Abstract
Reactive oxygen species (ROS) are key components of postreceptor intracellular signaling pathways; however, the role of ROS in signal initiation is uncertain. We discovered that receptor–ligand interaction caused the generation of hydrogen peroxide (H2O2). Using members of the hematopoietin receptor superfamily, as well as EGF receptor, we show that H2O2 is generated by specific receptor–ligand interaction in cells and in cell-free systems. With cognate ligand, the extracellular domain of the receptor was sufficient for H2O2 generation. We also found that production of H2O2 was diminished in a granulocyte–macrophage colony-stimulating factor receptor mutant unable to bind ligand. Exogenously added H2O2 induced signaling in the absence of ligand, whereas catalase and a membrane-bound peroxiredoxin inhibited ligand-dependent signaling. Our results suggest that H2O2 produced by receptor–ligand interaction is involved as a chemical mediator that facilitates cell signaling.
Keywords: reactive oxygen species, kinase, cytokine hematopoietin
Growth factors and cytokines achieve their effects by binding specific cell-surface receptors that initiate intracellular signaling cascades (1–3). Reactive oxygen species (ROS) are involved in modulating these signaling pathways (4, 5), but their precise role in signal initiation is unknown.
The granulocyte–macrophage colony-stimulating factor (GM-CSF) signaling pathway is known to involve ROS, and antioxidants inhibit early ligand-dependent responses (6, 7). Human GM-CSF is a hematopoietic growth factor that regulates the proliferation, maturation, and differentiation of myeloid progenitors, as well as the function of mature host defense cells (8). The GM-CSF receptor (GMR) is comprised of an 85-kDa α-subunit (αGMR) that binds ligand with low affinity and a 130-kDa β-subunit (βGMR) that cannot bind ligand alone but forms a high-affinity receptor with αGMR (9–11). The β-subunit is common to the GM-CSF, IL-3, and IL-5 receptors (12). GM-CSF initiates receptor-mediated transmembrane signaling events by means of the activation of the Janus family tyrosine kinase 2 (Jak2) (13) that then phosphorylates βGMR (14). The resultant phosphotyrosines on βGMR provide binding sites for signaling proteins with Src homology 2 domains, including signal transducers and activators of transcription (STAT) proteins (1, 15, 16). GM-CSF also activates the ras/raf signaling pathway, inducing mitogen-activated protein kinase (MAPK) phosphorylation and downstream transcription factors (8, 17, 18).
Receptors possessing intrinsic kinase domains, such as the EGF receptor (EGFR) and platelet-derived growth factor receptor (PDGFR), also signal by means of ROS-mediated mechanisms (19, 20). Hydrogen peroxide (H2O2) has been shown to be involved in the activation of EGFR and PDGFR (19, 20).
Our finding that activation of phosphatidylinositol 3-kinase by the isolated α-subunit of the GMR (αGMR) was inhibited by extracellular catalase (21) led to the hypothesis that H2O2 generated extracellularly by receptor–ligand interaction could permeate into the cell and facilitate signaling. It has been reported that normal and malignant cells, as well as isolated proteins, have the ability to generate H2O2 (22–24). We show in cells and cell-free systems that the extracellular domain of hematopoietin receptors interacting with cognate ligand is sufficient to generate H2O2. The EGFR also produced H2O2 when interacting with its ligand. These results suggest a general role of ROS in receptor-mediated facilitation of signaling.
Materials and Methods
Cells. A431 (human epidermoid carcinoma) and 293T (human kidney epithelial) cells were grown in DMEM-high glucose supplemented with 10% heat-inactivated FBS, 1% l-glutamine, and 1% penicillin/streptomycin. U937 (human histiocytic lymphoma) cells were obtained from the American Type Culture Collection (Manassas, VA) and grown in RPMI medium 1640 supplemented with 10% heat-inactivated FBS, 1% l-glutamine, and 1% penicillin/streptomycin. MO7e (human megakaryocytic leukemia, cytokine-dependent) cells were grown in Iscove's modified Dulbecco's medium with 10% heat-inactivated FBS, 1% l-glutamine, and 1% penicillin/streptomycin and supplemented with 30 pM GM-CSF.
Constructs. Human α- and β-subunits of GMR were in the pMX expression vector (21). Human EGFR was in the pUSE expression vector (Upstate Biotechnology, Lake Placid, NY). The αGMR and βGMR intracellular deletions in the pcDNA4/His-V5c expression vector (αΔ and βΔ) were provided by Jian Chen (Memorial Sloan–Kettering Cancer Center). The extracellular domain of αGMR (αGMRx) fused to a 6xHis tag was used to secrete receptors into media for isolation. Site-directed mutagenesis (QuikChange II, Stratagene) was used to create a cysteine-to-serine point mutation in αGMR (C136S) and αGMRx (C136Sx). The BD Advantage 2 PCR Enzyme System (BD Biosciences) was used to amplify peroxiredoxin-5 (Prdx5) from Marathon-Ready human lung cDNA (Clontech). The primers used were 5′-AGATCTCCCGGGATGGGACTAGCTGGCGTG-3′ and 3′-CTGCAGCCGCGGGAGCTGTGAGATGATATT-5′. Prdx5 was then cloned into the pDisplay expression vector (Invitrogen).
Transfections and Luciferase Assays. 293T cells were transiently transfected with cDNAs by using GeneJammer transfection reagent (Stratagene). STAT5-dependent signaling was performed with p8xGAS-luciferase reporter plasmid (25), and luciferase activity was quantitated with a luciferase reporter assay system (Promega). For experiments with fixed cells, transfected 293T cells were washed with PBS, fixed in 4% paraformaldehyde (pH 7.4) for 10 min, washed three times, and resuspended in PBS.
Receptor Binding Assay. Receptor binding assays were performed by using 293T cells (live or fixed) incubated overnight at 4°C with 125I-GM-CSF (PerkinElmer) in DMEM-high glucose serum-free medium. Cells were washed with FCS and then washed three times with PBS. Radioactivity of the cell pellets was measured by using a Cobra II AutoGamma (D5005) gamma counter (Packard).
Receptors and Ligands. αGMRx and C136Sx proteins were isolated from the supernatants of transfected 293T cells with Ni+-nitrilotriacetic acid (NTA) HisSorb Strips (Qiagen, Valencia, CA). Recombinant human ligands and receptors (R & D Systems) were >97% pure. Recombinant human soluble EGFR (Research Diagnostics, Flanders, NJ) was ≈80% pure. Commercially obtained proteins were carrier-free and reconstituted in PBS.
Immunoblotting. Full-length α- and β-subunits of GMR were detected with anti-αGMR (C18) and anti-βGMR (C20) antibodies, respectively (Santa Cruz Biotechnology). αΔ, αGMRx, C136Sx, and βΔ were detected with anti-αGMR (S50) and anti-βGMR (N20) antibodies (Santa Cruz Biotechnology). Receptor bound to Ni+-NTA plate was quantitated by using S50 antibody and rhodamine-conjugated secondary antibody (Santa Cruz Biotechnology). Expression of Prdx5 was detected with anti-HA antibody (clone 12CA5, Roche Diagnostics). Expression of p44/42-MAPK and phosphorylated p44/42-MAPK was detected as described in ref. 7. Phosphorylated Jak2 and unphosphorylated Jak2 were detected by using antibodies from BioSource International (Camarillo, CA). Expression of EGFR was determined by using anti-EGFR (sc-03) or anti-phospho-EGFR (sc-12531-R) (Santa Cruz Biotechnology) antibodies. The enhanced chemiluminescence Western blotting detection system was used (Amersham Biosciences).
Cellular Proliferation and Viability Assays. MO7e cells were incubated with 30 pM GM-CSF for 48 h with or without catalase. Cell proliferation and viability were determined by trypan blue exclusion. Viability of live and fixed cells was determined by WST-8-based colorimetric assay (CCK-8, Dojindo, Gaithersburg, MD).
Phosphorylation of βGMR. Cells (293T) expressing αGMR and βGMR were preincubated for 10 min with 5 × 103 units/ml bovine liver catalase (Sigma–Aldrich) and then treated with 1 nM GM-CSF for 15 min. The phosphorylation of βGMR was detected by immunoblotting with anti-phosphotyrosine (4G10 clone, Upstate Biotechnology) from extracts immunoprecipitated with anti-βGMR, as described in ref. 7. For in vitro phosphorylation of βGMR, the α/βGMR complex was immunoprecipitated with anti-βGMR antibody (S16, Santa Cruz Biotechnology) and Protein A beads. Beads were incubated for 15 min at 30°C in kinase reaction buffer (200 mM MgCl2/10 mM MnCl2/2 mM EGTA/80 mM β-glycerophosphate/80 mM imidazole HCl, pH 7.3) containing 15 μM ATP and 0.3 μCi/μL (1 Ci = 37 GBq) γ-32P ATP with or without GM-CSF, catalase, AG490 (Sigma–Aldrich), or H2O2. Beads were washed and resuspended in SDS-loading buffer, and βGMR phosphorylation was detected by autoradiography. Unphosphorylated βGMR was detected by immunoblotting.
H2O2 Detection and Quantitation. H2O2 was detected by using Amplex Red (A-22188, Molecular Probes) and using fluorescence microscopy and spectroscopy. Fluorescence microscopy was performed by using an Olympus BX60 microscope with NG filter, a QImaging CCD camera (Retiga 1300C), and qcapture software (QImaging, Burnaby, Canada). Exposure times varied between experiments but remained constant within each experiment. Fluorescence spectroscopy was performed by using a ThermoLab System Fluoroskan Ascent FL (Thermo Electron, Waltham, MA), and H2O2 was quantitated with standard curve. To detect extracellular H2O2 generation, supernatants (2 μl) from cells expressing αGMR, βGMR, or EGFR and treated with GM-CSF or EGF were applied to a nitrocellulose membrane (Bio-Rad), and fluorescence was detected by microscopy. Fluorescence was quantified from qcapture digital images with nih imagej software. To detect H2O2 generated by receptor–ligand interaction in cell-free systems, reactions using various combinations of IL-3, IL-5, human growth hormone (GH), and prolactin (ProL) ligands and receptors (final concentration of 3 μM) with or without 5 × 103 units/ml catalase were incubated at 37°C for 1 h with Amplex Red (final volume of 5 μl). Additionally, reactions using various combinations of EGF ligand and receptor (final concentration of 600 nM) were performed. Two microliters from each reaction was spotted onto a nitrocellulose membrane for H2O2 detection by fluorescence microscopy.
Phosphorylation of Jak2, MAPK, and EGFR. Cells were serum-starved overnight, washed in PBS, and preincubated for 15 min with or without catalase. Cells were treated with 1 nM GM-CSF or 500 μM H2O2 for 10 min, and phosphorylated Jak2 or MAPK was detected by immunoblotting (7). For activation of EGFR, cells were preincubated at 37°C for 30 min with or without catalase and then treated with or without EGF (10 ng/ml) for 3 min. EGFR-expressing 293T cells preincubated for 1 h at 37°C with the tyrosine kinase inhibitor AG1478 (Sigma–Aldrich) were treated with 50 ng/ml EGF for 5 min. Phosphorylated and unphosphorylated proteins were detected by immunoblotting.
Results
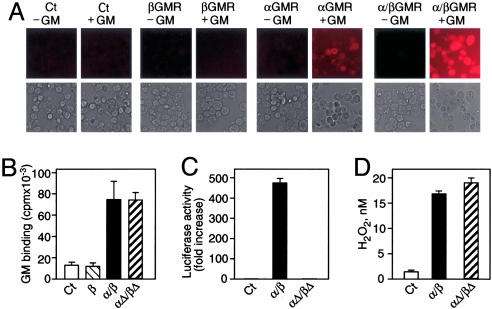
Receptor–Ligand Interaction Generates Extracellular H2O2 in Cells. To investigate the ability of the GMR to generate extracellular H2O2, 293T cells were transiently transfected with αGMR or βGMR or cotransfected with both subunits. The cellimpermeable reagent Amplex Red (10-acetyl-3,7-dihydroxyphenoxazine), which is converted into a fluorescent compound in the presence of H2O2, was used to detect extracellular H2O2. Cells expressing αGMR or the high-affinity α/βGMR generated an incremental increase in extracellular H2O2 over background when treated with GM-CSF (Fig. 1A). Neither untransfected cells nor cells expressing βGMR produced an increase in H2O2 in the presence of ligand, presumably because the βGMR subunit does not bind GM-CSF. We measured H2O2 in the supernatants of GM-CSF-treated 293T cells transfected with αGMR or with α/βGMR and found an almost 3-fold increase in generated H2O2 over control after subtracting background levels of H2O2 (data not shown). These results are consistent with the thesis that receptor–ligand interaction causes the generation of extracellular H2O2. However, the increase in H2O2 observed in this cellular system could include H2O2 resulting from signaling, as well as that generated by receptor–ligand interaction. To determine whether receptor–ligand interaction could generate extracellular H2O2 in the absence of signaling, we used intracellular deletion mutants (αΔ/βΔ). In an overexpression system, α/βGMR and αΔ/βΔ bound ligand with similar capacities (Fig. 1B). Compared with wild type, αΔ/βΔ was defective in STAT5-dependent signaling (Fig. 1C). In response to ligand, both α/βGMR and αΔ/βΔ produced a similar increase in H2O2 compared with untransfected cells after 2 min (Fig. 1D). These results suggest that the extracellular domains of α/βGMR are sufficient to generate a ligand-dependent increase in extracellular H2O2.
Fig. 1.
GM-CSF binding to its receptor generates extracellular H2O2.(A) 293T cells untransfected (Ct), transfected with βGMR or αGMR, or cotransfected with α/βGMR were incubated in Amplex Red for 1 h with or without 100 nM GM-CSF (GM) and analyzed by fluorescence microscopy. Below each fluorescent image is the corresponding light microscopy image. (B) GM binding to 293T cells untransfected (Ct) or transfected with βGMR (β), α/βGMR (α/β), or intracellular truncation mutants (αΔ/βΔ) using 1 nM 125I-GM. Error bars represent SDs. (C) STAT5-dependent transcriptional activation in 293T cells transfected with GMR (α/β or αΔ/βΔ) and p8xGAS-luc construct treated with 1 nM GM. (D) GM-dependent increase in H2O2 generation in 293T cells untransfected (Ct) or transfected with GMR (α/β or αΔ/βΔ) treated with 10 nM GM as quantitated by fluorescence spectroscopy.
Receptor–Ligand Interaction Produces H2O2 in Fixed Cells and Isolated Proteins. To further document that protein–protein interaction between receptor and ligand generated H2O2 extracellularly in the absence of signaling, we fixed with 4% paraformaldehyde 293T cells expressing GMRs. Fixed cells were able to retain the ability to bind ligand similar to live cells (Fig. 2A). Additionally, fixed cells were not viable, as determined by trypan blue exclusion (data not shown) or WST-8-based colorimetric assay (Fig. 2B). Compared with live cells expressing α/βGMR, fixed cells expressing α/βGMR failed to signal via STAT5 (Fig. 2C). As detected by fluorescence microscopy, supernatants from fixed cells expressing αGMR, but not βGMR, generated a GM-CSF-dependent increase in H2O2 (Fig. 2D). Fluorescence spectroscopy revealed that fixed cells expressing αGMR and α/βGMR generated a ligand-dependent increase in H2O2, whereas untransfected and βGMR-expressing cells did not (Fig. 2E). We confirmed the specificity of H2O2 production in fixed 293T cells expressing αGMR or EGFR. Only cognate receptor–ligand interaction generated an increase in H2O2 as detected by fluorescence microscopy (Fig. 2F). To demonstrate that H2O2 generation requires intact receptor–ligand binding, we created a binding-defective αGMR mutant (C136S) (Fig. 2G Left) (26). In a cell-free system, we found that αGMRx generated a GM-CSF-dependent increase in H2O2 production, whereas C136Sx did not (Fig. 2G Right). These results suggest that receptor–ligand interaction can generate an increase in H2O2 in the absence of signaling.
Fig. 2.
GMR–ligand interaction generates H2O2 in the absence of signaling. (A) GM-CSF (GM) binding using 1 nM 125I-GM in live and fixed 293T cells untransfected (Ct) or transfected with βGMR (β) or αGMR (α). (B) Cell viability of live or fixed 293T cells transfected with αGMR. (C) Luciferase activity of live and fixed 293T cells cotransfected with α/βGMR and STAT5-dependent luciferase reporter construct treated with or without 1 nM GM. (D) H2O2 generation in supernatants from fixed 293T cells transfected with αGMR or βGMR and treated with or without 10 nM GM. Digital images from Amplex Red-based fluorescence microscopy and H2O2 levels are shown. (E) GM-dependent H2O2 generation in fixed 293T cells untransfected (Ct) or transfected with αGMR, βGMR, or α/βGMR, as detected by fluorescence spectroscopy. (F) Extracellular H2O2 generation in fixed 293T cells transfected with αGMR or EGFR and then treated with 10 nM GM or EGF, as detected by fluorescence microscopy. (G) (Left) Relative 125I-GM binding (3 nM) in fixed 293T cells expressing wild type (αGMR) or mutant (C136S). (Right) GM-dependent (100 nM) H2O2 generation in vitro by αGMRx or mutant (C136Sx) bound to Ni+-nitrilotriacetic acid plates, as quantified by fluorescence spectroscopy. Error bars represent SDs.
We sought to determine whether extracellular H2O2 generation by receptor–ligand interaction was a general phenomenon in the hematopoietin receptor superfamily by studying purified proteins. Using human IL-3, IL-5, and respective soluble receptors (αIL-3Rs and αIL-5Rs), we found that ligand or receptor alone produced low levels of H2O2 (Fig. 3 A and B). However, cognate receptor–ligand pairs generated increases in H2O2 that exceeded additivity. To ensure specificity, we incubated αIL-3Rs with IL-5 and incubated αIL-5Rs with IL-3. Under these conditions, H2O2 production diminished to levels observed for receptor alone (Fig. 3 A and B). Using GH, ProL, and the extracellular domains of their respective receptors (GHRx and ProLRx), we found that cognate receptor–ligand combinations produced increased H2O2 (Fig. 3 C and D). Noncognate pairs also generated increases in H2O2 but at levels less than observed for cognate pairs. This finding likely reflects that GHR is known to bind ProL and vice versa (Fig. 3 C and D). Catalase abrogated H2O2 produced by cognate receptor–ligand interactions. These results indicate that the interaction between ligand and extracellular domain of receptor is sufficient to generate an increase in H2O2.
Fig. 3.
Receptor–ligand interactions generate H2O2, using purified proteins in vitro.(A) Purified human soluble αIL-3 receptor (αIL-3Rs) was incubated in Amplex Red with IL-3 or IL-5, and H2O2 was detected by fluorescence microscopy. (B) H2O2 generation using purified human soluble αIL-5 receptor (αIL-5Rs) incubated with IL-5 or IL-3 was detected as in A.(C) H2O2 generation using purified extracellular domain of the GH receptor (GHRx) incubated with GH or ProL was detected as in A. (D) H2O2 generation using purified extracellular domain of human ProL receptor (ProLRx) incubated with GH or ProL was detected as in A. cat, catalase.
Peroxidases Inhibit GM-CSF-Dependent Cellular Responses. We investigated the role of H2O2 in cellular responses induced by GM-CSF: specifically, survival of factor-dependent MO7e cells and STAT5 activation in α/βGMR-expressing 293T cells. MO7e cells were incubated in GM-CSF for 48 h with varying concentrations of catalase. Addition of extracellular catalase significantly decreased cell proliferation compared with control (Fig. 4A). Catalase did not affect viability in MO7e or U937 cells at 48 h as measured by trypan blue exclusion, suggesting that, at the concentration used, the enzyme was not cytotoxic (data not shown). Cells treated with catalase had a 45% reduction in cell number compared with control, suggesting that extracellular H2O2 plays a role in GM-CSF-supported growth. To assess the effect of extracellular catalase on GM-CSF-dependent STAT5 signaling, 293T cells were transfected with α/βGMR and p8xGAS-luciferase reporter construct. Cells treated with catalase and incubated for 5 h with GM-CSF had a nearly 40% reduction in STAT5-dependent transcriptional activation (Fig. 4B). We sought to confirm the catalase results by expressing human Prdx5 on the cell membrane (27). The effect of Prdx5 on GM-CSF-mediated STAT5-dependent signaling was examined in 293T cells transfected with α/βGMR and Prdx5. Cells expressing Prdx5 at the cell membrane exhibited a 45% reduction in STAT5-dependent luciferase activity (Fig. 4C). Catalase was also effective in diminishing GM-CSF-induced p44/42-MAPK phosphorylation in U937 and MO7e cell lines (Fig. 4D). These results suggest that extracellular H2O2 is involved in GM-CSF cell signaling. As assessed by immunoprecipitation and immunoblotting, the α/βGMR complex was unchanged in the presence of human or bovine catalase, suggesting that the enzyme does not have proteolytic activity toward the receptor, nor does it disrupt α/βGMR complex formation (data not shown).
Fig. 4.
GM-CSF-mediated signaling involves H2O2.(A) GM-CSF (GM)-dependent MO7e cells were incubated with 30 pM GM for 48 h with or without catalase. Cell number was determined by trypan blue exclusion (*, P = 0.04, one-tailed t test). (B) Luciferase activity of 293T cells transfected with α/βGMR and p8xGAS-luc construct and incubated for 5 h with 30 pM GM, with or without catalase (*, P < 0.005, one-tailed t test). (C) Luciferase activity of 293T cells transfected with α/βGMR, p8xGAS-luc, and either Prdx5 or pDisplay vector (Ct) incubated for 5 h with 30 pM GM (*, P < 0.0001, one-tailed t test). (Right) Protein expression was detected by immunoblotting. (D) GM-induced p44/42-MAPK (MAPK) phosphorylation in U937 and MO7e cells incubated with catalase. Phosphorylated (p-MAPK) and unphosphorylated MAPK were detected by immunoblotting. (E) Phosphorylation of MAPK and Jak2 in U937 cells treated with or without 500 μM H2O2, as detected by immunoblotting. (F) 293T cells expressing α/βGMR were incubated with 1 nM GM for 15 min, with or without catalase. After βGMR immunoprecipitation, p-βGMR was detected by using anti-phosphotyrosine antibody. Untransfected 293T cells were used as a control. (G) βGMR-associated proteins were immunoprecipitated from 293T cells expressing α/βGMR and in vitro kinase reactions performed with γ-[32P]ATP, 1 nM GM, and catalase (3 × 103units/ml) (Left) or Jak2 inhibitor AG490 (Right). p-βGMR was detected by autoradiography (Upper) and unphosphorylated βGMR was detected by immunoblotting (Lower). (H) In vitro kinase reactions were performed with βGMR-immunoprecipitated proteins incubated with γ-[32P]ATP, 50 μMH2O2, and catalase (3 × 103 units/ml). Proteins were detected as in G.
H2O2 Is Involved in GM-CSF-Induced Early Signaling Events. We reasoned that if H2O2 facilitates GM-CSF signaling after receptor–ligand interaction, then H2O2 should activate downstream signaling in the absence of ligand. We found that H2O2 was sufficient to induce phosphorylation of Jak2 and p44/42-MAPK in U937 cells (Fig. 4E). Additionally, in 293T cells expressing α/βGMR, GM-CSF induced phosphorylation of βGMR that was diminished by preincubation with catalase (Fig. 4F). Catalase alone appeared to increase phosphorylation of βGMR.
To address the role of H2O2 in the activation of kinases involved in the phosphorylation of βGMR, we developed a cell-free kinase assay based on βGMR-associated proteins. Using this in vitro system, GM-CSF induced βGMR phosphorylation that was inhibited by catalase and a Jak2 inhibitor, AG490 (Fig. 4G). The addition of H2O2 also resulted in βGMR phosphorylation (Fig. 4H).
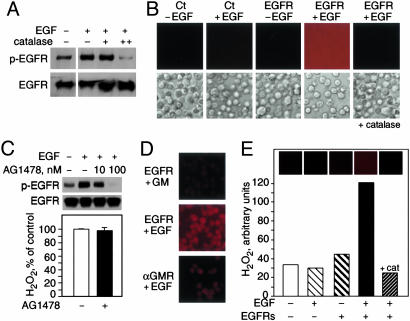
EGF–EGFR Interaction Generates H2O2 Independent of Intracellular Signaling. We investigated whether H2O2 consequent to receptor–ligand interaction could facilitate signaling mediated by receptors with intrinsic kinase domains. We used the EGFR as a model because it has been suggested that EGF-induced signaling involves the generation of H2O2 (19). Extracellular catalase inhibited EGF-induced phosphorylation of EGFR in A431 cells (Fig. 5A). These results support a role for H2O2 in early EGFR-mediated signaling events. We found that in 293T cells transiently transfected with EGFR, the addition of EGF resulted in extracellular H2O2 production (Fig. 5B). To assess whether receptor–ligand interaction could generate H2O2 in the absence of EGFR kinase function, 293T cells expressing EGFR were preincubated with a tyrosine kinase inhibitor, AG1478. At a concentration of AG1478 that completely inhibits EGFR phosphorylation by EGF, we found that AG1478 did not prevent EGFR–ligand-dependent H2O2 production (Fig. 5C). Additionally, we investigated the production of H2O2 in fixed 293T cells expressing EGFR or αGMR. We found that cognate ligand, but not GM-CSF, induced an increase in H2O2, indicating a requirement for receptor–ligand specificity (Fig. 5D). Membrane-based fluorescence assay with purified EGF and soluble EGFRs showed that receptor–ligand interaction using isolated proteins was sufficient to produce H2O2 (Fig. 5E).
Fig. 5.
EGFR–ligand interaction generates extracellular H2O2 in cells and with purified proteins. (A) EGF-induced phosphorylation of EGFR in A431 cells treated with catalase. Phosphorylated EGFR (p-EGFR) (Upper) and unphosphorylated EGFR (Lower) were detected by immunoblotting. (B) Fluorescence microscopy detection of extracellular H2O2 in 293T cells untransfected (Ct) or transfected with EGFR and incubated in Amplex Red for 30 min with or without 100 nM EGF. Corresponding light microscopy images are provided in Lower. (C) EGF-induced phosphorylation of EGFR-transfected 293T cells treated with AG1478. (Upper) Proteins were detected as in A.(Lower) EGFR–ligand-dependent H2O2 generation in 293T cells treated with or without AG1478. (D) Extracellular H2O2 generation by fixed 293T cells transfected with EGFR or αGMR and treated with EGF or GM, as detected by fluorescence microscopy. (E) Purified human soluble EGFR was incubated in Amplex Red with or without EGF and H2O2 detected by fluorescence microscopy. cat, catalase.
We propose that H2O2 generated extracellularly by receptor–ligand interaction can diffuse locally into the cell, where it may participate in the inactivation of phosphatases and/or the activation of kinases involved in signaling (Fig. 6).
Fig. 6.
Proposed role for H2O2 generated extracellularly by receptor–ligand interaction. Schematic representations for cytokine receptors (extrinsic kinase) and intrinsic kinase receptors are shown. Ligand binding to the receptor generates extracellular H2O2 that diffuses across the cell membrane to facilitate signaling.
Discussion
Whereas previous research has described H2O2 as an intracellular signaling molecule (4, 5, 28), we found H2O2 to be generated extracellularly by receptor–ligand interaction. We propose that H2O2 produced extracellularly at the receptor–ligand interface can permeate into the cell and facilitate activation of initiating kinases or inactivation of downstream phosphatases. H2O2 generation depended on ligand binding; only cognate receptor–ligand pairs resulted in substantial H2O2 production. Crossreacting pairs, such as GH and the ProL receptor, also produced an increase in H2O2 levels. Furthermore, an αGMR mutant unable to bind ligand abrogated H2O2 production, indicating a requirement for functional receptor–ligand binding. Our results with mutant receptor, kinase inhibitors, fixed cells, and purified proteins demonstrate that receptor–ligand binding, in the absence of cellular signaling, is sufficient to generate H2O2. Although this work does not illuminate the mechanism by which receptor–ligand interaction generates H2O2, it has been reported that some proteins and antibodies are capable of producing H2O2 (22, 24).
The molecular mechanism by which receptors lacking intrinsic kinase domains initiate signaling is not known. It is believed that ligand-induced receptor dimerization or oligomerization leads to activation of an “acquired” cytoplasmic tyrosine kinase, such as Jak2 in the case of the GMR (13, 29). The mechanism of Jak2 activation is not known in detail, although proximity-facilitated transphosphorylation is likely important. Our concept is that H2O2 generated by the ligand–receptor interaction facilitates activation of Jak2 or other initiating kinases. Because H2O2 is a small, diffusible molecule that can be generated and destroyed rapidly in response to cellular requirements, it has the properties of a signaling messenger (5). It is known that ROS can activate the Jak-STAT pathway (30, 31). We previously found that vitamin C prevented GM-CSF-dependent Jak2 activation and consequent βGMR phosphorylation, thus suggesting that a quenchable free radical may also be involved in early signaling events (7).
In the case of receptors with intrinsic kinases such as EGFR, it is uncertain that proximity alone is sufficient for EGFR kinase transphosphorylation. The EGFR kinase is known to be activated by H2O2 (19, 32, 33), and our data indicate that H2O2 is generated by the EGF–EGFR interaction and that catalase inhibits EGF-dependent phosphorylation of EGFR. Consistent with our data, a recent report indicated that EGF-induced phosphorylation of EGFR was not dependent on mitochondrial respiration (34). Our concept is that receptor oligomerization and H2O2 may act together to initiate receptor-mediated signaling in the hematopoietin superfamily, as well as in receptors with intrinsic kinases.
H2O2 can affect signaling through its ability to reversibly oxidize catalytic site cysteine residues on protein tyrosine phosphatases (PTPs), such as PTEN, that result in PTP inactivation (5, 35, 36). By inactivating phosphatases, H2O2 could increase the phosphorylation status of protein tyrosine kinases, resulting in increased signal transduction. H2O2 signaling can be modulated by peroxiredoxin enzymes that act as “floodgates” to maintain low resting levels of H2O2 while permitting higher levels during signal transduction (37). Both normal and malignant cells produce H2O2 (23), and H2O2 production could result as a consequence of signaling. Our in vitro data and experiments using nonsignaling mutant receptors indicate a separable source of H2O2 due to receptor–ligand interaction. The potential flux of H2O2 across the cell membrane may be part of an as yet unknown H2O2 cycling pathway.
It has been suggested that engagement of the B cell antigen receptor by antigen results in an increase in H2O2 production that could locally inhibit protein tyrosine phosphatases (38), which is in agreement with our proposed model (Fig. 6). Additionally, T cell receptor stimulation induces rapid H2O2 production independent of NADPH oxidase (39), which is consistent with our findings in α/βGMR-deletion constructs and in fixed cells lacking the capacity to signal. Our findings are also consistent with results from receptor chimera experiments in which ligand binding to the extracellular domain of the chimera provided an “on/off” signal, whereas the intracellular domain provided signaling instruction (40). In our model, the generation of H2O2 extracellularly and receptor oligomerization could act in concert to provide the on/off signal. We propose that H2O2 generated by receptor–ligand interaction diffuses across the plasma membrane, where it may locally activate kinases or inactivate phosphatases, thereby facilitating receptor-mediated signal transduction.
Acknowledgments
Dr. David W. Golde was our mentor and the inspiration behind this work. We thank Mr. Jeffrey R. Gardner for assistance with the MO7e proliferation assay and Dr. Jian Chen for the αGMR and βGMR truncation mutants. This work was supported by grants from the National Institutes of Health (CA30388), the New York State Department of Health, and the Lebensfeld and Schultz Foundations.
Author contributions: J.M.C. and D.W.G. designed research; G.J.D., O.B.-O., and C.C.S. performed research; G.J.D., O.B.-O., and C.C.S. contributed new reagents/analytic tools; G.J.D., J.M.C., and D.W.G. analyzed data; and G.J.D., J.M.C., and D.W.G. wrote the paper.
Abbreviations: ROS, reactive oxygen species; GM-CSF, granulocyte–macrophage colony-stimulating factor; GMR, GM-CSF receptor; EGFR, EGF receptor; Jak2, Janus family tyrosine kinase 2; STAT, signal transducers and activators of transcription; MAPK, mitogen-activated protein kinase; GH, human growth hormone; ProL, prolactin; Prdx5, peroxiredoxin-5; αGMRx, extracellular domain of αGMR.
References
- 1.Darnell, J. E., Jr. (1997) Science 277, 1630–1635. [DOI] [PubMed] [Google Scholar]
- 2.Brivanlou, A. H. & Darnell, J. E., Jr. (2002) Science 295, 813–818. [DOI] [PubMed] [Google Scholar]
- 3.Leaman, D. W., Leung, S., Li, X. & Stark, G. R. (1996) FASEB J. 10, 1578–1588. [PubMed] [Google Scholar]
- 4.Nathan, C. (2003) J. Clin. Invest. 111, 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhee, S. G., Chang, T. S., Bae, Y. S., Lee, S. R. & Kang, S. W. (2003) J. Am. Soc. Nephrol. 14, S211–S215. [DOI] [PubMed] [Google Scholar]
- 6.Sattler, M., Winkler, T., Verma, S., Byrne, C. H., Shrikhande, G., Salgia, R. & Griffin, J. D. (1999) Blood 93, 2928–2935. [PubMed] [Google Scholar]
- 7.Carcamo, J. M., Borquez-Ojeda, O. & Golde, D. W. (2002) Blood 99, 3205–3212. [DOI] [PubMed] [Google Scholar]
- 8.Gasson, J. C. (1991) Blood 77, 1131–1145. [PubMed] [Google Scholar]
- 9.Gearing, D. P., King, J. A., Gough, N. M. & Nicola, N. A. (1989) EMBO J. 8, 3667–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashida, K., Kitamura, T., Gorman, D. M., Arai, K., Yokota, T. & Miyajima, A. (1990) Proc. Natl. Acad. Sci. USA 87, 9655–9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasson, J. C., Kaufman, S. E., Weisbart, R. H., Tomonaga, M. & Golde, D. W. (1986) Proc. Natl. Acad. Sci. USA 83, 669–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitamura, T., Sato, N., Arai, K. & Miyajima, A. (1991) Cell 66, 1165–1174. [DOI] [PubMed] [Google Scholar]
- 13.Ihle, J. N., Witthuhn, B. A., Quelle, F. W., Yamamoto, K. & Silvennoinen, O. (1995) Annu. Rev. Immunol. 13, 369–398. [DOI] [PubMed] [Google Scholar]
- 14.Geijsen, N., Koenderman, L. & Coffer, P. J. (2001) Cytokine Growth Factor Rev. 12, 19–25. [DOI] [PubMed] [Google Scholar]
- 15.Zhao, Y., Wagner, F., Frank, S. J. & Kraft, A. S. (1995) J. Biol. Chem. 270, 13814–13818. [DOI] [PubMed] [Google Scholar]
- 16.Ihle, J. N. (2001) Curr. Opin. Cell Biol. 13, 211–217. [DOI] [PubMed] [Google Scholar]
- 17.Raines, M. A., Golde, D. W., Daeipour, M. & Nel, A. E. (1992) Blood 79, 3350–3354. [PubMed] [Google Scholar]
- 18.Suzuki, K., Hino, M., Hato, F., Tatsumi, N. & Kitagawa, S. (1999) Blood 93, 341–349. [PubMed] [Google Scholar]
- 19.Bae, Y. S., Kang, S. W., Seo, M. S., Baines, I. C., Tekle, E., Chock, P. B. & Rhee, S. G. (1997) J. Biol. Chem. 272, 217–221. [PubMed] [Google Scholar]
- 20.Bae, Y. S., Sung, J. Y., Kim, O. S., Kim, Y. J., Hur, K. C., Kazlauskas, A. & Rhee, S. G. (2000) J. Biol. Chem. 275, 10527–10531. [DOI] [PubMed] [Google Scholar]
- 21.Dhar-Mascareno, M., Chen, J., Zhang, R. H., Carcamo, J. M. & Golde, D. W. (2003) J. Biol. Chem. 278, 11107–11114. [DOI] [PubMed] [Google Scholar]
- 22.Wentworth, A. D., Jones, L. H., Wentworth, P., Jr., Janda, K. D. & Lerner, R. A. (2000) Proc. Natl. Acad. Sci. USA 97, 10930–10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szatrowski, T. P. & Nathan, C. F. (1991) Cancer Res. 51, 794–798. [PubMed] [Google Scholar]
- 24.Wentworth, P., Jr., Jones, L. H., Wentworth, A. D., Zhu, X., Larsen, N. A., Wilson, I. A., Xu, X., Goddard, W. A., III, Janda, K. D., Eschenmoser, A. & Lerner, R. A. (2001) Science 293, 1806–1811. [DOI] [PubMed] [Google Scholar]
- 25.Du, J., Alsayed, Y. M., Xin, F., Ackerman, S. J. & Platanias, L. C. (2000) J. Biol. Chem. 275, 33167–33175. [DOI] [PubMed] [Google Scholar]
- 26.Ronco, L. V., Silverman, S. L., Wong, S. G., Slamon, D. J., Park, L. S. & Gasson, J. C. (1994) J. Biol. Chem. 269, 277–283. [PubMed] [Google Scholar]
- 27.Knoops, B., Clippe, A., Bogard, C., Arsalane, K., Wattiez, R., Hermans, C., Duconseille, E., Falmagne, P. & Bernard, A. (1999) J. Biol. Chem. 274, 30451–30458. [DOI] [PubMed] [Google Scholar]
- 28.Reth, M. (2002) Nat. Immunol. 3, 1129–1134. [DOI] [PubMed] [Google Scholar]
- 29.Leonard, W. J. & O'Shea, J. J. (1998) Annu. Rev. Immunol. 16, 293–322. [DOI] [PubMed] [Google Scholar]
- 30.Al-Shami, A., Mahanna, W. & Naccache, P. H. (1998) J. Biol. Chem. 273, 1058–1063. [DOI] [PubMed] [Google Scholar]
- 31.Simon, A. R., Rai, U., Fanburg, B. L. & Cochran, B. H. (1998) Am. J. Physiol. 275, C1640–C1652. [DOI] [PubMed] [Google Scholar]
- 32.Kamata, H., Shibukawa, Y., Oka, S. I. & Hirata, H. (2000) Eur. J. Biochem. 267, 1933–1944. [DOI] [PubMed] [Google Scholar]
- 33.Peus, D., Beyerle, A., Vasa, M., Pott, M., Meves, A. & Pittelkow, M. R. (2004) Exp. Dermatol. 13, 78–85. [DOI] [PubMed] [Google Scholar]
- 34.Chen, K., Thomas, S. R., Albano, A., Murphy, M. P. & Keaney, J. F., Jr. (2004) J. Biol. Chem. 279, 35079–35086. [DOI] [PubMed] [Google Scholar]
- 35.Meng, T. C., Fukada, T. & Tonks, N. K. (2002) Mol. Cell 9, 387–399. [DOI] [PubMed] [Google Scholar]
- 36.Lee, S. R., Yang, K. S., Kwon, J., Lee, C., Jeong, W. & Rhee, S. G. (2002) J. Biol. Chem. 277, 20336–20342. [DOI] [PubMed] [Google Scholar]
- 37.Wood, Z. A., Poole, L. B. & Karplus, P. A. (2003) Science 300, 650–653. [DOI] [PubMed] [Google Scholar]
- 38.Rolli, V., Gallwitz, M., Wossning, T., Flemming, A., Schamel, W. W., Zurn, C. & Reth, M. (2002) Mol. Cell 10, 1057–1069. [DOI] [PubMed] [Google Scholar]
- 39.Jackson, S. H., Devadas, S., Kwon, J., Pinto, L. A. & Williams, M. S. (2004) Nat. Immunol. 5, 818–827. [DOI] [PubMed] [Google Scholar]
- 40.Socolovsky, M., Dusanter-Fourt, I. & Lodish, H. F. (1997) J. Biol. Chem. 272, 14009–14012. [DOI] [PubMed] [Google Scholar]