Abstract
In vertebrates, cranial placodes contribute to all sense organs and sensory ganglia and arise from a common pool of Six1/Eya2+ progenitors. Here we dissect the events that specify ectodermal cells as placode progenitors using newly identified genes upstream of the Six/Eya complex. We show in chick that two different tissues, namely the lateral head mesoderm and the prechordal mesendoderm, gradually induce placode progenitors: cells pass through successive transcriptional states, each identified by distinct factors and controlled by different signals. Both tissues initiate a common transcriptional state but over time impart regional character, with the acquisition of anterior identity dependent on Shh signalling. Using a network inference approach we predict the regulatory relationships among newly identified transcription factors and verify predicted links in knockdown experiments. Based on this analysis we propose a new model for placode progenitor induction, in which the initial induction of a generic transcriptional state precedes regional divergence.
KEY WORDS: Chick embryo, Quail graft, Cell fate, Sense organs, Sensory ganglia, Transcriptional networks, Signalling, Gene regulatory network
Summary: A new model is presented whereby multiple mesodermal tissues and signalling centres, rather than a single organizer, mediate the induction and patterning of chick sensory placodes.
INTRODUCTION
During early development, many fate decisions are controlled by inductive interactions, whereby an inducing cell population instructs responding cells to change their fate. Neural induction is perhaps the best-studied inductive event, and several models have been proposed to explain the induction and patterning of the nervous system by the organizer (Stern, 2001). The ʻmultiple organizer' model argues that different parts of the organizer induce distinct regions of the nervous system (Holtfreter, 1933a,b; Mangold, 1933; Saxen and Toivonen, 1962), whereas Waddington suggested a two-step model with ʻevocation' generating a generic, non-regionalised nervous system, followed by ʻindividuation' to impart regional character (Waddington and Needham, 1936). Nieuwkoop's ʻactivation-transformation' hypothesis suggests that initial induction generates anterior character, which is then posteriorised (Nieuwkoop and Nigtevecht, 1954). Currently, there is support for modified versions of both Nieuwkoop's and Mangold's models (Stern, 2001; Takemoto et al., 2006). However, the question remains whether these models could also explain the development of adjacent structures – the sensory placodes. Placodes arise in the head ectoderm in register with the neural tube and generate components of sense organs and cranial sensory ganglia (Baker and Bronner-Fraser, 2001; Grocott et al., 2011; Schlosser, 2010; Streit, 2008). Pushing the multiple organizer model to the extreme, it has been suggested that even placodes are induced by a subset of organizer cells (Mangold, 1933; Saxen and Toivonen, 1962; Spemann, 1938). Here we ask whether placodes are induced by a bona fide organizer that induces and patterns the placodal territory, and if so, which if any of these models explains this process.
Although contributing to diverse organs and ganglia, the cranial placodes arise from a common domain, termed the pre-placodal region (PPR), which is specified at head process stages as a strip of ectoderm surrounding the anterior neural plate (Baker and Bronner-Fraser, 2001; Grocott et al., 2012; Schlosser, 2010; Streit, 2008). PPR cells initially have the same developmental potential: they can give rise to any placode and express a common set of genes (Bailey et al., 2006; Bailey and Streit, 2006; Streit, 2008), among them Six and Eya factors, which impart PPR character to ectodermal cells (Brugmann et al., 2004; Chen et al., 2009; Christophorou et al., 2009; Laclef et al., 2003; Zheng et al., 2003; Zou et al., 2004, 2006). Surprisingly, few upstream regulators have been identified to explain how their expression is activated in, and confined to, sensory progenitors. Six1 transcription is directly activated by Dlx factors, but repressed by Msx1 (Sato et al., 2010). At gastrulation stages, BMP signalling is required for the PPR ‘competence factors’ Gata2/3, Tfap2a/c, Foxi1 and Dlx3, but must be inhibited later for Six1 activation (Ahrens and Schlosser, 2005; Brugmann et al., 2004; Kwon et al., 2010; Litsiou et al., 2005; Pieper et al., 2012). Subsequently, FGF signalling together with inhibition of BMP and Wnt is necessary for PPR formation and sufficient to induce sensory progenitors in non-placodal ectoderm (Brugmann et al., 2004; Litsiou et al., 2005). These signals emanate from the adjacent neural plate and the underlying head mesoderm (Ahrens and Schlosser, 2005; Litsiou et al., 2005), although their relative contribution to PPR induction remains unclear, as does the question of whether the signalling tissues are true organizers.
Using an established induction assay, we now dissect PPR induction and propose a new model. First, we design a molecular screen to identify new potential players and then use these factors to characterise the response to each PPR-inducing tissue and to establish a genetic hierarchy upstream of the Six/Eya complex. Combined with a network inference approach, our analysis proposes a new multistep model for PPR induction. We show that signals from the neural plate are unable to induce a PPR. Two different mesodermal populations, namely the lateral head mesoderm and the prechordal mesendoderm, initially induce a similar set of transcription factors (reminiscent of an ʻevocation'), but then gradually impart anterior and posterior bias to sensory progenitors (ʻindividuation'). We show that PPR induction is not mediated by an organizer, but instead involves multiple signalling centres, each inducing cells with distinct regional character.
RESULTS
A molecular screen reveals the complexity of PPR induction
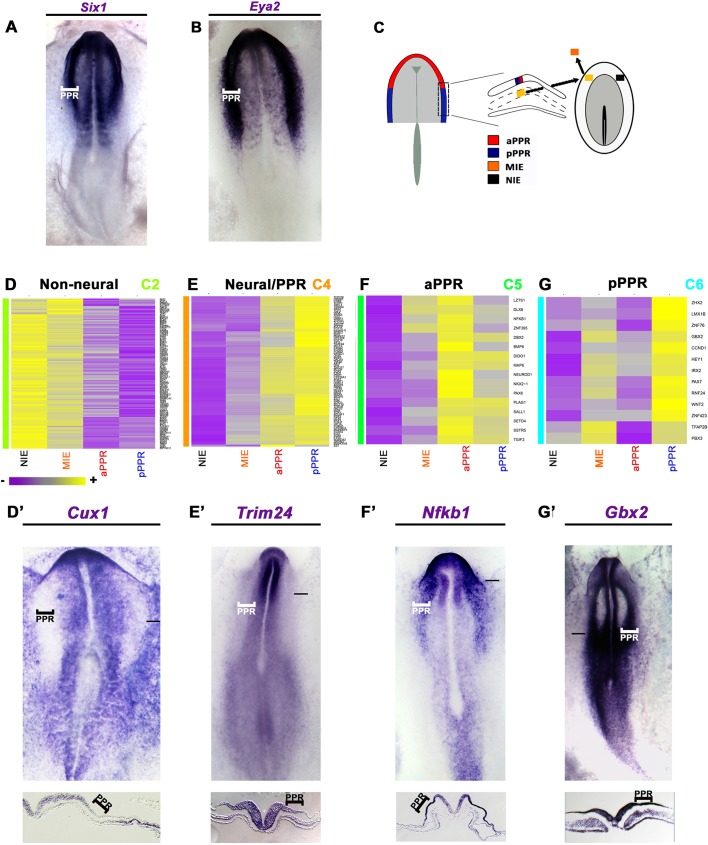
In chick, PPR induction is mediated by signals from the head mesoderm: when grafted next to competent epiblast this mesoderm induces a full set of bona fide PPR markers (Litsiou et al., 2005; see Fig. 2A,B). How long does this process take? In the same induction assay, quail lateral head mesoderm (lHM) requires at least 12 h of contact to induce the PPR markers Six1 and Eya2 in chick epiblast [Fig. 1A-B′; Six1: 4 h, 2/9; 8 h, 8/8; Eya2: 8 h, 0/7; 12 h, 3/10; Six4 (not shown): 4 h, 2/9; 8 h, 10/12].
Fig. 2.
New placode progenitor transcripts. (A,B) Six1 (A) and Eya2 (B) expression at HH7+/8−. (C) aPPR and pPPR from HH6 (Lleras-Forero et al., 2013) and mesoderm-induced (MIE) and non-induced ectoderm (NIE) were analysed by microarray. (D) Cluster C2 transcripts are excluded from the PPR, such as Cux1 (D′). (E) Cluster C4 includes aPPR- and pPPR-enriched factors, such as Trim24 (E′). (F) Cluster C5 includes aPPR factors, such as Nfkb1 (F′). (G) Cluster C6 includes pPPR factors, such as Gbx2 (G′). Bars in D′-G′ indicate level of sections shown beneath.
Fig. 1.
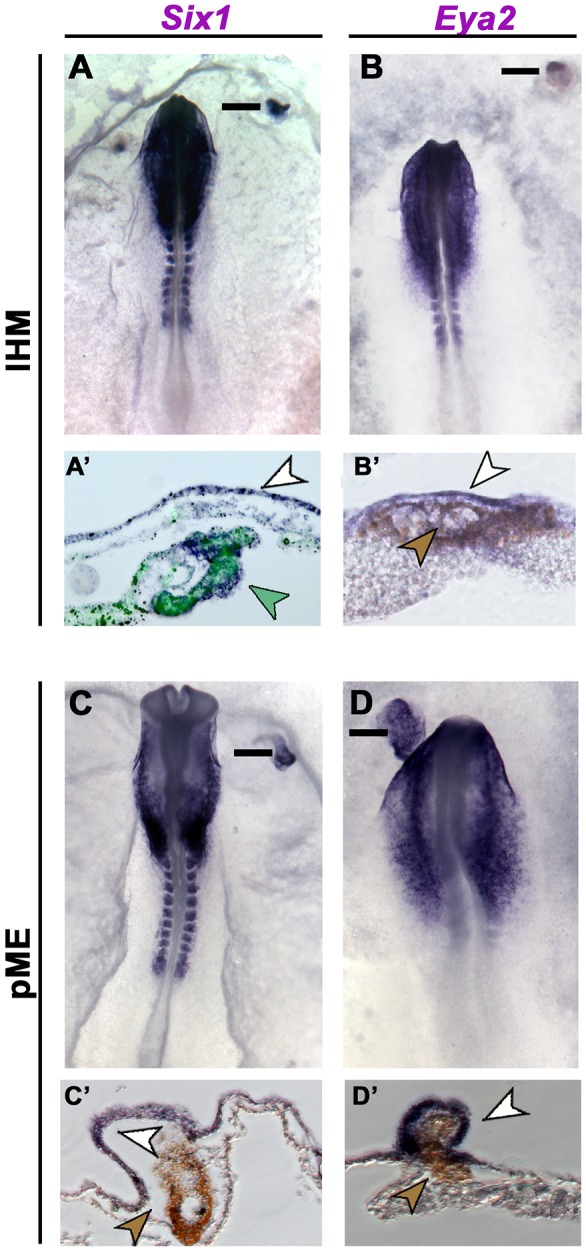
Induction of placode progenitors by the lateral head mesoderm (lHM) and prechordal mesendoderm (pME). (A-B′) CMFDA-labelled chick (green arrowhead in A′) or quail IHM (brown arrowhead in B′) from HH5/6 donors induces Six1 (A,A′, white arrowhead; 8 h, 8/8) and Eya2 (B,B′, white arrowhead; 12 h, 3/10) in host extraembryonic epiblast. (C-D′) pME grafts from HH5/6 quail donors (brown arrowheads) induce Six1 (C,C′, white arrowhead; 15-17 h, 5/8) and Eya2 (D,D′, white arrowhead; 15-17 h, 7/8). Bars in A-D indicate section levels in A′-D′.
To identify the components of the PPR induction cascade, we performed a microarray screen comparing the transcriptomes of epiblast from four conditions (Fig. 2C): mesoderm-induced epiblast (12 h; MIE), non-induced epiblast (control; NIE) from the contralateral side of the same embryos, and the normal anterior (aPPR) and posterior (pPPR) PPR at HH5/6 (Lleras-Forero et al., 2013). One-way ANOVA revealed 3475 probes with a significant change of more than 2-fold (P<0.05) in at least one cell population, representing 2868 unique transcripts including 206 known or putative transcription factors (Table S1, GSE81023). When compared with control epiblast, 1098 transcripts are mesoderm induced (>1.2-fold), whereas 1379 genes are repressed (Table S1). As expected, the PPR-enriched factors Eya2, Dach1 and Sox3 (Rex et al., 1977; Barembaum and Bronner-Fraser, 2007; Litsiou et al., 2005) are induced by the lHM, whereas neural crest (Snail2), neural plate (Sox2) and regionally restricted genes (Pax6) are not (Litsiou et al., 2005) (Fig. S1). Thus, the array screen replicates known changes in gene expression in response to mesodermal signals.
To ensure that the genes identified were induced by mesoderm rather than recruited from the host PPR, the screen was designed using extraembryonic tissue. However, for any gene to be relevant to PPR formation it should at some point be expressed in the prospective PPR. To identify synexpression groups we performed hierarchical clustering on all genes that changed significantly and in situ hybridisation to verify their spatiotemporal expression in normal embryos. In total, we assessed the in situ expression of 47 known or putative transcription factors, five chromatin modifiers and four signalling pathway components.
This analysis reveals seven major clusters with distinct profiles (C1-C7; Figs S1, S2 and S3, Table S4). C1 and C2 transcripts are largely absent from placode progenitors, like Cux1 (C2; Fig. 2D′), and repressed by the mesoderm, with C2 genes being enriched in the extraembryonic ectoderm (Fig. 2D). Cluster C5 transcripts (Fig. 2F) are strongly enriched in the aPPR, but only weakly mesoderm induced, and include aPPR-specific genes such as Pax6 and Sstr5 (Lleras-Forero et al., 2013), Nfkb1 (Fig. 2F′) and Sall1 (Fig. S2). By contrast, cluster C6 transcripts (Fig. 2G) are lHM induced, enriched in the pPPR and include Gbx2 (Fig. 2G′), Irx2 and Pax7 (Goriely et al., 1999; Khudyakov and Bronner-Fraser, 2009; Steventon et al., 2012) and novel transcripts such as Znf423, Znf76 and Rnf24. Finally, cluster C4 genes are mostly induced by mesodermal signals and are present throughout the entire PPR. In situ hybridisation reveals that most C4 transcripts are not restricted to sensory progenitors, but expressed broadly at primitive streak stages encompassing the future neural, neural crest and placode territories (Fig. 2E,E′, Figs S2 and S3), including Zic1 (Khudyakov and Bronner-Fraser, 2009), Otx2 (Bally-Cuif et al., 1995) and Fzd8 (Paxton et al., 2010) and many new genes (Figs S2 and S3). Only a few C4 transcripts are restricted to the PPR (Dmbx1, Homer2; Fig. S3). As development proceeds, some transcripts from cluster C4 remain expressed in both neural and placode cells, whereas others become confined to either tissue (Figs S2 and S3), suggesting that Six and Eya factors are among the few bona fide PPR markers.
Thus, the screen has identified many novel transcripts expressed in placode progenitors, revealing molecular similarity between precursors for the central and peripheral nervous systems. In addition, ultimately the lHM appears to induce posterior placode progenitors, but does not act as an organizer that induces and patterns the PPR. This implies that other PPR-inducing tissues exist, which should impart rostral PPR identity.
Prechordal mesendoderm induces placode progenitors
Endodermal signals have been implicated in anterior placode induction in amphibians (Henry and Grainger, 1990; Jacobson, 1963a,b) and we have recently shown that the anterior prechordal mesendoderm (pME) is required for Eya2, Pax6 and Pnoc expression in chick and zebrafish (Lleras-Forero et al., 2013). To test its PPR-inducing ability, we grafted quail HH5 pME next to chick extraembryonic epiblast (HH3+/4−). Both Six1 and Eya2 are induced after 15-17 h (Fig. 1C-D′). Thus, two different mesodermal tissues, namely lHM and pME, mediate PPR induction.
Lateral and axial mesoderm provide regional bias to sensory progenitors
Our results indicate that PPR induction is not mediated by an organizer, although it remains possible that lHM first induces aPPR, which is subsequently posteriorised. Alternatively, both mesoderms might first induce generic PPR character, which is then regionalised, or that each directly induces distinct rostrocaudal identity. To distinguish these possibilities and to capture the complexity of PPR induction, we compared the response to both tissues over time assessing 126 genes simultaneously. HH5/6 lHM or pME was grafted into the extraembryonic region of HH4− hosts and the underlying epiblast was collected after 3, 6 and 12 h, together with the contralateral control epiblast. Each experiment was performed in triplicate and gene expression was analysed by NanoString using a probe set containing known and new PPR transcripts, markers for different placodes, neural crest and neural plate (Table S2). Transcripts exhibiting a difference greater than 1.2-fold (P<0.05) between induced and time-matched control tissues were considered as differentially expressed.
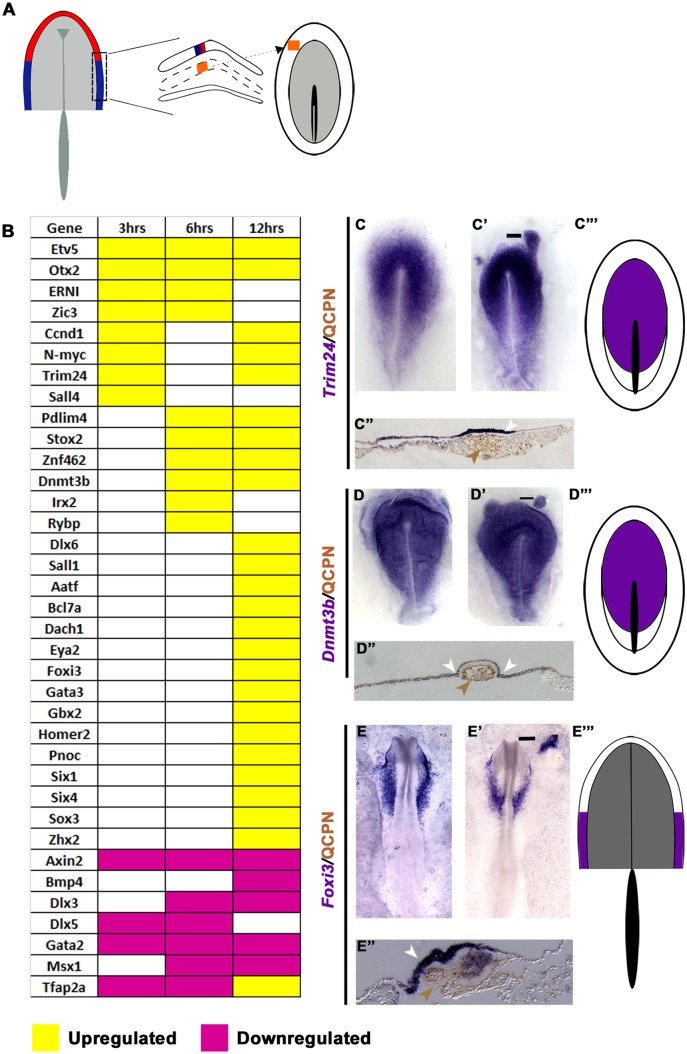
The transcriptional hierarchy in response to the lHM
lHM-derived signals initiate a dynamic response, with distinct groups of transcripts being regulated at each time point (Fig. 3A-E, Table S3). At 3 h, only a few factors are upregulated: Ccnd1, Etv5, ERNI, N-myc, Otx2, Sall4, Trim24 and Zic3 (Fig. 3B,C′,C″). Whereas some genes are maintained, others are only transiently induced: Otx2 expression levels are negligible (Fig. S4) and Sall4 disappears after 6 h, as do ERNI and Zic3 after 12 h (Fig. 3B). At 6 h, new transcripts appear in response to the mesoderm (Dnmt3b, Pdlim4, Irx2, Rybp, Stox2, Znf462; Fig. 3B,D′,D″, Table S3), whereas PPR genes (Six1/4, Eya2) are only upregulated after 12 h together with some of their known upstream regulators (Gata3, Foxi3, Tfap2a, Dlx6; Fig. 3B,E′,E″) (Kwon et al., 2010; Pieper et al., 2012; Qiao et al., 2012; Sato et al., 2010) and genes previously not associated with PPR induction (Aatf, Bcl7a, Zhx2; Fig. 3B, Table S3). By contrast, other genes are downregulated by the lHM (3 h), including Tfap2a, Dlx5, Gata2 and Axin2 (Fig. 3B, Table S3). At 12 h, most of these remain absent and other PPR repressors (Msx1, Bmp4) (Ahrens and Schlosser, 2005; Litsiou et al., 2005; Sato et al., 2010) are reduced; however, Tfap2a is now induced and Dlx5 is no longer repressed, consistent with their role as positive Six1 regulators (Kwon et al., 2010; Qiao et al., 2012) (Fig. 3B, Table S3).
Fig. 3.
Response to the lHM. (A) lHM (HH6; orange) was grafted into HH4− hosts; the adjacent ectoderm was analysed after 3, 6 and 12 h. (B) Transcripts induced or repressed (P<0.05; >1.2-fold change). (C-E‴) Expression patterns of 3 h (C-C‴), 6 h (D-D‴) and 12 h (E-E‴) induced genes. Quail lHM (brown arrowheads) induces Trim24 (C′,C″ white arrowhead), Dnmt3b (D′,D″ white arrowheads) and Foxi3 (E′,E″ white arrowhead). Bars in C′-E′ indicate the level of the sections shown in C″-E″. Area of overlap (purple) is summarised for 3 h (C‴), 6 h (D‴) and 12 h (E‴) induced transcripts.
To determine the character of the lHM-induced cells we examined the normal expression patterns of all genes in each cohort, considering their dynamic changes and that, at different times of development, they may characterise different cell populations (summarised in Fig. S2 and Table S4). This analysis reveals that 3 and 6 h induced transcripts are normally expressed early and broadly: at HH3-4 their domains encompass the presumptive neural plate and its border, including future neural crest and placodes (Fig. 3C,C‴,D,D‴, Figs S2 and S3, Table S4), except for ERNI and Otx2, which are already expressed in the pre-streak epiblast and then label much of the epiblast and the anterior ectoderm, respectively (Bally-Cuif et al., 1995). Most factors are maintained in the neural plate and/or PPR at head fold stages (HH6/7; Figs S2 and S3, Table S4). By contrast, most 12 h induced transcripts first appear at HH5/6 (Six1, Six4, Eya2) or begin broadly in the non-neural ectoderm (Gata3, Dlx6, Homer2, Foxi3) to become confined to, or upregulated in, the PPR (Fig. 3E,E‴, Table S4). Most mesoderm-induced genes are present in all placode progenitors; however, Foxi3 (Khatri et al., 2014) and Gbx2 (Steventon et al., 2012) are restricted to future otic and epibranchial cells by the time PPR markers are expressed; Otx2 remains present, although at extremely low levels (Fig. S4). Thus, at PPR stages the normal expression of all 12 h induced genes overlaps in the pPPR, suggesting that the induced tissue has acquired pPPR character.
Together, these data reveal that the lHM gradually induces PPR identity, with cells passing through sequential states. The first step does not generate anterior character but resembles pre-primitive streak or early streak stage epiblast, and a pPPR is established over time.
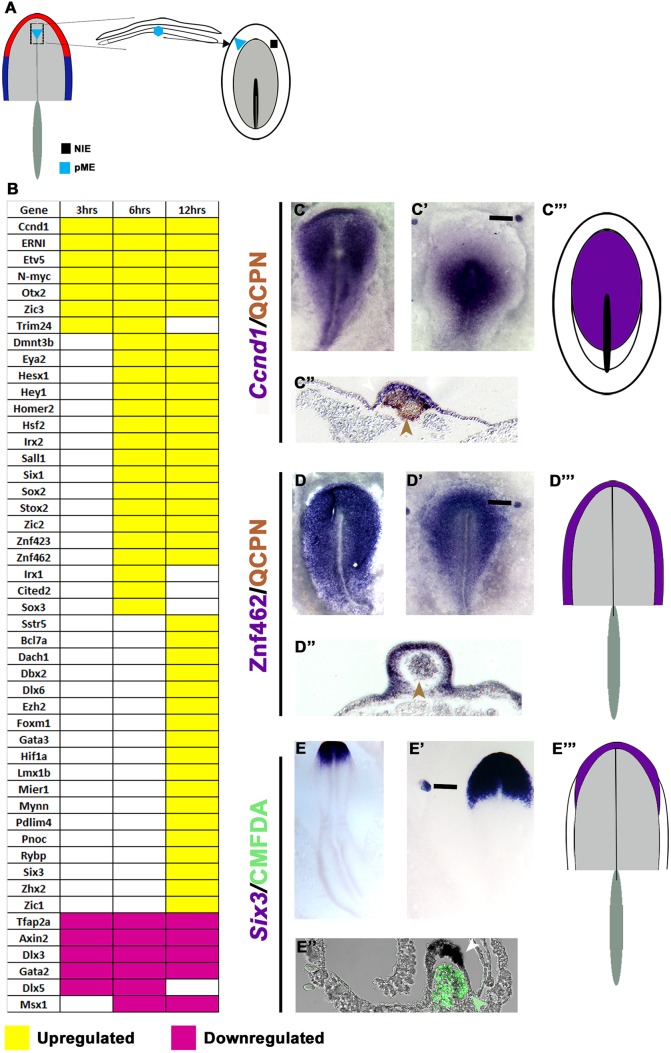
The transcriptional hierarchy in response to the pME
We observe a similar hierarchy in response to the pME. 3 h after grafting, the mesendoderm induces the same transcripts as the lHM (Fig. 4A,B,C′,C″, Table S3), with most genes being maintained until at least 12 h. The mesendoderm appears to induce Irx1, Gbx2 and Sstr5, albeit at extremely low levels (Fig. 4B, Figs S4 and S5, Table S3). This is followed by induction of 17 transcripts including PPR genes (Six1, Eya2, Irx1, Homer2; Fig. 4B,D′,D″) and, finally, by many genes at 12 h including the Six1 co-factor Dach1 (Fig. 4B,E′,E″, Table S3). The pME represses the same genes as the lHM, although the timing varies slightly (compare Fig. 3B with Fig. 4B).
Fig. 4.
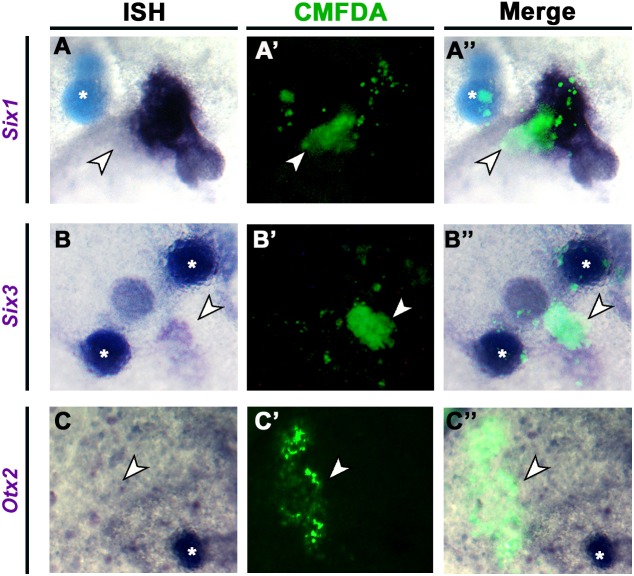
Response to the pME. (A) pME (HH6) was grafted into HH4− hosts; the adjacent ectoderm was analysed after 3, 6 and 12 h. (B) Transcripts induced or repressed (P<0.05; >1.2-fold change). (C-E‴) Expression patterns of 3 h (C-C‴), 6 h (D-D‴) and 12 h (E-E‴) induced genes. Quail pME (brown or green arrowheads) induces Ccnd1 (C′,C″ white arrowhead), Znf462 (D′,D″ white arrowhead) and Six3 (E′,E″ white arrowhead). Bars in C′-E′ indicate the level of the sections shown in C″-E″. Area of overlap (purple) is summarised for 3 h (C‴), 6 h (D‴) and 12 h (E‴) induced transcripts.
In normal embryos, 3 h pME-induced genes are expressed widely at HH3/4, labelling the future neural plate and its border (Fig. 4C,C‴,D, Figs S2 and S3, Table S4). 6 h induced genes overlap in the PPR (Fig. 4D,D‴), although two neural transcripts (Sox2, Znf423) are also present (Table S4). The expression domains of most 12 h induced genes continue to overlap in the PPR and the presence of Otx2, Pnoc, Sstr5 and Six3 suggests that cells have acquired anterior character (Fig. 4E,E‴). However, in addition, seven induced genes are expressed in the neural plate in normal embryos (Table S4). Thus, the pME rapidly induces aPPR, but over time generates tissue of mixed (neural/pre-placodal) anterior identity. The delayed induction of neural character excludes the possibility that an induced neural plate contributes to the accelerated induction of PPR transcripts by axial mesendoderm versus lHM. In summary, like the lHM, the pME initiates a sequence of transcriptional responses until PPR identity is established. Both tissues initially induce a small set of common transcription factors (Fig. S5), but subsequently impart anterior and posterior bias.
Neural plate signals are not sufficient to induce placode progenitors
The neural plate has been implicated in PPR induction in Xenopus (Ahrens and Schlosser, 2005), although this does not appear to be the case in chick (Litsiou et al., 2005). We now revisit this question using our newly identified genes. The future forebrain (aNP) or hindbrain (pNP) from HH5/6 donors was grafted into the extraembryonic region of HH4− hosts; epiblast exposed to neural plate signals was dissected together with non-induced epiblast from the contralateral side after 3, 6 and 12 h and processed for NanoString analysis (Fig. S6, Table S3).
After 3 h, the aNP induces many newly identified transcripts, as well as FGF mediators (Etv4, Etv5) and the neural crest marker Pax7 (Fig. S6). Except for Stox2 and Mynn, which are absent from the most central epiblast, in the embryo all other factors are expressed broadly at HH3/4 in the future neural plate and its border (Figs S2 and S3, Table S4). Most aNP-induced genes are not among those induced by mesoderm and are only transiently activated. Likewise, five 6 h induced transcripts are broadly expressed in the HH4 epiblast, while three (Hey1, Foxi3, Six1) are confined to the PPR. This is followed by many genes that, in the normal embryo, do not overlap in a single territory, including NP-specific transcripts (HH5/6; Sox2, Znf423) and non-neural ectoderm genes (Gata3, Pax6, Irx1, Eya2). The PPR marker Six1 is no longer induced, while other PPR-specific genes never respond to aNP grafts (Six4, Homer2). Finally, the aNP represses known Six1 regulators (Dlx5, Tfap2a) (Kwon et al., 2010; Qiao et al., 2012; Sato et al., 2010). Therefore, aNP-derived signals induce a mixture of neural and neural plate border cells rather than a unique territory and cannot initiate a complete PPR.
Similar results are observed in response to pNP-derived signals (Fig. S6). After 3 h, 28 genes are initiated, which are expressed widely in the normal embryo at HH3/4 (Table S4), except the neural plate border gene Irx1. Many transcripts continue to be present 6 and 12 h after pNP grafting, whereas 11 transcripts are only transiently induced. This is followed by neural plate (Sox2, Zic2, Znf423) and/or border genes (HH4+/5), with Eya2 being the only pNP-induced PPR marker. Simultaneously, non-neural markers and Six1 regulators (Gata2, Dlx3/5, Tfap2a) are repressed. Thus, the pNP induces a territory of mixed identity including many neural and non-neural transcripts, with Gbx2 and Irx2 indicating a posterior bias. Thus, like the aNP, the pNP induces different cell populations or cells with mixed identity (neural/placodal). Initially, aNP and pNP grafts induce distinct transcripts, whereas there is significant overlap later (Fig. S6). In summary, the neural plate induces many genes, but unlike the mesoderm it cannot induce the full transcriptional profile characteristic of placode progenitors.
Integration of signalling pathways during PPR induction
FGFs together with BMP and Wnt antagonists have previously been implicated in PPR induction (Brugmann et al., 2004; Kwon et al., 2010; Litsiou et al., 2005). Having identified distinct transcriptional states as cells adopt sensory precursor identity, we can now dissect the role of each signal at different time points by combining mesoderm grafts with pathway manipulation.
FGF initiates PPR induction
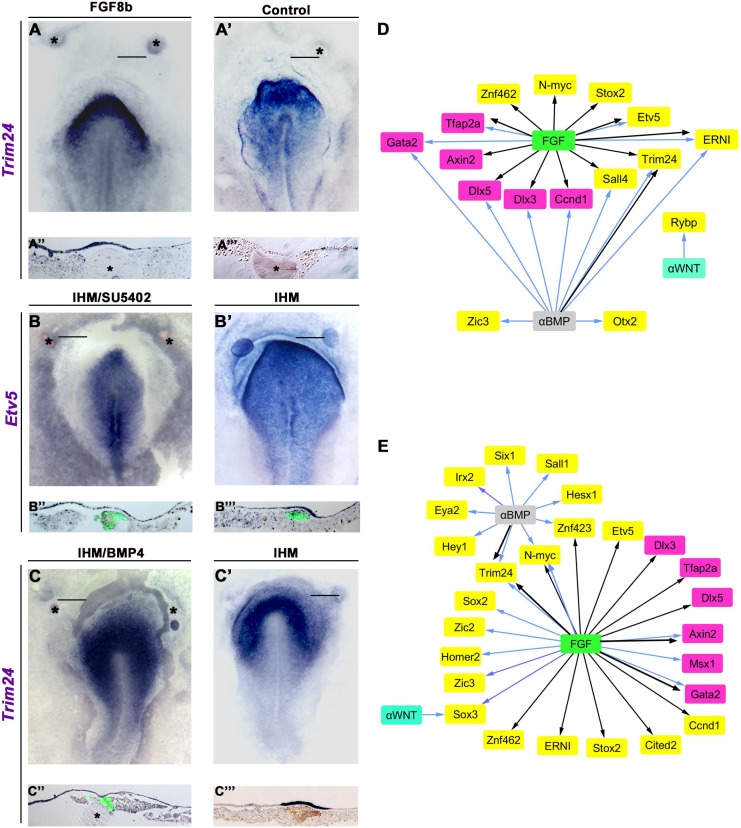
FGF signalling is required for PPR induction (Litsiou et al., 2005). To test which genes are FGF induced, we grafted FGF8-coated beads into the HH4− extraembryonic region and analysed FGF8-exposed and control epiblast gene expression after 3 and 6 h using NanoString. After 3 h, FGF8 induces a few genes (Etv5, Trim24, Ccnd1, ERNI, N-myc, Sall4; Fig. 5D, Fig. S7), largely overlapping with lHM-induced (6/8) and pME-induced (5/7) transcripts. ERNI is already known to be modulated by FGF8 (Streit et al., 2000). At 6 h, Etv5, Ccnd1, N-myc and Trim24 continue to be upregulated, as are some additional transcripts (Stox2, Cited2, Znf462; Fig. 5D, Fig. S7). We confirmed the induction of Trim24 (Fig. 5A,A″; 4/4) and N-myc (not shown; 6/9) by in situ hybridisation. By contrast, FGF8 – like the mesoderm – represses Tfap2a, Dlx3, Dlx5, Gata2 and Axin2 (Fig. 5D, Fig. S7, Table S3).
Fig. 5.
Signals controlling mesoderm-response genes. (A-A‴) Fgf8-coated (A,A″), but not control (A′,A‴), beads induce Trim24 after 3 h (4/4). (B-B‴) FGF signalling is required for the induction of Etv5 by lHM. lHM (green in B″,B‴) induces Etv5 in extraembryonic epiblast (B′,B‴; 5/6), and this is inhibited in the presence of SU5402-coated beads (B,B″; 4/4). (C-C‴) Trim24 is induced by lHM grafts (C′, brown in C‴; 3/3), and this induction is inhibited in the presence of BMP4-coated beads (C,C″; 4/4). Asterisks mark beads. Bars indicate level of sections shown beneath. (D,E) Different lHM (D) and pME (E) induced (yellow) or repressed (magenta) genes respond to modulation of FGF, BMP (αBMP, BMP antagonist) or Wnt (αWNT, Wnt antagonist). Black arrows indicate that signal is sufficient; blue arrows indicate that signal is required; see Table S3 and Fig. S7.
To assess the requirement of FGF signalling we combined lateral or axial mesoderm grafts with control beads or beads coated with the FGF receptor antagonist SU5402 (Fig. S7, Table S3). Both tissues induce Etv5 and N-myc; however, the requirement for FGF signalling differs: although FGF signalling is necessary for Etv5 induction by the lHM (Fig. 5B,B″) it is not required for its induction by the pME, whereas the opposite is true for N-myc (Fig. 5D,E, Fig. S7, Table S3). ERNI induction by the lHM requires FGF signalling as do pME-induced genes (Sox3, Homer2) and the neural genes Sox2, Zic2 and Zic3 (Fig. 5D,E, Fig. S7, Table S3). Finally, FGF signalling is necessary for Gata2 repression by both tissues, for Tfap2a suppression by the lHM, and for inhibition of Msx1 and Axin2 by the pME (Fig. 5D,E, Fig. S7, Table S3).
Together, these results show that FGF signalling is sufficient to induce or repress many genes that characterise the earliest response to mesoderm-derived signals (Fig. 5D,E, Fig. S7). However, only a few transcripts strictly depend on FGF, suggesting that each tissue contains other signals that can compensate for FGF loss.
BMP antagonism is required throughout PPR induction
The role of BMP signalling in PPR formation changes over time: during gastrulation it is required to induce Six/Eya regulators, but subsequently must be reduced to allow the emergence of sensory progenitors (Ahrens and Schlosser, 2005; Brugmann et al., 2004; Kwon et al., 2010; Litsiou et al., 2005). To test which mesoderm-response genes change after BMP pathway modulation, we reduced BMP signalling by growing primitive streak stage embryos in the BMP inhibitor dorsomorphin. After 3 and 6 h of culture, 11 transcripts are upregulated in PPR-competent epiblast when compared with stage-matched controls (Table S3). However, of these only Trim24 is induced by both mesodermal tissues (see Fig. 3C′).
To test which mesoderm-induced genes require BMP antagonism, we compared the inducing ability of mesoderm alone or mesoderm together with BMP4-coated beads after 3 and 6 h. BMP activation leads to the loss of Trim24, Ccnd1, Sall4, Otx2, ERNI and Zic3 induction by the lHM (Fig. 5D, Fig. S7, Table S3); we confirmed the reduction of Trim24 by in situ hybridisation (Fig. 5C,C″). Of the 23 pME-induced transcripts, ten are repressed after BMP activation, including Trim24, N-myc and the PPR genes Six1 and Eya2 (Fig. 5E, Fig. S7, Table S3). Likewise, the downregulation of some transcripts by the lHM is sensitive to BMP activation: Dlx3, Dlx5 and Gata2 increase when compared with control grafts (Fig. 5D, Fig. S7). By contrast, the pME continues to suppress Dlx genes even at elevated BMP levels, while Msx1 and Axin2 inhibition requires BMP antagonism (Fig. S7). Therefore, although BMP inhibition alone is sufficient to activate only a few mesoderm-induced genes, many transcripts require BMP antagonism, among them both early and late mesoderm-response genes. In addition, these findings highlight that although both mesoderm populations induce similar sets of transcription factors, each tissue provides a distinct signalling environment where other signals can compensate for the loss of a particular pathway.
Wnt antagonism is not required during early PPR induction
At the border of the neural plate increased levels of Wnt signalling promote neural crest formation at the expense of PPR, whereas Wnt antagonists have the opposite effect (Litsiou et al., 2005). To test which transcripts respond to Wnt modulation at different time points, we exposed the extraembryonic epiblast to the Wnt antagonist IWR and compared gene expression with control untreated epiblast using NanoString after 3 and 6 h. Wnt inhibition results in the upregulation of several genes, including Dlx5 and Nfkb1 as well as the Six1 co-factor Dach1 (Table S2). However, none of these is induced by mesoderm grafts, suggesting that antagonising Wnt signalling does not mimic the response to mesoderm signals.
To assess whether mesoderm-induced genes depend on Wnt antagonism, we combined tissue grafts with control DMSO-coated beads or beads coated with the Gsk3 inhibitor BIO to activate canonical Wnt signalling. Only a few genes, which are normally induced after 6 h, are affected: Rybp is no longer induced by the lHM, while Sox3 is no longer upregulated by the pME (Fig. 5D,E, Table S3, Fig. S7). Thus, modulation of Wnt signalling does not play a major role during the initial phase of PPR induction, but might be important for the decision between neural crest and placode progenitors (Litsiou et al., 2005; Villanueva et al., 2002).
Sonic hedgehog signalling is required for aPPR induction
The above experiments demonstrate that the axial mesendoderm is important to imbue sensory progenitor cells with anterior character. What is the nature of the anteriorising signal? Mesendoderm-derived somatostatin mediates PPR induction but does not impart regional identity (Lleras-Forero et al., 2013). Since sonic hedgehog (Shh) is prominently expressed in the pME (Dale et al., 1997), we tested whether it is required for aPPR formation. We grafted pME next to competent epiblast of HH4− hosts with or without cyclopamine, an Shh inhibitor (Fig. 6A-C). After 16 h, the general PPR markers Six1 (Fig. 6A) and Eya2 (not shown) continue to be induced, whereas the anterior markers Six3 and Otx2 are reduced (Fig. 6B,C). Shh-coated beads are not sufficient to induce PPR or anterior markers in the extraembryonic epiblast. We therefore tested whether Shh can anteriorise the lHM-induced PPR. When lHM grafts are combined with Shh-coated beads, the anterior marker Six3 is not induced (Fig. S11). Together, these data indicate that Shh alone is not sufficient to impart anterior identity or induce sensory progenitors, but plays a role in conferring rostral character to the PPR.
Fig. 6.
Sonic hedgehog is required for aPPR induction. CMFDA-labelled (green fluorescence) pME was grafted together with cyclopamine-coated beads into the extraembryonic region of chick hosts. After 16 h, Six1 is induced (A-A″; n=7/9 induced; no significant difference to controls, goodness of fit test). By contrast, Six3 (B-B″; 4/13 induced compared with 8/11 in controls; P=0.00166, goodness of fit test; see Fig. 4E′) and Otx2 (C-C″; 0/4 induced compared with 3/3 in controls; P=0.024, two-tailed goodness of fit test; see Fig. S11) induction is lost. Arrowheads indicate grafts; asterisks indicate beads. ISH, in situ hybridisation.
Network predictions support a transcriptional hierarchy during PPR induction
Our analysis suggests a gradual transition of epiblast cells towards placode progenitor fate, each step being defined by distinct transcriptional regulators and controlled by a combination of signals. Based on timecourse analysis we propose that at the top of the hierarchy a small set of transcription factors defines a ‘primed ectoderm state’ and controls the expression of a second tier of factors, which then directly regulate the Six/Eya cassette in the PPR. To test this model, we used an unbiased approach to investigate the topology of the PPR genetic network.
Using the NanoString datasets we inferred a regulatory network using the GENIE3 algorithm (Huynh-Thu et al., 2010). We created a network using all interactions with an importance measure of 0.02 or higher (Fig. S8A), which highlights important nodes (potential regulators) and weighted directional interactions. Visual inspection suggests the existence of three potential subclusters, which indeed emerge upon further dissection of the network using community clustering (Fig. S8, clusters G1-G3). Six1, Six4, Eya2 and other PPR-specific genes cluster together with known Six1 upstream regulators (Fig. S8C, cluster G2). In addition, G2 contains several early mesoderm-response genes including Otx2, ERNI, Sall1, Sall4, Ccnd1, Trim24, Zic3 and the FGF mediators Etv4 and Etv5 (Fig. S8C). Thus, the network predictions correlate well with our proposed model.
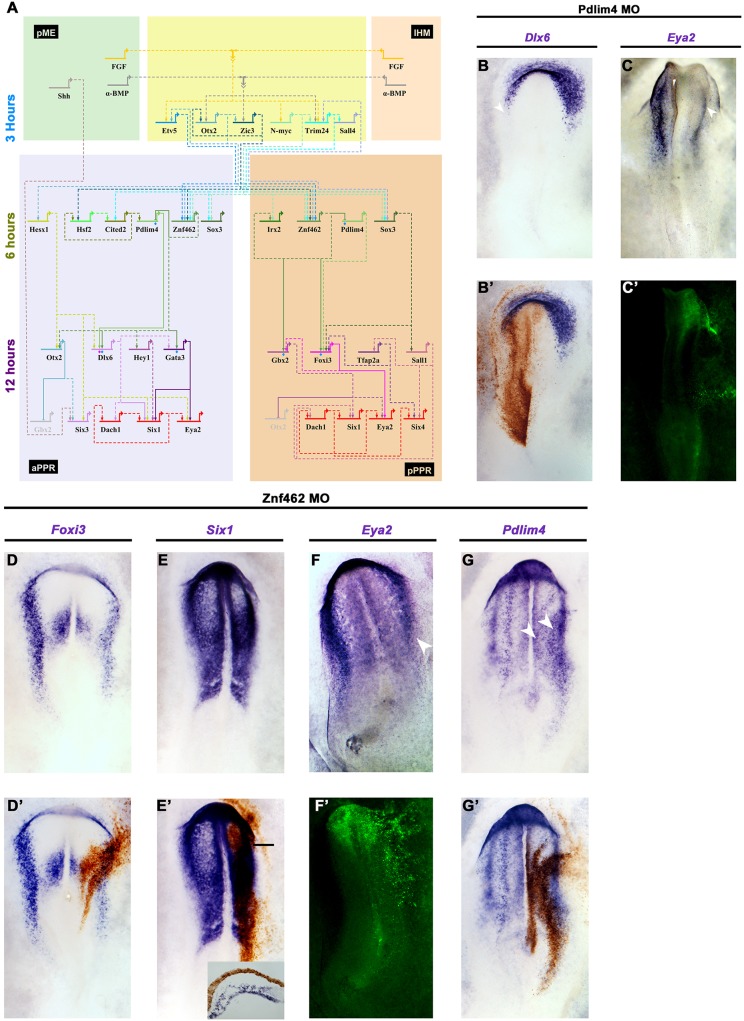
To assess potential direct regulators of Six1 and Eya2, we extracted their nearest neighbours from the network (Fig. S8A, Fig. S9). Using this information together with our induction timecourse and the spatial information from in situ hybridisation, we constructed a gene regulatory network in BioTapestry (Fig. 7A). Genes not induced by the mesoderm or not co-expressed with Six1 and Eya2 (e.g. Hey2, Irx1) were excluded from the BioTapestry network as potential direct interactors. Many first neighbours are shared by Six1 and Eya2, including known upstream regulators such as Gata3 and Dlx6 (Kwon et al., 2010; Sato et al., 2010). In addition, the network predicts novel regulatory relationships between the Six1/Eya2 complex and Sall1, Gbx2, Hey1 and Hesx1. To corroborate these predictions, we analysed the transcription factor-binding sites of the only PPR enhancer so far identified, Six1-14 (Sato et al., 2010), which directs Six1 expression in the aPPR. This analysis confirms the presence of Gata, Dlx, Gbx and Sall motifs (Table S5), indicating that these factors may indeed directly regulate Six1 (Fig. 7A, bottom, 12 h, purple genes).
Fig. 7.
A predicted gene network reveals new candidate Six1 and Eya2 regulators. (A) BioTapestry network that integrates time series of transcript induction and interactions predicted from GENIE3. Genes induced after 3 h, blue; 6 h, green; 12 h, purple; PPR genes, red. Dashed lines indicate predicted interactions; solid lines indicate known interactions and those experimentally verified in this study (blue diamonds for Znf462, pink diamonds for Pdlim4). (B-C′) Pdlim4 knockdown results in reduction of Dlx6 (B,B′; 5/5) and Eya2 (C,C′; 3/6). Arrowheads indicate changes in gene expression. (D-G′) Znf462 knockdown leads to reduction of Foxi3 (D,D′; 4/5), Six1 (E,E′; 4/5; inset, transverse section at level of bar) and Eya2 (F,F′; 4/6; arrowhead indicates changes in Eya2 expression), but to an expansion of Pdlim4 into the neural plate (G arrowheads, G′; 3/5). Morpholinos are visualised by DAB staining (brown) or by fluorescence (green).
By contrast, none of the early mesoderm-response genes defining the primed ectoderm state (Etv5, Otx2, Zic3, N-myc, Trim24; Fig. 7A, top, 3 h, blue genes) is predicted to interact with Six1 and Eya2 directly. We therefore analysed their first neighbours in the GENIE3 network as well as those of the predicted direct Six1/Eya2 regulators (Gata3, Dlx6, Gbx2, Foxi3, Sall1; Fig. S9). This analysis reveals a small set of intermediate transcripts downstream of the genes defining the primed ectoderm, among them Hesx1, Znf462, Hsf2, Pdlim4, Irx2, Cited2 and Sox3, which are all induced 6 h after exposure to mesodermal signals (Fig. 7A, middle 6h, green genes). In turn, they are connected to the Six1/Eya2 regulators suggesting that they provide the link between 3 and 12 h induced genes. We have termed the transcriptional state where 3 and 6 h induced genes are co-expressed the ‘PPR-primed’ state. In summary, our network inference approach supports a gradual transition of epiblast cells towards PPR fate and highlights the hierarchical nature of this process.
Confirming predicted interactions
To test whether these intermediate genes are regulators in the PPR gene network, we assessed the potential role of two transcription factors, Znf462 and Pdlim4. We designed two morpholinos for each factor to knockdown their expression. Experimental or control morpholinos were electroporated at primitive streak stages, the aPPR and pPPR were dissected at HH6/7 and then analysed by NanoString. Whereas reduction of Znf462 changes the expression of many transcripts when compared with controls (aPPR, 30; pPPR, 94), knockdown of Pdlim4 only affects nine genes (Table S3). This analysis verifies five of six predicted links downstream of Znf462 and one of two downstream of Pdlim4. To corroborate these findings we performed in situ hybridisation for selected genes after Znf462 and Pdlim4 knockdown. Whereas control morpholinos have no effect (Fig. S10), the reduction of either gene prevents Six1 and Eya2 expression (Fig. 7C,E,F). Furthermore, Znf462 regulates Foxi3 and Pdlim4 (Fig. 7D,G): although Znf462 is required for Foxi3, it seems to repress Pdlim4. Finally, Pdlim4 knockdown leads to a reduction in Dlx6 expression as predicted (Fig. 7B). Thus, these findings provide experimental support for the GENIE3-predicted gene regulatory network, and suggest that genes induced within 6 h after mesoderm grafting link early response genes and PPR competence factors.
DISCUSSION
Sensory placode progenitors are specified at head process stages in the ectoderm surrounding the anterior neural plate (Baker and Bronner-Fraser, 2001; Grocott et al., 2012; Schlosser, 2010; Streit, 2008). Initially, they are competent to give rise to all cranial placodes and share common features, but as individual placodes emerge their developmental potential becomes gradually restricted (Bailey et al., 2006; Bailey and Streit, 2006; Schlosser, 2010; Streit, 2008). Thus, induction of sensory progenitors is thought to be a common step that generates a non-regionalised placode territory, which is later subdivided along the rostrocaudal axis. Here, we provide evidence that PPR induction and regionalisation occur simultaneously, suggesting that a homogeneous PPR without anterior-posterior identity does not exist. Having identified new genes in the regulatory cascade that specifies sensory progenitors, we investigated their temporal hierarchy. This allowed us to dissect the events upstream of the PPR specifiers Six1 and Eya2 and revealed how signals from different tissues gradually induce placode precursors with distinct rostrocaudal identity.
A transcriptional hierarchy gradually specifies sensory placode progenitors with distinct rostrocaudal character
Three independent approaches – a PPR induction assay, gene expression during normal development, and network analysis – lead us to propose a new multistep model for PPR induction. Although we describe discrete steps as cells acquire PPR identity, it is likely that in reality the transitions between them are fluid and represent a continuum. In the first step, signals from the lHM and the pME rapidly induce a small set of transcription factors including ERNI, Etv5, N-myc, Otx2, Zic3 and Trim24 (akin to ‘evocation’ in Waddington's model). This set of transcription factors is largely identical irrespective of the signalling source, suggesting that both tissues initially promote a common transcriptional state. In the normal embryo these factors are expressed very early in development, with their domains encompassing the future neural plate, neural crest and placode territory, indicating that before acquiring PPR identity cells pass through a ‘primed ectoderm state’ that is common to progenitors for both the central and peripheral nervous systems. How this state is induced in the normal embryo remains to be elucidated; however, our results raise the possibility that FGF signalling might, at least in part, mediate this process. FGF8 largely mimics the response to mesodermal signals (see Fig. 5), is required for the early, but not the late, steps of PPR induction (Litsiou et al., 2005) and is known to initiate neural and neural crest formation (Delaune et al., 2005; Linker and Stern, 2004; Monsoro-Burq et al., 2003; Streit et al., 2000; Stuhlmiller and García-Castro, 2012; Wilson et al., 2000). It is thus possible that, like neural induction, PPR formation starts much earlier than previously thought and our induction assay rapidly recapitulates normal development.
The second phase of PPR induction is characterised by a set of new transcription factors (Fig. 7A, middle), the expression of which in the embryo starts after that of the genes defining the primed ectoderm, but is equally broad. Together, they characterise a transcriptional state that we have termed ‘PPR-primed’. Among these, the zinc-finger transcription factor Znf462, which is induced by both the lHM and axial mesendoderm, emerges as a potential integrator in the PPR gene network: several early response factors are predicted to provide input to Znf462, while Znf462 itself regulates Foxi3 and is predicted to control Gbx2, Gata3 and Dlx6, which in turn control Six1 and Eya2. In this phase, PPR regionalisation begins (akin to ‘individuation’ in Waddington's model) and, under the influence of lHM and axial mesendoderm, anterior-posterior markers are upregulated. Finally, the bona fide PPR markers Six1/4, Dach1 and Eya2 are induced, as are PPR competence factors (Bhat et al., 2013; Kwon et al., 2010; Litsiou et al., 2005; Pieper et al., 2012), which might regulate Six1 directly. Thus, as different populations of mesoderm emerge from the primitive streak, they transform primed ectoderm into PPR with regional character, without inducing definitive placode fates.
Although this hierarchical model emerges from an ectopic induction assay, epiblast cells go through the same transcriptional states as they gradually acquire PPR fate. Genes defining the primed ectoderm are present early, some before gastrulation (Grocott et al., 2012), followed by our newly identified PPR-primed factors. Where the latter overlap with PPR competence factors (Gata, Dlx, Tfap2a; Bhat et al., 2013; Kwon et al., 2010; Pieper et al., 2012), PPR specifiers are induced. Competence factors are initially expressed throughout the entire non-neural ectoderm at low levels (Bhat et al., 2013; Kwon et al., 2010; Pieper et al., 2012), but their expression increases in sensory progenitors as development proceeds. Our network analysis suggests that this enhanced expression might be controlled by the newly identified transcription factors, such as Znf462. Whether any of the competence or newly identified factors act as pioneer factors for the Six/Eya network remains to be elucidated.
This model for PPR induction somewhat resembles Waddington's model for neural induction, which proposes that the organizer initially induces a tissue of generic neural character and then imparts regional identity (Waddington and Needham, 1936). Although placode progenitor induction does not involve a bona fide organizer, but is instead mediated by different tissues, both initially induce a common transcriptional state (evocation) without rostrocaudal character, and subsequently impart regional bias (individuation). At this point PPR cells are specified as placode progenitors, but not yet committed to a specific placodal fate (reviewed by Bailey and Streit, 2006; Grocott et al., 2012; Streit, 2008). Together, our data suggest that during PPR induction cells initially pass through a primed ectoderm state, which is characterised by only a handful of genes, followed by a PPR-primed state defined by newly identified transcripts such as Znf462. Subsequently, mesoderm signals divert cells towards placode fate and simultaneously impart regional bias depending on their location along the rostrocaudal axis.
Tissues and signals in PPR initiation and regionalisation
Signals from the neural plate and the lHM have previously been implicated in placode progenitor induction (Ahrens and Schlosser, 2005; Litsiou et al., 2005). Here, we identify the pME as an inducer of aPPR fate. Assessing the behaviour of 126 transcripts simultaneously allows us to dissect the dynamic response to all three tissues and reveals that, although mesodermal signals initiate the full set of PPR transcripts in the induction assay, the neural plate alone is not sufficient to do so. It is likely that in normal development signals from the neural plate contribute to PPR induction. However, our findings suggest that they may play a less prominent role in initiating induction, but might be important to define regional character of the tissue induced.
FGF signalling mimics some of the earliest responses to signals from the mesoderm, although it is strictly required for only a few rapidly induced genes (Litsiou et al., 2005). Although BMP inhibition is sufficient to induce only two transcripts, namely Trim24 and Irx1, it is required for the induction of many more. Together, both pathways account for the majority of lHM-induced genes and for a considerable number of transcripts initiated by the pME. In particular, our findings confirm that antagonising BMP signalling is crucial for the expression of the PPR specifiers Six1 and Eya (Ahrens and Schlosser, 2005; Brugmann et al., 2004; Litsiou et al., 2005). Together, these results suggest that the cooperation of FGF activation and BMP antagonism is crucial during the early phase of PPR induction and that both contribute to establishing a PPR-primed state, but are not sufficient for PPR specification.
By contrast, Wnt signalling only appears to play a minor role during the first step of PPR specification. Wnt antagonism does not mimic any activity of the mesoderm and only very few genes depend on Wnt inhibition. This suggests that modulation of canonical Wnt signalling is important during the late phase of PPR induction, where it might mediate different processes. On the one hand, Wnt antagonists mediate the decision between neural crest and placode precursors at the border of the neural plate and protect PPR cells from Wnts emanating from surrounding tissues (Brugmann et al., 2004; Litsiou et al., 2005). On the other hand, Wnt antagonism might also be important to promote pPPR identity. Although active Wnt signalling is generally considered to posteriorise neural and non-neural ectoderm (Wilson and Houart, 2004), Wnt inhibition might be required to fine-tune pre-placodal fate. Finally, while studies in zebrafish have implicated Shh in the decision between adenohypophysis and lens character (Dutta et al., 2005), our results suggest an even earlier role at pre-placodal stages. Shh and somatostatin (Lleras-Forero et al., 2013) from the axial mesendoderm may cooperate to initiate Pnoc expression in aPPR cells, which in turn is required for the expression of aPPR transcription factors.
Conclusions
The combination of embryological timecourse experiments analysing more than 100 genes and network analysis reveals a new multistep model for PPR induction. We demonstrate that the acquisition of placode progenitor fate occurs gradually and that cells transit through different transcriptional states, each defined by distinct factors. The first step generates primed ectoderm cells, followed by a PPR-primed state, which might be shared with neural and neural crest induction. Subsequently, two different tissues, namely the pME and the lHM, gradually impart anterior and posterior PPR character, respectively. Thus, PPR induction and regionalisation occur simultaneously, suggesting that, once induced, PPR cells do not share the same transcriptional profile. However, since both tissues initially elicit an almost identical response and cells only diverge later, PPR induction is somewhat reminiscent of Waddington's evocation-individuation model for neural induction.
MATERIALS AND METHODS
Embryo techniques and in situ hybridisation
Fertile hens' (Winter Farm) and quails' (Potter Farm) eggs were incubated at 38°C to obtain embryos of appropriate Hamburger and Hamilton (HH) stages (Hamburger and Hamilton, 1951). Chick embryos at stage 3+/4− were cultured according to New (1955) as modified by Stern and Ireland (1981). To isolate quail and chick tissues for grafting, embryos were collected in Tyrode's saline. lHM and pME were dissected from HH5/6 donors using fine steel needles and small amounts of dispase (1 mg/ml). Neural plate from different rostrocaudal levels was obtained from HH6 donors after removal of the endoderm and mesoderm. For fluorescently labelled grafts, tissue was collected as above and incubated for 45 min at 38°C in DMEM containing 10 µM 5-chloromethylfluorescein diacetate (CMFDA; Thermofisher). Tissues were kept on ice in Tyrode's saline until grafted into the inner margin of the extraembryonic area opaca of HH4− hosts.
To inhibit Wnt or BMP signalling, whole embryos (HH4−) were cultured in modified New culture (Stern and Ireland, 1981) with albumen containing 30 µM IWR-1 (Sigma Aldrich) or 20 µM dorsomorphin (Tocris Bioscience), respectively. Heparin beads were coated in 50 µg/ml FGF8 (R&D Systems) in PBS containing 0.1% BSA on ice for 1 h, washed briefly in Tyrode's saline and grafted into the inner third of the area opaca of HH4− stage chick embryos. Heparin beads were coated with 100 µg/ml Shh (R&D Systems) in PBS containing 0.1% BSA for 1 h on ice, washed in Tyrode's saline and grafted into the area opaca alone or in combination with lHM. To modulate different signalling pathways, AG1X2 beads (Sigma Aldrich) were coated with 1 µg/ml BMP4 (R&D Systems) in PBS containing 0.1% BSA for 1 h on ice, with DMSO (control), 25 µM SU5402 (Tocris Bioscience) or 2.5 µM BIO (Sigma Aldrich) in DMSO at room temperature for 2 h. Affi-Gel blue beads (Bio-Rad) were coated with 1 µM cyclopamine (Sigma Aldrich) in DMSO for 2 h. Beads were then washed in PBS and grafted together with lHM or pME. SU5402 is yellow, and beads retain their colour for at least 6 h after grafting, but have released all inhibitor after 15 h and appear white. To ensure that BMP4 beads are effective over the 6 h timecourse, we grafted beads next to the neural tube and assessed Sox2 expression (Fig. S11). The effectiveness of IWR and dorsomorphin was assessed after growing embryos from HH5/6 in the presence of these drugs and assessing heart and somite formation (Fig. S11). As positive controls for Shh and cyclopamine we grafted beads next to the node at HH4 and assayed for Pitx2 expression after 12 h of culture (Fig. S11).
For whole-mount in situ hybridisation, embryos were harvested in PBS, fixed in 4% paraformaldehyde and processed for hybridisation using DiG-labelled antisense mRNA as previously described (Streit and Stern, 2001). EST clones are listed in Table S6. To reveal quail tissue, QCPN antibody was used (1:5; AB 531886, Developmental Studies Hybridoma Bank).
Electroporation and morpholino knockdown experiments
Two independent fluorescein-labelled morpholinos were designed by GeneTools targeting Pdlim4 (MO1, 3′-CGACACCACGTGCACCATACC-5′; MO2, 3′-CATCCACTTAAAGCGGCTCCGAGGC-5′) and Znf462 (MO1, 3′-AGACACACAGATCCTTACCCTCTCT-5′; MO2, 3′-TGCAGCACCTCCATGGTTCAAGGAT-5′). These are splice-blocking morpholinos; blastN does not reveal any other potential targets and their effectiveness was assessed by RT-PCR. Control morpholino was 3′-CCTCTTACCTCAGTTACAATTTATA-5′. For electroporation, each morpholino (1 µg/µl) was mixed with plasmid DNA (0.5 µg/µl) as a carrier and 0.01% Fast Green, and injected between the vitelline membrane and the epiblast. Primitive streak stage embryos (HH3+/4−) were prepared for New culture; morpholinos were transferred into epiblast cells using four 5 mV pulses for 50 ms each, with an interval of 500 ms. Targeted cells are visualised by fluorescence or by immunostaining with anti-fluorescein antibodies. For NanoString analysis, electroporated aPPR and pPPR was dissected from HH6/7 after electroporation.
Microarray analysis
To identify differentially regulated transcripts, lHM was grafted into the area opaca of HH4− chick hosts. After 12 h, mesoderm grafts were removed using small amounts of dispase and the underlying area opaca epiblast was collected together with control epiblast from the contralateral side. Tissue collection was repeated on three independent occasions, with 35-40 explants collected for each sample. Using 5 ng total RNA, labelled cell extracts were prepared and hybridised to Affymetrix Chick GeneChip according to Chambers and Lumsden (2008). Probe level values were derived from the raw data using the MAS5 algorithm (version 1.2, Affymetrix). Data were analysed using the GeneSpring package (version 7.3.1, Agilent Technologies). The suitability of the datasets for further analysis and the relationship between and within the biological replicates was determined using principal components analysis and hierarchical clustering. Differential expression between the conditions under investigation was determined by a stepwise process. Samples were first normalised to the 50th percentile across the whole expression dataset and then each gene was normalised to the median of its own expression across each cell type. Prior to statistical analysis, genes classed as not expressed (absent in biological replicates) or not varying in expression above a 2-fold threshold in any of the cell types were removed from the analysis. From the remaining set of genes, those with expression levels that differ significantly (P≤0.05) between each cell type were determined by one-way ANOVA. Microarray data generated in this study were deposited in Gene Expression Omnibus (GSE81023). Transcripts expressed in the aPPR and pPPR have been identified previously (Lleras-Forero et al., 2013; GEO accession GSE48116). For hierarchical clustering and heatmap generation, the R statistical packages Hclust and Heatmap.2 were used.
NanoString nCounter
Area opaca exposed to signals from different tissues and non-induced area opaca explants were collected at 3, 6 and 12 h after grafting. Three independent replicates of 5-10 explants (5000-10,000 cells) per condition were collected on ice in RNase-free PBS, which was then replaced by lysis buffer (Ambion) and the lysed tissues were quickly spun (13,000 rpm in an Eppendorf 5415R) before being snap frozen on dry ice. Samples were stored at −80°C until required. RNA lysates were hybridised at 65°C overnight, eluted according to the nCounter manual and counted by the nCounter digital analyser. Counts were normalised to the positive controls present in each hybridisation mix. Subsequently, the negative control probe values were used to create a background threshold level; transcripts with expression values below the threshold were removed from further analysis. Counts were then normalised to the total amount of mRNA counted in each sample. Differential expression of transcripts between different conditions was determined using an unpaired two-tailed Student's t-test comparing the average of three biological replicates (P<0.05, >1.2-fold change).
GENIE3-inferred network analysis
NanoString data from tissue grafting timecourses and from signalling experiments that did not use DMSO (i.e. lHM/BMP4 for 3 and 6 h; FGF8 for 3 and 6 h) were used to generate a predicted gene regulatory network. The mean expression values for each gene under each condition were analysed by the GENIE3 algorithm (Huynh-Thu et al., 2010). In brief, this algorithm decomposes n genes into n different regression problems. For each regression problem, the expression profile of one gene (target gene) is predicted from the expression profiles of all other genes (input genes) using tree-based ensemble methods. Within this, the importance of a single input gene in explaining the profile of the target gene is assessed and an importance measure is generated. This importance measure is then used to predict the regulatory links and their direction within the network. Following analysis, interactions above an importance measure of 0.02 were isolated, based on the strength of their importance measure, and the network was viewed using Cytoscape (www.cytoscape.org/). For subsequent analysis, genes of interest were highlighted with their first neighbours (putative regulators and targets) and small networks were created. To identify more closely related genes, community clustering was performed using the GLay plugin in Cytoscape (Su et al., 2010).
Acknowledgements
We are grateful to Ewa Kolano for excellent technical assistance, Ramya Ranganathan for Cux1 in situ hybridisation in Fig. 2D′, Laura Lleras-Forero for in situ hybridisation of HH4 Ece1 in Fig. S2 and Dmbx1 in Fig. S3, Claudio D. Stern for comments on the manuscript, and the A.S. laboratory for discussions. QCPN antibody was obtained from the Developmental Studies Hybridoma Bank, maintained by the Department of Pharmacology and Molecular Sciences, The Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA and the Department of Biological Sciences, University of Iowa, Iowa City 52242, USA under contract N01-HD-2-3144 from the NICHD.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.H., A.S.; Formal analysis: M.H., A.S.; Investigation: M.H., R.S.P., M.T., N.A.D.C., M.A., T.G.; Writing - original draft: M.H., A.S.; Writing - review & editing: A.S.; Visualization: M.H., R.S.P., M.T.; Supervision: A.S.; Project administration: A.S.; Funding acquisition: A.S.
Funding
This work was supported by a Diana Trebble Studentship from King's College London Dental Institute, a Studentship from Defeating Deafness UK (now Action on Hearing Loss) to A.S. and by project grants to A.S. from the National Institutes of Health (DE022065) and the Biotechnology and Biological Sciences Research Council (BB/I021647/1). Deposited in PMC for release after 6 months.
Data availability
Microarray data generated in this study have been deposited in Gene Expression Omnibus under accession number GSE81023.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.147942.supplemental
References
- Ahrens K. and Schlosser G. (2005). Tissues and signals involved in the induction of placodal Six1 expression in Xenopus laevis. Dev. Biol. 288, 40-59. 10.1016/j.ydbio.2005.07.022 [DOI] [PubMed] [Google Scholar]
- Bailey A. P. and Streit A. (2006). Sensory organs: making and breaking the pre-placodal region. Curr. Top. Dev. Biol. 72, 167-204. 10.1016/S0070-2153(05)72003-2 [DOI] [PubMed] [Google Scholar]
- Bailey A. P., Bhattacharyya S., Bronner-Fraser M. and Streit A. (2006). Lens specification is the ground state of all sensory placodes, from which FGF promotes olfactory identity. Dev. Cell 11, 505-517. 10.1016/j.devcel.2006.08.009 [DOI] [PubMed] [Google Scholar]
- Baker C. V. H. and Bronner-Fraser M. (2001). Vertebrate cranial placodes I. Embryonic induction. Dev. Biol. 232, 1-61. 10.1006/dbio.2001.0156 [DOI] [PubMed] [Google Scholar]
- Bally-Cuif L., Gulisano M., Broccoli V. and Boncinelli E. (1995). C-Otx2 is expressed in two different phases of gastrulation and is sensitive to retinoic acid treatment in chick embryo. Mech. Dev. 49, 49-63. 10.1016/0925-4773(94)00301-3 [DOI] [PubMed] [Google Scholar]
- Barembaum M. and Bronner-Fraser M. (2007). Spalt4 mediates invagination and otic placode gene expression in cranial ectoderm. Development 134, 3805-3814. 10.1242/dev.02885 [DOI] [PubMed] [Google Scholar]
- Bhat N., Kwon H.-J. and Riley B. B. (2013). A gene network that coordinates preplacodal competence and neural crest specification in zebrafish. Dev. Biol. 373, 107-117. 10.1016/j.ydbio.2012.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugmann S. A., Pandur P. D., Kenyon K. L., Pignoni F. and Moody S. A. (2004). Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development 131, 5871-5881. 10.1242/dev.01516 [DOI] [PubMed] [Google Scholar]
- Chambers D. and Lumsden A. (2008). Profiling gene transcription in the developing embryo: microarray analysis on gene chips. Methods Mol. Biol. 461, 631-655. 10.1007/978-1-60327-483-8_42 [DOI] [PubMed] [Google Scholar]
- Chen B., Kim E.-H. and Xu P.-X. (2009). Initiation of olfactory placode development and neurogenesis is blocked in mice lacking both Six1 and Six4. Dev. Biol. 326, 75-85. 10.1016/j.ydbio.2008.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou N. A. D., Bailey A. P., Hanson S. and Streit A. (2009). Activation of Six1 target genes is required for sensory placode formation. Dev. Biol. 336, 327-336. 10.1016/j.ydbio.2009.09.025 [DOI] [PubMed] [Google Scholar]
- Dale J. K., Vesque C., Lints T. J., Sampath T. K., Furley A., Dodd J. and Placzek M. (1997). Cooperation of BMP7 and SHH in the induction of forebrain ventral midline cells by prechordal mesoderm. Cell 90, 257-269. 10.1016/S0092-8674(00)80334-7 [DOI] [PubMed] [Google Scholar]
- Delaune E., Lemaire P. and Kodjabachian L. (2005). Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development 132, 299-310. 10.1242/dev.01582 [DOI] [PubMed] [Google Scholar]
- Dutta S., Dietrich J.-E., Aspöck G., Burdine R. D., Schier A., Westerfield M. and Varga Z. M. (2005). Pitx3 defines an equivalence domain for lens and anterior pituitary placode. Development 132, 1579-1590. 10.1242/dev.01723 [DOI] [PubMed] [Google Scholar]
- Goriely A., Diez del Corral R. and Storey K. G. (1999). c-Irx2 expression reveals an early subdivision of the neural plate in the chick embryo. Mech. Dev. 87, 203-206. 10.1016/S0925-4773(99)00149-5 [DOI] [PubMed] [Google Scholar]
- Grocott T., Johnson S., Bailey A. P. and Streit A. (2011). Neural crest cells organize the eye via TGF-β and canonical Wnt signalling. Nat. Commun. 2, 265-265 10.1038/ncomms1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grocott T., Tambalo M. and Streit A. (2012). The peripheral sensory nervous system in the vertebrate head: a gene regulatory perspective. Dev. Biol. 370, 3-23. 10.1016/j.ydbio.2012.06.028 [DOI] [PubMed] [Google Scholar]
- Hamburger V. and Hamilton H. L. (1951). A series of normal stages in the development of the chick embryo. J. Morphol. 88, 49-92. 10.1002/jmor.1050880104 [DOI] [PubMed] [Google Scholar]
- Henry J. J. and Grainger R. M. (1990). Early tissue interactions leading to embryonic lens formation in Xenopus laevis. Dev. Biol. 141, 149-163. 10.1016/0012-1606(90)90110-5 [DOI] [PubMed] [Google Scholar]
- Holtfreter J. (1933a). Der Einfluss von Wirtsalter und verschiedenen Organbezirken auf die Differenzierung von angelagertem Gastrulaektoderm. Roux's Arch. Entw. Mech. 127, 619-775. 10.1007/BF01380476 [DOI] [PubMed] [Google Scholar]
- Holtfreter J. (1933b). Eigenschaften und Verbreitung induzierender Stoffe. Naturwissenschaften 21, 766-770. 10.1007/BF01503741 [DOI] [Google Scholar]
- Huynh-Thu V. A., Irrthum A., Wehenkel L. and Geurts P. (2010). Inferring regulatory networks from expression data using tree-based methods. PLoS ONE 5, e12776 10.1371/journal.pone.0012776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A. G. (1963a). The determination and positioning of the nose, lens and ear. I. Interactions within the ectoderm, and between the ectoderm and underlying tissues. J. Exp. Zool. 154, 273-283. 10.1002/jez.1401540303 [DOI] [PubMed] [Google Scholar]
- Jacobson A. G. (1963b). The Determination and positioning of the nose, lens and ear. II. The role of the endoderm. J. Exp. Zool. 154, 285-291. 10.1002/jez.1401540304 [DOI] [PubMed] [Google Scholar]
- Khatri S. B., Edlund R. K. and Groves A. K. (2014). Foxi3 is necessary for the induction of the chick otic placode in response to FGF signaling. Dev. Biol. 391, 158-169. 10.1016/j.ydbio.2014.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khudyakov J. and Bronner-Fraser M. (2009). Comprehensive spatiotemporal analysis of early chick neural crest network genes. Dev. Dyn. 238, 716-723. 10.1002/dvdy.21881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H.-J., Bhat N., Sweet E. M., Cornell R. A. and Riley B. B. (2010). Identification of early requirements for preplacodal ectoderm and sensory organ development. PLoS Genet. 6, e1001133 10.1371/journal.pgen.1001133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laclef C., Souil E., Demignon J. and Maire P. (2003). Thymus, kidney and craniofacial abnormalities in Six 1 deficient mice. Mech. Dev. 120, 669-679. 10.1016/S0925-4773(03)00065-0 [DOI] [PubMed] [Google Scholar]
- Linker C. and Stern C. D. (2004). Neural induction requires BMP inhibition only as a late step, and involves signals other than FGF and Wnt antagonists. Development 131, 5671-5681. 10.1242/dev.01445 [DOI] [PubMed] [Google Scholar]
- Litsiou A., Hanson S. and Streit A. (2005). A balance of FGF, BMP and WNT signalling positions the future placode territory in the head. Development 132, 4051-4062. 10.1242/dev.01964 [DOI] [PubMed] [Google Scholar]
- Lleras-Forero L., Tambalo M., Christophorou N., Chambers D., Houart C. and Streit A. (2013). Neuropeptides: developmental signals in placode progenitor formation. Dev. Cell 26, 195-203. 10.1016/j.devcel.2013.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold O. (1933). Über die Induktionsfähigkeit der verschiedenen Bezirke der Neurula von Urodelen. Naturwissenschaften 21, 761-766. 10.1007/BF01503740 [DOI] [Google Scholar]
- Monsoro-Burq A.-H., Fletcher R. B. and Harland R. M. (2003). Neural crest induction by paraxial mesoderm in Xenopus embryos requires FGF signals. Development 130, 3111-3124. 10.1242/dev.00531 [DOI] [PubMed] [Google Scholar]
- New D. A. T. (1955). A new technique for the cultivation of the chick embryo in vitro. J. Embryol. Exp. Morph. 3, 326-331. [Google Scholar]
- Nieuwkoop P. D. and Nigtevecht G. V. (1954). Neural activation and transformation in explants of competent ectoderm under the influence of fragments of anterior notochord in Urodeles. J. Embryol. Exp. Morph. 2, 175-193. [Google Scholar]
- Paxton C. N., Bleyl S. B., Chapman S. C. and Schoenwolf G. C. (2010). Identification of differentially expressed genes in early inner ear development. Gene Expr. Patterns 10, 31-43. 10.1016/j.gep.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper M., Ahrens K., Rink E., Peter A. and Schlosser G. (2012). Differential distribution of competence for panplacodal and neural crest induction to non-neural and neural ectoderm. Development 139, 1175-1187. 10.1242/dev.074468 [DOI] [PubMed] [Google Scholar]
- Qiao Y., Zhu Y., Sheng N., Chen J., Tao R., Zhu Q., Zhang T., Qian C. and Jing N. (2012). AP2[gamma] regulates neural and epidermal development downstream of the BMP pathway at early stages of ectodermal patterning. Cell Res. 22, 1546-1561. 10.1038/cr.2012.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex M., Orme A., Uwanogho D., Tointon K., Wigmore P. M., Sharpe P. T. and Scotting P. J. (1997). Dynamic expression of chicken Sox2 and Sox3 genes in ectoderm induced to form neural tissue. Dev. Dyn. 209, 323-332. [DOI] [PubMed] [Google Scholar]
- Sato S., Ikeda K., Shioi G., Ochi H., Ogino H., Yajima H. and Kawakami K. (2010). Conserved expression of mouse Six1 in the pre-placodal region (PPR) and identification of an enhancer for the rostral PPR. Dev. Biol. 344, 158-171. 10.1016/j.ydbio.2010.04.029 [DOI] [PubMed] [Google Scholar]
- Saxen L. and Toivonen S. (1962). Primary Embryonic Induction. London: Logos/Academic. [DOI] [PubMed] [Google Scholar]
- Schlosser G. (2010). Making senses development of vertebrate cranial placodes. Int. Rev. Cell Mol. Biol. 283, 129-234. 10.1016/S1937-6448(10)83004-7 [DOI] [PubMed] [Google Scholar]
- Spemann H. (1938). Embryonic Development and Induction. New Haven: Yale University Press. [Google Scholar]
- Stern C. D. (2001). Initial patterning of the central nervous system: how many organizers? Nat. Rev. Neurosci. 2, 92-98. 10.1038/35053563 [DOI] [PubMed] [Google Scholar]
- Stern C. D. and Ireland G. W. (1981). An integrated experimental study of endoderm formation in avian embryos. Anat. Embryol. 163, 245-263. 10.1007/BF00315703 [DOI] [PubMed] [Google Scholar]
- Steventon B., Mayor R. and Streit A. (2012). Mutual repression between Gbx2 and Otx2 in sensory placodes reveals a general mechanism for ectodermal patterning. Dev. Biol. 367, 55-65. 10.1016/j.ydbio.2012.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A. (2008). The cranial sensory nervous system: specification of sensory progenitors and placodes. StemBook, doi/10.3824/stembook.1.31.1 10.3824/stembook.1.31.1 [DOI] [PubMed] [Google Scholar]
- Streit A. and Stern C. D. (2001). Combined whole-mount in situ hybridization and immunohistochemistry in avian embryos. Methods 23, 339-344. 10.1006/meth.2000.1146 [DOI] [PubMed] [Google Scholar]
- Streit A., Berliner A. J., Papanayotou C., Sirulnik A. and Stern C. D. (2000). Initiation of neural induction by FGF signalling before gastrulation. Nature 406, 74-78. 10.1038/35017617 [DOI] [PubMed] [Google Scholar]
- Stuhlmiller T. J. and García-Castro M. I. (2012). FGF/MAPK signaling is required in the gastrula epiblast for avian neural crest induction. Development 139, 289-300. 10.1242/dev.070276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G., Kuchinsky A., Morris J. H., States D. J. and Meng F. (2010). GLay: community structure analysis of biological networks. Bioinformatics 26, 3135-3137. 10.1093/bioinformatics/btq596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto T., Uchikawa M., Kamachi Y. and Kondoh H. (2006). Convergence of Wnt and FGF signals in the genesis of posterior neural plate through activation of the Sox2 enhancer N-1. Development 133, 297-306. 10.1242/dev.02196 [DOI] [PubMed] [Google Scholar]
- Villanueva S., Glavic A., Ruiz P. and Mayor R. (2002). Posteriorization by FGF, Wnt, and retinoic acid is required for neural crest induction. Dev. Biol. 241, 289-301. 10.1006/dbio.2001.0485 [DOI] [PubMed] [Google Scholar]
- Waddington C. H. and Needham J. (1936). Evocation, individuation, and competence in amphibian organizer action. Proc. Kon. Akad. Wetensch. Amsterdam 39, 887-891. [Google Scholar]
- Wilson S. W. and Houart C. (2004). Early steps in the development of the forebrain. Dev. Cell 6, 167-181. 10.1016/S1534-5807(04)00027-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S. I., Graziano E., Harland R., Jessell T. M. and Edlund T. (2000). An early requirement for FGF signalling in the acquisition of neural cell fate in the chick embryo. Curr. Biol. 10, 421-429. 10.1016/S0960-9822(00)00431-0 [DOI] [PubMed] [Google Scholar]
- Zheng W., Huang L., Wei Z.-B., Silvius D., Tang B. and Xu P.-X. (2003). The role of Six1 in mammalian auditory system development. Development 130, 3989-4000. 10.1242/dev.00628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D., Silvius D., Fritzsch B. and Xu P.-X. (2004). Eya1 and Six1 are essential for early steps of sensory neurogenesis in mammalian cranial placodes. Development 131, 5561-5572. 10.1242/dev.01437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D., Silvius D., Rodrigo-Blomqvist S., Enerbäck S. and Xu P.-X. (2006). Eya1 regulates the growth of otic epithelium and interacts with Pax2 during the development of all sensory areas in the inner ear. Dev. Biol. 298, 430-441. 10.1016/j.ydbio.2006.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]