Abstract
Oral squamous cell carcinoma (OSCC) is a common malignant tumor with high metastatic potential. However, no good biomarker has been identified to refine which subtype is of high metastatic potential to make decisions regarding the elective and therapeutic management of lymphatic metastases. In this study, we investigated the role of the metabolic enzyme phosphoglycerate mutase 1 (PGAM1) in OSCC. PGAM1 expression was examined in tissue samples of 122 OSCC patients using immunohistochemistry, and the correlation between clinicopathological expression and PGAM1 expression was determined. Survival curves were generated using the Kaplan-Meier method, and multivariate analysis was performed by the Cox proportional hazards model. Moreover, PGAM1 was knocked down in the OSCC cell lines Cal27 and HN12, followed by determination of the change in cell migration and signaling pathways. PGAM1 expression is correlated with age, lymphatic metastasis and tumor recurrence and is closely associated with poor overall survival (OS) and disease-free survival (DFS). Intriguingly, PGAM1 is an independent risk factor for OS and DFS. After knocking down PGAM1 in Cal27 and HN12 cells, cell migration was remarkably decreased along with signaling pathway molecules, such as proto-oncogene c-SRC (SRC), Focal adhesion kinase (FAK) and Paxillin. The effect on cell migration was abolished following pretreatment with an SRC inhibitor. This study suggested that PGAM1 is a poor prognostic biomarker of OSCC and may be used to select patients of high metastatic potential in the clinic, and PGAM1 promotes the migration of OSCC cells is associated with the SRC pathway.
Keywords: Oral cancer, Phosphoglycerate mutase 1, Migration, Prognosis, SRC
Introduction
Oral squamous cell carcinoma (OSCC) is one of the most prevalent tumor types in the head and neck region around the world 1. OSCC is characterized by severe progression with a high potential for both lymphatic metastasis and locoregional invasion 2. Despite advances in surgery, radiotherapy, and chemotherapy in the past 30 years, the 5-year survival rate of this disease remains less than 50% 3. The unfavorable clinical outcome is closely associated with its high lymphatic and local metastatic potential 4. Thus far, decisions regarding the elective and therapeutic management of lymphatic metastases are made mainly on clinical grounds, which are not very precise because we cannot make predictions from the size and extent of invasion of the primary tumors 5. Therefore, new biomarkers are needed to refine which subtype of primary OSCC is of high metastatic character, and could be tremendously beneficial regarding how to manage OSCC in the clinic.
Phosphoglycerate mutase 1 (PGAM1) is an essential metabolic enzyme in glycolysis that catalyzes the conversion of 3-phosphoglycerate (3-PG) into 2-phosphoglycerate (2-PG) 6. Several studies have shown that PGAM1 is highly expressed in various human cancers, including lung cancer, liver cancer and colon cancer 7-10. Additionally, much attention has been paid to the impact of PGAM1 on tumor growth 11-13 and that the inhibition of PGAM1 activity results in the inhibition of tumor growth and enhancement of cell apoptosis. Moreover, the phosphorylation of PGAM1 (Y26) induced by oncogenic tyrosine kinases, such as FGFR1, EGFR, FLT3 and JAK2, has been shown to activate its activity and promote tumor growth 14. These studies revealed PGAM1 as an important contributor to the tumor development and potentially as a therapeutic target 15, 16.
Thus far, limited studies have focused on the role of PGAM1 in OSCC, and it is unclear whether PGAM1 also plays a critical role in OSCC development. In this study, we analyzed the expression of PGAM1 in clinical samples and performed the correlation analysis of its expression to clinicopathological parameters and found that PGAM1 is closely correlated with poor overall survival (OS) and disease-free survival (DFS). Additionally, high expression of PGAM1 is correlated with lymphatic metastasis and tumor occurrence of OSCC. Intriguingly, we found that PGAM1 is an independent factor for the poor prognosis of patients with OSCC. Mechanistically, we found that PGAM1 could affect the migration of OSCC cells associated with the SRC pathway, which seems to be important for the high metastatic potential of OSCC. Therefore, this study widens the current understanding of the functions of PGAM1 and suggests that PGAM1 may be a biomarker for the management of OSCC for selecting patients with high metastatic potential.
Material and Methods
Patients and specimens
Human paraffin-embedded tissue samples were collected from 122 patients (67 were male and 55 were female), who were examined and treated for OSCC at the Stomatological Hospital of Jiangsu Province, Nanjing, China. Written informed consent from these patients was obtained to use the tissue samples and for follow-up interviews in research. The study was conducted in accordance with the guidelines in the Declaration of Helsinki. The ethical review board (Committee of Ethics of Nanjing Medical University) approved the use of human paraffin-embedded tissues. All of the patients underwent radical resection without any previous radiation or chemotherapy. The tumor grade was classified as poor, moderate, or well differentiated. The pathological stage was defined according to the American Joint Committee on Cancer (AJCC) TNM staging system. Both the tumor grade and pathological stage were evaluated by two pathologists independently. The primary tumor sites included the tongue (n=46), gingiva (n=25), buccal mucosa (n=30), mouth floor (n=5), palate (n=11), and jaw (n=5). The follow-up data were collected through direct interviews with patients or their relatives. At the time of data collection, 28 patients (23%) showed evidence of disease recurrence, and 39 patients (32%) had died of the disease.
Immunohistochemistry (IHC)
The PGAM1 antibody (ab129191, abcam, MA, USA) was used for IHC detection. Briefly, the universal R.T.U. Vectastain Elite ABC Kit (Vector Laboratories, Burlingame, CA, USA) was used. Endogenous peroxidase activity was blocked by incubation with 0.3% hydrogen peroxide. The antigen was retrieved by heating sections in 10 mM sodium citrate (pH 6.0) at 95°C for 30 minutes. The primary antibody PGAM1 was diluted at 1:200 and incubated at 4°C overnight, followed by avidin-biotin-peroxidase complex staining according to the manufacturer's instructions. Negative controls were included by incubating tissue sections with normal mouse serum or phosphate-buffered saline. All of the sections were counterstained with hematoxylin, and the tissue image was captured with a digital camera. PGAM1 expression was evaluated semi-quantitatively as the total immunostaining score, which was calculated as the product of the proportion score and intensity score. Briefly, the proportion score was defined as the fraction of positively stained cells: 1, <5%; 2, 5-10%; 3, 10-50%; 4, 50-75%; 5, >75% of cells stained. The staining intensity was evaluated as follows: 0, no staining signal; 1, weak positive signal; 2, moderate positive signal; 3, strong positive signal. Thus, the total expression score could range from 0 to 15. The classification of high or low expression of PGAM1 is based on regular definition as reported before 17, that is high PGAM1 expression was defined as a total expression score ≥7, and low PGAM1 expression was defined as a total expression score <7. The evaluation was performed independently by two observers, and the average of two readings was used for statistical analysis. The histological images were taken under the OLYMPUS microscope (BX41).
Cell culture
The human oral squamous cell carcinoma cell lines Cal27 and HN12 were generous gifts from Dongxia Ye's Lab in Shanghai Ninth People's Hospital (Shanghai, China). Cal27 and HN12 cell lines were cultured in Dulbecco's modified Eagle medium supplemented with 10% FBS under a humidified atmosphere of 5% CO2 at 37°C.
RNA interference
Short interfering RNAs (siRNAs) were synthesized by Ruibo (Guangzhou, China), and the sequences specifically targeting human PGAM1 were as follows: siRNA1 (5′-CGACUGGUAUUCCCAUUGU TT-3′), siRNA2 (5′-GUCCUGUCCAAGUGUAUCUTT-3′). The non-targeting scrambled siRNA sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. For siRNA delivery, Cal27 or HN12 cells were seeded at 30-50% confluency in 6-cm dishes and were transfected with 20 nM of siRNAs using 5 μL of RNAiMAX (Invitrogen, CA, USA) according to the manufacturer's instructions.
CCK8 assay
The cells were transfected with scrambled siRNA and PGAM1 siRNAs for 48 h, and then aliquots of cells (10,000/well) were seeded into 96-well plates. After 24 h, the cell proliferation rate was measured by adding 10 μl of CCK8 solution (Dojindo, Tokyo, Japan) to each well, followed by incubation at 37°C for 2 h. The absorbance at 450 nm was measured by SPECTRA MAX190 spectrophotometry (Sunnyvale, CA, USA). In each assay, six parallel wells were included, and the results were collected as the mean of three independent experiments.
Cell migration assay and wound-healing assay
For cell migration, Cal27 (2 × 105 cells) and HN12 (1 × 105 cells) were starved in serum- and growth factor-free medium for 12 hours, and aliquots of cells were added to the upper chambers of 24-well Transwell plates (8 μm; Corning Costar Corp, Corning, NY). The bottom chambers were filled with growth medium supplemented with 10% FBS. After incubation for 24 hours at 37°C, non-migrating cells were removed from the upper chamber with a cotton swab. Migrated cells were fixed in ethanol (90%) and were stained with 1% crystal violet solution for 15 minutes at room temperature. Each well was photographed under a light microscope at a magnification of 100×. Fixed cells were solubilized by the addition of 100 μL of 33% acetic acid, and the absorbance was measured at 570 nm using a multiwell spectrophotometer (VERSAmax; Molecular Devices, Sunnyvale, CA). For the transwell assay with compound AZD0530 (Selleckchem, TX, USA), 1 µM AZD0530 was added to the medium before the assay, and PGAM1 was knocked down for 48 h and migrated for 24 h.
For the wound-healing assays, Cal27 and HN12 cells were grown to full confluence in 24-well plates, and the wound was induced by scraping the cell monolayer with a pipette tip of 200 µL and left to heal for 24 h. The assay was performed in triplicate and was repeated twice.
Western blotting
Cells were collected and lysed in RIPA (Beyotime, Jiangsu, China) supplemented with protease inhibitor cocktail (Roche, IN, USA) and then were incubated on ice for 30 min, followed by centrifugation at 12,000 g at 4°C for 30 min. Next, the BCA protein assay kit (Beyotime, Jiangsu, China) was used to determine the concentration of proteins. Equal total protein was loaded into each well for separation. After electrophoresis, the proteins were transferred to a nitrocellulose membrane and were blocked with 5% non-fat milk in TBST (25 mM Tris, 150 mM NaCl, 2 mM KCl, pH 7.4, supplemented with 0.1% Tween 20) for 1 h. Next, primary antibodies (Src (#2018), p-Src (Tyr416) (#6943), FAK (#3285), p-FAK (Tyr397) (#8556), Paxillin (#2542), p-Paxillin (Tyr118) (#2541), GAPDH (#5174)) (Cell Signaling Technology Danvers, MA, USA) and Beta-actin (Abmart) were added and incubated at 4°C overnight. After washing with TBST, horseradish peroxidase-conjugated anti-rabbit IgG or anti-mouse IgG antibody (KangCheng Bio-tech, Shanghai, China) was added and incubated for 1 h at room temperature. The membranes were washed and developed with enhanced chemiluminescence (ECL) (Pierce) and were subsequently exposed to KODAK X-OMAT BT Film (Kodak, Rochester, NY).
Statistical Analysis
The clinicopathological and immunohistochemistry data were analyzed using SPSS statistical software (version 13.0; SPSS Inc, Chicago, IL, USA). The statistical significance among all of these comparisons was examined using Chi-squared analysis or Fisher's exact test. Survival curves were made using the Kaplan-Meier method and were compared by the long-rank test. Additionally, the significance of variables for the survival was analyzed using the Cox proportional hazards model in multivariate analysis. In vitro data were analyzed by One-Way ANOVA, and a value of P<0.05 was considered to be statistically significant.
Results
PGAM1 is associated with lymphatic metastasis and the poor prognosis of OSCC
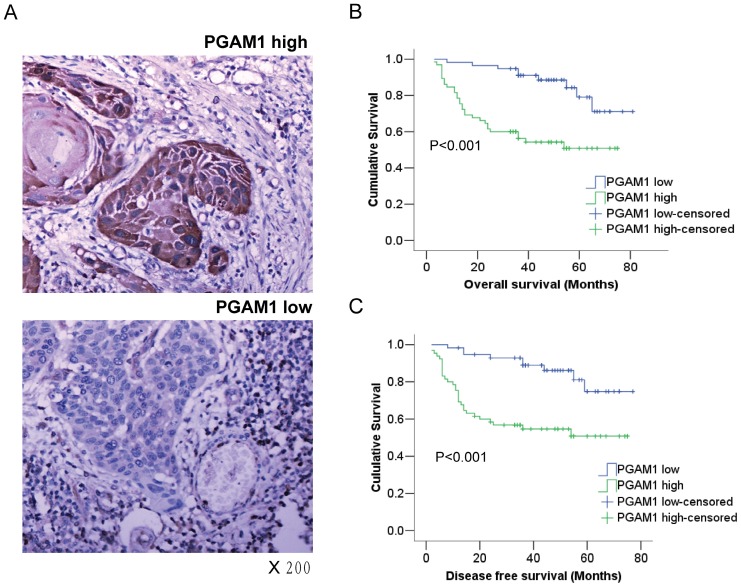
The expression of PGAM1 in OSCC from the tissue samples of 122 patients was examined using immunohistochemistry. According to the immunostaining score, the samples were classified into the PGAM1 high-expression group or the PGAM1 low-expression group (Figure 1A), and PGAM1 was predominantly expressed in the cytosol as expected. The correlation of PGAM1 expression with various clinicopathological features was analyzed, as shown in Table 1, and PGAM1 was highly expressed in 65 (53%) OSCC samples compared with 57 (47%) low-expressed samples. Interestingly, PGAM1 expression was significantly correlated with lymphatic metastasis (P=0.001), tumor recurrence (P=0.028) and age (P=0.029), but it was not correlated with other clinicopathological factors, such as gender, alcohol and cigarette abuse, tumor size, histological grade, TNM classification and distant metastasis.
Figure 1.
PGAM1 expression is closely associated with the poor prognosis of oral squamous cell carcinoma. A) Representative images of high-expression sample and low expression sample stained for PGAM1 (brown) using immunohistochemistry. The sections were counterstained with hematoxylin (blue). B) PGAM1 expression is significantly correlated with poor overall survival in OSCC patients. C) PGAM1 expression is significantly correlated with poor disease-free survival in OSCC patients. The data were analyzed by Kaplan-Meier analysis.
Table 1.
Correlation between PGAM1 expression and clinicopathological parameters (n=122)
| Category | Subcategory | PGAM1 low expression (n=57) | PGAM1 high expression (n=65) | P value |
|---|---|---|---|---|
| Gender | Male | 31 | 36 | 0.913 |
| Female | 26 | 29 | ||
| Age (y) | ≤ 60 | 35 | 27 | 0.029* |
| > 60 | 22 | 38 | ||
| Alcohol and cigarette abuse | Positive | 14 | 11 | 0.301 |
| Negative | 43 | 54 | ||
| Tumor size (cm) | ≤4 | 39 | 48 | 0.310 |
| >4 | 18 | 17 | ||
| Histological grade | Well | 33 | 37 | 0.550 |
| Moderate | 19 | 23 | ||
| Poor | 3 | 5 | ||
| TNM classification (1-4) | Class 1 | 5 | 8 | 0.185 |
| Class 2 | 28 | 18 | ||
| Class 3 | 16 | 25 | ||
| Class 4 | 8 | 14 | ||
| Lymphatic metastasis | Positive | 14 | 36 | 0.001* |
| Negative | 43 | 29 | ||
| Distant metastasis | Positive | 3 | 8 | 0.178 |
| Negative | 54 | 57 | ||
| Tumor recurrence | Positive | 8 | 20 | 0.028* |
| Negative | 49 | 45 |
*Statistically significant difference.
Kaplan-Meier survival analysis was conducted to determine the prognostic significance of PGAM1 expression. Intriguingly, it showed that PGAM1 high-expression patients had a worse overall survival (OS) and disease-free survival (DFS) than PGAM1 low-expression patients (P<0.001, Figure 1B, 1C). Furthermore, multivariate Cox regression analysis was performed to evaluate the independent prognostic factors of OS and DFS, including gender, age, histological grade, tumor size and PGAM1 expression. As shown in Table 2, PGAM1 expression was an independent factor of OS (HR, 3.427; 95%CI, 1.579-7.440; P=0.002) and DFS (HR, 3.264; 95%CI, 1.509-7.057; P=0.003), while other factors were not, such as gender, age, histological grade and tumor size. These data demonstrated that the high expression of PGAM1 is strongly associated with the poor prognosis of patients with OSCC.
Table 2.
Multivariate analysis of factors with OS and DFS
| Parameter | Overall survival | Disease-free survival | |||||
|---|---|---|---|---|---|---|---|
| Category | Subcategory | P | HR | 95% CI | P | HR | 95% CI |
| Gender | Male vs Female | 0.147 | 0.597 | 0.298-1.198 | 0.187 | 0.628 | 0.315-1.254 |
| Age (y) | ≤ 60 vs > 60 | 0.849 | 0.938 | 0.485-1.812 | 0.913 | 0.964 | 0.498-1.865 |
| Histological grade | Well vs Poor | 0.259 | 1.306 | 0.822-2.076 | 0.320 | 1.263 | 0.797-2.002 |
| Tumor size (cm) | ≤4 vs>4 | 0.104 | 0.507 | 0.223-1.151 | 0.114 | 0.514 | 0.225-1.174 |
| PGAM1 expression | Low vs high | 0.002* | 3.427 | 1.579-7.440 | 0.003* | 3.264 | 1.509-7.057 |
*Statistically significant difference.
Knocking down of PGAM1 decreased the mobility of OSCC cells
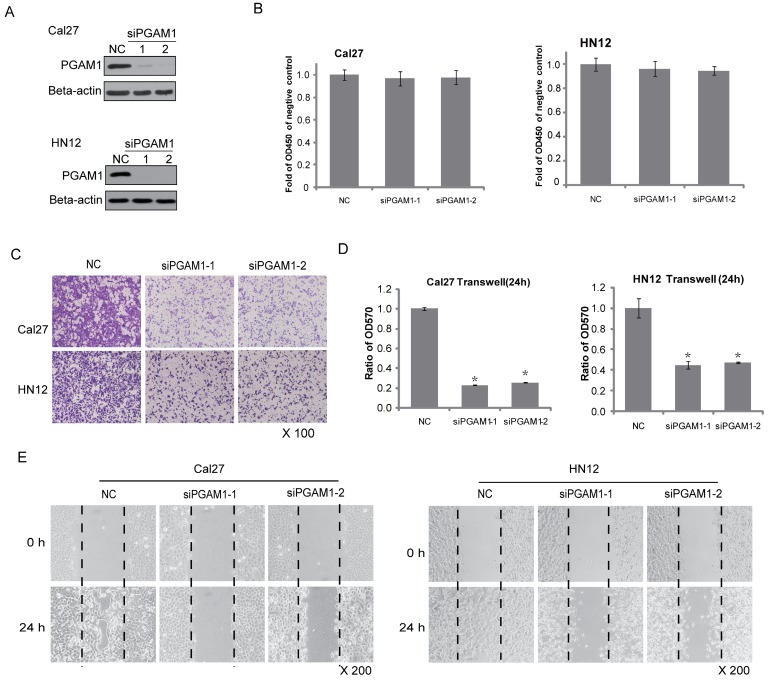
Because PGAM1 is correlated with lymphatic metastasis, it might participate in cancer cell migration. To test this hypothesis, two OSCC cell lines, Cal27 and HN12, were used. PGAM1 was knocked down by siRNA for 48 h in both Cal27 and HN12 cells (Figure 2A), and cell proliferation was determined by plating aliquots of cells from each group on 96-well plates for an additional 24 h. No difference was found in the cell proliferation between the scrambled group and the PGAM1-knocked down group in both Cal27 and HN12 cells (Figure 2B). In the same setting, the cell migration was determined in parallel using the transwell assay. A remarkable decrease in cell migration was shown after knocking down PGAM1 (Figure 2C), and the ratios of the migrated cells in the PGAM1-knocked down groups versus that in the control groups were significantly reduced as only approximately 20% of Cal27 cells and 40% of HN12 cells (Figure 2D). Moreover, the wound healing assay was also exploited to test the cell mobility, showing that the migration of both Cal27 cells and HN12 cells was dramatically decreased after knocking down PGAM1 (Figure 2E), a finding that is consistent with the above result. Collectively, these data demonstrated that PGAM1 plays a critical role in migration of OSCC cells, but not cell proliferation.
Figure 2.
Decreased cell migration after knocking down PGAM1. A) Verification of the knock down efficiency of PGAM1 (48 h) by western blotting in both Cal27 and HN12 cells. B) Cell proliferation (24 h) does not show significant alteration after knocking down PGAM1 in both Cal27 and HN12 cells. C) Cell migration (24 h) was dramatically decreased after knocking down PGAM1 in both Cal27 and HN12 cells. D) Relative quantification of the above images by OD measurement of solubilized crystal violet dye. *P<0.05, as compared to NC group. E) Cell mobility was decreased after knocking down of PGAM1 in both Cal27 and HN12, as determined by the wound-healing assay. NC represents scrambled siRNA, and siPGAM1-1 and siPGAM1-2 represent two different PGAM1 siRNAs. The data were from three independent experiments.
Knocking down of PGAM1 decreased the phosphorylation of SRC, FAK and Paxillin
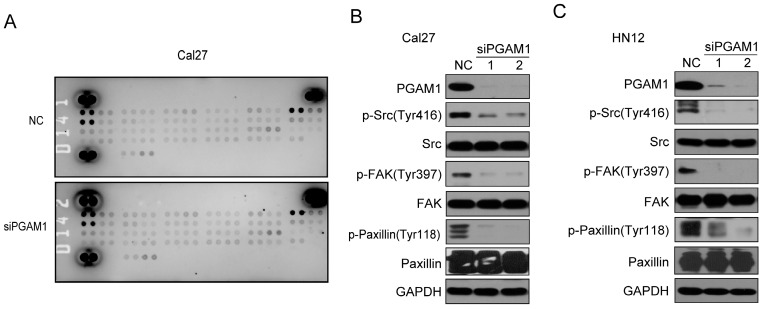
To study the mechanism by which PGAM1 regulates the migration of OSCC cells, the Human Phospho-RTK Array was applied to explore the alteration of signaling pathways, including the EGFR, FGFR, HGFR, PDGFR, IGF1R, IR, FLT-3, ROR, ALK, EphA and EphB (Supplementary material); unfortunately, we found no changes in all these molecules (Figure 3A). Thus, we suppose that some classical signaling pathways might change, such as the Paxillin/FAK/SRC signaling pathway, which is critical for the acquisition of metastatic potential in cancer cells. To this end, we knocked down the expression of PGAM1 in Cal27 cells and found a remarkable decrease in the phosphorylation of SRC, FAK and Paxillin after the downregulation of PGAM1 (Figure 3B). To determine whether this is a unique phenomenon that occurs in Cal27 cells, we also knocked down the expression of PGAM1 in another OSCC cell line, HN12, and found that the phosphorylation of SRC, FAK and Paxillin were also decreased after the depletion of PGAM1 (Figure 3C). These results suggested that the Paxillin/FAK/SRC signaling pathway may be involved in PGAM1-mediated cell migration.
Figure 3.
Phosphorylation of SRC, FAK and Paxillin is decreased after knocking down PGAM1. A) Analysis of the phosphorylation of receptor tyrosine kinase using the proteome array profiler, and no obvious alteration was detected between siPGAM1 and NC. B) Phosphorylation of SRC, FAK and Paxillin was decreased after knocking down PGAM1 for 72 h in Cal27 cells. C) Phosphorylation of SRC, FAK and Paxillin was decreased after knocking down PGAM1 for 72 h in Cal27 cells.
SRC inhibitor abolished the effect of PGAM1 on cell migration
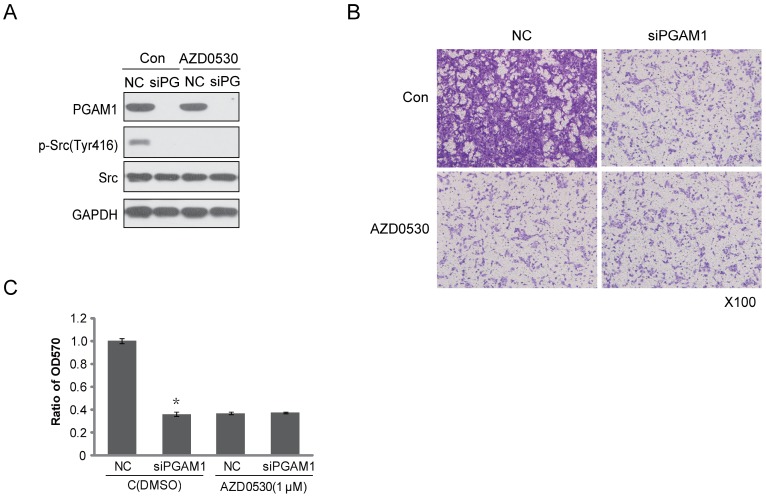
To identify the effect of PGAM1 on migration through the Paxillin/FAK/SRC pathway, the SRC inhibitor AZD0530 was used to test whether it could block the PGAM1-mediated cell migration. To this end, Cal27 cells were pretreated with AZD0530 to block SRC phosphorylation and then PGAM1 siRNA was used to knock down the expression of PGAM1 and determine whether cell migration was further decreased. As shown in Figure 4A, SRC phosphorylation was inhibited by AZD0530, and knocking down PGAM1 could not further decrease of the migration of Cal27 cells (Figure 4B); thus, knocking down PGAM1 did not have an additional effect on cell migration after pretreatment with the SRC inhibitor (Figure 4C). This result suggests that the SRC pathway may involve in PGAM1-mediated cell migration.
Figure 4.
SRC phosphorylation is required for PGAM1-mediated cell migration. A) Verification of the inhibition effect of AZD0530 and PGAM1 on SRC phosphorylation in Cal27 cells. B) Cell migration was decreased by AZD0530 or PGAM1 knock down, while pretreatment with AZD0530 abolished a further decrease in cell migration when PGAM1 was knocked down. C) Relative quantification of the above images by OD measurement of solubilized crystal violet dye. *P<0.05, as compared to NC group.
Discussion
In this study, we investigated the role of PGAM1 in OSCC and showed that PGAM1 is an adverse indicator of OS and DFS. Moreover, high expression of PGAM1 was closely correlated with the lymphatic metastasis and tumor occurrence of OSCC, which might account for the poor prognosis of patients. Intriguingly, multivariate Cox regression analysis showed that PGAM1 was an independent factor of OS and DFS but that age, gender, histological grade and tumor size were not. Collectively, these clinical data suggested that PGAM1 may play an important role in the tumor metastasis of OSCC. As such, we used the OSCC cell lines Cal27 and HN12 to investigate whether PGAM1 could affect the cell migration. Indeed, we found that PGAM1 is essential for the migration of both Cal27 and HN12 cells.
This finding suggested that cell migration may be more critical for PGAM1 involved in the development of OSCC, which is different from previous reports that PGAM1 mainly participates in tumor growth in other tumor types because it provides a metabolic advantage for cancer cell growth. For instance, PGAM1 could coordinate glycolysis and anabolic biosynthesis by controlling the intracellular levels of 3-PG and 2-PG in lung cancer 13. Additionally, both 3-PG and 2-PG have additional biological functions, and 3-PG could inhibit glucose-6-phosphate dehydrogenase activity and consequently the decrease in the oxidative pentose phosphate pathway and anabolic biosynthesis. The decrease in 2-PG leads to decreased PHGDH activity, which facilitates 3-PG accumulation and, in turn, permits high levels of PPP and biosynthesis. PGAM1 was also identified as a novel therapeutic target in hepatocellular carcinoma using quantitative proteomic analysis of human hepatocellular samples, and the shRNA-mediated repression of PGAM1 resulted in the inhibition of liver cancer cell growth both in vitro and in vivo 12. By contrast, our study suggested that cell migration may be more critical for PGAM1 involved in OSCC, but not cell proliferation.
Furthermore, we investigated the mechanism by which PGAM1 regulates cancer cell migration. We screened a panel of receptor tyrosine kinases, EGFR, FGFR, HGFR, PDGFR, IGF1R, IR, FLT-3, ROR, ALK, EphA and EphB, however, unfortunately none were found to be changed after knocking down PGAM1. Thus, we supposed that classical signaling pathways that are involved in cell migration may change, and we found that the phosphorylation of the Paxillin/FAK/SRC signaling pathway was decreased after knocking down PGAM1, and pretreatment with a SRC inhibitor could block the effect of PGAM1 on cell migration. Taken together, these results suggested that the SRC pathway mediates the effect of PGAM1 on cell migration, which might account for the lymphatic metastasis and poor prognosis of patients with OSCC.
PGAM1 is known as a metabolic enzyme in glycolysis, but how could it affect the migration of OSCC cells? There may be several factors involved. First, PGAM1 may have non-metabolic functions that regulate critical proteins involved in promoting cell migration. As an increasing number of discoveries of non-metabolic functions of metabolic enzymes, such as pyruvate kinase M2, which works as a protein kinase, and protein-protein interactions that regulate chromosome segregation, the cell cycle, gene transcription and tumorigenesis 18-21. Indeed, as shown in previous study, PGAM1 interacts with α-smooth muscle actin (ACTA2) to promote the migration of breast cancer cells independent of its metabolic activity 22, which needs to be further identified in this system. Second, there may be interactions between PGAM1 and upstream signaling proteins such as membrane proteins or structural proteins or transcription factors, which regulate downstream mobility-related proteins. It is possible that several factors may contribute to the migration of oral cancer cells, which depends on the cellular context. Although the direct mechanism by which PGAM1 regulates cell migration in OSCC was not disclosed in this study, the signaling pathway in OSCC induced by PGAM1 occurs, at least in part, through the SRC pathway. Besides, other protein-protein interactions or upstream signaling of SRC may also be affected, while these hypotheses need to be further tested. We have some limitations in this study, such as the prognostic value of PGAM1 in OSCC has not yet been validated in independent clinical samples, and the mechanistic insight of PGAM1 in cell migration was not fully demonstrated. Therefore, further studies are required to validate the prognostic value of PGAM1 in prospective studies, as well as the in-depth study of the unique mechanism of PGAM1 in the migration of OSCC, and which would provide full understanding of its role in OSCC, and it is possible PGAM1 would be used as a biomarker in selection of OSCC patients of high metastatic potential in future.
Conclusions
This study demonstrated that the high expression of PGAM1 is closely correlated with lymphatic metastasis and the poor prognosis of OSCC, and it is an independent risk factor for OS and DFS. Knocking down of PGAM1 decreased the migration of OSCC cells, and the mechanism by which PGAM1 regulates cell migration is associated with the SRC pathway. Thus, we displayed a novel function of PGAM1 in regulating cell migration in OSCC which is different from the known function as a metabolic enzyme reported previously. Besides, these findings also suggest that PGAM1 may be useful as a biomarker for the management of OSCC in the clinic in future.
Supplementary Material
Supplementary information.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81202549) and the “Personalized Medicines——Molecular Signature-based Drug Discovery and Development”, Strategic Priority Research Program of the Chinese Academy of Sciences, Grant No. XDA12020341.
Conceived and designed the experiments: ZQX, HWL, MH and MYG. Performed the experiments:
DDZ, HMW and XMZ. Analyzed the data: ZQX, HMW and XD. Wrote the paper: ZQX, HMW and HWL.
Abbreviations
- OSCC
Oral squamous cell carcinoma
- OS
overall survival
- DFS
disease free survival
- 3-PG
3-phosphoglycerate
- 2-PG
2-phosphoglycerate.
References
- 1.Chen YJ, Lin SC, Kao T, Chang CS, Hong PS, Shieh TM. et al. Genome-wide profiling of oral squamous cell carcinoma. J Pathol. 2004;204:326–32. doi: 10.1002/path.1640. [DOI] [PubMed] [Google Scholar]
- 2.Woolgar JA, Rogers S, West CR, Errington RD, Brown JS, Vaughan ED. Survival and patterns of recurrence in 200 oral cancer patients treated by radical surgery and neck dissection. Oral Oncol. 1999;35:257–65. doi: 10.1016/s1368-8375(98)00113-4. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 4.Sasahira T, Ueda N, Yamamoto K, Kurihara M, Matsushima S, Bhawal UK. et al. Prox1 and FOXC2 act as regulators of lymphangiogenesis and angiogenesis in oral squamous cell carcinoma. PLoS One. 2014;9:e92534. doi: 10.1371/journal.pone.0092534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sano D, Gule MK, Rosenthal DI, Bell D, Yates J, El-Naggar AK. et al. Early postoperative epidermal growth factor receptor inhibition: safety and effectiveness in inhibiting microscopic residual of oral squamous cell carcinoma in vivo. Head Neck. 2013;35:321–8. doi: 10.1002/hed.22961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fothergill-Gilmore LA, Watson HC. The phosphoglycerate mutases. Adv Enzymol Relat Areas Mol Biol. 1989;62:227–313. doi: 10.1002/9780470123089.ch6. [DOI] [PubMed] [Google Scholar]
- 7.Buhrens RI, Amelung JT, Reymond MA, Beshay M. Protein expression in human non-small cell lung cancer: a systematic database. Pathobiology. 2009;76:277–85. doi: 10.1159/000245893. [DOI] [PubMed] [Google Scholar]
- 8.Shen J, Wang W, Wu J, Feng B, Chen W, Wang M. et al. Comparative proteomic profiling of human bile reveals SSP411 as a novel biomarker of cholangiocarcinoma. PLoS One. 2012;7:e47476. doi: 10.1371/journal.pone.0047476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li C, Xiao Z, Chen Z, Zhang X, Li J, Wu X. et al. Proteome analysis of human lung squamous carcinoma. Proteomics. 2006;6:547–58. doi: 10.1002/pmic.200500256. [DOI] [PubMed] [Google Scholar]
- 10.Durany N, Joseph J, Campo E, Molina R, Carreras J. Phosphoglycerate mutase, 2,3-bisphosphoglycerate phosphatase and enolase activity and isoenzymes in lung, colon and liver carcinomas. Br J Cancer. 1997;75:969–77. doi: 10.1038/bjc.1997.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans MJ, Saghatelian A, Sorensen EJ, Cravatt BF. Target discovery in small-molecule cell-based screens by in situ proteome reactivity profiling. Nat Biotechnol. 2005;23:1303–7. doi: 10.1038/nbt1149. [DOI] [PubMed] [Google Scholar]
- 12.Ren F, Wu H, Lei Y, Zhang H, Liu R, Zhao Y. et al. Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma. Mol Cancer. 2010;9:81. doi: 10.1186/1476-4598-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hitosugi T, Zhou L, Elf S, Fan J, Kang HB, Seo JH. et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell. 2012;22:585–600. doi: 10.1016/j.ccr.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hitosugi T, Zhou L, Fan J, Elf S, Zhang L, Xie J. et al. Tyr26 phosphorylation of PGAM1 provides a metabolic advantage to tumours by stabilizing the active conformation. Nat Commun. 2013;4:1790. doi: 10.1038/ncomms2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang X, Sun Q, Li H, Li K, Ren X. The role of phosphoglycerate mutase 1 in tumor aerobic glycolysis and its potential therapeutic implications. Int J Cancer. 2014;135:1991–6. doi: 10.1002/ijc.28637. [DOI] [PubMed] [Google Scholar]
- 16.Guo S, Zou J, Wang G. Advances in the proteomic discovery of novel therapeutic targets in cancer. Drug Des Devel Ther. 2013;7:1259–71. doi: 10.2147/DDDT.S52216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun W, Zhang X, Ding X, Li H, Geng M, Xie Z. et al. Lactate dehydrogenase B is associated with the response to neoadjuvant chemotherapy in oral squamous cell carcinoma. PLoS One. 2015;10:e0125976. doi: 10.1371/journal.pone.0125976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Y, Li X, Yang W, Hawke DH, Zheng Y, Xia Y. et al. PKM2 Regulates Chromosome Segregation and Mitosis Progression of Tumor Cells. Mol Cell. 2014;53:75–87. doi: 10.1016/j.molcel.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase m2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W. et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–22. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R. et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–44. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang D, Jin N, Sun W, Li X, Liu B, Xie Z. et al. Phosphoglycerate mutase 1 promotes cancer cell migration independent of its metabolic activity. Oncogene. 2017;36:2900–9. doi: 10.1038/onc.2016.446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.