Abstract
We recently discovered it was possible to physically magnify preserved biological specimens by embedding them in a densely crosslinked polyelectrolyte gel, anchoring key labels or biomolecules to the gel, mechanically homogenizing the specimen, and then swelling the gel-specimen composite by ~4.5x in linear dimension, a process we call expansion microscopy (ExM). Here we describe iterative expansion microscopy (iExM), in which a sample is expanded, then a second swellable polymer mesh is formed in the space newly opened up by the first expansion, and finally the sample is expanded again. iExM expands biological specimens ~4.5 × 4.5 or ~20x, and enables ~25 nm resolution imaging of cells and tissues on conventional microscopes. We used iExM to visualize synaptic proteins, as well as the detailed architecture of dendritic spines, in mouse brain circuitry.
INTRODUCTION
We recently discovered that preserved biological specimens, embedded in a swellable polymer gel with key biomolecules or labels anchored to the gel, and then mechanically homogenized, could be isotropically swelled ~4.5x in linear dimension by immersion in water – a process we call expansion microscopy (ExM)1. Since our original paper on ExM, we have developed variants that anchor proteins or RNA directly to the gel, enabling ease of use in a diversity of scientific and clinical contexts2,3. However, all ExM variants published to date expand biological specimens by ~4.5x in linear dimension, resulting in an effective resolution for a ~300 nm diffraction limited objective lens of ~60–70 nm (~300/4.5), raising the question of whether it might be possible to expand biological specimens by greater expansion factors, resulting in better resolution.
In our original ExM protocol1, biological molecules of interest were first labeled with a primary antibody, followed by a secondary antibody bearing an oligonucleotide. Then, a second oligonucleotide bearing a gel-anchoring moiety (a 5′ acrydite group) and a fluorophore was applied and anchored to a swellable polyelectrolyte gel synthesized evenly throughout the specimen, using a crosslinker to insure an interconnected topology for the polymer strands to support isotropic expansion. After mechanical homogenization with strong protease treatment, the polymer-specimen composite could then be expanded in water1. Expanding a gel > 4.5-fold was possible1 but resulted in fragile gels. We here explored whether it would be possible to synthesize, post-expansion, a second gel that could expand the specimen further, that would provide sufficient mechanical support (Fig. 1a–e). For such an iterative ExM (iExM) protocol, we must transfer the information from the first gel to the second, then disrupt the first gel, and finally expand the second gel; to make the first gel disruptible, we used a chemically cleavable crosslinker for its synthesis.
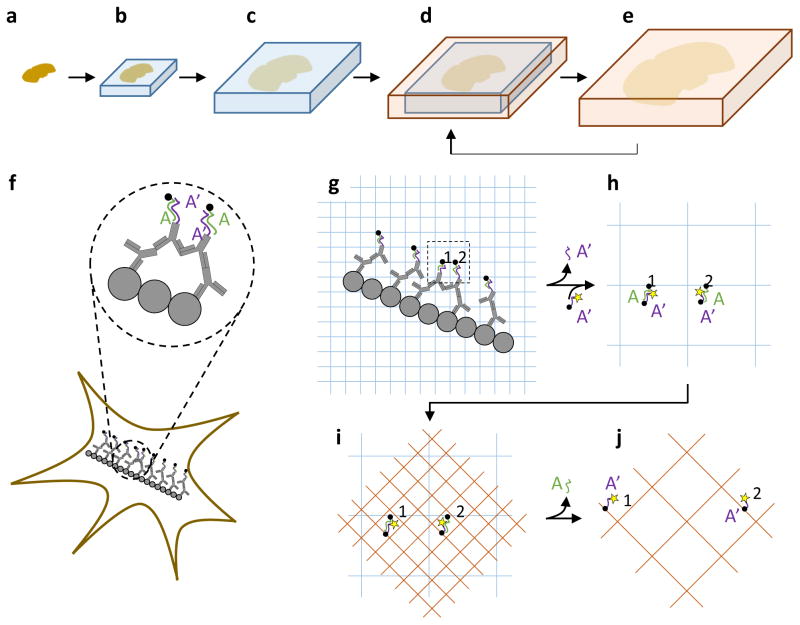
Figure 1. Iterative expansion microscopy (iExM) concept.
(a–e) Schematic of iterative expansion, showing how a brain slice can be expanded multiple times. First, a swellable polyelectrolyte gel network containing a cleavable crosslinker is formed throughout a brain slice (b), then mechanically homogenized and expanded (c), as in our original ExM protocol1. After expansion, a second swellable polyelectrolyte gel network is formed throughout the first (d), and then expanded after dissolving the first gel (e). This process (d,e) can be applied repeatedly to increase the physical magnification still further, if desired. (f–j), Molecular view of the iExM process. (f) Biomolecules of interest (gray circles) are first labeled with a primary antibody (shown also in gray) followed by a DNA (purple, sequence A′)-conjugated secondary antibody, then a complementary DNA (green, sequence A) bearing a gel-anchoring moiety (acrydite; black dot), as in our original ExM procedure1. (g) The sample is then embedded in a swellable polyelectrolyte gel (blue mesh), which critically involves a chemically cleavable crosslinker. This gel incorporates the DNA of sequence A at the gel-anchoring site, and is expanded. The sample is re-embedded in a charge-neutral backbone polymer (not shown, for simplicity) with a cleavable crosslinker to enable new electrolyte monomers to be infused, and to support DNA hybridization without shrinkage3. (h) In order to enable a second round of expansion, a DNA oligo with the original A′ sequence (purple strand), but now bearing a fluorophore (yellow star) and a new gel-anchoring moiety (acrydite; black dot), is hybridized to the anchored A-sequence DNA (green). (i) A second swellable gel (orange mesh) is formed, this time with an uncleavable crosslinker. This gel incorporates the final fluorophore-bearing DNA oligo (sequence A′, purple). (j) The gel expands the labels away from each other after digesting the first and re-embedding gel through crosslinker cleavage.
RESULTS
Design of iExM chemistry
We implemented iExM by first taking a sample and expanding it using ExM as in our original paper1, but with the second oligonucleotide applied (green in Fig. 1f) without a fluorophore, and using a cleavable crosslinker (e.g., the commercially available crosslinker N,N′-(1,2-dihydroxyethylene) bisacrylamide (DHEBA), whose diol bond can be cleaved at high pH4) for gel synthesis (Fig. 1g). We then embedded the expanded sample in an uncharged polyacrylamide gel prepared with a cleavable crosslinker (the re-embedding gel3) so that it could be held in the expanded state during subsequent steps. In particular, this re-embedding gel allowed us to apply a third oligonucleotide (Fig. 1h), bearing a gel-anchoring moiety and fluorophore, which hybridized to the oligonucleotide anchored to the first polymer. We then formed a second polyacrylate gel, made with a conventional crosslinker (e.g., N,N′-methylenebis(acrylamide) (BIS)), which incorporated the third oligonucleotide (and thus the fluorophore, Fig. 1i), and then we dissolved the original gels by cleaving their crosslinkers before expanding the fluorophores away from each other through immersion in water (Fig. 1j). We found that iExM would typically result in expansion ratios of ~4.5–5.5x in the first round, and ~4x in the second round, for a total increase of ~16x–22x (see Supplementary Note 1 for details). In addition to this implementation of iExM, we also explored a second strategy, namely using the re-embedding gel as the final gel, and hydrolyzing the side groups at high pH into carboxyl groups5 (dissolving the first gel’s cross-linkers simultaneously), a process we call “high pH” iterative expansion microscopy (hp-iExM). hp-iExM resulted in expansion ratios slightly smaller than that of iExM (Supplementary Note 1), so in the main text we focus on iExM (but will mention data from hp-iExM in the supplementary information throughout).
Validation of iExM resolution and distortion
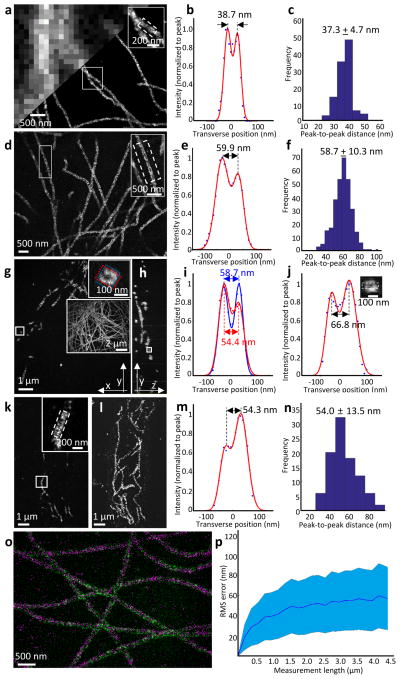
To validate iExM, we imaged the configuration of biomolecules of known organization, analyzing both the resolution obtained, as well as distortion over various length scales. We analyzed microtubules, hollow tubes with an outer diameter of ~25 nm as determined by transmission electron microscopy (TEM)6, due to their small size and stereotyped appearance in BS-C-1 cells (Fig. 2a, upper left). When imaged with stochastic optical reconstruction microscopy (STORM), hollow microtubule structures were clearly resolved (Fig. 2a, lower right; Fig. 2b). When the cross-section was fit with a sum of Gaussians, the peak-to-peak distance between the sidewalls was 37.3 ± 4.7 nm (mean ± standard deviation throughout; Fig. 2c), similar to previous super-resolution microscopy data7,8. When these cells were expanded via iExM (Fig. 2d,e; ~20x physical magnification), such hollow structures could be resolved with confocal microscopy (or widefield microscopy; Supplementary Fig. 1a), which was not possible with earlier ~4.5x expansion factor forms of ExM1. The average distance between the sidewall peaks, for iExM-expanded samples, was 58.7 ± 10.3 nm (Fig. 2f; see Supplementary Fig. 1b–d for hp-iExM processed cells). In 3-D confocal z-stacks of such cells (Fig. 2g for a single xy-plane image; Fig. 2h for a single yz-plane image reconstructed from the z-stack image shown in Fig. 2g), tubular cross-sections of microtubules could be easily seen and characterized (Fig. 2i).
Figure 2. Validation of the nanoscale precision of iterative expansion microscopy.
(a-c) STORM imaging of cultured BS-C-1 cells after microtubules were labeled with an anti-tubulin antibody. (a) Epifluorescence image (upper left) and STORM image (lower right) of microtubules before expansion. The inset in upper right zooms in on the small box at center. (b) Transverse profile of microtubules in the boxed region (dotted lines) of the inset of a after averaging down the long axis of the box and then normalizing to the peak value (blue dots), with superimposed fit with a sum of two Gaussians (red lines). (c) Population data for 110 microtubule segments from two samples (mean ± standard deviation), showing a histogram of peak-to-peak distances. (d–j) Confocal imaging of cultured BS-C-1 cells with labeled microtubules, after ~20-fold expansion via iExM. (d) Single xy-plane image at the bottom of the cell. The inset in upper right zooms in on the small box at left. (e) As in b, but for the inset of d. (f) As in c, but for iExM-processed BS-C-1 cells. n=307 microtubule segments from one expanded sample. (g) Single xy-plane image 1.6 μm above the bottom of the cell. The inset in upper right zooms in on the small box indicated at left, highlighting the circular cross-section of the microtubule (blue and red boxes are used to calculate the profile of i). The large inset at right shows the entire cellular context, as a maximum intensity projection of the sample. (h) Single yz-plane within the volume imaged in g; the small box is highlighted in the inset of j. (i) Transverse profiles (i.e., plotting along the long axis of the highlighting box) of the microtubule in the upper right inset of g, with color corresponding to that of the highlighting box in the inset. (j) Transverse profile of the microtubule in the small box of h. Inset, zoomed-in image of the box of h, showing the cross-section of the microtubule being resolved along the optical axis. (k) Confocal image of a 100-μm thick slice of mouse cortex with microtubules labeled, after ~18-fold expansion via iExM, and imaged at a single xy-plane. (l) Maximum-intensity projection of the sample shown in k. (m) As in e, but for the inset of k. (n) Population data for 96 microtubule segments from one expanded sample, showing a histogram of the peak-to-peak distances. (o) Overlay, using only a rigid registration, of a STORM image (magenta) of cultured BS-C-1 cells stained with anti-tubulin pre-expansion, with a confocal image (green) of the same sample post-expansion. (p) RMS length measurement error of biological measurements, calculated using the distortion vector field method9, using STORM microscopy pre-expansion followed by confocal imaging of iExM-processed samples (~20x expanded) (blue line, mean; shaded area, ± 1 standard deviation; n = 3 samples).
To understand the peak-to-peak distances measured by iExM vs. STORM, we took into account the size of the probes used to stain the microtubules in each case. We simulated iExM images of microtubules labeled with DNA-conjugated secondary antibodies (description of simulation in Supplementary Figure 2 and Supplementary Note 2; simulator code contained in Supplementary Software 1). Using this software, we calculated the inner and outer radii of a cylinder that would contain the ends of DNA oligos borne by secondary antibodies, which corresponds to a specific actual dataset (Supplementary Fig. 2; see Supplementary Fig. 3 for a sketch of how a typical microtubule equipped with antibodies and DNA might appear). The DNA-equipped antibodies of iExM, we calculated, may shift the appearance of target proteins up to ~4.6 nm relative to the position that would be obtained via classical super-resolution microscopy using antibodies lacking DNA (modeled in Supplementary Fig. 4; see also Supplementary Note 3). Such positional errors could be reduced in the future by using different DNA-antibody conjugation strategies (schematized in Supplementary Fig. 5; see also Supplementary Note 4).
Using these models, we quantitatively estimated the resolution of the overall iExM process. First, we measured the full width at half maximum (FWHM) of single microtubule sidewalls, deriving a value of 25.8 ± 7.7 nm for the point spread function (PSF) of the overall iExM process, from staining to gelation and expansion to optical imaging (Supplementary Fig. 6a). To attempt to estimate the resolution of the iExM process independent of the label (e.g., focusing on the optical, gelation and expansion components), we deconvolved actual images of microtubule sidewalls by a simulated structure of an idealized DNA/antibody-labeled microtubule sidewall (generated according to the model of Supplementary Fig. 3), resulting in the slightly smaller value of 22.3 ± 5.3 nm (Supplementary Fig. 6b). Finally, we attempted to isolate just the amount of PSF broadening due to the gelation and expansion steps specifically. We simulated (Supplementary Fig. 6a) how microtubules would be expected to look, after staining and optical imaging, assuming that gelation and expansion induced zero error. The resultant PSF was ~6 nm smaller than the actual PSF obtained for the entire iExM process, suggesting that the processes of forming and expanding the multiple gels involved with iExM introduced ~6 nm of additional resolution error, beyond the effects of the antibodies, DNA, and optics (see Supplementary Note 5). Such a PSF broadening does not greatly alter the mean peak-to-peak distance between target proteins arranged in a complex (Supplementary Fig. 6c), but instead widens the appearance of small things via broadening the PSF of iExM.
In expansion microscopy, physical expansion occurs in axial as well as lateral directions, and thus magnifies specimens along the optical axis as well as in the focal plane1. When a yz-plane (Fig. 2h) was reconstructed from the z-stack image shown in Fig. 2g, the circular cross-section of a microtubule was resolvable (Fig. 2j, inset). The nanoscale axial resolution of iExM enabled clear visualization of microtubules of BS-C-1 cells in 3-D (Supplementary Videos 1 and 2).
We applied iExM to preserved mouse tissues, including brain, liver, and lung, to determine whether iExM could resolve ~20-nm biological structures in intact tissues. As shown in the inset of Fig. 2k (single z-plane image; see Fig. 2l for the entire cellular context), the sidewalls of microtubules in mouse brain slices were resolvable on a confocal microscope. The distance between the two peaks of the fitted Gaussians was similar to that obtained in the cultured cell case (Fig. 2m; population data in Fig. 2n). The sidewalls of microtubules in cells of mouse lung and liver tissue slices were also easily resolved on confocal microscopes (Supplementary Fig. 7a–h). In addition to the visualization of the sidewalls of microtubules in tissues, we found that individual components of microtubule bundles in the mouse cortex could be resolved after 18-fold expansion (Supplementary Fig. 7i–l).
In addition to resolution, the ability to tell finely spaced objects apart, another optical parameter of interest is distortion across more extended length scales. Accordingly, we quantified the distortion caused by iExM over various length scales that correspond to feature sizes of interest in cell biology. To measure distortion over scales of several microns, we compared pre-expansion images taken on a super-resolution microscope to post-expansion images taken on a conventional diffraction-limited microscope1,2,9. We prepared samples with secondary antibodies labeled with STORM dyes, and simultaneously applied DNA-conjugated secondary antibodies so that they could be processed for iExM and visualized post-expansion. We co-registered the pre-expansion STORM image and post-expansion confocal image via a rigid transformation (Fig. 2o), and then calculated the deformation vector field between the two images1,2,9. Although the image qualities enabled by STORM and iExM were compromised in this specific experiment because of the special requirements involved in imaging the same sample with both methods (e.g., each label will occur at half the antibody labeling density of a typical experiment, since we are dual labeling), the root-mean-square (RMS) alignment error between iExM and STORM was nonetheless small, about 2.5% of measurement length (Fig. 2p) over scales of several microns, similar to the 1%–4% range of alignment errors previously determined for ExM1,2,9. We estimated the distortion of iExM across length scales of tens to hundreds of nanometers by examining the variation of microtubule diameter along 400 nm distances down the long axis of the microtubule. The estimated distortion was found to be 9 nm for cells and 13 nm for tissues (see Supplementary Note 6 for details).
Nanoscale imaging of synapses
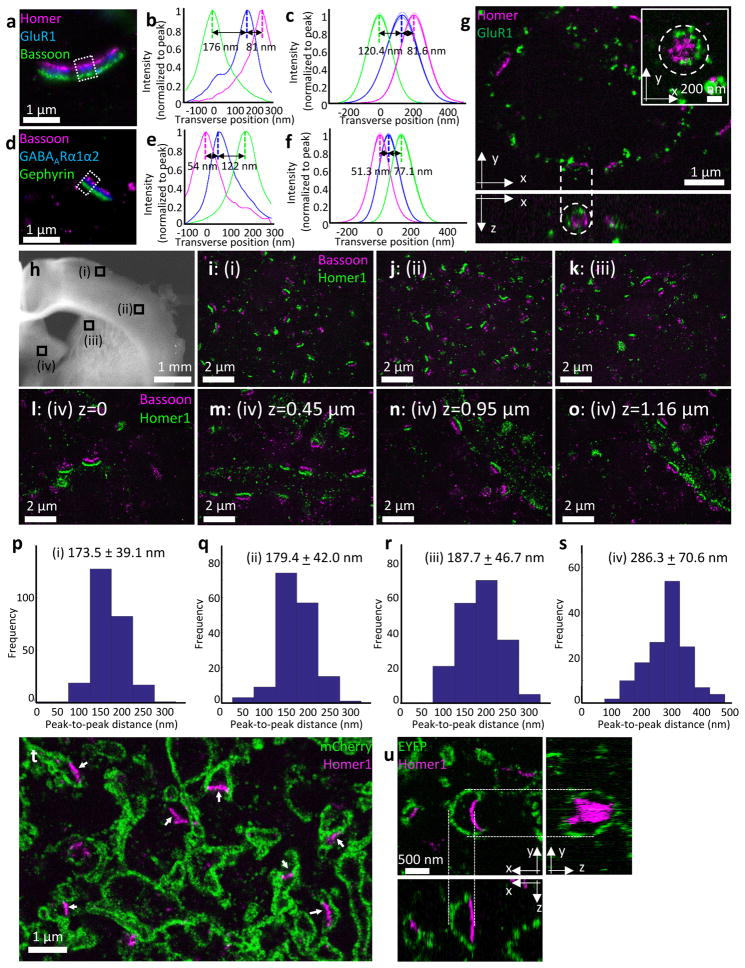
We next explored the utility of iExM in the context of resolving proteins within synapses. To improve brightness of expanded specimens, we pursued two signal amplification methods, using either DNA or locked nucleic acid (LNA) probes to increase the number of fluorophores associated with a single gel-anchored oligo (see Supplementary Fig. 8 for schematic; see also Supplementary Note 7). We first examined synapses of cultured mouse hippocampal neurons. We labeled synapses with sets of antibodies that indicate putative excitatory (Fig. 3a–c) or inhibitory (Fig. 3d–f) synapses – anti-Homer1, anti-Bassoon, and anti-Glutamate receptor 1 (GluR1) for the former, and anti-Gephyrin, anti-Bassoon, and anti-Gamma-aminobutyric acid receptor Aα1/anti-Gamma-aminobutyric acid receptor Aα2 (GABAARα1/α2; labeled with the same oligonucleotide strand) for the latter. It was possible not only to resolve the presynaptic scaffolding protein Bassoon from the post-synaptic scaffolding proteins Homer1 and Gephyrin, but also possible to resolve proteins within a synaptic compartment – resolving the neurotransmitter receptors GluR1 and GABAARα1/α2 from their respective post-synaptic scaffolding proteins as well (Fig. 3c, f). We could observe the geometric organization of proteins within synapses, for example seeing how GluR1 proteins sometimes formed ring structures around Homer1 proteins (Fig. 3g, dotted circle in the upper right inset), as has been previously reported using STORM10. The isotropic 3-D nature of iExM expansion allowed us to resolve structures organized along the optical axis of the microscope, for example resolving ring structures of GluR1 when the synaptic cleft was parallel to the microscope’s optical axis (Fig 3g, dotted circle in the bottom).
Figure 3. Nanoscale resolution imaging of synapses using iExM.
(a) Epifluorescence image of cultured hippocampal neurons stained with antibodies against Homer1 (magenta), Glutamate receptor 1 (GluR1, blue), and Bassoon (green), after ~13-fold expansion via iExM and DNA hybridization-based signal amplification. Boxed regions are analyzed further in b. (b) Transverse profile of the three proteins imaged in the sample of a (in the boxed region), after normalizing to the peak (Homer1 in magenta, GluR1 in blue, Bassoon in green). (c) Sum of Gaussian functions fitted to curves as in b, for 10 synapses from one sample, normalized to peak (thick lines, mean; thin lines, ± 1 standard deviation). (d) As in a, but stained with antibodies against Bassoon (magenta), GABAARα1/α2 (blue), and Gephyrin (green) (e) As in b, but for the boxed region in d (Bassoon in magenta, GABAARα1/α2 in blue, Gephyrin in green). (f) As in c, but for the labels of d; 14 synapses from one sample. (g) Confocal z-stack (top, a single xy-plane; bottom, a single xz-plane; dotted lines connect corresponding points in the two cross-sections) of cultured hippocampal neurons with labeled Homer1 (magenta) and Glutamate receptor 1 (GluR1, green), after ~20-fold expansion via iExM. Inset of upper panel shows a zoomed-in image of a synapse (from another field of view) showing the circular distribution of GluR1 around Homer1. (h) low magnification widefield image of a mouse brain slice (corresponding to slide 57 of the Allen Brain Reference Atlas, P56 mouse, coronal sections) showing four regions i–iv that were imaged after expansion in i–o: (i,ii) primary somatosensory cortex, (iii) dorsal striatum, (iv) medial pallidum. (i–k) Confocal images of three regions (i), (ii), (iii) highlighted in h after labeling with anti-Bassoon (magenta) and anti-Homer1 (green), and 16-fold expansion via iExM. (l–o) Single xy-plane imaged at iv in h, at different z-heights. (p–s) Population data of the Homer1-Bassoon separation (mean ± standard deviation) measured in the four regions shown in h. The number of Homer1-Bassoon pairs analyzed was p, 248 pairs from one specimen; q, 159 pairs from one specimen; r, 189 pairs from one specimen; s, 147 pairs from one specimen. (t,u) Confocal images of motor cortex areas (t, slide 57 of the Allen Brain Reference Atlas P56 mouse coronal sections; u, slide 47 of the same Atlas) after immunostaining and expansion. (t) Confocal image of the specimen after immunostaining with antibodies against Homer1 (magenta) and mCherry (green) and 16-fold expansion via iExM. (u) Z-stack confocal image of the specimen after immunostaining with antibodies against Homer1 (magenta) and EYFP (green) and 20-fold expansion via iExM. Upper left shows a single xy-plane image; right shows a single yz-plane image reconstructed from the z-stack image; bottom shows a single xz-plane image reconstructed from the z-stack image.
We demonstrated the utility of iExM to resolve synaptic structures in the mouse brain. We immunostained a mouse brain slice with antibodies against Bassoon and Homer1, expanded the brain slice 16-fold with iExM, and then imaged putative synapses within four different brain regions (overview in Fig. 3h; iExM images taken on a confocal microscope in Fig. 3i–o; see Supplementary Fig. 9 for additional images taken with epifluorescence microscopy). The average distances between Bassoon and Homer1 observed in two regions within primary somatosensory cortex (indicated with Roman numerals i and ii in Fig. 3h, and highlighted in Fig. 3i and 3j) were similar to each other (Fig. 3p,q) and to Bassoon-Homer distances observed in the dorsal striatum (indicated with Roman numerals iii in Fig. 3h, and highlighted in Fig. 3k; Fig. 3r for the population data). However, a fourth region - the medial pallidum (indicated with Roman numeral iv in Fig. 3h, and highlighted in Fig. l–o), exhibited distances between Bassoon and Homer1 that were 50% larger (Fig. 3s), suggestive of a different synaptic architecture; furthermore, although putative synapses were evenly distributed in cortex and striatum, in the pallidum, synapses were arranged in regularly spaced patterns as if they were tiling a cylindrical target (Fig. 3l–o; Supplementary Video 3). We used iExM to explore other regional heterogeneities in localization of pre- and post-synaptic proteins (Supplementary Fig. 10 and Supplementary Video 4). Thus, iExM may be useful for analyzing the varying nanoscale configurations of proteins across brain circuits and regions, because it can support large volume imaging with nanoscale precision.
We applied Brainbow adeno-associated viruses (AAVs)11, in which Cre-expressing neurons are virally transduced to express, in random combinations, subsets of four fluorescent proteins (TagBFP, mTFP, mCherry, and EYFP), each fused to a farnesylation tag for membrane targeting. When such mouse brain slices (focusing on the motor and somatosensory cortices) were immunostained with antibodies against Homer1, as well as against fluorescent proteins (mCherry for Fig. 3t; EYFP for Fig. 3u), we found that postsynaptic proteins and membrane outlines could be co-visualized (Fig. 3t, 3u). At synaptic contacts (as indicated by Homer1 staining) we observed less membrane-bound fluorophore (e.g., arrows of Fig. 3t, Fig. 3u), perhaps because the density of proteins at the synaptic cleft prevents the inward diffusion of membrane-anchored fluorophores. Thus iExM may be useful for mapping out how proteins are arranged in small, even nanoscale, compartments of neurons.
Nanoscale imaging of 3-D mouse brain circuitry
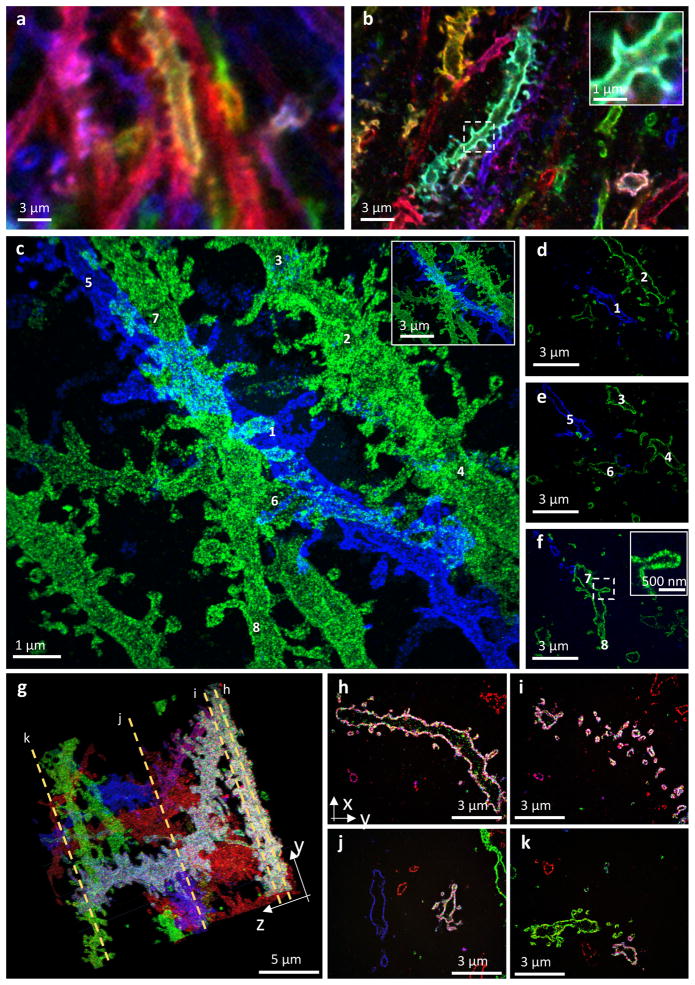
We prepared Brainbow AAV-labeled mouse brain samples as above, and performed iExM with LNA hybridization-based signal amplification. Brainbow-labeled dendritic spines in the molecular layer of the mouse hippocampal dentate gyrus are hard to resolve without expansion (Fig. 4a). In such samples processed with ~4.5x expansion factor protein-retention expansion microscopy (proExM, in which antibodies, genetically encoded fluorophores, or other proteins within a specimen are anchored to the swellable gel, and then expanded2), dendritic spines could be identified and even sometimes distinguished from one another, but not in all cases, and their shapes were difficult to analyze (Fig. 4b). After iExM, the number, size, position, and shapes of dendritic spines were easily visualized, as shown in the maximum intensity projection of Fig. 4c (see Supplementary Video 5 for 3-D visualization; note that as in Fig. 3t, 3u, membrane-anchored fluorophores are less dense or even absent at the tips of spines, consistent with membrane-anchored fluorophore exclusion by postsynaptic proteins as hypothesized above; see Supplementary Fig. 11). In particular, the hollow space within neurons (Fig. 4d–f) and spines was easily visualized when we used membrane-localized fluorescent proteins (Fig. 4f; for more examples, see Supplementary Fig. 12 and Supplementary Video 6). With iExM, it is possible to visualize structures such as spines along neural processes that extend over large 3-D volumes, for example along branching dendrites (shown in four sections of Fig. 4g in Fig. 4h–k; see also Supplementary Video 7; further examples in Supplementary Fig. 13 and Supplementary Videos 8, 9). Neuronal geometries could even to some extent be resolved with epifluorescence microscopy (mouse cortex, 16-fold expansion via hp-iExM; Supplementary Fig. 14). Thus, iExM can be used to explore neural connectivity in 3-D with spatial precision sufficient to resolve individual synaptic connections.
Figure 4. Nanoscale imaging of mouse hippocampal brain circuitry.
(a) Confocal image of immunostained Emx1-Cre mouse hippocampus with neurons expressing membrane-bound fluorescent proteins (Brainbow AAVs) before expansion. Blue: EYFP, Red: TagBFP, and Green: mTFP. (b) As in a, but expanded 4.5-fold by the antibody anchoring form of the ProExM protocol2. Blue: EYFP, Red: TagBFP, Green: mTFP. Inset shows a magnified image of a spine in the dotted box of b. (c–f) Confocal z-stack image of 20-fold expanded mouse hippocampal circuitry with labeled EYFP (blue) and mCherry (green). (c) Maximum intensity projection of the stack shown in (d–f); numbers refer to neural processes that are highlighted within individual z-stacks in (d–f). Inset shows a de–magnified view of the image of (c), with the same scale bar with a and b. (d–f) Single xy-plane images at different z-heights from the bottom of the specimen. (d) z=1.9 μm; (e) z=2.4 μm; (f) z=3.2 μm. See Supplementary Video 5 for 3-D video and surface rendering. Inset of f shows a magnified view of a spine in the dotted box of f. (g–k) Confocal z-stack image of 20-fold expanded mouse hippocampal circuitry with labeled EYFP and mTFP (blue; both EYFP and mTFP were labeled in a same color), mCherry (green), and tagBFP (red). (g) Maximum intensity projection of the stack; dotted orange lines highlight four z-planes which yielded the images of h–k. (h–k) Single z-plane images of the stack of (g). See Supplementary Video 7 for 3-D video.
Can iExM be applied beyond two rounds? In principle we could perform the second round of expansion so that a third round would be possible, by using a crosslinker whose cleaving chemistry is orthogonal to that of the first crosslinker. We found that it was possible to magnify a sample by 4.6 × 3.2 × 3.6 ~53-fold (see Supplementary Note 8 for details; see Supplementary Fig. 15 for 53-fold expanded BS-C-1 cells after antibody labeling tubulin). Although this might seem to imply an effective resolution of 300 nm/53 = 5.7 nm, the actual resolution is limited by the size of antibodies, the use of DNA anchors (additional ~4.6-nm positional errors, as estimated above), and the broadening of PSF by the gelation and expansion process (additional ~6 nm errors, as estimated above). However, with nanobody-based12 or small molecule tags13 compatible with iExM, or if direct protein anchoring2,9 versions of iExM are developed, iterated expansion strategies may be able to further improve the resolution beyond 25 nm.
DISCUSSION
iExM achieves resolutions comparable to those of the highest-performing forms of super-resolution light microscopy. Although expanded samples prepared with iExM can be quite large, they are transparent and homogeneous in refractive index (since they are 99.99% polymer and water, and less than 0.01% original biomaterial), analogous to previous ExM versions1–3,9, and thus may be amenable to fast, large volume imaging modalities compatible with transparent tissues, such as light-sheet microscopy14. Indeed, light-sheet imaging of ExM-processed tissues has recently been shown to be feasible3. With objective lenses of working distance ~8 mm available (e.g., Olympus 25x 0.9NA15), ~400 micron thick slices could be expanded by ~20x and imaged without further sectioning. iExM-processed samples are stiff enough to support post-expansion sectioning (e.g., with a vibratome); any sectioning error is effectively divided by the expansion factor in terms of impact on the biological information, and thus iExM could in principle help support the mapping of neural circuitry over large volumes, e.g. entire neural circuits or even entire brains. The volumetric dilution of iExM results in a lower density of biomolecules and labels, but the additional room created by expansion can support amplification chemistries such as those used here, or other variants of hybridization-based fluorescence amplification such as the hybridization chain reaction (HCR)16. In fact, we recently utilized HCR in the context of expanded brain tissues to visualize single RNAs within synaptic compartments of neurons in intact mouse brain circuits, taking advantage of the room made by expansion to append on the order of perhaps several dozen fluorophores to a single RNA strand3.
iExM is a strategy, not a single chemistry, and thus could be applied to other fundamental ExM chemistries, e.g. cleavable monomers that could support iterative removal of previous gels, as well as alternative polymer systems17. Since iExM concludes with nucleic acid strands, whose sequences code for protein identity, anchored throughout a polymer network at locations determined by the original protein locations, iExM may be able to support multiplexed in situ proteomics through serial hybridization of fluorescent strands as done in DNA-PAINT18. We recently demonstrated serial hybridization readout of multiple RNAs using our ExFISH variant of ExM3. Because iExM decrowds protein labels to the point of appearing punctate, coded hybridization strategies where the same strand is imaged many times with different sets of probes may allow an exponential number of proteins to be probed given a linear number of hybridization rounds – as has been previously done for RNA19,20. Finally, the additional room around biomolecules created by expansion could enable potentially complex reactions, including sequencing21, to be conducted on expanded tissues, furthering the ability to read out the molecular composition of complex biological systems in a multiplexed, yet scalable, way.
ONLINE METHODS
A Step-by-step protocol of this method can be found in Protocol Exchange22 and the Supplementary Protocol document. A table of all chemicals can be found in Supplementary Table 1.
DNA, LNA, primary antibody, and secondary antibody preparation
Oligonucleotides were purchased from Integrated DNA technologies (IDT) with standard desalting purification (see Supplementary Tables 2–8 for the sequences). Locked nucleic acids (LNAs) were purchased from Exiqon with high performance liquid chromatography (HPLC) purification (see Supplementary Table 9 for the sequences). Primary and secondary antibodies were purchased from multiple vendors (see Supplementary Tables 10–12). Oligonucleotides with a 5′ amine modification (see Supplementary Table 2 for the sequences) were conjugated to secondary antibodies using a modified protocol from a commercial kit (Solulink, Antibody-Oligonucleotide All-in-One conjugation kit; please visit http://expansionmicroscopy.org/ to find step-by-step instructions for the DNA-antibody conjugation).
Cultured BS-C-1 cell preparation
BS-C-1 cells (American Type Culture Collection, product number CCL-26) were cultured in Nunc Lab-Tek II chambered coverglasses (ThermoFisher, 155409) with Eagle’s Minimum Essential Medium supplemented with 10% heat inactivated fetal bovine serum and 1% penicillin-streptomycin, incubated at 37°C in 5% CO2.
Cultured hippocampal neuron preparation
Hippocampal neurons were prepared from postnatal day 0 or day 1 Swiss Webster (Taconic) mice as previously described23,24, but with the following modifications. Hippocampal tissues were isolated and digested with 50 units of papain for 6–8 minutes, and then the digestion was stopped with ovomucoid trypsin inhibitor. 10,000 – 20,000 cells were plated in Matrigel (BD Biosciences)-coated 96-well glass-bottom plates with 100 μL of plating medium containing MEM (Life Technologies), glucose (33 mM, Sigma), transferrin (0.01%, Sigma), Hepes (10 mM), Glutagro (2 mM, Corning), insulin (0.13%, Millipore), B27 supplement (2%, Gibco), and heat inactivated fetal bovine serum (7.5%, Corning). AraC (0.002 mM, Sigma) was added when glial density reached 50–70% of confluence. Neurons were cultured at 37°C in humidified 5% CO2.
Brainbow AAV injection and brain preparation
All the following procedures involving animals were approved by the Massachusetts Institute of Technology Committee on Animal Care and were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. 4 Emx1-Cre mice ages ~3–5 months old were used. Mice were used without regard for sex. Brainbow rAAV (AAV9.hEF1a.lox.TagBFP.lox.eYFP.lox.WPRE.hGH-InvBYF and AAV9.hEF1a.lox.mCherry.lox.mTFP1.lox.WPRE.hGH-InvCheTF; University of Pennsylvania, Penn Vector Core) was injected into Emx1-Cre mice11. Adult Emx1-Cre mice were first head-fixed to a stereotaxic apparatus and a small (~0.5 mm2) craniotomy was performed under continuous isoflurane anesthesia. A 34-gauge injection needle pre-loaded with the AAV solution (7.5 × 1012 genome copy/mL) was then inserted into the brain to a depth of ~500 μm from the cortical surface, and the virus infused at a rate of 0.2 μL/min. After injecting 2 μL of the virus solution, the needle was left at the injection site for additional 5 minutes to allow for viral diffusion. Mice were allowed to recover from surgery and express virus for 3–4 weeks before transcardial perfusion. Using isoflurane, mice were deeply anesthetized and perfused with 30 mL room temperature 1x PBS, and then 30 mL room temperature fixative solution (4% paraformaldehyde in 1x phosphate buffered saline (PBS)). Brains were then harvested and stored in the same fixative at 4°C for 24 hours. 100-μm or 150-μm thick brain slices were prepared by slicing brains with 100 mM glycine in 1x PBS on a vibratome (Leica VT1000s). The slices were stored in 1x PBS at 4°C until staining.
Immunostaining of tissues (except the microtubule staining of mouse tissue slices)
All following steps were conducted at room temperature with gentle shaking, unless otherwise noted. To stain Brainbow slices, two different conditions were used. To stain only Brainbow AAV fluorescent proteins (FPs), Brainbow mouse brain slices were first permeabilized and blocked in “0.5T” blocking buffer (0.5% Triton X-100, 5% normal donkey serum (NDS), 1x PBS) for two hours. Slices were then incubated with primary antibodies (see Supplementary Tables 10 and 11 for details) in “0.25T” blocking buffer (0.25% Triton X-100, 5% NDS, 1x PBS) for 2–3 days at 4°C with gentle shaking. Slices were washed in 0.25T blocking buffer four times, for thirty minutes each time. Slices were incubated with DNA-conjugated secondary antibodies in hybridization buffer (2x SSC buffer, 10% dextran sulfate, 1 mg/mL yeast tRNA, 5% NDS, 0.1% Triton X-100) overnight and washed in 0.25T blocking buffer four times, for thirty minutes each time. Slices were then incubated with DNAs with a 5′acrydite modification at a concentration of 1 ng/μL in hybridization buffer overnight, and then washed in 0.25T blocking buffer four times, for thirty minutes each time.
To stain synaptic proteins, or synaptic proteins and Brainbow fluorescent proteins, Brainbow slices were first permeabilized and blocked in “0.1T” blocking buffer (0.1% Triton X-100, 1x PBS, 5% NDS) for two hours. Primary antibody staining and subsequent washing steps were identical to the FP staining protocol described above, but conducted in 0.1T blocking buffer. DNA-conjugated antibody and DNA staining steps were identical to the FP staining protocol. Subsequent washing steps were conducted in 0.1T blocking buffer. To stain FPs for proExM, the permeabilization, primary antibody staining, washing steps, and secondary antibody staining were conducted in 0.1T blocking buffer.
Immunostaining of tubulin in cultured cells and tissue slices
All following steps were conducted at room temperature, unless otherwise noted. Cells were first washed in 1x PBS three times, and then extracted in cytoskeleton extraction buffer25 (0.2% Triton X-100, 0.1 M 1,4-piperazinediethanesulfonic acid (PIPES), 1 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA), 1 mM magnesium chloride, pH 7.0) for 1 min, and then fixed in tubulin fixation solution (3% formaldehyde, 0.1% glutaraldehyde, 1x PBS) for 10 minutes, followed by reduction with 0.1% sodium borohydride in 1x PBS for 7 minutes and washing with 100mM glycine in 1x PBS three times, for 5 minutes each time. Cells were permeabilized and blocked in 0.2T blocking buffer (0.2% Triton X-100, 1x PBS, 5% NDS) for ten minutes and incubated with rabbit anti-beta tubulin antibody in “0.2T” blocking buffer at a concentration of 10 μg/mL for one hour, and then washed in 1x PBS three times. Cells were incubated with DNA-conjugated anti-rabbit secondary antibody (RbA1′ in Supplementary Table 12) in hybridization buffer at a concentration of 10 μg/mL for one hour with gentle shaking, then washed in 1x PBS three times. Cells were incubated with DNA (A1′ 5′acrydite 3′ Alexa 488 in Supplementary Table 3) in hybridization buffer at a concentration of 0.5 ng/μL for one hour with gentle shaking, then washed three times in 1x PBS.
To stain microtubules of mouse tissue slices, Thy1-YFP mice were deeply anesthetized using isoflurane and perfused with 30 mL room temperature 1x PBS. Brains, livers, and lungs were then harvested and sliced on a vibratome (Leica VT1000s) to a thickness of 100 μm in 1x PBS. Slices were extracted in cytoskeleton extraction buffer25 for 5 min with gentle shaking, and then fixed in tubulin fixation solution for 30 minutes with gentle shaking, followed by reduction with 0.1% sodium borohydride in 1x PBS for 7 minutes with gentle shaking and washing with 100mM glycine in 1x PBS three times with gentle shaking, for 10 minutes each time.
Slices were permeabilized and blocked in 0.2T blocking buffer (0.2% Triton X-100, 5% NDS, 1x PBS) for two hours with gentle shaking. Primary antibody staining and all washing steps were identical to the synaptic protein staining protocol, but conducted in 0.2T blocking buffer. DNA-conjugated antibody and DNA staining steps were identical to the synaptic protein staining protocol.
Immunostaining of synaptic proteins in cultured neurons
All following steps were conducted at room temperature, unless otherwise noted. Cultured neurons were fixed two weeks after initial plating. Cultured neurons were first washed in 1x PBS three times, and then fixed with 4% formaldehyde in 1x PBS for 10 minutes and washed with 100 mM glycine in 1x PBS three times, for 5 minutes each time. Subsequent procedures were identical to the microtubule staining of cultured cells.
First round expansion (except Supplementary Fig. 15)
After immunostaining, cultured cells, neurons, and tissue slices were first incubated in pre-gel incubation solution (see Supplementary Table 13 for details) overnight at 4°C. After the incubation, specimens were incubated in 1st gelation solution (Supplementary Table 13) twice, for 30 minutes each at 4°C. For cultured cells and neurons, 200 μL of 1st gelation solution was added to each well, and then incubated at 37°C for three hours. For tissue slices, slices were placed between two pieces of #1 coverglass separated by another #1 coverglass, and then incubated at 37°C for 3 hours.
After the incubation, gels (including cultured cells, neurons, and tissue slices) were incubated with Proteinase K at a concentration of 8 units/mL (1:100 dilution) in digestion buffer (50 mM Tris pH8, 1 mM EDTA, 0.5% Triton-X100, 0.8 M guanidine HCl) overnight at room temperature with gentle shaking. Digested gels were next placed in an excess volume of fresh distilled water for three periods (2 hours, 2 hours, overnight) at room temperature with gentle shaking.
Re-embedding and DNA hybridization (except Supplementary Fig. 15)
All following steps were conducted at room temperature with gentle shaking, unless otherwise noted. Expanded gels were incubated in a freshly prepared re-embedding solution (Supplementary Table 13) twice, for thirty minutes each. After the incubation, gels were placed between two pieces of #1 coverglass, and then incubated at 37°C for 1.5 hours in a nitrogen-filled chamber. Following the incubation, gels were detached from the coverglass, and then washed in DNA hybridization buffer (20% (v/v) formamide in 4x saline-sodium citrate (SSC) buffer) for thirty minutes to remove any unreacted monomers from gels.
Gels that would not undergo signal amplification were incubated with DNAs (see Supplementary Tables 4 and 10 for details) at a concentration of 0.5 ng/μL in DNA hybridization buffer overnight, and then washed in DNA hybridization buffer three times, for 2 hours, 2 hours, respectively.
Gels that would undergo DNA or LNA hybridization-based signal amplification were incubated with linker DNAs (see Supplementary Table 4 and 10 for details) at a concentration of 2 ng/μL in DNA hybridization buffer overnight, and then washed in DNA hybridization buffer three times, for 2 hours, 2 hours, and overnight, respectively.
Second round expansion (except Supplementary Fig. 15)
All following steps were conducted at room temperature with gentle shaking, unless otherwise noted. For hp-iExM, gels were incubated in a freshly prepared hp-iExM 2nd gel solution (Supplementary Table 13) twice, for thirty minutes each. After the incubation, gels were placed between two pieces of #1 coverglass, and then incubated in a nitrogen-filled chamber at 37 °C for 1.5 hours. After the incubation, gels were incubated in 0.2 M NaOH for overnight and washed in DI water multiple times until the size of the gels plateaued.
For iExM, gels were incubated in a freshly prepared iExM 2nd gel solution (Supplementary Table 13) twice, for thirty minutes each. After the incubation, gels were placed between two pieces of #1 coverglass, and then incubated in a nitrogen-filled chamber at 37°C for 1.5 hours. After the incubation, gels were incubated in 0.2 M NaOH for 1 hour. Gels were washed in DNA hybridization buffer twice, for thirty minutes each time, and then incubated with fluorophore-tagged DNAs for DNA hybridization-based signal amplification and fluorophore-tagged LNAs for LNA hybridization-based signal amplification at a concentration of 0.5 ng/μL in DNA hybridization buffer overnight, and then washed in DNA hybridization buffer three times (2 hours, 2 hours, and overnight). Gels were then washed in 0.2x PBS multiple times for DNA-hybridization based signal amplification, and in DI water for LNA-hybridization based signal amplification, until the size of the gels plateaued.
ProExM
Immunostained brain slices were first incubated in 6-((acryloyl)amino)hexanoic acid, succinimidyl ester (AcX; resuspended in anhydrous DMSO at a concentration of 10 mg/mL and then diluted in 1x PBS at a concentration of 0.1 mg/mL) at room temperature overnight with gentle shaking. Slices were then incubated in monomer solution (1x PBS, 2 M NaCl, 8.625% (w/w) sodium acrylate, 2.5% (w/w) acrylamide, 0.15% (w/w) N,N′-methylenebisacrylamide (BIS), 0.2% (w/w) ammonium persulfate (APS), 0.2% (v/v) tetramethylethylenediamine (TEMED), 0.01% (w/w) 4-hydroxy-2,2,6,6-tetramethylpiperidin-1-oxyl (H-TEMPO)) twice, for thirty minutes each at 4°C and placed between two pieces of #1 coverglass separated by another #1 coverglass and then incubated in a humidified 37°C incubator for two hours. Following the incubation, gels were digested in Proteinase K at a concentration of 8 units/mL in 50 mM Tris (pH 8) with 1 mM EDTA, 0.5% Triton X-100, and 1 M NaCl overnight at room temperature with gentle shaking and then expanded in distilled (DI) water several times until the size of gels plateaued.
Imaging
Imaging was performed on an Andor spinning disk confocal microscope with a 40x 1.15 NA water immersion objective (Fig. 2d, 2g, 2k, 2l, 2o, Fig. 3g, 3i–o, 3t, 3u, Fig. 4, Supplementary Fig. 1b, 2b, 7, 10–13) or Nikon Eclipse Ti inverted microscope with the same objective (Fig. 3a, 3d, 3h, Supplementary Fig. 1a, 9, 14, 15). Background of images was corrected by using the ‘Subtract background’ function implemented in Fiji with a 50-pixel wide ‘rolling ball’ algorithm.
Expansion factor measurement
To determine the expansion factors for each round of expansion, we imaged whole specimens (tissues and cultured cells) with a widefield microscope before vs. after the expansion of the first gel. The expansion factor for the first round was then determined by measuring the distance between two landmarks in the specimen before vs. after the first round of expansion. The expansion factor of the second round was determined in a same way.
RMS error measurement
RMS error measurement was performed in a similar way as in previous studies1,9. Briefly, STORM images before expansion and confocal images after expansion were registered using rigid body registration as implemented in Fiji (Plugins → Registration → TurboReg → Rigid Body/Accurate/Manual)1. After the registration, deformation vector fields were calculated by using Elastix and Transformix as in ref.9 (see Supplementary Protocol 1 of ref.9 for details).
Deconvolution and de-noising
Images shown in Fig. 2g were deconvolved using custom-written MATLAB code that uses the Richardson-Lucy algorithm with wavelet regularization and a theoretical point-spread function. The deconvolution was performed with a GPU (NVidia, Tesla K40c). For Fig. 4c–k, Supplementary Fig. 10–13, Supplementary Video 5–9, the images were first deconvolved, and then the background and signals from non-specifically bound fluorophores were removed by using connected component analysis26.
STORM imaging
BS-C-1 cells were cultured, extracted, fixed, and stained with a primary antibody as in ‘Immunostaining of tubulin of cultured cells and tissue slices’. For Fig. 2a–c, primary antibody-stained cells were incubated with Alexa 647-conjugated anti-rabbit antibody (10 μg/μL) in 0.2T blocking buffer for thirty minutes, and washed in 1x PBS three times. STORM imaging was performed in STORM imaging buffer (100 mM Tris (pH 8.0), 50 mM NaCl, 1% β-Mercaptoethanol, 5% glucose, 1 μg/μL glucose oxidase, 40 μg/mL catalase) on a custom-built STORM microscope using the oblique-incidence geometry. For Fig. 2o, primary antibody-stained cells were incubated with a mixture of Alexa 647-conjugated anti-rabbit secondary antibody (13.3 μg/mL) and DNA-conjugated anti-rabbit secondary antibody (13.3 μg/mL, RbA1′) in hybridization buffer for one hour at room temperature with gentle shaking, and then washed in 1x PBS three times. After the wash, STORM imaging was performed on a commercial Nikon N-STORM microscope in total internal reflection fluorescence (TIRF) mode in STORM imaging buffer (1 M Tris (pH 8.0), 50 mM NaCl, 1% β-Mercaptoethanol, 5% glucose, 1 μg/μL glucose oxidase, 40 μg/mL catalase). After STORM imaging, cells were washed in 1x PBS, and then incubated with DNA (A1′ 5′acrydite 3′ Alexa 488 in Supplementary Table 3) in hybridization buffer at a concentration of 0.5 ng/μL for 1 hour at room temperature with gentle shaking, then washed three times in 1x PBS.
Statistics
In this study, the sample sizes were decided not based upon a power analysis, since the goal was to develop a new technology. As noted in ref. 27, recommended by the NIH, “In experiments based on the success or failure of a desired goal, the number of animals required is difficult to estimate…” As noted in the aforementioned paper, “The number of animals required is usually estimated by experience instead of by any formal statistical calculation, although the procedures will be terminated [when the goal is achieved].”27 The sample sizes of this study reflect our past experience in developing ExM technologies1–3. For animal studies, sample-size estimation was not performed. Exclusion, randomization, and blinding of samples were not performed.
Triple round expansion
All following steps were conducted at room temperature with gentle shaking, unless otherwise noted. Immunostaining of BS-C-1 cells, pre-gel incubation, first gel synthesis, re-embedding, and second gel synthesis steps were identical to the iExM procedure for BS-C-1 cells with labeled tubulin, but with the following modifications. RbB1′ and DNA B1 5′acrydite were used during the staining step. Pre-gel incubation solution, 1st gel solution, and 1st re-embedding solution shown in Supplementary Table 14 were used to form a 1st swellable gel and re-embedding gel. After the re-embedding, gels were incubated with DNA B1′ A2 5′acrydite (Supplementary Table 8). 2nd gel solution shown in Supplementary Table 14 was used to form a 2nd swellable gel.
After the 2nd gel formation, gels were incubated in 0.25 M tris(2-carboxyethyl)phosphine (TCEP; 1 M stock solution of TCEP diluted in 1 M Tris-HCl pH 8.0) overnight, and then expanded in DI water three times. Expanded gels were re-embedded again in 2nd re-embedding solution (Supplementary Table 14), and then incubated with a linker DNA (A2′ 4LNA-A1′ 5′acrydite) in DNA hybridization buffer for overnight at a concentration of 2 ng/μL, and then washed in DNA hybridization buffer three times for two hours, two hours, and overnight, respectively.
Gels were then embedded in 3rd gel solution (Supplementary Table 14) and digested in 0.2 M NaOH for 1 hour. Gels were then washed in DNA hybridization buffer and incubated with fluorophore-tagged LNA (LNA-A1 3′atto 565) at a concentration of 0.5 ng/μL in DNA hybridization buffer overnight, and then washed in DNA hybridization buffer three times, for 2 hours, 2 hours, and overnight respectively. Gels were then washed in DI water multiple times.
MATLAB simulation of iExM images
We developed a simulator of iExM images of microtubules labeled with DNA-conjugated secondary antibodies (code contained in Supplemental Software 1, and described in Supplementary Fig. 2). Simulation of iExM was performed by first creating a cylinder with an inner radius of Ri and outer radius of Ro. 5′acrydite moieties were randomly assigned to voxels within the cylindrical volume to simulate the stochastic staining of a microtubule. To gauge the impact of the broadening of the PSF on the simulation (Supplementary Fig. 6c), the positions of the 5′acrydite moieties were randomly perturbed with a standard deviation Ep (parameter ‘PositionE’ in the MATLAB code). Then, the cylindrical volume was projected onto a 3-D image stack by convolving the volume with the 3-D point-spread-function (PSF) of a confocal microscope with an objective lens of 40x magnification and 1.15 numerical aperture. Then the volume was down-sampled by pixel-binning in the lateral dimension (with a pixel size of 6 nm) and sub-sampled in the axial dimension to incorporate the pixel pitch and the z-step size of the microscope. The simulation was performed multiple times with varying Ri and Ro.
Once the microtubule profiles with various combinations of Ri and Ro were generated, we then fitted the simulated profiles with a sum of two Gaussians and the peak-to-peak distances of the fitted sum of two Gaussians were measured. The measured peak-to-peak distances were compared to the peak-to-peak distances of each experimental microtubule profile. If the difference between these two distances was smaller than a single pixel size of the simulation (6 nm), then the Ri and Ro value of the corresponding simulated profile was retained for further analysis. The collected Ri and Ro values were averaged to find an average inner and outer radius of the DNA layer best fitted to experimental microtubule profiles. For example, the average Ri and Ro was 30.6 nm and 34.8 nm respectively, for the experimental microtubule profile shown in Supplementary Fig. 2c.
Code availability
iExM image simulator was contained in Supplemental Software 1.
Supplementary Material
Acknowledgments
E.S.B. was funded by the HHMI-Simons Faculty Scholars Program, the NIH Director’s Pioneer Award 1DP1NS087724, the New York Stem Cell Foundation-Robertson Award, U. S. Army Research Laboratory and the U. S. Army Research Office under contract/grant number W911NF1510548, US-Israel Binational Science Foundation Grant 2014509, the Picower Institute Innovation Fund, NIH grants IARPA D16PC00008, 1R01MH110932, 1R43MH101943, 1R01MH103910, 1R01EY023173, and 2R01DA029639, the IET A. F. Harvey Prize, the Open Philanthropy Project, and the MIT Media Lab. J.-B.C was supported by the Simons Postdoctoral Fellowship. F.C. was supported by the NSF Fellowship and Poitras Fellowship. Y.-G.Y., J.S.K., and H.-J.S. were supported by Samsung Scholarships. P.W.T. and A.T.W. were supported by Hertz Foundation fellowships. Confocal imaging was performed in the W. M. Keck Facility for Biological Imaging at the Whitehead Institute for Biomedical Research. We acknowledge W. Salmon for her assistance with confocal imaging. STORM imaging shown in Fig. 2o was performed in the Center for Brain Science at Harvard University. We acknowledge E. Garner, C. Wivagg, and S. Turney for allowing us to use the N-STORM microscope and their assistance with STORM imaging. We acknowledge D. Park for assistance with the preparation of cultured neurons. We also acknowledge for helpful discussion, S. Shim, Y. Sigal, C. Speer, M. Thanawala, D. Kim, M. Sauer, and S. Alon.
Footnotes
AUTHOR CONTRIBUTIONS
E.S.B. and J.-B.C. conceived the main idea and designed experiments. P.W.T. conceived hp-iExM strategy. J.-B.C. performed immunostaining and expanded specimens. J.-B.C., F.C., and A.T.W. developed re-embedding process. J.-B.C. calculated RMS error of iExM. F.C. conceived signal amplification methods. E.J. performed immunostaining. Y.-G.Y. performed deconvolution and de-noising and developed the iExM simulator. H.-J.S. and N.P performed the brainbow virus injection and perfusion. Y.-G.Y. and S.A. created 3-D videos. H.B. contributed STORM data in Fig. 2a–c. J.-B.C. performed STORM imaging for Fig. 2o,p. D.C. provided antibodies against Brainbow fluorescent proteins. J.-B.C. and J.S.K. imaged samples and performed image processing. J.-B.C. performed statistical analysis. E.S.B. and J.-B.C. wrote the paper. All authors contributed to editing of the paper. E.S.B. supervised this work.
COMPETING FINANCIAL INTEREST STATEMENT
E.S.B., J.-B.C., F.C., and P.W.T. have applied for a patent on iExM. E.S.B. is co-founder of a company, Expansion Technologies, that aims to provide expansion microscopy kits and services to the community.
Supplementary Information is available in the online version of the paper.
DATA AVAILABLITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon request.
REFERENCES (FOR MAIN TEXT ONLY)
- 1.Chen F, Tillberg P, Boyden ES. Expansion Microscopy. Science (80- ) 2015;347:543. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tillberg PW, et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat Biotechnol. 2016;34:987–992. doi: 10.1038/nbt.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen F, et al. Nanoscale Imaging of RNA with Expansion Microscopy. Nat Methods. 2016;13:679–684. doi: 10.1038/nmeth.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Connell PBH, Brady CJ. Polyacrylamide gels with modified cross-linkages. Anal Biochem. 1976;76:63–73. doi: 10.1016/0003-2697(76)90264-5. [DOI] [PubMed] [Google Scholar]
- 5.Kurenkov VF, Hartan HG, Lobanov FI. Alkaline Hydrolysis of Polyacrylamide. Russ J Appl Chem. 2001;74:543–554. [Google Scholar]
- 6.Weber K, Rathke PC, Osborn M. Cytoplasmic microtubular images in glutaraldehyde-fixed tissue culture cells by electron microscopy and by immunofluorescence microscopy. Proc Natl Acad Sci. 1978;75:1820–1824. doi: 10.1073/pnas.75.4.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aquino D, et al. Two-color nanoscopy of three-dimensional volumes by 4Pi detection of stochastically switched fluorophores. Nat Methods. 2011;8:353–359. doi: 10.1038/nmeth.1583. [DOI] [PubMed] [Google Scholar]
- 9.Chozinski TJ, et al. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat Methods. 2016;13:485–488. doi: 10.1038/nmeth.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dani A, Huang B, Bergan J, Dulac C, Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–56. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Improved tools for the Brainbow toolbox. Nat Methods. 2013;10:540–547. [PubMed] [Google Scholar]
- 12.Olivier N, Keller D, Gönczy P, Manley S. Resolution doubling in 3D-STORM imaging through improved buffers. PLoS One. 2013;8:e69004. doi: 10.1371/journal.pone.0069004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wombacher R, Cornish VW. Chemical tags: Applications in live cell fluorescence imaging. J Biophotonics. 2011;4:391–402. doi: 10.1002/jbio.201100018. [DOI] [PubMed] [Google Scholar]
- 14.Chen B-C, et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science (80- ) 2014:346. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ke MT, Fujimoto S, Imai T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci. 2013;16:1154–1161. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- 16.Choi HMT, et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat Biotech. 2010;28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cipriano BH, et al. Superabsorbent Hydrogels That Are Robust and Highly Stretchable. Macromolecules. 2014;47:4445–4452. [Google Scholar]
- 18.Jungmann R, et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat Methods. 2014 doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed]
- 19.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science (80- ) 2015:348. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JH, et al. Highly Multiplexed Subcellular RNA Sequencing in Situ. Science (80- ) 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Protocol exchange, iExM.
- 23.Klapoetke NC, et al. Independent optical excitation of distinct neural populations. Nat Methods. 2014;11:338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chow BY, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaughan JC, Dempsey GT, Sun E, Zhuang X. Phosphine Quenching of Cyanine Dyes as a Versatile Tool for Fluorescence Microscopy. J Am Chem Soc. 2013;135:1197–1200. doi: 10.1021/ja3105279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legant WR, et al. High-density three-dimensional localization microscopy across large volumes. Nat Methods. 2016;13:359–365. doi: 10.1038/nmeth.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dell RB, Holleran S, Ramakrishnan R. Sample Size Determination. ILAR J. 2002;43:207–213. doi: 10.1093/ilar.43.4.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.