Abstract
Bacteriophages are the most abundant life forms in the biosphere. They play important roles in bacterial ecology, evolution, adaptation to new environments, and pathogenesis of human bacterial infections. Here, we report the complete genomic sequences, and predicted proteins of 27 bacteriophages of the Gram-positive bacterium Staphylococcus aureus. Comparative nucleotide and protein sequence analysis indicates that these phages are a remarkable source of untapped genetic diversity, encoding 2,170 predicted protein-encoding ORFs, of which 1,402 cannot be annotated for structure or function, and 522 are proteins with no similarity to other phage or bacterial sequences. Based on their genome size, organization of their gene map and comparative nucleotide and protein sequence analysis, the S. aureus phages can be organized into three groups. Comparison of their gene maps reveals extensive genome mosaicism, hinting to a large reservoir of unidentified S. aureus phage genes. Among the phages in the largest size class (178–214 kbp) that we characterized is phage Twort, the first discovered bacteriophage (responsible for the Twort-D'Herelle effect). These phage genomes offer an exciting opportunity to discern molecular mechanisms of phage evolution and diversity.
Keywords: genomics
Bacteriophages play a critical role in bacterial biology, diversity, and evolution. They represent an important force in microbial evolution, including their capacity to transduce host genes and mediate the acquisition of novel genetic information. The ability of bacteriophages to impart novel biochemical and physiological properties not only provides the host with the opportunity to adapt to new environments, but, in some instances, confers novel virulence properties associated with pathogenesis in human bacterial infections.
Bacteriophages usually have a narrow host range, and the global bacteriophage population has been estimated to be on the order of 1031 (1), with the majority of this population turning over every few weeks (2, 3). Additionally, bacteriophages appear to represent an enormous and unique untapped source of protein sequence diversity. Recent sequence analyses of bacteriophages from Mycobacterium tuberculosis and other bacterial species indicates that between 50% and 75% of ORFs predicted from the phage genomes have no match in GenBank (4–6). An independent estimation of the global phage metagenome using nonparametric estimation predicts that <0.0002% of the global metagenome has been sampled, with ≈2 billion different phage-encoded ORFs remaining to be discovered (7). Thus, the systematic characterization of bacteriophage genomes represents a unique opportunity to increase the size and knowledge of both the global proteome and overall genetic diversity. In addition, the comparative analysis of multiple bacteriophages from a single bacterial species offers a unique opportunity to study the mechanisms driving prokaryotic genetic diversity, including lateral gene transfer and illegitimate recombination (4, 8–10).
Lytic bacteriophages have shown clinical promise as therapeutic agents for topical or systemic treatments of bacterial infections (11) or for their ability to block and subvert essential host metabolic pathways (12). The latter point makes them attractive tools to discover and validate essential bacterial genes targeted during the phage replicative cycle. Using this strategy, we have previously shown that several Staphylococcus aureus bacteriophages encode proteins that target components of the DNA replication and RNA transcription machinery (12). As part of this ongoing effort, we now report the complete genomes and predicted proteins of a group of 27 S. aureus bacteriophages. The proteome of these phages was annotated by comparative analyses within the phage group itself and with the known sequences of three S. aureus lytic phages, 44AHJD, P68, and K (13, 14). Among the phages we characterized is Twort, the first bacteriophage described in the literature and responsible for the lysin activity reported by Twort in 1915 (15), later known as the Twort–D'Herelle effect upon publication of a more detailed description of phage growth by d'Herelle in 1917 (16). The phage sequence information reported herein constitutes a valuable resource to better study the mechanism of genome and proteome diversity in phages.
Materials and Methods
Sources of Phages. S. aureus bacteriophages were obtained from the following sources: phages 3A, 47, 29, 77, 42e, 55, 52A, 53, 71, 85, and 96 were from the Laboratory Centre for Disease Control (Ottawa); phages 44AHJD, P68, 187, 2638A, and Twort were from H.-W. Ackermann (Felix d'Hérelle Reference Center for Bacterial Viruses, Québec City); phages 66, 69, X2, EW, 37, and ROSA were from the National Collection of Type Cultures (London); phages 92 and 88 were from the American Type Culture Collection (Manassas, VA); and phage G1 was isolated from a mixture of S. aureus bacteriophages (Bacteriophagum staphylococcum liquidum, lot no. 361098, BioPharm, Tbilisi, Republic of Georgia). Phage PT1028 was isolated from a mitomycin C-treated culture of S. aureus strain NY940 (Mount Sinai Hospital, Toronto).
Phage Propagation and Preparation of Phage DNA. Isolation and propagation of phages, preparation of phage genomic DNA, and restriction enzyme digests followed published protocols (17). Individual plaques were twice purified, and large-scale infections were performed by using the agar plate method, followed by phage elution and high-speed centrifugation (40,000 × g for 2.5 h at 4°C in a JA-20 Beckman rotor). Two successive cesium chloride (CsCl) gradients were performed to isolate phage particles. Genomic DNA was isolated by treatment with Proteinase K (50 μg/ml) for 1 h at 65°C, extracted with phenol, phenol-chloroform, and chloroform, and dialyzed overnight at 4°C against 10 mM Tris·HCl, pH 8.0/1 mM EDTA.
Phage Genome Sequencing. Approximately 10 μg of phage DNA was sheared by sonication, repaired, size-selected (1–2 kbp), and cloned into pKSII+ (Stratagene). Individual clones were sequenced by using ABI Prism technology and assembled using sequencher 3.1 (Gene Codes, Ann Arbor, MI) or phred-phrap/consed 12.0 (CodonCode, Dedham, MA) software. Generally, the assembled phage genome sequences had at least 2-fold coverage in both orientations. Primer walking was sometimes used to close gaps in the sequence. The quality of the assembled sequences was independently verified by sequencing >1,000 ORFs amplified directly from phage DNA (12).
Phage ORF Prediction and Analysis. ORFs were predicted by using a customized in-house software package. Briefly, the primary nucleotide sequence was scanned in all reading frames for one of the following start codons: AUG (methionine), UUG and CUG (leucine), GUG (valine), and AUA (isoleucine), starting from the first position of the genomic sequence. Once a start codon is identified, the number of in-frame codons is counted until a termination codon is reached. A minimum threshold of 33 codons defines this bounded sequence as an ORF. Putative genes were predicted based on the presence of a Shine–Dalgarno (S.D.) sequence optimally centered between 8 and 12 bp upstream of the start codon. A position-specific scoring matrix was calculated in two independent experiments to determine the optimal S.D. sequence (5′-AGGAGG-3′) for known protein coding sequences in S. aureus. The training set consisted of sequences from 500 5′ UTRs (spanning 30 bp upstream of the AUG initiation codon) from the S. aureus genome to determine the consensus ribosomal binding sequence. The program consensus (18, 19) was used to determine the weight matrix from this training set, and a 6-bp matrix was chosen because it provided optimal information content. All phage ORFs are designated with a unique Targanta ID number.
Results and Discussion
General Features of Staphylococcal Phage Genomes. The complete genomes of 27 double-stranded DNA S. aureus bacteriophages, including 24 previously uncharacterized bacteriophages, were sequenced and analyzed by a variety of bioinformatics tools. The phages could be arranged into three classes based on genome sizes of <20 kbp (class I), ≈40 kbp (class II), and >125 kbp (class III) (Table 1). Three of the four class I phages (P68, 66, and 44AHJD) show C1 morphotype characteristics with a short, noncontractile tail and an isometric head (20), whereas the morphology of PT1028 remains to be established. The majority of class II phages (187, 69, 53, 85, 2638A, 77, 37, 96, 71, 55, 29, 52A, 88, 92, and X2) show a B1 morphotype with a long, noncontractile tail and an isometric head. In this group, phages 42e, 3A, and 47 display a B2 morphotype with a long, noncontractile tail and elongated head, whereas EW is classified as morphotype A, and phage ROSA has yet to be classified. The three phages from class III (Twort, K, and G1) are structurally defined as members of the Myoviridae family. The clustering of these phages into three discrete genome-size classes is not observed in the phages of Mycobacterium where 14 recently reported phages displayed heterogeneous genome sizes ranging from 49 to 156 kbp (4), nor in Pseudomonas aeruginosa where analysis of 10 phages reveals diverse genome sizes ranging from 36 to 281 kbp (data not shown). Combined sequence analysis of the 27 phage genomes show a GC content of 33.7%, which is similar to that of S. aureus (32.9%) (Table 1). This similarity is in agreement with that previously reported for coliphages and Escherichia coli (20), and those of Mycobacterium [63.6% (phage) vs. 65.6% (host) for M. tuberculosis] (4) and Streptococcus pneumoniae [39.8% (phage) vs. 39.7% (host)] (data not shown). As opposed to Mycobacterium phages (4), there is no relationship between GC content and genome size in S. aureus phages.
Table 1. Summary of genome size, gene content, gene density, and proteome content for 27 S. aureus bacteriophages.
| Phage | Genome Size, bp | Percent GC | Predicted genes | Percent coding | Gene density, genes per kbp | Putative function | Unknown function | NDM | Average gene size, aa |
|---|---|---|---|---|---|---|---|---|---|
| PT1028 | 15,603 | 31.4 | 22 | 77.4 | 1.41 | 9 | 13 | 2 | 187 |
| 66 | 18,199 | 29.3 | 27 | 91.8 | 1.48 | 14 | 13 | 6 | 216 |
| 44AHJD | 16,668 | 29.7 | 32 | 93.6 | 1.92 | 9 | 23 | 13 | 182 |
| P68 | 18,221 | 29.3 | 29 | 93.6 | 1.59 | 13 | 16 | 9 | 210 |
| 187 | 39,620 | 34.3 | 77 | 93.5 | 1.94 | 32 | 45 | 14 | 170 |
| 69 | 42,732 | 34.3 | 76 | 93.0 | 1.78 | 26 | 50 | 9 | 180 |
| 53 | 43,883 | 34.1 | 79 | 93.4 | 1.80 | 30 | 49 | 14 | 179 |
| 85 | 44,283 | 34.6 | 78 | 91.8 | 1.76 | 30 | 48 | 15 | 180 |
| 2638A | 41,318 | 36.9 | 57 | 92.7 | 1.38 | 19 | 38 | 17 | 224 |
| 77 | 41,708 | 33.5 | 69 | 92.1 | 1.65 | 25 | 44 | 0 | 186 |
| 42e | 45,861 | 33.7 | 79 | 93.4 | 1.72 | 33 | 46 | 14 | 186 |
| 3A | 43,095 | 33.5 | 67 | 93.9 | 1.55 | 27 | 40 | 7 | 206 |
| 47 | 44,777 | 33.5 | 72 | 94.0 | 1.61 | 28 | 44 | 11 | 199 |
| 37 | 43,681 | 35.1 | 77 | 95.6 | 1.76 | 31 | 46 | 26 | 188 |
| EW | 45,286 | 38.0 | 77 | 92.0 | 1.70 | 30 | 47 | 24 | 186 |
| 96 | 43,576 | 35.0 | 79 | 92.0 | 1.81 | 26 | 53 | 15 | 179 |
| ROSA | 43,155 | 35.1 | 74 | 92.9 | 1.71 | 31 | 43 | 8 | 183 |
| 71 | 43,114 | 35.2 | 72 | 94.6 | 1.67 | 27 | 45 | 11 | 193 |
| 55 | 41,902 | 35.7 | 77 | 93.4 | 1.84 | 31 | 46 | 14 | 180 |
| 29 | 42,802 | 35.4 | 75 | 92.2 | 1.75 | 29 | 46 | 16 | 185 |
| 52A | 41,690 | 35.5 | 65 | 91.6 | 1.56 | 30 | 35 | 11 | 202 |
| 88 | 43,231 | 35.5 | 72 | 94.3 | 1.67 | 34 | 38 | 7 | 194 |
| 92 | 42,431 | 35.7 | 74 | 95.2 | 1.74 | 30 | 44 | 13 | 192 |
| X2 | 43,440 | 36.0 | 77 | 92.8 | 1.77 | 28 | 49 | 16 | 181 |
| K | 127,395 | 30.6 | 178 | 88.7 | 1.40 | 44 | 134 | 52 | 213 |
| G1 | 138,715 | 30.4 | 214 | 88.5 | 1.54 | 50 | 164 | 69 | 193 |
| Twort | 130,706 | 30.3 | 195 | 88.0 | 1.49 | 52 | 143 | 89 | 200 |
| Total | 1,327,092 | 2,170 | 768 | 1402 | 522 | ||||
| Average | 33.7 | 92.1 | 1.67 | 192 |
The phage genomes were translated and a list of predicted protein-encoding ORFs (genes) is tabulated in Table 1, with their relative position on circular maps of the genomes shown in Fig. 6, which is published as supporting information on the PNAS web site. A total of 2,170 genes are predicted from the 27 phages. The gene maps illustrate that the coding regions are tightly packed, with very few intergenic spaces between them (Fig. 6). On average, the gene-coding potential of each phage genome is 92.1% (Table 1), with 1.67 genes per kilobase pair of nucleotide sequence, a number similar to that reported for Mycobacterium phages (1.69 genes per kbp) (4). Bacteriophage 187 shows the highest gene density (1.94 genes per kbp) and phage 2638A shows the lowest gene density (1.38 genes per kbp). As expected, the number of genes was proportional to the phage genome size, with PT1028 containing the fewest number (22 genes) and G1 containing the largest (214 genes).
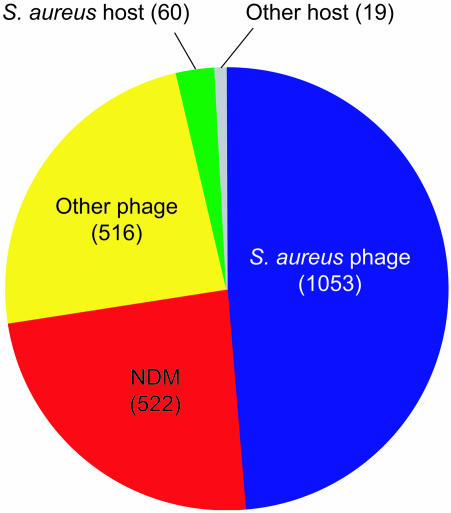
All predicted proteins were examined for similarity to known bacterial and bacteriophage sequences deposited in public databases. Positive hits were identified by using the blast server with a cutoff E value of 10–4 (Table 1 and Fig. 1), and this information was used to provide detailed annotations of the phage proteomes (Fig. 6). Several important points emerge. First, the phage proteomes appear to be a rich source of untapped protein-sequence diversity. Indeed, a large proportion of predicted proteins (1,402 genes; 65% of the proteome) show no obvious biological function compared with only 768 (35%) that can be structurally or functionally annotated (Fig. 1). Second, 1,053 ORFs (49%) show sequence similarity only to ORFs encoded by other S. aureus phage genomes (Table 1). This number is higher than the proportion of genes with identifiable homologs among other phages (516 genes; 24% of the proteome) or within the S. aureus host genome (60 genes; 3% of the proteome), which is consistent with a process of gene transfer among S. aureus phages being more predominant than recombination between S. aureus phages and phages of other species or between S. aureus phages and their host (Fig. 1). Third, a significant proportion of predicted ORFs (522 genes; 24% of the proteome) are unique and show no database match (NDM) to any publicly available prokaryotic sequence.
Fig. 1.
Bacteriophage genomics. Distribution illustrating relationship of S. aureus bacteriophage proteome to current entries in GenBank Bacteria and Phage databases. The number of bacteriophage proteins with homology (blast E value cutoff = 10–4) to another S. aureus bacteriophage (blue), other bacteriophages (yellow), to the S. aureus genome (green), to other hosts (gray), or NDM (red) is indicated in parentheses.
Analysis of the global gene organization of the phage genomes revealed several key features (Fig. 6). First, the majority of S. aureus bacteriophage genes are transcribed from one strand, although small clusters of genes are transcribed from the other strand in a seemingly mutually exclusive fashion (Fig. 6). Second, when the phage gene set is functionally classified into six arbitrary categories (DNA replication, integration, packaging, head, tail, and lysis), it is clear that predicted genes are not randomly distributed, but, rather, map to discrete modules. This modular organization and their respective order is shared by many phages in the same genome-size class. Third, genes involved in genome integration are almost always located on the least ORF-populated strand and are transcribed in the opposite direction, relative to the majority of other genes. Finally, in the three largest phages, there is a second DNA-replication module, located on the minus strand, with duplicated associated lytic functions (amidase and holin). These additional functions may be responsible for the increased host range of some members of this class of phages, infecting both coagulase-positive and -negative staphylococci (20).
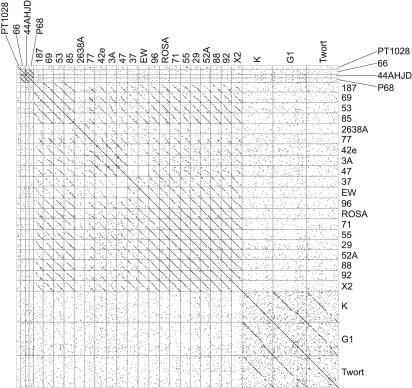
Comparative Genomic Analysis. A pairwise comparison of the nucleotide sequence of the 27 phages was carried out and is shown as a dot matrix (Fig. 2). This analysis shows that, in general, phages of the same genome-size class often demonstrate nucleotide sequence relatedness among each other, but not to phages from other genome size classes. This analysis identifies further diversity among the three genome-size classes. For class I (≈20 kb), phages 66, 44AHJD, and P68 are clearly related, whereas phage PT1028 is unique and not related to the other three, despite having a similar genome size. Class II phages can be further organized into three clades: clade A (phages 69, 53, and 85), clade B (phages 42e, 3A, and 47), and clade C (phages 37, EW, 96, ROSA, 71, 55, 29, 52A, 88, 92, and X2). Class II phage 2638A is unique and does not belong to any of the three clades. In addition, phages 187 and 77 are difficult to classify because they share similarities to several members of the different clades. This clustering may reflect some sampling bias, in particular, for some of the clade C phages. Finally, class III phages G1, K, and Twort are clearly related to each other (in particular, G1 and K), but share no similarity to any other S. aureus phages, which is consistent with their classification in the Myoviridae family. Indeed, at the nucleotide level, phages K and G1 are 90% identical, whereas the relationship between Twort and G1 or K is not as high (51–52% identity).
Fig. 2.
Comparative nucleotide sequence analysis of S. aureus bacteriophages genomes. Shown is a dot matrix comparing the relatedness of the nucleotide sequences of each phage genome that were generated with the software program dotter (32) by using a sliding window of 25 bp.
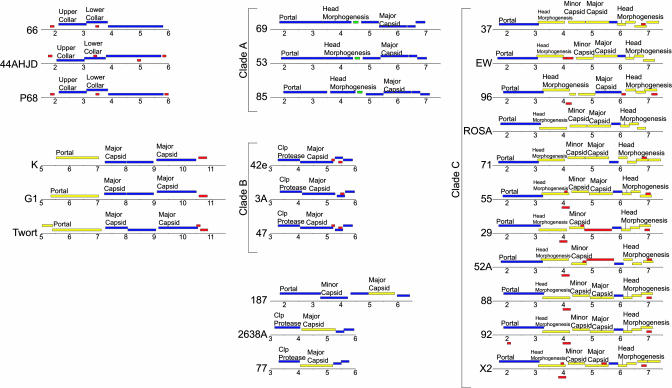
Annotated functional modules in the 27 phages were further examined with respect to number, position, and reading frame of individual ORFs, superimposed on potential similarity to genes from S. aureus phages, other phages, the S. aureus host genome, and no NDMs. This analysis provided an additional measure of structural conservation amongst the phage genomes and proteomes, and allowed confirmation of the grouping described above. There is a particularly high degree of conservation of the structural ORF map in the region encoding head proteins of different phages (Fig. 3). For class I, this analysis verified the uniqueness of phage PT1028 and the close similarity among phages 66, 44AHJD, and P68. For class II, this analysis further validated the classification of clades A, B, and C, and the presence of unique phages in this group (2638A, 187, and 77), as noted above in the nucleotide sequence analyses (Fig. 2).
Fig. 3.
Comparative gene arrangements among S. aureus bacteriophage head regions. The gene content of each phage head region and identity of the encoded proteins are indicated by colored boxes, using the same color key as described for Fig. 1. The map location of each head region is indicated and the annotation is aligned such that the leftmost part of the gene is directly below the start of the text. Genes lacking annotation have no predicted function. Note that for PT1028, no head region was identified from the structural ORF map.
A pairwise comparison of the proteomes of the 27 phages was carried out and is presented in Table 2, which is published as supporting information on the PNAS web site. This analysis confirms the results of nucleotide sequence comparisons, with phages from the different size classes being more related to each other than to other classes (Table 2). This relationship is maintained whether moderately stringent criteria (E > 10–20) or more relaxed parameters (E > 10–4) are used in the analysis. This analysis extended to ORFs showing NDMs (522 genes), indicating that phages within a given genome-size class share a small number of NDM genes, but this relationship does not extend to phages between classes (Table 3, which is published as supporting information on the PNAS web site).
Although space limitations preclude a detailed review of individual phage proteomes, comparison of the 27 annotated genomes (Fig. 6 and Table 4, which is published as supporting information on the PNAS web site) identifies several interesting features. First, PT1028 is dissimilar in gene content, compared with other phages of the ≈20-kbp size class. There appears to be no lysis functions. It is interesting to note the phage was isolated after mitomycin C treatment of an S. aureus clinical isolate, suggesting that it may be a relatively small and highly evolved temperate phage (Fig. 6). Second, the overall gene organization (clockwise from 0°) of packaging, head, tail, lysis, integration, and DNA-replication functions appear generally conserved among the majority of phages (although there are clear outliers, such as PT1028 and the class III phages, which lack specific functions or contain duplicated regions, respectively). Third, phages G1, K, and Twort contain a second DNA-replication module that encodes a greater number of DNA-replication proteins (Fig. 6). Unique to these three phages are components of the ribonucleoside-diphosphate reductase complex, which are responsible for catalyzing de novo reductive synthesis of deoxyribonucleotides from their corresponding ribonucleotides, and necessary for providing DNA synthesis precursors (21). Also, they contain: (i) histone-like bacterial DNA-binding proteins, (ii) RecA, which plays a role in homologous DNA recombination and DNA repair, and (iii) DNA primase, a nucleotidyl-transferase responsible for synthesis of oligonucleotide primers required for DNA replication on the lagging strand of the replication fork. These functions indicate that class III phages have evolved a larger set of DNA-replication functions derived from their hosts (because these exhibit homology to bacterial not phage proteins), a result observed with other large members of the Myoviridae family.
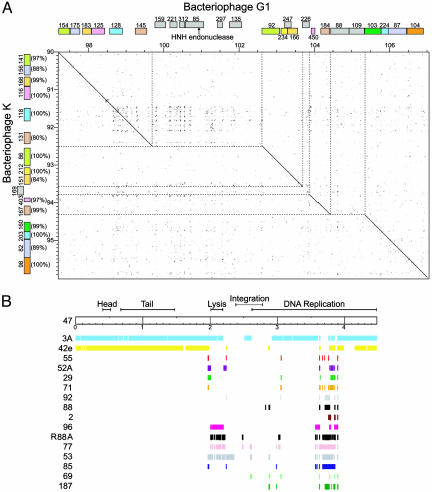
Extensive Mosaicism in the Phage Genomes. The S. aureus phages show widespread insertions/deletions within their genomes. This finding is exemplified by examination of a segment of the DNA-replication module of phages G1 and K (Fig. 4A). First, the overall gene organization within this region is conserved with many of the genes between the two phages being highly related. Second, there are unique insertions/deletions restricted to one phage. Phage G1 ORFs 159, 221, 312, 85, 297, and 135 are not found in phage K and are located between two ORFS (145 and 92), that have homologous counterparts in phage K (ORFs 131 and 86). Third, unrelated ORFs can be found inserted between two conserved genes, such as phage G1 ORF 226 and phage K ORF 169. With the exception of the ORF encoding an HNH endonuclease-related function, none of the genes unique to either phage G1 or K have a homologue in the GenBank Bacteria and Phage database. This large amount of mosaicism found among phages support the idea of large-scale genetic exchange in prokaryotic viruses (22–25).
Fig. 4.
Mosaicism in S. aureus phages. (A) A highly mosaic segment of one of the DNA-replication modules of phages G1 and K. Related ORFs are identified by using a color code, with the percent identity shown to the left of bacteriophage K ORFs. Nonhomologous regions are evident on the dot matrix because they result in discontinuity of the diagonal plot and are shaded gray. None of the ORFs within this region, with the exception of G1 ORF 85 (HNH endonuclease) encode proteins with known function. (B) Mosaic nature of phage 47. The nucleotide sequence of phage 47 was scanned for conserved blocks of ≥50 bp showing≥98% identity with the other 26 S. aureus phages (identified to the left). Identified regions are aligned below the white box that schematically represents phage 47.
A separate comparative analysis was performed by using phage 47 (a clade B phage) as an example, in which genomic regions (≥50 bp) from other S. aureus phages having ≥98% identity were aligned. The head and tail regions, and part of the region encoding DNA-replication functions of phage 47 show the greatest similarity to the corresponding regions of phages 3A and 42e, which is consistent with these phages being categorized as B2 morphotypes (see above). The lysis function of phage 47 shows the greatest similarity to the corresponding region from phages 3A, 96, ROSA, 77, and 53. The nucleotide sequence of phage 47 spanning the integration functions appears as a patchwork of nucleotide sequences similar to a number of other phages, with a large block related to the corresponding region in phage 3A. Part of the DNA-replication module of phage 47 is more homologous to the corresponding region of phages ROSA, 77, 53, and 85, although a number of other phages have related blocks of nucleotide sequence that span this region as well. These examples illustrate the mosaic nature of the S. aureus phage genomes (Fig. 4B), which is consistent with previous reports documenting mosaicism among phages (4, 26–29). Our results support the idea that taxonomical classification of phages based on sequence similarity is inadequate (9, 23), and that alternative methods such as those based on the phage proteome may better describe phage relationships (24).
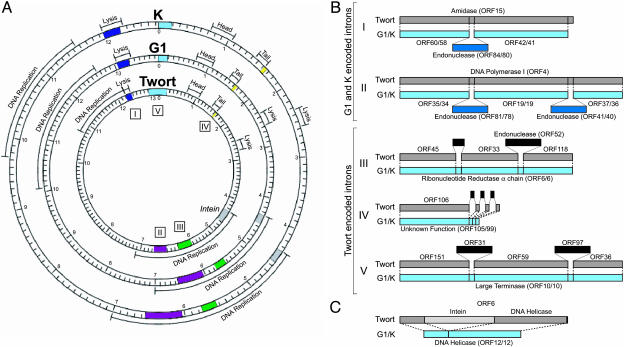
Splicing. The presence of group I introns have been previously documented in phages K and Twort (14, 30, 31). Five genomic regions implicated in splicing were analyzed in detail (Fig. 5A). Introns within the amidase and DNA polymerase genes of phage K (that encode endonucleases) have been previously characterized (14), and are also present within their respective homologues within phage G1, but are absent in Twort (Fig. 5B, clusters I and II). Previously documented introns within the ribonuclease reductase gene of Twort (30, 31) and Twort ORF 106 [corresponds to Twort ORF 142 documented by Landthaler and Shub (30)] are not present within homologues in phages G1 or K (Fig. 5B, clusters III and IV). We performed a genome-wide comparison between Twort and G1, searching for relatedness between two genes of one phage (separated by one ORF) to one complete ORF of a second phage. The complete ORF had to score for homology that spanned multiple genes from the first phage. This analysis identified two introns within the large terminase gene of Twort, both absent in G1 and K (Fig. 5, cluster V). The intron encoding ORF 31 is 1,545 bp in length and likely corresponds to the unassigned ≈1,400-nucleotide intron detected by Landthaler et al. (31).
Fig. 5.
Splicing within Twort, G1, and K. (A) Circular map of phages K, G1, and Twort, illustrating the relative location of the regions involved in splicing. (B) Introns present in the G1, K, and Twort genomes. Intervening sequences are indicated above or below the denoted ORFs in dark blue. The five regions compared for conservation of splicing events are numbered I–V. (C) Schematic representation illustrating the predicted presence of an intein within Twort ORF 6. The intein is denoted by a lightly shaded gray box. The predicted downstream extein has putative DNA helicase activity (blast E value = 10–34).
An intein-encoding gene within Twort was identified during a proteome comparative analysis (Fig. 5C). The predicted inteincoding region is not present in phages G1 or K, and interrupts two exteins, one of which has predicted DNA helicase activity. The intein also harbors a DOD endonuclease motif that confers genetic mobility to the intein (data not shown). Differences in intron and intein content among these three phages are other examples of lateral gene-transfer events, leading to phage mosaicism.
Conclusion
The isolation and characterization of a large set of Staphylococcal phages indicates a clear distribution of the phage genomes within three size classes. These phages contain a large gene set with uncharacterized function. Diversity within the Staphylococcus phage population further occurs through a number of mechanisms, including apparent recombination among other phages, and, less frequently, between the phages and the S. aureus host. The mosaic nature of the phages reported herein is evident from comparative genomics analysis and highlights the dynamic nature of this process, which is consistent with what has been previously reported (4, 22, 25). By providing primary sequence information from a large group of phages from a single organism, this report not only provides a compendium of protein sequences but also allows a systematic approach to study phage evolution and diversity (25).
Supplementary Material
Acknowledgments
We thank the scientific and support personnel of Targanta Therapeutics for their contribution to this project over the last 7 years. We thank the National Research Council (Canada) Industrial Research Assistance Program for partial support of our research program. T.K. and J.L. are recipients of the Natural Sciences and Engineering Research Council of Canada Industrial Research Fellowship.
Author contributions: M.D., P.G., and J.P. designed research; T.K. and J.L. performed research; T.K. analyzed data; and T.K., P.G., and J.P. wrote the paper.
Abbreviation: NDM, no database match.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology (NCBI) database (accession nos. AY954948–AY954970).
References
- 1.Wommack, K. E. & Colwell, R. R. (2000) Microbiol. Mol. Biol. Rev. 64, 69–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilhelm, S. W., Brigden, S. M. & Suttle, C. A. (2002) Microb. Ecol. 43, 168–173. [DOI] [PubMed] [Google Scholar]
- 3.Breitbart, M., Wegley, L., Leeds, S., Schoenfeld, T. & Rohwer, F. (2004) Appl. Environ. Microbiol. 70, 1633–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedulla, M. L., Ford, M. E., Houtz, J. M., Karthikeyan, T., Wadsworth, C., Lewis, J. A., Jacobs-Sera, D., Falbo, J., Gross, J., Pannunzio, N. R., et al. (2003) Cell 113, 171–182. [DOI] [PubMed] [Google Scholar]
- 5.Breitbart, M., Salamon, P., Andresen, B., Mahaffy, J. M., Segall, A. M., Mead, D., Azam, F. & Rohwer, F. (2002) Proc. Natl. Acad. Sci. USA 99, 14250–14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breitbart, M., Hewson, I., Felts, B., Mahaffy, J. M., Nulton, J., Salamon, P. & Rohwer, F. (2003) J. Bacteriol. 185, 6220–6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao, A. (1984) Scand. J. Stat. 11, 783–791. [Google Scholar]
- 8.Proux, C., van Sinderen, D., Suarez, J., Garcia, P., Ladero, V., Fitzgerald, G. F., Desiere, F. & Brussow, H. (2002) J. Bacteriol. 184, 6026–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendrix, R. W. (2000) Theor. Popul. Biol. 61, 471–480. [DOI] [PubMed] [Google Scholar]
- 10.Datta, A., Hendrix, M., Lipsitch, M. & Jinks-Robertson, S. (1997) Proc. Natl. Acad. Sci. USA 94, 9757–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chibani-Chennoufi, S., Sidoti, J., Bruttin, A., Kutter, E., Sarker, S. & Brussow, H. (2004) Antimicrob. Agents Chemother. 48, 2558–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, J., Dehbi, M., Moeck, G., Arhin, F., Bauda, P., Bergeron, D., Callejo, M., Ferretti, V., Ha, N., Kwan, T., et al. (2004) Nat. Biotechnol. 22, 185–191. [DOI] [PubMed] [Google Scholar]
- 13.Vybiral, D., Takac, M., Loessner, M., Witte, A., von Ahsen, U. & Blasi, U. (2003) FEMS Microbiol. Lett. 219, 275–283. [DOI] [PubMed] [Google Scholar]
- 14.O'Flaherty, S., Coffey, A., Edwards, R., Meaney, W., Fitzgerald, G. F. & Ross, R. P. (2004) J. Bacteriol. 186, 2862–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Twort, F. W. (1915) Lancet 189, 1241–1243. [Google Scholar]
- 16.d'Herelle, F. (1917) C. R. Acad. Sci. Paris 165, 373–375. [Google Scholar]
- 17.Sambrook, J. & Russell, D. W. (2001) Molecular Cloning. A Laboratory Manual (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 18.Hertz, G. Z., Hartzell, G. W., III, & Stormo, G. D. (1990) Comput. Appl. Biosci. 6, 81–92. [DOI] [PubMed] [Google Scholar]
- 19.Stormo, G. D. & Hartzell, G. W., III. (1989) Proc. Natl. Acad. Sci. USA 86, 1183–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ackermann, H. W. & DuBow, M. (1987) Viruses of Prokaryotes (CRC, Boca Raton, FL).
- 21.Nilsson, O., Lundqvist, T., Hahne, S. & Sjoberg, B. M. (1988) Biochem. Soc. Trans. 16, 91–94. [DOI] [PubMed] [Google Scholar]
- 22.Hendrix, R. W. (2003) Cur. Opin. Microbiol. 6, 506–511. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence, J. G., Hatfull, G. F. & Hendrix, R. W. (2002) J. Bacteriol. 184, 4891–4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohwer, F. & Edwards, R. (2002) J. Bacteriol. 184, 4529–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendrix, R. W., Hatfull, G. F. & Smith, M. C. (2003) Res. Microbiol. 154, 253–257. [DOI] [PubMed] [Google Scholar]
- 26.Juhala, R. J., Ford, M. E., Duda, R. L., Youlton, A., Hatfull, G. F. & Hendrix, R. W. (2000) J. Mol. Biol. 299, 27–51. [DOI] [PubMed] [Google Scholar]
- 27.Brussow, H., Bruttin, A., Desiere, F., Lucchini, S. & Foley, S. (1998) Virus Genes 16, 95–109. [DOI] [PubMed] [Google Scholar]
- 28.Desiere, F., Lucchini, S. & Brussow, H. (1999) Virology 260, 244–253. [DOI] [PubMed] [Google Scholar]
- 29.Lucchini, S., Desiere, F. & Brussow, H. (1999) J. Virol. 73, 8647–8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landthaler, M. & Shub, D. A. (1999) Proc. Natl. Acad. Sci. USA 96, 7005–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landthaler, M., Begley, U., Lau, N. C. & Shub, D. A. (2002) Nucleic Acids Res. 30, 1935–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sonnhammer, E. L. & Durbin, R. (1995) Gene 167, GC1–GC10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.