Abstract
The B cell developmental pathway represents a leading system for the analysis of regulatory circuits that orchestrate cell fate specification and commitment. Considerable progress has been achieved within the past decade in the identification and genetic analysis of various regulatory components. These components include the transcription factors PU.1, Ikaros, Bcl11a, E2A, EBF, and Pax-5, as well as the cytokine receptors Flk2 and IL-7R. Experimental evidence of connectivity among the regulatory components is used to assemble sequentially acting and contingent gene regulatory networks. Transient signaling inputs, self-sustaining positive feedback loops, and crossantagonism among alternate cell fate determinants are key features of the proposed networks that instruct the development of B lymphocyte precursors from hematopoietic stem cells.
Keywords: B lymphopoiesis, cell fate determination, hematopoiesis, transcription factors, cytokine signaling
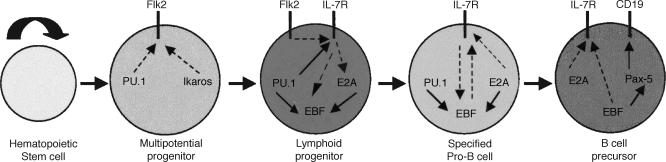
The hematopoietic system represents an excellent developmental model for exploring how a well defined multipotential stem cell gives rise to various cell types of the blood and immune system, including erythrocytes, megakaryocytes, myeloid cells (macrophages and granulocytes), and lymphocytes. Reverse genetics targeting transcription factors and signaling proteins, as well as the characterization of mutations in genes that result in leukemias, have enabled the analysis of a large set of regulatory molecules that control the generation of one or more lineages of the hematopoietic system. These regulators can be viewed as components of complex gene regulatory networks, which orchestrate cell fate specification, commitment, and differentiation. This article attempts to assemble sequentially acting and contingent gene regulatory networks, which instruct the development of B lymphocyte precursors from hematopoietic stem cells (HSCs). The B lineage is particularly well suited for such a synthesis of sequentially acting regulatory circuits. The B cell pathway can be modeled as a series of discrete developmental states (Fig. 1) that correspond to isolatable progenitors or precursors (1). Each state can be described on the basis of a unique constellation of cell surface markers and/or gene expression patterns, as well as Ig gene rearrangements. Finally, sufficient numbers of regulatory components have been analyzed by loss- and gain-of-function experiments (2) that it is now possible to examine their connectivity so as to link them as components of coherent regulatory networks.
Fig. 1.
Gene regulatory networks that direct the generation of a B cell precursor from a HSC. Four successive, interdependent developmental states are depicted. Each transition involves distinct combinations of regulatory molecules: gene regulatory proteins (e.g., PU.1) and signaling receptors (e.g., IL-7R). Gene regulators activate or repress target genes, whereas signaling receptors induce or modify the activities of gene regulators. CD19 is a B lineage-specific cell surface protein (Ig coreceptor component) that signifies commitment to the B cell fate. Regulatory connections are shown as dashed or solid arrows, depending on the strength of the supportive experimental evidence.
Transcriptional Control of Hematopoiesis: Rules of the Game
Before detailing gene regulatory networks that underlie B lymphocyte development, we elaborate a general framework by which transcriptional regulatory proteins specify distinct cell fates within the hematopoietic system. This framework is inferred from numerous experiments involving the manipulation of individual regulatory proteins and the analysis of gene expression patterns within single hematopoietic cells representing distinct multipotential progenitors (MPPs). (i) Each cell fate is specified by a unique combinatorial code of transcription factors (e.g., PU.1, Ikaros, Bcl11a, E2A, EBF, and Pax-5) that determine the B cell fate (3). The combinatorial code includes transcription factors that function in a hierarchical as well as combinatorial manner (see below). The level and activity state of a transcription factor is assigned a unique value within the combinatorial code; e.g., the B cell fate depends on low level/activity of PU.1, whereas the macrophage fate requires high level/activity of PU.1 (4). (ii) Individual HSCs and MPPs exhibit low levels of mixed lineage patterns of gene expression (5). It follows that cell fate specification involves the activation of lineage-appropriate subsets of genes and the concerted repression of lineage-inappropriate subsets (6). (iii) The mixed lineage developmental states may be dictated by heterogeneous sets of primary cell fate determinants (transcriptional regulators), which are simultaneously active (low expression/activity states) in individual MPPs. Cell fate specification is likely initiated by the induction of a primary cell fate determinant (induced expression/activity) and its antagonism of alternate lineage-determining transcription factors (7), which enables the eventual resolution of a mixed lineage pattern of gene expression into one specific for a given lineage.
Regulatory Circuits Promoting Specification of the B Cell Fate
Two cytokine receptors (Flk2/Flt3 and IL-7R) and six transcription factors (PU.1, Ikaros, E2A, Bcl11a, EBF, and Pax-5) are critical for the development of B cell precursors (8–16). These proteins represent key components of sequentially acting regulatory circuits detailed in Fig. 1. The circuits are shown to be operative in discrete developmental states that correspond to defined cellular intermediates. Such cells can be resolved and isolated by flow cytometry from the fetal liver or bone marrow, which are the sites of embryonic and adult B lymphopoiesis, respectively (1).
Specification of the B cell fate involves the expression of a set of B lineage-specific genes, including mb-1, B29, λ5, and VpreB, and the onset of ordered DNA rearrangements of the Ig heavy chain (IgH) locus catalyzed by the recombinase complex consisting of Rag-1 and Rag-2 (1). The mb-1, B29, λ5, and VpreB genes encode components of the preB cell antigen receptor (preBCR), which is assembled upon productive rearrangement and expression of the IgH locus. The mb-1 and B29 gene products represent signaling subunits of the preB and B cell receptors (BCRs). The λ5 and VpreB proteins are surrogate light chains, which are transiently expressed during B cell development and associate with the heavy chain protein to form the preBCR. The preBCR executes a crucial developmental checkpoint by promoting the survival and expansion of B cell precursors that have productively rearranged an IgH locus. It also regulates the differentiation of preB cells into mature B cells, which entails rearrangement and expression of conventional Ig light chain loci. As a consequence, B cells assemble and express the Ig molecule on their cell surface and use it as an antigen receptor (BCR).
B lineage and all other immune cells are derived from a rare population of HSCs. Considerable progress is being made in analyzing the molecular circuitry that instructs stem cell self-renewal versus differentiation. Upon exiting the stem cell niche, HSCs give rise to MPPs (Fig. 1). Although MPPs can generate all of the blood lineages, they are unable to self-renew. Expression of the cell surface protein CD27 on MPPs permits their separation from HSCs by flow cytometry (17). Intriguingly, HSCs express at low levels many lineage-specific genes reflecting a developmentally poised state (18). Single-cell RT-PCR analyses have demonstrated that HSCs and MPPs exhibit broader multilineage gene expression patterns than their progeny, the lymphoid and myeloid progenitors, which express more restricted sets of genes (19). The molecular mechanisms by which MPPs restrict and refine their gene expression patterns to generate lineage-specified progenitors appear to involve crossantagonism between opposing subsets of lineage-determining transcription factors (see below).
The earliest regulatory event that appears to trigger B cell development is the expression of the receptor tyrosine kinase Flk2/Flt3 within a subset of MPPs (Fig. 1). Expression of Flk2/Flt3 within the MPP population is associated with loss of stem cell and myeloid lineage potential but sustained lymphoid reconstitution in vivo (20). Furthermore, targeted inactivation of the Flk2/Flt3 gene results in a severe deficiency in the generation of B lineage progenitors (15). Consistent with the requirement for Flk2/Flt3 signaling in the development of B lineage progenitors is the significant decrease in common lymphoid progenitors (CLPs) observed in mice deficient in the Flk2/Flt3 ligand (21). Taken together, these results strongly suggest that specification of the B lymphoid cell fate initiates within the MPP population as a consequence of expression of Flk2/Flt3.
The signaling pathway through which Flk2/Flt3 selectively favors the generation of B lineage progenitors is unknown, but in vitro data suggest that activation of this receptor promotes expression of the IL-7 receptor (IL-7R) (22). The development of pro-B cells in the bone marrow requires signaling through IL-7R (23). Furthermore, in a culture system, IL-7R signaling is sufficient to induce the differentiation of CLPs into pro-B cells. Importantly, two recent studies have shown that combined loss of Flk2/Flt3 and IL-7R results in a complete failure to develop B lineage cells during both fetal and adult hematopoiesis (24, 25). These results raise the strong possibility that signaling through these two cytokine receptors may activate the expression or modulate the activity of key transcriptional regulators such as E2A and/or EBF, which are required for specification of the B cell fate (see below).
Analysis of the regulation of Flk2/Flt3 and IL-7Rα gene expression is likely to provide insight into the earliest circuitry underlying B cell development. The transcription factors PU.1 and Ikaros appear to regulate expression of these two cytokine receptor genes. Ikaros null or Ikaros DN–/– hematopoietic progenitors are deficient in expression of Flk2/Flt3 (26). PU.1–/– fetal liver hematopoietic progenitors also exhibit reduced Flk2/Flt3 transcripts (27). In addition to the defect in Flk2/Flt3 expression, PU.1–/– fetal liver hematopoietic progenitors are impaired in expression of IL-7R. PU.1 is implicated in directly regulating the transcription of the IL-7Rα-chain gene (27). Thus, the severe reduction in B lymphoid progenitors caused by the PU.1–/– mutation is likely due to failed expression of both the Flk2/Flt3 receptor and IL-7R (3). Collectively, these data suggest that expression of Flk2/Flt3 in MPPs may depend on the concerted activities of Ikaros and PU.1, whereas PU.1 and Flk2 signaling may drive the subsequent expression of IL-7R in CLPs (Fig. 1).
The transcription factors E2A and EBF are required for specification of the B cell fate and therefore are designated as primary cell fate determinants. They regulate the early program of B lineage gene expression (mb-1, B29, λ5, VpreB, and Rag-1,2) and DNA rearrangements of the IgH locus (Fig. 2). The E2A gene encodes two basic helix–loop–helix proteins, E12 and E47, generated by differential splicing, whose expression and activities are induced during early B cell development (28, 29). EBF is an atypical helix–loop–helix zinc finger protein that is expressed exclusively in the B lineage within the hematopoietic system (30). Targeted inactivation of the E2A or EBF gene results in a block in B cell development before the onset of early B lineage gene expression and the initiation of D-J rearrangements at the IgH locus (9, 11, 14). E2A/EBF compound-mutant heterozygotes display a more severe defect in B lymphopoiesis than the single-heterozygous animals, suggesting that the two regulators may function synergistically to activate transcription of B lineage genes (31). Consistent with this possibility, ectopic expression of E2A and EBF results in activation of the λ5 and VpreB genes. E2A or EBF can also induce D-JH DNA rearrangements in a non-B cell line when expressed with the recombinase proteins Rag-1 and Rag-2, suggesting direct roles for these transcription factors in IgH recombination (32). Regulation of D-JH rearrangements in the IgH locus likely depends on E2A binding sites in the intron enhancer. This enhancer regulates transcription from the constant region of the unrearranged IgH locus, as well as D-JH rearrangements (33). Loss of E2A activity results in severely diminished transcription initiated within the IgH intron enhancer and D-JH recombination (9). EBF appears to directly regulate the transcription of the B lineage genes, mb-1, λ5, and VpreB, by binding to functionally important sites in their promoter regions (31). Thus, the specification of the B cell fate appears to be regulated by two primary cell fate determinants, E2A and EBF. These regulatory proteins not only promote B cell differentiation, but also appear to be important in restricting alternate developmental fates. E2A–/– hematopoietic cells can be expanded in culture by using the cytokines SCF (stem cell factor), FL, and IL-7. They phenotypically resemble early B lineage progenitors but retain multilineage developmental potential (34). As is the case for the E2A mutation, EBF–/– early B lineage progenitors can be stably propagated on stromal cells in the presence of SCF, FL, and IL-7 (K.L.M. and H.S., unpublished data). These EBF–/– progenitors retain the ability to generate T-lymphoid and myeloid progeny. Cell fate restriction by E2A and EBF is likely initiated within the context of a CLP and culminated in a pro-B cell by the transcription factor Pax-5 (see below).
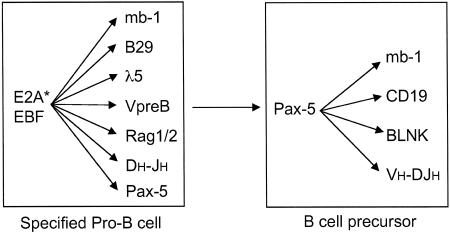
Fig. 2.
Gene expression programs activated in specified pro-B cells and their differentiated progeny (B cell precursors). E2A-, EBF-, and Pax-5-regulated genes and IgH locus DNA rearrangements are shown. Pax-5 expression is contingent on E2A and EBF. Pax-5 reinforces and expands the B lineage program of gene expression. E2A* indicates E2A activation.
From a regulatory standpoint, it is of considerable interest to determine the means by which the expression and/or activity of E2A and EBF are induced in CLPs. E2A transcripts are expressed in the stem cell compartment, although transcriptionally active complexes of E47 homodimers or E47/E12 heterodimers are detected primarily in B lineage progenitors (33). Analysis of mice expressing an E2A-GFP knock-in allele has shown that E2A expression in B lineage cells is up-regulated concomitant with induction of EBF and initiation of D-JH recombination (35). At present, it remains to be determined how E2A expression and activity are induced. We suggest that signaling through the IL-7R induces the activity of E2A proteins (Fig. 1). It should be noted that Notch signaling and inhibitory basic helix–loop–helix proteins have been suggested to antagonize E2A activity (33). The Notch pathway may inhibit transactivation by E2A proteins (36) and/or target them for degradation (37). Thus, relief from these inhibitory pathways may also contribute to the developmental induction of E2A activity (Fig. 1). We propose that induction of E2A activity is initiated in the CLP, thereby enabling Rag and EBF gene expression as well as D-JH gene rearrangements. EBF transcripts are first detectable in a subset of CLPs that express both Rag-1 and Rag-2 and contain D-JH rearrangements (38). Although EBF transcript levels are low in this population, they likely enable sufficient levels of protein accumulation to promote Rag gene expression and IgH rearrangement in conjunction with E2A. A functional binding site for E2A has been found in the EBF promoter, suggesting that E2A is directly involved in regulation of EBF expression (39). Recently, it was shown that EBF expression is impaired in E47–/– fetal liver hematopoietic progenitors (40). EBF expression is also compromised in PU.1–/– fetal liver hematopoietic progenitors, suggesting that PU.1 also participates in regulation of the EBF gene. PU.1 binds in vitro and in vivo to a conserved site within the first intron of the EBF gene (3). Thus, we suggest that induction of the EBF gene depends on activation of E2A and a regulatory input from PU.1 (Fig. 1). Finally, the developmental induction of E2A gene expression, detected by the E2A-GFP knock-in allele, is compromised in EBF–/– mice, raising the possibility that EBF may be a component of a feedback loop that is used to sustain higher levels of E2A expression in pro-B cells (35).
Given that EBF expression in vivo is contingent on PU.1 and E2A, functional bypass experiments have been pursued to determine whether restoration of EBF expression enables B cell development in the absence of PU.1 or E2A. Retroviral transduction of EBF can rapidly restore the generation of B cell precursors from PU.1–/– fetal liver hematopoietic progenitors (3). Surprisingly, restoration of EBF expression in E47–/– fetal liver hematopoietic progenitors also promotes the generation of B cell precursors (40). This result suggests that EBF can function synergistically with low levels of alternative E-proteins to regulate the early B lineage program of gene expression.
EBF in concert with E2A regulates expression of a secondary B cell fate determinant, Pax-5 (41) (Fig. 1). In contrast to E2A and EBF, the transcription factor Pax-5 is not required for specification of the B cell fate but is essential for commitment. Pax-5–/– pro-B cells express early B lineage genes and undergo DH-JH and proximal VH-DJH gene rearrangements (12). However, unlike their wild-type counterparts, Pax-5–/– pro-B lines exhibit extensive developmental plasticity. They can give rise to T-lymphoid, myeloid, and even erythroid progeny upon transplantation (42, 43). Pax-5 antagonizes the T cell fate by down-regulating the expression of Notch-1 (44). Notch signaling is required for the induction of T cell development in the thymus (45). Not only is Pax-5 required for commitment to the B lineage and suppression of alternative cell fates, but it is also needed for maintenance of B cell identity (46). Pax-5 actively and continuously represses the expression of various myeloid genes, including M-CSFR (macrophage colony-stimulating factor receptor) in B lineage cells. The conditional deletion of Pax-5 in B lineage cells results in the misexpression of myeloid genes. The mechanistic basis of Pax-5-mediated inhibition of myeloid gene expression remains to be elucidated but could involve antagonism of the myeloid transcription factor C/EBPα (see below). Pax-5 also plays an essential role in reinforcing and expanding the early program of B lineage gene expression (Fig. 2). Pax-5 target genes include mb-1, CD19, and BLNK, all of which encode lineage-specific antigen receptor or coreceptor signaling components (43). In addition to regulating B lineage gene expression, Pax-5 also promotes distal VH-DJH gene rearrangements (47). Thus, Pax-5 is dispensable for specification of the B cell fate but is required for elaboration of the B cell developmental program, as well as lineage commitment and maintenance of identity. On this basis, it is proposed to represent a secondary cell fate determinant.
We note that in addition to the aforementioned regulators, another transcription factor, Bcl11a, is required for the generation of B lineage progenitors. Bcl11a is a Kruppel-related zinc finger protein expressed in multiple lineages, including hematopoietic progenitors (48). The Bcl11a gene was originally identified as an oncogene, which is translocated in a variety of B cell malignancies (16, 49). Targeted inactivation of Bcl11a shows that it is dispensable for the generation of myeloid or erythroid lineages but is required for the development of B lineage progenitors and thymocyte maturation (16). Consistent with the profound block to B cell development in Bcl11a–/– embryos, analysis of fetal liver hematopoietic progenitors revealed the absence of EBF, Pax-5, and IL-7R transcripts. Conversely, the expression of genes required for early T cell development was not impaired in Bcl11a–/– fetal thymocytes, including the IL-7R. The Bcl11a mutant phenotype in the lymphoid compartment is similar to the Flk2/Flt3 receptor or Flk2/Flt3 ligand knockout phenotypes. It remains to be determined whether Bcl11a participates indirectly (by means of the Flk2 receptor and/or IL-7R signaling) or directly in the regulation of the EBF gene.
Taken together, these studies reveal that the molecular circuits driving specification of the B cell fate involve the contingent as well as combinatorial activities of the transcription factors PU.1, Ikaros, E2A, Bcl11a, EBF, and Pax-5 and the cytokine receptors Flk2/Flt3 and IL-7R (Fig. 1). PU.1 and Ikaros are required for expression of Flk2/Flt3. Flk2/Flt3 signaling in concert with PU.1 induces IL-7R. Signaling through the IL-7R is proposed to induce E2A, which, in turn, is directly implicated in regulating expression of the EBF gene along with PU.1. Once expressed, EBF may reinforce E2A expression. EBF in concert with E2A activates the early program of B lineage gene expression, as well as DH-JH recombination, specifying the B cell fate. In addition to promoting specification of the B cell fate, EBF induces the expression of Pax-5, which culminates a gradual process of restriction of alternative lineage options, thereby ensuring commitment to the B cell fate. Pax-5 also functions coordinately with EBF and E2A to reinforce and expand the B lineage program of gene expression. Importantly, Pax-5 function, in the differentiation of pro-B cells, is contingent on EBF. Expression of the mb-1 gene is coordinately regulated by EBF and Pax-5. EBF is required for chromatin remodeling and DNA demethylation of the mb-1 promoter, which in turn regulates the ability of Pax-5 to activate the mb-1 promoter (50). This scenario provides an attractive molecular explanation for the contingency of Pax-5 function on EBF and may apply to other B lineage-specific genes that are codependent on EBF and Pax-5 for their activities.
Network Architecture
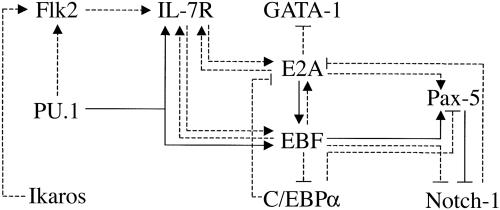
Given the extensive set of regulatory links that are being revealed among the aforementioned components, it is useful to attempt a depiction of the network architecture, which provides insight into its structure and dynamics (Fig. 3). Both stimulatory and inhibitory inputs are shown, indicated as solid or dashed lines, depending on the strength of the experimental evidence. Much of this evidence has been described above, and the remaining links are detailed below. IL-7 signaling appears to regulate EBF expression, because IL-7–/– CLPs have greatly diminished B cell developmental potential (51) and express considerably lower levels of EBF and Pax-5 transcripts when compared with wild-type CLPs (58). EBF can up-regulate IL-7Rα gene expression upon its precocious expression in MPPs (J.M.R.P. and H.S., unpublished data). E2A can also induce the expression of IL-7Rα upon ectopic expression in a macrophage cell line (52). Thus, IL-7R, EBF, and E2A, through positive feedback loops, may generate a self-sustaining regulatory network. The establishment of this network would depend on transient signals and inputs from the cytokine receptor Flk2 and the transcription factors PU.1 and Ikaros, respectively. Pax-5 expression depends on EBF (3) and can also be induced upon expression of EBF in MPPs (J.M.R.P. and H.S., unpublished data). Curiously, Pax-5 appears to induce EBF expression when ectopically expressed in developing T cells in the thymus (53). Thus, Pax-5 could reinforce and stabilize EBF expression, thereby further consolidating the self-sustaining network. We note that because EBF can complement E47–/– progenitors (40), resulting in the generation of B cell precursors, the proposed network appears to be sustainable by low levels of alternative E-proteins.
Fig. 3.
A self-sustaining regulatory network established in a B cell precursor. It is suggested that the establishment of this network depends on transient signaling and inputs from the cytokine receptor Flk2 and the transcription factors PU.1 and Ikaros, respectively. Positive feedback loops involving the cytokine receptor IL-7R and the transcription factors EBF, E2A, and Pax-5 may generate a self-sustaining circuit. The network architecture also features crossantagonism with alternate cell fate-determining transcription factors such as GATA-1, C/EBPα, and Notch-1. Stimulatory and inhibitory inputs are indicated as solid or dashed lines, depending on the strength of the experimental evidence.
The network architecture also features crossantagonism among various cell fate-determining transcription factors (Fig. 3). From the standpoint of specification of the B cell fate, E2A antagonizes the expression of the erythroid transcription factor GATA-1 (34). Furthermore, EBF appears to antagonize the expression of the myeloid transcription factor C/EBPα and can also override Notch signaling (J.M.R.P. and H.S., unpublished data). Conversely, Notch-1 and C/EBPα are able to counteract E2A and Pax-5 (36, 37, 54), respectively. Some of these mutually antagonistic interactions are likely to play a crucial role in dictating alternate cell fate choices. They may also be involved in lineage reprogramming (see below).
Reversibility of the Committed State and Network Reconfiguration
According to our model, lineage commitment is the culmination of contingent gene regulatory networks, which promote lineage-specific gene expression and repress alternate lineage programs. Lineage commitment is generally considered to be an irreversible process or, at the very least, one that cannot be overridden readily. This concept has been challenged by the demonstration that enforced expression of C/EBPα/β transcription factors in bone marrow-derived CD19+ B cells in culture leads to their reprogramming into macrophages (54). This “lineage conversion” is accompanied by rapid and efficient down-regulation of CD19 (a Pax-5-dependent B lineage gene) and up-regulation of the macrophage marker Mac-1. It was suggested that C/EBP factors antagonize Pax-5 activity, thereby reversing the committed state. We note that C/EBP factors appear to also regulate the Id1 gene (55). Id1 negatively regulates E2A DNA-binding activity through heterodimerization, thus inhibiting the expression of E2A target genes. Expression of an Id1 transgene driven by Ig regulatory elements has been shown to induce a profound block to early B cell development (56). Thus, we propose that reprogramming of B lineage cells into macrophages observed after enforced expression of C/EBPα/β may also involve dysregulated Id1 expression. Therefore, C/EBP factors may reprogram B lineage cells into macrophages by antagonizing primary (E2A) as well as secondary (Pax-5) B cell fate determinants.
Although expression of C/EBP factors in committed preB and B cells efficiently induces CD19 down-regulation and upregulation of Mac-1, the aforementioned study did not unambiguously identify, through clonogenic assays, the precursors that give rise to macrophages upon ectopic expression of C/EBP factors. Such cells could be the CD19+B220– bipotent B/macrophage progenitors previously demonstrated in adult bone marrow (57). Thus, it remains possible that C/EBP factors divert bipotential progenitors along the macrophage differentiation pathway rather than reprogram differentiated B cells into macrophages. The latter process would necessarily entail disassembly of a complex gene regulatory network specific to B cells (Fig. 3) and its reconfiguration into one specific for macrophages. The stability and potential reversibility of gene regulatory networks that are operative in committed precursors and differentiated cell types need to be explored further.
Perspective
The B cell developmental pathway represents a leading system for the analysis of regulatory circuits that orchestrate cell fate specification and commitment. Considerable progress has been achieved within the past decade in the identification and characterization of various regulatory components, including transcription factors and signaling molecules. It is now possible to initiate the assembly of molecular circuits that underlie cell fate choice and specific developmental transitions. We have attempted to assemble such control circuits, which operate in MPPs, CLPs, pro-B, and preB cells. At present, the circuits are rudimentary and tentative in structure, because additional components remain to be identified and analyzed. Moreover, the connectivity of various components needs to be rigorously tested and extended. Nevertheless, the proposed circuit architecture is foreshadowing design principles that include transient signaling inputs, self-sustaining positive feedback loops, and crossantagonism among alternate cell fate determinants. The elaboration of similar circuits in a variety of cellular systems should consolidate general principles of network architecture and dynamics, which underlie a wide variety of biological patterning and differentiation processes. Detailed knowledge of regulatory networks that specify and sustain diverse mammalian cell fates should also facilitate the directed and efficient generation of lineage-specific progenitors from embryonic stem cells for therapeutic purposes.
Acknowledgments
We thank members of the H.S. laboratory for stimulating discussions. H.S. is an Investigator with the Howard Hughes Medical Institute, and K.L.M. is a Fellow of the Irvington Institute.
Author contributions: H.S., K.L.M., and J.M.R.P. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HSC, hematopoietic stem cell; IL-7R, IL-7 receptor; MPP, multipotential progenitor; IgH, Ig heavy chain; CLP, common lymphoid progenitor.
References
- 1.Hardy, R. R. & Hayakawa, K. (2001) Annu. Rev. Immunol. 19, 595–621. [DOI] [PubMed] [Google Scholar]
- 2.Busslinger, M. (2004) Annu. Rev. Immunol. 22, 55–79. [DOI] [PubMed] [Google Scholar]
- 3.Medina, K. L., Pongubala, J. M., Reddy, K. L., Lancki, D. W., Dekoter, R., Kieslinger, M., Grosschedl, R. & Singh, H. (2004) Dev. Cell 7, 607–617. [DOI] [PubMed] [Google Scholar]
- 4.DeKoter, R. P. & Singh, H. (2000) Science 288, 1439–1441. [DOI] [PubMed] [Google Scholar]
- 5.Hu, M., Krause, D., Greaves, M., Sharkis, S., Dexter, M., Heyworth, C. & Enver, T. (1997) Genes Dev. 11, 774–785. [DOI] [PubMed] [Google Scholar]
- 6.Enver, T. & Greaves, M. (1998) Cell 94, 9–12. [DOI] [PubMed] [Google Scholar]
- 7.Cantor, A. B. & Orkin, S. H. (2002) Oncogene 21, 3368–3376. [DOI] [PubMed] [Google Scholar]
- 8.Georgopoulos, K., Bigby, M., Wang, J. H., Molnar, A., Wu, P., Winandy, S. & Sharpe, A. (1994) Cell 79, 143–156. [DOI] [PubMed] [Google Scholar]
- 9.Bain, G., Maandag, E. C., Izon, D. J., Amsen, D., Kruisbeek, A. M., Weintraub, B. C., Krop, I., Schlissel, M. S., Feeney, A. J., van Roon, M., et al. (1994) Cell 79, 885–892. [DOI] [PubMed] [Google Scholar]
- 10.Scott, E. W., Simon, M. C., Anastasi, J. & Singh, H. (1994) Science 265, 1573–1577. [DOI] [PubMed] [Google Scholar]
- 11.Zhuang, Y., Soriano, P. & Weintraub, H. (1994) Cell 79, 875–884. [DOI] [PubMed] [Google Scholar]
- 12.Nutt, S. L., Urbanek, P., Rolink, A. & Busslinger, M. (1997) Genes Dev. 11, 476–491. [DOI] [PubMed] [Google Scholar]
- 13.Peschon, J. J., Morrissey, P. J., Grabstein, K. H., Ramsdell, F. J., Maraskovsky, E., Gliniak, B. C., Park, L. S., Ziegler, S. F., Williams, D. E., Ware, C. B., et al. (1994) J. Exp. Med. 180, 1955–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, H. & Grosschedl, R. (1995) Nature 376, 263–267. [DOI] [PubMed] [Google Scholar]
- 15.Mackarenhtschian, K., Hardin, J. D., Moore, K. A., Boast, S., Goff, S. P. & Lemischka, I. R. (1995) Immunity 3, 147–161. [DOI] [PubMed] [Google Scholar]
- 16.Liu, P., Keller, J. R., Ortiz, M., Tessarollo, L., Rachel, R. A., Nakamura, T., Jenkins, N. A. & Copeland, N. G. (2003) Nat. Immunol. 4, 525–532. [DOI] [PubMed] [Google Scholar]
- 17.Wiesmann, A., Phillips, R. L., Mojica, M., Pierce, L. J., Searles, A. E., Spangrude, G. J. & Lemischka, I. (2000) Immunity 12, 193–199. [DOI] [PubMed] [Google Scholar]
- 18.Cross, M. A. & Enver, T. (1997) Curr. Opin. Genet. Dev. 7, 609–613. [DOI] [PubMed] [Google Scholar]
- 19.Miyamoto, T., Iwasaki, H., Reizis, B., Ye, M., Graf, T., Weissman, I. L. & Akashi, K. (2002) Dev. Cell 3, 137–147. [DOI] [PubMed] [Google Scholar]
- 20.Adolfsson, J., Borge, O. J., Bryder, D., Theilgaard-Monch, K., Astrand-Grundstrom, I., Sitnicka, E., Sasaki, Y. & Jacobsen, E. W. (2001) Immunity 15, 659–669. [DOI] [PubMed] [Google Scholar]
- 21.Sitnicka, E., Bryder, D., Theilgaard-Monch, K., Buza-Vidas, N., Adolfsson, J. & Jacobsen, S. E. (2002) Immunity 17, 463–472. [DOI] [PubMed] [Google Scholar]
- 22.Borge, O. J., Adolfsson, J., Martensson, A., Martensson, I. & Jacobsen, S. E. W. (1999) Blood 94, 3781–3790. [PubMed] [Google Scholar]
- 23.Miller, J. P., Izon, D., DeMuth, W., Gerstein, R., Bhandoola, A. & Allman, D. (2002) J. Exp. Med. 196, 705–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sitnicka, E., Brakebusch, C., Martensson, I. L., Svensson, M., Agace, W. W., Sigvardsson, M., Buza-Vidas, N., Bryder, D., Cilio, C. M., Ahlenius, H., et al. (2003) J. Exp. Med. 198, 1495–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vosshenrich, C. A., Cumano, A., Muller, W., Di Santo, J. P. & Vieira, P. (2003) Nat. Immunol. 4, 773–779. [DOI] [PubMed] [Google Scholar]
- 26.Nichogiannopoulou, A., Trevisan, M., Neben, S., Friedrich, C. & Georgopoulos, K. (1999) J. Exp. Med. 190, 1201–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeKoter, R. P., Lee, H. J. & Singh, H. (2002) Immunity 16, 297–309. [DOI] [PubMed] [Google Scholar]
- 28.Murre, C., McCaw, P. S. & Baltimore, D. (1989) Cell 56, 777–783. [DOI] [PubMed] [Google Scholar]
- 29.Murre, C., Bain, G., van Dijk, M. A., Engel, I., Furnari, B. A., Massari, M. E., Matthews, J. R., Quong, M. W., Rivera, R. R. & Stuiver, M. H. (1994) Biochim. Biophys. Acta 1218, 129–135. [DOI] [PubMed] [Google Scholar]
- 30.Hagman, J., Belanger, C., Travis, A., Turck, C. W. & Grosschedl, R. (1993) Genes Dev. 7, 760–773. [DOI] [PubMed] [Google Scholar]
- 31.O'Riordan, M. & Grosschedl, R. (1999) Immunity 11, 21–31. [DOI] [PubMed] [Google Scholar]
- 32.Romanow, W. J., Langerak, A. W., Goebel, P., Wolvers-Tettero, I. L., van Dongen, J. J., Feeney, A. J. & Murre, C. (2000) Mol. Cell 5, 343–353. [DOI] [PubMed] [Google Scholar]
- 33.Quong, M. W., Romanow, W. J. & Murre, C. (2002) Annu. Rev. Immunol. 20, 301–322. [DOI] [PubMed] [Google Scholar]
- 34.Ikawa, T., Kawamoto, H., Wright, L. Y. & Murre, C. (2004) Immunity 20, 349–360. [DOI] [PubMed] [Google Scholar]
- 35.Zhuang, Y., Jackson, A., Pan, L., Shen, K. & Dai, M. (2004) Mol. Immunol. 40, 1165–1177. [DOI] [PubMed] [Google Scholar]
- 36.Ordentlich, P., Lin, A., Shen, C. P., Blaumueller, C., Matsuno, K., Artavanis-Tsakonas, S. & Kadesch, T. (1998) Mol. Cell. Biol. 18, 2230–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nie, L., Xu, M., Vladimirova, A. & Sun, X. H. (2003) EMBO J. 22, 5780–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Igarashi, H., Gregory, S. C., Yokota, T., Sakaguchi, N. & Kincade, P. W. (2002) Immunity 17, 117–130. [DOI] [PubMed] [Google Scholar]
- 39.Smith, E. M., Gisler, R. & Sigvardsson, M. (2002) J. Immunol. 169, 261–270. [DOI] [PubMed] [Google Scholar]
- 40.Seet, C. S., Brumbaugh, R. L. & Kee, B. L. (2004) J. Exp. Med. 199, 1689–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schebesta, M., Heavey, B. & Busslinger, M. (2002) Curr. Opin. Immunol. 14, 216–223. [DOI] [PubMed] [Google Scholar]
- 42.Nutt, S. L., Heavey, B., Rolink, A. G. & Busslinger, M. (1999) Nature 401, 556–562. [DOI] [PubMed] [Google Scholar]
- 43.Nutt, S. L., Eberhard, D., Horcher, M., Rolink, A. G. & Busslinger, M. (2001) Int. Rev. Immunol. 20, 65–82. [DOI] [PubMed] [Google Scholar]
- 44.Souabni, A., Cobaleda, C., Schebesta, M. & Busslinger, M. (2002) Immunity 17, 781–793. [DOI] [PubMed] [Google Scholar]
- 45.Maillard, I., Adler, S. H. & Pear, W. S. (2003) Immunity 19, 781–791. [DOI] [PubMed] [Google Scholar]
- 46.Mikkola, I., Heavey, B., Horcher, M. & Busslinger, M. (2002) Science 297, 110–113. [DOI] [PubMed] [Google Scholar]
- 47.Hesslein, D. G., Pflugh, D. L., Chowdhury, D., Bothwell, A. L., Sen, R. & Schatz, D. G. (2003) Genes Dev. 17, 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saiki, Y., Yamazaki, Y., Yoshida, M., Katoh, O. & Nakamura, T. (2000) Genomics 70, 387–391. [DOI] [PubMed] [Google Scholar]
- 49.Satterwhite, E., Sonoki, T., Willis, T. G., Harder, L., Nowak, R., Arriola, E. L., Liu, H., Price, H. P., Gesk, S., Steinemann, D., et al. (2001) Blood 98, 3413–3420. [DOI] [PubMed] [Google Scholar]
- 50.Maier, H., Ostraat, R., Gao, H., Fields, S., Shinton, S. A., Medina, K. L., Ikawa, T., Murre, C., Singh, H., Hardy, R. R. & Hagman, J. (2004) Nat. Immunol. 5, 1069–1077. [DOI] [PubMed] [Google Scholar]
- 51.Carvalho, T. L., Mota-Santos, T., Cumano, A., Demengeot, J. & Vieira, P. (2001) J. Exp. Med. 194, 1141–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kee, B. L. & Murre, C. (1998) J. Exp. Med. 188, 699–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fuxa, M., Skok, J., Souabni, A., Salvagiotto, G., Roldan, E. & Busslinger, M. (2004) Genes Dev. 18, 411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie, H., Ye, M., Feng, R. & Graf, T. (2004) Cell 117, 663–676. [DOI] [PubMed] [Google Scholar]
- 55.Saisanit, S. & Sun, X. H. (1997) Mol. Cell. Biol. 17, 844–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun, X. H. (1994) Cell 79, 893–900. [DOI] [PubMed] [Google Scholar]
- 57.Montecino-Rodriguez, E., Leathers, H. & Dorshkind, K. (2001) Nat. Immunol. 2, 83–88. [DOI] [PubMed] [Google Scholar]
- 58.Dias, S., Silva, H., Jr., Cumano, A. & Vieira, P. (March 15, 2005) J. Exp. Med., 10.1084/jem.20042393. [DOI] [PMC free article] [PubMed]