Abstract
In skeletal muscle cells, voltage-dependent potentiation of Ca2+ channel activity requires phosphorylation by cAMP-dependent protein kinase (PKA) anchored via an A-kinase anchoring protein (AKAP15), and the most rapid sites of phosphorylation are located in the C-terminal domain. Surprisingly, the site of interaction of the complex of PKA and AKAP15 with the α1-subunit of CaV1.1 channels lies in the distal C terminus, which is cleaved from the remainder of the channel by in vivo proteolytic processing. Here we report that the distal C terminus is noncovalently associated with the remainder of the channel via an interaction with a site in the proximal C-terminal domain when expressed as a separate protein in mammalian nonmuscle cells. Deletion mapping of the C terminus of the α1-subunit using the yeast two-hybrid assay revealed that a distal C-terminal peptide containing amino acids 1802–1841 specifically interacts with a region in the proximal C terminus containing amino acid residues 1556–1612. Analysis of the purified α1-subunit of CaV1.1 channels from skeletal muscle by saturation sequencing of the intracellular peptides by tandem mass spectrometry identified the site of proteolytic processing as alanine 1664. Our results support the conclusion that a noncovalently associated complex of the α1-subunit truncated at A1664 with the proteolytically cleaved distal C-terminal domain, AKAP15, and PKA is the primary physiological form of CaV1.1 channels in skeletal muscle cells.
Keywords: calcium channels, contraction coupling, excitation, protein kinase, proteolysis
In skeletal muscle, voltage-gated Ca2+ channels localized in the transverse tubule membrane play a pivotal role in excitation–contraction (E–C) coupling (1). After membrane depolarization during an action potential, these Ca2+ channels function as voltage sensors to initiate E–C coupling and as a slowly activating Ca2+ entry pathway that regulates contractile force (2, 3). Trains of high-frequency depolarizing stimuli that mimic action potentials or single long depolarizing pulses greatly increase the activity of these Ca2+ channels (4, 5). This “potentiation” of Ca2+ channel activity is strongly voltage-dependent and, in skeletal muscle, requires phosphorylation by cAMP-dependent protein kinase (PKA) anchored via an AKAP (AKAP15; refs. 4 and 6–8).
The molecular mechanism underlying the targeting of PKA to the CaV1 family of Ca2+ channels has recently emerged. AKAP15 directly targets PKA to the distal C terminus of cardiac and skeletal muscle CaV1 channels through a modified leucine zipper interaction (9, 10). Mutation of this motif prevents PKA anchoring, and disruption of this interaction with dominant-negative peptides prevents PKA-dependent potentiation of Ca2+ channel activity in skeletal and cardiac myocytes (9, 10). Thus, anchoring of PKA to CaV1.1 channels via AKAP15 favors rapid phosphorylation and modulation of these channels as part of the overall regulation of contractile force in response to motor nerve stimulation (11, 12).
Two size forms of the α1-subunit of CaV1.1 channels, ≈175 and 212 kDa, exist in membrane preparations from skeletal muscle (13–16). The predominant (>90%) short form results from in vivo proteolytic processing of the C terminus near amino acid residue 1685, approximately halfway through the C-terminal domain (14). Similarly, the related CaV1.2 channels in heart and brain are also truncated by in vivo proteolysis at a similar position in their C termini (17–20). Surprisingly, the site of interaction with AKAP15 and PKA is located beyond the point of truncation in the C terminus (9). Because PKA anchored at this site is required for effective regulation of skeletal muscle Ca2+ channels by voltage-dependent potentiation and PKA, these results raise the possibility that the proteolytically cleaved distal C terminus remains associated with the remainder of the skeletal muscle Ca2+ channel to allow this regulation to occur. In the experiments reported here, we have demonstrated that the distal C terminus associates with the remainder of the channel when coexpressed as a separate protein in nonmuscle cells, mapped the site of interaction between the distal and proximal C-terminal domains, and defined the point of proteolytic truncation of the CaV1.1 channel precisely by mass spectrometric analysis. Our results provide biochemical evidence for the formation of a specific complex of the α1-subunit of the CaV1.1 channel truncated at alanine 1664 with its distal cleaved C-terminal peptide beginning at asparagine 1665, which has specifically bound AKAP15 and PKA.
Experimental Procedures
Antibodies, Peptides, and cDNA Constructs. Rabbit polyclonal anti-CP1 and anti-CP11 antibodies were generated against peptides corresponding to residues (1857–1873) and (1601–1618) of the rabbit skeletal muscle α1 sequence (21) and characterized as described (14). Monoclonal anti-myc antibody was purchased from Invitrogen. CaV1.1 Δ1684 was constructed by amplifying a BamHI/NotI fragment with primers incorporating a stop codon after position 1684 and subcloning the truncated fragment into α1.1 pCDNA3. CaV1.1 1685–1873 was amplified by PCR using specific primers and cloned in-frame into pCDNA3mycHisA (Invitrogen). Sequences corresponding to the proximal C-terminal fragments (1382–1698, 1382–1556, 1518–1698, 1518–1572, 1556–1612, and 1606–1656) of the α1-subunit of the rabbit skeletal Ca2+ channel were amplified by PCR and cloned in frame into the Gal4 DNA binding domain vector, pAS2-1 (Clontech). Distal C-terminal constructs (1753–1873, 1774–1841, and 1802–1841) were generated in the same manner and cloned in-frame into the Gal4 activation domain vector, pACT2 (Clontech). N-terminally acylated C-terminal fragments of CaV1.1 were constructed by incorporating the sequence (MGQLCC) that directs N-terminal myristoylation and palmitoylation of AKAP15 onto the N terminus of the C-terminal fragments by PCR and cloned in-frame into pcDNA3mycHisA (Invitrogen). The orientation and reading frame of all constructs were confirmed by DNA sequencing.
Coimmunoprecipitation Experiments. TsA-201cells were cultured in DMEM F12 supplemented with 10% FBS and 100 units/ml penicillin and streptomycin and plated on 15-cm dishes. Cells were transfected with 50 μg of an equimolar ratio of expression plasmid cDNA using the calcium phosphate method. Forty-eight hours after transfection, cells were washed in PBS, solubilized in ice-cold bRIA (50 mM Tris·HCl, pH 7.4/150 mM NaCl/1 mg·ml–1 BSA/50 mM NaF/5 mM EGTA/5 mM EDTA/1% Triton X-100, plus protease inhibitors) and rotated at 4°C for 30 min. Unsolubilized material was removed by centrifugation, and lysates were precleared with protein A Sepharose. Precleared lysates were incubated with 10 μg of anti-CP11, anti-CP1, or control nonimmune IgG for 2 h at 4°C, followed by the addition of protein A Sepharose (50 μl) for an additional 2 h. Immune complexes bound to the Sepharose beads were washed extensively, and proteins were separated by SDS/PAGE, transferred to nitrocellulose, and analyzed by immunoblotting. CaV1.1 Δ1684 was detected by anti-CP11. CaV1.1 1685–1873 and the Myc-tagged N-acyl C-terminal fragments were detected by using monoclonal anti-myc antibody. Each immunoprecipitation experiment was repeated at least three times.
Yeast Two-Hybrid Assay. cDNAs encoding different regions of the proximal and distal C terminus of CaV1.1 were cotransformed into the Y190 yeast strain, containing two Gal4-inducible reporter genes, HIS3 and LacZ, by using the TRAFCO LiAc method. Cotransformants were first plated onto synthetic dropout medium lacking tryptophan and leucine (trp–leu–) to select for colonies containing both hybrid plasmids and then transferred onto medium that also lacked histidine (trp–leu–his–) to select for protein–protein interactions. The media was supplemented with 30 mM 3-amino-1,2,4-triazole to suppress background growth of the Y190 yeast strain. Only those transformants that grew on trp–leu–his– media at 30°C were assayed for β-galactosidase activity by using the colony filter lift assay. Data shown are representative of at least four separate experiments.
Purification and Mass Spectrometric Analysis of Skeletal Muscle CaV1.1 Channels. [3H]PN200-110, endoproteinase Asp-N and endoproteinase Glu-C were purchased from Roche Applied Sciences. Trypsin Gold, Mass Spectrometry Grade, was from Promega. Wheat germ agglutinin (WGA)–Sepharose and N-acetylglucosamine were from Sigma. CNBr-Activated Sepharose 4B and DEAE Sephadex A-25 were obtained from Amersham Pharmacia. Skeletal muscle CaV1.1 channels were purified by chromatography on WGA–Sepharose and DEAE ion exchange chromatography (22) from rabbit skeletal muscle transverse tubule membranes prepared according to Florio et al. (23). All buffers contained 35 μg/ml phenylmethanesulfonyl fluoride, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin A, 1 μg/ml aprotinin, and 200 μg/ml o-phenanthroline. The purity and quantity of the CaV1.1 channel preparation were analyzed by SDS/PAGE and Coomassie Blue staining, and the moles of purified Ca2+ channel were estimated assuming a molecular mass of 429 kDa for the channel complex. The α1-subunit band was excised from the gel and subjected to “in-gel” proteolysis with three different proteases: trypsin to cleave at the carboxyl side of lysine and arginine residues; Asp-N to cleave at the amino side of asparagine residues; and Glu-C to cleave at the carboxyl side of glutamate residues. In-gel digested peptide samples were cleaned before tandem mass spectrometry (MS/MS) analyses by application to 1-ml SPE C18 columns (Discovery DSC-18, Supelco, Bellefonte, PA) and eluted with 80% acetonitrile/0.1% trifluoroacetic acid. The eluted peptides were concentrated by using a Speed-Vac SC 250 Express (ThermoSavant, Holbrook, NY). BCA protein assays were performed to determine the final peptide concentrations, and samples were snap frozen and stored at –80°C. The peptides were analyzed by high-resolution reversed-phase capillary liquid chromatography coupled to an electrospray ionization interface with a ThermoFinnigan LCQ ion trap mass spectrometer (ThermoFinnigan, San Jose, CA) to perform MS/MS measurements as described (24). To enhance detection of short peptides down to ≈7 amino acid residues in length, analyses employing narrow segmented mass ranges (m/z windows) were performed. SEQUEST (ThermoFinnigan) matching analysis of the MS/MS spectra was performed against the NCBI Oryctolagus cuniculus (rabbit) protein FASTA database (6,110 proteins) applying stringent filtering guidelines. Using an approach similar to that described by Qian et al. (25), confident tryptic peptide identifications were obtained by the application of the following scoring criteria: (i) SEQUEST DelCN value of ≥0.10 and (ii) SEQUEST XCorr (correlation score) ≥1.5 for charge state 1+ and full tryptic peptides, ≥3.1 for partial tryptic peptides; ≥1.9 for charge state 2+ and full tryptic peptides, ≥3.8 for partial tryptic; and ≥2.9 for charge state 3+ and full tryptic peptides, ≥4.5 for partial tryptic peptides. Confident peptide identifications for Asp-N and Glu-C protease digests were obtained by applying the following scoring criteria: (i) SEQUEST DelCN value ≥0.05 and (ii) SEQUEST XCorr ≥1.8 for charge state 1+; ≥1.9 for charge state 2+; and ≥3.75 for charge state 3+.
Results and Discussion
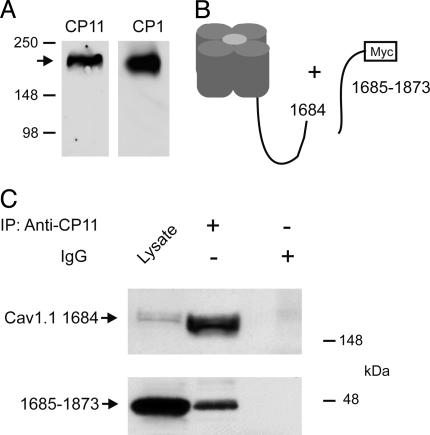
Association of the Distal C Terminus with the Cleaved CaV1.1 Channel. Expression of CaV1.1 channels by transfection of full-length cDNA in tsA-201 cells yields only full-length α1-subunit proteins, as detected by anti-peptide antibodies against the final C-terminal peptide (Fig. 1A). This finding is in marked contrast to skeletal muscle, where C-terminal truncated α1-subunit proteins are the predominant form (14). These results suggest that the proteolytic enzyme necessary for processing CaV1 channels is not present in these nonmuscle cells. To examine the association of the cleaved distal C terminus with the remainder of the CaV1.1 channel, we expressed the distal C terminus in tsA-201 cells as a separate protein, consisting of amino acid residues 1685–1873 and a C-terminal myc epitope tag, in combination with the remainder of the CaV1.1 channel (amino acid residues 1–1684; Fig. 1B). Analysis of cell lysates by immunoblotting revealed robust expression of both proteins (Fig. 1C, lane 1). Immunoprecipitation of the CaV1.1Δ1684 α1-subunit with anti-CP11 antibodies resulted in coimmunoprecipitation of the distal C terminus as detected with antibodies against the myc epitope (Fig. 1C Lower, lane 2). No coimmunoprecipitation was observed with nonimmune IgG (Fig. 1C Lower, lane 3). These results demonstrate formation of a specific complex between the cleaved distal C-terminal domain of CaV1.1 and the remainder of the channel protein when expressed in nonmuscle cells.
Fig. 1.
Coimmunoprecipitation of the separately expressed distal C terminus with truncated CaV1.1 channels in tsA-201 cells. (A) Immunoblot showing that expression of CaV1.1 channels by transfection of full-length cDNA in tsA-201 cells yields only full-length α1-subunit proteins, as detected by an anti-peptide antibody against the final C-terminal peptide (anti-CP1). (B) Schematic diagram of truncated CaV1.1 channels and the separately expressed distal C terminus used in coimmunoprecipitation experiments. (C) Lysates of tsA-201 cells transfected with CaV1.1 1684 and the distal C terminus (1685–1873) were immunoprecipitated with anti-CP11 (lane 2) or control IgG (lane 3). Immunoblots were probed with anti-CP11 (Upper) or anti-myc (Lower). Positive control for immunoblotting was 20 μl of lysate (lane 1).
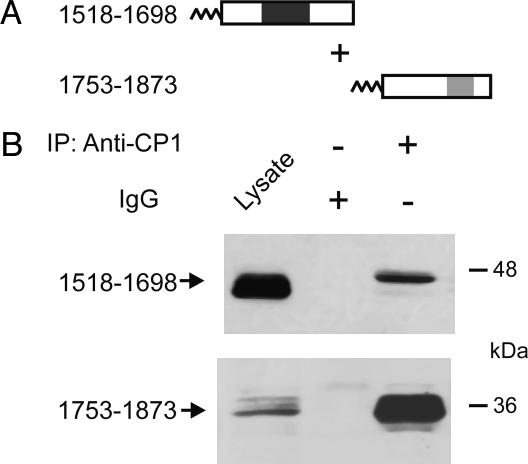
Specific Association of Peptides from the Distal and Proximal C-Terminal Domains in Mammalian Cells. We hypothesized that the distal C terminus may remain in noncovalent association with the proximal C terminus after proteolytic cleavage in vivo. To examine the interaction between the proximal and distal C-terminal domains in intact cells, immunoprecipitation experiments were performed in the tsA-201 line of human embryonic kidney cells. The N-terminal acylation sequence of AKAP15 was engineered onto the N terminus of the distal and proximal CaV1.1 C-terminal fusion proteins to target them to the plasma membrane as described (9). Cell lysates were immunoprecipitated with anti-CP1 to immunoprecipitate CaV1.1 1753–1873 or control IgG, and the immunoprecipitated CaV1.1 1518–1698 protein was detected with anti-CP11. Immunoblotting of cell lysates with anti-CP11 or anti-CP1 demonstrated robust expression of the CaV1.1 1518–1698 and CaV1.1 1753–1873 proteins, respectively (Fig. 2, lane 1). CaV1.1 1518–1698 specifically coimmunoprecipitated with CaV1.1 1753–1873 (Fig. 2B Upper, lane 3) but not control IgG (Fig. 2B Upper, lane 2). These results demonstrate that the proximal C-terminal domain directly binds to the distal C-terminal domain through noncovalent protein–protein interactions.
Fig. 2.
Coimmunoprecipitation of the distal and proximal C-terminal domains of CaV1.1 channels expressed in tsA-201 cells. (A) Schematic map of the lipid-anchored CaV1.1 C-terminal constructs examined in coimmunoprecipitation experiments. (B) TsA-201 cells were cotransfected with the constructs depicted in A, and cell lysates were immunoprecipitated with anti-CP1 or control IgG. Immunoblots were probed with anti-CP11 (Upper) or anti-CP1 (Lower) to detect immunoprecipitated CaV1.1 1518–1698 and CaV1.1 1753–1873 protein, respectively.
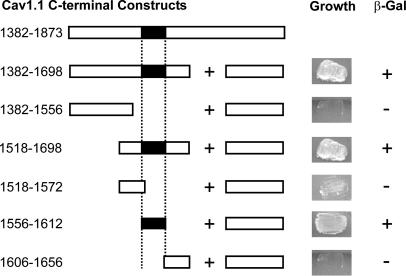
Mapping the Interaction Site for the Distal C Terminus Within the Proximal C Terminus of the CaV1.1 α1-Subunit. To define the points of interaction between the distal and proximal C-terminal domains, we used the yeast two-hybrid assay. We found that a full-length proximal C-terminal peptide consisting of amino acid residues 1382–1698 interacted effectively with a distal C-terminal peptide consisting of amino acid residues 1753–1873 to reconstitute an active gal-4 transcription factor and allow transcription of nutritional markers in intact yeast cells (Fig. 3). Analysis of the interactions of five deletion constructs revealed that a peptide consisting of amino acid residues 1556–1612 was sufficient to interact with the distal C terminus (1753–1873; Fig. 3). Thus, this 56-aa segment of the proximal C terminus is likely to form an interaction site for the distal C terminus.
Fig. 3.
Amino acid residues 1556–1612 in the proximal C terminus of CaV1.1 bind the distal C terminus. (Left) Schematic map of the proximal C-terminal constructs of CaV1.1 that were cloned into the pAS2.1 vector and cotransformed with the distal C terminus (1753–1873 pACT2) or pACT2 alone into the Y190 yeast strain. Numbers correspond to the amino acid residues in CaV1.1. (Right) Representative growth of yeast cotransformed with the CaV1.1 proximal and distal C-terminal plasmids depicted on the left and the resulting β-galactosidase assay (+ or –).
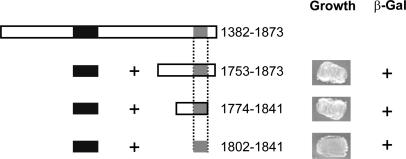
Mapping the Interaction Site for the Proximal C Terminus Within the Distal C Terminus of the CaV1.1 α1-Subunit. We performed a complementary set of analyses using the yeast two-hybrid system to identify the amino acid sequence in the distal C terminus that is responsible for its interaction with the proximal C terminus (Fig. 4). Analysis of three deletion constructs showed that a peptide consisting of amino acid residues 1802–1841 in the distal C terminus was sufficient to interact with the proximal C-terminal interaction domain (PCID, residues 1556–1612; Fig. 4). Taken together, these data suggest that this 39-amino-acid-residue peptide forms a distal C-terminal interaction domain (DCID) for binding of the distal C terminus to the PCID.
Fig. 4.
Amino acid residues 1802–1841 in the distal C terminus of CaV1.1 bind the proximal C terminus. (Left) Schematic map of the distal C-terminal constructs of CaV1.1 that were cloned into the pACT2 vector and cotransformed with the proximal C terminus (1556–1612 pAS2.1) or pAS2.1 alone into the Y190 yeast strain. Numbers correspond to the amino acid residues in CaV1.1. (Right) Representative growth of yeast cotransformed with the CaV1.1 proximal and distal C-terminal plasmids depicted on the left and the resulting β-galactosidase assay (+ or –).
Identification of the C-Terminal Amino Acid Residue of the α1-Subunit of CaV1.1 Channels. Our results defining the sites of specific interaction of the cleaved distal C terminus with the proximal C-terminal domain of CaV1.1 emphasize the importance of identifying the precise point of proteolytic truncation of CaV1.1 to fully characterize the molecular properties of the components of this complex. In previous studies, we mapped the C terminus of the α1-subunit of CaV1.1 channels by using an overlapping series of anti-peptide antibodies and concluded that it was near amino acid residues 1685–1699 (14). The development of high-resolution mass spectrometry techniques now allows precise determination of the C-terminal amino acid residue by a saturation sequencing strategy in which the hydrophilic peptides in the intracellular domains of the α1-subunit are identified and sequenced as completely as possible. We purified the CaV1.1 channel from rabbit skeletal muscle and isolated the α1-subunit by SDS-PAGE as described in Experimental Procedures (Fig. 5A). The α1-subunit band was excised from the gel and subjected to in-gel proteolysis with three different proteases: trypsin to cleave at the carboxyl side of lysine and arginine residues; Asp-N to cleave at the amino side of asparagine residues; and Glu-C to cleave at the carboxyl side of glutamate residues. The resulting peptides were analyzed by MS/MS combined with high-resolution liquid chromatography methods that led to confident identification of peptides as small as seven amino acid residues in length. In this analysis, all internal peptides should end in the amino acid residue characteristic of the protease used for cleavage in vitro, except for the C-terminal peptide, which should end in its natural terminal amino acid residue when cleaved with all three proteases.
Fig. 5.
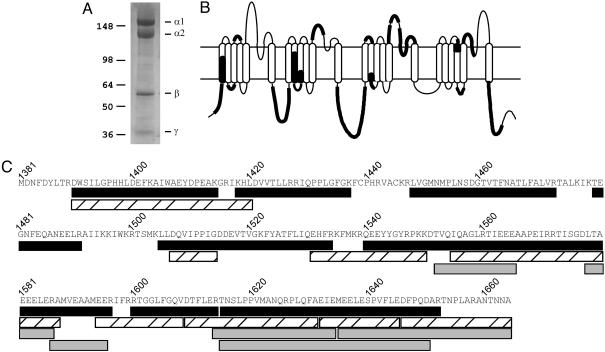
Identification of the C terminus of proteolytically processed CaV1.1 by mass spectrometry. (A) Coomassie blue staining of the CaV1.1 complex. The α1-subunit band was excised from the gel and subjected to “in-gel” proteolysis with three different proteases. The other bands on the gel correspond to the α2-, β-, and γ-subunits that copurify in the CaV1.1 channel complex. (B) Schematic diagram showing the coverage of proteolytic peptides of the α1-subunit of CaV1.1 channels identified by LC/MS/MS. (C) Deduced amino acid sequence of the cleaved CaV1.1 C terminus. The location of the proteolytic peptides obtained from digests with trypsin (black bar), endoproteinase Asp-N (striped bar) and endoproteinase Glu-C (gray bar) are shown.
As illustrated in Fig. 5B, we achieved near-saturation sequencing of the proteolytic peptides derived from the N and C termini and the large hydrophilic intracellular loops of the CaV1.1 channel up to residue 1664 in the C-terminal domain (see Table 1, which is published as supporting information on the PNAS web site). In the C-terminal domain, nearly all peptides up to position 1664 were identified that had predicted length >6 aa after cleavage with one of the three proteases (Fig. 5C). In the analysis of tryptic peptides, the final peptide ended at arginine 1652, a residue expected from tryptic cleavage. In contrast, the final Asp-N peptide ended at alanine 1664 (A1664), an amino acid residue not expected from Asp-N cleavage. Similarly, the final Glu-C peptide also ended at A1664, which is not expected from Glu-C cleavage (Fig. 5C). These results implicate A1664 as the final amino acid residue in the in vivo truncated form of CaV1.1. The results of the tryptic peptide analysis are also consistent with this conclusion because the final predicted peptide from tryptic cleavage would be the hexapeptide A1659–A1664, which would not be readily detected because it is <7 aa.
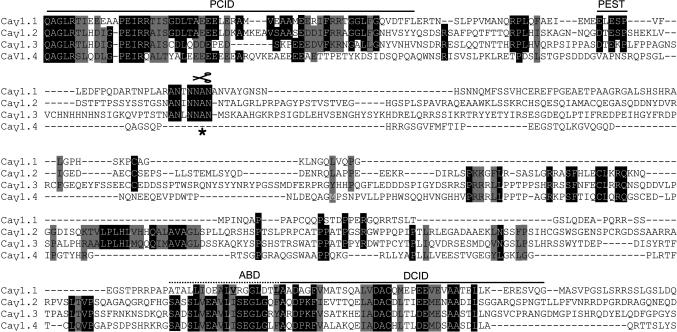
Conserved Amino Acid Sequences of CaV1.1 and CaV1.2 Channels at the Sites of C-Terminal Interaction and Proteolytic Cleavage. It is interesting to compare the amino acid sequences of the CaV1 family of channels in the sites of interaction between the proximal and distal C-terminal domains and in the region surrounding the site of proteolytic cleavage (Fig. 6), because both CaV1.1 and CaV1.2 channels are subject to in vivo proteolytic processing (14, 19, 20, 26). Alignment of the C-terminal domains of CaV1.1 and CaV1.2 channels shows striking conservation of the amino acid sequences in the PCID, the AKAP15-binding domain (ABD), and DCID (Fig. 6) compared to the poorly conserved amino acid sequences between them. Interestingly, the ABD and DCID are immediately adjacent in the distal C terminus. Similarly, comparison of the aligned amino acid sequences around the site of proteolytic cleavage also reveals striking conservation of amino acid sequence (Fig. 6). In fact, this sequence was proposed as a possible site of proteolytic recognition or functional regulation based on its high conservation of amino acid sequence in our previous antibody-mapping studies (14). The high level of amino acid sequence conservation is consistent with the conclusions that both CaV1.1 and CaV1.2 channels are cleaved at the equivalent of A1664 in vivo and that the resulting distal C-terminal domain remains associated with the remainder of the channels through noncovalent interactions of analogous sites in the proximal and distal C-terminal domains.
Fig. 6.
Conserved amino acid sequences of CaV1.1 and CaV1.2 channels at the sites of C-terminal interaction and proteolytic cleavage. Amino acid sequence alignment of the PCID, DCID, PEST motif, the proteolytic cleavage site (asterisk and scissors), and the AKAP15-binding domain (ABD; critical residues shown in bold) identified in CaV1.1 with CaV1.2, CaV1.3, and CaV1.4. Conserved amino acids are shaded in black, and similar amino acids are shaded in gray.
Comparison of the amino acid sequences of CaV1.3 and CaV1.4 channels with the PCID, proteolytic cleavage site, ABD, and DCID from CaV1.1 channels reveals an interesting dichotomy. The amino acid sequence of CaV1.3 channels is well conserved at all four of these sites (Fig. 6). Therefore, it is likely that this channel is also proteolytically processed, its distal C-terminal domain remains associated with the channel through interactions of its DCID and PCID, and a complex of PKA and AKAP15 binds to the ABD and regulates channel function. In marked contrast, the ABD of CaV1.4 channels is well conserved, especially the heptad repeat of hydrophobic residues that binds AKAP15 via a modified leucine zipper, but the proteolytic cleavage site is not well conserved. This striking difference in amino acid sequence raises the interesting possibility that CaV1.4 channels are not proteolytically processed so that the distal C-terminal domain is not required to associate noncovalently with the proximal C-terminal domain. Nevertheless, CaV1.4 channels, which are highly expressed in the retina, may be regulated by PKA specifically targeted to them through interaction with AKAP15 bound to the conserved ABD.
Possible Mechanism of Proteolytic Cleavage. Indirect evidence suggests that a calpain-like protease may be responsible for in vivo proteolytic cleavage of CaV1 channels. The C-terminal domain of CaV1.2 channels in brain is truncated (26). In intact hippocampal neurons in brain slices, regulated proteolytic cleavage of the C terminus of CaV1.2 channels is induced by Ca2+ influx through NMDA-activated glutamate receptors, and proteolytic cleavage of α11.2 in these intact cells can be prevented by calpain inhibitors (27). Purified calpain 1 can cleave the C-terminal domain of purified CaV1.1 channels in vitro, but the site of cleavage is not identical to the in vivo site based on SDS/PAGE analysis (28). Cleavage at A1664 is also consistent with the catalytic specificity of calpain proteases (29). Moreover, a conserved PEST sequence rich in proline, glutamate, serine, and threonine is located ≈27 aa upstream of A1664 (PEST, Fig. 6) in position to serve as a substrate recognition site for calpains (30). Further experiments with specific members of the family of 12 calpain isozymes are needed to confirm that a calpain protease is indeed involved in the in vivo proteolytic processing of this family of Ca2+ channels.
Potential Physiological Significance of Proteolytic Processing. The conserved proteolytic processing of CaV1.1 and CaV1.2 channels suggests that this cleavage has physiological significance. What physiological role might be served by this proteolytic processing event? Expression of CaV1.1 and CaV1.2 channels with truncated C termini in Xenopus oocytes or tsA-201 cells increases the functional activity of these channels (31–34), suggesting that the distal C terminus has an autoinhibitory effect. Short peptides derived from the distal C-terminal domain inhibit CaV1.2 channels (33). Moreover, coexpression of the distal C-terminal domain as a separate protein with the remainder of CaV1.2 channels in tsA-201 cells gives potent inhibition, decreasing channel activity below the level of the full-length α1-subunit.∥ In light of these results with CaV1.2 channels, the cleaved distal C-terminal domain of CaV1.1 channels may also serve an autoinhibitory function. In skeletal muscle cells, the level of Ca2+ channel ion conductance activity is low compared to the large number of CaV1.1 channels present (3, 34), and the principal function of the CaV1.1 channel is voltage-dependent activation of the ryanodine-sensitive sarcoplasmic reticulum Ca2+ channel, which releases Ca2+ into the cytosol to initiate contraction (1). Inhibition of Ca2+ conductance by the cleaved, noncovalently associated C-terminal autoinhibitory domain may serve to reduce unwanted Ca2+ entry through CaV1.1 channels unless that inhibition is relieved by cAMP-dependent phosphorylation. Consistent with that idea, disruption of the leucine zipper interaction between AKAP15 and the distal C terminus of CaV1.1 is effective in blocking voltage-dependent potentiation, which depends on PKA activity (4, 6, 9). Evidently, a noncovalent complex composed of CaV1.1 channels truncated at A1664, the cleaved distal C-terminal domain, AKAP15, and PKA is likely to be the primary physiological form of CaV1.1 channels in skeletal muscle cells. Recent studies indicate that Ca2+ channel regulation is impaired in dystrophic mouse muscle cells (35), suggesting that dysfunction of this signaling complex may be important in neuromuscular disease.
Supplementary Material
Acknowledgments
This work was supported by Muscular Dystrophy Association research grants and National Institutes of Health (NIH) Grant PO1 HL 44948 (to W.A.C.), by a research grant from the American Heart Association (to J.T.H.), and by the NIH National Center for Research Resources (RR18522) and the Laboratory Directed Research and Development Program, Pacific Northwest National Laboratory (Richland, WA), under Department of Energy Contract DE-AC06-76RL01830 (to M.A.G., D.G.C., and D.J.B). High-resolution LC/MS/MS analyses were performed by Ronald J. Moore, David J. Anderson, and Therese R. W. Clauss in the Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, which is sponsored by the Office of Biological and Environmental Research, U.S. Department of Energy.
Author contributions: J.T.H., K.K., D.G.C., D.J.B., and W.A.C. designed research; J.T.H., K.K., T.W.-C.L., and M.A.G. performed research; D.G.C. contributed new reagents/analytic tools; J.T.H., K.K., T.W.-C.L., M.A.G., D.G.C., D.J.B., and W.A.C. analyzed data; and J.T.H., K.K., and W.A.C. wrote the paper.
Abbreviations: MS/MS, tandem MS; PCID, proximal C-terminal interaction domain; DCID, distal C-terminal interaction domain; ABD, AKAP15-binding domain.
Footnotes
Hulme, J. T., Scheuer, T. & Catterall, W. A. (2004) Biophys. J. 86, 185a (abstr.).
References
- 1.Catterall, W. A. (1991) Cell 64, 871–874. [DOI] [PubMed] [Google Scholar]
- 2.Adams, B. A. & Beam, K. G. (1990) FASEB J. 4, 2809–2816. [DOI] [PubMed] [Google Scholar]
- 3.Rios, E. & Pizarro, G. (1991) Physiol. Rev. 71, 849–908. [DOI] [PubMed] [Google Scholar]
- 4.Sculptoreanu, A., Scheuer, T. & Catterall, W. A. (1993) Nature 364, 240–243. [DOI] [PubMed] [Google Scholar]
- 5.Fleig, A. & Penner, R. (1995) J. Physiol. (London) 489, 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson, B. D., Scheuer, T. & Catterall, W. A. (1994) Proc. Natl. Acad. Sci. USA 91, 11492–11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson, B. D., Brousal, J. P., Peterson, B. Z., Gallombardo, P. A., Hockerman, G. H., Lai, Y., Scheuer, T. & Catterall, W. A. (1997) J. Neurosci. 17, 1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray, P. C., Johnson, B. D., Westenbroek, R. E., Hays, L. G., Yates, I. J., Scheuer, T., Catterall, W. A. & Murphy, B. J. (1998) Neuron 20, 1017–1026. [DOI] [PubMed] [Google Scholar]
- 9.Hulme, J. T., Ahn, M., Hauschka, S. D., Scheuer, T. & Catterall, W. A. (2002) J. Biol. Chem. 277, 4079–4087. [DOI] [PubMed] [Google Scholar]
- 10.Hulme, J. T., Lin, T. W., Westenbroek, R. E., Scheuer, T. & Catterall, W. A. (2003) Proc. Natl. Acad. Sci. USA 100, 13093–13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kernell, D., Eerbeek, O. & Verhey, B. A. (1983) Exp. Brain Res. 50, 220–227. [DOI] [PubMed] [Google Scholar]
- 12.Arreola, J., Calvo, J., Garcia, M. C. & Sánchez, J. A. (1987) J. Physiol. (London) 393, 307–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Jongh, K. S., Merrick, D. K. & Catterall, W. A. (1989) Proc. Natl. Acad. Sci. USA 86, 8585–8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Jongh, K. S., Warner, C., Colvin, A. A. & Catterall, W. A. (1991) Proc. Natl. Acad. Sci. USA 88, 10778–10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai, Y., Seagar, M. J., Takahashi, M. & Catterall, W. A. (1990) J. Biol. Chem. 265, 20839–20848. [PubMed] [Google Scholar]
- 16.Brawley, R. M. & Hosey, M. M. (1992) J. Biol. Chem. 267, 18218–18223. [PubMed] [Google Scholar]
- 17.Chang, F. C. & Hosey, M. M. (1988) J. Biol. Chem. 263, 18929–18937. [PubMed] [Google Scholar]
- 18.Schneider, T. & Hofmann, F. (1988) Eur. J. Biochem. 174, 369–375. [DOI] [PubMed] [Google Scholar]
- 19.Gao, T. Y., Puri, T. S., Gerhardstein, B. L., Chien, A. J., Green, R. D. & Hosey, M. M. (1997) J. Biol. Chem. 272, 19401–19407. [DOI] [PubMed] [Google Scholar]
- 20.De Jongh, K. S., Murphy, B. J., Colvin, A. A., Hell, J. W., Takahashi, M. & Catterall, W. A. (1996) Biochemistry 35, 10392–10402. [DOI] [PubMed] [Google Scholar]
- 21.Ellis, S. B., Williams, M. E., Ways, N. R., Brenner, R., Sharp, A. H., Leung, A. T., Campbell, K. P., McKenna, E., Koch, W. J., Hui, A., et al. (1988) Science 241, 1661–1664. [DOI] [PubMed] [Google Scholar]
- 22.Curtis, B. M. & Catterall, W. A. (1984) Biochemistry 23, 2113–2118. [DOI] [PubMed] [Google Scholar]
- 23.Florio, V., Striessnig, J. & Catterall, W. A. (1992) Methods Enzymol. 207, 529–546. [DOI] [PubMed] [Google Scholar]
- 24.Shen, Y., Zhao. R., Belov, M. E., Conrads. T. P., Anderson, G. A., Tang, K., Pasa-Tolic, L., Veenstra, T. D., Lipton, M. S., Udseth, H. R. & Smith, R. D. (2001) Anal. Chem. 73, 1766–1775. [DOI] [PubMed] [Google Scholar]
- 25.Qian, W. J., Liu, T., Monroe, M. E., Strittmatter, E. F., Jacobs, J. M., Kangas, L. J., Petritis, K., Camp, D. G., II, & Smith, R. D. (2005) J. Proteome. Res. 4, 53–62. [DOI] [PubMed] [Google Scholar]
- 26.Hell, J. W., Yokoyama, C. T., Wong, S. T., Warner, C., Snutch, T. P. & Catterall, W. A. (1993) J. Biol. Chem. 268, 19451–19457. [PubMed] [Google Scholar]
- 27.Hell, J. W., Westenbroek, R. E., Breeze, L. J., Wang, K. K. W., Chavkin, C. & Catterall, W. A. (1996) Proc. Natl. Acad. Sci. USA 93, 3362–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Jongh, K. S., Colvin, A. A., Wang, K. K. W. & Catterall, W. A. (1994) J. Neurochem. 63, 1558–1564. [DOI] [PubMed] [Google Scholar]
- 29.Tompa, P., Buzder-Lantos, P., Tantos, A., Farkas, A., Szilagyi, A., Banoczi, Z., Hudecz, F. & Friedrich, P. (2004) J. Biol. Chem. 279, 20775–20785. [DOI] [PubMed] [Google Scholar]
- 30.Wang, K. K., Villalobo, A. & Roufogalis, B. D. (1989) Biochem. J. 262, 693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrill, J. A. & Cannon, S. C. (2000) J. Gen. Physiol. 116, 341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei, X. N. A., Lacerda, A. E., Olcese, R., Stefani, E., Perez-Reyes, E. & Birnbaumer, L. (1994) J. Biol. Chem. 269, 1635–1640. [PubMed] [Google Scholar]
- 33.Gao, T., Cuadra, A. E., Ma, H., Bunemann, M., Gerhardstein, B. L., Cheng, T., Eick, R. T. & Hosey, M. M. (2001) J. Biol. Chem. 276, 21089–21097. [DOI] [PubMed] [Google Scholar]
- 34.Schwartz, L. M., McCleskey, E. W. & Almers, W. (1985) Nature 314, 747–751. [DOI] [PubMed] [Google Scholar]
- 35.Johnson, B. D., Scheuer, T. & Catterall, W. A. (2005) Proc. Natl. Acad. Sci. USA 102, in press. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.