Abstract
Many stem cell niches contain support cells that increase contact with stem cells by enwrapping them in cellular processes. One example is the germ stem cell niche in C. elegans, which is composed of a single niche cell termed the distal tip cell (DTC) that extends cellular processes, constructing an elaborate plexus that enwraps germ stem cells. To identify genes required for plexus formation and to explore the function of this specialized enwrapping behavior, a series of targeted and tissue-specific RNAi screens were performed. Here we identify genes that promote stem cell enwrapment by the DTC plexus, including a set that specifically functions within the DTC, such as the chromatin modifier lin-40/MTA1, and others that act within the germline, such as the 14-3-3 signaling protein par-5. Analysis of genes that function within the germline to mediate plexus development reveal that they are required for expansion of the germ progenitor zone, supporting the emerging idea that germ stem cells signal to the niche to stimulate enwrapping behavior. Examination of wild-type animals with asymmetric plexus formation and animals with reduced DTC plexus elaboration via loss of two candidates including lin-40 indicate that cellular enwrapment promotes GLP-1/Notch signaling and germ stem cell fate. Together, our work identifies novel regulators of cellular enwrapment and suggests that reciprocal signaling between the DTC niche and the germ stem cells promotes enwrapment behavior and stem cell fate.
Keywords: niche, distal tip cell, germ cell, stem cell, somatic gonad
Introduction
Stem cells require a specialized microenvironment, or niche, comprised of support cells and signaling factors to coordinate stem cell maintenance with production of new cells for tissue renewal, organ regeneration, and tissue expansion (Mesa et al., 2015). Niche dysregulation is implicated in diseases, including leukemia (Wiseman, 2011; Zambetti et al., 2016), myelodysplastic syndrome (Hoggatt et al., 2016), Alzheimer’s disease (Hamilton et al., 2015), medulloblastoma (Bassett et al., 2016), and testicular germ cell tumors (Silván et al., 2013). Thus, understanding mechanisms that regulate stem cell-niche interactions is important for our basic understanding of tissue maintenance, growth, regeneration, and numerous human diseases.
Niche cues are often restricted to stem cells in direct contact with niche cells (Inaba et al., 2016). In many niches, support cells extend cellular processes that enwrap stem cells. For example, in the mouse testis, Sertoli cells contribute to the niche and extend elaborate cytoplasmic processes around spermatogonial stem cells and progenitors (Oatley et al., 2011; Smith et al., 2012). Regulated niche-germ cell contact is also seen in the Drosophila testis, where germ stem cells extend projections termed nanotubes into the niche (Inaba et al., 2015). In the Drosophila intestinal niche, the peripheral cell extends processes surrounding intestinal progenitors (Mathur et al., 2010). Cellular extensions have also been observed in the Drosophila lymph gland niche (Mandal et al., 2007). Cellular enwrapment of hematopoietic stem cells by the perivascular niche of zebrafish has been referred to as “endothelial cuddling” (Tamplin et al., 2015). It has been speculated that enwrapment of stem cells might facilitate stem cell-niche signaling (Buszczak et al., 2016; Kornberg and Roy, 2014) or adhesion (S. Chen et al., 2013) to promote stem cell fate. Direct evidence for a role for enwrapment of stem cells, however, is lacking. Further, the mechanisms that promote this specialized behavior are poorly understood.
In C. elegans, germline stem cells are maintained by a single-cell niche, called the distal tip cell (DTC) (Kimble and White, 1981). The DTC extends elaborate processes in young adults that contact germ progenitor cells (stem cells and their proliferative and early-differentiating progeny) (Byrd and Kimble, 2009; Byrd et al., 2014; Cinquin et al., 2015; Crittenden et al., 2017; Lee et al., 2016; B. G. Wong et al., 2013). In the distal-most region of the gonad where the germ stem cells reside, the DTC extends elaborate processes that enwrap these cells, forming what has been termed the DTC plexus (Byrd et al., 2014; Lee et al., 2016). The correlation of the plexus with the stem cell pool suggests that enwrapment might promote stem cell fate, repress differentiation, or both. Intriguingly, previous work showed that genetically forcing germ cells to differentiate caused loss of DTC elaboration (Byrd et al., 2014), suggesting that germ progenitor cells might be required to stimulate enwrapping behavior to construct the DTC plexus. Together these observations suggest possible reciprocal signaling interactions between the DTC and progenitor cells that promote stem cell fate.
To identify regulators of germ stem cell enwrapment by the C. elegans DTC niche, a targeted RNAi screen of over 700 genes required for robust fecundity and cell communication was performed. Using germline- and DTC-specific RNAi strains, the screen isolated genes that function directly in the DTC to promote plexus formation. Reduction of the plexus correlated with a decrease in the stem cell pool, indicating that the enwrapment of germ cells within the plexus supports germ stem cell fate. The screen also identified genes that promote enwrapment and function within the germline. All of the germline-functioning genes analyzed were required for expansion of the germ progenitor zone, supporting the idea that germ stem cells foster enwrapping behavior to form the DTC plexus. Together, these studies reveal numerous genes that regulate germ stem cell enwrapment. Further, our results reveal an apparent positive feedback loop between the niche and the germ stem cells: enwrapment by the DTC niche promotes germ stem cell fate and germ stem cells signal back to the niche and stimulate enwrapment.
Results
A targeted RNAi screen identifies genes required for DTC plexus formation
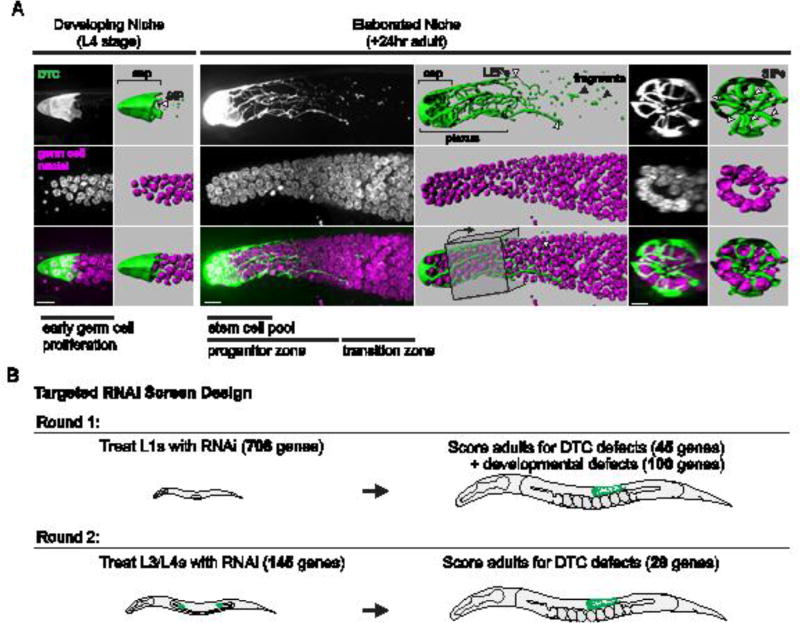
The DTC niche in C. elegans hermaphrodites elaborates at the L4 larval stage from a cap-like structure with a few short intercalating processes (SIPs) that extend between germ cells to a dramatically elaborated plexus, composed of extensive processes that enwrap germ stem cells by the early adult stage (Fig. 1A). As germ stem cells divide and are displaced from the niche, they differentiate into gametes, producing sperm during the L4 larval stage and oocytes in adult worms. We reasoned that genes promoting germ stem cell niche formation might impact fecundity. Additionally, we hypothesized that dynamic DTC enwrapment of germ cells would involve genes mediating cell-cell communication. We thus designed a targeted RNAi screen of 708 genes known to be required for robust fecundity and cell communication (see Methods). RNAi knockdown was performed for these genes by first treating L1 worms containing markers for fluorescently-labeled DTC membranes and germ cell nuclei (Fig. 1B and Table S1). DTC morphology was then evaluated in 1- to 2-day-old adult animals. We used a >50% reduction in DTC plexus length as the criteria for a DTC plexus defect (the plexus surrounds one to four rows of germ cells instead of eight to nine rows as in wild-type animals (Byrd et al., 2014)). Importantly, the plexus is defined as the portion of the DTC (including the cap) with short intercalating processes (SIPs) that enwrap germ cells and is distinct from the few long processes and fragments of DTC membrane that extend beyond the SIPs (Byrd et al., 2014) (Fig. 1A). These long processes do not correlate with the extent of the germ progenitor zone (Cinquin et al., 2015; Crittenden et al., 2006; 1994; Fitzgerald and Greenwald, 1995; Hall et al., 1999; Lee et al., 2016; Pepper et al., 2003), whereas the plexus region overlaps with the stem cell pool (Byrd et al., 2014). In our initial screen of 708 genes by L1 RNAi treatment, we found that RNAi-mediated loss of 45 genes produced a DTC plexus defect in 15% or more of treated animals (Table S2; see Methods for rationale for determining the threshold for plexus defects). Knockdown of an additional 100 genes resulted in developmental defects, which precluded scoring of adults (Table S3).
Figure 1. Enwrapment in the C. elegans germ stem cell niche and screen design.
(A) The distal tip cell niche (DTC, green, lag-2 > GFP::CAAX) elaborates from a simple cap in the L4 larval stage (left) to form a plexus of processes enwrapping germ stem cells (magenta, mex-5 > H2B::mCh) in the adult C. elegans gonad 24 hours later (right). The germ stem cell pool (the distal-most 5 – 8 rows of germ cells), the progenitor zone (stem cells and their proliferative and early-differentiating progeny), and the meiotic transition zone (marked by entry into meiotic prophase) are labeled (bottom panel, third column). The DTC plexus region including the cap and short intercalating processes (SIPs) is labeled, and long external processes (LEPs) that extend farther than the plexus (white arrowheads) and fragments of processes are also indicated (black arrowheads, top panel, fourth column). Fluorescence images are full projections of the DTC from confocal z slices (see Methods). Lateral views of 3D isosurface renderings and transverse views of a 30-µm region of the DTC plexus rendering (boxed) (top panel, fifth and sixth columns) are shown indicating short intercalating processes (SIPs, arrowheads). Adult gonad image is comprised of two tiled images. Scale bars, 10 µm. (B) A targeted RNAi screen for failure of germ cell enwrapment identified 45 genes required early in development following L1 exposure to RNAi (at least 15% of examined animals had a 50% reduction in DTC plexus length), along with 100 genes producing developmental defects that precluded DTC scoring. A second round of screening identified 29 genes required specifically during the L4 and young adult stage, the critical period of DTC elaboration.
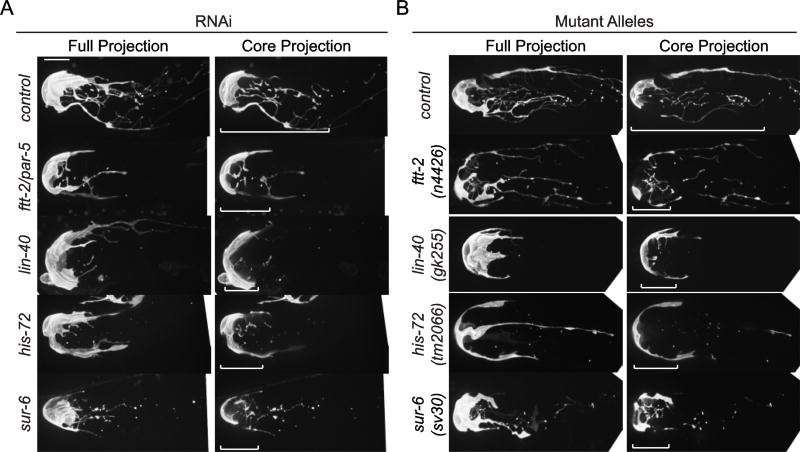
To more selectively identify genes that act during the critical period of DTC plexus formation and avoid developmental perturbations that might indirectly affect DTC enwrapping behavior, we performed an additional L3/L4 RNAi screen, exposing worms to RNAi approximately 12 hours prior to DTC plexus formation (Dalfó et al., 2012; Kamath et al., 2001). With this temporally restricted knockdown strategy, we rescreened the 45 genes required for DTC plexus formation. We also rescreened the 100 genes that were required for developmental progression after L1 RNAi treatment to avoid early developmental defects associated with loss of these genes. With this secondary screen, we identified 29 genes whose RNAi-mediated loss caused a DTC plexus defect (Table 1, Table S4). These genes represent multiple functional classes (Table 1), including protein synthesis and degradation (hars-1, rpl-11.1, F23B12.7, W07E6.2/Notchless, ppp-1, uba-1, dre-1), mitochondrial function (nuo-1, let-754, C16A3.5, rpom-1, dld-1), cell signaling (clr-1, lin-3/EGF, sur-6), transport and vesicle trafficking (dyn-1, sec-10, emo-1, copa-1, sar-1), RNA regulation (rnp-7, snr-2), chromatin and transcription (lin-40/MTA1, pab-1, his-72), cytoskeleton (unc-112), and nuclear transport and signaling (ran-1/Ran, ftt-2). Four genes associated with the screen for which putative null alleles were available (ftt-2, lin-40, his-72, sur-6) showed a reduction in the DTC plexus. We also examined animals harboring a null mutation of exoc-8, a regulatory subunit of the exocyst complex (Jiu et al., 2012), as a surrogate for the exocyst component sec-10, whose null allele is lethal (Zou et al., 2015). Loss of exoc-8 also showed a reduced DTC plexus. Together, the reduction of DTC elaboration in these null mutants confirms the specificity of their RNAi knockdown phenotypes and the fidelity of the screen (Fig. 2 and Table S5).
Table 1.
Genes required for DTC plexus formation in the L3/L4 stage.
| Gene Public Name (Reference) |
Sequence Name (Gene) |
CDS Description |
DTC Plexus Defect |
PercentC Defect |
Tissue-specific RNAi Site of Action |
|---|---|---|---|---|---|
| Mitochondria and energy metabolism | |||||
| nuo-1 | C09H10.3 | NADH ubiquinone oxidoreductase | 7/11 | 64% | DTC |
| let-754 | C29E4.8 | Adenylate kinase | 4/16 | 25% | DTC, GCs |
| C16A3.5 | C16A3.5 | NADH:ubiquinone oxidoreductase | 4/20 | 20% | DTC |
| rpom-1 | Y105E8A.23 | Mitochondrial RNA polymerase | 4/20 | 20% | |
| dld-1 | LLC1.3 | Dihydrolipoamide dehydrogenase | 3/19 | 16% | GCs |
| Transport and vesicle trafficking | |||||
| sec-10 | C33H5.9 | Exocyst subunit | 7/18 | 39% | DTC |
| emo-1 | F32D8.6 | Sec61p gamma subunit | 4/15 | 27% | DTC, GCs |
| copa-1 | Y71F9AL.17 | COPI complex alpha subunit | 3/9 | 33% | DTC, GCs |
| sar-1 | ZK180.4 | SAR (Secretion Associated, Ras-related) COPII vesicle coat protein | 3/19 | 16% | |
| dyn-1 | C02C6.1 | Dynamin GTPase | 5/24 | 21% | |
| RNA Regulation | |||||
| rnp-7 | K04G7.10 | RNP (RRM RNA binding domain)-containing | 4/20 | 20% | DTC |
| snr-2 | W08E3.1 | Small nuclear ribonucleoprotei n polypeptide | 3/16 | 19% | GCs |
| Protein Synthesis and Degradation | |||||
| dre-1 | K04A8.6 | F-box domain-containing protein | 9/29 | 31% | GCs |
| hars-1 | T11G6.1 | Histidyl tRNA synthetase | 8/10 | 80% | DTC |
| rpl-11.1 | T22F3.4 | Large ribosomal subunit L11 | 5/17 | 29% | |
| F23B12.7 | F23B12.7 | Constituent of 66S pre-ribosomal particles | 7/19 | 37% | |
| W07E6.2 | W07E6.2 | WD-repeat protein RSA4/Notchless | 4/26 | 15% | GCs |
| ppp-1 | C15F1.4 | Translation initiation factor eIF2B gamma subunit | 3/19 | 16% | |
| uba-1 | C47E12.5 | Ubiquitin-activating enzyme | 6/18 | 33% | GCs |
| Chromatin and transcription | |||||
| lin-40 | T27C4.4 | Histone deacetylase complex, chromatin binding | 9/19 | 47% | DTC |
| pab-1 | Y106G6H.2 | Poly(A)-binding protein | 7/29 | 24% | DTC, GCs |
| his-72 | Y49E10.6 | H3 histone | 3/20 | 15% | GCs |
| Cell signaling | |||||
| clr-1 | F56D1.4 | Receptor tyrosine phosphatase | 5/21 | 24% | |
| sur-6 | F26E4.1 | Regulatory subunit of serine/ threonine protein phosphatase 2A | 3/19 | 16% | |
| lin-3 | F36H1.4 | EGF family ligand | 4/27 | 15% | |
| Nuclear Transport and signaling | |||||
| ran-1 | K01G5.4 | Ran GTPase | 5/15 | 33% | DTC, GCs |
| ftt-2/par-5a | F52D10.3 | 14-3-3 signaling and scaffolding protein | 5/18 | 28% | DTC (ftt-2), GCs (par-5) |
| Focal adhesion | |||||
| unc-112 | C47E8.7 | Fermitin, PH domain-containing protein | 9/17 | 53% | DTC |
| Uncharacterized nematode-specific gene | |||||
| R08C7.1 | R08C7.1 | Protein coding gene | 6/25 | 24% | DTC, GCs |
Genes are grouped by reported function or predicted function based on homologs. DTC plexus defect is reported as the number of animals with a DTC defect out of total number of animals examined for that treatment. The negative control (empty RNAi vector L4440) showed a DTC plexus defect in 8/270 animals scored. Site of action determined by testing RNAis on tissue-specific RNAi strain is listed for distal tip cell (DTC)-specific RNAi and germ cell (GCs)-specific RNAi. For the site of action screen, defects >15% penetrance are indicated by bolding.
ftt-2 RNAi also targets the close paralog par-5; thus, gene-specific RNAi was performed to determine site of action (Table S5).
Figure 2. DTC plexus defects after RNAi depletion validated with null alleles.
In wild-type animals or worms fed control RNAi (L4440 empty vector), DTCs (lag-2 > 2x mKate2::PH) display an elaborate plexus of processes by the one-day-adult stage. In RNAi-treated animals (A) and putative null alleles (B) for ftt-2, lin-40, his-72, and sur-6, DTC plexus defects of > 50% reduction in plexus length were observed. ftt-2 RNAi also targets a close paralog, par-5. Confocal images are displayed as full projections of the DTC (left) and core 10 µm-thick projections of the DTC plexus from central confocal z slices (right; see Methods). Brackets indicate the length of the elaborated DTC plexus. Scale bar, 10 µm.
Identification of genes that act in germ cells to promote plexus formation
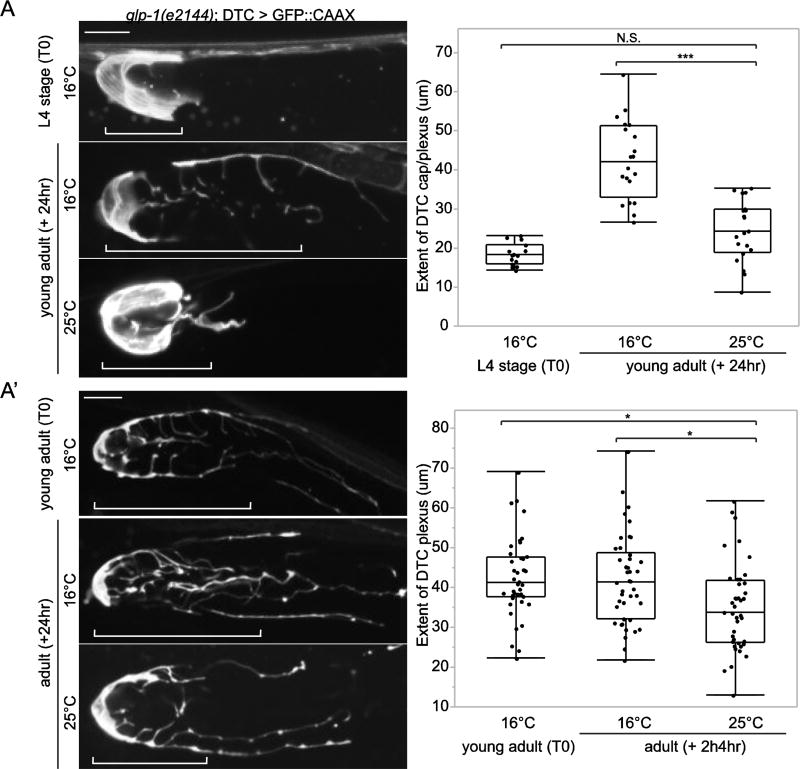
Germ progenitor cells may signal to the DTC to foster enwrapment during plexus formation. Forcing premature differentiation of germ cells in young adult animals has been reported to result in a failure of DTC elaboration, suggesting that germ progenitor cells are required for plexus formation (Byrd et al., 2014). This defect, however, was not quantified. We thus repeated the experiment, utilizing the GLP-1/Notch temperature-sensitive allele glp-1(e2144) that causes germ cell differentiation (adoption of meiotic fate) at 25°C (Austin and Kimble, 1987; Priess et al., 1987). Quantitative analysis revealed that the plexus completely failed to elaborate in glp-1(e2144) animals after shifting L4 animals to the restrictive temperature (25°C) (Fig. 3A), suggesting that the DTC does not enwrap meiotic germ cells. This analysis confirms that initial DTC elaboration requires GLP-1/Notch signaling, presumably due to a requirement for germ progenitor cells, as previous studies have shown that restoration of germ progenitor cells in a GLP-1/Notch-deficient background restores the DTC plexus (Byrd et al., 2014). To determine whether the DTC plexus continues to depend on germ progenitor cells after it forms in young adults, we next performed a temperature shift after DTC elaboration occurred. Young adult animals exposed to the restrictive temperature showed reduced plexus length compared to control animals (Fig. 3A’), suggesting that germ progenitor cells help maintain the DTC plexus.
Figure 3. Germ progenitor cells are required for initiation and maintenance of the DTC plexus.
(A) Forcing germ cell differentiation in temperature-sensitive glp-1(e2144) L4 larval animals at the restrictive temperature resulted in failure of DTC plexus initiation compared to control animals at the permissive temperature (***p < 0.0005, N.S., not significant (p > 0.05), n > 15 for each group, ANOVA followed by Tukey-Kramer HSD test). (A’) Forcing germ cell differentiation in glp-1(e2144) young adults at the restrictive temperature resulted in failure of DTC plexus maintenance compared to control animals at the permissive temperature (*p < 0.05, n > 23 for each group, ANOVA followed by Tukey-Kramer HSD test). Confocal images are displayed as 6 µm-thick core projections of the DTC plexus or cap. Brackets indicate the length of the DTC plexus. Scale bars, 10 µm.
We next hypothesized that some of the genes identified in our screen might function indirectly in the germ cells to promote enwrapment, since the DTC appears to require a robust germ progenitor cell population to form and maintain the plexus. Thus, compromising germ progenitor cells may indirectly affect plexus formation. To determine if genes identified in our screen function cell-autonomously within the germline but not the DTC, we used the rrf-1(pk1417) background, which reduces somatic but not germline RNAi efficiency (Sijen et al., 2001). This strain has been widely used for germline-specific RNAi, although it has been reported to have RNAi activity in the intestine (Kumsta and Hansen, 2012). Importantly, the rrf-1 strain does not appear to be sensitive to RNAi in the DTC (Kumsta and Hansen, 2012), making it suitable for testing germline versus DTC site of action.
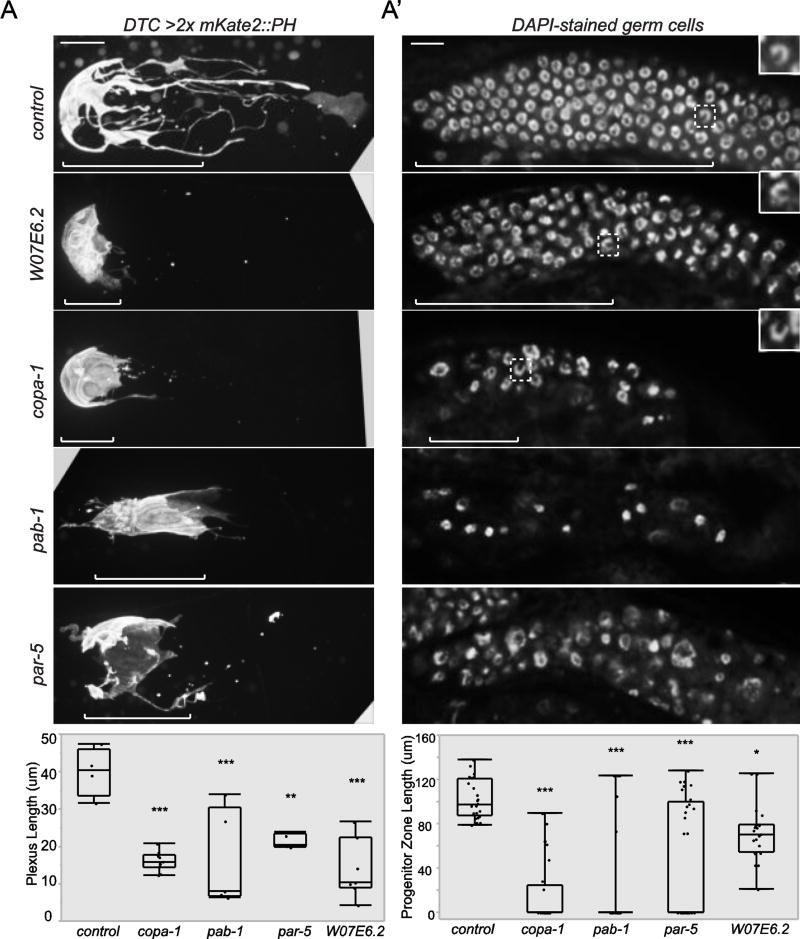
Using the rrf-1 germline-specific RNAi strain, we identified a set of genes whose loss in the germline indirectly caused DTC plexus defects (Table 2). DTC defects were observed in > 15% of animals after germline RNAi knockdown of the histone variant his-72 (Ooi et al., 2006), the polyA-binding protein pab-1, a subunit of the COPI Golgi-to-ER (endoplasmic reticulum) vesicle coat protein complex copa-1, the ER protein translocator Sec61p γ homolog emo-1, the GTPase ran-1/Ran, a regulator of ribosomal biogenesis W07E6.2/Notchless, and the 14-3-3 signaling protein par-5 (Fig. 4A). SAGE analysis has indicated that these genes are expressed in dissected gonads, though multiple somatic gonad cells including the DTC are present in this sample (Xin Wang et al., 2009). Notably, pab-1, W07E6.2, and par-5 have reported roles in germ cell proliferation (Aristizábal-Corrales et al., 2012; Ko et al., 2010; Voutev et al., 2006). Germline-expression profiling previously showed that his-72, ran-1, W07E6.2, and snr-2 are enriched in wild-type animals compared to germline-depleted mutants (Reinke et al., 2004), suggesting that these genes are expressed in the germline. Additionally, a single copy transgenic reporter for par-5 (Mikl and Cowan, 2014) is expressed in germ cells (Fig. 5). Though we did not observe expression of a copa-1 reporter in germ cells, germline silencing of multicopy transgenic arrays has been frequently observed (Praitis et al., 2001), and in situ hybridization for copa-1 from the NEXTDB database showed strong staining throughout the gonad (Antoshechkin and Sternberg, 2007).
Table 2.
Genes that function in the germ cells to indirectly promote plexus formation.
| Germline-specific RNAi (L3/L4 plating)
| |||
|---|---|---|---|
| Gene Name |
defect | total | % defect |
| his-72 | 13 | 23 | 57% |
| pab-1 | 8 | 23 | 35% |
| copa-1 | 10 | 29 | 34% |
| emo-1 | 6 | 25 | 24% |
| ran-1 | 8 | 39 | 21% |
| W07E6.2 | 10 | 50 | 20% |
| ftt-2/par-5a | 8 | 42 | 19% |
| uba-1 | 3 | 33 | 9% |
| dld-1 | 2 | 29 | 7% |
| snr-2 | 1 | 21 | 5% |
| let-754 | 1 | 27 | 4% |
| dre-1 | 1 | 44 | 2% |
| R08C7.1 | 1 | 66 | 2% |
| L4440 | 0 | 24 | 0% |
DTC plexus defects were scored as >50% reduction in plexus length. The number of animals with DTC defects and the total number of animals examined are shown. All 29 genes identified in whole-body RNAi screens were scored in the germline-specific rrf-1(pk1417); RNAi knockdown of genes not shown had no observable DTC defects.
par-5 and ftt-2 are close paralogs targeted by the same RNAi construct.
Figure 4. The DTC plexus depends on the germ cell progenitor zone.
(A) Full projections of confocal z slices of DTCs (lag-2 > 2x mKate2::PH) show that RNAi depletion of W07E6.2, copa-1, par-5, and pab-1 result in a reduction of plexus length compared to control RNAi (L4440 empty vector) (**p < 0.005, ***p < 0.0005, n > 4, ANOVA followed by Tukey-Kramer HSD test). Brackets indicate the length of the DTC plexus. (A’) RNAi knockdown of W07E6.2, copa-1, par-5, and pab-1 resulted in loss or reduction of the germ progenitor zone compared to empty vector control (*p < 0.05, **p < 0.005, ***p < 0.0005, n > 22 for all groups, ANOVA followed by Tukey-Kramer HSD test). A single superficial confocal z slice of DAPI-stained germ cells is shown. Brackets indicate the length of the progenitor zone. Dashed boxes surround meiotic nuclei identified by their crescent shape (inset magnification). Scale bars, 10 µm.
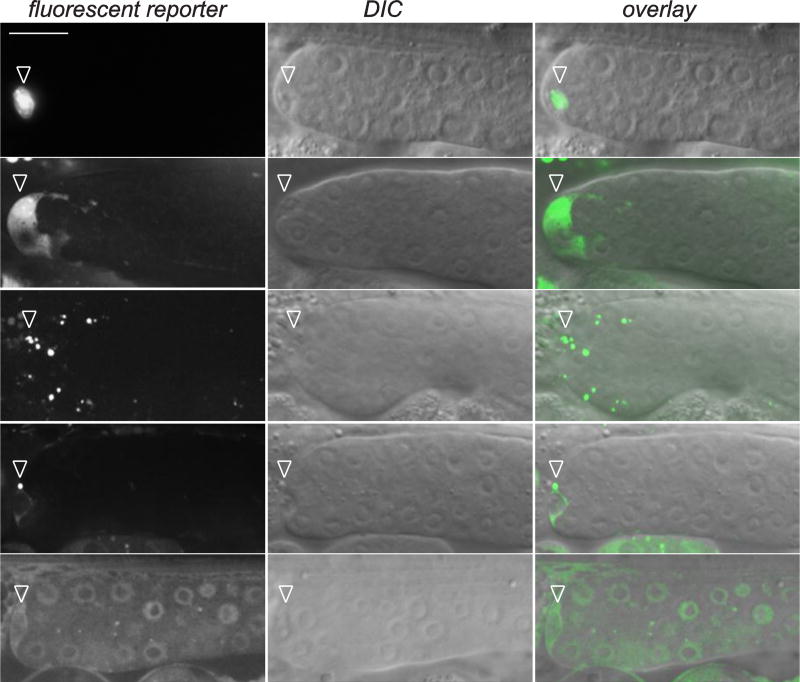
Figure 5. Transgene reporter expression of genes that cell-autonomously promote DTC enwrapping behavior.
Fluorescence image, left, corresponding differential interference contrast (DIC) image, middle, and overlay, right. Transcriptional (>) and translational (::) reporter construct expression was observed in the DTC in one-day-old adult animals. A translational reporter for the exocyst component sec-10 showed a punctate pattern in the DTC cell body (n = 11/11 animals observed). PAR-5::GFP was also expressed in germ cells. Arrowheads indicate DTC expression, and brackets indicate germ cell expression. copa-1 > GFP and sec-10 > SEC-10::GFP images are displayed as 8 µm-thick projections from central confocal z slices. A single confocal z slice is shown for ftt-2 > FTT-2::mCh and par-5 > PAR-5::GFP. A wide-field fluorescence image of lin-40 > GFP is shown. Scale bar, 10 µm.
To determine if loss of these germline-functioning genes might alter the DTC plexus indirectly by affecting germ progenitor cells, we next examined the germ cells after knockdown of selected genes. Short gonad arms with fewer germ cells were observed in germline-specific L1 knockdown of W07E6.2 (observed in 7/22 animals), copa-1 (21/25), pab-1 (23/28), and par-5 (7/29), compared to the RNAi control (L4440 empty vector, 0/24). Reduced progenitor zone length was observed in W07E6.2/Notchless, copa-1, pab-1, and par-5 (Fig. 4A’). Large nuclei suggestive of premature differentiation or cell division defects in the distal gonad were also observed for par-5 (16/29 animals), copa-1 (21/25), and pab-1 (10/28), compared to the empty vector control (0/24). Taken together, these observations support the idea that while the DTC promotes germ stem cell fate (Byrd and Kimble, 2009; Kimble and White, 1981), the progenitor cells provide a reciprocal cue back to the DTC to promote its enwrapping behavior during plexus formation.
Identification of genes that function in the DTC to promote plexus formation
We next wanted to determine if any genes identified in our screen promote DTC enwrapping behavior directly (cell-autonomously) in the DTC. To accomplish this, we generated a DTC-specific, RNAi-hypersensitized strain with fluorescently labeled DTC membranes to highlight the DTC plexus [(lag-2>mNeonGreen::PH::F2A::rde-1; rrf-3(pk1426); rde-1(ne219)]; see Methods for construction and validation of DTC specificity and Fig. S1). DTC plexus defects were observed in >15% of animals after DTC knockdown of the 14-3-3 proteins ftt-2 and par-5, the sole C. elegans Ran GTPase ortholog ran-1, the NuRD nucleosome remodeling complex subunit lin-40/MTA1, the polyA-binding protein pab-1, the COPI subunit copa-1, the histidyl tRNA synthetase hars-1, the focal adhesion and muscle dense body component unc-112, and the spliceosome component rnp-7 (Table 3). Importantly, DTC knockdown of these eight genes did not grossly perturb the germline, consistent with specific knockdown and function in the DTC (n = 20/20 animals per treatment showed wild-type size and regionalization of the germline) (Fig. S2). Three of these genes – ran-1, copa-1, and pab-1 – were required in both the DTC and germ cells, consistent with their essential cellular functions. Available expression reporters for lin-40 (McKay et al., 2003), sec-10, and copa-1 were expressed in the DTC (Fig. 5), consistent with a direct role in promoting DTC enwrapping behavior.
Table 3.
Genes that function in the DTC to promote plexus formation.
| DTC-specific RNAi (L3/L4 plating)
| |||
|---|---|---|---|
| Gene Name | defect | total | % defect |
| ftt-2/par-5a | 14 | 21 | 67% |
| ran-1 | 9 | 20 | 45% |
| hars-1 | 9 | 20 | 45% |
| lin-40 | 10 | 29 | 34% |
| pab-1 | 9 | 28 | 32% |
| copa-1 | 8 | 47 | 17% |
| rnp-7 | 3 | 20 | 15% |
| unc-112 | 3 | 20 | 15% |
| nuo-1 | 4 | 32 | 13% |
| sec-10 | 3 | 26 | 12% |
| emo-1 | 3 | 26 | 12% |
| C16A3.5 | 3 | 27 | 11% |
| let-754 | 2 | 30 | 7% |
| R08C7.1 | 2 | 33 | 6% |
DTC plexus defects were scored as >50% reduction in plexus length. The number of animals with DTC defects and the total number of animals examined are shown. All 29 genes identified in whole-body RNAi screens were scored in the DTC-specific lag-2>RDE-1 strain; RNAi knockdown of genes not shown had no observable DTC defects.
par-5 and ftt-2 are close paralogs targeted by the same RNAi construct.
Another gene that appeared to be required in both the DTC and germline to promote DTC plexus formation was par-5. However, the RNAi clone that targets par-5 is also predicted to knock down its close paralog ftt-2 (Li et al., 2007; Rual et al., 2004). To definitively determine the site of action of par-5 and ftt-2, we designed off target-optimized RNAi constructs for par-5 and ftt-2 with the dsCheck program (Naito et al., 2005). Gene-specific knockdown of par-5 yielded strong DTC plexus defects in the germline-specific RNAi strain (Table S5 and Fig. S3) but not in the DTC-specific RNAi strain. These observations are consistent with the reported role of par-5 in directly promoting germ cell proliferation (Aristizábal-Corrales et al., 2012). Consistent with reports that par-5 is the sole germline C. elegans 14-3-3 protein (Aristizábal-Corrales et al., 2012), a par-5 reporter transgene was expressed in germ cells (Fig. 5) and knockdown of ftt-2 in the germline did not affect DTC plexus formation. In contrast to par-5, loss of ftt-2 in the DTC resulted in a plexus defect (Table S5), and a reporter for ftt-2 was expressed in the DTC (Fig. 5). Together these data suggest that this pair of 14-3-3 proteins in C. elegans act in distinct tissues to affect the niche: par-5 promotes DTC elaboration indirectly via its role in germ cell proliferation, and ftt-2 functions in the DTC to promote its enwrapping behavior.
DTC enwrapment of germ stem cells promotes stem cell fate
The DTC acts as the germline stem cell niche and expresses transmembrane Notch ligands that activate GLP-1/Notch signaling in germ cells and promote stem cell fate through direct transcriptional targets including sygl-1 (Kershner et al., 2014). As sygl-1 is a direct Notch target and stem cell regulator, sygl-1 expression allows more precise assessment of germ stem cells in contrast to measurements of the progenitor zone, which include some early differentiating cells that do not yet show overt features of differentiation (Crittenden et al, 2017). Single molecule FISH for the stem cell marker sygl-1 showed that germ stem cells actively transcribing sygl-1 are more likely to contact DTC processes (Lee et al., 2016), suggesting that DTC contact is required for sygl-1 expression. We hypothesized that the function of DTC enwrapment of germ cells is to present a localized source of Notch ligands to a larger pool of cells than an unelaborated cell could contact, thereby allowing a single niche cell to maintain a robust stem cell population. To assess the consequences of reduced DTC enwrapment on germ stem cells, we examined the robustness of niche signaling by utilizing a transcriptional reporter for sygl-1 (sygl-1 > H2B::GFP) (Kershner et al., 2014). The sygl-1 > H2B::GFP-positive cell population expanded 2.5-fold from the L4 to adult stages, correlating with the time window of plexus formation (Fig. S4). This might seem surprising given the reported similar lengths of the sygl-1 active transcription zone measured in germ cell diameters (Lee et al., 2016). However, the circumferential width of the distal gonad dramatically increases from L4 to adult stages (from ~3 germ cell diameters wide to ~5 germ cell diameters wide, Fig. 1A), which might account for the increased sygl-1 > H2B::GFP-positive cell population.
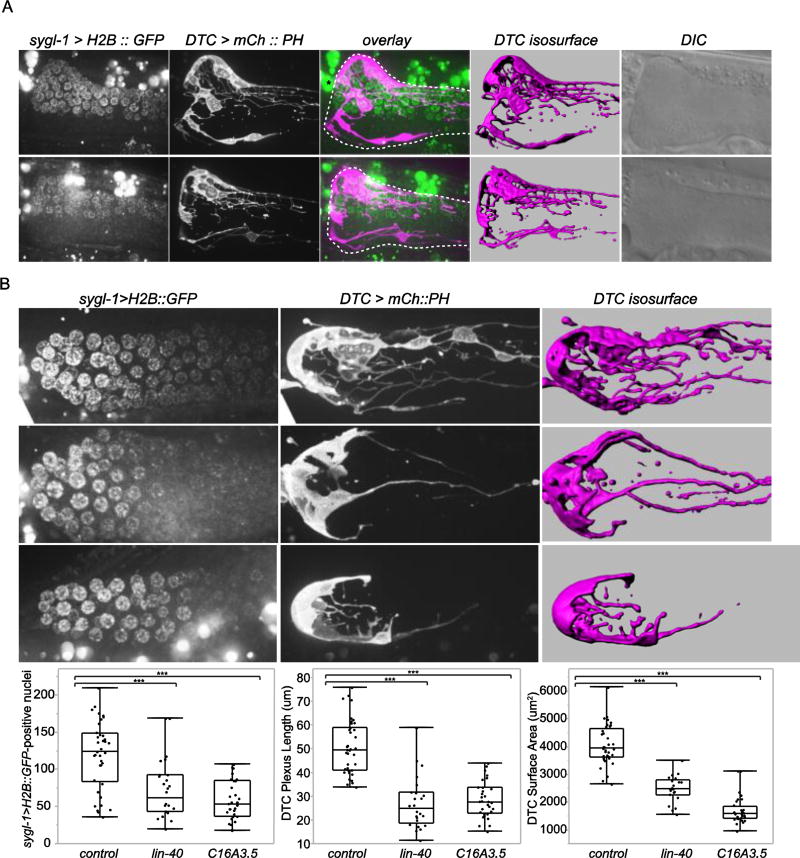
Strikingly, in rare cases of wild-type animals with the dense DTC plexus biased to one side of the gonad, sygl-1 > H2B::GFP was exclusively expressed in germ cells within the asymmetric plexus and was absent in distal germ cells on the other side of the gonad (n = 4/4 cases of asymmetric plexus formation and sygl-1 expression observed in 688 animals) (Fig. 6A). Notably in two of these animals (shown in Fig. 6A), there were unbranched superficial DTC processes opposite the asymmetric plexus and no sygl-1 reporter expression was observed. This is likely because the long superficial processes are not highly branched and do not intercalate between germ cells to effectively activate Notch expression.
Figure 6. Enwrapment by the DTC promotes germ stem cell fate.
(A) Expression of the GLP-1/Notch transcriptional target sygl-1 (sygl-1 > H2B::GFP, green) was observed specifically in germ stem cells enwrapped by the DTC plexus in two cases of rare wild-type animals with the DTC plexus (magenta, lag-2 > mCh::PH) formed on one side of the gonad arm. 15 or 20 µm-thick projections of confocal z slices of sygl-1 > H2B::GFP-expressing cells, the DTC, and fluorescent overlay, isosurface renderings, and DIC images, from left to right, are shown. Dashed white lines indicate the gonad, and an asterisk marks autofluorescent gut granules. (B) lin-40 and C16A3.5(RNAi) animals showed a reduced number of sygl-1 > H2B::GFP-expressing cells, left, reduced DTC plexus, middle, and reduced surface area of the DTC assessed by 3D isosurface renderings, right (***p < 0.0005; > 24 animals per category; ANOVA followed by Tukey-Kramer HSD test). Full projections of fluorescent images and isosurface renderings of the DTC are shown. All scale bars, 10 µm.
The distribution of sygl-1 > H2B::GFP-positive nuclei during the wild-type developmental progression of the L4-stage DTC cap to the adult plexus, as well as in the rare asymmetric adult plexus, suggest that DTC plexus-germ cell contact leads to sygl-1 reporter activation and the normal expansion of the germ stem cell pool in the adult. We hypothesized that RNAi causing plexus reduction would therefore cause a reduction in the number of sygl-1-positive cells. Indeed, whole body RNAi knockdown of the DTC-autonomous genes lin-40/MTA1 and C16A3.5, which specifically affected the DTC and did not perturb gonad development (Table S2, Fig. S2), resulted in reduction of DTC plexus length and surface area and a reduced number of sygl-1 > H2B::GFP-positive nuclei (Fig. 6B). Notably, the number of sygl-1-reporter positive nuclei was similar to the number of cells observed in wild-type L4 animals with unelaborated DTCs (Fig. S4). Taken together, these observations offer compelling evidence that germ cell enwrapment within the adult DTC plexus extends Notch signaling to additional germ cells to promote germ stem cell fate.
Discussion
Niche cells transmit cues to promote stem cell fate. Cellular enwrapment of stem cells by their niche is commonly observed for a wide range of stem cell systems and is a potential mechanism for mediating signaling specificity. Despite its widespread occurrence, the regulation and function of enwrapment of stem cells by their niche support cells is poorly understood. We performed the first screen for regulators of enwrapment in the C. elegans germ stem cell niche and identified 29 genes required during the critical period when the distal tip cell (DTC) niche forms the DTC plexus by enwrapping germ stem cells. We further investigated whether these genes were required in the DTC niche or the stem cells. We confirmed that germ progenitor cells induce enwrapment by their niche and demonstrated that niche elaboration promotes germ stem cell fate. This reciprocal signaling between niche and stem cell may comprise a positive feedback loop that expands and maintains the stem cell pool.
To detect genes required for DTC plexus formation, we performed an RNAi screen for severe loss of the DTC plexus. We stringently defined a DTC plexus defect as a 50% reduction in plexus length, which is not observed in wild-type animals. Thus we are confident that even low penetrance defects by RNAi are informative. As knockdown of genes by feeding animals bacteria expressing dsRNA is often incompletely penetrant and variable (Simmer et al., 2003), a low penetrance defect by feeding RNAi can detect genes with significant roles. As a positive control, we performed RNAi to the key niche regulator lag-2 and observed a low penetrance defect by our stringent criteria. Finally, we validated our RNAi screen candidates with available mutant alleles, which showed higher penetrance defects and confirmed our screening approach.
Previous work has suggested that there might be a dynamic interplay between the germ progenitor cells and the DTC to promote its enwrapping behavior during plexus formation. For example, an apparent dependence of the DTC processes on germ progenitor cells has been observed (Byrd et al., 2014), indicating that genes might affect DTC elaboration indirectly through their roles in the germline. To identify genes that promote DTC plexus formation and determine their site of action, we thus used targeted and tissue-specific RNAi to define sets of genes functioning specifically in the germline and DTC. Such temporally- and tissue-specific strategies enable dissection of the chicken-and-egg relationship of the niche and stem cells.
DTC enwrapping behavior initiates plexus formation between the late L4 and young adult stages, a period of dramatic morphological change in the distal gonad. Careful examination of RNAi knockdown of several germline-functioning genes (copa-1, par-5, W07E6.2, and pab-1) revealed that all led to a reduced germ progenitor zone and to plexus reduction. Further, premature differentiation of distal germ cells in the glp-1(ts) mutant similarly causes plexus reduction (Byrd et al., 2014). Thus, we hypothesize that the germ progenitor cells generate a cue to stimulate DTC enwrapping behavior to form the plexus in the late L4 and the early adult. As only the germ stem cells seem to be enwrapped (Byrd et al., 2014, and see below), it is possible that only the stem cells generate a cue that promotes enwrapment. Alternatively, all of the mitotic cells at the L4 – adult transition may send a signal that stimulates enwrapment, and only those that then become enwrapped retain stemness through adulthood. This enwrapment-promoting cue may be generated specifically during the L4 – adult transition when the DTC plexus elaborates. Alternatively, the cue might be produced earlier and the DTC becomes receptive to the signal during the late L4 stage. Notably, the late L4 is the time when DTC migration to form the gonad arm is completed, suggesting that plexus formation might require cessation of migration (Byrd et al., 2014). The germ cell-autonomous genes we identified may act upstream of a potential germ progenitor cell cue, as none are secreted or transmembrane and thus are not likely candidates to be the cue itself. Further screens that identify germline-expressed secreted or transmembrane ligands required for DTC enwrapment may identify our hypothesized germ progenitor cell cue. In addition to a role for the germ progenitor cells in the L4 stage in promoting plexus formation, we also found that differentiation of germ progenitor cells in the one-day-old adult reduced the plexus. Thus the germ progenitor cells appear to stimulate enwrapping behavior during plexus formation and are also required for its maintenance.
Our screen was biased towards identifying genes required for fecundity, which may include genes that promote germ progenitor cell fate and thus act to support the process of stem cell enwrapment. This is likely the reason this screen did not reveal any genes whose reduction increased the DTC plexus. Notably, genes that function to repress DTC plexus length do exist, as loss of the mitochondrial 2-oxoglutarate carrier misc-1 increases the length of DTC processes and the germ progenitor zone (Gallo et al., 2011).
As our RNAi screen focused on genes with annotated roles in fecundity, it is possible that we failed to identify genes that promote enwrapment by the DTC but that do not dramatically affect fecundity. Indeed, while we show that a reduced plexus results in a smaller stem cell pool, it is not yet clear if this reduction in germ stem cells dramatically affects fecundity or whether the strong fecundity defects caused by these genes are due to pleiotropic roles in germline development. However, the observations that a number of the genes we screened function in the DTC to promote enwrapment and that this enwrapment regulates the stem cell pool are consistent with the idea that our screen of genes involved in fecundity likely enriched for direct regulators of germ stem cell enwrapment. It will be interesting to see in future screens if genes that do not alter fecundity reduce DTC elaboration.
We favor the idea that the germ progenitor cells generate a cue that stimulates enwrapment, rather than acting to stretch DTC processes passively due to adhesion to dividing germ cells. One important piece of evidence for this is that DTC elaboration initiates at a specific developmental stage, the L4-adult transition, when germ cell divisions have slowed (Roy et al., 2015). In addition, previous studies performed live imaging of DTC process growth and observed that DTC processes grow not only with the distal-to-proximal progression of dividing germ cells but also in the opposite direction (Wong et al, 2013). These studies also observed that processes form transient structures including rings and branching points and can grow in spurts, all dynamic behaviors that suggest that DTC elaboration is at least in part an active, DTC-autonomous process.
Using our newly developed DTC-specific RNAi strain, our studies identified genes that are required in the DTC for plexus formation. One gene that is necessary for DTC elaboration is lin-40/MTA1, a chromatin regulator, which is part of the nucleosome remodeling and histone deacetylation (NuRD) complex (Solari et al., 1999). Formation of other stem cell niches, including in the murine testis (Wu et al., 2017) and in the Drosophila ovary (Hitrik et al., 2016; Upadhyay et al., 2016), also requires specific chromatin regulators. As the DTC actively remodels from a simple cap to a highly branched and elaborate structure, energetically intensive and dynamic membrane addition and remodeling may also be key components underlying plexus formation. Consistent with this idea, our screen implicated genes required for mitochondria (NADH:ubiquinone oxidoreductases nuo-1/NDUFV1 and C16A3.5/NDUFB9), the cytoskeleton (focal adhesion component unc-112/FERMT2), and exocytosis (exocyst complex component sec-10/EXOC-5). Interestingly, lin-3/EGF was identified in our initial whole body RNAi screen. The role of EGF signaling in other stem cell niches including in the Drosophila testis (H. Chen et al., 2013; Kitadate and Kobayashi, 2013; Sarkar et al., 2007) and intestinal stem cell niches (Biteau and Jasper, 2011) suggests an interesting parallel between signaling pathways required in the C. elegans and Drosophila stem cell niches.
Niches precisely regulate delivery of stemness signals by various mechanisms including cell-contact based signaling (Anklesaria et al., 1990; Ding et al., 2012), interactions with extracellular matrix (Xiaomeng Wang et al., 2008), and polarized signaling protrusions (Inaba et al., 2015; Mandal et al., 2007; Rojas-Ríos et al., 2012). The cellular enwrapment observed in the DTC plexus was previously hypothesized to facilitate juxtacrine cell-cell signaling, to anchor stem cells in the niche, or to impact the microenvironment (Byrd et al., 2014; Crittenden et al., 2006). The GLP-1/Notch ligands LAG-2 and APX-1 are both expressed in the DTC, and LAG-2 localizes to the entire DTC surface including its processes (Crittenden et al., 2006). Though we excluded the possibility that our candidates regulate lag-2 expression, some of the DTC-autonomous genes identified in our screen could regulate the expression of APX-1 or posttranslational modifications or localization of these ligands.
We examined a direct target of Notch signaling, sygl-1 (Kershner et al., 2014), to dissect the relationship of germ stem cells and the DTC plexus. By examining wild-type animals with asymmetric plexus formation and identifying genes that cell-autonomously promote DTC plexus formation, we found that enwrapment by the plexus promotes expression of the stem cell marker sygl-1. The expansion of DTC processes between L4 and adult stages forms the plexus and encompasses a larger number of sygl-1-reporter-positive germ stem cells, potentially establishing the adult germ stem cell pool. While strong sygl-1 transcription overlaps with the extent of the DTC cap in the L4 stage, some sygl-1-positive nuclei appear to reside out of contact with the DTC cap in L4 animals (Lee et al. 2016). Possibly, DTC contact may not be required in the L4 stage for GLP-1/Notch activation during larval germline expansion. We also examined rare wild-type adult animals with asymmetric plexus formation, allowing us to observe that distal germ cells without DTC plexus contacts do not express a sygl-1 reporter. These data show that all sygl-1 > GFP::H2B+ cells specifically reside within the asymmetric plexus, strongly suggesting that DTC-germ cell interactions in the adult directly promote Notch activation. Interestingly, the border of sygl-1 expression was sharply tied to the side of gonad with the asymmetrically localized DTC plexus. This contrasts to the more graded distal-proximal decrease in sygl-1 expression at the end of the plexus observed in a typical distal-localized plexus (Lee et al., 2016). It is possible that cytoplasmic streaming within the syncytial gonad occurs most strongly along the distal-to-proximal axis, thus spreading sygl-1 mRNA (Lee et al., 2016) or the intracellular domain of GLP-1/Notch after interaction with its DTC-expressed ligands to proximal germ cell nuclei (but not nuclei orthogonal to this axis) in a graded manner.
Extending the idea that the plexus is tied to the germ stem cell pool, knockdown of DTC-functioning genes lin-40 and C16A3.5 showed a shorter DTC plexus phenotype and displayed reduced numbers of sygl-1 > GFP::H2B-expressing cells. We do not believe this decrease in Notch activation is caused by the failure of the DTC to produce or localize Notch ligands in these experiments because sygl-1 > GFP::H2B is still activated within the plexus, and germ cells do not differentiate in the distal zone, a hallmark phenotype of Notch signaling defects (Austin and Kimble, 1987). These observations support the idea that one key function of the DTC plexus — and perhaps other enwrapping stem cell niches — is to foster contact-mediated signaling in stem cells, extending the range of niche signaling to promote stem cell fate in a larger pool of cells.
Taken together, our studies have identified new regulators of germ stem cell enwrapment by the DTC niche in C. elegans and have revealed a probable positive feedback loop between the enwrapping DTC and germ stem cells that reinforces and extends the germ stem cell pool. Our dissection of stem cell enwrapment provides reagents and assays to deepen our understanding of DTC plexus-germ cell interactions and identify and examine additional possible functions of the DTC plexus. As enwrapment of stem cells by niche support cells is broadly observed, we expect these studies will have wide-ranging significance to understanding stem cell regulation and human health.
Materials and Methods
Strains and culture
Worm strains were maintained on NGM plates at 16, 18, or 20°C (Brenner, 1974). In strain descriptions, we designate linkage to a promoter with a greater-than sign (>) and designate in-frame protein fusions with a double semicolon (::). The following transgenes and alleles were used in this study: qyIs353(lag-2>GFP::CAAX), qyIs382(lag-2>mCh::PH), stIs11646(egr-1a::H1-wCherry), sEx10059(Y71F9AL.17::GFP), crc102(ftt-2>ftt-2::Spep-TEV-mCherry); LGI, cpIs121(lag-2>mNG::PH::F2A::rde-1), rrf-1(pk1417), exoc-8(ok2523), sur-6(sv30)/hT2; LGII, qSi26(sygl-1>H2B::GFP::sygl-1 3' UTR), oaSi10(par-5>par-5::GFP), cpIs91(lag-2>2x mKate2::PH), cpIs122(lag-2>mNeonGreen::PH), naSi2(mex-5>H2B::mCh), rrf-3(pk1426); LGIII, his-72(tm2066), glp-1(e2144); LGIV, lin-40(gk255)/nT1 [qIs51], teIs1(oma-1::GFP); LGV, qIs19(lag-2>GFP::unc-54), qIs56(lag-2>GFP), rde-1(ne219); LGX, ftt-2(n4426). Mutations were followed in crosses by PCR, sequencing, or previously reported phenotypes. naSi2 was a kind gift from Jane Hubbard.
Molecular biology and strain construction
We utilized the DTC-specific expression of the lag-2 promoter to drive membrane-targeted GFP::CAAX and mCherry::PH. The 2.8 kb region upstream of the lag-2 ATG start codon was amplified by PCR and cloned into the pPD95.75 vector at HindIII and BamHI sites. The lag-2 promoter region was then amplified and fused to GFP::CAAX (pSA129) or mCh::PH (pAA173 (Ziel et al., 2009)) by fusion PCR and coinjected with transformation marker pPD#MM016B (unc-119) as well as EcoRI-digested salmon sperm DNA and pBluescript II as carrier DNA at 50 ng/uL into unc-119(ed4) animals. pPD95.75 was a gift from Andrew Fire (Addgene plasmid # 1494), and pSA129 was a gift of Scott Alper. Integrated lines were obtained by standard protocol (Inoue et al., 2002). Other DTC marker strains were generated via CRISPR-Cas9 mediated recombination to insert lag-2 > mNG::PH or lag-2 > 2x mKate2::PH near the standard MoSCI insertion site ttTi5605 on chromosome II (Dickinson and Goldstein, 2016). (Guide sequences for targeting the MoSCI sites are: chromosome I, GAAATCGCCGACTTGCGAGG; chromosome II, ATATCAGTCTGTTTCGTAA.)
To develop a RNAi-hypersensitized DTC-specific RNAi strain that labels the DTC plexus, a single copy of lag-2>mNG::PH::F2A::rde-1 was inserted into the MoSCI site ttTi4348 on chromosome I in the rrf-3(pk1426); rde-1(ne219) background. In contrast to a previously generated DTC-specific RNAi strain (Martynovsky et al., 2012), this new strain contains the RNAi-hypersensitive mutation rrf-3 to facilitate screening (Simmer et al., 2003). Additionally, this new DTC-specific strain labels the DTC cell membrane instead of predominantly labeling the cell body with cytoplasmic GFP. Finally, in contrast to the previous strain which contains two transgenic arrays, this new DTC-specific strain contains one single copy transgene, lag-2>mNG::PH::F2A::rde-1, labeling all rde-1-rescued cells with NeonGreen. This single copy strategy also reduces background DTC migration defects present in the previous strain; in contrast, our strain shows wild-type migration (Table S6). The lag-2 promoter is specific to the DTC in L4 and adult stages (Fig. S1) and has been previously used for DTC-specific RNAi (Martynovsky et al., 2012; Tannoury et al., 2010; Wong et al., 2014). We observed broader lag-2 expression in the L1 stage in the somatic gonad precursors Z1 and Z4 and in the L2 stage in the seam cells. To avoid knockdown in these other tissues, we utilized L3/L4 platings on RNAi for scoring of DTC plexus defects, thus allowing DTC-specific knockdown.
To confirm that the DTC-specific strain was sensitive to RNAi in the DTC, worms were treated with RNAi to genes known to regulate DTC migration cell-autonomously (gon-1 (Blelloch and Kimble, 1999), mig-38 (Martynovsky et al., 2012)), and penetrant DTC migration defects were observed (Table S6). To confirm that the DTC-specific RNAi strain was insensitive to RNAi in other tissues, worms were treated with RNAi to germline- and muscle-expressed genes. First, worms were treated with RNAi for four essential germline genes (gld-1 (Francis et al., 1995), glh-1 (Gruidl et al., 1996), fbf-2 (Crittenden et al., 2002), fog-3 (Ellis and Kimble, 1995)), and expected phenotypes were observed in the germline-specific rrf-1(pk1417) strain but not in the DTC-specific lag-2>RDE-1 strain (Table S6). Next, worms were treated with RNAi to the muscle-expressed collagen emb-9 (Graham et al., 1997), and worm rupture was observed in the whole-body RNAi-sensitized strain rrf-3(pk1426) but not in the DTC-specific lag-2>RDE-1 strain (Table S6).
Off-target optimized RNAi constructs including 200 or 600 bp of coding sequence were designed using the Web portal dsCheck (Naito et al., 2005), transformed into the RNAi vector HT115 (Timmons 2001), and retested in tissue-specific RNAi strains to confirm DTC plexus defects (Table S5). The redesigned ran-1 clone caused a highly penetrant DTC plexus defect (100%, n = 30). The redesigned W07E6.2 clone did not show a penetrant defect, so we cannot rule out off-target effects; alternatively, the shorter RNAi construct may be ineffective. Primers are available upon request.
L1 plating RNAi screen
We carried out a targeted RNAi screen of genes known to affect fecundity and cell communication utilizing the C. elegans ORF-RNAi feeding RNAi library (Rual et al., 2004) (Table S1). Specifically, we selected genes with reported RNAi phenotypes of ‘reduced brood size’ (649 genes) or ‘fewer germ cells’ (31 genes) utilizing the WormMart tool on WormBase.org. We also selected genes annotated with the cell communication GO term (GO:0007154) (74 genes) (Ashburner et al., 2000), utilizing http://go.princeton.edu/cgi-bin/GOTermFinder. We then added a short list of additional candidate genes based on literature searches of other cases of cell enwrapping behaviors: actin regulation (9 genes), germline ensheathment in Drosophila, engulfment and glial cell ensheathment in C. elegans (8 genes), and entosis of cancer cells (3 genes). We performed L1 RNAi treatments targeting 708 total genes and L3/L4 RNAi treatments targeting 145 genes according to standard protocols (Dalfó et al., 2012; Kamath et al., 2001). The L1 stage was chosen for the initial high throughput screen, as synchronized worms can be rapidly manipulated at this stage (Rual et al., 2004). To determine a biologically relevant cutoff and as a positive control, we performed RNAi to the key niche regulator lag-2, a DTC-expressed Notch ligand (Henderson et al., 1994), and observed a low penetrance defect (7% of animals scored, Table S2). To confidently identify genes of interest, we set a more stringent threshold of 15%, which is statistically significant for a sample size of 40 animals (close to the number that can be scored in one experiment) in a Fisher’s exact test. Since we also stringently defined a DTC plexus defect as a 50% reduction in plexus length, which is not observed in wild-type animals, we are confident that even low penetrance defects by RNAi suggest genes with significant roles in DTC plexus formation. Knockdown of genes by feeding animals bacteria expressing dsRNA is often incompletely penetrant and variable (Simmer et al., 2003), such that a low penetrance defect by feeding RNAi can detect genes with significant roles. The known stem cell regulator fbf-2 was identified in this screen. fbf-2 RNAi may knock out both fbf-1 and fbf-2 (Crittenden et al., 2002) and serves as a control for known genes affecting the stem cell niche.
L3/L4 RNAi plating followup screens
To assay the effect of RNAi knockdown specifically during the time window of DTC elaboration, RNAi clones resulting in a DTC plexus defect or developmental defects in the L1 screen were repeated in a second screen with a later RNAi exposure. We exposed worms to RNAi approximately 12 hours prior to DTC plexus formation in the L3/L4 stage (Byrd et al., 2014; B. G. Wong et al., 2013) to target the developmental window of DTC elaboration. Synchronized L1s were plated on OP50 and then transferred to RNAi plates at the L3/early L4 stages, determined by observing vulval invagination (approximately 47 hours post L1 plating at 18°C). L3/L4 animals were transferred to RNAi plates by washing five times in M9 to remove OP50 bacteria. Adults were scored after 1.5 – 2 days of RNAi feeding. Twenty-nine RNAi treatments resulted in DTC plexus defects in 15% or more of animals exposed (Table 1, Table S4) and were sequenced to confirm the correct insert. We then knocked down these candidate genes in the germline-specific rrf-1 strain (Kumsta and Hansen, 2012) and our newly developed DTC-specific RNAi strain. We expected variability in gene knockdown by RNAi given these different strain backgrounds, particularly for the DTC-specific line containing the RNAi-hypersensitizing mutation rrf-3 (Simmer et al., 2003) and rescuing the RNAi response in the DTC with lag-2 > RDE-1. Due to this variability, while we focus on genes whose loss results in > 15% penetrance defects, we report all scoring data for these screens (Table 2, Table 3).
Scoring DTC Morphology
DTC plexus defects were scored as >50% reduction in DTC plexus length at the one-day-adult stage, staged as 24 hours after the midL4 stage at 20°C as previously described (Byrd et al., 2014). We retained genes whose knockdown produced DTC plexus defects in > 15% of animals exposed to RNAi in initial screens. Animals that bagged, displayed incorrect gonad migration, failed to reach the egg-laying adult stage, or showed reduced lag-2 reporter expression were excluded from scoring. Scoring was performed blind to the identity of the RNAi clone.
Quantification of DTC morphology and germline characteristics
Measurements of germline progenitor zone length, which has been previously referred to as the proliferative or mitotic zone, and length of the DTC plexus were performed as previously described (Byrd et al., 2014; Crittenden et al., 2017). Briefly, the progenitor zone was measured from the distal end of the gonad to the first row of germ cells containing multiple crescent-shaped nuclei indicating meiotic prophase in DAPI-stained animals. The DAPI staining method was modified from a previous protocol (Hanazawa et al., 2004). Animals were collected in M9, pelleted and fixed in ice-cold methanol for one minute, washed in PBS-TX100 three times, then stained with 1 µg/mL DAPI (diamidino-2-phenylindole, Thermo Scientific #62248) in PBS-TX100 for 30 min, followed by three PBS-TX100 washes prior to imaging.
DTC plexus length was determined as the extent of short intercalating DTC processes (SIPs) along the gonad arm, excluding single SIPs separated by more than 25 µm (approximately 5 germ cell diameters). These criteria allowed the measurement of the plexus region of densely-packed intercalating processes.
The number of sygl-1-expressing cells was determined by counting the number of nuclei with detectable nuclear sygl-1>H2B::GFP (Kershner et al., 2014) in the distal gonad. This sygl-1 reporter does not capture the difference between sygl-1 active transcription and the diffusion of mRNA between syncytial germ cells as in (Lee et al., 2016). As germline-expressed transgenes are commonly silenced in C. elegans (Frøkjær-Jensen et al., 2016; Praitis et al., 2001), we utilized the oma-1::GFP transgene to reduce silencing (Shirayama et al., 2012). Animals that showed germline silencing were excluded from analysis. As a control for the experiment reducing lin-40 and C16A3.5 expression and examining DTC plexus formation and GLP-1/Notch signaling, we found that lag-2 expression was not altered by lin-40 or C16A3.5 RNAi treatment (data not shown). LAG-2 protein levels and localization were not assessed, nor was expression of another DTC-expressed Notch ligand, apx-1 (Nadarajan et al., 2009).
Additionally, we confirmed that knockdown of DTC-autonomous genes including lin-40 and C16A3.5 did not cause gross germline defects (Fig. S4). Wild-type gonad length was determined by migration of the gonad arm adjacent to or past the uterus. Wild-type proliferation and differentiation of germ cells was assayed by scoring for the presence of the progenitor, pachytene, and oocyte zones by examining chromatin structure in DAPI-stained or mCherry::histone labeled germ cells at timepoints matching previous assays. All quantification was performed blind.
Germ cell fate manipulation
glp-1(e2144) animals were maintained at 16°C. Early L4 animals or young adult animals (28 hours past the mid-L4 stage) were transferred to new plates, some of which were imaged (T0) and some of which were transferred to the restrictive temperature (25°C) or returned to the permissive temperature (16°C) to be imaged 24 hours later. For comparisons of DTC elaboration between L4 stage T0 and upshifted or control glp-1(e2144) adult animals, the extent of the SIPs under the cap or plexus was measured (Fig. 3A).
Statistical Analysis
Fisher’s exact tests, ANOVA followed by the Tukey-Kramer HSD test, or two-tailed unpaired Student’s t tests were performed in JMP Pro 13 as noted in figure legends and text. Power calculations performed in JMP Pro 13 were used to determine sample size.
Microscopy and image acquisition, processing, and analysis
Confocal DIC and fluorescent images were acquired on an AxioImager A1 microscope (Carl Zeiss) equipped with an EMCCD camera (Hamamatsu Photonics), a 100x or 40x Plan-Apochromat (1.4 NA) objective, and a spinning disc confocal scan head (CSU-10; Yokogawa Electric Corporation) driven by µManager software (Edelstein et al., 2010) at 20°C. Widefield DIC and fluorescent images were acquired with an AxioImager A1 microscope (Carl Zeiss) equipped with a CCD camera (AxioCam MRm; Carl Zeiss) and 100x Plan-Apochromat objective (1.4 NA) driven by Axiovision software (Carl Zeiss). Worms were mounted on 5% noble agar pads containing 0.01 M sodium azide for imaging.
Images were processed with FIJI 2.0 and Photoshop CS6 (Adobe Systems Inc.). Images of entire germlines were comprised of multiple images stitched with the FIJI pairwise stitching method (Preibisch et al., 2009). Confocal images were displayed as full projections of the entire DTC or core projections of the central z slices as previously described, as the outer surface of the DTC obscures viewing the plexus (Byrd et al., 2014). Core projections were generated in FIJI from a stack of z slices acquired at intervals of 0.5 or 1 µm and are 10 µm-thick projections unless otherwise noted in figure legends. Isosurface renderings of the DTC cell membrane were built in Imaris 7 (Bitplane) using a threshold that captured the DTC plexus, and the total surface area of the DTC was measured blind. Figures and graphs generated by JMP Pro 13 were constructed using Illustrator CS6 (Adobe Systems Inc.).
Supplementary Material
The lag-2 promoter is specific to DTCs (arrowheads) in the L4 and adult stages targeted in our screen. Early in development, in the L1 stage, expression of lag-2>NeonGreen::rde-1 was observed in the somatic gonad precursors Z1 and Z4, and in the L2, the seam cells expressed lag-2>NeonGreen::rde-1. All images are full projections of tiled images of the entire animal from confocal z slices. Only one DTC is sharply in focus in the L3 – adult stages during and after gonad migration. Five or more animals were examined per developmental stage. Scale bars, 10 µm.
(A) Loss of genes detected by the DTC-specific RNAi screen (hars-1, par-5, rnp-7, unc-112, C16A3.5, copa-1, pab-1, lin-40, and ran-1) did not grossly perturb germlines after L3/L4 plating in the DTC-specific strain, paralleling Table 3. Superficial 2.5 µm-thick projections of DAPI-stained germlines are shown. 20 or more animals per treatment were examined for presence of germ progenitor cells, the pachytene and oocyte zones, and complete gonad migration. (B) Likewise, loss of lin-40 and C16A3.5 by whole body RNAi treatment from the L1 stage did not grossly perturb germlines (mex-5>H2B::mCherry), as a control for Figure 6. Scale bars, 20 µm.
Using gene-specific RNAi constructs for the paralogs par-5 and ftt-2 revealed that par-5(RNAi) knockdown results in loss of expression of the translational reporter par-5 > PAR-5::GFP but does not reduce expression of the translational reporter ftt-2 > FTT-2::mCherry. Conversely, ftt-2(RNAi) knocks down expression of ftt-2 > FTT-2::mCherry but does not reduce expression of par-5 > PAR-5::GFP. All images are single confocal z slices. Scale bar, 10 µm.
The number of germ cell nuclei expressing a reporter for the GLP-1/Notch transcriptional target sygl-1 (sygl-1 > H2B::GFP, green) increases 2.5-fold from L4 to adult stages (L4 stage, 48 cells on average; adult stage, 120 cells on average; n > 10 animals per stage; p < 0.0001, Student’s t test) during the time window of DTC plexus formation (magenta, lag-2 > mCh::PH). Adult control data from Figure 6B is shown for reference. Scale bar, 10 µm.
Highlights.
RNAi screens identify regulators of germ stem cell enwrapment by its niche.
LIN-40/MTA1 (chromatin remodeling factor) acts in the niche to promote enwrapment.
Germ progenitor cells stimulate enwrapping behavior by the niche.
Enwrapment of germ cells extends Notch signaling and the stem cell pool.
Acknowledgments
We thank Judith Kimble and Jane Hubbard for strains and discussions; we thank Jeremy Nance and Christian Riedel for strains; and we thank R. Jayadev and E. Hastie for comments on the manuscript and members of the Sherwood lab for input on experimental approaches. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). L.M.L. is supported by graduate research fellowship DGE-1106401 from the National Science Foundation. K.L.G. is supported by postdoctoral fellowship GM121015-01 from the National Institutes of Health. A.M.P. was supported by postdoctoral fellowship F32 GM115151 from the National Institutes of Health. This work was supported by NIGMS R01 GM083071 to B.G. and a Pew Scholars Award and MIRA Award (GM118049-01) to D.R.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anklesaria P, Teixidó J, Laiho M, Pierce JH, Greenberger JS, Massagué J. Cell-cell adhesion mediated by binding of membrane-anchored transforming growth factor alpha to epidermal growth factor receptors promotes cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 1990;87:3289–3293. doi: 10.1073/pnas.87.9.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoshechkin I, Sternberg PW. The versatile worm: genetic and genomic resources for Caenorhabditis elegans research. Nature Reviews Genetics. 2007;8:518–532. doi: 10.1038/nrg2105. [DOI] [PubMed] [Google Scholar]
- Aristizábal-Corrales D, Fontrodona L, Porta-de-la-Riva M, Guerra-Moreno A, Cerón J, Schwartz S. The 14-3-3 gene par-5 is required for germline development and DNA damage response in Caenorhabditis elegans. J. Cell. Sci. 2012;125:1716–1726. doi: 10.1242/jcs.094896. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin J, Kimble JE. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987;51:589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- Bassett EA, Tokarew N, Allemano EA, Mazerolle C, Morin K, Mears AJ, McNeill B, Ringuette R, Campbell C, Smiley S, Pokrajac NT, Dubuc AM, Ramaswamy V, Northcott PA, Remke M, Monnier PP, Potter D, Paes K, Kirkpatrick LL, Coker KJ, Rice DS, Perez-Iratxeta C, Taylor MD, Wallace VA. Norrin/Frizzled4 signalling in the preneoplastic niche blocks medulloblastoma initiation. Elife. 2016:5. doi: 10.7554/eLife.16764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Jasper H. EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development. 2011;138:1045–1055. doi: 10.1242/dev.056671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blelloch R, Kimble J. Control of organ shape by a secreted metalloprotease in the nematode Caenorhabditis elegans. Nature. 1999;399:586–590. doi: 10.1038/21196. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Inaba M, Yamashita YM. Signaling by Cellular Protrusions: Keeping the Conversation Private. Trends Cell Biol. 2016;26:526–534. doi: 10.1016/j.tcb.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd DT, Kimble J. Scratching the niche that controls Caenorhabditis elegans germline stem cells. Semin. Cell Dev. Biol. 2009;20:1107–1113. doi: 10.1016/j.semcdb.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd DT, Knobelb K, Affeldt K, Crittenden SL, Kimble Judith. A DTC Niche Plexus Surrounds the Germline Stem Cell Pool in Caenorhabditis elegans. PLoS ONE. 2014:9. doi: 10.1371/journal.pone.0088372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chen X, Zheng Y. The Nuclear Lamina Regulates Germline Stem Cell Niche Organization via Modulation of EGFR Signaling. Cell Stem Cell. 2013;13:73–86. doi: 10.1016/j.stem.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Lewallen M, Xie T. Adhesion in the stem cell niche: biological roles and regulation. Development. 2013;140:255–265. doi: 10.1242/dev.083139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinquin A, Zheng L, Taylor PH, Paz A, Zhang L, Chiang M, Snow JJ, Nie Q, Cinquin O. Semi-permeable Diffusion Barriers Enhance Patterning Robustness in the C. elegans Germline. Developmental Cell. 2015;35:405–417. doi: 10.1016/j.devcel.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R, Wickens M, Kimble J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- Crittenden SL, Leonhard KA, Byrd DT, Kimble J. Cellular Analyses of the Mitotic Region in the Caenorhabditis elegans Adult Germ Line. Molecular Biology of the Cell. 2006;17:3051–3061. doi: 10.1091/mbc.E06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden SL, Seidel HS, Kimble J. Analysis of the C. elegans Germline Stem Cell Pool. Methods Mol. Biol. 2017;1463:1–33. doi: 10.1007/978-1-4939-4017-2_1. [DOI] [PubMed] [Google Scholar]
- Crittenden SL, Troemel ER, Evans TC, Kimble J. GLP-1 is localized to the mitotic region of the C. elegans germ line. Development. 1994;120:2901–2911. doi: 10.1242/dev.120.10.2901. [DOI] [PubMed] [Google Scholar]
- Dalfó D, Michaelson D, Hubbard EJA. Sensory regulation of the C. elegans germline through TGF-β-dependent signaling in the niche. Curr. Biol. 2012;22:712–719. doi: 10.1016/j.cub.2012.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Goldstein B. CRISPR-Based Methods for Caenorhabditis elegans Genome Engineering. Genetics. 2016;202:885–901. doi: 10.1534/genetics.115.182162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using µManager. Curr Protoc Mol Biol Chapter. 2010;14 doi: 10.1002/0471142727.mb1420s92. Unit14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RE, Kimble J. The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics. 1995;139:561–577. doi: 10.1093/genetics/139.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K, Greenwald I. Interchangeability of Caenorhabditis elegans DSL proteins and intrinsic signalling activity of their extracellular domains in vivo. Development. 1995;121:1–8. doi: 10.1242/dev.121.12.4275. [DOI] [PubMed] [Google Scholar]
- Francis R, Barton MK, Kimble J, Schedl T. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics. 1995;139:579–606. doi: 10.1093/genetics/139.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C, Jain N, Hansen L, Davis MW, Li Y, Zhao D, Rebora K, Millet JRM, Liu X, Kim SK, Dupuy D, Jorgensen EM, Fire AZ. An Abundant Class of Non-coding DNA Can Prevent Stochastic Gene Silencing in the C. elegans Germline. Cell. 2016;166:343–357. doi: 10.1016/j.cell.2016.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo M, Park D, Luciani DS, Kida K, Palmieri F, Blacque OE, Johnson JD, Riddle DL. MISC-1/OGC links mitochondrial metabolism, apoptosis and insulin secretion. PLoS ONE. 2011;6:e17827–e17827. doi: 10.1371/journal.pone.0017827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PL, Johnson JJ, Wang S, Sibley MH, Gupta MC, Kramer JM. Type IV collagen is detectable in most, but not all, basement membranes of Caenorhabditis elegans and assembles on tissues that do not express it. The Journal of Cell Biology. 1997;137:1171–1183. doi: 10.1083/jcb.137.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruidl ME, Smith PA, Kuznicki KA, McCrone JS, Kirchner J, Roussell DL, Strome S, Bennett KL. Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13837–13842. doi: 10.1073/pnas.93.24.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DH, Winfrey VP, Blaeuer G, Hoffman LH, Furuta T, Rose KL, Hobert O, Greenstein D. Ultrastructural Features of the Adult Hermaphrodite Gonad of Caenorhabditis elegans: Relations between the Germ Line and Soma. Developmental Biology. 1999:1–23. doi: 10.1006/dbio.1999.9356. [DOI] [PubMed] [Google Scholar]
- Hamilton LK, Dufresne M, Joppé SE, Petryszyn S, Aumont A, Calon F, Barnabé-Heider F, Furtos A, Parent M, Chaurand P, Fernandes KJL. Aberrant Lipid Metabolism in the Forebrain Niche Suppresses Adult Neural Stem Cell Proliferation in an Animal Model of Alzheimer’s Disease. Cell Stem Cell. 2015;17:397–411. doi: 10.1016/j.stem.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Hanazawa M, Kawasaki I, Kunitomo H, Gengyo-Ando K, Bennett KL, Mitani S, Iino Y. The Caenorhabditis elegans eukaryotic initiation factor 5A homologue, IFF-1, is required for germ cell proliferation, gametogenesis and localization of the P-granule component PGL-1. Mech. Dev. 2004;121:213–224. doi: 10.1016/j.mod.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Gao D, Lambie EJ, Kimble J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development. 1994;120:2913–2924. doi: 10.1242/dev.120.10.2913. [DOI] [PubMed] [Google Scholar]
- Hitrik A, Popliker M, Gancz D, Mukamel Z, Lifshitz A, Schwartzman O, Tanay A, Gilboa L. Combgap Promotes Ovarian Niche Development and Chromatin Association of EcR-Binding Regions in BR-C. PLoS Genet. 2016;12:e1006330. doi: 10.1371/journal.pgen.1006330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggatt J, Kfoury Y, Scadden DT. Hematopoietic Stem Cell Niche in Health and Disease. Annu Rev Pathol. 2016;11:555–581. doi: 10.1146/annurev-pathol-012615-044414. [DOI] [PubMed] [Google Scholar]
- Inaba M, Buszczak M, Yamashita YM. Nanotubes mediate niche-stem-cell signalling in the Drosophila testis. Nature. 2015;523:329–332. doi: 10.1038/nature14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba M, Yamashita YM, Buszczak M. Keeping stem cells under control: New insights into the mechanisms that limit niche-stem cell signaling within the reproductive system. Mol. Reprod. Dev. 2016;83:675–683. doi: 10.1002/mrd.22682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Sherwood DR, Aspöck G, Butler JA, Gupta BP, Kirouac M, Wang M, Lee P-Y, Kramer JM, Hope I, Bürglin TR, Sternberg PW. Gene expression markers for Caenorhabditis elegans vulval cells. Mech. Dev. 2002;119(Suppl 1):S203–9. doi: 10.1016/s0925-4773(03)00117-5. [DOI] [PubMed] [Google Scholar]
- Jiu Y, Jin C, Liu Y, Holmberg CI, Jäntti J. Exocyst subunits Exo70 and Exo84 cooperate with small GTPases to regulate behavior and endocytic trafficking in C. elegans. PLoS ONE. 2012;7:e32077. doi: 10.1371/journal.pone.0032077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0002. RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershner AM, Shin H, Hansen TJ, Kimble J. Discovery of two GLP-1/Notch target genes that account for the role of GLP-1/Notch signaling in stem cell maintenance. PNAS. 2014;111:3739–3744. doi: 10.1073/pnas.1401861111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble JE, White JG. On the Control of Germ Cell Development in Caenorhabditis elegans. Developmental Biology. 1981:1–12. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- Kitadate Y, Kobayashi S. Notch and Egfr signaling act antagonistically to regulate germ-line stem cell niche formation in Drosophila male embryonic gonads. PNAS. 2010;107:14241–14246. doi: 10.1073/pnas.1003462107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko S, Park J-H, Lee A-R, Kim E, Jiyoung-Kim Kawasaki I, Shim Y-H. Two mutations in pab-1 encoding poly(A)-binding protein show similar defects in germline stem cell proliferation but different longevity in C. elegans. Mol. Cells. 2010;30:167–172. doi: 10.1007/s10059-010-0103-2. [DOI] [PubMed] [Google Scholar]
- Kornberg TB, Roy S. Communicating by touch--neurons are not alone. Trends Cell Biol. 2014;24:370–376. doi: 10.1016/j.tcb.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumsta C, Hansen M. C. elegans rrf-1 mutations maintain RNAi efficiency in the soma in addition to the germline. PLoS ONE. 2012;7:e35428. doi: 10.1371/journal.pone.0035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Sorensen EB, Lynch TR, Kimble J. C. elegans GLP-1/Notch activates transcription in a probability gradient across the germline stem cell pool. Elife. 2016:5. doi: 10.7554/eLife.18370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tewari M, Vidal M, Lee SS. The 14-3-3 protein FTT-2 regulates DAF-16 in Caenorhabditis elegans. Developmental Biology. 2007;301:82–91. doi: 10.1016/j.ydbio.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal L, Martinez-Agosto JA, Evans CJ, Hartenstein V, Banerjee U. A Hedgehog- and Antennapedia-dependent niche maintains Drosophila haematopoietic precursors. Nature. 2007;446:320–324. doi: 10.1038/nature05585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynovsky M, Wong M-C, Byrd DT, Kimble J, Schwarzbauer JE. mig-38, a novel gene that regulates distal tip cell turning during gonadogenesis in C. elegans hermaphrodites. Developmental Biology. 2012;368:404–414. doi: 10.1016/j.ydbio.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur D, Bost A, Driver I, Ohlstein B. A transient niche regulates the specification of Drosophila intestinal stem cells. Science. 2010;327:210–213. doi: 10.1126/science.1181958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, Chan S, Dube N, Fang L, Goszczynski B, Ha E, Halfnight E, Hollebakken R, Huang P, Hung K, Jensen V, Jones SJM, Kai H, Li D, Mah A, Marra M, McGhee J, Newbury R, Pouzyrev A, Riddle DL, Sonnhammer E, Tian H, Tu D, Tyson JR, Vatcher G, Warner A, Wong K, Zhao Z, Moerman DG. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb. Symp. Quant. Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- Mesa KR, Rompolas P, Greco V. The Dynamic Duo: Niche/Stem Cell Interdependency. Stem Cell Reports. 2015;4:961–966. doi: 10.1016/j.stemcr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikl M, Cowan CR. Alternative 3’ UTR selection controls PAR-5 homeostasis and cell polarity in C. elegans embryos. Cell Rep. 2014;8:1380–1390. doi: 10.1016/j.celrep.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Nadarajan S, Govindan JA, McGovern M, Hubbard EJA, Greenstein D. MSP and GLP-1/Notch signaling coordinately regulate actomyosin-dependent cytoplasmic streaming and oocyte growth in C. elegans. Development. 2009;136:2223–2234. doi: 10.1242/dev.034603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y, Yamada T, Matsumiya T, Ui-Tei K, Saigo K, Morishita S. dsCheck: highly sensitive off-target search software for double-stranded RNA-mediated RNA interference. Nucleic Acids Res. 2005;33:W589–91. doi: 10.1093/nar/gki419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley MJ, Racicot KE, Oatley JM. Sertoli cells dictate spermatogonial stem cell niches in the mouse testis. Biol. Reprod. 2011;84:639–645. doi: 10.1095/biolreprod.110.087320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SL, Priess JR, Henikoff S. Histone H3.3 variant dynamics in the germline of Caenorhabditis elegans. PLoS Genet. 2006;2:e97. doi: 10.1371/journal.pgen.0020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper AS-R, Lo TW, Killian DJ, Hall DH, Hubbard EJA. The establishment of Caenorhabditis elegans germline pattern is controlled by overlapping proximal and distal somatic gonad signals. Developmental Biology. 2003;259:336–350. doi: 10.1016/s0012-1606(03)00203-3. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess JRJ, Schnabel HH, Schnabel RR. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell. 1987;51:601–611. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- Rojas-Ríos P, Guerrero I, González-Reyes A. Cytoneme-mediated delivery of hedgehog regulates the expression of bone morphogenetic proteins to maintain germline stem cells in Drosophila. PLoS Biol. 2012;10:e1001298. doi: 10.1371/journal.pbio.1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Michaelson D, Hochman T, Santella A, Bao Z, Goldberg JD, Hubbard EJA. Cell cycle features of C. elegans germline stem/progenitor cells vary temporally and spatially. Developmental Biology. 2015 doi: 10.1016/j.ydbio.2015.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J-F, Cerón J, Koreth J, Hao T, Nicot A-S, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, Vidal M. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Research. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Parikh N, Hearn SA, Fuller MT, Tazuke SI, Schulz C. Antagonistic roles of Rac and Rho in organizing the germ cell microenvironment. Curr. Biol. 2007;17:1253–1258. doi: 10.1016/j.cub.2007.06.048. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee H-C, Gu W, Ishidate T, Conte D, Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Silván U, Díez-Torre A, Moreno P, Arluzea J, Andrade R, Silió M, Aréchaga J. The spermatogonial stem cell niche in testicular germ cell tumors. Int. J. Dev. Biol. 2013;57:185–195. doi: 10.1387/ijdb.130068ja. [DOI] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PVE, Kamath RS, Fraser AG, Ahringer J, Plasterk RHA. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LB, Milne L, Nelson N, Eddie S, Brown P, Atanassova N, O’Bryan MK, O’Donnell L, Rhodes D, Wells S, Napper D, Nolan P, Lalanne Z, Cheeseman M, Peters J. KATNAL1 regulation of sertoli cell microtubule dynamics is essential for spermiogenesis and male fertility. PLoS Genet. 2012;8:e1002697. doi: 10.1371/journal.pgen.1002697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solari F, Bateman A, Ahringer J. The Caenorhabditis elegans genes egl-27 and egr-1 are similar to MTA1, a member of a chromatin regulatory complex, and are redundantly required for embryonic patterning. Development. 1999;126:2483–2494. doi: 10.1242/dev.126.11.2483. [DOI] [PubMed] [Google Scholar]
- Tamplin OJ, Durand EM, Carr LA, Childs SJ, Hagedorn EJ, Li P, Yzaguirre AD, Speck NA, Zon LI. Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell. 2015;160:241–252. doi: 10.1016/j.cell.2014.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannoury H, Rodriguez V, Kovacevic I, Ibourk M, Lee M, Cram EJ. CACN-1/Cactin interacts genetically with MIG-2 GTPase signaling to control distal tip cell migration in C. elegans. Developmental Biology. 2010;341:176–185. doi: 10.1016/j.ydbio.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay M, Martino Cortez Y, Wong-Deyrup S, Tavares L, Schowalter S, Flora P, Hill C, Nasrallah MA, Chittur S, Rangan P. Transposon Dysregulation Modulates dWnt4 Signaling to Control Germline Stem Cell Differentiation in Drosophila. PLoS Genet. 2016;12:e1005918. doi: 10.1371/journal.pgen.1005918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voutev R, Killian DJ, Ahn JH, Hubbard EJA. Alterations in ribosome biogenesis cause specific defects in C. elegans hermaphrodite gonadogenesis. Developmental Biology. 2006;298:45–58. doi: 10.1016/j.ydbio.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Wang Xiaomeng, Harris RE, Bayston LJ, Ashe HL. Type IV collagens regulate BMP signalling in Drosophila. Nature. 2008;455:72–77. doi: 10.1038/nature07214. [DOI] [PubMed] [Google Scholar]
- Wang Xin, Zhao Y, Wong K, Ehlers P, Kohara Y, Jones SJ, Marra MA, Holt RA, Moerman DG, Hansen D. Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics. 2009;10:213. doi: 10.1186/1471-2164-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman DH. Donor cell leukemia: a review. Biol. Blood Marrow Transplant. 2011;17:771–789. doi: 10.1016/j.bbmt.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Wong BG, Paz A, Corrado MA, Ramos BR, Cinquin A, Cinquin O, Hui EE. Live imaging reveals active infiltration of mitotic zone by its stem cell niche. Integr Biol (Camb) 2013;5:976–982. doi: 10.1039/c3ib20291g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M-C, Kennedy WP, Schwarzbauer JE. Transcriptionally regulated cell adhesion network dictates distal tip cell directionality. Dev. Dyn. 2014;243:999–1010. doi: 10.1002/dvdy.24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R-C, Zeng Y, Chen Y-F, Lanz RB, Wu M-Y. Temporal-spatial Establishment of Initial Niche for the Primary Spermatogonial Stem Cell Formation is Determined by an ARID4B Regulatory Network. Stem Cells. 2017 doi: 10.1002/stem.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambetti NA, Ping Z, Chen S, Kenswil KJG, Mylona MA, Sanders MA, Hoogenboezem RM, Bindels EMJ, Adisty MN, Van Strien PMH, van der Leije CS, Westers TM, Cremers EMP, Milanese C, Mastroberardino PG, van Leeuwen JPTM, van der Eerden BCJ, Touw IP, Kuijpers TW, Kanaar R, van de Loosdrecht AA, Vogl T, Raaijmakers MHGP. Mesenchymal Inflammation Drives Genotoxic Stress in Hematopoietic Stem Cells and Predicts Disease Evolution in Human Pre-leukemia. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.08.021. [DOI] [PubMed] [Google Scholar]
- Ziel JW, Hagedorn EJ, Audhya A, Sherwood DR. UNC-6 (netrin) orients the invasive membrane of the anchor cell in C. elegans. Nature Publishing Group. 2009;11:183–189. doi: 10.1038/ncb1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Yadav S, DeVault L, Nung Jan Y, Sherwood DR. RAB-10-Dependent Membrane Transport Is Required for Dendrite Arborization. PLoS Genet. 2015;11:e1005484. doi: 10.1371/journal.pgen.1005484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The lag-2 promoter is specific to DTCs (arrowheads) in the L4 and adult stages targeted in our screen. Early in development, in the L1 stage, expression of lag-2>NeonGreen::rde-1 was observed in the somatic gonad precursors Z1 and Z4, and in the L2, the seam cells expressed lag-2>NeonGreen::rde-1. All images are full projections of tiled images of the entire animal from confocal z slices. Only one DTC is sharply in focus in the L3 – adult stages during and after gonad migration. Five or more animals were examined per developmental stage. Scale bars, 10 µm.
(A) Loss of genes detected by the DTC-specific RNAi screen (hars-1, par-5, rnp-7, unc-112, C16A3.5, copa-1, pab-1, lin-40, and ran-1) did not grossly perturb germlines after L3/L4 plating in the DTC-specific strain, paralleling Table 3. Superficial 2.5 µm-thick projections of DAPI-stained germlines are shown. 20 or more animals per treatment were examined for presence of germ progenitor cells, the pachytene and oocyte zones, and complete gonad migration. (B) Likewise, loss of lin-40 and C16A3.5 by whole body RNAi treatment from the L1 stage did not grossly perturb germlines (mex-5>H2B::mCherry), as a control for Figure 6. Scale bars, 20 µm.
Using gene-specific RNAi constructs for the paralogs par-5 and ftt-2 revealed that par-5(RNAi) knockdown results in loss of expression of the translational reporter par-5 > PAR-5::GFP but does not reduce expression of the translational reporter ftt-2 > FTT-2::mCherry. Conversely, ftt-2(RNAi) knocks down expression of ftt-2 > FTT-2::mCherry but does not reduce expression of par-5 > PAR-5::GFP. All images are single confocal z slices. Scale bar, 10 µm.
The number of germ cell nuclei expressing a reporter for the GLP-1/Notch transcriptional target sygl-1 (sygl-1 > H2B::GFP, green) increases 2.5-fold from L4 to adult stages (L4 stage, 48 cells on average; adult stage, 120 cells on average; n > 10 animals per stage; p < 0.0001, Student’s t test) during the time window of DTC plexus formation (magenta, lag-2 > mCh::PH). Adult control data from Figure 6B is shown for reference. Scale bar, 10 µm.