Abstract
In eukaryotes, the primary components of the ribosome are encoded by multicopy nuclear ribosomal RNA (rRNA) genes: 28/26S, 18S, 5.8S, and 5S. Copies of these genes are typically localized within tandem arrays and homogenized within a genome. As a result, nuclear rRNA gene families have become a paradigm of concerted evolution. In filamentous fungi of the subphylum Pezizomycotina, 5S rRNA genes exist as a large and dispersed multigene family, with between 50 and 100 copies per genome. To determine whether these genes defy the concerted evolution paradigm, we examined the patterns of evolution of these genes by using sequences from the complete genomes of four species. Analyses of these sequences revealed (i) multiple 5S gene types within a genome, (ii) interspecies clustering of gene types, (iii) multiple identical gene types shared among species, (iv) multiple pseudogenes within a genome, and (v) presence/absence variation of individual 5S copies in comparisons of closely related species. These results demonstrate that the 5S family in these species is characterized by birth-and-death evolution under strong purifying selection. Furthermore, our results suggest that birth-and-death evolution occurs at different rates in the genera examined, and that the multiplication and movement of 5S genes across the genome are highly dynamic. As such, we hypothesize that a mechanism resembling retroposition controls 5S rRNA gene amplification, dispersal, and integration in the genomes of filamentous fungi.
Keywords: 5S rRNA, retroposition
Eukaryotes possess four nuclear ribosomal RNA (rRNA) genes: 28/26S, 18S, 5.8S, and 5S. These genes are repeated several hundred to a few thousand times in the genome and are nearly always arranged either in a single large tandem repeat array or in multiple tandem arrays found on one or a few chromosomes. Brown et al. (1) first proposed the idea that rRNA gene copies evolve “horizontally,” meaning that a mutation that arises in one copy spreads to all other copies, eventually homogenizing all of them. Later, this process was called “concerted evolution” (2). Because concerted evolution theory could explain the observed lack of genetic variability among rRNA gene copies in many different species, it eventually became dogma that it was the fundamental mode of rRNA multigene family evolution (3–5).
Although the overwhelming majority of studies have shown that most rRNA multigene families evolve in a strictly concerted manner (meaning that all gene copies in the family are homogenized), some exceptional cases have been identified in actinobacteria, oak trees, flatworms, and apicomplexan protists (6–9). In the case of actinobacteria, errors during DNA replication are assumed to be the cause when horizontal transfer can be ruled out (10). In the flatworm (7, 8) and oak tree (11) species, there apparently have been “escapees” from concerted evolution, in which a few distinct rRNA types are maintained within the genome. Still, all copies of each distinct type are homogenized by concerted evolution in these exceptional cases. Apicomplexans present a rather interesting case of birth-and-death evolution of a small rRNA gene family (9). In these organisms, rRNA genes are dispersed throughout the genome and are not organized in a tandem array. This dispersed gene organization apparently facilitates birth-and-death evolution wherein rRNA genes diversify from one another, some of which are unique to a given species, whereas others are shared among species (9).
Unlike the other nuclear rRNA genes, the 5S genes of most eukaryotes are dispersed throughout the genome [e.g., as in Schizosaccharomyces pombe (12)] or found in their own distinct tandem arrays [e.g., as in soybeans (13)]. In some species, both types of gene arrangement are found [e.g., as in humans (14)]. This flexibility of 5S rRNA gene organization in different genomes is rather unusual and reminiscent of what is observed in some large multigene families that undergo birth-and-death evolution (Fig. 1), such as histones (15). Therefore, the purpose of this study was to investigate 5S rRNA multigene family evolution in filamentous fungi, because the genomes of four relatively closely related species from this group have been completely sequenced, are reasonably well assembled, and contain 5S rRNA gene families with 50–100 copies.
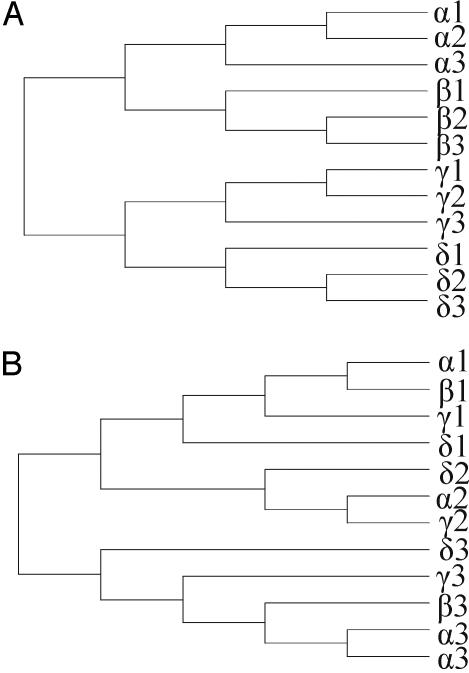
Fig. 1.
Expected gene relationships under the concerted evolution model (A) and the birth-and-death evolution model (B). Species are denoted by Greek letters, and genes are denoted by numbers. Under the concerted evolution model, genes cluster according to species; however, they do not under the birth-and-death model, except in cases of recent gene duplication (e.g., the α3 sequences in B). The figure was adapted from Rooney (9).
Materials and Methods
Genomic blast searches were used to mine 5S gene copies from the complete genomes of Aspergillus nidulans FGSC A4, Fusarium graminearum NRRL 31084 (also known as Gibberella zeae PH-1), Magnaporthe grisea 70–15, and Neurospora crassa OR74A (www.ncbi.nlm.nih.gov/genomes/FUNGI/funtab.html). Initial searches in each genome were performed by using a 5S rRNA gene sequence from A. nidulans (GenBank accession no. K03162). Subsequent searches were performed by using genome-specific sequences obtained from the initial search. This process was repeated with different sequences until no new gene copies were found. Genomic blast search results occasionally included partial sequences of ≈40–50% of the complete sequence length (119–120 nt). In all cases, the regions immediately flanking each partial sequence were directly examined to determine whether the sequence was truly incomplete or whether it merely contained large numbers of substitutions relative to the query sequence. Once all of the 5S genes were extracted from each of the above genomes, an alignment was constructed by using the computer program sequencher 4.1.2 (Gene Codes, Ann Arbor, MI). The resulting alignment was subsequently inspected for errors but required virtually no editing because of the highly conserved nature of the sequences.
For each genome, we randomly chose several loci to screen for the presence of orthologous 5S genes in closely related species. Oligonucleotide primers in predicted protein-coding sequences flanking each selected 5S gene copy were used to amplify the corresponding region in related species. If a locus failed to amplify with primers set in protein-coding regions, we designed alternative primer sets in which one of the primers was located in the 5S rRNA gene itself. PCR amplifications were performed with Platinum TaqDNA polymerase high fidelity (Invitrogen) under standard reaction conditions. The oligonucleotide primer sequences and specific PCR reaction conditions are available upon request. Amplification products were purified by using Montage PCR cleanup filter plates (Millipore). Sequencing reactions were conducted as described in Ward et al. (16) by using bigdye 3.0 sequencing chemistry (Applied Biosystems). Reaction products were purified by ethanol precipitation and run on an ABI3100 or ABI3730 genetic analyzer (Applied Biosystems). DNA sequences were edited by using sequencher 4.1.2 and added to the alignment containing the genomic sequences. The sequences have been deposited in GenBank under accession nos. AY924880–AY924946.
The computer program mega 2 (17) was used to conduct all phylogenetic and nucleotide sequence analyses. Kimura two-parameter distances (18) were computed and used to generate phylogenetic trees by using the neighbor-joining method (19). The statistical reliabilities of the internal branches were assessed for all trees by using 1,000 bootstrap pseudoreplicates.
Results
The 5S Gene Complement and Sequence Divergence Levels. The size of the 5S rRNA gene complement in the A. nidulans, F. graminearum, M. grisea, and N. crassa genomes is consistent with expectations based on genome size (Table 1). Accordingly, N. crassa, which has the largest genome size (43 Mb), possesses the greatest number of total 5S gene copies (92), and A. nidulans, which has the smallest genome size (31 Mb), possesses the least number of total gene copies (52). The F. graminearum and M. grisea genomes are approximately the same size (40 Mb) and have similar numbers of 5S gene copies (66 and 71, respectively). Of the total 5S gene copy number in each genome, a portion was comprised of truncated 5S gene copies (Table 1), but the majority consisted of intact gene copies. There are a total of 36 intact copies in the A. nidulans genome but only 16 distinct copy types, because 20 copies were identical to one or more of the others (Table 1). Similarly, the M. grisea genome contains 38 intact copies but only 17 distinct types. In contrast, the F. graminearum genome has 59 intact copies and 24 distinct types, whereas the N. crassa genome has 79 intact copies and 29 distinct types.
Table 1. Frequency and diversity of 5S rRNA gene copies in the complete filamentous fungal genomes studied here.
| Genome size, Mb | Total number of gene copies | Number of intact gene copies* | Number of distinct intact copy types* | Number of pseudogene copies | Proportion of intact copies/Mb | Proportion of pseudogene copies/Mb† | |
|---|---|---|---|---|---|---|---|
| A. nidulans | 31 | 52 | 36 (0.03) | 16 (0.03) | 16 | 1.16 | 0.52 |
| M. grisea | 40 | 71 | 38 (0.01) | 17 (0.02) | 33 | 0.95 | 0.83 |
| F. graminearum | 40 | 66 | 59 (0.09) | 24 (0.12) | 7 | 1.48 | 0.18 |
| N. crassa | 43 | 92 | 78 (0.07) | 29 (0.11) | 14 | 1.81 | 0.33 |
Average p̄ distance among sequences is given in parentheses.
Results for comparisons of pseudogene frequency by using a two-tailed G test were significant for A. nidulans vs. F. graminearum (P = 0.05), M. grisea vs. F. graminearum (P = 0.007), and M. grisea vs. N. crassa (P = 0.02).
When the distances between the intact copies within each of the four genomes were analyzed, we found that 5S genes in the A. nidulans and M. grisea genomes are much more closely related than those of the F. graminearum and N. crassa genomes. For example, the average distance between the 16 distinct types in the A. nidulans genome is p̄ = 0.03. Similarly, the 17 distinct types in the M. grisea genome differed by p̄ = 0.02. In contrast, the F. graminearum genome contains 24 distinct types that show an average divergence of p̄ = 0.12, and the N. crassa genome contains 29 distinct types that show an average divergence of p̄ = 0.11. When these distance estimates were compared to the estimates generated for all intact types, we found that they were either the same or highly similar (Table 1). In contrast, the estimates differed substantially between the N. crassa and F. graminearum genomes vs. the A. nidulans and M. grisea genomes (Table 1). Thus, 5S gene sequences are much more closely related in the A. nidulans and M. grisea genomes than in the F. graminearum and N. crassa genomes.
The above patterns can be explained by one of the following three possibilities: (i) the 5S gene copies of all four species evolve in concert, but some of the gene copies in the F. graminearum and N. crassa genomes evolve distinctly from the others; (ii) the 5S gene copies of all four species undergo birth-and-death evolution, but A. nidulans and M. grisea undergo a faster rate, resulting in a higher turnover frequency in 5S gene copies; or (iii) the 5S gene copies of A. nidulans and M. grisea are evolving in concert, whereas the 5S gene copies of F. graminearum and N. crassa undergo birth-and-death evolution. To determine which of these hypotheses is correct, we conducted the analyses described below.
Pseudogenes and Birth-and-Death Evolution. As mentioned previously, in all four genomes, we found that a portion of the total gene number was comprised of truncated 5S gene sequences. These truncated sequences are ≈40–50% of the intact gene sequence length. We consider these truncated sequences to be pseudogenes because their lack of an intact coding sequence effectively destroys the secondary structure of the 5S rRNA molecule that they would have otherwise encoded. The presence of pseudogenes in a multigene family strongly suggests that the family evolves under a birth-and-death process (9, 15, 20). Under this process, a multigene family both expands because of gene duplication and contracts because of gene loss (e.g., as a result of unequal crossover). Thus, some family members exist in the genome for only short periods of evolutionary time, whereas others persist for much longer periods. Eventually, distinct gene copies accumulate differences, leading some of them to degenerate into pseudogenes. Birth-and-death evolution can be detected through two means: (i) a phylogenetic analysis of multigene family members, whereby a between-species gene clustering pattern will be evident; and (ii) through an examination of gene sequence divergence levels, in which case a relatively high proportion of changes between gene family members indicates a lack of homogenization (9, 15, 20) (Fig. 1). In contrast, sequences evolving in a concerted fashion will show evidence of extensive homogenization due to gene conversion or unequal crossover, resulting in a pattern of within-species gene clustering accompanied by very small distance estimates in comparisons between gene copies of the same organism (9, 15, 20).
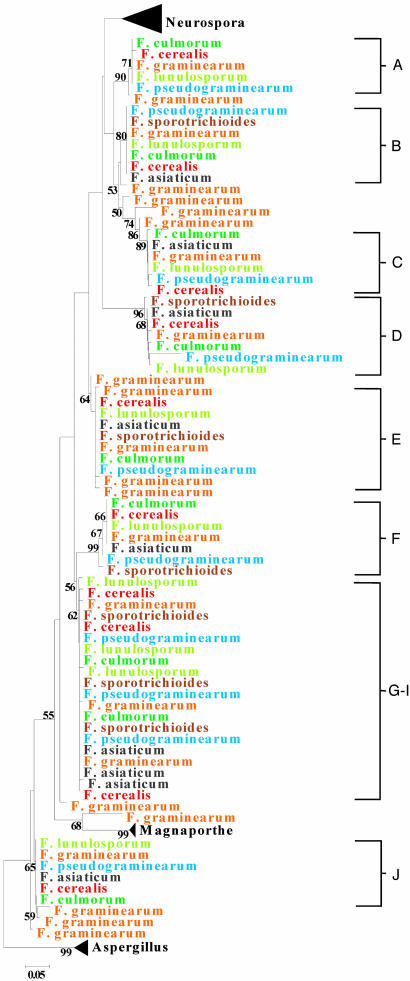
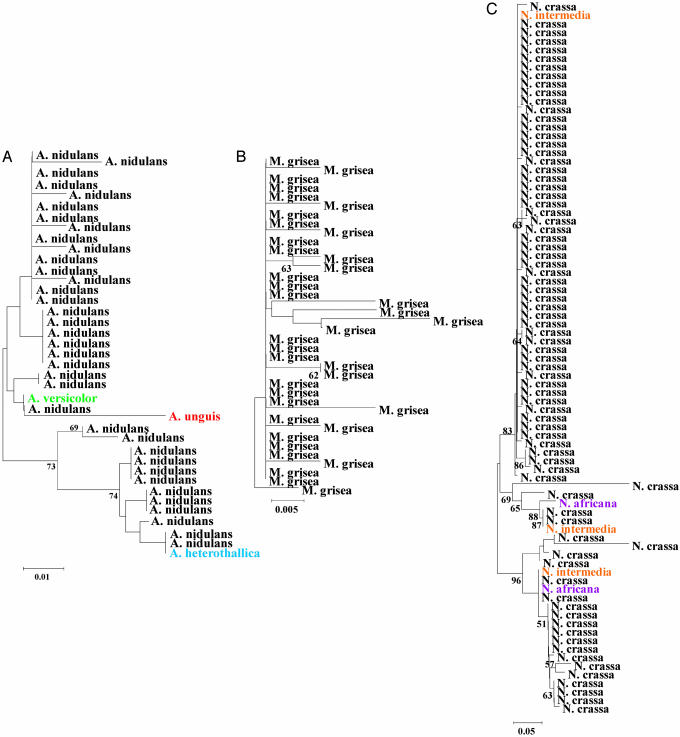
We analyzed the phylogenetic relationships between the 5S genes from all four genomes along with sequences from closely related taxa that we obtained from our own laboratory work (Figs. 2 and 3). Comparison of the gene copies from the four complete genome sequences provided little evidence of interspecies clustering of 5S types. All 5S copies from the N. crassa, M. grisea, and A. nidulans genome sequences formed individual monophyletic groups, with 5S copies from the F. graminearum genome more broadly distributed in a weakly supported, but paraphyletic arrangement (Fig. 2). However, comparisons involving closely related species revealed extensive interspecies clustering and the presence of identical 5S types shared among species (Figs. 2 and 3). This is best seen in the case of F. graminearum (Fig. 2), for which we had samples of several closely related species. The 5S rRNA genes are split into several multispecies clades, some of which are well supported by relatively high bootstrap values. Because these clades represent orthologous loci (Fig. 2), the phylogeny is characterized by locus-specific clustering, as genes that were amplified and sequenced from a particular locus cluster together to the exclusion of genes from other loci regardless of the species from which they came (Fig. 2). The one exception involves a cluster in which the genes present at three loci were identical in all species examined (Fig. 2, G–I). However, this observation is not unexpected if the genes originated from a recent duplication event and/or if purifying selection is strong. In fact, the important observations that can be made from examining this phylogeny are that (i) 5S gene sequences form several clusters composed of different species and (i) the gene sequences in these clusters are either identical to or display only one or two differences from each other. Taken together, these observations indicate that 5S nucleotide sequence homogeneity is maintained by purifying selection, not concerted evolution, and that the gene copies evolve under a birth-and-death process. The same patterns were found for Aspergillus and Neurospora (Fig. 3), although we screened fewer sister species and 5S gene loci in these cases.
Fig. 2.
Phylogeny of 5S rRNA genes from the filamentous fungal species examined in this study. Because of the large number of sequences analyzed, we show only the Fusarium gene sequences. The gene sequences from Aspergillus, Magnaporthe, and Neurospora are shown in Fig. 3. Each Fusarium species is shown in a different color. The clusters labeled A–J represent distinct loci.
Fig. 3.
Phylogeny of the 5S rRNA genes from Aspergillus (A), Magnaporthe (B), and Neurospora (C). Each species is shown in a different color.
Evidence for birth-and-death evolution was also found as a result of identifying instances in which specific 5S copies from the complete genomes were absent from the genomes of closely related species or strains. For example, at the top of Fig. 2, one clade represents a locus that contains sequences from only five Fusarium species; apparently, the other two (F. asiaticum and F. sporotrichioides) did not possess a 5S sequence at that locus. This suggests that the 5S gene was lost at that locus in the latter two taxa. We made similar observations for the Neurospora and Aspergillus species that we examined (Fig. 3). For instance, for one of the N. crassa loci that we examined, N. crassa and N. intermedia had a 5S gene at one locus but N. Africana did not (Fig. 3). We also saw evidence of gene “birth” in our laboratory analyses. This was shown best in our analysis of M. grisea and its relatives. Here, we looked at one locus in three M. grisea genome strain relatives: (i) a strain of M. grisea that was not the genome strain, (ii) M. salvinii (a close relative of M. grisea), and (iii) Gaeumannomyces graminis var. tritici (a member of a genus closely related to Magnaporthe). None of these isolates possessed a 5S gene at that locus. The most parsimonious explanation is that the 5S gene was inserted in the M. grisea genome strain after it had diverged from these other genomes.
Discussion
For several decades now, researchers have been interested in understanding how rRNA genes and gene families diversify and evolve (e.g., see refs. 1, 3, 5, 7–9, and 21–27). Brown et al. (1) first proposed that rRNA genes evolve “horizontally” or in a concerted fashion on the basis of restriction digest patterns. Since then, rRNA multigene families have come to be viewed as a paradigm of concerted evolution (4, 28–31). Only rarely have any departures from this model been found, the most notable of which were mentioned at the beginning of this paper. Moreover, virtually all of these departures from the rRNA concerted evolution paradigm are limited in scope and usually involve the existence of one or a few divergent types (e.g., see refs. 7, 8, and 11). In addition, these departing types encompass many gene copies that are highly homogenized. So far, the only identified case of an rRNA multigene family undergoing birth-and-death evolution is the apicomplexan rRNA multigene family (9). This family encompasses a small number of copies (usually five to seven) that have a dispersed genomic organization, which is quite unlike what is seen in most other eukaryotes. In this paper, we identify an example of a large dispersed rRNA multigene family undergoing birth-and-death evolution: the 5S rRNA genes of filamentous fungi within the ascomycete subphylum Pezizomycotina, as represented by the genera Aspergillus, Fusarium, Magnaporthe, and Neurospora.
Previous studies claimed to show that the 5S genes of A. nidulans (25) and N. crassa (26) undergo concerted evolution. However, we did not find this to be true for these species or for M. grisea or the Fusarium species on the basis of nucleotide sequence divergence levels, phylogenetic clustering patterns, and the existence of 5S pseudogenes. It is true that we observed multiple copies of identical gene sequences. However, the reasons for this are (i) strong purifying selection and (ii) frequent 5S gene copying and multiplication resulting in rapid gene turnover. When combined, these processes will curtail the chances of extensive divergence between genes. In a study on 5S rRNA gene evolution in the filamentous fungi Epichloë and Neotyphodium, which are also members of the subphylum Pezizomycotina, Ganley and Scott (32) claimed that the 5S genes of these species evolve only partially in concert, with limited divergence between genes. After examining their results, we conclude that a more likely explanation is that the genes undergo birth-and-death evolution with strong purifying selection and rapid gene turnover.
Depending on how rapid the gene turnover rate is, the distinct gene types produced through birth-and-death evolution will show varying degrees of divergence from each other. In our study, we found that the gene types of A. nidulans and M. grisea were much more closely related than were the gene types of F. graminearum and N. crassa (Table 1), suggesting that rates of gene turnover are higher in the latter two genomes than in the former two. This conclusion is consistent with the finding that the genomes possessing very closely related gene types (A. nidulans and M. grisea) had a higher frequency of pseudogenes than the genomes whose gene types were more divergent (F. graminearum and N. crassa) (Table 1). Moreover, there was a strong, although not statistically significant (P = 0.088), negative correlation (r = –0.912) between the number of pseudogenes per megabase and the average level of nucleotide sequence divergence among 5S gene types (Table 1). Overall, our results show that there are high levels of gene amplification and dispersal in all of the species studied, indicating that their genomes are highly dynamic. However, the rate of birth and death of 5S copies appears to be different in the four pezizomycotine genera examined, with Magnaporthe having the fastest rate and Fusarium having the slowest rate. More generally, our results suggest that pseudogene formation is linked to rates of gene turnover in this large multigene family.
Given the prevalence of concerted evolution among tandemly arrayed rRNA gene families, some might hypothesize that the dispersed nature of the apicomplexan and ascomycete rRNA gene families is responsible for their exceptional patterns of evolution. Although this might be true of apicomplexan rRNA genes (9), the dispersed organization of pezizomycotine 5S genes is more a result of the birth-and-death process rather than a facilitator of it. For the most part, a dispersed genomic organization of multigene family members ordinarily arises through a process involving gene duplication followed by insertion of copies into other areas of the genome via unequal crossover, inversion and/or translocation. Mobile elements, on the other, hand, use a process of transposition or retrotransposition to amplify, disperse, and subsequently integrate copies into the genome. Our findings appear to be more in line with this type of process for best explaining the pattern of 5S gene organization and dispersal observed in this study. Gene amplification involves the copying of a progenitor gene or genetic element and subsequent insertion of the daughter copies into other areas of the genome. The retrotransposition of long interspersed elements (LINEs) and short interspersed elements (SINEs) are classic examples of amplification (28–30, 33). The amplification of a LINE occurs when its primary transcript is reverse transcribed, copied, and then integrated into another area of the genome. SINEs operate essentially the same way but rely on LINEs, at least in part, for their own retroposition, amplification, and genomic integration (34–36).
We hypothesize that 5S genes are capable of multiplying and integrating into other areas of the genome through a process the same as, or akin to, retroposition. The 5S rRNA genes and SINEs have the same type of internal RNA polymerase III promoter (33, 37). Furthermore, Kapitonov and Jurka (38) have identified an unique class of SINEs that are derived from the fusion of a 5S rRNA gene and a LINE, showing that 5S rRNA genes and retroelements can indeed interact with one another. A number of retroelements, including SINEs, LINEs, and related retrotransposons, have been identified in A. nidulans (39), F. graminearum (40), M. grisea (41, 42), N. crassa (43), and a number of other related fungal species (44–49). Interestingly, Esnault et al. (50) have shown that LINEs are capable of amplifying and integrating non-LINE mRNA transcripts by turning them into processed pseudogenes and then integrating them into other parts of the genome. Apparently, the mRNA transcripts redirect the LINE enzymatic machinery to accomplish this. As such, it is interesting to consider the possibility that the 5S rRNA gene of filamentous fungi might also be capable of redirecting the enzymatic machinery of LINEs, LINE-like elements, or even some other retroelement for their own amplification, dispersal, and genomic integration.
Acknowledgments
We thank J. R. Dettman (University of California, Berkeley) and S. B. Goodwin, K. O'Donnell, and S. W. Peterson (Agricultural Research Service, U.S. Department of Agriculture) for providing fungal strains. We are grateful to T. Usgaard and J. Robinson for laboratory support.
Author contributions: A.P.R. and T.J.W. designed research; A.P.R. and T.J.W. performed research; A.P.R. and T.J.W. analyzed data; and A.P.R. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: rRNA, ribosomal RNA; LINE, long interspersed element; SINE, short interspersed element.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY924880–AY924946).
References
- 1.Brown, D. D., Wensink, P. C. & Jordan, E. (1972) J. Mol. Biol. 63, 57–73. [DOI] [PubMed] [Google Scholar]
- 2.Zimmer, E. A., Martin, S. L., Beverley, S. M., Kan, Y. W. & Wilson, A. C. (1980) Proc. Natl. Acad. Sci. USA 77, 2158–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coen, E., Strachan, T. & Dover, G. (1982) J. Mol. Biol. 158, 17–35. [DOI] [PubMed] [Google Scholar]
- 4.Liao, D. (1999) Am. J. Hum. Genet. 64, 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao, D. (2000) J. Mol. Evol. 51, 305–317. [DOI] [PubMed] [Google Scholar]
- 6.Gunderson, J. H., Sogin, M. L., Wollett, G., Hollingdale, M., de la Cruz, V. F., Waters, A. P. & McCutchan, T. F. (1987) Science 238, 933–937. [DOI] [PubMed] [Google Scholar]
- 7.Carranza, S., Giribet, G., Ribera, C., Baguñà, J. & Riutort, M. (1996) Mol. Biol. Evol. 13, 824–832. [DOI] [PubMed] [Google Scholar]
- 8.Carranza, S., Baguñà, J. & Riutort, M. (1999) J. Mol. Evol. 49, 250–259. [DOI] [PubMed] [Google Scholar]
- 9.Rooney, A. P. (2004) Mol. Biol. Evol. 21, 1704–1711. [DOI] [PubMed] [Google Scholar]
- 10.Ueda, K., Seki, T., Kudo, T., Yoshida, T. & Kataoka, M. (1999) J. Bacteriol. 181, 78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muir, G., Fleming, C. C. & Schlötterer, C. (2001) Mol. Biol. Evol. 18, 112–119. [DOI] [PubMed] [Google Scholar]
- 12.Wood, V., Gwilliam, R., Rajandream, M. A., Lyne, M., Lyne, R., Stewart, A., Sgouros, J., Peat, N., Hayles, J., Baker, S., et al. (2002) Nature 415, 871–880. [DOI] [PubMed] [Google Scholar]
- 13.Gottlob-McHugh, S. G., Levesque, M., MacKenzie, K., Olson, M., Yarosh, O. & Johnson, D. A. (1990) Genome 33, 486–494. [DOI] [PubMed] [Google Scholar]
- 14.Little, R. D. & Braaten, B. C. (1989) Genomics 4, 376–383. [DOI] [PubMed] [Google Scholar]
- 15.Rooney, A. P., Piontkivska, H. & Nei, M. (2002) Mol. Biol. Evol. 19, 68–75. [DOI] [PubMed] [Google Scholar]
- 16.Ward, T. J., Gorski, L., Borucki, M. K., Mandrell, R. E., Hutchins, J. & Pupedis, K. (2004) J. Bacteriol. 186, 4994–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- 18.Kimura, M. (1980) J. Mol. Evol. 16, 111–120. [DOI] [PubMed] [Google Scholar]
- 19.Saitou, N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 20.Nei, M. Gu, X. & Sitnikova, T. (1997) Proc. Natl. Acad. Sci. USA 94, 7799–7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore, R. L. & McCarthy, B. J. (1968) Biochem. Genet. 2, 75–86. [DOI] [PubMed] [Google Scholar]
- 22.Pinder J. C., Gould, H. J. & Smith, I. (1969) J. Mol. Biol. 40, 289–298. [DOI] [PubMed] [Google Scholar]
- 23.Perry, R. P., Cheng, T. Y., Freed, J. J., Greenberg, J. R., Kelley, D. E. & Tartof, K. D. (1970) Proc. Natl. Acad. Sci. USA 65, 609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown, D. D. & Sugimoto, K. (1974) Cold Spring Harbor Symp. Quant. Biol. 38, 501–505. [DOI] [PubMed] [Google Scholar]
- 25.Borsuk, P., Gniadkowski, M., Bartnik, E. & Stepien, P. P. (1988) J. Mol. Evol. 28, 125–130. [DOI] [PubMed] [Google Scholar]
- 26.Morzycka-Wroblewska, E., Selker, E. U., Stevens, J. N. & Metzenberg, R. L. (1985) Mol. Cell. Biol. 5, 46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drouin, G. & de Sá, M. M. (1995) Mol. Biol. Evol. 12, 481–493. [DOI] [PubMed] [Google Scholar]
- 28.Nei, M. (1987) Molecular Evolutionary Genetics (Columbia Univ. Press, New York).
- 29.Li, W.-H. (1997) Molecular Evolution (Sinauer, Sunderland, MA).
- 30.Graur, D. & Li, W.-H. (2000) Fundamentals of Molecular Evolution (Sinauer, Sunderland, MA).
- 31.Ohta, T. (2000) Gene 259, 45–52.11163960 [Google Scholar]
- 32.Ganley, A. R. & Scott, B. (2002) Fungal Genet. Biol. 35, 39–51. [DOI] [PubMed] [Google Scholar]
- 33.Weiner, A. M. (2002) Curr. Opin. Cell Biol. 14, 343–350. [DOI] [PubMed] [Google Scholar]
- 34.Oshima, K., Hamada, M., Terai, Y. & Okada, N. (1996) Proc. Natl. Acad. Sci. USA 90, 6260–6264. [Google Scholar]
- 35.Okada, N., Hamada, M., Ogiwara, I. & Ohshima, K. (1997) Gene 205, 229–243. [DOI] [PubMed] [Google Scholar]
- 36.Terai, Y., Takahashi, K. & Okada, N. (1998) Mol. Biol. Evol. 15, 1460–1471. [DOI] [PubMed] [Google Scholar]
- 37.Paule, M. R. & White, R. J. (2000) Nucleic Acids Res. 28, 1283–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapitonov, V. V. & Jurka, J. (2003) Mol. Biol. Evol. 20, 694–702. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen, M. L., Hermansen, T. D. & Aleksenko, A. (2001) Mol. Genet. Genomics 265, 883–837. [DOI] [PubMed] [Google Scholar]
- 40.Mes, J. J., Haring, M. A. & Cornelissen, B. J. (2000) Mol. Gen. Genet. 263, 271–280. [DOI] [PubMed] [Google Scholar]
- 41.Kachroo, P., Leong, S. A. & Chattoo, B. B. (1995) Proc. Natl. Acad. Sci. USA 92, 11125–11129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamer, J. E., Farrall, L., Orbach, M. J., Valent, B. & Chumley, F. G. (1989) Proc. Natl. Acad. Sci. USA 86, 9981–9985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cambareri, E. B., Helber J. & Kinsey, J. A. (1994) Mol. Gen. Genet. 242, 658–665. [DOI] [PubMed] [Google Scholar]
- 44.Rasmussen, M., Rossen, L. & Giese, H. (1993) Mol. Gen. Genet. 239, 298–303. [DOI] [PubMed] [Google Scholar]
- 45.Kim, H. G., Meinhardt, L. W., Benny, U. & Kistler, H. C. (1995) Mol. Plant–Microbe Interact. 8, 524–531. [DOI] [PubMed] [Google Scholar]
- 46.Eto, Y., Ikeda, K., Chuma, I., Kataoka, T., Kuroda, S., Kikuchi, N., Don, L. D., Kusaba, M., Nakayashiki, H., Tosa Y., et al. (2001) Mol. Gen. Genet. 264, 565–577. [DOI] [PubMed] [Google Scholar]
- 47.Goyon, C., Rossignol, J. L. & Faugeron, G. (1996) Nucleic Acids Res. 24, 3348–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He, C., Nourse, J. P., Kelemu, S., Irwin, J. A. & Manners, J. M. (1996) Mol. Gen. Genet. 252, 320–331. [DOI] [PubMed] [Google Scholar]
- 49.Luderer, R., Takken, F. L., de Wit, P. J. & Joosten, M. H. (2002) Mol. Microbiol. 45, 875–884. [DOI] [PubMed] [Google Scholar]
- 50.Esnault, C., Maestre, J. & Heidmann, T. (2000) Nat. Genet. 24, 363–367. [DOI] [PubMed] [Google Scholar]