Abstract
Congenital left-sided cardiac lesions (LSLs) are a significant contributor to the mortality and morbidity of congenital heart disease (CHD). Structural copy number variants (CNVs) have been implicated in LSL without extra-cardiac features; however, non-penetrance and variable expressivity have created uncertainty over the use of CNV analyses in such patients. High-density SNP microarray genotyping data was used to infer large, likely-pathogenic, autosomal CNVs in a cohort of 1,139 probands with LSL and their families. CNVs were molecularly confirmed and the medical records of individual carriers reviewed. The gene content of novel CNVs was then compared with public CNV data from CHD patients. Large CNVs (> 1 MB) were observed in 33 probands (~3%). Six of these were de novo and 14 were not observed in the only available parent sample. Associated cardiac phenotypes spanned a broad spectrum without clear predilection. Candidate CNVs were largely non-recurrent, associated with heterozygous loss of copy number, and overlapped known CHD genomic regions. Novel CNV regions were enriched for cardiac development genes, including seven that have not been previously associated with human CHD. CNV analysis can be a clinically useful and molecularly informative tool in LSLs without obvious extra-cardiac defects, and may identify a clinically-relevant genomic disorder in a small but important proportion of these individuals.
INTRODUCTION
DNA copy number variants (CNVs) have been associated with a variety of human diseases or susceptibility to diseases [Feuk et al 2006]. Population-based studies have demonstrated that large CNVs greater than 250 kilobases (kb) in size are significantly enriched among individuals with developmental disorders [Cooper et al 2011] and birth defects [Southard et al 2012], and CNVs greater than 1 megabase (Mb) in size carry an even higher risk of clinical abnormalities [Girirajan et al 2011]. Therefore, large CNVs, particularly those that occur de novo, are more likely to be pathogenic and have an important clinical impact. The role of pathogenic CNVs in developmental delays/intellectual disabilities, dysmorphic features, autism spectrum disorder, and multiple congenital anomalies is now well established, with an estimated test yield of 5–15% [Ellison et al 2012].
CNVs are also important genetic contributors to congenital heart disease (CHD) in the presence of additional birth defects and developmental delays. Multiple studies have identified CNV susceptibility loci in CHD [Glessner et al 2014; Goldmuntz et al 2011; Greenway et al 2009; Hitz et al 2012; Lalani et al 2013; Payne et al 2012; Thienpont et al 2007; Tomita-Mitchell et al 2012; Warburton et al 2014], but lack of controls and conflation of cardiac phenotypes in several previous studies limit their clinical applicability to LSLs. Many of the CNVs have also been observed among apparently unaffected population-based controls, implying that penetrance is incomplete, making genetic counselling of families with such CNVs more difficult. Likely pathogenic CNVs have been observed more often in individuals with CHD but without developmental delays, dysmorphic features, or other birth defects (hereafter called isolated CHDs) than in children without CHD [Kim et al 2016], but the utility of CNV analysis in this group of individuals has remained equivocal; in turn, this has meant that genetic testing for CNVs in isolated CHDs controversial [Connor et al 2014].
Grouping of CHDs together in studying CNVs also potentially dilutes important developmental and clinical categorizations. Left sided lesions (LSLs) constitute 14–20% of all CHDs, and include hypoplastic left heart syndrome (HLHS), Shone complex, mitral valve atresia, coarctation of the aorta (CoA), aortic valve stenosis (AS) and bicuspid aortic valve (BAV)[Lalani and Belmont 2014]. These lesions have a particularly high morbidity and mortality, consuming a high proportion of resources in pediatric cardiology centers [Czosek et al 2013]. LSLs occur in the context of chromosomal disorders, such as Turner and Jacobsen syndrome, as well as in monogenic disorders where there is often CHD concurrent with additional clinical complications (syndromic CHD), evident in Kabuki and Adams-Oliver syndromes. Isolated LSLs appear to have a more complex origin, although familial clustering of cases and an increased risk of LSL in first degree relatives strongly supports a significant genetic contribution [Kerstjens-Frederikse et al 2011; McBride et al 2005; McBride et al 2009]. Few previous papers on the role of CNVs in this category of CHDs have been published (Table 1). Most have not distinguished between isolated and syndromic cases [Geng et al 2014; Glessner et al 2014; Hitz et al 2012; Payne et al 2012; Warburton et al 2014; White et al 2014] and have typically included broad case descriptions and CNVs with variable levels of presumed pathogenicity.
Table 1.
LSLs with CNVs from literature; HLHS – Hypoplastic Left Heart Syndrome; CoA – Coarctation of the Aorta; AVS – Aortic Valve Stenosis; BAV – bicuspid Aortic Valve; ASD – Autism Spectrum Disorder
| Source | Method | Group | Totals | HLHS | CoA | AVS/BAV | LSL + ASD |
|---|---|---|---|---|---|---|---|
| Hitz 2012 [12] (a) | Affymetrix SNP | Multiplex families | 59 | ||||
| Array 6.0 | Trios | 8 | |||||
|
| |||||||
| Payne 2012 [14] (a) | All Cases | 43 | 43 | ||||
| Syndromic cases | 10 | 10 | |||||
|
| |||||||
| Geng 2014 [25] (b) | Agilent 244K | CHD Cases (multiple types) | 410 | 47 | 74 | ||
| Isolated CHD cases | 31 | 26 | |||||
| Syndromic CHD cases | 248 | 16 | 48 | ||||
| Pathogenic CNV Isolated cases | 162 | 5 | 5 | ||||
| Pathogenic CNV Isolated and Syndromic Cases | 8 | 19 | |||||
|
| |||||||
| White 2014 [25] | HumanHap550 V3 | CHD Cases (multiple types) | 257 | 122 | 83 | 51 | |
| Pathogenic Inherited CNV All cases (c) | 68 | ||||||
| Pathogenic de novo CNV All Cases (c) | 5 | ||||||
|
| |||||||
| Glessner 2014 [10](d) | Omni 1M | CHD cases SNP genotype array (multiple types) | 415 | 60 | 97 | 204 | |
| WES | CHD cases WES | 358 | 46 | 65 | 146 | ||
| CHD Cases by Both methods | 233 | ||||||
| Pathogenic de novo CNV All Cases | 11 | 6 | 5 | ||||
| Pathogenic de novo CNV cases (extracardiac lesions) | 3 | 3 | 3 | ||||
|
| |||||||
| Warburton 2014 [18] | Nimblegen HD2 | CHD Cases (Conotruncal and HLHS) | 223 | 71 | |||
| Pathogenic de novo CNV All Cases | 9 | ||||||
- CNV pathogenicity not defined, therefore CNVs not included
- No parental testing; CoA group includes CoA, AS, and "other"
- large CNVs (>269 kb)
– No differentiation between isolated and extracardiac; de novo events in isolated stated at 6% and in extracardiac at 31%
The primary purpose of this study was to identify large de novo genomic events of potential clinical impact in a large cohort of individuals with LSLs. At the time of recruitment all probands had what was considered to be isolated LSL. We catalogued the clinical presentations of the probands with large events and compared the regions and genes identified with CNVs observed in a large database of controls and with previous reports. Finally, we established the rate of CNVs among individuals with isolated LSLs, demonstrating that CNVs represent a clinically important cause of these defects.
MATERIALS AND METHODS
Samples and Subjects
Probands and their family members were enrolled through three sites: a) Texas Children’s Hospital (TCH) in Houston, Texas, b) Nationwide Children’s Hospital (NCH) in Columbus, Ohio c) Primary Children’s Hospital (PCH), Salt Lake City, Utah, under the respective IRB approved protocols at each institution. In addition, 49 parent-proband trios with HLHS were recruited at Children’s Hospital in Linz, Austria (CHL) and processed alongside the TCH cohort. Families enrolled through TCH, NCH, CHL are extensions of previously reported cohorts [Hanchard et al 2016; Lewin et al 2004; McBride et al 2009]. Patients were eligible for the study if they were willing to have DNA taken and banked, and had a characteristic LSL lesion without overt extra-cardiac involvement; i.e. non-dysmorphic/non-syndromic in the opinion of the treating physician, and no overt suspicion of other birth defects at the time of enrollment. The primary inclusion criterion was a congenital LSL cardiac defect; therefore, all age-groups were eligible for enrollment, although the vast majority were recruited in infancy and early childhood (PCH median age at recruitment 9.25 years, range: birth – 46 years; TCH median age 3.7 years, range: birth – 45 years; NCH median age 9.1 years, range: birth – 46 years; 67% male). Recruitment started in 2001 at TCH, 2006 at NCH, and 2009 at PCH and is ongoing at all three sites. Several recruited individuals were followed clinically as part of their standard of care, and in some cases the full clinical phenotype further evolved over time; nonetheless, these individuals were retained in the study based on their evaluation at the time of enrollment. Eligible LSLs were defined as: congenital AS – including BAV; COA; HLHS – mitral valve atresia or stenosis and aortic valve atresia or stenosis with hypoplasia of the left ventricle and aortic arch; Shone complex; and mitral atresia or stenosis (MA/MS). We also included individuals if they had a BAV in addition to their primary malformation. Diagnoses were confirmed by echocardiography, cardiac catheterization, or direct observation at cardiac surgery. These studies were approved by the Institutional Review Boards of Baylor College of Medicine and Nationwide Children’s Hospital under the auspices of the National Heart Lung and Blood Institute of the National Institutes of Health (for CHL samples). Subjects from PCH were enrolled under Utah Institutional Review Board (Protocol #00021080) and cases were defined in the same manner as the other recruited cohorts. After obtaining informed consent in writing (from parents/guardians for patients less than 18 years of age), either saliva (for geographically distant relatives) or blood samples were obtained. Lymphoblastic transformation by Ebstein-Barr Virus was undertaken to establish immortal cell lines in all NCH and ~90% of TCH proband samples. Remaining TCH proband DNA samples were from saliva. DNA from all samples was extracted, processed, and quantified using standard protocols. We did not separate our analyses on the basis of specific LSL lesions or geographical ascertainment; epidemiologically, the phenotypic and developmental overlap between LSLs justifies a combined analysis [Botto et al 2007] and the case definition used was uniform across included sites. Details of the LSL diagnosis demography and family structures for all recruited probands are given in (Supplementary Table 1).
SNP array genotyping of LSL families
Genotyping was undertaken in two phases, each on a separate, but related microarray platform. In the first phase (hereafter referred to as the primary cohort 797 probands and 1047 parents and siblings were genotyped using an iScan system for HumanOmniExpress-12 v1.0 BeadChips (Illumina, CA) according to manufacturer’s instructions. These microarrays include ~750,000 markers and are designed to provide genome-wide SNP coverage (LD > 0.85) in persons of European ancestry. Resulting iDAT files were uploaded to GenomeStudio software (Illumina, CA) and were used to interpret normalized fluorescent intensities as genotypes. SNPs with genotyping efficiency < 95% and samples with < 98% genotypes were removed.
The second phase of genotyping consisted of 342 probands genotyped on the HumanOmniExpressExome chip v 1.2, which includes SNPs found on the OmniExpress chip as well as rare variants discovered from recent population-based exome sequencing. For this analysis, samples with genotyping efficiency < 95% were excluded and the two cohorts were analyzed separately. The final analysis incorporated data for a total of 1139 affected cases genotyped at ~733,202 SNPs in the family-based cohort and ~961,073 SNPs in the proband-only cohort.
CNV analysis
In the family-based dataset, an annotated file of the LogR Ratio, B-allele frequency and corresponding genotype for each SNP was output and adapted for use with PennCNV [Wang et al 2007]. All chromosome positions were translated to build hg19 using the UCSC lift-over tool (https://genome.ucsc.edu/cgi-bin/hgLiftOver). CNV analysis was subsequently performed using GC wave adjustment and automatic quality control using a standard deviation for LogR Ratio of 0.2. Pericentromeric, telomeric and immunoglobulin regions were omitted from analysis. Similarly, we also omitted CNV calls with fewer than 50 SNPs in the intervening region, or data from the sex chromosomes. Fewer than 10% of samples failed sample QC in PennCNV. Probands with multiple CNV calls (> 10 of any size) were removed from analysis as possible cell line artifacts. Where a proband had multiple CNV calls, the chromosome and genomic coordinates were used to resolve the difference between multiple unrelated CNVs and boundary effects in which one large CNV may have been called as several smaller CNVs. For trio analysis, we restricted our analysis to complete trios where all members of the trio passed sample QC as implemented above. PLINK [Purcell et al 2007] was used to identify non-Mendelian inheritance and gender discrepancies that might suggest a sample swap or mislabeling. The updated trios were then passed to PennCNV. CNV partition v3.1.6 was used to visually validate large de novo CNVs called from PennCNV. We used parameter settings of a confidence of 50 and minimum of 5 SNPs to call any CNV. Large CNVs were confirmed by visual inspection of the chromosome plots of BAF and LRR generated in the Chromosome Browser of GenomeStudio, and by comparing the CNV call from PennCNV in the same region. These large event CNVs were of high confidence and were easily detectable using either software. In phase 2 (proband-only dataset), CNV detection was limited to CNVpartition using the same parameters as above. In our analyses we observed a large number of uncertain duplication (gain) events between 1 Mb and 1.5 Mb upon visual inspection and so limited our analysis to deletions (losses) larger than 1 Mb and duplications (gains) > 1.5 Mb.
These large CNVs were validated using a quantitative PCR assay in the affected proband and tested in the available parents using the qBiomarker Copy Number PCR Assay system from Qiagen, following kit protocol. This included families where parental samples were available, but had not undergone genome-wide SNP genotyping. Briefly, for each individual, reactions were assembled using a multi-copy reference assay (cat. No. VPH000-0000000A) and a site-specific copy number PCR assay (see Supplemental Data). For each assay, reactions were completed in quadruplicate. Calibrator DNA was also included on each tray as a control. Each ten-microliter reaction contained 8ng genomic DNA and the recommended volumes of qBiomarker SYBR Master Mix (cat. no. 337820) and PCR Assay, either multi-reference or site specific. Thermal cycling was completed on an Applied Biosystems 7500 FAST Real-Time PCR System using recommended conditions (FAST conditions were not used). For NCH and Utah samples, if EBV-transformed lymphoblasts were used for the microarray-based analyses, primary white blood cell DNA was used for molecular confirmation. We also confirmed large CNV calls by comparing them with calls performed on the same samples using a different pipeline [Prakash et al 2016] as part of another study of CNVs in bicuspid aortic valve. All large CNVs reported here were also observed and reported by the BAV pipeline.
Control Datasets
Datasets from the Database of Genotypes and Phenotypes (dbGAP) were used as comparison control datasets and consisted of two datasets – the first has been previously used in a study of thoracic aortic aneurysm [Prakash et al 2010]; the other was matched for the OmniExpress platform. Data was derived from Genetic Epidemiology of Refractive Error in the KORA (Kooperative Gesundheitsforschung in der Region Augsburg) Study, dbGaP accession phs000303.v1.p1 (n=1865); The Genetic Architecture of Smoking and Smoking Cessation, phs000404.v1.p1 (n=935); A Genome-Wide Association Study of Fuchs' Endothelial Corneal Dystrophy (FECD), phs000421.v1.p1 (n=3218); and Health and Retirement Study (HRS), phs000428.v1.p1 (n=9428) for a total control set of n=15446. CNVs in both datasets were called using PennCNV and CNV partition.
Bioinformatics
Gene lists were generated by importing the coordinates of the CNV regions into Biomart (http://www.ensembl.org/biomart/martview). Gene Ontology terms GO: 0048598 “embryonic morphogenesis, and GO:0072358 “cardiovascular development” were used in Amigo (http://amigo.geneontology.org/amigo) to create a subset of genes important in heart development from the list of all genes in the CNV intervals. PhenoGram (http://visualization.ritchielab.psu.edu/) was used to illustrate overlapping CNVs.
Pathway and network analysis was performed using two groups of data. The first group consisted of CNVs associated with CHDs as described by [Thorsson et al 2015], which was used as a reference framework. Genes within CNV intervals between 1Mb and 10Mb in size and also present in at least five described cases were chosen for analysis. The second group consisted of the CNVs identified in this study which included de novo or transmitted deletion and duplication CNVs > 1Mb, with the exception of the complex chromosome 9 event which was excluded (Supplementary Table 3). The generated gene list was then imported into Ingenuity Pathway Analysis web server (IPA, Ingenuity Systems). The reference set for comparison was the Ingenuity Knowledgebase gene set. Default analysis settings were used, including: generation of interaction networks (maximum 35 molecules per network and 25 networks per analysis); all available data sources; consider only experimentally observed relationships; and all mammalian species. The significance of biological pathways was assessed by Fisher’s exact test, and was adjusted for multiple testing via Benjamini-Hochberg analysis, with a false discovery rate of less than 0.05. Network scores were obtained using the hypergeometric distribution, with one-sided Fisher’s exact test.
RESULTS
Large genomic events in LSL
CNVs were classified as likely pathogenic if they were observed de novo in a proband from a trio, or if they were not present in our control dataset. We initially focused on de novo CNV events as those most likely to be pathogenic. We confirmed large de novo CNVs in 6 of the 379 parent-trio probands (~1.6%) and at least 14 potential de novo events (absent in the only available parent) among the remaining 760 probands (~1.8%) (Table 2). The majority of copy number losses were heterozygous and included a number of complex rearrangements, including two unbalanced translocations. We also observed 13 large events (1–2.7 Mb) that were transmitted from apparently unaffected parents. None of these large events were noted in our control datasets. In general, transmitted events were smaller than de novo events and were usually heterozygous copy number gains (duplication). We observed multiple instances of copy number change at five chromosomal regions – 11q24.2 to 11q25; 1q21.1, 15q11.2, 16p13 and 21q21.3 - with both gains and losses of chromosomal material being observed at all of these regions except 21q21.3 (two gains) (Table 2).
Table 2.
Large De Novo, Potential De Novo, and transmitted large CNVs in LSL. Shaded CNVs represent unbalanced translocations in a single individual. BAV - Bicuspid Aortic Valve; CoA - coarctation of the Aorta; MS- Mitral Stenosis; AVS - aortic valve Stenosis; AR - Aortic regurgitation; AO - Aortic obstruction; HLHS - Hypoplastic Left Heart Syndrome; SC- Shone Complex; TGA – Transposition of the Great Arteries; VSD – ventricular septal defect; Gend. – Gender; Ethn. – Self-reported ethnicity; Age – Age at recruitment.
| ID | Gend. | Ethn. | Age | Chromosome Location | Size (bp) | Cytoband | Copy No. |
LSL | Other | |
|---|---|---|---|---|---|---|---|---|---|---|
| De Novo | LO0607 | M | Ca | 8mo | chr10:81577613-88945341 | 7,367,728 | q23.1 - q23.2 | 1 | CoA, AO | |
|
| ||||||||||
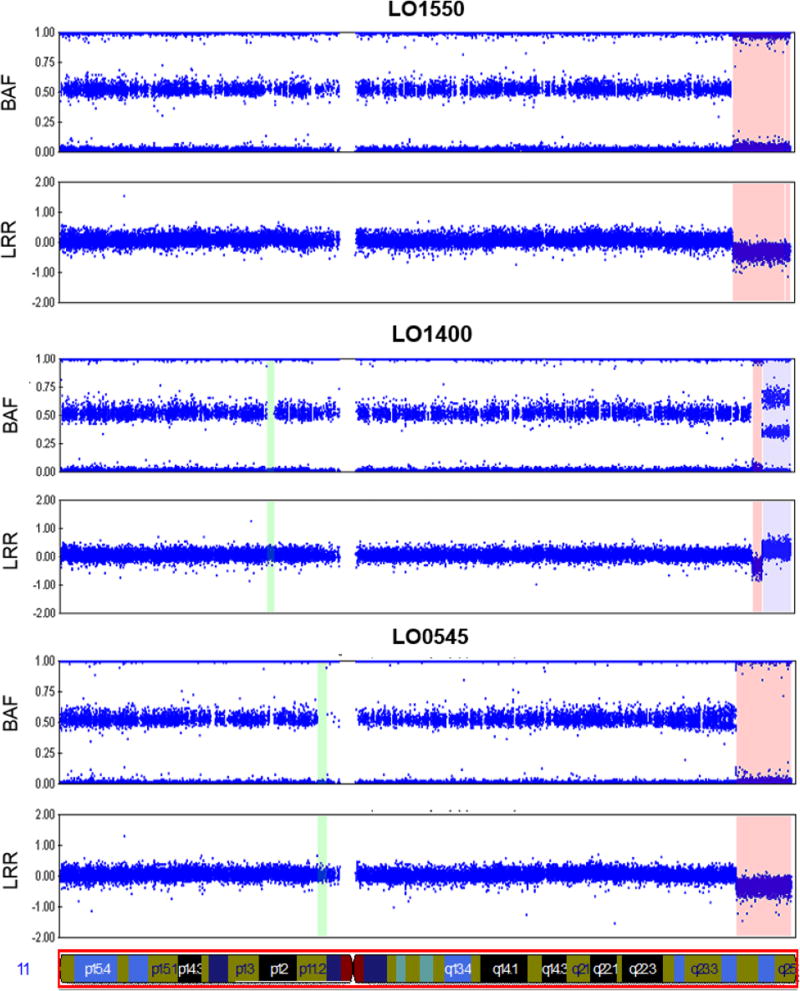
| LO1550 | M | H | 5dys | chr11:124323660-134934063 | 10,610,403 | q24.2 - q25 | 1 | CoA | Deceased; hypoplastic digits, bilateral renal cysts, single umbilical artery, cryptorchidism, del 11q24.1 detected at autopsy | |
|
| ||||||||||
| LO1400 | F | H | 9dys | chr11:124870136-134934063 | 10,063,927 | q24.2 - q25 | 1 | HLHS | Deceased; unusual facial features, del 11q24.1 detected at autopsy | |
|
| ||||||||||
| LO1705 | F | M | 1mo | chr18:75380175-78015180 | 2,635,005 | q23 | 1 | MS | Sib with AVS | |
| chr20:50494953-62801181 | 12,306,228 | q13.2 - q13.33 | 3 | |||||||
|
| ||||||||||
| OS0200 | M | Ca | 10yrs | chr19:509569-1539267 | 1,029,698 | p13.3 | 3 | BAV | Irregular facial mass (resected) with soft-tissue calcification | |
|
| ||||||||||
| LO1397 | F | Ca | 4mo | chr20:6981282-8343543 | 1,362,261 | p12.3 | 1 | SC | ||
|
| ||||||||||
| One parent available | OS0491 | M | Ca | 10yrs | chr3:67615531-69019978 | 1,404,447 | p14.1 | 1 | CoA | |
|
| ||||||||||
| UT0139 | M | Ca | 39 mo | chr4:177310481-188590745 | 11,280,264 | q34.3 - q35.2 | 1 | BAV, VSD | Had formal genetics consultation | |
|
| ||||||||||
| UT1758 | F | Ca | 7 dys | chr6:148690764-151430051 | 2,739,287 | q24.3 - q25.1 | 1 | CoA, HLHS | ||
|
| ||||||||||
| UT1089 | M | Ca | 12 yrs | chr8:7308659-11898209 | 4,589,550 | p23.1 | 3 | CoA | Autism Spectrum Disorder | |
|
| ||||||||||
| OS0920b | F | Ca | 5mo | chr9:71301557-141066491 | 69,764,934 | q21.11 - q34.3 | 3 | HLHS, PS | Karyotype 46, XX; nrml FISH for 22q11.2 | |
|
| ||||||||||
| LO0253* | F | Ca | 3mo | chr9:95580375-115042805 | 19,462,430 | q22.31 - q31.3 | 1 | CoA, TGA | Deceased - congenital abnormalities of kidneys and brain | |
|
| ||||||||||
| UT0372 | M | Ca | 8 yrs | chr10:46202216-47703869 | 1,501,653 | q11.22 | 3 | CoA, BAV, AVS | ||
|
| ||||||||||
| LO0545a | M | H | 6dys | chr11:129769925-134934063 | 5,164,138 | q24.3 - q25 | 3 | HLHS | ||
| chr11:127900579-129737988 | 1,837,409 | q24.3 | 1 | |||||||
|
| ||||||||||
| OS0010a | M | Ca | 15 yrs | chr14:83028831-87815467 | 4,786,636 | q31.1 - q31.3 | 3 | BAV | Aortic root dilation, Pulmonary insufficiency | |
| chr7:118777401-153071114 | 34,293,713 | q31.31 - q36.2 | 1 | |||||||
|
| ||||||||||
| OS0199 | M | Ca | 4 yrs | chr15:32922947-34807851 | 1,884,904 | q13.3 - q14 | 1 | BAV | Tourette's, ADHD | |
|
| ||||||||||
| UT2188 | M | Ca | 4 mo | chr16:14975292-16363239 | 1,387,947 | p13.11 | 3 | CoA | Clinical microarray showed 16p13.11 dup; Laryngomalacia, Hemivertebra, FH of CHD | |
|
| ||||||||||
| LO0445 | F | H | 5yrs | chr19:20788625-22175566 | 1,386,941 | p12 | 3 | CoA, AO | ||
|
| ||||||||||
| UT0240 | F | Ca | 4 yrs | chr20:7533180-9196090 | 1,662,910 | p12.3 | 3 | AVS, BAV | Post-operative choreiform movements; Dysarthria | |
|
| ||||||||||
| UT0690 | M | Ca | 6 yrs | chr22:18886915-21463730 | 2,576,815 | q11.2 | 3 | CoA | ||
|
| ||||||||||
| Transmitted | LO0821 | M | Ca | NR | chr1:146152553-147784656 | 1,632,103 | q21.1 | 3 | MA, AVS | |
|
| ||||||||||
| LO1821 | - | NR | chr1:146089254-147826789 | 1,737,535 | q21.1 -q21.2 | 1 | - | |||
|
| ||||||||||
| OS0007 | M | Ca | 9yrs | chr2:111392259-112816047 | 1,423,788 | q13 | 3 | HLHS | Anxiety, depression, Post-traumatic stress Disorder | |
|
| ||||||||||
| LO2313 | Ca | 18yrs | chr4:186366615-188924979 | 2,558,364 | q35.1 - q35.2 | 1 | AVS, AR, PVCs | |||
|
| ||||||||||
| OS1625 | F | Ca | 1yr | chr6:149609-1533447 | 1,383,838 | p25.3 | 1 | AVS, MS | Platelet deficiency, sleep apnea, Chiari malformation type I, Epilepsy; sub-aortic membrane, tortuous aortic arch | |
|
| ||||||||||
| OS0782 | M | Ca | 18yrs | chr15:20612840-22576118 | 1,963,278 | q11.2 | 3 | CoA | Anxiety | |
|
| ||||||||||
| UT2628 | M | Ca | 4 yrs | chr15:22652330-23656946 | 1,004,616 | q11.2 | 1 | BAV, PDA | Uncle with Congenital Heart Disease | |
|
| ||||||||||
| OS0939 | M | Ca | 9yrs | chr16:15092778-16303388 | 1,210,610 | p13.11 | 1 | CoA | Wolf-Parkinson-White syndrome, idiopathic epilepsy | |
|
| ||||||||||
| UT0573 | F | Ca | 1 mo | chr16:15369019-18164698 | 2,795,679 | p13.11 - p12.3 | 3 | CoA, BAV | ||
|
| ||||||||||
| LO0668 | M | H | 9.5yrs | chr18:6936251-8080357 | 1,144,106 | p11.31 - p11.23 | 3 | AS, BAV | ||
|
| ||||||||||
| LO0210 | M | H | 1yr | chr19:27795706-29612355 | 1,816,649 | q12 | 3 | CoA, AO | Hemiparesis, developmental delay, seizures, microcephaly | |
|
| ||||||||||
| LO0735 | M | H | 1.5yrs | chr21:28240392-29417762 | 1,177,370 | q21.3 | 3 | HLHS | Deceased; Scoliosis, microcephaly, neurogenic bladder | |
|
| ||||||||||
| LO1186 | M | H | 15yrs | chr21:28244672-29422978 | 1,178,306 | q21.3 | 3 | CoA, AO | ||
The clinical features of probands with large events ranged from uncomplicated LSL lesions to the subsequent revelation of a syndromic diagnosis (Table 2). Three samples had evidence for CNVs at chromosome 11q24q25 - two individuals had large deletions of the region and a third had a complex deletion-duplication-deletion variant (Figure 1). The 11q23q24 region is associated with Jacobsen syndrome (OMIM #147791), and although both of the neonates with the distal deletion of the region subsequently died because of their complex CHD, the genetic syndromic diagnosis was made at autopsy. Similarly, the two individuals with unbalanced translocations (LO1705; OS0010) were not suspected to be syndromic at the time of recruitment, despite having very large events. At the other end of the spectrum, the majority of individuals with large events were non-syndromic with relatively uncomplicated and unambiguous LSLs. In between these extremes were individuals with more complex LSLs and/or neurodevelopmental co-morbidities detected over time. We found these general observations to be independent of gender and ancestry (Table 2).
Figure 1.
Large CNVs on chromosome 11q24-q25 detected by SNP microarray. Figure shows the logR Ratio (LRR) and B-allele frequency (BAF) of SNPs genotyped across chromosome 11 in three LSL probands. Loss of single copy number is shaded in orange; gain of a single copy is shaded in purple.
Overlap between large genomic events in LSL and cardiac developmental loci
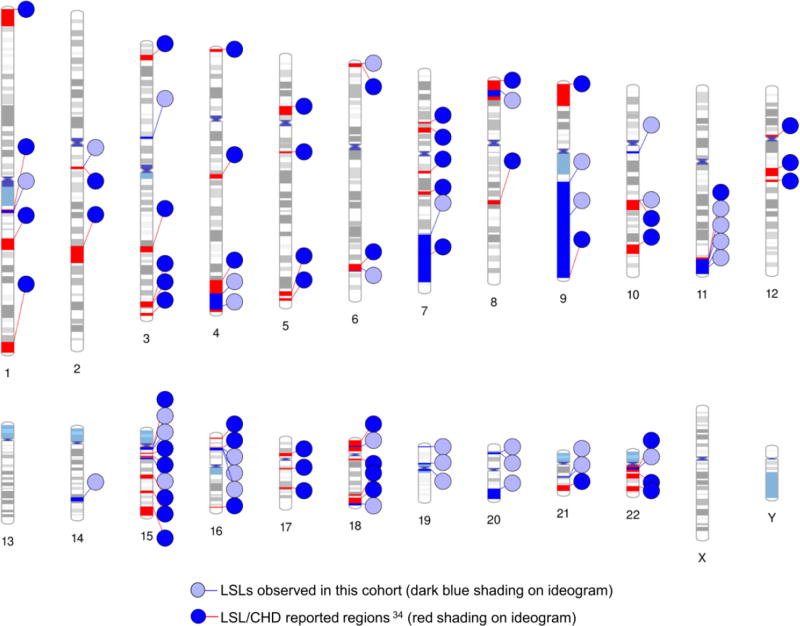
Most of the large genomic events we observed were singleton events; therefore, we attempted to place the observed events into a broader gene-based context. When we analyzed the genes in all identified CNV regions we found that 120 of the 818 contained genes (~15%) were associated with the biologic process GO:0048598 “embryonic morphogenesis”, and 36 (4.4%) were associated with GO:0072358 “cardiovascular development” (Supplementary Table 3). Repeating the GO term query without limiting the search space to the genes in our identified CNV regions identified 919 genes (from a total of 45,493 genes (2%) – 20,313 protein coding genes and 25,180 non-coding genes in Ensembl) associated with “cardiovascular development” (GO:0072358). Comparison of the proportion of “cardiovascular development” genes in the CNV regions versus those in the entire genome demonstrated a statistically significant enrichment in our CNV regions (Fisher’s exact 2-sided p<0.001). The Ingenuity Pathway Analysis (Supplementary Table 2) corroborated these findings, demonstrating a statistically significant excess of genes and processes involved in embryonic development and cardiac development or cardiac defects in multiple groups.
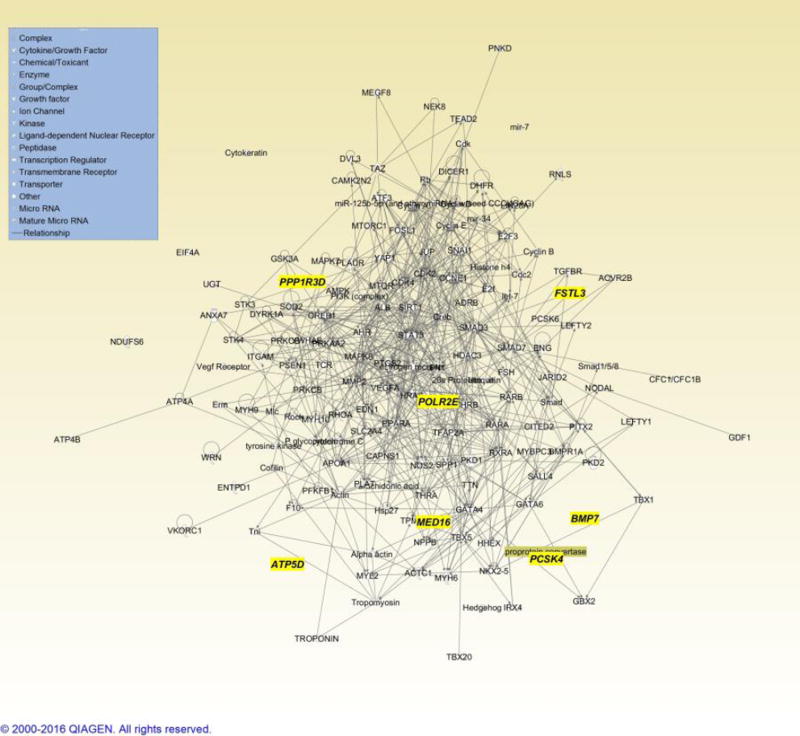
Half of the large events observed in our cohort (14/33) occurred outside of previous established CHD genomic regions [Thorsson et al 2015] (Figure 2). Given the enrichment for cardiac developmental genes among our CNVs, we considered whether regions outside of established regions might provide insight to new CHD genes. In order to provide a framework of reference genes that would be relevant to CHD, we first interrogated the gene content of more established CHD CNVs, proxied by recurrent events recorded by [Thorsson et al 2015](see Methods and Supplemental Table 3). In this analysis, genes and processes involved in embryonic development and cardiac development or cardiac defects were represented in multiple groups that contained other genes not relevant to heart development; therefore we generated a secondary list by merging networks with cardiovascular development. The resulting network contained genes important in heart development, including transcription factors (GATA4, TBX1, NKX2-5), cell signaling pathways (Wnt, Tgf beta), laterality, extra-cellular matrix (integrins), and a variety of mediators (Supplementary Table 3 and 4). We then overlaid genes contained in non-overlapping CNV intervals from this study (Supplementary Table 3) onto this merged network (Figure 3). Seven genes (ATP5D, BMP7, FSTL3, MED16, PCSK4, POLR2E, and PPP1R3D) in the CNV intervals from this study intersected this network, and are involved in key developmental processes, including signaling in development and chromatin remodeling.
Figure 2.
CNV loci in CHD patients. The overlap of large CNV events observed in this study (light blue circles, blue lines, dark blue shading) with large (1–10 mb) CNV regions reported in more than five CHD cases by Thorsson et. al.36 (dark blue circles, red lines, red shading).
Figure 3.
Cardiac development candidate genes (yellow labels) inferred from ‘unique’ large CNV regions identified. Inferred cardiac development genes and corresponding network are shown in grey shading.
DISCUSSION
We undertook CNV analysis in a cohort of 1,139 LSL patients and their families, with the express purpose of assessing the contribution of large autosomal CNVs of likely pathogenicity. This represents one of the largest LSL cohorts so far reported. Despite focusing exclusively on large copy number events, we found likely-pathogenic, clinically relevant, de novo (or potentially de novo) autosomal CNVs in ~1.8% of individuals (20/1139). This figure is congruent with the 2% de novo mutation rate recently reported for single nucleotide variants (SNV) identified by whole exome sequencing in isolated CHD [Homsy et al 2015], and suggests that the contribution of de novo CNV formation is likely as important to the pathogenesis of non-syndromic LSLs as SNVs. The CNV events observed in this study affected probands with a variety of cardiac phenotypes spanning LSLs with complex CHD to isolated LSLs, and included individuals of both Hispanic and European ancestry. Some of the individuals recruited as infants or neonates were subsequently noted to have additional neurodevelopmental features - in the neonatal period, particularly when children with CHD are critically ill and require cardiac support, syndromic genetic diagnoses can be challenging to make for treating physicians, especially in the context of pressing clinical priorities (e.g. cardiac instability); nonetheless, our data would suggest that in the absence of clear syndromic, dysmorphic, or extra-cardiac features, as assessed by primary physicians, a genomic and genetic evaluation is still warranted, especially in cases where a full clinical genetic evaluation is not readily available. The potential clinical yield of assessing large CNVs appears to be applicable to most cases of LSL with or without overt extra-cardiac features, a finding consistent with a recent study of CNVs in isolated CHD [Kim et al 2016].
Most of the large genomic events involved copy number loss. This is consistent with the few previous assessments of CNVs in CHD, and LSLs specifically and is congruent with copy number losses being more associated with a clinical phenotype. More than half of the regions implicated in our study have been previously reported as being associated with CHD or LSLs [Thorsson et al 2015] and, also consistent with previous reports, most events were seen once in the cohort. Recurrent events included gains and losses on 1q21.1 and 15q11.2. Haploinsufficiency at both loci has been associated with a clinical phenotype that includes CHD, although both non-penetrance and/or variable expressivity have been frequently observed [Brunetti-Pierri et al 2008; Glessner et al 2014; Rosenfeld et al 2013]; this is consistent with our observation of inheritance from apparently unaffected parents. Visual inspection of these CNVs in parents did not suggest mosaicism. We did not, however, systematically evaluate parents for phenotypically milder cardiac phenotypes. As such the designation of ‘unaffected’ does not exclude variable expressivity of the observed CNVs nor does it preclude ascribing pathogenicity to these CNVs. These observations are consistent with recent studies of isolated CHD in which rare loss-of-function variants in cardiac genes were also often inherited [Sifrim et al 2016].
We observed three overlapping large events at chromosome 16p13.11 – two of which were inherited from overtly unaffected parents. Smaller CNVs in both the 16p13.11 and 15q11 regions have been noted in datasets not ascertained by cardiovascular phenotype. 16p13.11 includes the MYH11 gene (MIM #160745), which has been implicated in Familial Thoracic Aortic Aneurysms and Dissections (FTAAD) [Kuang et al 2011], a diagnosis typically arising at or beyond the 5th decade of life, as a well as patent ductus arteriosus (PDA) – diagnosed in the neonatal period. None of the patients with CNVs in this region had a PDA; however, all three had coarctation of the aorta, underscoring the importance of MYH11 to vascular smooth muscle. Our results imply that MYH11 variation has highly variable and pleiotropic expressivity, contributing to both an increased susceptibility to FTAAD among adult carriers and a congenital LSL aortopathy phenotype in children and neonates. These observations further highlight the complex genetics underlying LSLs and may underlie the increased incidence of milder LSL lesions seen in parents of affected offspring, for whom parental echocardiograms are recommended [Kerstjens-Frederikse et al 2011; Lewin et al 2004] but seldom obtained.
We also observed three events at chromosome 11q24 to 11qter – two de novo deletions and one complex unbalanced translocation of uncertain inheritance. This region is at the distal end of the chromosome 11q23 region that is associated with Jacobsen syndrome (MIM # 147791), in which LSLs are well described [Grossfeld et al 2004]. A consistent clinical challenge is the diagnostic evaluation of newborns and infants with apparently isolated CHD; features of neurodevelopmental delay (NDD) may only be recognized later in infancy or childhood, and clinically-relevant dysmorphism may erroneously be thought to be related to medical, surgical or anesthetic intervention or obscured in early evaluations. The classical presenting features of Jacobsen syndrome include developmental delay and a variety of non-diagnostic dysmorphic features that can be difficult to appreciate in newborns. This is particularly true in cardiac or neonatal ICUs where the acuity of clinical care can limit physical examination. Among our three cases, two infants died as a result of their cardiac lesion. It may be that distal CNVs in this region are particularly important to the cardiac phenotype of Jacobsen syndrome as opposed to the cognitive and developmental features. This distal region includes the ETS1 gene that has been recently postulated as a candidate gene for CHD in Jacobsen syndrome [Ye et al 2010].
Aside from CNVs affecting known CHD-genes and loci in our cohort, a collective view of the novel regions implicated in our cohort (i.e. not overlapping previously described regions) also suggested enrichment for genes known to be involved in cardiac development or CHD. For instance, our analysis implicated seven genes, including BMP7 - a member of the TGF-beta signaling network, which is known to play a substantial role in cardiac development. In murine models, Bmp7 and Bmp6 appear to interact in the formation in the endocardial cushions of the developing cardiac outflow tract[Kim et al 2001], and in humans, BMP7 is implicated in congenital abnormalities of the kidney and urinary tract (CAKUT) , which shares substantial overlap with cardiac developmental genetics [San Agustin et al 2016]. The genes identified by this analysis therefore represent candidates for gene-based surveys of pathogenic variation in CHDs [Homsy et al 2015; Zaidi et al 2013], and could aid in the interpretation of smaller CNVs among CHD cases, which would, in turn, further improve the clinical yield of copy number testing in CHD.
We chose to focus strictly on large events for both clinical and technical reasons. In general, large events typically encompass a large number of genes, and thus are likely to be clinically relevant from a diagnostic standpoint [Cooper et al 2011; Girirajan et al 2011]. Large pathogenic CNVs may also have implications for medical management; for example, the post-operative prognosis (and implications) for patients with cyto-genomic disorders may be different from cases with other causes. Second, calling algorithms for CNVs using SNP genotyping arrays are known to be imprecise for smaller events. Concordance between calling algorithms on SNP microarray data is low – 50% or less in identifying deletions or duplications[Eckel-Passow et al 2011] - thus most studies employ multiple algorithms and report overlap, as we did here. In comparison of SNP genotype data against gold-standard (but more expensive) aCGH, there are high false positive and false negative rates, with many events under 1 Mb missed. The array platform we chose allowed collection of data from a large number of individuals at low cost, providing a reasonable estimate of pathogenic and likely pathogenic CNVs in isolated LSLs. Future studies of these individuals by whole-genome sequencing technology will allow a more accurate and much finer resolution of CNVs.
Our report suggests that including an assessment for large, de novo, autosomal CNVs in congenital LSL can uncover variants of clinical importance; this is consistent with similar smaller surveys[Bachman et al 2015]. Such evaluations also have the potential to uncover novel genomic regions that are important to cardiovascular development and pathology. Studies of larger CHD and cardiovascular cohorts across a multiple of modalities are likely to bear fruit for enumerating the full spectrum of CHD genes and the mechanisms underlying their pathogenesis.
Supplementary Material
Supplemental Table S1 – a. Demography of PCH case cohort; CoA – coarctation of the aorta; AS = Aortic Stenosis; HLHS= Hypoplastic Left Heart Syndrome; b. Summary of genotyped family pedigrees for all cohorts.
Supplemental Table S2 – Ingenuity Pathway Analysis summary
Supplemental Table S3 – Gene Lists, GO Terms used in pathway analyses
Supplemental Table S4 – IPA top networks
Acknowledgments
The authors would like to acknowledge the many families and auxiliary staff who participated in the study. This work was funded in part by National Institutes of Health (NIH) grants to JWB (1U54 HD083092, 5RO1 HD039056, 5RO1 HL090506, 5RO1 HL091771) and KM (5 R01 HL109758 and 1R21 HL106549-01). NH is funded by a Clinical Scientist Development Award from the Doris Duke Charitable Foundation (Grant #:2013096). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Conflicts of Interest Statement
J.W.B. is a fulltime employee of Illumina Inc., but all work was performed under the listed affiliation. The remaining authors have no conflicts of interest to declare.
References
- Bachman KK, DeWard SJ, Chrysostomou C, Munoz R, Madan-Khetarpal S. Array CGH as a first-tier test for neonates with congenital heart disease. Cardiol Young. 2015;25:115–122. doi: 10.1017/S1047951113001868. [DOI] [PubMed] [Google Scholar]
- Botto LD, Lin AE, Riehle-Colarusso T, Malik S, Correa A, National Birth Defects Prevention S Seeking causes: Classifying and evaluating congenital heart defects in etiologic studies. Birth Defects Res A Clin Mol Teratol. 2007;79:714–727. doi: 10.1002/bdra.20403. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, Lalani SR, Graham B, Lee B, Shinawi M, Shen J, Kang SH, Pursley A, Lotze T, Kennedy G, Lansky-Shafer S, Weaver C, Roeder ER, Grebe TA, Arnold GL, Hutchison T, Reimschisel T, Amato S, Geragthy MT, Innis JW, Obersztyn E, Nowakowska B, Rosengren SS, Bader PI, Grange DK, Naqvi S, Garnica AD, Bernes SM, Fong CT, Summers A, Walters WD, Lupski JR, Stankiewicz P, Cheung SW, Patel A. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor JA, Hinton RB, Miller EM, Sund KL, Ruschman JG, Ware SM. Genetic testing practices in infants with congenital heart disease. Congenit Heart Dis. 2014;9:158–167. doi: 10.1111/chd.12112. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, Abdel-Hamid H, Bader P, McCracken E, Niyazov D, Leppig K, Thiese H, Hummel M, Alexander N, Gorski J, Kussmann J, Shashi V, Johnson K, Rehder C, Ballif BC, Shaffer LG, Eichler EE. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czosek RJ, Anderson JB, Heaton PC, Cassedy A, Schnell B, Cnota JF. Staged palliation of hypoplastic left heart syndrome: trends in mortality, cost, and length of stay using a national database from 2000 through 2009. Am J Cardiol. 2013;111:1792–1799. doi: 10.1016/j.amjcard.2013.02.039. [DOI] [PubMed] [Google Scholar]
- Eckel-Passow JE, Atkinson EJ, Maharjan S, Kardia SL, de Andrade M. Software comparison for evaluating genomic copy number variation for Affymetrix 6.0 SNP array platform. BMC Bioinformatics. 2011;12:220. doi: 10.1186/1471-2105-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison JW, Ravnan JB, Rosenfeld JA, Morton SA, Neill NJ, Williams MS, Lewis J, Torchia BS, Walker C, Traylor RN, Moles K, Miller E, Lantz J, Valentin C, Minier SL, Leiser K, Powell BR, Wilks TM, Shaffer LG. Clinical utility of chromosomal microarray analysis. Pediatrics. 2012;130:e1085–1095. doi: 10.1542/peds.2012-0568. [DOI] [PubMed] [Google Scholar]
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Geng J, Picker J, Zheng Z, Zhang X, Wang J, Hisama F, Brown DW, Mullen MP, Harris D, Stoler J, Seman A, Miller DT, Fu Q, Roberts AE, Shen Y. Chromosome microarray testing for patients with congenital heart defects reveals novel disease causing loci and high diagnostic yield. BMC Genomics. 2014;15:1127. doi: 10.1186/1471-2164-15-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Brkanac Z, Coe BP, Baker C, Vives L, Vu TH, Shafer N, Bernier R, Ferrero GB, Silengo M, Warren ST, Moreno CS, Fichera M, Romano C, Raskind WH, Eichler EE. Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet. 2011;7:e1002334. doi: 10.1371/journal.pgen.1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Bick AG, Ito K, Homsy JG, Rodriguez-Murillo L, Fromer M, Mazaika E, Vardarajan B, Italia M, Leipzig J, DePalma SR, Golhar R, Sanders SJ, Yamrom B, Ronemus M, Iossifov I, Willsey AJ, State MW, Kaltman JR, White PS, Shen Y, Warburton D, Brueckner M, Seidman C, Goldmuntz E, Gelb BD, Lifton R, Seidman J, Hakonarson H, Chung WK. Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circ Res. 2014;115:884–896. doi: 10.1161/CIRCRESAHA.115.304458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmuntz E, Paluru P, Glessner J, Hakonarson H, Biegel JA, White PS, Gai X, Shaikh TH. Microdeletions and microduplications in patients with congenital heart disease and multiple congenital anomalies. Congenit Heart Dis. 2011;6:592–602. doi: 10.1111/j.1747-0803.2011.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway SC, Pereira AC, Lin JC, DePalma SR, Israel SJ, Mesquita SM, Ergul E, Conta JH, Korn JM, McCarroll SA, Gorham JM, Gabriel S, Altshuler DM, Quintanilla-Dieck Mde L, Artunduaga MA, Eavey RD, Plenge RM, Shadick NA, Weinblatt ME, De Jager PL, Hafler DA, Breitbart RE, Seidman JG, Seidman CE. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet. 2009;41:931–935. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossfeld PD, Mattina T, Lai Z, Favier R, Jones KL, Cotter F, Jones C. The 11q terminal deletion disorder: a prospective study of 110 cases. Am J Med Genet A. 2004;129A:51–61. doi: 10.1002/ajmg.a.30090. [DOI] [PubMed] [Google Scholar]
- Hanchard NA, Swaminathan S, Bucasas K, Furthner D, Fernbach S, Azamian MS, Wang X, Lewin M, Towbin JA, D'Alessandro LC, Morris SA, Dreyer W, Denfield S, Ayres NA, Franklin WJ, Justino H, Lantin-Hermoso MR, Ocampo EC, Santos AB, Parekh D, Moodie D, Jeewa A, Lawrence E, Allen HD, Penny DJ, Fraser CD, Lupski JR, Popoola M, Wadhwa L, Brook JD, Bu'Lock FA, Bhattacharya S, Lalani SR, Zender GA, Fitzgerald-Butt SM, Bowman J, Corsmeier D, White P, Lecerf K, Zapata G, Hernandez P, Goodship JA, Garg V, Keavney BD, Leal SM, Cordell HJ, Belmont JW, McBride KL. A genome-wide association study of congenital cardiovascular left-sided lesions shows association with a locus on chromosome 20. Hum Mol Genet. 2016;25:2331–2341. doi: 10.1093/hmg/ddw071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitz MP, Lemieux-Perreault LP, Marshall C, Feroz-Zada Y, Davies R, Yang SW, Lionel AC, D'Amours G, Lemyre E, Cullum R, Bigras JL, Thibeault M, Chetaille P, Montpetit A, Khairy P, Overduin B, Klaassen S, Hoodless P, Awadalla P, Hussin J, Idaghdour Y, Nemer M, Stewart AF, Boerkoel C, Scherer SW, Richter A, Dube MP, Andelfinger G. Rare copy number variants contribute to congenital left-sided heart disease. PLoS Genet. 2012;8:e1002903. doi: 10.1371/journal.pgen.1002903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, Jin SC, Deanfield J, Giardini A, Porter GA, Jr, Kim R, Bilguvar K, Lopez-Giraldez F, Tikhonova I, Mane S, Romano-Adesman A, Qi H, Vardarajan B, Ma L, Daly M, Roberts AE, Russell MW, Mital S, Newburger JW, Gaynor JW, Breitbart RE, Iossifov I, Ronemus M, Sanders SJ, Kaltman JR, Seidman JG, Brueckner M, Gelb BD, Goldmuntz E, Lifton RP, Seidman CE, Chung WK. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstjens-Frederikse WS, Du Marchie Sarvaas GJ, Ruiter JS, Van Den Akker PC, Temmerman AM, Van Melle JP, Hofstra RM, Berger RM. Left ventricular outflow tract obstruction: should cardiac screening be offered to first-degree relatives? Heart. 2011;97:1228–1232. doi: 10.1136/hrt.2010.211433. [DOI] [PubMed] [Google Scholar]
- Kim DS, Kim JH, Burt AA, Crosslin DR, Burnham N, Kim CE, McDonald-McGinn DM, Zackai EH, Nicolson SC, Spray TL, Stanaway IB, Nickerson DA, Heagerty PJ, Hakonarson H, Gaynor JW, Jarvik GP. Burden of potentially pathologic copy number variants is higher in children with isolated congenital heart disease and significantly impairs covariate-adjusted transplant-free survival. J Thorac Cardiovasc Surg. 2016;151:1147–1151. doi: 10.1016/j.jtcvs.2015.09.136. e1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RY, Robertson EJ, Solloway MJ. Bmp6 and Bmp7 are required for cushion formation and septation in the developing mouse heart. Dev Biol. 2001;235:449–466. doi: 10.1006/dbio.2001.0284. [DOI] [PubMed] [Google Scholar]
- Kuang SQ, Guo DC, Prakash SK, McDonald ML, Johnson RJ, Wang M, Regalado ES, Russell L, Cao JM, Kwartler C, Fraivillig K, Coselli JS, Safi HJ, Estrera AL, Leal SM, Lemaire SA, Belmont JW, Milewicz DM, Gen TACI. Recurrent chromosome 16p13.1 duplications are a risk factor for aortic dissections. PLoS Genet. 2011;7:e1002118. doi: 10.1371/journal.pgen.1002118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani SR, Belmont JW. Genetic basis of congenital cardiovascular malformations. Eur J Med Genet. 2014;57:402–413. doi: 10.1016/j.ejmg.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani SR, Shaw C, Wang X, Patel A, Patterson LW, Kolodziejska K, Szafranski P, Ou Z, Tian Q, Kang SH, Jinnah A, Ali S, Malik A, Hixson P, Potocki L, Lupski JR, Stankiewicz P, Bacino CA, Dawson B, Beaudet AL, Boricha FM, Whittaker R, Li C, Ware SM, Cheung SW, Penny DJ, Jefferies JL, Belmont JW. Rare DNA copy number variants in cardiovascular malformations with extracardiac abnormalities. Eur J Hum Genet. 2013;21:173–181. doi: 10.1038/ejhg.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin MB, McBride KL, Pignatelli R, Fernbach S, Combes A, Menesses A, Lam W, Bezold LI, Kaplan N, Towbin JA, Belmont JW. Echocardiographic evaluation of asymptomatic parental and sibling cardiovascular anomalies associated with congenital left ventricular outflow tract lesions. Pediatrics. 2004;114:691–696. doi: 10.1542/peds.2003-0782-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KL, Pignatelli R, Lewin M, Ho T, Fernbach S, Menesses A, Lam W, Leal SM, Kaplan N, Schliekelman P, Towbin JA, Belmont JW. Inheritance analysis of congenital left ventricular outflow tract obstruction malformations: Segregation, multiplex relative risk, and heritability. Am J Med Genet A. 2005;134A:180–186. doi: 10.1002/ajmg.a.30602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KL, Zender GA, Fitzgerald-Butt SM, Koehler D, Menesses-Diaz A, Fernbach S, Lee K, Towbin JA, Leal S, Belmont JW. Linkage analysis of left ventricular outflow tract malformations (aortic valve stenosis, coarctation of the aorta, and hypoplastic left heart syndrome) Eur J Hum Genet. 2009;17:811–819. doi: 10.1038/ejhg.2008.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne AR, Chang SW, Koenig SN, Zinn AR, Garg V. Submicroscopic chromosomal copy number variations identified in children with hypoplastic left heart syndrome. Pediatr Cardiol. 2012;33:757–763. doi: 10.1007/s00246-012-0208-9. [DOI] [PubMed] [Google Scholar]
- Prakash S, Kuang SQ, Gen TACRI, Regalado E, Guo D, Milewicz D. Recurrent Rare Genomic Copy Number Variants and Bicuspid Aortic Valve Are Enriched in Early Onset Thoracic Aortic Aneurysms and Dissections. PLoS One. 2016;11:e0153543. doi: 10.1371/journal.pone.0153543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash SK, LeMaire SA, Guo DC, Russell L, Regalado ES, Golabbakhsh H, Johnson RJ, Safi HJ, Estrera AL, Coselli JS, Bray MS, Leal SM, Milewicz DM, Belmont JW. Rare copy number variants disrupt genes regulating vascular smooth muscle cell adhesion and contractility in sporadic thoracic aortic aneurysms and dissections. Am J Hum Genet. 2010;87:743–756. doi: 10.1016/j.ajhg.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld JA, Coe BP, Eichler EE, Cuckle H, Shaffer LG. Estimates of penetrance for recurrent pathogenic copy-number variations. Genet Med. 2013;15:478–481. doi: 10.1038/gim.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Agustin JT, Klena N, Granath K, Panigrahy A, Stewart E, Devine W, Strittmatter L, Jonassen JA, Liu X, Lo CW, Pazour GJ. Genetic link between renal birth defects and congenital heart disease. Nat Commun. 2016;7:11103. doi: 10.1038/ncomms11103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifrim A, Hitz MP, Wilsdon A, Breckpot J, Turki SH, Thienpont B, McRae J, Fitzgerald TW, Singh T, Swaminathan GJ, Prigmore E, Rajan D, Abdul-Khaliq H, Banka S, Bauer UM, Bentham J, Berger F, Bhattacharya S, Bu'Lock F, Canham N, Colgiu IG, Cosgrove C, Cox H, Daehnert I, Daly A, Danesh J, Fryer A, Gewillig M, Hobson E, Hoff K, Homfray T, Study I, Kahlert AK, Ketley A, Kramer HH, Lachlan K, Lampe AK, Louw JJ, Manickara AK, Manase D, McCarthy KP, Metcalfe K, Moore C, Newbury-Ecob R, Omer SO, Ouwehand WH, Park SM, Parker MJ, Pickardt T, Pollard MO, Robert L, Roberts DJ, Sambrook J, Setchfield K, Stiller B, Thornborough C, Toka O, Watkins H, Williams D, Wright M, Mital S, Daubeney PE, Keavney B, Goodship J, Consortium UK, Abu-Sulaiman RM, Klaassen S, Wright CF, Firth HV, Barrett JC, Devriendt K, FitzPatrick DR, Brook JD, Deciphering Developmental Disorders S, Hurles ME. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet. 2016;48:1060–1065. doi: 10.1038/ng.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southard AE, Edelmann LJ, Gelb BD. Role of copy number variants in structural birth defects. Pediatrics. 2012;129:755–763. doi: 10.1542/peds.2011-2337. [DOI] [PubMed] [Google Scholar]
- Thienpont B, Mertens L, de Ravel T, Eyskens B, Boshoff D, Maas N, Fryns JP, Gewillig M, Vermeesch JR, Devriendt K. Submicroscopic chromosomal imbalances detected by array-CGH are a frequent cause of congenital heart defects in selected patients. Eur Heart J. 2007;28:2778–2784. doi: 10.1093/eurheartj/ehl560. [DOI] [PubMed] [Google Scholar]
- Thorsson T, Russell WW, El-Kashlan N, Soemedi R, Levine J, Geisler SB, Ackley T, Tomita-Mitchell A, Rosenfeld JA, Topf A, Tayeh M, Goodship J, Innis JW, Keavney B, Russell MW. Chromosomal Imbalances in Patients with Congenital Cardiac Defects: A Meta-analysis Reveals Novel Potential Critical Regions Involved in Heart Development. Congenit Heart Dis. 2015;10:193–208. doi: 10.1111/chd.12179. [DOI] [PubMed] [Google Scholar]
- Tomita-Mitchell A, Mahnke DK, Struble CA, Tuffnell ME, Stamm KD, Hidestrand M, Harris SE, Goetsch MA, Simpson PM, Bick DP, Broeckel U, Pelech AN, Tweddell JS, Mitchell ME. Human gene copy number spectra analysis in congenital heart malformations. Physiol Genomics. 2012;44:518–541. doi: 10.1152/physiolgenomics.00013.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D, Ronemus M, Kline J, Jobanputra V, Williams I, Anyane-Yeboa K, Chung W, Yu L, Wong N, Awad D, Yu CY, Leotta A, Kendall J, Yamrom B, Lee YH, Wigler M, Levy D. The contribution of de novo and rare inherited copy number changes to congenital heart disease in an unselected sample of children with conotruncal defects or hypoplastic left heart disease. Hum Genet. 2014;133:11–27. doi: 10.1007/s00439-013-1353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PS, Xie HM, Werner P, Glessner J, Latney B, Hakonarson H, Goldmuntz E. Analysis of chromosomal structural variation in patients with congenital left-sided cardiac lesions. Birth Defects Res A Clin Mol Teratol. 2014;100:951–964. doi: 10.1002/bdra.23279. [DOI] [PubMed] [Google Scholar]
- Ye M, Coldren C, Liang X, Mattina T, Goldmuntz E, Benson DW, Ivy D, Perryman MB, Garrett-Sinha LA, Grossfeld P. Deletion of ETS-1, a gene in the Jacobsen syndrome critical region, causes ventricular septal defects and abnormal ventricular morphology in mice. Hum Mol Genet. 2010;19:648–656. doi: 10.1093/hmg/ddp532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, Carriero NJ, Cheung YH, Deanfield J, DePalma S, Fakhro KA, Glessner J, Hakonarson H, Italia MJ, Kaltman JR, Kaski J, Kim R, Kline JK, Lee T, Leipzig J, Lopez A, Mane SM, Mitchell LE, Newburger JW, Parfenov M, Pe'er I, Porter G, Roberts AE, Sachidanandam R, Sanders SJ, Seiden HS, State MW, Subramanian S, Tikhonova IR, Wang W, Warburton D, White PS, Williams IA, Zhao H, Seidman JG, Brueckner M, Chung WK, Gelb BD, Goldmuntz E, Seidman CE, Lifton RP. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1 – a. Demography of PCH case cohort; CoA – coarctation of the aorta; AS = Aortic Stenosis; HLHS= Hypoplastic Left Heart Syndrome; b. Summary of genotyped family pedigrees for all cohorts.
Supplemental Table S2 – Ingenuity Pathway Analysis summary
Supplemental Table S3 – Gene Lists, GO Terms used in pathway analyses
Supplemental Table S4 – IPA top networks