Classical paradigms of prokaryotic transcriptional regulation are simple and elegant. In some cases, modification of an activator leads to DNA binding that enables initiation of transcription by an RNA polymerase. In others, interaction with an inducer relieves inhibition by a DNA-binding repressor. Alternative sigma factors activated under specific stress conditions may compete with the housekeeping sigma factor to direct a core RNA polymerase in the transcription of specific gene subsets. However, as the control of individual genes is examined in greater detail, the situation often becomes more complex. Transcription of individual genes may reflect input from multiple DNA-binding proteins, regulatory cascades, RNA-binding proteins, redox-sensing motifs, and small RNAs or metabolites (1), as well as factors influencing genome structure and local DNA superhelicity (2). One example, the Escherichia coli sodA gene encoding manganese superoxide dismutase, is controlled by at least five transcriptional regulators: SoxS, Fur, ArcA, Fnr, and IHF (3). In this respect, sodA is by no means unusual. Genes encoded by the SPI-1 pathogenicity island required for invasion of host cells by Salmonella are controlled by four transcriptional regulators and modulated by as many as a dozen others (4). Expression of the alternative sigma factor σS is controlled by as many as two dozen regulatory factors at the level of transcription, translation, or proteolysis (5). Microbiologists might be forgiven for wondering whether transcriptional regulatory networks were designed by Rube Goldberg (1883–1970), a cartoonist famous for his invention of inordinately complicated schemes to achieve simple tasks.
Microarray technology has revolutionized the ability to comprehensively examine bacterial transcriptional responses to environmental changes and to examine the contribution of specific regulatory loci (6). Such analyses have revealed unanticipated complexity of regulatory phenomena: in E. coli, a simple transition from growth on glucose to lactose was shown by microarray analysis to alter the expression of hundreds of genes in addition to the lactose operon (7). Moreover, microarrays have facilitated the study of gene expression in organisms in which conventional genetic approaches are difficult and tools are limited. In this issue of PNAS, Fisher et al. (8) report the use of microarrays to identify genes controlled by the alternative sigma factors σN (σ54) and σS in the Lyme disease spirochete, Borrelia burgdorferi. The authors confirm a regulatory cascade involving σN and σS, as shown earlier by others (9), and identify hundreds of genetic loci whose expression depends on either or both sigma factors. In contrast to the earlier authors (9), Fisher et al. did not find σS expression to depend absolutely on σN, so that some genes appeared to be regulated by σS alone.
Alternative sigma factors play a critical role in the complex life cycle of B. burgdorferi, in which the spirochetes encounter many different environmental conditions. Both σN- and σS-deficient B. burgdorferi mutants are avirulent in mice (8, 10), and σN was shown in the present study to be required for the entry of the spirochete into tick salivary glands as well, implying a role in the transmission of Lyme disease. Neither sigma factor was essential for growth in culture medium, consistent with the notion that these regulons are primarily involved in stress responses. Fisher et al. observed up-regulation as well as down-regulation of genes in response to the loss of σN or σS, which the authors attribute to indirect regulatory actions or sigma competition. Another possibility is that some genes whose expression is elevated in sigma factor mutants might belong to stress responses compensating for the loss of the alternative sigma factor, analogous to the phage shock protein operon that is induced to compensate for the loss of σE in Salmonella (11).
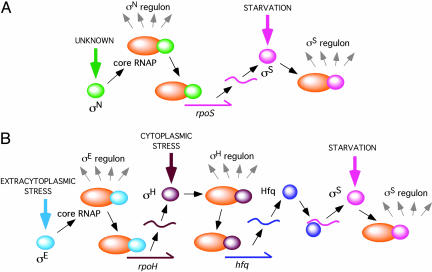
The activity of alternative sigma factors is typically controlled by proteolytic turnover or inactivation of cognate anti-sigma factors (12), which, in turn, are triggered by environmental conditions such as the presence of unfolded proteins or nutrient deprivation. In Bacillus sp., sigma factor cascades provide temporal and spatial compartmentalization of gene expression critical for the sporulation process (13). However, in other bacteria, alternative sigma factors are usually considered to independently control discrete subsets of genes in response to different environmental stimuli (14). Although alternative sigma factors can act independently and are likely to do so in many instances, the observations in B. burgdorferi (8, 9) demonstrate that there can be considerable overlap between the genes controlled by alternative sigma factors as a consequence of regulatory interactions. Recent observations demonstrating a sigma cascade linking σE, σH, and σS in Salmonella (15) suggest that such cascades may be more widespread than has been previously appreciated (Fig. 1). In this instance, σE and σH enhance translation of σS by increasing expression of the RNA-binding protein Hfq (HF-1). The activation of one sigma factor by another allows the integration of diverse environmental signals to result in expression of a common stress response. As the data of Fisher et al. demonstrate, such activation may not necessarily result in expression of an entire regulon but could nevertheless play an important role for specific loci or conditions in which levels of the downstream sigma factor are limiting.
Fig. 1.
Sigma factor cascades in B. burgdorferi and Salmonella typhimurium. Alternative sigma factors can direct core RNA polymerase to transcribe specific gene subsets. (A)In B. burgdorferi, the alternative sigma factor σN appears to directly stimulate transcription of the rpoS gene encoding the sigma factor σS (9). Thus, activation of σN can enhance the expression of a large number of both σN- and σS-regulated genes (8). (B)In S. typhimurium, the alternative sigma factors σE, σH, and σS are linked by a more complex mechanism (15). The sigma factor σE stimulates transcription of the rpoH gene encoding the sigma factorσH which, in turn, stimulates transcription of the hfq gene encoding the RNA-binding protein Hfq. Binding of Hfq to the rpoS mRNA facilitates translation of the sigma factor σS. Regulatory interactions between sigma factors permit the integration of diverse environmental stimuli to trigger expression of a common stress response (in these instances, the σS regulon).
In addition to Borrelia, alternative sigma factors have been found to play a crucial role in the virulence of pathogenic bacteria including Salmonella, Mycobacterium, Pseudomonas, Listeria, Legionella, Vibrio, and Staphylococcus sp. (16–22). This important role in pathogenesis reflects the diversity of environmental conditions faced by pathogenic microbes as they travel through their life cycles of transmission and replication. In some cases, the role of alternative sigma factors in pathogenesis may simply reflect the need for a microbe to withstand specific stress conditions encountered in the host environment [e.g., σE-mediated resistance to antimicrobial peptides and reactive oxygen species (23–25)], whereas, in other cases, specific virulence determinants appear to have coopted preexisting regulatory systems that match their temporal and spatial requirements [e.g., regulation of the Salmonella spv plasmid virulence genes by σS (17)].
Activation of one sigma factor by another allows the integration of diverse environmental signals.
New investigative approaches and mathematical analyses are revealing that regulatory complexity has important functional consequences, which include asymmetric kinetics (26), robust control (27), graded responses (28), and the generation of noise (29). As Emerson observed (30), “The highest simplicity of structure is produced, not by few elements, but by the highest level of complexity.” In analogy to neural and other networks, the whole is greater than the sum of the parts, and complexity gives rise to novel emergent properties. Such complexity must be both embraced in its entirety as well as deconstructed (6) if the true nature of regulatory networks is to be understood.
Prokaryotic regulatory networks have evolved to respond with exquisite precision to environmental changes of significance to the organism. Overlapping and interacting circuits maintain gene expression in an equipoise that is stable yet able to respond appropriately to new stimuli. In that light, what may have first appeared to be an unnecessarily complex and ungainly invention can be seen as a work of art. That is surely something Rube Goldberg would have appreciated.
Acknowledgments
I thank L. Kenney, I. S. Bang, and S. Libby for critical reading of this Commentary. This work was supported by National Institutes of Health Grant AI44486.
See companion article on page5162.
References
- 1.Wick, L. M. & Egli, T. (2004) Adv. Biochem. Eng. Biotechnol. 89, 1–45. [DOI] [PubMed] [Google Scholar]
- 2.Dorman, C. J. (1996) Trends Microbiol. 4, 214–216. [DOI] [PubMed] [Google Scholar]
- 3.Compan, I. & Touati, D. (1993) J. Bacteriol. 175, 1687–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lostroh, C. P. & Lee, C. A. (2001) Microbes Infect. 3, 1281–1291. [DOI] [PubMed] [Google Scholar]
- 5.Hengge-Aronis, R. (2002) Microbiol. Mol. Biol. Rev. 66, 373–395, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conway, T. & Schoolnik, G. K. (2003) Mol. Microbiol. 47, 879–889. [DOI] [PubMed] [Google Scholar]
- 7.Chang, D. E., Smalley, D. J. & Conway, T. (2002) Mol. Microbiol. 45, 289–306. [DOI] [PubMed] [Google Scholar]
- 8.Fisher, M. A., Grimm, D., Henion, A. K., Elias, A. F., Stewart, P. E., Rosa, P. A. & Gherardini, F. C. (2005) Proc. Natl. Acad. Sci. USA 102, 5162–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hubner, A., Yang, X., Nolen, D. M., Popova, T. G., Cabello, F. C. & Norgard, M. V. (2001) Proc. Natl. Acad. Sci. USA 98, 12724–12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caimano, M. J., Eggers, C. H., Hazlett, K. R. & Radolf, J. D. (2004) Infect. Immun. 72, 6433–6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becker, L. A., Bang, I. S., Crouch, M.-L. & Fang, F. C. (2005) Mol. Microbiol., in press. [DOI] [PubMed]
- 12.Hughes, K. T. & Mathee, K. (1998) Annu. Rev. Microbiol. 52, 231–286. [DOI] [PubMed] [Google Scholar]
- 13.Stragier, P. & Losick, R. (1990) Mol. Microbiol. 4, 1801–1806. [DOI] [PubMed] [Google Scholar]
- 14.Gruber, T. M. & Gross, C. A. (2003) Annu. Rev. Microbiol. 57, 441–466. [DOI] [PubMed] [Google Scholar]
- 15.Bang, I. S., Frye, J. G., McClelland, M., Velayudhan, J. & Fang, F. C. (March 9, 2005) Mol. Microbiol., 10.1111/j.1365-2958.2005.04580.x. [DOI] [PubMed]
- 16.Totten, P. A., Lara, J. C. & Lory, S. (1990) J. Bacteriol. 172, 389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang, F. C., Libby, S. J., Buchmeier, N. A., Loewen, P. C., Switala, J., Harwood, J. & Guiney, D. G. (1992) Proc. Natl. Acad. Sci. USA 89, 11978–11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachman, M. A. & Swanson, M. S. (2001) Mol. Microbiol. 40, 1201–1214. [DOI] [PubMed] [Google Scholar]
- 19.Kovacikova, G. & Skorupski, K. (2002) Infect. Immun. 70, 5355–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazmierczak, M. J., Mithoe, S. C., Boor, K. J. & Wiedmann, M. (2003) J. Bacteriol. 185, 5722–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manganelli, R., Provvedi, R., Rodrigue, S., Beaucher, J., Gaudreau, L. & Smith, I. (2004) J. Bacteriol. 186, 895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonsson, I. M., Arvidson, S., Foster, S. & Tarkowski, A. (2004) Infect. Immun. 72, 6106–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humphreys, S., Stevenson, A., Bacon, A., Weinhardt, A. B. & Roberts, M. (1999) Infect. Immun. 67, 1560–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Testerman, T. L., Vazquez-Torres, A., Xu, Y., Jones-Carson, J., Libby, S. J. & Fang, F. C. (2002) Mol. Microbiol. 43, 771–782. [DOI] [PubMed] [Google Scholar]
- 25.Crouch, M.-L., Becker, L. A., Bang, I. S., Tanabe, H., Ouellette, A. J. & Fang, F. C. (March 7, 2005) Mol. Microbiol., 10.1111/j.1365-2958.2005.04578.x. [DOI] [PubMed]
- 26.Mangan, S. & Alon, U. (2003) Proc. Natl. Acad. Sci. USA 100, 11980–11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batchelor, E. & Goulian, M. (2003) Proc. Natl. Acad. Sci. USA 100, 691–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Batchelor, E., Silhavy, T. J. & Goulian, M. (2004) J. Bacteriol. 186, 7618–7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korobkova, E., Emonet, T., Vilar, J. M., Shimizu, T. S. & Cluzel, P. (2004) Nature 428, 574–578. [DOI] [PubMed] [Google Scholar]
- 30.Emerson, R. W. (1850) Representative Men (Phillips and Sampson, Boston).