Abstract
Regulatory T cells are critical for maintaining self-tolerance and to negatively regulate immune responses. Foxp3 is a regulatory T cell-specific transcription factor that functions as the master regulator of the development and function of regulatory T cells. Here, we report the generation of a mouse model, in which a bicistronic reporter expressing a red fluorescent protein has been knocked into the endogenous Foxp3 locus. Using this mouse model, we assessed Foxp3 expression in various lymphocyte compartments and identified previously unreported Foxp3-expressing cells. In addition, we showed that de novo Foxp3 expression along with suppressive function were induced by TGF-β in activated CD4 T cells in vitro. Finally, we demonstrated that non-Foxp3-expressing CD4 T cells could not be converted into Foxp3-expressing cells upon adoptive transfer into immunodeficient hosts. This Foxp3 bicistronic reporter knockin mouse model should greatly enhance the study of regulation and function of Foxp3-expressing regulatory T cells.
Keywords: regulatory T cells, TGF-β
How the body maintains immunological self-tolerance and negative control of physiological and pathological immune responses has been a long-standing question in immunology. Thirty years ago, it was proposed that subsets of thymus-derived CD4 T cells are essential to actively suppress immune responses and maintain self-tolerance (1–3). Recent studies have identified specific regulatory T cells that likely explain these phenomena (4–7). Subsets of regulatory T cells have been described, and they share common features of being hypoproliferative to T cell antigen receptor (TCR) stimulation in vitro and immunosuppressive in vitro and in vivo. Among regulatory T cells, naturally occurring regulatory T (Treg) cells and antigen-induced IL-10 producing regulatory T cells (Tr1) are the best characterized. Naturally occurring CD4+CD25+ Treg cells comprise 5–10% of peripheral CD4+ T cells and exert suppressive function through cell–cell contact and cytokine secretion, whereas induced CD4+CD25– Tr1 cells mediate immunosuppression through IL-10 and TGF-β (8–10).
Many attempts in recent times have been made to identify a reliable surface marker for naturally occurring Treg cells. CD5high, CD45RBlow, GITRhigh, and CD25+ have all been used to enrich for the Treg population (5, 11–14). CD25 has been the most prominent marker used for identifying Treg cells. However, the expression of CD25 on Treg cells is unstable. CD25 is down-regulated when Treg cells are transferred into severe combined immunodeficient mice, whereas the suppressive function of Treg cells is maintained (15). In addition, non-Tr1 CD4+CD25– peripheral T cells have been found to possess regulatory activity (16, 17). Furthermore, CD25 is ubiquitously expressed by activated T cells, which makes it a marginally useful marker in identifying Treg cells from activated T cells in vitro or in vivo. Thus, a more reliable and unambiguous marker for Treg cells is needed.
Foxp3 is a transcription factor belonging to the forkhead family. In humans, mutation in Foxp3 results in immunodysregulation, polyendocrinopathy, enteropathy, X linked syndrome, an X linked immunodeficiency syndrome associated with autoimmune disease in multiple endocrine organs (18–20). A frameshift mutation of Scurfin, the mouse orthologue of Foxp3, results in early lethality due to hyperactivation of T cells in Scurfy mice (21). Recent studies have unveiled a specific role of Foxp3 in the development and function of Treg cells. Foxp3 is predominantly expressed in CD4+CD25+ thymocytes and CD4+CD25+ peripheral T cells (22, 23). Retroviral-mediated Foxp3 expression in CD4+CD25– T cells converts them into Treg-like cells phenotypically and functionally (22, 23). Analysis of Foxp3-transgenic or -knockout mice has further established an essential role for Foxp3 in regulatory T cell development. In mice transgenic for ≈16 copies of cosmid containing endogenous Foxp3 gene, the number of CD4+CD25+ Treg cells is increased. Furthermore, in these mice, both CD4+CD25 and CD8+ T cells that express Foxp3 exhibit immunosuppressive activity (21, 24, 25). Like Scurfy mice, Foxp3-deficient mice show hyperreactivity of T cells. This finding is due to a deficiency in the development of CD4+CD25+ Treg cells (23). Thus, Foxp3 appears to be a reliable marker for Treg cells as it has been shown to be the master regulator specific for the development and function of these cells. However, as an intracellular protein, Foxp3 cannot be easily detected, which has hampered the study of the biology of Foxp3-expressing cells. Quantitation of Foxp3 mRNA has been used to estimate Treg frequency. There is, however, no guarantee that Foxp3 mRNA levels faithfully reflect Foxp3 protein, although a recent study suggests this method may be reliable (23).
Using a gene targeting approach, we have generated a mouse in which we knocked-in a bicistronic fluorescent reporter into the endogenous Foxp3 locus, allowing us to identify Foxp3 expressing cells from different lymphocyte lineages and lymphoid organs. Our results indicate that, as expected, Foxp3 is predominantly expressed in CD4+CD25+ peripheral T cells. In addition, we detected Foxp3 expression in a subset of CD4+CD25– peripheral T cells, a subset of CD4+ thymocytes and CD4+TCR+ bone marrow cells. Moreover, we show that de novo Foxp3 expression in activated CD4 T cells was potently induced by TGF-β and conferred suppressive function on these cells in vitro. Finally, our results demonstrated that Foxp3– CD4 T cells could not be converted into Foxp3+ cells in immunodeficient hosts.
Materials and Methods
Mice. BALB/c mice (blastocyst donors), CD1 mice (foster mothers), C57BL/6 (B6) mice, Tet-Cre transgenic mice (“deletor” mice, C57BL/6 background), Rag-deficient mice (C57BL/6 background) and Foxp3-IRES-mRFP (FIR) mice (C57BL/6 background) were kept under specific pathogen-free conditions in the animal care facility at the Yale University. All mouse experiments were approved by Institutional Animal Care and Use Committee of Yale University.
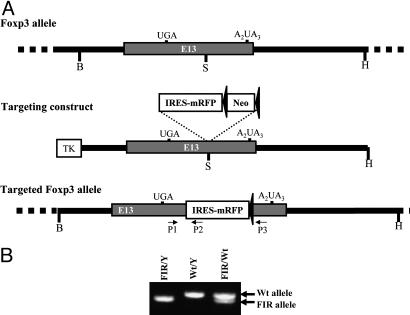
Generation of FIR Mice. A BAC clone (RP23–446O15) consisting of Foxp3 genomic DNA derived from C57BL/6 mice was purchased from BacPac (Oakland, CA). An 11-kb BstZ17I/HpaI fragment comprising exon 13 for Foxp3 gene was cloned into pEasy-Flox vector adjacent to the thymidine kinase selection marker. The internal ribosomal entry site-linked monomeric red fluorescent protein (IRES-mRFP) cassette was generated by substituting eGFP cDNA in MSCV-IRES-eGFP with mRFP cDNA (kindly provided by Roger Tsien, University of California at San Diego, La Jolla, CA). The IRES-mRFP cassette was linked to a LoxP-flanked neomycin (Neo) selection marker to obtain the IRES-mRFP-Neo cassette. The targeting construct was generated by cloning the IRES-mRFP-Neo cassette into an SspI site between the translation stop codon (UGA) and the polyadenylation signal (A2UA3) of the Foxp3 gene. The targeting construct was linearized by ClaI cleavage and subsequently electroporated into Bruce4 C57BL/6 ES cells. Transfected ES cells were selected in the presence of 300 μg/ml G418 and 1 μM ganciclovir. Drug resistant ES cell clones were screened for homologous recombination by Southern blot. To obtain chimeric mice, correctly targeted ES clones were injected into BALB/c blastocysts, which were then implanted in CD1 pseudopregnant foster mothers. Male chimeras were bred with C57BL/6 to screen for germ-line-transmitted offspring. Germ-line-transmitted mice were bred with Tet-Cre transgenic mice (deletor mice) to remove the neomycin gene. Mice bearing targeted Foxp3 allele were screened by PCR.
Antibodies and PCR Primers. Anti-B220 (FITC-conjugated, no. 553088) anti-CD4 [FITC-conjugated, no. 553651; phycoerythrin (PE)-Cy5-conjugated, no. 553654], anti-CD8a (FITC-conjugated, no. 553031; PE-Cy5 conjugated, no. 553034), anti-CD25 (FITC-conjugated, no. 553071), anti-CD45.1 (PE-conjugated, no. 553776), anti-TCR (FITC-conjugated, no. 01304D), and anti-IFN-γ (allo-phycocyanin-conjugated, no. 554413) antibodies were purchased from Becton Dickinson Pharmingen. PCR primers for genotyping FIR mice (P1: 5′-CAAAAC CAAGAAAAGGTGGGC-3′; P2: 5′-GGAATGCTCGTCAAGAAGACAGG-3′; P3: 5′-CAGTGCTGTTGCTGTGTAAGGGTC-3′) and TaqMan real-time PCR primers for Foxp3 (Forward: 5′-GGCCCTTCTCCAGGACAGA-3′ and Reverse 5′-GCTGATCATGGCTGGGTTGT-3′) were synthesized by the Yale Keck facility. TaqMan real-time probe (5′-6-FAM-ACTTCATGCATCAGCTCTCCACTGTGGAT-BHQ-1–3′) for Foxp3 was purchased from Biosearch.
Flow Cytometry and FACS. Harvested lymphocytes were treated with an ammonium, chloride, potassium lysis buffer (BioSource International, Caramillo, CA, no. P304) to remove red blood cells and washed with PBS containing 1% FBS (Gemini Biological Products, Calabasas, CA). Cells were then stained with 1:400 dilution of the indicated antibodies together with 5 μg/ml anti-Fc-Receptor blocking antibody (2.4G2) (American Type Culture Collection) in PBS containing 1% FBS and then washed twice with PBS. Before cell sorting, CD4 T cells were enriched by magnetic-activated cell sorting beads per instructions (Miltenyi Biotec, Auburn, CA) and then stained with the indicated antibodies. The Becton Dickinson FACSVantage system was used for fluorescence detection and cell sorting.
T Cell Culture and Stimulation. T cells were cultured under tissue culture conditions (37°C and 5% CO2) in Bruff's media (GIBCO/BRL) supplemented with 10% FBS/1% penicillin and streptomycin/1% glutamine (GIBCO/BRL). T cells were stimulated with 2 μg/ml soluble anti-CD3 (2C11) and 2 μg/ml soluble anti-CD28 (American Type Culture Collection) in the presence of irradiated antigen-presenting cells (APCs). The ratio between T cells and APCs was 1:4 unless stated otherwise. To prepare APCs, splenocytes depleted of CD4 and CD8 T cells were irradiated with 3,000 rad by using a Pantak RAD 320 (AGFA NDT, Lewistown, PA).
T Cell Proliferation, Suppression, and Cytokine Assays. T cell proliferation was assayed by [3H]thymidine incorporation assay, in which 2 × 104 FACS-purified CD4 T cells were stimulated in the presence of 8 × 104 irradiated APCs. Seventy-two hours after stimulation, 1 μCi (1 Ci = 37 GBq) of [3H]thymidine was added into the T cell culture. Eight hours later, cells were recovered by cell harvestor (Tomtec, Orange, CT), and the amounts of incorporated 3H were measured by β-counter (PerkinElmer). For T cell suppression assay, 2 × 104 purified responder T cells were combined with FACS-purified suppressor T cells at different ratios in the presence of 8 × 104 irradiated APCs. The T cell mixture was stimulated and T cell proliferation was measured by [3H]thymidine incorporation assay as above. For 5-,6-carboxyfluorescein diacetate succinimidyl ester (CFSE) staining, FACS-purified CD4 T cells were labeled with 5 μM CFSE (Molecular Probes) per manufacturer's protocols. To measure intracellular IFN-γ production, previously stimulated CD4 T cells were restimulated for 4 h with 50 ng/ml phorbol 12-myristate 13-acetate (Sigma) and 1 μM ionomycin (Sigma) in the presence of BD-GolgiStop. Cells were stained with different fluorophore-conjugated antibodies and assayed per protocols by BD Biosciences.
Adoptive Transfer Assay and Bone Marrow Transplantation. FACS-purified peripheral Foxp3–CD25–CD4+ cells or Foxp3– bone marrow cells (1 × 106) from FIR mice were transferred into sublethally irradiated (600 rad) Rag-deficient recipients through retroorbital injection. At different time points after transfer, recipients were killed and Foxp3 expressing cells were monitored by flow cytometry.
Results and Discussion
Generation of FIR Mice. To mark Foxp3-expressing cells with fluorescent protein, we decided to use mRFP. mRFP is a derivative of Discosoma red fluorescent protein (DsRed) that was developed through serial mutagenesis (26). Unlike DsRed, which has to form tetramers to be fluorescent, mRFP is monomeric. Thus, compared with DsRed, mRFP protein matures faster, does not form aggregates inside cells and, thus, is less toxic to cells. In addition, mRFP and eGFP can be readily distinguished optically allowing the labeling of one cell with two fluorescent proteins.
To trace Foxp3-expressing cells, an IRES-mRFP was inserted into the 3′ untranslated region of the endogenous Foxp3 locus (Fig. 1A) by gene targeting. The IRES we used, a DNA sequence derived from Encephalomyocarditis virus (27), is transcribed and recruits cellular translational machinery to reinitiate translation of the downstream second gene (mRFP) on the bicistronic mRNA independent of the upstream sequence (Foxp3). Thus, in targeted Foxp3-expressing cells, mRFP is transcribed along with the endogenous Foxp3 gene under the control of the endogenous Foxp3 promoter, and mRFP is translated independently under IRES control. Mice bearing the Foxp3-IRES-mRFP allele (referred to as FIR mice below) were confirmed and genotyped by PCR (Fig. 1B).
Fig. 1.
Targeting IRES-mRFP reporter into the mouse Foxp3 locus. (A) Maps for mouse Foxp3 locus, targeting DNA construct, and the targeted Foxp3 locus. An 11-kb mouse genomic DNA, including exon 13 of Foxp3 gene, was excised by using BstZ17I (B) and HpaI (H) (Top) and cloned into pEasy-Flox vector adjacent to the thymindine kinase (TK) selection marker. A cassette containing IRES-mRFP and LoxP-flanked neomycin (Neo) selection marker was inserted into an SspI (S) site between the translation stop codon (UGA) and the polyadenylation signal (A2UA3) of Foxp3 gene (Middle). A correctly targeted ES cell was used to create chimeras and germ-line-transmitted mice. The Neo gene was removed in vivo by using deletor mice transgenic for Cre recombinase to generate mice bearing targeted Foxp3 locus (Lower). (B) PCR geno-typing FIR mice. Three primers (P1 to P3 as indicated) were designed to genotype FIR mice. PCR yielded 517-bp product for the wild-type (Wt) Foxp3 allele and 470-bp product for targeted Foxp3 allele.
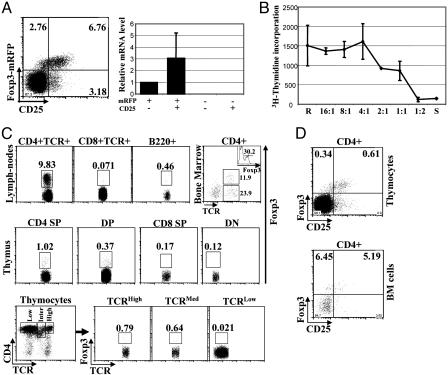
mRFP Faithfully Marks the Expression of Foxp3 in FIR Mice, and Foxp3-Expressing Cells Were Detected in CD4 T Cells from Different Lymphoid Organs. Mice bearing the FIR knockin allele develop normally and are fertile. FIR mice displayed normal development of different lymphocyte lineages, such as thymocytes, mature T cells, and B cells (data not shown). To assess whether mRFP expression reflects Foxp3 expression in FIR mice, T cells from FIR mice were stained with fluorophore-conjugated CD4 and CD25 antibodies and analyzed by flow cytometry (Fig. 2A Left). As expected, most CD4 T cells (≈90%) were negative for CD25 and mRFP, and ≈7% of CD4 T cells were positive for both CD25 and mRFP. In addition, small populations of cells were CD25+mRFP– or CD25–mRFP+ (≈3%). These four populations of CD4 T cells, namely CD25–mRFP+, CD25+mRFP+, CD25–mRFP– and CD25+mRFP– were purified by FACS, and the steady-state mRNA levels of Foxp3 were measured by real-time quantitative PCR (Fig. 2 A Right). Foxp3 mRNA was detected only in CD25+mRFP+ and CD25–mRFP+ populations, although lower levels of Foxp3 mRNA were detected in CD25–mRFP+ CD4 T cells. Interestingly, the geometric mean fluorescence intensity of mRFP as measured by flow cytometry was also lower in the CD25–mRFP+ (≈37) as compared with the CD25+mRFP+ (≈59) population. Thus, in FIR mice, mRFP expression faithfully marks Foxp3 expression and can be used as a reliable marker for detecting Foxp3-expressing cells.
Fig. 2.
mRFP expression faithfully marks Foxp3-expressing CD4 T cells without compromising their regulatory activity, and Foxp3 expression was detected in different lymphocyte compartments. (A) Peripheral lymphocytes from FIR mice were harvested and stained with fluorophore-conjugated anti-CD4 and anti-CD25 antibodies. mRFP expression in CD4 T cells was monitored by flow cytometry (Left). RNA was extracted from different populations of peripheral CD4 T cells (as indicated) purified from FIR mice by FACS. Relative mRNA levels of Foxp3 were determined by TaqMan real-time quantitative PCR, and combined results of two experiments were plotted. (B) CD4+mRFP+ suppressor (S) and CD4+CD25–mRFP– responder (R) T cells were purified by FACS. Suppressor and responder cells were either cultured alone or mixed at indicated ratios (R:S), whereas the number of responder cells remained the same (2 × 104). T cells were activated by soluble anti-CD3 and anti-CD28 antibodies in the presence of irradiated APCs. Three days after stimulation, T cell proliferation was measured by a [3H]thymidine incorporation assay. Combined results from two experiments are shown. (C) In FIR mice, cells from peripheral lymph nodes, bone marrow, and thymus were harvested and stained with fluorophore-conjugated anti-CD4, anti-CD8, anti-B220, and anti-TCR antibodies. By detecting mRFP, the expression of Foxp3 was assessed in CD4 T cells (CD4+TCR+), CD8 T cells (CD8+TCR+), and B cells (B220+) from peripheral lymphocytes (Top Left, Top Left Center, and Top Right Center); in CD4+ cells from bone marrow cells (Top Right with inserted histogram showing the percentage of Foxp3+ cells in CD4+TCR+ population); and in CD4 SP, double-positive (DP), CD8 SP, double-negative (DN) thymocytes (Middle). In addition, based on the expression levels of TCR, CD4+ thymocytes were divided into three groups, TCRHigh, TCRMed, and TCRLow, (Lower Left) and Foxp3 expression in each population was evaluated (Lower Left Center, Lower Right Center, and Lower Right). Typical results of three experiments are shown. (D) Thymocytes and bone marrow cells from FIR mice were stained with fluorophore-conjugated anti-CD4 and anti-CD25 antibodies and analyzed by flow cytometry. Results typical of three experiments are shown.
To test whether mRFP expression perturbed the immunological function of Treg cells, T cell proliferation and suppression assays were performed by using FACS-purified CD4+mRFP+ and CD4+CD25–mRFP– T cells as suppressor cells and responder cells, respectively (Fig. 2B). Suppressor and responder T cells were activated with soluble anti-CD3 and anti-CD28 antibodies in the presence of irradiated APCs either alone or as a combination of the two populations mixed at different ratios. T cell proliferation was measured by the incorporation of [3H]thymidine. In contrast to CD4+CD25–mRFP– T cells, which exhibited extensive proliferation (Fig. 2B, label R), CD4+mRFP+ T cells did not proliferate (Fig. 2B, label S). Furthermore, CD4+mRFP+ T cells inhibited the proliferation of CD4+CD25–mRFP– T cells in a dose-dependent manner. Therefore, the immunosuppresive function of Foxp3-expressing cells is not impaired when mRFP is coexpressed. Thus, in FIR mice, mRFP marks Foxp3-expressing cells with high fidelity and without compromising their immunosuppressive functions.
By measuring mRFP, we assessed the expression of Foxp3 in different lymphocyte lineages from different lymphoid organs (Fig. 2C). In peripheral lymph nodes, very few B cells (B220+) or CD8 T cells (CD8+TCR+) expressed Foxp3 (<1%), whereas Foxp3 expression was detected in ≈10% of CD4+ T cells (CD4+TCR+) (Fig. 2C Top Left, Top Left Center, and Top Right Center). Immunosuppressive activity in the bone marrow has been documented in ref. 28. We therefore investigated whether Foxp3-expressing cells could be found in the bone marrow (Fig. 2C Top Right). Indeed, ≈12% of CD4+ cells residing in the bone marrow expressed Foxp3. Based on TCR expression, ≈35% of CD4+ bone marrow cells were also TCR positive (CD4+TCR+), and Foxp3 expression was detected only in the CD4+TCR+ population in the bone marrow. Thus, interestingly, >30% of the CD4+TCR+ bone marrow cells expressed Foxp3, which was a much higher percentage than that of peripheral CD4+TCR+ cells (Fig. 2C Top Left). The origin and the property of Foxp3 expressing CD4+TCR+ cells in the bone marrow remain to be elucidated. A small percentage of CD4+ thymocytes have been shown to express Foxp3 and possess immunosuppressive activities (16, 22). This result prompted us to investigate Foxp3 expression during T cell thymic development. In the thymus, Foxp3 was expressed in ≈1% of CD4 single positive and ≈0.37% of CD4CD8 double-positive thymocytes. This result is in accordance with recent work done by Tai et al. (29) showing that Foxp3 expression is induced in double-positive (DP) thymocytes upon TCR engagement in vitro. Neither CD8 SP nor CD4CD8 double-negative thymocytes expressed Foxp3 (Fig. 2C Middle). CD4+ thymocytes were divided into three groups based on the expression levels of TCR, TCRHigh, TCRMed, and TCRLow. It appeared that Foxp3 expression levels correlated with TCR expression levels (Fig. 2C Lower). This finding agrees with the reports showing that, in the thymus, TCR signaling is essential for Treg cell development, and the generation of Treg cells directly correlates with TCR signaling strength (30, 31). We also assessed CD25 expression on Foxp3-expressing cells in the thymus and bone marrow. Like CD4+ T cells in the peripheral lymph nodes, a significant fraction of Foxp3-expressing CD4+ cells in the thymus and the bone marrow were negative for CD25 (Fig. 2D).
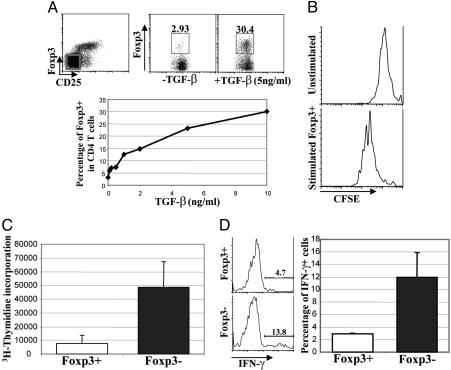
TGF-β Induces de Novo Foxp3 Expression and Regulatory Activity in CD4 T Cells After Antigenic Stimulation in Vitro. It has been suggested that TGF-β induces Foxp3 expression in CD4 T cells under in vitro and in vivo conditions (32–34). However, the question still remains as to whether TGF-β does so by inducing de novo Foxp3 expression in these cells. To address this question, CD4+CD25– T cells that did not express Foxp3 were purified by FACS. Cells were activated by soluble anti-CD3 and anti-CD28 with irradiated APCs in the absence or presence of increasing amounts of TGF-β. Three days later, Foxp3 expression was analyzed by flow cytometry. Foxp3 was highly induced in a substantial portion (up to 30%) of CD4 T cells treated with TGF-β in a dose-dependent manner after TCR engagement (Fig. 3A). Approximately 2–3% of cells expressed Foxp3 in the absence of exogenous TGF-β. We speculated this finding was due to the effects of endogenous TGF-β produced by cultured cells. Thus, TGF-β potently induces de novo Foxp3 expression in activated CD4 T cells. Furthermore, this result suggests that the expression of Foxp3 is not limited to naturally occurring regulatory T cells; rather, it can also be induced in a high percent of CD4 T cells. To further investigate whether this induction can occur in dividing CD4 T cells, CD4+Foxp3– T cells were purified by FACS and labeled with CFSE. CFSE is a fluorescent intracellular protein dye, whose fluorescent intensity is reduced to half after every cell division, allowing us to monitor cell division through flow cytometry. In the presence of exogenous TGF-β, CFSE-labeled cells were either left inactivated or activated with soluble anti-CD3 and anti-CD28 in the presence of irradiated APCs. After 3 days of stimulation, CD4+Foxp3+ cells were gated and the cell divisions were measured by flow cytometry (Fig. 3B). Our results demonstrate that Foxp3 expression can be induced in dividing CD4 T cells in the presence of TGF-β.
Fig. 3.
TGF-β induces de novo Foxp3 expression and regulatory function in CD4 T cells after antigenic stimulation. (A) CD4+CD25–Foxp3– T cells were purified by FACS (Upper Left). With or without TGF-β treatment, purified T cells were activated with soluble anti-CD3 and anti-CD28 in the presence of irradiated APCs. Three days after stimulation, cells were stained with fluorophore-conjugated anti-CD4, and the percentage of Foxp3 expressing CD4 T cells were assessed by flow cytometry (Upper Center and Upper Right). In similar experiments, titrated amount of TGF-β were used, and the correlation between the percentage of Foxp3+ cells and the amount of TGF-β used was plotted (Lower). Typical results of two experiments are shown. (B) FACS-purified CD4+CD25–Foxp3– cells were labeled with CFSE. Under the treatment of 5 ng/ml TGF-β, purified T cells were either left inactivated or activated by soluble anti-CD3 and anti-CD28 antibodies in the presence of irradiated APCs. Three days after stimulation, cells were stained with fluorophore-conjugated anti-CD4 antibodies and analyzed by flow cytometry. CD4+Foxp3+ cells were gated, and the CFSE fluorescence intensity of gated cells was assessed and plotted as histograms. Results typical of two experiments are shown. (C) After 3 days of activation in the presence of 5 ng/ml TGF-β, CD4+Foxp3+, and CD4+Foxp3– cells from the experiments described in A were purified by FACS and restimulated with soluble anti-CD3 and anti-CD28 in the presence of irradiated APCs. Three days after restimulation, T cell proliferation was measured by [3H]thymindine incorporation. Combined results from three experiments are shown. (D) FACS-purified CD4+Foxp3+ (Foxp3+) and CD4+Foxp3– (Foxp3–) T cells as described in A were combined with equal amounts of FACS-purified CD4+CD25– T cells bearing the CD45.1 congenic marker. Cocultured T cells were stimulated with soluble anti-CD3 and anti-CD28 in the presence of irradiated APCs. Four days later, T cells were restimulated with phorbol 12-myristate 13-acetate and ionomycin, and IFN-γ production in CD45.1+CD4+ T cells were monitored by intracellular staining (Left). Combined results from two experiments were plotted (Right).
Foxp3 expressing CD25+CD4+ naturally occurring Treg cells are hyporesponsive toward TCR stimulation and suppress T cell effector functions (22, 23). To address whether TGF-β-induced Foxp3-expressing T cells generated in vitro share these characteristics of naturally occurring Treg cells, we first purified Foxp3+ and Foxp3– CD4 T cells after 3 days of activation in the presence of TGF-β. Purified T cells were restimulated by soluble anti-CD3 and anti-CD28 in the presence of irradiated APCs, and T cell proliferation was assayed 3 days later by [3H]thymidine incorporation. TGF-β-induced Foxp3-expressing CD4 T cells proliferated poorly upon TCR engagement compared with Foxp3 negative counterparts (Fig. 3C). To assess whether TGF-β-induced Foxp3-expressing CD4 T cells possess immunosuppressive function, purified Foxp3+ and Foxp3– CD4 T cells were cocultured with CD4+CD25– T cells bearing the CD45.1 congenic marker. Cells were activated by soluble anti-CD3 and anti-CD28 in the presence of irradiated APCs. Four days later, cells were restimulated with phorbol 12-myristate 13-acetate and ionomycin, and IFN-γ production in CD45.1+ CD4 T cells was measured by intracellular staining (Fig. 3D). TGF-β-induced Foxp3-expressing CD4 T cells inhibited effector function of other T cells as measured by the decrease in IFN-γ production of T cells cultured with Foxp3+ as compared with Foxp3– T cells. Thus, like naturally occurring Treg cells, TGF-β-induced Foxp3-expressing CD4 T cells exhibit immunosuppressive activities, and this result reinforces the notion that Foxp3 is a reliable marker for suppressor T cells.
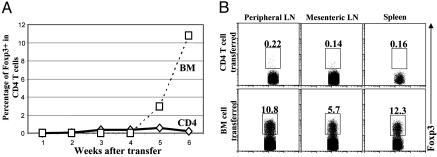
Non-Foxp3-Expressing CD4 T Cells Are Not Converted into Foxp3-Expressing Cells in Immunodeficient Hosts. It has been suggested that in sublethally irradiated wild-type hosts, CD4+CD25– T cells can be converted into CD4+CD25+ Foxp3-expressing cells with potent immunosuppression function (35, 36). However, severe combined immunodeficient mice develop inflammatory bowel disease upon receipt of transferred CD4+CD25– T cells (37). CD25 is expressed by activated T cells and, thus, it is not a reliable marker for Foxp3-expressing regulatory CD4 T cells in a chronically inflamed host. Moreover, CD4+CD25– T cells, which convert into CD4+CD25+ Foxp3-expressing cells, might either initiate as Foxp3– or Foxp3+CD4+CD25– cells because our data show that some CD4+CD25– T cells are Foxp3+. Therefore, it still remains unclear whether CD4+CD25–Foxp3– T cells are capable of developing into CD4+Foxp3+ cells in immunodeficient hosts. To investigate whether CD4+Foxp3– T cells can be converted into CD4+Foxp3+ T cells in the immunodeficient host, the FIR mice were used in adoptive transfer experiments. CD4+CD25–Foxp3– T cells were purified by FACS, and then transferred into sublethally irradiated syngeneic Rag-deficient hosts. FACS-purified Foxp3– bone marrow cells were transplanted into Rag-deficient hosts in parallel to show that transferred hematopoietic cells can express Foxp3. Peripheral lymphocytes were collected and Foxp3-expressing CD4 T cells were monitored by flow cytometry at various time points after transfer (Fig. 4A). In the hosts that received CD4+CD25–Foxp3– T cells, no significant numbers of CD4+Foxp3+ T cells were detected in different lymphoid organs at any given time point tested (Fig. 4). However, substantial amounts (≈3%) of CD4 T cells expressed Foxp3 in bone marrow-transplanted hosts 5 weeks after transfer, and the percentage of Foxp3-expressing CD4 T cells reached normal levels (≈10%) by 6 weeks after transfer (Fig. 4). In addition, 2 weeks after transfer, CD4+CD25–Foxp3– T cell transferred hosts developed IBD as manifested by morbidity, hunched back, diarrhea, and an enlarged spleen, intestine, and colon (data now shown), whereas bone marrow transplanted hosts showed no sign of any disease even 6 weeks after transfer. Thus, in the immunodeficient hosts, mature non-Foxp3 expressing CD4 T cells cannot be converted into Foxp3-expressing CD4 T cells, and the failure to generate Foxp3+ Treg cells may contribute to the development of IBD in this model. It would be interesting to know whether CD4+Foxp3– T cells can become CD4+Foxp3+ T cells in the sublethally irradiated wild-type hosts.
Fig. 4.
CD4+Foxp3– T cells are not converted into CD4+Foxp3+ cells in the immunodeficient hosts after adoptive transfer. (A) From FIR mice, CD4+CD25–Foxp3– (CD4) peripheral T cells and Foxp3– bone marrow cells (BM) were purified by FACS and then transferred into multiple sublethally irradiated (600 rad) Rag-deficient syngeneic hosts with 1 × 106 cells per recipient. At various time points after transfer, recipient mice were killed, and the peripheral T cells were harvested and stained with fluorophore-conjugated anti-CD4 antibodies; the percentage of Foxp3+ cells among CD4 T cells was determined by flow cytometry, and combined results were plotted with each dot represents one recipient mouse. (B) Six weeks after transfer, hosts that received CD4+CD25–Foxp3– peripheral T cells or Foxp3– bone marrow cells were killed. Lymphocytes from peripheral lymph nodes, mesenteric lymph nodes, and the spleens were harvested and stained with fluorophore-conjugated CD4 antibodies. Foxp3-expressing CD4 T cells were detected by flow cytometry.
The FIR Mouse Is a Promising Model to Study the Biology of Foxp3 Expressing Treg Cells. It has been widely accepted that Treg cells are critical for maintaining self-tolerance and actively suppressing immune responses. Attempts have been made to manipulate Treg populations to treat various human pathologies, enhance antitumor immunity, and maintain allograft tolerance after organ transplantation (38–40). Further understanding the biology of Treg cells clearly has important clinical implications. However, the lack of reliable markers has been a major hindrance to the study of Treg cells. With few exceptions, e.g., Tr1 cells (41–43), the expression of Foxp3 goes hand in hand with suppressive T cell function. Ample studies had been done to establish Foxp3 as an essential gene that plays specific roles in regulating the development and function of Treg cells. Thus, the expression of Foxp3 appears to specifically mark Treg cells. By a knockin approach, we have inserted a bicistronic red fluorescent reporter into the endogenous Foxp3 locus to generate FIR mice without compromising the expression of Foxp3 and the function of Foxp3-expressing Treg cells. Using this FIR mouse, we were able to identify previously reported and some unreported Foxp3-expressing cells in mouse. Using FIR cells, we isolated TGF-β-induced Foxp3-expressing functional suppressor T cells after TCR stimulation in vitro, which could have not been done by relying on other traditional markers for Treg cells. Furthermore, we demonstrated that this model is of value to study the generation of Foxp3-expressing Treg cells in vivo. The fact that these mice were generated on the inbred B6 background will substantially facilitate their further use particularly for adoptive transfer studies to other mice. In summary, FIR mice greatly enhanced the ability to study the biology of Foxp3-expressing Treg cells and will be a valuable tool to test both scientific hypotheses and clinical applications of Treg cells in mice.
Acknowledgments
We thank Klaus Rajewsky (Harvard Medical School, Boston) for the gift of Bruce4 ES cells (44), Roger Tsien for providing mRFP cDNA, Linda Evangelisti and Cindy Hughes for generating ES cells and chimeric mice, respectively, Judy Stein for the Southern screening, Elizabeth Eynon for managing the mouse program, and Fran Manzo for help with manuscript preparation. Y.Y.W. was supported by a grant from the Cancer Research Institute. This work was supported by a grant from the National Institutes of Health (RO1 DK51665) and the American Diabetes Association. R.A.F. is an investigator of the Howard Hughes Medical Institute.
Abbreviations: APC, antigen-presenting cell; CFSE, 5-,6-carboxyfluorescein diacetate succinimidyl ester; IRES-mRFP, internal ribosomal entry site-linked monomeric red fluorescent protein; TCR, T cell antigen receptor; Treg, regulatory T.
References
- 1.Nishizuka, Y. & Sakakura, T. (1969) Science 166, 753–755. [DOI] [PubMed] [Google Scholar]
- 2.Penhale, W. J., Farmer, A., McKenna, R. P. & Irvine, W. J. (1973) Clin. Exp. Immunol. 15, 225–236. [PMC free article] [PubMed] [Google Scholar]
- 3.Penhale, W. J., Irvine, W. J., Inglis, J. R. & Farmer, A. (1976) Clin. Exp. Immunol. 25, 6–16. [PMC free article] [PubMed] [Google Scholar]
- 4.Powrie, F. & Mason, D. (1990) J. Exp. Med. 172, 1701–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakaguchi, S., Fukuma, K., Kuribayashi, K. & Masuda, T. (1985) J. Exp. Med. 161, 72–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugihara, S., Izumi, Y., Yoshioka, T., Yagi, H., Tsujimura, T., Tarutani, O., Kohno, Y., Murakami, S., Hamaoka, T. & Fujiwara, H. (1988) J. Immunol. 141, 105–113. [PubMed] [Google Scholar]
- 7.McKeever, U., Mordes, J. P., Greiner, D. L., Appel, M. C., Rozing, J., Handler, E. S. & Rossini, A. A. (1990) Proc. Natl. Acad. Sci. USA 87, 7618–7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maloy, K. J. & Powrie, F. (2001) Nat. Immunol. 2, 816–822. [DOI] [PubMed] [Google Scholar]
- 9.Sakaguchi, S. (2004) Annu. Rev. Immunol. 22, 531–562. [DOI] [PubMed] [Google Scholar]
- 10.Roncarolo, M. G., Bacchetta, R., Bordignon, C., Narula, S. & Levings, M. K. (2001) Immunol. Rev. 182, 68–79. [DOI] [PubMed] [Google Scholar]
- 11.Powrie, F., Leach, M. W., Mauze, S., Caddle, L. B. & Coffman, R. L. (1993) Int. Immunol. 5, 1461–1471. [DOI] [PubMed] [Google Scholar]
- 12.Morrissey, P. J., Charrier, K., Braddy, S., Liggitt, D. & Watson, J. D. (1993) J. Exp. Med. 178, 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimizu, J., Yamazaki, S., Takahashi, T., Ishida, Y. & Sakaguchi, S. (2002) Nat. Immunol. 3, 135–142. [DOI] [PubMed] [Google Scholar]
- 14.Sakaguchi, S., Sakaguchi, N., Asano, M., Itoh, M. & Toda, M. (1995) J. Immunol. 155, 1151–1164. [PubMed] [Google Scholar]
- 15.Gavin, M. A., Clarke, S. R., Negrou, E., Gallegos, A. & Rudensky, A. (2002) Nat. Immunol. 3, 33–41. [DOI] [PubMed] [Google Scholar]
- 16.Stephens, L. A. & Mason, D. (2000) J. Immunol. 165, 3105–3110. [DOI] [PubMed] [Google Scholar]
- 17.Apostolou, I., Sarukhan, A., Klein, L. & von Boehmer, H. (2002) Nat. Immunol. 3, 756–763. [DOI] [PubMed] [Google Scholar]
- 18.Chatila, T. A., Blaeser, F., Ho, N., Lederman, H. M., Voulgaropoulos, C., Helms, C. & Bowcock, A. M. (2000) J. Clin. Invest. 106, R75–R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wildin, R. S., Ramsdell, F., Peake, J., Faravelli, F., Casanova, J. L., Buist, N., Levy-Lahad, E., Mazzella, M., Goulet, O., Perroni, L., et al. (2001) Nat. Genet. 27, 18–20. [DOI] [PubMed] [Google Scholar]
- 20.Bennett, C. L., Christie, J., Ramsdell, F., Brunkow, M. E., Ferguson, P. J., Whitesell, L., Kelly, T. E., Saulsbury, F. T., Chance, P. F. & Ochs, H. D. (2001) Nat. Genet. 27, 20–21. [DOI] [PubMed] [Google Scholar]
- 21.Brunkow, M. E., Jeffery, E. W., Hjerrild, K. A., Paeper, B., Clark, L. B., Yasayko, S. A., Wilkinson, J. E., Galas, D., Ziegler, S. F. & Ramsdell, F. (2001) Nat. Genet. 27, 68–73. [DOI] [PubMed] [Google Scholar]
- 22.Hori, S., Nomura, T. & Sakaguchi, S. (2003) Science 299, 1057–1061.12522256 [Google Scholar]
- 23.Fontenot, J. D., Gavin, M. A. & Rudensky, A. Y. (2003) Nat. Immunol. 4, 330–336. [DOI] [PubMed] [Google Scholar]
- 24.Khattri, R., Cox, T., Yasayko, S. A. & Ramsdell, F. (2003) Nat. Immunol. 4, 337–342. [DOI] [PubMed] [Google Scholar]
- 25.Khattri, R., Kasprowicz, D., Cox, T., Mortrud, M., Appleby, M. W., Brunkow, M. E., Ziegler, S. F. & Ramsdell, F. (2001) J. Immunol. 167, 6312–6320. [DOI] [PubMed] [Google Scholar]
- 26.Campbell, R. E., Tour, O., Palmer, A. E., Steinbach, P. A., Baird, G. S., Zacharias, D. A. & Tsien, R. Y. (2002) Proc. Natl. Acad. Sci. USA 99, 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jang, S. K. & Wimmer, E. (1990) Genes Dev. 4, 1560–1572. [DOI] [PubMed] [Google Scholar]
- 28.Muraoka, S. & Miller, R. G. (1980) J. Exp. Med. 152, 54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tai, X., Cowan, M., Feigenbaum, L. & Singer, A. (2005) Nat. Immunol. 6, 152–162. [DOI] [PubMed] [Google Scholar]
- 30.Jordan, M. S., Boesteanu, A., Reed, A. J., Petrone, A. L., Holenbeck, A. E., Lerman, M. A., Naji, A. & Caton, A. J. (2001) Nat. Immunol. 2, 301–306. [DOI] [PubMed] [Google Scholar]
- 31.Suto, A., Nakajima, H., Ikeda, K., Kubo, S., Nakayama, T., Taniguchi, M., Saito, Y. & Iwamoto, I. (2002) Blood 99, 555–560. [DOI] [PubMed] [Google Scholar]
- 32.Chen, W., Jin, W., Hardegen, N., Lei, K. J., Li, L., Marinos, N., McGrady, G. & Wahl, S. M. (2003) J. Exp. Med. 198, 1875–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng, Y., Laouar, Y., Li, M. O., Green, E. A. & Flavell, R. A. (2004) Proc. Natl. Acad. Sci. USA 101, 4572–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green, E. A., Gorelik, L., McGregor, C. M., Tran, E. H. & Flavell, R. A. (2003) Proc. Natl. Acad. Sci. USA 100, 10878–10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang, S., Alard, P., Zhao, Y., Parnell, S., Clark, S. L. & Kosiewicz, M. M. (2005) J. Exp. Med. 201, 127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Curotto de Lafaille, M. A., Lino, A. C., Kutchukhidze, N. & Lafaille, J. J. (2004) J. Immunol. 173, 7259–7268. [DOI] [PubMed] [Google Scholar]
- 37.Singh, B., Read, S., Asseman, C., Malmstrom, V., Mottet, C., Stephens, L. A., Stepankova, R., Tlaskalova, H. & Powrie, F. (2001) Immunol. Rev. 182, 190–200. [DOI] [PubMed] [Google Scholar]
- 38.Baecher-Allan, C. & Hafler, D. A. (2004) J. Exp. Med. 200, 273–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutmuller, R. P., van Duivenvoorde, L. M., van Elsas, A., Schumacher, T. N., Wildenberg, M. E., Allison, J. P., Toes, R. E., Offringa, R. & Melief, C. J. (2001) J. Exp. Med. 194, 823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edinger, M., Hoffmann, P., Ermann, J., Drago, K., Fathman, C. G., Strober, S. & Negrin, R. S. (2003) Nat. Med. 9, 1144–1150. [DOI] [PubMed] [Google Scholar]
- 41.Groux, H., O'Garra, A., Bigler, M., Rouleau, M., Antonenko, S., de Vries, J. E. & Roncarolo, M. G. (1997) Nature 389, 737–742. [DOI] [PubMed] [Google Scholar]
- 42.Levings, M. K., Sangregorio, R., Sartirana, C., Moschin, A. L., Battaglia, M., Orban, P. C. & Roncarolo, M. G. (2002) J. Exp. Med. 196, 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levings, M. K., Gregori, S., Tresoldi, E., Cazzaniga, S., Bonini, C. & Roncarolo, M. G. (2005) Blood 105, 1162–1169. [DOI] [PubMed] [Google Scholar]
- 44.Kontgen, F., Suss, G., Stewart, C., Steinmetz, M. & Bluethmann, H. (1993) Int. Immunol. 5, 957–964. [DOI] [PubMed] [Google Scholar]