Abstract
Background
Understanding natural HIV control may lead to new preventative or therapeutic strategies. Several protective MHC genotypes were found in humans and rhesus macaques. Here, we report a SIV controller MHC genotype in Mauritian cynomolgus macaques (MCMs).
Methods
Twelve MHC-genotyped MCMs were infected with SIVmac251 and monitored for viral loads and CD4+ T cell counts.
Results
Two macaques with M3M4 genotype exhibited the lowest peak viral loads (log plasma SIV RNA copies/ml), nearly 3 logs lower than those in most macaques with other MHC haplotype combinations, and set point viral loads below the level of detection limit by RT-qPCR (< 2 log RNA copies/ml). They maintained healthy CD4+ T cell counts of > 500 cells/μl blood, while CD4 counts in the vast majority of other macaques were below this level.
Conclusions
The M3M4 MHC genotype may confer enhanced control of SIV replication in MCMs.
Keywords: HIV, cynomolgus macaque, SIV, MHC, haplotype, M3/M4 genotype
Introduction
The natural course of human immunodeficiency virus (HIV) infection progresses through 3 typical phases [1, 2]. During the acute phase, viral load (plasma HIV RNA concentration) peaks, up to > 106 copies/ml, and CD4+ cell count declines. In a few weeks, CD4 count partially recovers and viral load decreases to a steady state known as the set point. This is the chronic phase that can last for months or years. Without antiretroviral therapy (ART), eventually viral loads increases and CD4 levels decline, leading to acquired immunodeficiency syndrome (AIDS). However, a small subset of HIV infected individuals, the HIV controllers, maintain normal CD4 counts (> threshold of 500 cells/μl blood) and control viral loads to low (< 2000 copies/ml) or undetectable level (< 50 copies/ml) by conventional assays, in the absence of ART [2]. This natural form of HIV control is widely considered an ideal model to inform the development of new vaccines or therapeutics [2].
While there are several proposed mechanisms accounting for the level of viral containment in HIV controllers, recent studies suggested that host genetics and immunological responses play an important role [3]. Several genotypes of the major histocompatibility complex (MHC), or human leukocyte antigen (HLA) in humans, including HLA-B*57 and HLA-B*27, appear to be the dominant genetic correlate of protection [4-7]. Accumulating evidence suggested that protective HLA molecules may contribute to viral control by enhancing CD8+ T cell antiviral efficacy in HIV controllers [4]. Several mechanisms have been proposed based on the abilities of the HLA molecules to: present a broad repertoire of viral peptides to induce a greater breadth of CD8+ T cell responses [8, 9], target conserved viral antigens [8, 10], more efficiently prime CD8+ T cell cytotoxic responses [11], enhance the expansion ability of CD8+ T cells [12], mediate higher CD8+ T cell cytotoxic capacity [13], or induce early CD8+ T cell responses [14].
An additional, but less characterized mechanism is that protective HLA molecules may mediate innate immunity for early/initial HIV control through interacting with killer immunoglobulin-like receptors (KIR) on natural killer (NK) cells [4, 7, 15-17]. HLA-B*57 is a natural ligand of the NK cell receptor, KIR3DL1. A stronger effect of HLA-B*57 on HIV control was observed in the presence of higher level of KIR3DL1 [8].
While protective MHC genotypes are widely observed as a major determinant of HIV control, not all the individuals with these genotypes showed the same phenotype [4]. The effect of protective MHC genotypes seems to be subject to modulations by other host-specific genetic and immunological factors in the context of complex interaction between the host and virus [18-20].
Simian immunodeficiency virus (SIV) infection of nonhuman primates (NHPs) is currently the best animal model to study HIV pathogenesis, vaccines or therapeutics [21-26]. Traditionally, rhesus macaques (Macaca mulatta) are the favorite choice among NHPs. A wealth of knowledge has been accumulated for this species regarding SIV-host interaction, viral and cellular dynamics following SIV infection, genetics and physiology [22, 27]. SIV-controller MHC genotypes have been identified in rhesus macaques including Mamu-A*01 [28-31], Mamu-B*08 [32] and Mamu-B*17 [33] MHC class I alleles. Mamu-B*08 and the human HLA-B*27 were found to bind peptides with sequence similarity [34].
The availability of rhesus macaques has been greatly reduced due to a ban of their export from India (the major source) and most other south Asian countries [35]. Cynomolgus macaques (Macaca fascicularis) have now become by far the most internationally traded NHP for laboratory experiments [22]. The largest laboratory supply of cynomolgus macaques is available from the island of Mauritius. The Mauritian cynomolgus macaques (MCM) descended from a small group of founder animals and are characterized by limited MHC diversity, with only seven haplotypes, named M1 through M7 [36, 37]. This helps reduce variability between animals and thus reduce animal numbers needed to achieve statistically significant results, making them a practically useful animal model in HIV studies [27].
SIV control phenotypes have also been observed in MCMs, however the underlying factors are poorly understood. A major study in this field suggested that the M1 or M7 MHC haplotypes may be correlates of protection [38]. Here, we report that the combination of M3 and M4 MHC haplotypes may be a SIV control genotype in MCMs.
Materials and Methods
Humane care guidelines
14 female Mauritian cynomolgus macaques (Macaca facicularis) from Bioculture (Mauritius) Ltd were used in a pre-study to optimize immunization condition, MHC haplotype selection, and titration of SIVmac251 for repeated low dose intravaginal challenge for a large vaccine efficacy study. The vaccine candidate was SIV peptides derived from the sequences of the protease cleavage sites, delivered by recombinant vesicular stomatitis virus and nanoparticles (the PCS vaccine). Eight monkeys received immunization and six monkeys were controls (Table S1). The animal work was performed in accordance with Canadian Council on Animal Care guidelines and the Animal Use Document was approved by the Canadian Sciences Centre for Human and Animal Health Animal Care Committee (protocol number: H-12-014R2). Animals were double housed in standard non-human primate cages, received standard primate feed as well as fresh fruit and enrichment daily, and had continual access to water. Temperature (19-24 °C), humidity (45-60%) and light (approximately 323 lux) were monitored and maintained within recommended limits, the light/dark cycle was maintained at 12 hour split. Environmental enrichment was provided. Animals were observed twice daily by the PI, a co-investigator, or the veterinary staff for signs of clinical illness.
SIV challenge
The 14 animals were intravaginally challenged with 1000 TCID50 SIVmac251 (Desrosiers” 2010-Day 8 viral stock, provided by Drs. Jon Warren, and Nancy Miller, Vaccine Research Program, NIH). The challenge stock titer was based on a previous publication [39]. The challenge was repeatedly carried out until positive plasma viral load (VL) was detected. Two animals that did not show any detectable viral load after four challenges were excluded from study.
Viral load assay
Nucleic acids were extracted from 1.0 ml plasma using the EasyMag system and reagents (bioMerieux Canada, St Laurent, QC), and eluted into 110 μl buffer. Viral quantitation was performed by qPCR using the ABI 7900HT (Applied Biosystems, Streetsville, Ontario, Canada) and the QuantiTect Probe RT-PCR kit (Qiagen, Toronto, Ontario, Canada) and primers and probe previously described by Horton et al [40]. All plasma viral loads were converted to log10 values prior to further analysis. The viral load assay was validated by direct comparisons with an established assay in the lab of Jonathan Heeney (Biomedical Primate Research Center, The Netherlands).
CD4+ T cell quantification
An aliquot of nonhuman primate (NHP) whole blood (100 μl) collected in a heparinized vacutainer tube was transferred into a BD round-bottom FACS tube. The whole blood was stained using 10 μl of NHP T lymphocyte cocktail (BD Biosciences) in the dark. The mixture was vortexed lightly and incubated at 4 °C for 30 minutes. The labelled cells were then fixed using 200 μl of 4% paraformaldehyde. 200μL of Flow Cytometry Absolute Count beads (Bang Laboratories Inc.) were added prior to acquisition of 1000 beads using LSR II flow cytometry (BD). A blank tube containing only 200 μL Flow Cytometry Absolute Count beads was used as a negative control. Flow data acquisition was done using FACS DIVATM software (BD).
MHC genotyping
The cynomolgus macaque MHC haplotype typing was conducted by Wisconsin Nonhuman Primate Research Centre Genetics Services [41, 42].
Results
Viral load profiles of SIV-infected macaques with different MHC haplotypes
We conducted a pre-study with Mauritian cynomolgus macaques (MCM) to optimize the study design of a large project for evaluation of a new HIV vaccine strategy. The vaccine of interest was based on SIV peptides derived from the sequences of the viral protease cleavage sites (the PCS vaccine). The primary purposes of the pilot study were to screen MHC haplotypes that can generate immune responses to the PCS vaccine peptides and test dose of SIVmac251 intravaginal challenges in female MCMs. Fourteen MHC-genotyped female MCMs were involved, among which eight were immunized with the PCS vaccine (Table S1).
Using this monkey cohort, we took the opportunity to investigate patterns of disease progression in SIV-infected MCMs in the context of various MHC haplotypes. To mimic natural route of vaginal HIV infection in humans, repeated low-dose SIVmac251 [43, 44] intravaginal challenges were conducted until infection was established at detectable level. The course of infection was monitored by quantification of plasma viral load (SIV RNA copy number/ml) over extended period of time (Fig. 1). After four challenges 12 monkeys were infected. Among them, two were of M3/M4 haplotypes. For simplicity, these were termed M3M4 monkeys and the others non-M3M4 monkeys. Among the ten non-M3M4 monkeys, eight showed infection profiles typical of SIV non-controllers (Non-M3M4 subgroup 1, Fig. 1A). They were characterized by high peak viral loads ranging from 5.694 to 6.832 (Log10 SIV RNA copies/ml plasma) with a mean of 6.439, as well as by high set point viral loads ranging from 2.830 to 5.227 with a mean of 3.757 (Figs. 1 and 2). The other two non-M3M4 monkeys (Non-M3M4 subgroup 2, Fig. 1B) displayed a pattern of viral control. Compared to the non-controllers (Fig. 1A), they had lower peak viral loads, 5.632 and 5.065, with a mean of 5.344, and lower set point viral loads, 1.281 and 1.776 with a mean of 1.529 (Fig. 2). Both animals shared a common MHC haplotype, M1. It is consistent with previous report that monkeys of M1 haplotype can better control SIV infection [38]. The two M3M4 monkeys (M3M4 group, Fig. 1C) exhibited a strong SIV control phenotype. They had the lowest peak viral loads among the 12 monkeys, 2.989 and 3.900 (Figs. 1C and 2A), with a mean of 3.445, which is nearly 3 and 2 logs lower than those of the non-controllers and M1 controllers, respectively. The two M3M4 monkeys had low set point viral loads, 1.633 and 1.013 (Figs. 1C and 2A), with a mean of 1.323, the lowest among all the 3 sub-groups of monkeys. Since both monkeys exhibited similar viral load profile between the one (BM795) that received the vaccine and the one (BM796) that did not, immunization with the PCS vaccine could not be the major factor influencing viral load of the M3M4 monkeys. These results suggest a possible role of the M3M4 MHC genotype in SIV control.
Figure 1. Viral load profiles of individual SIV-infected macaques.
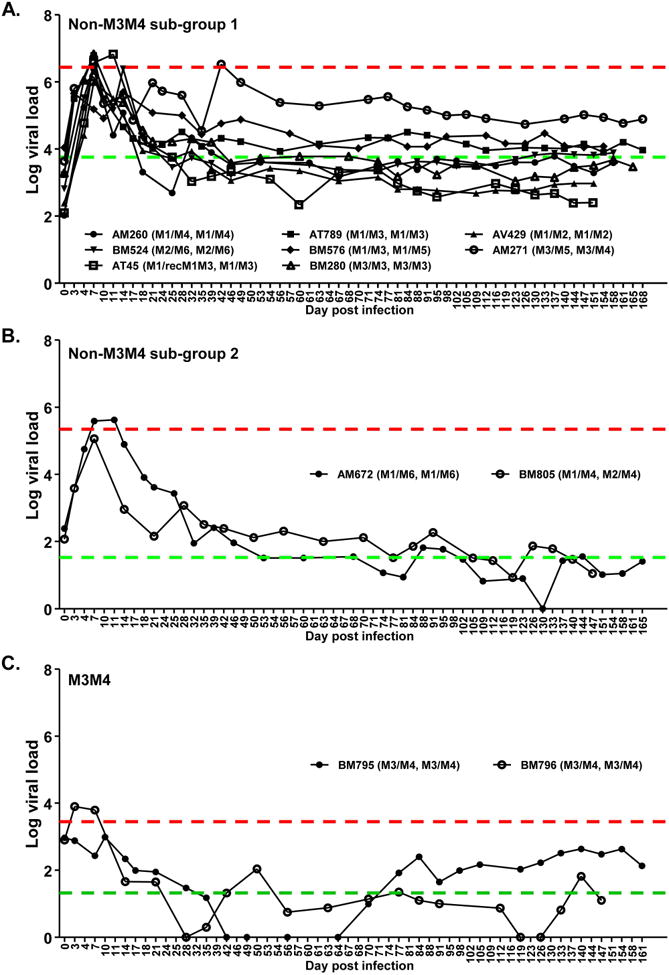
Twelve female Mauritian cynomolgus macaques were vaginally infected with SIVmac251 and monitored for viral load (Log10 SIV RNA copy number/ml plasma) over time. Animal ID (MHC I haplotype, MHC II haplotype) is shown for each individual monkey. Red line: mean peak viral load of monkey group. Green line: Mean set point viral load of monkey group. (A) Non-M3M4 sub-group 1: Monkeys of non-M3M4 genotypes with poor SIV control. (B) Non-M3M4 sub-group 2: Monkeys of non-M3M4 genotypes showing SIV control. (C) M3M4: Monkey group of M3M4 genotype. Note that two more monkeys (with M3M4 and non-M3M4 genotypes, respectively) were challenged in the same experiments, but did not show detectable viral load and were excluded from study (See Table S1).
Figure 2. Quantitative analysis of viral loads and CD4 counts in Non-M3M4 versus M3M4 monkeys.
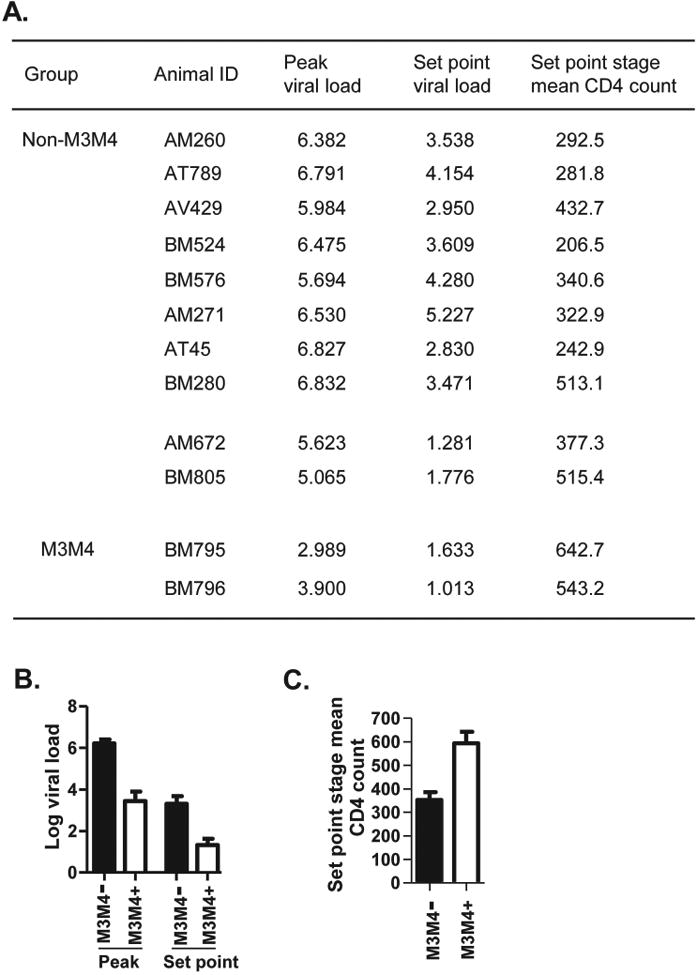
(A) List of peak and set point viral loads (Log SIV RNA copies/ml plasma) and set point stage mean CD4 counts (cell number/μl blood) in SIV-infected monkeys from Fig. 1. (B) Peak and set point viral loads were compared between Non-M3M4 monkeys (M3M4-, all animals from Fig. 1A and B combined) and M3M4 monkeys (M3M4+, animals from Fig.1C). Data are mean ± SEM. (C) Set point stage mean CD4 count (CD4+ cell number/μl blood) was compared between the two groups.
The M3M4 genotype demonstrates enhanced control of SIV infection
We compared the SIV infection profiles of the two M3M4 monkeys versus all the ten non-M3M4 monkeys (Fig. 2). The M3M4 group showed trends of lower peak and set point viral loads than the non-M3M4 group, as well as higher CD4 counts, which were above the healthy threshold of 500 cells/μl blood, as opposed to the CD4 counts lower than the threshold in non-M3M4 group (Fig. 2). Our observation is consistent with a previous study on SIV infection in MCMs. The only M3M4 monkey in that study population was found to control SIV, with a viral load profile similar to the two SIV-infected M3M4 monkeys in our study [45]. Altogether, these findings strongly suggest that the M3M4 MHC genotype may play a protective role in controlling SIV infection in MCMs and is worthy of further exploration.
M3 or M4 haplotype alone is not sufficient to confer SIV control
Among the SIV-infected monkeys, the M3M4 monkeys showed SIV control, but such phenotype was not seen in monkeys with either M3 or M4 haplotype alone (Fig. 3). It is interesting to note that a monkey with “M3/M5/4” hybrid haplotypes, AM271, which had the MHC I region of M5 and MHC II region of M4 (Fig. 3A left panel), failed to control SIV infection (Fig. 2B). It suggests that both M3 and M4 class I regions contributed to the SIV control. This is consistent with the importance of MHC class I restricted CD8+ T cells in SIV control [20, 46, 47]. The better control of SIV infection by the combination of M3 and M4 is also in agreement with the previously reported MHC heterozygosity advantage over homozygosity in SIV-infected MCMs [45].
Figure 3. M3 or M4 haplotype alone is not sufficient for the SIV control phenotype found in M3M4 monkeys.
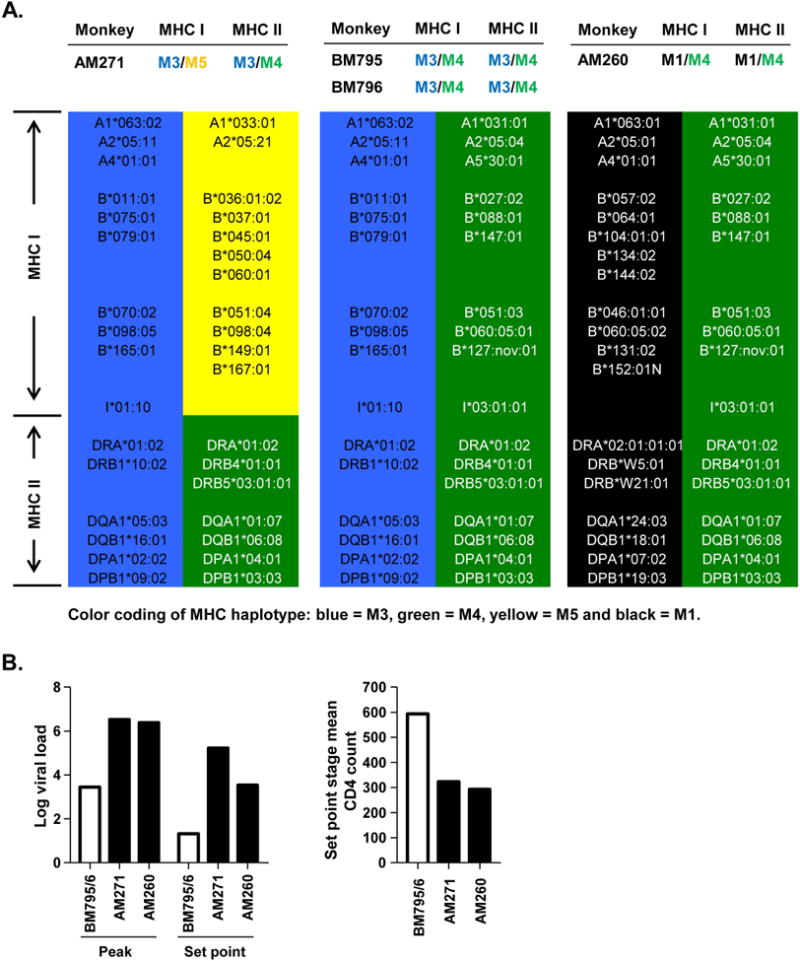
(A) MHC haplotypes with alleles are illustrated for: the two monkeys with a combination of complete M3 and M4 haplotypes (BM795 and BM796, middle panel), a monkey with M3 but no M4 for MHC I (AM271, left panel), and a monkey with M4 but no M3 (AM260, right panel). (B) Viral loads and CD4 counts of the monkeys in A. In contrast to M3M4 monkeys (white bars), M3 or M4 monkeys (black bars) showed high viral loads and low CD4 counts. Two other monkeys with M3 but not M4, AT789 (M1/M3, M1/M3) and BM280 (M3/M3, M3/M3), had similar disease profiles (not illustrated here).
Discussion
Understanding the natural control of HIV and SIV infection will help to develop better preventative and therapeutic strategies. While the mechanisms of viral control remain to be fully understood, several host MHC genotypes have been extensively described as protective factors in humans and rhesus macaques against pathogenic HIV/SIV infection.
Mauritian cynomolgus macaques (MCM) are an increasingly important NHP model for HIV research, due to their availability and simple MHC haplotypes. We know much less about the protective MCM MHC genotypes in SIV control than we know about Indian origin rhesus monkeys. Previous studies have shown that MCMs with M1 or M7 haplotypes can control SIV infection [38] and M3 homozygotes had higher viral loads than M1/M3 heterozygotes [38, 45]. In this study, we observed SIV control in some, but not all monkeys possessing the protective haplotype, M1 [38]. It could be that the protective effect also depends on additional factors, such as combination with other MHC haplotypes [20, 38, 45].
Our observation that MCMs with M3M4 genotype can control SIV infection added new dimension of understanding about immunologic control of viral infection in MCMs. This observation is consistent with the data from a previous study [45]. Because CD8+ T cells are primary mediators of sustained viral control [20, 46, 47], the M3M4 MHC genotype may lead to more effective anti-SIV CD8+ T cell responses. Since the presence of either M3 or M4 haplotype alone is not sufficient to control SIVmac251, it is likely that CD8+ T cell responses mediated by these two haplotypes complement each other, leading to enhanced viral targeting. An important part of the CD8+ T cell responses may be mediated by the MHC I region of the M4 haplotype, since a monkey missing this part could not control SIVmac251 infection.
In conclusion, this study identified the combination of M3 and M4 MHC haplotypes as a novel SIV control genotype in MCMs. It may help to better understand natural control of SIV infection. The information will help for future studies using MCMs as an AIDS model, in animal selection, experimental design and result interpretation.
Supplementary Material
Acknowledgments
Funding: The study was supported by an NIH grant (R01AI111805), a CIHR/CHVI bridging grant and funding from National Microbiology Laboratory of Canada.
We would like to thank the VTS staff at Canadian Science Centre for Human and Animal Health, Christine De Graff, Julie Kubay, Michelle French, Stephanie Kucas, Kimberly Azaransky, Carissa EmburyHyatt and Valerie Smid, for tremendous technical support. We would also like to thank Dr. Jon Warren and Dr. Nancy Miller, NIH Vaccine Research Program, for providing the SIVmac251 Desrosiers” 2010-Day 8 viral stock, and Wisconsin Nonhuman Primate Research Centre Genetics Services for MHC genotyping. We recognize Dr. Stuart Shapiro, NIH Vaccine Research Program, and Dr. Matthew Gilmour, National Microbiology Laboratory of Canada, for their important support. This work was supported by a CIHR Operating Grant - PA: Bridge Funding - CHVI Vaccine Discovery and Social Research, a NIH grant (R01AI111805) and funding from National Microbiology Laboratory of Canada.
References
- 1.O'Connell KA, Bailey JR, Blankson JN. Elucidating the elite: mechanisms of control in HIV-1 infection. Trends Pharmacol Sci. 2009;30:631–637. doi: 10.1016/j.tips.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Saag M, Deeks SG. How do HIV elite controllers do what they do? Clin Infect Dis. 2010;51:239–241. doi: 10.1086/653678. [DOI] [PubMed] [Google Scholar]
- 3.Gaardbo JC, Hartling HJ, Gerstoft J, Nielsen SD. Thirty Years with HIV Infection-Nonprogression Is Still Puzzling: Lessons to Be Learned from Controllers and Long-Term Nonprogressors. AIDS Res Treat. 2012;2012:161584. doi: 10.1155/2012/161584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Poropatich K, Sullivan DJ., Jr Human immunodeficiency virus type 1 long-term non-progressors: the viral, genetic and immunological basis for disease non-progression. J Gen Virol. 2011;92:247–268. doi: 10.1099/vir.0.027102-0. [DOI] [PubMed] [Google Scholar]
- 6.Dominguez-Molina B, Tarancon-Diez L, Hua S, Abad-Molina C, Rodriguez-Gallego E, Machmach K, Vidal F, Tural C, Moreno S, Goni JM, Ramirez de Arellano E, Del Val M, Gonzalez-Escribano MF, Del Romero J, Rodriguez C, Capa L, Viciana P, Alcami J, Yu XG, Walker BD, Leal M, Lichterfeld M, Ruiz-Mateos E. HLA-B*57 and IFNL4-related polymorphisms are associated with protection against HIV-1 disease progression in controllers. Clin Infect Dis. 2016 doi: 10.1093/cid/ciw833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaunders J, van Bockel D. Innate and Adaptive Immunity in Long-Term Non-Progression in HIV Disease. Front Immunol. 2013;4:95. doi: 10.3389/fimmu.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altfeld M, Addo MM, Rosenberg ES, Hecht FM, Lee PK, Vogel M, Yu XG, Draenert R, Johnston MN, Strick D, Allen TM, Feeney ME, Kahn JO, Sekaly RP, Levy JA, Rockstroh JK, Goulder PJ, Walker BD. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS. 2003;17:2581–2591. doi: 10.1097/00002030-200312050-00005. [DOI] [PubMed] [Google Scholar]
- 9.Jansen CA, Kostense S, Vandenberghe K, Nanlohy NM, De Cuyper IM, Piriou E, Manting EH, Miedema F, van Baarle D. High responsiveness of HLA-B57-restricted Gag-specific CD8+ T cells in vitro may contribute to the protective effect of HLA-B57 in HIV-infection. Eur J Immunol. 2005;35:150–158. doi: 10.1002/eji.200425487. [DOI] [PubMed] [Google Scholar]
- 10.Altfeld M, Allen TM. Hitting HIV where it hurts: an alternative approach to HIV vaccine design. Trends Immunol. 2006;27:504–510. doi: 10.1016/j.it.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Migueles SA, Rood JE, Berkley AM, Guo T, Mendoza D, Patamawenu A, Hallahan CW, Cogliano NA, Frahm N, Duerr A, McElrath MJ, Connors M. Trivalent adenovirus type 5 HIV recombinant vaccine primes for modest cytotoxic capacity that is greatest in humans with protective HLA class I alleles. PLoS Pathog. 2011;7:e1002002. doi: 10.1371/journal.ppat.1002002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez M, Peris A, Soriano V, Lozano S, Vicario JL, Rallon NI, Restrepo C, Benito JM. The expansion ability but not the quality of HIV-specific CD8(+) T cells is associated with protective human leucocyte antigen class I alleles in long-term non-progressors. Immunology. 2011;134:305–313. doi: 10.1111/j.1365-2567.2011.03490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Migueles SA, Mendoza D, Zimmerman MG, Martins KM, Toulmin SA, Kelly EP, Peterson BA, Johnson SA, Galson E, Poropatich KO, Patamawenu A, Imamichi H, Ober A, Rehm CA, Jones S, Hallahan CW, Follmann DA, Connors M. CD8(+) T-cell Cytotoxic Capacity Associated with Human Immunodeficiency Virus-1 Control Can Be Mediated through Various Epitopes and Human Leukocyte Antigen Types. EBioMedicine. 2015;2:46–58. doi: 10.1016/j.ebiom.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennan CA, Ibarrondo FJ, Sugar CA, Hausner MA, Shih R, Ng HL, Detels R, Margolick JB, Rinaldo CR, Phair J, Jacobson LP, Yang OO, Jamieson BD. Early HLA-B*57-restricted CD8+ T lymphocyte responses predict HIV-1 disease progression. J Virol. 2012;86:10505–10516. doi: 10.1128/JVI.00102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malnati MS, Ugolotti E, Monti MC, Battista D, Vanni I, Bordo D, Sironi F, Larghero P, Marco ED, Biswas P, Poli G, Vicenzi E, Riva A, Tarkowski M, Tambussi G, Nozza S, Tripodi G, Marras F, Maria A, Pistorio A, Biassoni R. Activating Killer Immunoglobulin Receptors and HLA-C: a successful combination providing HIV-1 control. Sci Rep. 2017;7:42470. doi: 10.1038/srep42470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Y, Chen O, Cui C, Zhao B, Han X, Zhang Z, Liu J, Xu J, Hu Q, Liao C, Shang H. KIR3DS1/L1 and HLA-Bw4-80I are associated with HIV disease progression among HIV typical progressors and long-term nonprogressors. BMC Infect Dis. 2013;13:405. doi: 10.1186/1471-2334-13-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grifoni A, Montesano C, Palma P, Salerno A, Colizzi V, Amicosante M. Role of HLA-B alpha-3 domain amino acid position 194 in HIV disease progression. Mol Immunol. 2013;53:410–413. doi: 10.1016/j.molimm.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Salgado M, Simon A, Sanz-Minguela B, Rallon NI, Lopez M, Vicario JL, Benito JM, Rodes B. An additive effect of protective host genetic factors correlates with HIV nonprogression status. J Acquir Immune Defic Syndr. 2011;56:300–305. doi: 10.1097/QAI.0b013e3182036f14. [DOI] [PubMed] [Google Scholar]
- 19.Casado C, Colombo S, Rauch A, Martinez R, Gunthard HF, Garcia S, Rodriguez C, Del Romero J, Telenti A, Lopez-Galindez C. Host and viral genetic correlates of clinical definitions of HIV-1 disease progression. PLoS One. 2010;5:e11079. doi: 10.1371/journal.pone.0011079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ericsen AJ, Starrett GJ, Greene JM, Lauck M, Raveendran M, Deiros DR, Mohns MS, Vince N, Cain BT, Pham NH, Weinfurter JT, Bailey AL, Budde ML, Wiseman RW, Gibbs R, Muzny D, Friedrich TC, Rogers J, O'Connor DH. Whole genome sequencing of SIV-infected macaques identifies candidate loci that may contribute to host control of virus replication. Genome Biol. 2014;15:478. doi: 10.1186/s13059-014-0478-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitz JE, Korioth-Schmitz B. Immunopathogenesis of simian immunodeficiency virus infection in nonhuman primates. Curr Opin HIV AIDS. 2013;8:273–279. doi: 10.1097/COH.0b013e328361cf5b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antony JM, MacDonald KS. A critical analysis of the cynomolgus macaque, Macaca fascicularis, as a model to test HIV-1/SIV vaccine efficacy. Vaccine. 2015;33:3073–3083. doi: 10.1016/j.vaccine.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Xu H, Wang X, Malam N, Aye PP, Alvarez X, Lackner AA, Veazey RS. Persistent Simian Immunodeficiency Virus Infection Drives Differentiation, Aberrant Accumulation, and Latent Infection of Germinal Center Follicular T Helper Cells. J Virol. 2015;90:1578–1587. doi: 10.1128/JVI.02471-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gill AF, Ahsan MH, Lackner AA, Veazey RS. Hematologic abnormalities associated with simian immunodeficieny virus (SIV) infection mimic those in HIV infection. J Med Primatol. 2012;41:214–224. doi: 10.1111/j.1600-0684.2012.00543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasegawa A, Liu H, Ling B, Borda JT, Alvarez X, Sugimoto C, Vinet-Oliphant H, Kim WK, Williams KC, Ribeiro RM, Lackner AA, Veazey RS, Kuroda MJ. The level of monocyte turnover predicts disease progression in the macaque model of AIDS. Blood. 2009;114:2917–2925. doi: 10.1182/blood-2009-02-204263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breed MW, Jordan AP, Aye PP, Sugimoto C, Alvarez X, Kuroda MJ, Pahar B, Keele BF, Hoxie JA, Lackner AA. A single amino acid mutation in the envelope cytoplasmic tail restores the ability of an attenuated simian immunodeficiency virus mutant to deplete mucosal CD4+ T cells. J Virol. 2013;87:13048–13052. doi: 10.1128/JVI.02126-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sui Y, Gordon S, Franchini G, Berzofsky JA. Nonhuman primate models for HIV/AIDS vaccine development. Curr Protoc Immunol. 2013;102(Unit 12):14. doi: 10.1002/0471142735.im1214s102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muhl T, Krawczak M, Ten Haaft P, Hunsmann G, Sauermann U. MHC class I alleles influence set-point viral load and survival time in simian immunodeficiency virus-infected rhesus monkeys. J Immunol. 2002;169:3438–3446. doi: 10.4049/jimmunol.169.6.3438. [DOI] [PubMed] [Google Scholar]
- 29.Zhang ZQ, Fu TM, Casimiro DR, Davies ME, Liang X, Schleif WA, Handt L, Tussey L, Chen M, Tang A, Wilson KA, Trigona WL, Freed DC, Tan CY, Horton M, Emini EA, Shiver JW. Mamu-A*01 allele-mediated attenuation of disease progression in simian-human immunodeficiency virus infection. J Virol. 2002;76:12845–12854. doi: 10.1128/JVI.76.24.12845-12854.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W, Cong Z, Liu X, Tong W, Qiao H, Jiang H, Wei Q, Qin C. Frequency of the major histocompatibility complex Mamu-A*01 allele in experimental rhesus macaques in China. J Med Primatol. 2010;39:374–380. doi: 10.1111/j.1600-0684.2010.00420.x. [DOI] [PubMed] [Google Scholar]
- 31.Lim SY, Chan T, Gelman RS, Whitney JB, O'Brien KL, Barouch DH, Goldstein DB, Haynes BF, Letvin NL. Contributions of Mamu-A*01 status and TRIM5 allele expression, but not CCL3L copy number variation, to the control of SIVmac251 replication in Indian-origin rhesus monkeys. PLoS Genet. 2010;6:e1000997. doi: 10.1371/journal.pgen.1000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, Bean AT, Beal DR, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol. 2007;81:8827–8832. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, Lifson JD, O'Connor DH, Carrington M, Watkins DI. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2006;80:5074–5077. doi: 10.1128/JVI.80.10.5074-5077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loffredo JT, Sidney J, Bean AT, Beal DR, Bardet W, Wahl A, Hawkins OE, Piaskowski S, Wilson NA, Hildebrand WH, Watkins DI, Sette A. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J Immunol. 2009;182:7763–7775. doi: 10.4049/jimmunol.0900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan G, Zhang G, Fang X, Zhang Y, Li C, Ling F, Cooper DN, Li Q, Li Y, van Gool AJ, Du H, Chen J, Chen R, Zhang P, Huang Z, Thompson JR, Meng Y, Bai Y, Wang J, Zhuo M, Wang T, Huang Y, Wei L, Li J, Wang Z, Hu H, Yang P, Le L, Stenson PD, Li B, Liu X, Ball EV, An N, Huang Q, Fan W, Zhang X, Wang W, Katze MG, Su B, Nielsen R, Yang H, Wang X. Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nat Biotechnol. 2011;29:1019–1023. doi: 10.1038/nbt.1992. [DOI] [PubMed] [Google Scholar]
- 36.Wiseman RW, Wojcechowskyj JA, Greene JM, Blasky AJ, Gopon T, Soma T, Friedrich TC, O'Connor SL, O'Connor DH. Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J Virol. 2007;81:349–361. doi: 10.1128/JVI.01841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mee ET, Badhan A, Karl JA, Wiseman RW, Cutler K, Knapp LA, Almond N, O'Connor DH, Rose NJ. MHC haplotype frequencies in a UK breeding colony of Mauritian cynomolgus macaques mirror those found in a distinct population from the same geographic origin. J Med Primatol. 2009;38:1–14. doi: 10.1111/j.1600-0684.2008.00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Budde ML, Greene JM, Chin EN, Ericsen AJ, Scarlotta M, Cain BT, Pham NH, Becker EA, Harris M, Weinfurter JT, O'Connor SL, Piatak M, Jr, Lifson JD, Gostick E, Price DA, Friedrich TC, O'Connor DH. Specific CD8+ T cell responses correlate with control of simian immunodeficiency virus replication in Mauritian cynomolgus macaques. J Virol. 2012;86:7596–7604. doi: 10.1128/JVI.00716-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Del Prete GQ, Scarlotta M, Newman L, Reid C, Parodi LM, Roser JD, Oswald K, Marx PA, Miller CJ, Desrosiers RC, Barouch DH, Pal R, Piatak M, Jr, Chertova E, Giavedoni LD, O'Connor DH, Lifson JD, Keele BF. Comparative characterization of transfection- and infection-derived simian immunodeficiency virus challenge stocks for in vivo nonhuman primate studies. J Virol. 2013;87:4584–4595. doi: 10.1128/JVI.03507-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horton H, Vogel TU, Carter DK, Vielhuber K, Fuller DH, Shipley T, Fuller JT, Kunstman KJ, Sutter G, Montefiori DC, Erfle V, Desrosiers RC, Wilson N, Picker LJ, Wolinsky SM, Wang C, Allison DB, Watkins DI. Immunization of rhesus macaques with a DNA prime/modified vaccinia virus Ankara boost regimen induces broad simian immunodeficiency virus (SIV)-specific T-cell responses and reduces initial viral replication but does not prevent disease progression following challenge with pathogenic SIVmac239. J Virol. 2002;76:7187–7202. doi: 10.1128/JVI.76.14.7187-7202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Budde ML, Wiseman RW, Karl JA, Hanczaruk B, Simen BB, O'Connor DH. Characterization of Mauritian cynomolgus macaque major histocompatibility complex class I haplotypes by high-resolution pyrosequencing. Immunogenetics. 2010;62:773–780. doi: 10.1007/s00251-010-0481-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiseman RW, Karl JA, Bohn PS, Nimityongskul FA, Starrett GJ, O'Connor DH. Haplessly hoping: macaque major histocompatibility complex made easy. ILAR J. 2013;54:196–210. doi: 10.1093/ilar/ilt036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitney JB, Hill AL, Sanisetty S, Penaloza-MacMaster P, Liu J, Shetty M, Parenteau L, Cabral C, Shields J, Blackmore S, Smith JY, Brinkman AL, Peter LE, Mathew SI, Smith KM, Borducchi EN, Rosenbloom DI, Lewis MG, Hattersley J, Li B, Hesselgesser J, Geleziunas R, Robb ML, Kim JH, Michael NL, Barouch DH. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature. 2014;512:74–77. doi: 10.1038/nature13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitney JB, Luedemann C, Bao S, Miura A, Rao SS, Mascola JR, Letvin NL. Monitoring HIV vaccine trial participants for primary infection: studies in the SIV/macaque model. AIDS. 2009;23:1453–1460. doi: 10.1097/QAD.0b013e32832b43d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Connor SL, Lhost JJ, Becker EA, Detmer AM, Johnson RC, Macnair CE, Wiseman RW, Karl JA, Greene JM, Burwitz BJ, Bimber BN, Lank SM, Tuscher JJ, Mee ET, Rose NJ, Desrosiers RC, Hughes AL, Friedrich TC, Carrington M, O'Connor DH. MHC heterozygote advantage in simian immunodeficiency virus-infected Mauritian cynomolgus macaques. Sci Transl Med. 2010;2:22ra18. doi: 10.1126/scitranslmed.3000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 47.Friedrich TC, Valentine LE, Yant LJ, Rakasz EG, Piaskowski SM, Furlott JR, Weisgrau KL, Burwitz B, May GE, Leon EJ, Soma T, Napoe G, Capuano SV, 3rd, Wilson NA, Watkins DI. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J Virol. 2007;81:3465–3476. doi: 10.1128/JVI.02392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


