SUMMARY
The ring-shaped cohesin complex regulates transcription, DNA repair, and chromosome segregation by dynamically entrapping chromosomes to promote chromosome compaction and sister-chromatid cohesion. The cohesin ring needs to open and close to allow its loading to and release from chromosomes. Cohesin dynamics are controlled by the releasing factors Pds5 and Wapl and the cohesin stabilizer Sororin. Here, we report the crystal structure of human Pds5B bound to a conserved peptide motif found in both Wapl and Sororin. Our structure establishes the basis for how Wapl and Sororin antagonistically influence cohesin dynamics. The structure further reveals that Pds5 can bind inositol hexakisphosphate (IP6). The IP6-binding segment of Pds5B is shaped like the jaw of a plier lever and inhibits the binding of Scc1 to Smc3. We propose that Pds5 stabilizes a transient, open state of cohesin to promote its release from chromosomes.
INTRODUCTION
Cohesin regulates diverse cellular processes, including chromosome segregation and compaction, DNA repair, and transcription (Haarhuis et al., 2014; Nasmyth and Haering, 2009; Onn et al., 2008; Peters et al., 2008). All functions of cohesin likely involve its unique ability to topologically entrap chromosomes within its ring (Haering et al., 2008). Dynamic entrapment of distal elements in the same chromosome by cohesin in G1 produces chromosome loops and impacts transcription (Merkenschlager and Odom, 2013). Stable cohesin entrapment of sister chromatids coupled to DNA replication or induced by DNA damage establishes sister-chromatid cohesion, which is critical for accurate chromosome segregation and homology-directed DNA repair (Rolef Ben-Shahar et al., 2008; Strom et al., 2007; Unal et al., 2008; Wu et al., 2012). Mutations of cohesin and its regulators perturb many facets of chromosome biology, and are linked to human cancers and developmental diseases (Bose and Gerton, 2010; Solomon et al., 2011).
Chromosome entrapment by cohesin is regulated during the cell cycle. Chromosome-bound cohesin is highly dynamic in G1 (Gerlich et al., 2006). The Huntingtin-Elongation factor 3-A subunit-TOR (HEAT) repeat proteins, Pds5 and Wapl, release cohesin from chromosomes, in a reaction that requires the opening of the cohesin ring at the Smc3–Scc1 interface (termed the DNA exit gate) (Buheitel and Stemmann, 2013; Chan et al., 2012; Gandhi et al., 2006; Gligoris et al., 2014; Huis in’t Veld et al., 2014; Kueng et al., 2006; Murayama and Uhlmann, 2015; Rowland et al., 2009). During S phase, DNA replication-coupled Smc3 acetylation by Esco1/2 inhibits Pds5–Wapl-dependent cohesin-releasing activity and establishes sister-chromatid cohesion (Chan et al., 2012; Rolef Ben-Shahar et al., 2008; Rowland et al., 2009; Sherwood et al., 2010; Unal et al., 2008). In metazoans, cohesion establishment also requires Sororin, which competes with Wapl for binding to Pds5 (Nishiyama et al., 2010; Rankin, 2005). In mitosis, cohesin is removed from chromosomes by Pds5–Wapl-dependent release and separase-dependent cleavage (Gandhi et al., 2006; Kueng et al., 2006; Shintomi and Hirano, 2009; Uhlmann et al., 2000), leading to chromosome segregation.
Paradoxically, Pds5 has both positive and negative functions in cohesion regulation (Chan et al., 2012; Losada et al., 2005; Rowland et al., 2009; Shintomi and Hirano, 2009; Sutani et al., 2009). Pds5 promotes Smc3 acetylation through recruiting Esco1 to cohesin (Minamino et al., 2015; Vaur et al., 2012). In vertebrates, Pds5 interacts with Sororin (Nishiyama et al., 2010). These Pds5-dependent events are expected to stabilize cohesin on chromosomes and strengthen sister-chromatid cohesion. On the other hand, Pds5 engages and collaborates with Wapl to promote the release of cohesin from chromosomes (Murayama and Uhlmann, 2015; Nishiyama et al., 2010; Shintomi and Hirano, 2009).
To better define the dual functions of Pds5, we determined the crystal structure of human Pds5B bound to a conserved tyrosine-serine-arginine (YSR) motif found in both Wapl and Sororin. The YSR motifs of Wapl and Sororin bind to the same conserved site on Pds5B and compete for Pds5B binding, helping to explain the well-established Wapl-Sororin antagonism in cohesin regulation. Our structure unexpectedly reveals inositol hexakisphophate (IP6) as a tightly bound cofactor of Pds5. The IP6-binding segment of Pds5 and its flanking region form a jaw-like structure to engage the N-terminal region of Scc1 and inhibit its binding to Smc3. These findings suggest a rather direct role of Pds5 in cohesin release from chromosomes, possibly through stabilizing a fleeting, open state of cohesin during its ATPase cycle.
RESULTS
Identification of a Conserved Pds5-binding Motif in Human Wapl and Sororin
We characterized the binding of bacterially expressed human Wapl and Sororin to human Pds5B expressed and purified from insect cells. Through systematic deletion mutagenesis, we mapped the Pds5B-binding region of Wapl to residues 1–33 (Figure S1A and S1B). Similarly, we mapped the Pds5B-binding region of Sororin to residues 131–171 (Figure S1C). A synthetic Sororin peptide (residues 132–171) competed effectively with GST-Wapl1–33 for binding to Pds5B (Figure 1A). As measured by isothermal titration calorimetry (ITC), a C-terminal fragment of Sororin containing this region bound to Pds5B with a dissociation constant (Kd) of 3.0 μM (Figure 1B). We could not detect the binding of Wapl1–33 to Pds5B using ITC, suggesting that Wapl1–33 bound more weakly to Pds5B. The Pds5B-binding sequences of both Wapl and Sororin contained a previously unknown YSR motif (with the consensus of [K/R][S/T]YSR) conserved in vertebrates (Figure 1C). Mutation of the YSR motif to ASE (YSR>ASE) in Wapl or Sororin reduced their binding to Pds5 in vitro (Figure S1D and S1E) and in human cells (Figure 1D and 1E). Thus, Wapl and Sororin compete for Pds5 binding through a similar conserved motif.
Figure 1. Identification of a Functional Pds5-binding Motif in Human Wapl and Sororin.
(A) Coomassie-stained SDS-PAGE gel of recombinant human Pds5B1–1120 bound to GST-Wapl1–33, in the absence or presence of increasing concentrations of a Sororin peptide. Asterisk indicates a proteolytic fragment of GST-Wapl1–33.
(B) Isothermal titration calorimetry (ITC) curves of the binding between purified recombinant human Pds5B1–1120 and Sororin91–252, with Kd and binding stoichiometry (N) indicated. DP, differential power.
(C) Schematic drawing of domains and motifs of human Wapl and Sororin, and sequence alignment of the YSR motifs of Wapl and Sororin from human (Hs), mouse (Mm), zebrafish (Dr), and Xenopus (Xl). CBM, cohesin-binding motif.
(D) Anti-GFP, anti-Pds5B, and anti-SA2 blots of anti-GFP immunoprecipitates (IP) of HeLa cells transfected with the indicated GFP-Wapl plasmids. WT, wild type; ASE, Y9A/R11E; AGA, F73A/F75A.
(E) Anti-Myc and anti-GFP blots of lysates and anti-Myc IP of HeLa cells transfected with plasmids encoding GFP-Pds5B and the indicated Myc-Sororin proteins. WT, wild type; ASE, Y146A/R148E; AGA, F166A/F168A.
(F) Anti-Wapl and anti-β-tubulin immunoblots of lysates of HeLa cells that were transfected with Wapl siRNA and increasing amounts of the indicated GFP-Wapl plasmids. Endo, endogenous. The increase of the untagged Wapl band intensity in GFP-Wapl samples was due to proteolysis of GFP-Wapl proteins or internal translation start of the transgene.
(G) Representative metaphase spreads of cells in (E) with arm-resolved or arm-unresolved chromosomes. Spreads were stained with DAPI (gray) and the kinetochore marker CREST (red). Selected sister chromatids are magnified in inset. Scale bar, 5 μm. Quantification of the percentages of mitotic cells in (E) that had arm-unresolved chromosomes. Error bars, SD (n = 3 independent experiments) (see also Figure S1).
Both Wapl and Sororin also contain phenylalanine-glycine-phenylalanine (FGF) motifs, which have been implicated in their antagonism (Nishiyama et al., 2010; Shintomi and Hirano, 2009). The first FGF motif of Wapl and the only FGF motif in Sororin are located in close proximity to their YSR motif (Figure 1C). We tested the contributions of FGF motifs of Wapl and Sororin to Pds5 binding. Mutation of FGF to AGA (FGF>AGA) in Wapl slightly weakens the binding of Wapl1–100 to Pds5 in vitro, whereas mutation of FGF in Sororin had no effect on Pds5 binding (Figure S1D and S1E). In contrast, these FGF mutations substantially reduced the binding of Wapl or Sororin to Pds5B or cohesin in human cells (Figure 1D and 1E). Thus, the FGF motif of Wapl and Sororin might primarily compete for binding to cohesin, and cohesin might strengthen Pds5–Wapl and Pds5–Sororin interactions in vivo.
To verify the functional importance of the YSR and FGF motifs, we depleted endogenous Wapl from human cells with RNA interference (RNAi), complemented them with RNAi-resistant Wapl wild type (WT), the YSR>ASE mutant, or the FGF>AGA mutant, and performed chromosome spreads of these cells enriched in mitosis (Figure 1F and 1G). Depletion of Wapl hindered the resolution of chromosome arms in early mitosis, presumably due to defective cohesin removal through the prophase pathway. Expression of GFP-Wapl WT restored arm resolution in Wapl RNAi cells in a dose-dependent manner. Wapl YSR>ASE or FGF>AGA was less efficient in rescuing the arm resolution defect caused by Wapl depletion. Thus, both the YSR and FGF motifs of Wapl are functionally important.
Structure of Human Pds5B Bound to the Wapl YSR motif
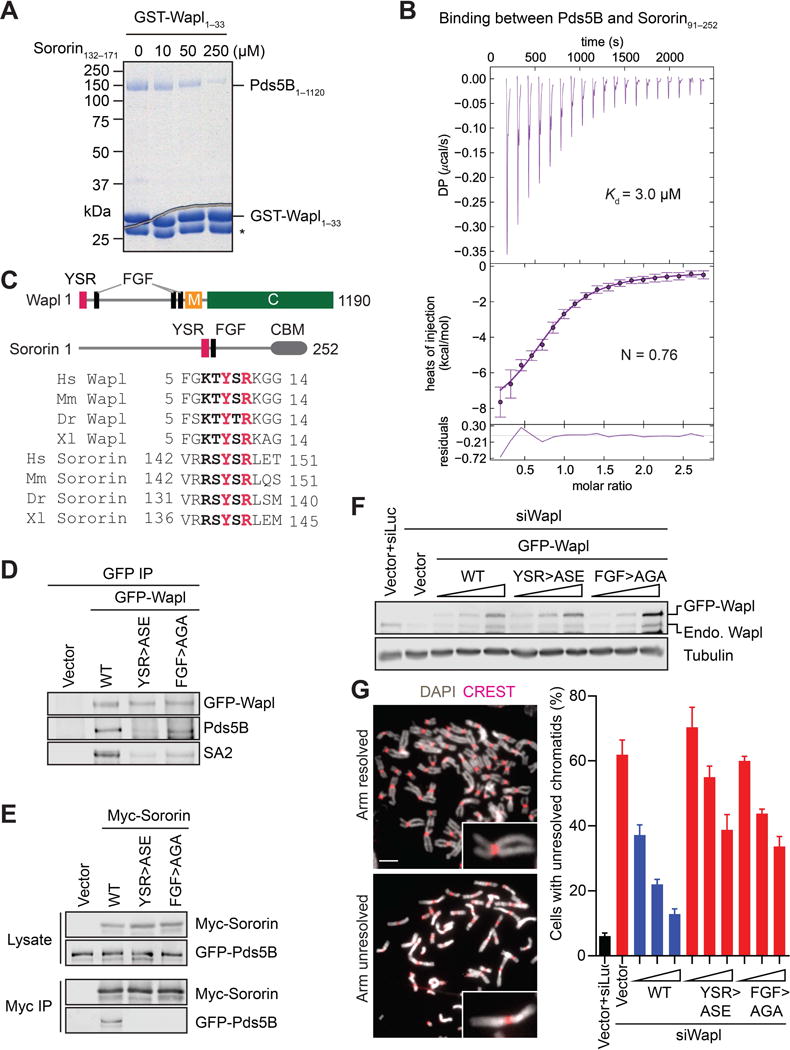
We next crystallized human Pds5B (residues 21–1120) bound to Wapl1–33 and determined the structure using X-ray crystallography to 2.7 Å resolution (Table 1). Pds5B consists of 20 HEAT repeats and a helical insert domain (HID), and folds into a structure shaped like a big dipper or a plier lever (Figure 2A and Figure S2). Although the boundaries of some HEAT repeats were correctly predicted in previous studies (Huis in’t Veld et al., 2014; Panizza et al., 2000), the curvature of these repeats was not correctly modeled. The N-terminal 8 HEAT repeats form the handle of the lever. The HID resembles the pivot, where the two levers of a plier connect. The HEAT repeats H9–H20 and the HID form the jaw. An extra N-terminal helix and a C-terminal extension pack against the first and last HEAT repeat, respectively. There is a sharp bend at HEAT repeats 15 and 16. At this bend lies an unanticipated cofactor of Pds5, inositol hexakisphosphate (IP6). The curved structure of Pds5B is reminiscent of that of SA2 or its yeast ortholog Scc3 (Hara et al., 2014; Roig et al., 2014). Interestingly, the sharp bend in SA2 forms a critical Scc1-binding site (Hara et al., 2014).
Table 1.
Data Collection, Structure Determination, and Refinement Statistics of Human Pds5B Bound to Wapl1–33
| Data collection | |
|---|---|
| Space group | P212121 |
| Cell constants a, b, c (Å) | 120.76, 162.37, 173.06 |
| Wavelength (Å) | 0.97918 |
| Resolution range (Å) | 40.6 – 2.70 (2.75 – 2.70)a |
| Unique reflections | 92,470 (4,540) |
| Multiplicity | 8.1 (6.6) |
| Data completeness (%) | 99.9 (99.4) |
| Rmerge (%)b,c | 9.5 (100) |
| Rpim(%)c,d | 4.7 (76.5) |
| I/σ(I) | 18.4 (1.2) |
| Wilson B-value (Å2) | 38.1 |
| Phase determination | |
| Anomalous scatterers | Se, 73 out of 66 possible sites |
| Refinement statistics | |
| Resolution range (Å) | 40.6 – 2.71 (2.78 – 2.71) |
| No. of reflections Rwork/Rfree | 81,101/1,991 (2,181/61) |
| Data completeness (%) | 87.4 (34.0) |
| Atoms (non-H protein/peptide/IP6) | 17,387/45/72 |
| Rwork (%) | 21.6 (32.9) |
| Rfree (%) | 25.3 (43.3) |
| R.m.s.d. bond length (Å) | 0.003 |
| R.m.s.d. bond angle (°) | 0.62 |
| Mean B-value (Å2) (protein, chain A/IP6, chain A/protein, chain B/IP6, chain B/peptide, chain C) | 41.4/33.4/63.4/46.4/91.6 |
| Ramachandran plot (%) (favored/additional/disallowed)e | 97.2/2.6/0.2 |
| Missing residues | A: 589–594, 1102–1107, 1117–1120. B: 46–48, 91–94, 539–543, 587–595, 1101–1107. C: 1–6, 12–33. |
Data for the outermost shell are given in parentheses.
Rmerge = 100 ΣhΣi|Ih,i—〈Ih〉|/ΣhΣi 〈Ih,i〉, where the outer sum (h) is over the unique reflections and the inner sum (i) is over the set of independent observations of each unique reflection.
Bijvoet-pairs were kept separate for data processing.
Rpim = 100 ΣhΣi [1/(nh − 1)]1/2|Ih,i—〈Ih〉|/ΣhΣi 〈Ih,i〉, where nh is the number of observations of reflections h (Evans, 2011).
As defined by the validation suite MolProbity (Chen et al., 2010).
Figure 2. Crystal Structure of Human Pds5B Bound to YSR motif of Wapl.
(A) Cartoon drawing of the crystal structure of human Pds5B in complex with the YSR motif of Wapl in two different orientations. The HEAT repeats and the helical insert domain (HID) are colored teal and gray, respectively. The Wapl peptide and IP6 are shown as sticks. The N- and C-termini and the 20 HEAT repeats (H1–H20) are labeled. Pds5B is shaped like a plier lever, with H1–H8 resembling the handle, the HID resembling the pivot, and H9–H20 forming the jaw. All structure figures are made with PyMOL (www.pymol.org).
(B) Zoomed in view of the Pds5B–Wapl interface. Pds5B and Wapl residues are shown as gray and yellow sticks, respectively.
(C) Quantification of the relative 35S-Pds5B intensities bound to GST-Wapl1–150. Error bars, SD (n = 3 independent experiments) (see also Figures S2 and S3).
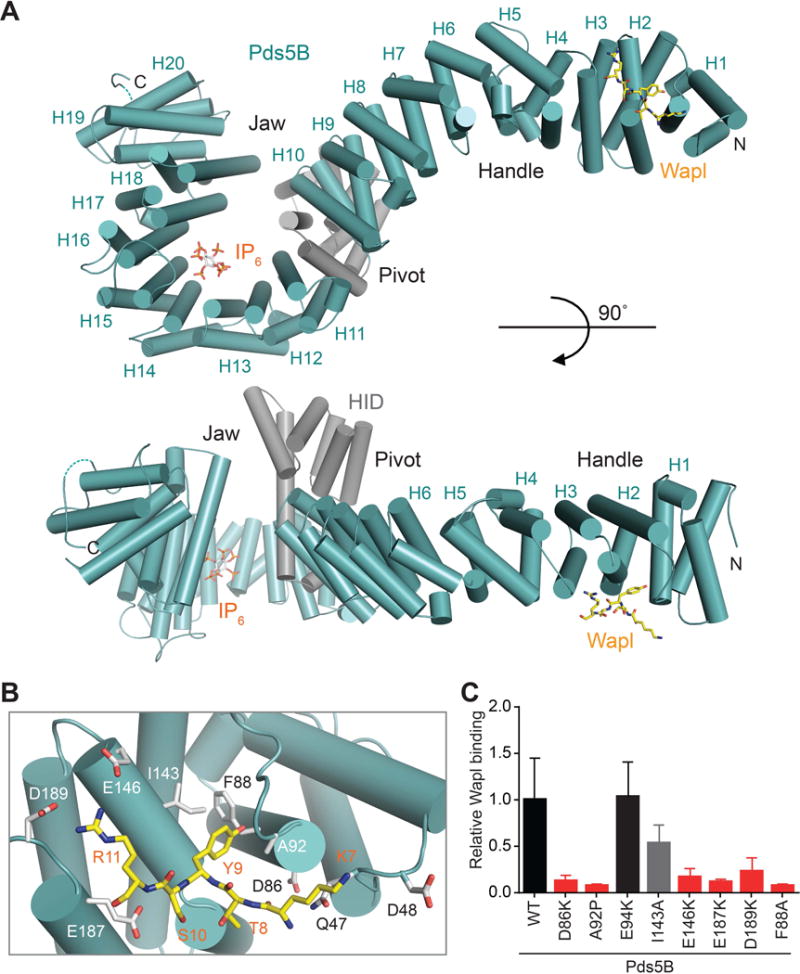
Only one of two Pds5B molecules in one asymmetric unit bound to Wapl. The electron density corresponding to Wapl was weak, with only that belonging to residues 7–11 (KTYSR) being visible (Figure S3A). Wapl binds along the ridge of HEAT repeats 1–3 at the tip of the handle (Figure 2A). K7, Y9, S10, and R11 of Wapl form polar, electrostatic, and hydrophobic interactions with Q47, A92, F88, I143, E146, E187, and D189 of Pds5B (Figure 2B), consistent with the importance of the YSR motif in Pds5 binding. Most of the Wapl-binding residues of Pds5 are conserved among various organisms (Figure S2). Importantly, single point mutations of several corresponding Wapl-binding residues in the budding yeast Pds5 suppress the cohesion defects of eco1–1 mutant (Rowland et al., 2009).
To validate this Wapl-binding surface of Pds5, we mutated the hydrophobic or polar residues to alanine and introduced charge-reversal mutations to charged residues. We also created the A92P mutant, based on a yeast Pds5 mutation that suppressed eco1–1 phenotypes (Heidinger-Pauli et al., 2010). All mutations, except I143A, greatly reduced the binding of Wapl1–100 to Pds5B in vitro (Figure 2C and S3B). These mutations also reduced the binding of Sororin to Pds5B (Figure S3B and S3C), suggesting that Sororin and Wapl bound to a similar site on Pds5B. As a negative control, the E94K mutation targeting an adjacent residue that did not contact Wapl had no effect on Wapl or Sororin binding to Pds5B.
The YSR motifs of vertebrate Wapl and Sororin thus compete for binding to a similar site on Pds5 that is conserved from yeast to man. Furthermore, residues 2–8 (RAYGKRG) of the budding yeast Wapl (Wpl) partially conform to the consensus of the YSR motif, and may bind to the same site on yeast Pds5. It is possible that an unidentified YSR-containing factor in yeast may counteract the binding of Wapl to Pds5 in that organism.
Mutual Wapl-Sororin Antagonism at the YSR-binding Site of Pds5
We expressed a subset of Wapl-binding-deficient Pds5B mutants in human cells and tested their binding to endogenous Wapl, Sororin, and cohesin. The D86K, A92P, and E187K mutations weakened or abolished binding of Pds5B to Wapl or Sororin, but had no effect on its binding to Smc1, a core subunit of cohesin (Figure 3A and 3B). Thus, the cohesin-binding surface of Pds5B is distinct from its Wapl- and Sororin-binding site.
Figure 3. Biochemical and functional analyses of the YSR-binding site of Pds5B.

(A) Immunoblots of the lysates and anti-Wapl immunoprecipitates (IP) of HeLa cells transfected with the indicated plasmids and siRNAs. WT, wild type.
(B) Anti-Myc, anti-Smc1 and anti-Sororin blots of anti-Myc IP of HeLa cells transfected with the indicated Myc-Pds5B plasmids. The two gel panels were from two different experiments. The mutants can only be compared to the WT control in the same experiment.
(C) Representative metaphase spreads of cells with arm-resolved or arm-unresolved chromosomes (upper panels). Spreads were stained with DAPI (gray) and the kinetochore marker CREST (red). Selected sister chromatids are magnified in inset. Scale bar, 5 μm. Quantification of the percentages of mitotic HeLa cells (transfected with the indicated Myc-Pds5B plasmids and Pds5A/B siRNAs) that had arm-unresolved chromosomes (lower panel). WT, wild type. Error bars, SD (n ≥ 3 independent experiments for all samples except those of K795E and R932E, which were performed only once; >100 cells were counted for each sample in each experiment).
(D) Recovery curves of normalized Smc1-GFP intensity of cells transfected with the indicated mCherry-Pds5B plasmids followed by Luciferase (Luc) or Pds5A/B siRNAs. Vec, vector. WT, wild type. Ipre, intensity before bleaching; I0, intensity immediately after bleaching; It, intensity at each time point after bleaching. Error bars, SEM (Vec+siLuc, n = 15 cells; Vec+siPds5A/B, n = 18; WT+siPds5A/B, n = 12; D86K+siPds5A/B, n = 15). The plateau and half-life of recovery for each sample are tabulated below (see also Figures S4 and S5).
To identify Pds5 residues required for cohesin binding, we systematically mutated conserved surface-exposed residues, and tested the binding of these mutants to cohesin, Wapl, and Sororin in human cells (Figure 3A and 3B). Among them, the K400E/R401E and Y445A/N447A mutants exhibited reduced binding to Smc1, with Y445A/N447A being more deficient. Both mutants were also deficient in binding to Wapl or Sororin, suggesting that cohesin was required for the association of Wapl and Sororin with Pds5 in vivo. This finding is consistent with the fact that other regions of Wapl and Sororin have been implicated in cohesin binding (Hara et al., 2014; Ouyang et al., 2013; Wu et al., 2011). Thus, the YSR motif is required, but not sufficient, to mediate the binding of Wapl or Sororin to Pds5 in human cells. Additional interfaces between Wapl/Sororin and cohesin are needed to form detectable cohesin–Pds5–Wapl or cohesin–Pds5–Sororin ternary interactions. We note, however that the Y445A/N447A mutant retains cohesin-independent binding to GST-Wapl or GST-Sororin in vitro (see Figure S5 below), indicating that this mutant is not globally unfolded.
Consistent with a previous report (Shintomi and Hirano, 2009), depletion of both Pds5A and Pds5B in HeLa cells caused arm resolution defects in mitotic chromosome spreads (Figure 3C and Figure S4A), a phenotype similar to that seen with Wapl depletion (Gandhi et al., 2006; Kueng et al., 2006). Like Wapl inactivation (Kueng et al., 2006; Tedeschi et al., 2013), depletion of Pds5A/B caused worm-like cohesin assembly on interphase chromatin (Figure S4B), and as revealed by fluorescence recovery after photobleaching (FRAP), reduced the kinetics and extent of cohesin turnover on chromatin (Figure S4C and S4D). For unknown reasons, the kinetics of cohesin turnover in control and Wapl RNAi cells in this study was faster than that reported previously (Kueng et al., 2006). Co-depletion of Pds5A/B and Wapl did not further impede the kinetics of cohesin exchange, but slightly decreased the pool of dynamic cohesin (Figure S4C). Collectively, these results confirm that Pds5 collaborates with Wapl to promote cohesin release from chromosomes in both mitosis and interphase.
Expression of Pds5B WT restored arm resolution in Pds5A/B-depleted cells (Figure 3C). In contrast, expression of Wapl-binding-deficient Pds5B mutants, including D86K, E146K, E187K, and D189K, failed to rescue the arm resolution defects. Likewise, the cohesin-binding-deficient Pds5B Y445A/N447A mutant also failed to complement. In addition, as revealed by FRAP, expression of Pds5B WT, but not the Wapl-binding-deficient D86K mutant, restored the kinetics of cohesin turnover on interphase chromatin (Figure 3D and S4E). In fact, Pds5B D86K appeared to further slow down cohesin dynamics, suggesting that it might dominant-negatively inhibit the residual, endogenous Pds5B. Taken together, these results establish the functional importance of the Wapl-binding site in Pds5 in cohesin release.
Pds5 plays both negative and positive roles in sister-chromatid cohesion in diverse organisms (Chan et al., 2012; Losada et al., 2005; Rowland et al., 2009; Shintomi and Hirano, 2009; Sutani et al., 2009). Unlike Wapl depletion (Ouyang et al., 2013), depletion of Pds5A/B in human cells did not rescue the cohesion defects and the resulting spindle checkpoint-dependent mitotic arrest caused by Sgo1 or Sororin depletion (Figure S5A and S5B). Furthermore, depletion of Pds5A/B inhibited two well-established cohesin stabilization mechanisms: Smc3 acetylation and subsequent Sororin association with cohesin (Figure S5C and S5D). Although depletion of Pds5A or Pds5B or both did not produce cohesion defects in metaphase spreads (Figure S5E), depletion of either protein weakened sister-chromatid cohesion in interphase cells, consistent with the molecular defects (Figure S5F and S5G). Therefore, even though depletion of Pds5A/B ostensibly produces phenotypes similar to depletion of Wapl, Pds5 actually plays dual roles in cohesin dynamics.
Inositol Hexakisphosphate (IP6) as a Structural Cofactor of Pds5
IP6 is an abundant lipid-derived metabolite in eukaryotic cells (Monserrate and York, 2010). As opposed to lower inositol polyphosphates with signaling functions (e.g. IP3), IP6 and other higher inositol polyphosphates (e.g. IP5 and IP4) have been shown to be structural cofactors for the human RNA-editing enzyme ADAR2 (Macbeth et al., 2005), the plant hormone receptors (Sheard et al., 2010; Tan et al., 2007), the yeast mRNA export helicase Dbp5 complex (Montpetit et al., 2011), among other proteins. In many cases, these inositol polyphosphates directly participate in protein–protein or protein–ligand interactions.
During the refinement of the Pds5B structure, we noticed an electron density located near the sharp bend of Pds5B at HEAT repeats 13–17. Both Pds5B molecules in the asymmetric unit contained this density, which fitted well with IP6 (Figure 4A). The 1D 1H nuclear magnetic resonance (NMR) spectrum of the cofactor isolated from purified Pds5B from insect cells matched perfectly with that of authentic IP6 (Figure 4B), confirming its identity. IP6 binds at a positively charged surface at the bottom of the jaw of Pds5B (Figure 4C). Several basic IP6-binding residues, including K727, K830, K888, and K925, are conserved from yeast to man (Figure 4A and S2), suggesting that IP6 binding is a conserved feature of Pds5.
Figure 4.
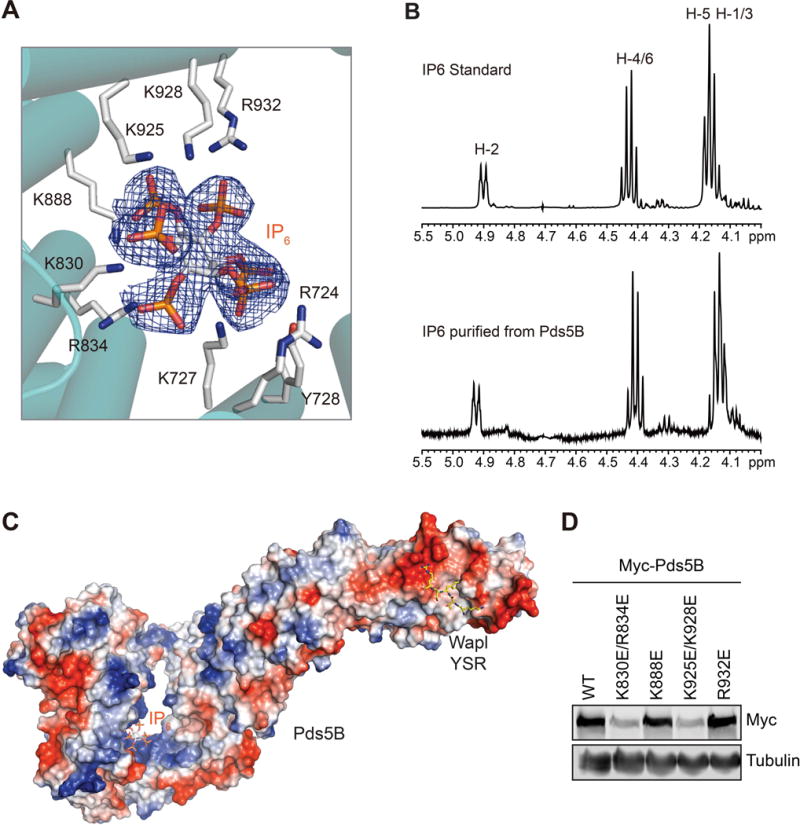
IP6 as a Structural Cofactor of Pds5B
(A) Zoomed in view of the IP6-binding site of Pds5B. IP6 is shown as stick, along with its 2Fo-Fc electron density map (blue mesh) contoured at 1.0 σ. IP6-binding residues are shown as sticks and labeled.
(B) 1D 1H NMR spectra of authentic IP6 standard (top) and IP6 isolated from recombinant human Pds5B, with the 1H assignment indicated.
(C) Surface drawing of human Pds5B colored with its electrostatic potential (blue, positive; red, negative). IP6 and the Wapl peptide are shown in sticks.
(D) Anti-Myc and anti-β-tubulin immunoblots of lysates of HeLa cells transfected with the same amount of the indicated Myc-Pds5B plasmids. WT, wild type (see also Figure S6).
We then mutated the IP6-binding residues in Pds5B. When the same amounts of plasmids were used in transient transfection of HeLa cells, the protein levels of Pds5B K727E/Y728A, K830E/R834E, and K925E/K928E mutants were much lower than that of the wild type, whereas the levels of K888E and R932E were similar to wild-type levels (Figure 4D). Similar patterns were observed with the expression of these Pds5B mutants in insect cells. These findings suggest that IP6 binding is required for the structural integrity of Pds5B. The single mutants presumably retain IP6 binding and are thus stable. Unfortunately, our NMR-based assay required milligram amounts of Pds5 protein, and we lacked a sensitive IP6 detection assay that could directly verify IP6 binding by human Pds5B wild type (or the lack of IP6 binding by Pds5B mutants) in human cells.
Contributions of the IP6-binding Segment of Pds5B to Cohesin Binding
We have identified four conserved residues in Pds5B that are required for cohesin binding (Figure 3B). K400 and R401 are located in the loop connecting the two helices in HEAT repeat H9, whereas Y445 and N447 reside in the loop of H10 (Figure S2). They define a critical cohesin-binding site of Pds5B on one side of the jaw, which is near the pivot of the lever and connects to the handle (Figure 5A). This pivot site, however, was not sufficient to mediate Pds5B binding to cohesin, as a Pds5B mutant with the C-terminal 8 HEAT repeats (H13–H20) deleted (Δ722–1116) was deficient in binding to cohesin, Wapl, or Sororin in human cells (Figure 5B). Consequently, Pds5B Δ722–1116 failed to support the arm resolution of sister chromatids during early mitosis of Pds5A/B-depleted HeLa cells (Figure 5C). Therefore, the C-terminal region, including the IP6-binding segment, contributes to cohesin binding.
Figure 5. Requirement for IP6 in Cohesin Binding by Pds5B.
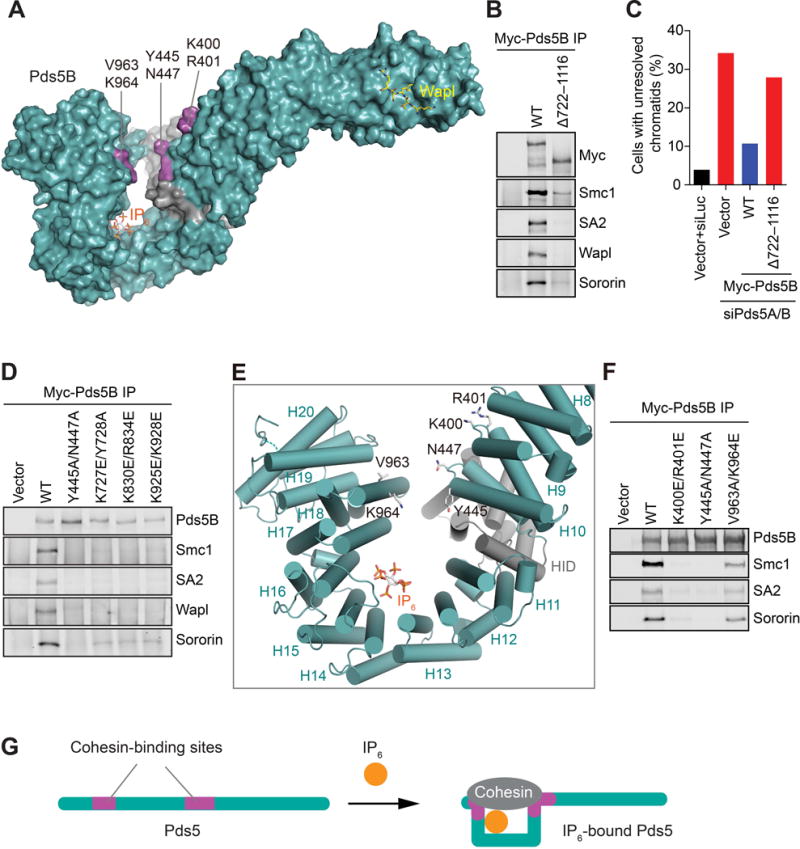
(A) Surface drawing of Pds5B, with the cohesin-binding residues colored purple and labeled. IP6 and the Wapl peptide are shown in sticks.
(B) Immunoblots of anti-Myc immunoprecipitates (IP) of HeLa cells that were transfected with the indicated Myc-Pds5B plasmids.
(C) Quantification of the percentages of mitotic HeLa cells (transfected with the indicated plasmids and siRNAs) that had arm-unresolved chromosomes.
(D) Immunoblots of anti-Myc immunoprecipitates (IP) of HeLa cells transfected with the indicated Myc-Pds5B plasmids. WT, wild type.
(E) Cartoon diagram of the jaw of Pds5B, with IP6 and cohesin-binding residues shown in sticks. HID, helical insert domain.
(F) Immunoblots of anti-Myc immunoprecipitates (IP) of HeLa cells transfected with the indicated Myc-Pds5B plasmids. WT, wild type.
(G) Model for IP6-dependent cohesin binding by Pds5 (see also Figure S6).
To test whether IP6 was required for the Pds5B–cohesin interaction in human cells, we normalized the expression of Pds5B mutants deficient for IP6 binding to that of wild type by using more mutant plasmids in transfections. Even when expressed at similar levels, the IP6-binding-deficient Pds5B mutants, including K727E/Y728A, K830E/R834E, and K925E/K928E, were deficient in binding to cohesin, Wapl, or Sororin (Figure 5D). Because cohesin was required for the binding of Pds5B to Wapl or Sororin in human cells, the defects of these Pds5B mutants in Wapl or Sororin binding could be an indirect consequence of their inability to bind cohesin. Consistent with this notion, the same Pds5B mutants retained binding to recombinant Wapl1–100 and Sororin131–252 in vitro (Figure S6). This finding also indicated that the IP6-binding-deficient Pds5B mutants were still folded in the N-terminal region. These results suggest that IP6 is required for cohesin binding by Pds5B.
Both the N-terminal side of the jaw and IP6 at the bottom of jaw are required for Pds5B to bind cohesin. The C-terminal side of the jaw, especially the αB helix of HEAT repeat H18, is in spatial proximity to the cohesin-binding site near the pivot (Figure 5E). We tested whether this helix was involved in cohesin binding. Indeed, mutation of two surface-exposed residues on this helix, V963 and K964, reduced Pds5B binding to cohesin, albeit to a lesser degree than the K400E/R401E and Y445A/N447A mutations did (Figure 5E and 5F). Thus, the HEAT repeats C-terminal to the IP6-binding segment contribute an auxiliary cohesin-contacting site. We propose that one mechanism by which IP6 contributes to cohesin binding is to sharply bend Pds5B at repeat H15, juxtaposing two cohesin-contacting sites that are otherwise spatially separated (Figure 5G).
Inhibition of the Binding of Scc1 to Smc3 by Pds5B
Several recent studies have implicated the Smc3–Scc1 interface as an evolutionarily conserved DNA exit gate of the cohesin ring (Buheitel and Stemmann, 2013; Chan et al., 2012; Gligoris et al., 2014; Huis in’t Veld et al., 2014). At this interface, the N-terminal helical domain (NHD) of Scc1 bind to the coiled coil region of Smc3 that is adjacent to its ATPase domain, forming a four-helix bundle (Gligoris et al., 2014). Human cohesin with four conserved hydrophobic Scc1 residues at this interface mutated to glutamate (Scc1 4E) is loaded on chromatin, but dissociates from chromatin with abnormally fast kinetics in a Wapl-independent manner, suggesting that the cohesin-releasing activity might act through disrupting the Smc3–Scc1 interface (Huis in’t Veld et al., 2014).
We next defined the element within cohesin that interacted with the IP6-dependent cohesin-binding surface of Pds5B. The Scc11–210-Myc fragment and its N-terminally truncated species bound efficiently to GFP-Pds5B in HeLa cells (Figure 6A). An Scc176–150 fragment immediately C-terminal to the Smc3-binding NHD retained partial binding to Pds5B. Mutations of cohesin-binding residues located at either side of the jaw in Pds5B weakened its binding to both Scc1 fragments. Recombinant purified Scc176–150 bound to Pds5B surprisingly tightly, with a Kd of 4.3 nM, as measured by ITC (Figure 6B). Purified recombinant Pds5B V963/K964E and Y445A/N447A mutants also bound weaker to Scc176–150 in vitro (Figure 6C). Because mutations of the corresponding region in yeast Scc1 are known to disrupt Pds5 binding, Scc176–150 contains a conserved Pds5-binding element that binds at the IP6-dependent cohesin-binding site in Pds5. Scc176–150 bound to Pds5B less tightly than Scc11–210 in human cells, indicating that other Scc1 elements, possibly the NHD, contribute to Pds5 binding. Full-length Scc1 or any fragments containing the NHD were, however, either not expressed or insoluble in bacteria or insect cells, preventing us from measuring the affinity between Pds5B and larger Scc1 fragments.
Figure 6. Identification of the Pds5B-binding region in Scc1.
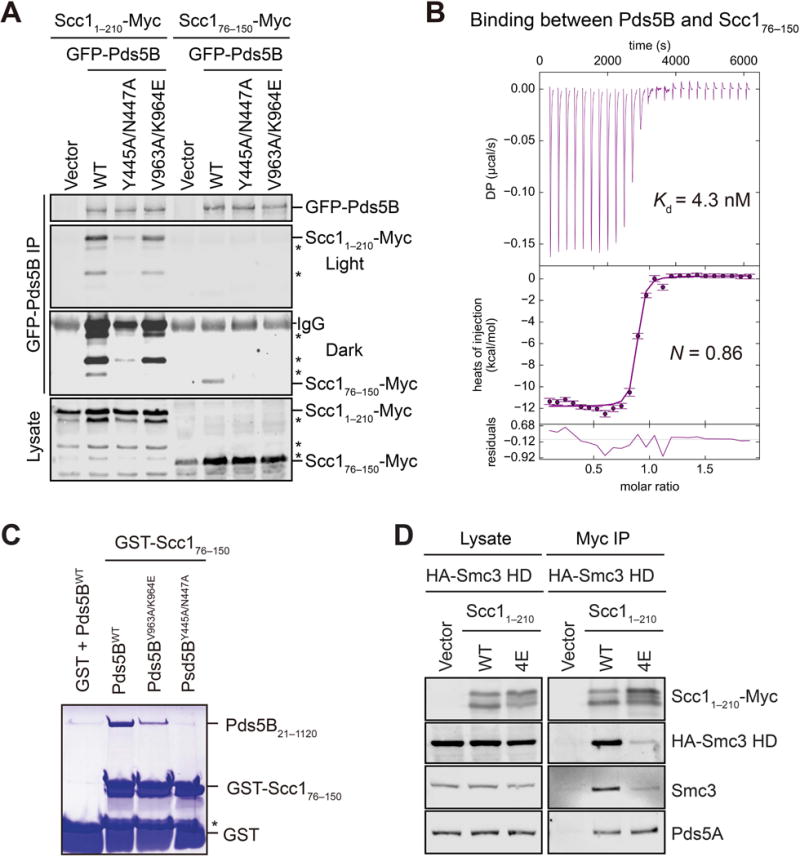
(A) Immunoblots of lysate and anti-GFP immunoprecipitates (IP) of HeLa cells transfected with the indicated Scc1-Myc and GFP-Pds5B plasmids. Asterisks indicate N-terminally truncated forms of Scc11–210-Myc. WT, wild type.
(B) ITC curves of the binding between purified recombinant Pds5B21–1120 and Scc176–150, with the dissociation constant (Kd) and binding stoichiometry (N) indicated. DP, differential power.
(C) Coomassie-stained SDS-PAGE gel of recombinant human Pds5B21–1120 wild type (WT) or mutant proteins bound to beads containing GST or GST-Scc176–150. Asterisk indicates a proteolytic fragment of GST-Scc176–150.
(D) Anti-Myc, anti-HA, anti-Smc3, and anti-Pds5A blots of lysates and anti-Myc immunoprecipitates (IP) of HeLa cells that were transfected with plasmids encoding HA-Smc3 HD and the indicated Scc11–210-Myc plasmids. WT, wild type; 4E, L53E/L59E/Y67E/L74E.
The fact that Pds5B bound with high affinity to an Scc1 region bordering the Smc3-binding NHD promoted us to test whether Pds5B could disrupt the Smc3–Scc1 interface. An Smc3 head domain (HD) containing the ATPase domain and an adjacent coiled coil segment bound strongly to Scc11–210, but only weakly to Scc11–210 4E, in human cells (Figure 6D). Likewise, GST-Smc3 HD bound efficiently to in vitro translated Scc11–210, and weakly to Scc1 4E (Figure 7A). These results indicate that we can construct an isolated DNA exit gate of cohesin with a functional Smc3–Scc1 interface. Unlike the uncleaved Scc11–210, the N-terminally truncated species of Scc11–210 4E present completely lost binding to Smc3 HD (Figure 7A). Furthermore, deletion of the N-terminal 20 residues of Scc1 on its own did not reduce binding to Smc3 HD, but when combined with the 4E mutation, completely abolished Smc3 binding (Figure S7). Thus, the N-terminal tail of Scc1 can mediate weak binding to Smc3 HD when the Scc1 NHD–Smc3 helical interactions are disrupted. The weak binding of Scc1 4E to Smc3 might help to explain the apparently normal loading of Scc1 4E-containing cohesin onto chromatin (Huis in’t Veld et al., 2014).
Figure 7. Inhibition of DNA exit gate closure by Pds5B.

(A) Beads bound to GST or GST-Smc3 head domain (HD) were incubated with in vitro translated 35S-labeled Scc11–210 or Scc11–210 4E and washed. The beads were then incubated with the indicated combinations of Pds5B (WT or Y445A/N447A), full-length Wapl, Wapl501–1190, and SA2. After washing, the bound proteins were separated on SDS-PAGE and analyzed with a phosphoimager (top panel) and Coomassie staining (bottom panel). Asterisk indicates N-terminally truncated forms of Scc11–210. The schematic drawing on the right shows that Pds5B does not disrupt the pre-formed Smc3–Scc1 complex.
(B) Binding between GST-Smc3 head domain (HD) and in vitro translated 35S-Scc11–210 (WT or 4E) that had been pre-incubated with the indicated combinations of Pds5B (WT or Y445A/N447A), Wapl501–1190, and SA2. Autoradiograph (top) and Coomassie stained gel (bottom) of 10% input proteins and proteins bound to GST or GST-Smc3 HD beads are shown. Asterisk indicates N-terminally truncated forms of Scc11–210.
(C) Quantification of the relative Scc1 intensities of the indicated lanes in (B). Error bars, SD (n = 3 independent experiments). The schematic drawing on top shows that Pds5-bound Scc1 is deficient in Smc3 binding.
(D) A speculative model for Pds5-dependent cohesin release from chromosomes. Pds5 bridges the interaction between cohesin and the YSR motif of Wapl to strengthen binding of Wapl–Pds5 to cohesin. This function of Pds5 is antagonized by the YSR motif of Sororin. Pds5 inhibits the formation of the Smc3–Scc1 interaction, suggesting that Pds5 might also promote cohesin release through stabilizing a transient open state of cohesin (see also Figure S7).
Consistent with our previous studies (Ouyang et al., 2013), addition of Pds5B either alone or together with SA2 or Wapl did not reduce the amount of Scc1 already bound to Smc3 HD (Figure 7A), indicating that these releasing factors cannot disrupt the preformed DNA exit gate in vitro. This finding is rather expected, as the release of cohesin from chromatin likely requires its ATPase activity, and the isolated Smc3 HD is not a functional ATPase in the absence of Smc1. Indeed, when Scc1 in the context of intact cohesin is cleaved, Wapl–Pds5 can release the N-terminal fragment Scc1 from Smc3.
Interestingly, a pre-incubation of Pds5B with Scc11–210 or Scc121–210 (prior to the addition of Smc3 HD) greatly reduced the Smc3–Scc1 interaction (Figure 7B, 7C, and S7). As an important control, pre-incubating Scc1 with Pds5B Y445A/N447A deficient in Scc1 binding did not have any effect. SA2 and the C-terminal fragment of Wapl (Wapl501–1190) did not prevent Scc1 binding to Smc3, although addition of both appeared to slightly enhance the effects of Pds5B. Pds5B also reduced the residual binding of Scc1 4E to Smc3 mediated by the N-tail of Scc1. This result suggests that Pds5B might hinder the formation of the DNA exit gate of cohesin, but cannot disrupt the preformed one. We infer from this finding that the half-life of the isolated Smc3–Scc1 complex must be exceedingly long in vitro. There is little exchange between free and Smc3-bound states of Scc1 during the time course of our experiments.
DISCUSSION
Our results presented herein confirm and extend the current paradigm that Pds5 has both positive and negative roles in cohesin regulation. Although we could not observe cohesion defects in Pds5A/B-depleted cells using metaphase spreads, we have provided evidence to suggest that Pds5A/B depletion caused phenotypes congruent with cohesion establishment defects. Conversely, we have clearly established a requirement for Pds5 in cohesin release in human cells. One mechanism by which Pds5 promotes cohesin release is to promote the productive association of Wapl with cohesin, through engaging the YSR motif of Wapl with the N-terminal handle and binding to Scc1 through the C-terminal IP6-bound jaw (Figure 7D). This mechanism is antagonized by the YSR motif of Sororin and possibly other unidentified factors in yeast and human.
One possible explanation for why we could not observe cohesion defects in metaphase spreads of Pds5 RNAi cells is that a small amount of Pds5 suffices to perform the positive functions in cohesion establishment, whereas a larger amount of Pds5 is needed to perform its Wapl-dependent cohesin release functions. A partial depletion of Pds5 is expected to more severely cripple the cohesin-release function of Pds5. This notion is consistent with the role of Pds5 in promoting Smc3 acetylation by Esco1, which is a catalytic process. In contrast, the cohesin-release function of Pds5 involves its stoichiometric binding to Wapl and cohesin.
The dual roles of Pds5 in cohesin dynamics may underlie the peculiar behavior of the Pds5B A92P mutant. Unlike other Wapl-binding-deficient mutants, Pds5B A92P supported chromosome-arm resolution (Figure 3C). We propose that Wapl and Sororin are mutually antagonistic in sister-chromatid cohesion. The net balance of their activities determines cohesion status. Pds5 recruits both Wapl and Sororin to cohesin. Most Pds5B mutations destabilize Wapl binding more severely than Sororin binding, thus displaying cohesin-release defects. Pds5B A92P disrupts Sororin binding more thoroughly than Wapl binding (Figure 3A and 3B), and is thus still functional in supporting cohesin release from chromatin.
Mutation of FGF motif in Sororin weakens sister-chromatid cohesion in Xenopus egg extracts (Nishiyama et al., 2010). In contrast, a previous study has shown that deletion of a region encompassing both YSR and FGF motifs of Sororin did not cause overt cohesion defects in human cells (Wu et al., 2011), a finding we could reproduce. Because both motifs of Wapl are important for cohesin release in human cells, we suspect that another protein might act redundantly with the YSR and FGF motifs of Sororin to antagonize these motifs of Wapl in human cells.
Unexpectedly, we have identified IP6 as a structural cofactor of Pds5. Because IP6 and other inositol polyphosphates often reside at functional interfaces and directly contribute to protein–protein interactions, we suspect that IP6 in Pds5 might also form direct contact with cohesin. Because IP6 is an abundant metabolite and is required for the structural integrity of Pds5, IP6 is likely a constitutive structural cofactor of Pds5. On the other hand, we cannot exclude possible regulatory roles of IP6. In the future, it will be interesting to test whether the levels of IP6 fluctuate during the cell cycle.
In addition to the scaffolding role of Pds5, Scc1 binding by the Pds5 jaw appears to directly counteract the formation of the Smc3–Scc1 interface, which is a DNA exit gate of cohesin. The mechanism by which Pds5 inhibits the binding of Scc1 to Smc3 remains to be established. Because Scc1 4E deficient in Smc3 binding can still interact with Pds5 (Figure 6D), Pds5 is unlikely to compete with Smc3 for the same binding interface on Scc1. It is possible that Pds5 molds the N-terminal region of free Scc1 into an alternative conformation, which is not compatible for Smc3 binding.
A recent study by Uhlmann and coworkers has provided key insight into Wapl–Pds5-mediated cohesin release (Murayama and Uhlmann, 2015). Wapl–Pds5 does not stimulate the ATPase activity of cohesin, but requires the re-binding of ATP to nucleotide-free cohesin to open the cohesin ring. Based on this important insight and our results presented here, we propose the following speculative mechanism to explain the opening of cohesin ring by Wapl–Pds5 (Figure 7D). Cohesin contains two interlocked gates: an inner gate formed by the ATPase heads of Smc1 and Smc3 and stabilized by ATP; and an outer gate formed by the Scc1–Smc3 interface. ATP hydrolysis opens the inner gate, allowing the entrapped DNA to move freely in the large ring whose outer gate is closed. We speculate that Wapl–Pds5 might preferably recognize the inner-gate-open conformation of cohesin. Binding of ATP to the nucleotide-free cohesin–Wapl–Pds5 complex closes the inner gate, but produce a fleeting outer-gate-open state of cohesin, in which the N-tail and NHD of Scc1 are detached from Smc3 HD. This open state during the ATPase cycle might be too transient to allow cohesin release from chromatin. The IP6-bound jaw of Pds5 binds to the N-terminal region of Scc1 and inhibits its re-association with Smc3. In effect, Pds5 stabilizes a transient, open state of cohesin, and prolongs its lifetime, thereby promoting cohesin release from chromosomes. Even though Pds5 is likely the primary regulator of the latter step, Wapl might also be a facilitator, as previously proposed (Gandhi et al., 2006).
By virtue of its ability to topologically embrace chromosomes, the ring-shaped cohesin complex mediates diverse, fundamental cellular processes, including transcription, DNA repair, and chromosome segregation. Our findings reveal the structural basis of the Wapl–Sororin antagonism in cohesin dynamics, provide rationales for the dual functions of Pds5 in cohesin dynamics, and suggest a testable model for Pds5-dependent cohesin release from chromosomes.
EXPERIMENTAL PROCEDURES
Protein Purification, Crystallization, and Structure Determination
Recombinant human Pds5B proteins were expressed in insect cells using the Bac-to-Bac system (Invitrogen) and purified with a combination of affinity and conventional chromatography. Selenomethionine-containing Pds5B21–1120 was concentrated to 11.2 mg/ml, and a synthetic Wapl1–33 peptide was added to a 1:5 molar ratio. Crystals were grown with the sitting drop vapor diffusion method, optimized with seeding, and cryoprotected. Diffraction data were collected at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL) and processed with HKL3000 (Minor et al., 2006). Phases were initially obtained with single-wavelength selenium anomalous dispersion, and were subsequently refined. Iterative model building and refinement produced the final structure. The parameters of data collection, phasing, and refinement statistics of the final model are shown in Table 1. See Supplemental Information (SI) for more details.
Protein Binding Assays
Standard GST pull-down assays were used to assess the binding of Pds5B to Wapl, Sororin, and Scc1 in vitro and test the effects of Pds5B and Wapl on the Scc1–Smc3 interaction. The affinity between purified recombinant Pds5B1–1120 and Sororin91–252 was measured with Isothermal Titration Calorimetry (ITC). See SI for details.
Isolation and NMR Analysis of IP6 from Recombinant Pds5B
The isolation of IP6 from human Pds5B expressed and purified from insect cells was performed essentially as described (Sheard et al., 2010). The 1D 1H NMR spectra were acquired on both the isolated IP6 and purchased IP6 standard (Sigma) and compared. See SI for details.
Mammalian Cell Culture, Transfection, and Synchronization
HeLa Tet-On cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS) and 2 mM L-glutamine. Plasmid and siRNA transfections were performed using the Effectene reagent (Qiagen) and Lipofectamine RNAiMAX (Invitrogen), respectively. See SI for details and siRNA sequences. For mitotic synchronization, cells were treated with 2 mM thymidine for 16–18 h, released into fresh medium for 9 h, and blocked at mitosis with the addition of 5 μM nocodazole (Sigma) for 2 h.
Antibodies, Immunoblotting, and Immunoprecipitation
The anti-Wapl antibody was generated against a C-terminal fragment of human Wapl (residues 601–1190) as described previously (Wu et al., 2012). Rabbit polyclonal antibodies against eGFP, human Sororin91–252, and human Pds5B1140–1310 were raised at Yenzym Antibodies with purified recombinant proteins as antigens. A list of commercial antibodies is provided in SI. For immunoblotting, the blots were incubated first with primary antibodies at 1 μg/ml and then with either fluorescently labeled or horseradish peroxidase-linked secondary antibodies. The blots were scanned with an Odyssey Infrared Imaging System (LI-COR) or developed with the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). For immunoprecipitation, antibodies were coupled to Affi-Prep Protein A beads (Bio-Rad) at a concentration of 1 mg/ml. Cleared cell lysates were incubated with antibody beads for 3 h at 4°C. Proteins bound to beads were dissolved in SDS sample buffer, separated by SDS-PAGE, and blotted with the appropriate antibodies. See SI for details.
Flow Cytometry
Cells were harvested with trypsinization and fixed in 70% ice-cold ethanol overnight. After being washed with PBS, cells were permeabilized with PBS containing 0.25% Triton X-100 on ice for 5 min. Then, cells were incubated with the antibody to MPM2 in PBS containing 1% BSA for 3 h at room temperature, followed by an incubation with a fluorescent secondary antibody (Invitrogen) for 30 min. After being washed with PBS, cells were resuspended in PBS containing 0.1% Triton X-100, RNase A, and propidium iodide, and analyzed with a flow cytometer. Data were processed with FlowJo.
Immunofluorescence
Mitotic HeLa Tet-On cells were treated with 55 mM KCl hypotonic solution at 37°C for 15 min, and spun onto microscope slides with a Shandon Cytospin centrifuge. Cells on the slides were permeabilized, fixed in 4% paraformaldehyde for 10 min, and stained with CREST and DAPI. For fluorescence in situ hybridization (FISH), fluorescently labeled probes derived from the BAC clone RP11-466L19 were hybridized to interphase HeLa Tet-On cells. Slides were viewed with a DeltaVision fluorescence microscope (GE Healthcare). Image processing and quantification were performed with Image J. See SI for details.
Fluorescence Recovery after Photobleaching (FRAP)
HeLa cells stably expressing Smc1-GFP were transfected with empty vector or RNAi-resistant mCherry-Pds5B WT or mutant plasmids, and then transfected with siPds5A and siPds5B. FRAP was performed using a custom built spinning disk confocal microscope (BioVision). Single-stack images were captured with a 100× objective. A circular region in each cell was bleached. Images were acquired at 30 sec intervals for 60 min after bleaching. Data analysis and fitting were performed with ImageJ and Prism. See SI for details.
Supplementary Material
Acknowledgments
We thank Susannah Rankin and Prasad Jallepalli for reagents, Haydn Ball for peptide synthesis, Chad Brautigam for help with isothermal titration calorimetry, and members of the Yu laboratory for helpful discussions. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. H.Y. is an investigator with the Howard Hughes Medical Institute. This work is supported in part by grants from the Cancer Prevention and Research Institute of Texas (RP110465-P3 to H.Y.) and the Welch Foundation (I-1441 to H.Y.).
Footnotes
AUTHOR CONTRIBUTIONS
Z.O. performed the structure determination and in vitro protein binding experiments. G.Z. performed all cellular experiments. D.R.T. assisted in X-ray diffraction data collection and structure determination. X.L. performed the NMR experiment on IP6. H.Y. supervised the project, and participated in experimental design and data analysis. Z.O., G.Z., and H.Y. wrote the paper with input from all authors.
ACCESSION NUMBERS
The coordinates of human Pds5B bound to the Wapl YSR peptide have been deposited to the Protein Data Bank under the code 5HDT.
References
- Bose T, Gerton JL. Cohesinopathies, gene expression, and chromatin organization. J Cell Biol. 2010;189:201–210. doi: 10.1083/jcb.200912129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buheitel J, Stemmann O. Prophase pathway-dependent removal of cohesin from human chromosomes requires opening of the Smc3-Scc1 gate. EMBO J. 2013;32:666–676. doi: 10.1038/emboj.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Roig MB, Hu B, Beckouet F, Metson J, Nasmyth K. Cohesin’s DNA exit gate is distinct from its entrance gate and is regulated by acetylation. Cell. 2012;150:961–974. doi: 10.1016/j.cell.2012.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol. 2006;16:2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol. 2006;16:1571–1578. doi: 10.1016/j.cub.2006.06.068. [DOI] [PubMed] [Google Scholar]
- Gligoris TG, Scheinost JC, Burmann F, Petela N, Chan KL, Uluocak P, Beckouet F, Gruber S, Nasmyth K, Lowe J. Closing the cohesin ring: structure and function of its Smc3-kleisin interface. Science. 2014;346:963–967. doi: 10.1126/science.1256917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis JH, Elbatsh AM, Rowland BD. Cohesin and its regulation: on the logic of X-shaped chromosomes. Dev Cell. 2014;31:7–18. doi: 10.1016/j.devcel.2014.09.010. [DOI] [PubMed] [Google Scholar]
- Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- Hara K, Zheng G, Qu Q, Liu H, Ouyang Z, Chen Z, Tomchick DR, Yu H. Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion. Nat Struct Mol Biol. 2014;21:864–870. doi: 10.1038/nsmb.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Onn I, Koshland D. Genetic evidence that the acetylation of the Smc3p subunit of cohesin modulates its ATP-bound state to promote cohesion establishment in Saccharomyces cerevisiae. Genetics. 2010;185:1249–1256. doi: 10.1534/genetics.110.116871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huis in’t Veld PJ, Herzog F, Ladurner R, Davidson IF, Piric S, Kreidl E, Bhaskara V, Aebersold R, Peters JM. Characterization of a DNA exit gate in the human cohesin ring. Science. 2014;346:968–972. doi: 10.1126/science.1256904. [DOI] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci. 2005;118:2133–2141. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M, Odom DT. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 2013;152:1285–1297. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- Minamino M, Ishibashi M, Nakato R, Akiyama K, Tanaka H, Kato Y, Negishi L, Hirota T, Sutani T, Bando M, et al. Esco1 Acetylates Cohesin via a Mechanism Different from That of Esco2. Curr Biol. 2015;25:1694–1706. doi: 10.1016/j.cub.2015.05.017. [DOI] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Monserrate JP, York JD. Inositol phosphate synthesis and the nuclear processes they affect. Curr Opin Cell Biol. 2010;22:365–373. doi: 10.1016/j.ceb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472:238–242. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama Y, Uhlmann F. DNA Entry into and Exit out of the Cohesin Ring by an Interlocking Gate Mechanism. Cell. 2015;163:1628–1640. doi: 10.1016/j.cell.2015.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K, et al. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell. 2010;143:737–749. doi: 10.1016/j.cell.2010.10.031. [DOI] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Zheng G, Song J, Borek DM, Otwinowski Z, Brautigam CA, Tomchick DR, Rankin S, Yu H. Structure of the human cohesin inhibitor Wapl. Proc Natl Acad Sci U S A. 2013;110:11355–11360. doi: 10.1073/pnas.1304594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol. 2000;10:1557–1564. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- Rankin S. Sororin, the cell cycle and sister chromatid cohesion. Cell Cycle. 2005;4:1039–1042. doi: 10.4161/cc.4.8.1926. [DOI] [PubMed] [Google Scholar]
- Roig MB, Lowe J, Chan KL, Beckouet F, Metson J, Nasmyth K. Structure and function of cohesin’s Scc3/SA regulatory subunit. FEBS Lett. 2014;588:3692–3702. doi: 10.1016/j.febslet.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouet F, Underwood P, Metson J, Imre R, et al. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell. 2009;33:763–774. doi: 10.1016/j.molcel.2009.02.028. [DOI] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood R, Takahashi TS, Jallepalli PV. Sister acts: coordinating DNA replication and cohesion establishment. Genes Dev. 2010;24:2723–2731. doi: 10.1101/gad.1976710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K, Hirano T. Releasing cohesin from chromosome arms in early mitosis: opposing actions of Wapl-Pds5 and Sgo1. Genes Dev. 2009;23:2224–2236. doi: 10.1101/gad.1844309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, Toretsky JA, Rosenberg SA, Shukla N, Ladanyi M, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjogren C. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–245. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K. Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr Biol. 2009;19:492–497. doi: 10.1016/j.cub.2009.01.062. [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- Tedeschi A, Wutz G, Huet S, Jaritz M, Wuensche A, Schirghuber E, Davidson IF, Tang W, Cisneros DA, Bhaskara V, et al. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature. 2013;501:564–568. doi: 10.1038/nature12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- Vaur S, Feytout A, Vazquez S, Javerzat JP. Pds5 promotes cohesin acetylation and stable cohesin-chromosome interaction. EMBO Rep. 2012;13:645–652. doi: 10.1038/embor.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FM, Nguyen JV, Rankin S. A conserved motif at the C terminus of sororin is required for sister chromatid cohesion. J Biol Chem. 2011;286:3579–3586. doi: 10.1074/jbc.M110.196758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Kong X, Ji Z, Zeng W, Potts PR, Yokomori K, Yu H. Scc1 sumoylation by Mms21 promotes sister chromatid recombination through counteracting Wapl. Genes Dev. 2012;26:1473–1485. doi: 10.1101/gad.193615.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


