Abstract
We report locus-specific disintegration of megabase-scale chromosomal conformations in brain after neuronal ablation of Kmt1e/Setdb1 histone H3-lysine 9 methyltransferase, including a large topologically associated 1.2Mb domain conserved in human and mouse and encompassing >70 genes at the clustered Protocadherin (cPcdh) locus. TADcPcdh in mutant neurons showed abnormal accumulations of CTCF transcriptional regulator and 3D genome organizer at cryptic binding sites, converted into permissive state with DNA cytosine hypomethylation and histone hyperacetylation. Broadly upregulated expression across cPcdh included defective S-type Protocadherin single-cell stochastic constraint. Setdb1-dependent loop formations, bypassing 0.2–1Mb of linear genome, radiated from TADPcdh fringes towards cPcdh cis-regulatory sequences, counterbalanced shorter-range facilitative promoter-enhancer contacts and carried loop-bound polymorphisms associated with genetic risk for schizophrenia. We show that KRAB-zinc finger Setdb1 repressor complex, shielding neuronal 3D genomes from excess CTCF binding, is critically required for structural maintenance of TADcPcdh.
Introduction
A significant portion of the chromosomal material is compartmentalized into ‘topologically associated domains’ (TADs), typically encompassing several hundred kilobases of linear genome folded upon itself with regulatory proteins including cohesion and the multifunctional CCCTC-binding factor CTCF constraining expression of TAD-associated genes1–4. TAD-like structures in brain5, 6 were implicated in the genetic risk architecture of psychiatric disease7 but regulatory mechanisms remain unexplored. Here, we report that neuronal maintenance of a subset of very large ‘superTADs’ critically requires Set-domain-bifurcated 1 (Setdb1/Eset/Kmt1e) histone H3-lysine 9 methyltransferase8. Kmt1e/Setdb1 is important for prenatal development and pup survival9 and broadly regulates retroelement suppression and transcriptional silencing in stem cells10–12. Little is known about its essential functions in differentiated cells including neurons. Cell type-specific 3D genome, CTCF, DNA methylation and histone modification profilings, in conjunction with targeted epigenomic editing and conditional mutagenesis uncovered a Setdb1-dependent ‘shield’ protecting genomes from excess CTCF binding and unique locus-specific epigenomic vulnerabilities triggering higher order chromatin collapse on a megabase-scale.
Results
Locus-specific TAD disintegration in Setdb1 mutant neurons
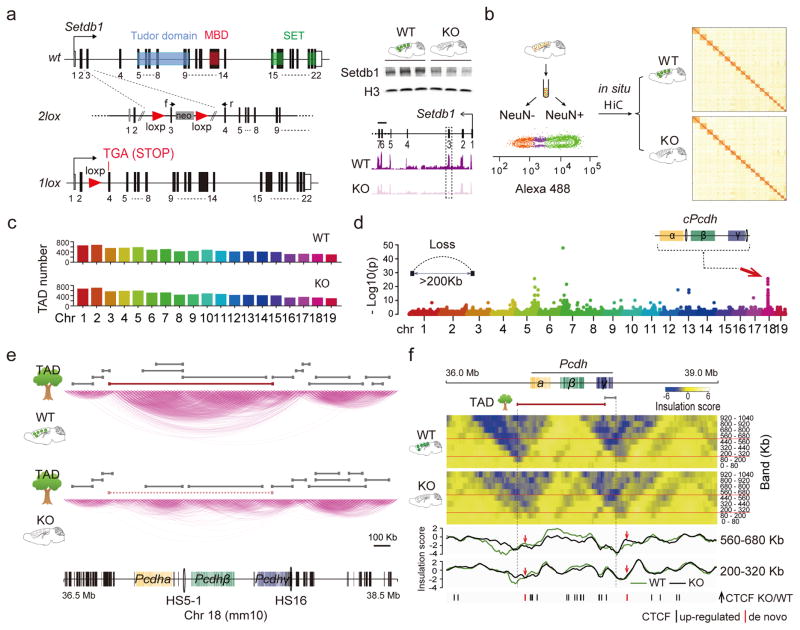
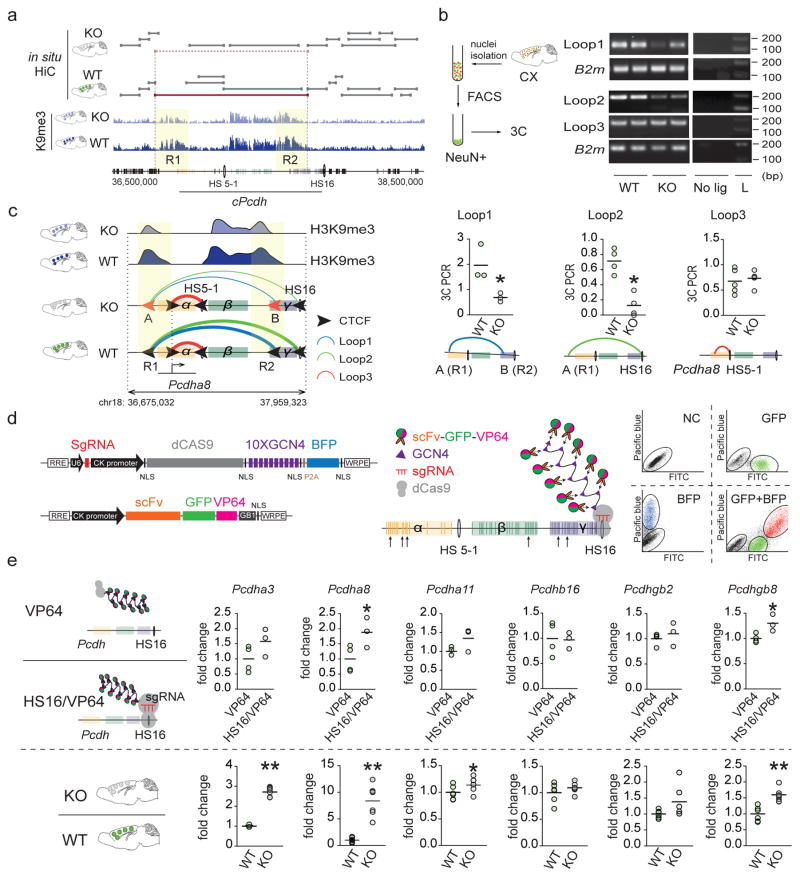
To explore higher order chromatin in Setdb1-deficient brain, we generated a mouse line for CK-Cre driven recombination in postnatal forebrain neurons with exon 3 deletion, frameshift and premature stop upstream of the critical Tudor, methyl-CpG binding (MBD) and catalytic SET domains. CK-Cre+ Setdb12lox/2lox mutant, in comparison to CK-Cre−Setdb12lox/2lox control mice showed grossly normal brain cytoarchitecture. However, adult mutants consistently showed a reduction in brain weight, without premature death or neuronal loss as assessed by flow cytometry-based nuclei counts and COMET nuclear DNA damage assays (Figure 1a, Supplementary Figure 1). We conducted in situ Hi-C1, or genome-scale DNA-DNA proximity mappings in formalin-fixed, restriction-digested, religated NeuN+ neuronal nuclei collected by fluorescence-activated sorting (to avoid signal contribution from non-neuronal nuclei) in adult CK-Cre+Setdb12lox/2lox and CK-Cre−Setdb12lox/2lox cortex (Figure 1B). However, chromosomal contact mappings at 40kb resolution (N=2 in situ Hi-C libraries/genotype, with 250–300M aligned reads/library, Figure 1b and Supplementary Figure 2a) showed that mutant neurons were not affected by a generalized disorganization of the 3D genome. For example, length distributions and numbers of autosomal TADs, assessed by TADtree1, were indistinguishable and minimally different between genotypes, with ~200kb median length as expected for mammalian genomes1 (Figure 1c, Supplementary Figure 2b). We then assessed longer-range chromosomal contacts, spanning >200kb of linear genome. We identified genome-wide 110 long-range loop contacts affected in mutant neurons (DESeq2 P<0.05). Unexpectedly, the large majority, 84/110 or 76%, represented clustered, locus-specific ‘loop aggregates’ showing massive weakening, or complete loss, after neuronal Setdb1 ablation (Figure 1d, Supplementary Figure 2c). These included a singular hotspot on chromosome 18 with remarkable chromosome-wide enrichment (1Mb sliding window (1Mbsw) Poisson test P=1.2×10−24, Figure 1d,e), fully engulfing the clustered Protocadherin (cPcdh) locus harboring 77 genes including 58 cell adhesion molecules linearly arranged as three gene clusters (Pcdh α, β,γ), regulating neuronal connectivity13, 14. Closer inspection of the wildtype cPcdh domain revealed multiple small (~100kb length) cluster-specific subTADs nested into a massive superTAD encompassing at least 1.2Mb of linear genome. TADtree analyses confirmed that this superTAD completely disintegrated after Setdb1 ablation, leaving behind only subTAD remnants in mutant neurons (Figure 1e, Supplementary Figure 2d). Additional loci, including distal portions of chromosomes 5 and 7, showed partial loss of large-sized TADs (Figure 1d, Supplementary Figure 2e). Insulation, measuring the strength of physical segregation of neighboring chromosomal sequences, informs about functional compartmentalization of chromatin15. We quantified contact insulation across multiple DNA-DNA contact scales or ‘bands’ (Figure 1F) defined by increasing genomic distance16. Indeed, wildtype neurons showed very strong insulation scores at the fringes of the cPcdh locus. However, insulation of corresponding sequences in mutant neurons was dramatically weakened across multiple bands (Figure 1f). Therefore, multiple computational approaches, including (i) TADtree, (ii) long-range contact mapping and (iii) insulation analysis reveal structural disintegration of the superTADcPcdh after Setdb1 ablation.
Figure 1. 3D genomes in Setdb1-deficient cortical neurons.
(a) (Left) Conditional Setdb1 ablation with loxP sites surrounding exon 3. Recombination results in frame shift and premature stop (TGA) upstream of Tudor, methyl-CpG-binding (MBD) and catalytic SET domains. (Right) Setdb1 immunoblot (histone H3 loading control) (complete blot shown in Supplementary Figure 1c) and RNA-seq from adult CK-Cre+ Setdb12lox/2lox mutant (K), in comparison to CK-Cre−Setdb12lox/2lox (WT) cortex. (b) (Left) Flow cytometry-based sorting of adult cortex NeuN immunotagged nuclei. (Right) genome-scale in situ Hi-C contact matrix from WT and KO NeuN+ nuclei. (c) TAD numbers per autosome for mutant (KO) and wildtype (WT) NeuN+, (N=2/genotype). (d) Manhattan plot summarizing loss of long-range DNA loop contacts bypassing >200kb linear genome in KO compared to WT NeuN+. Notice localized aggregates of densely spaced loop losses on chromosomes 5, 7 and 18. (e) in situ HiC 2Mb window showing chromosome 18 conformations in KO and WT at position marked by red arrow in Manhattan blot in panel D, with TADs called (TADtree) in both genotypes marked gray. Large ‘superTAD’ called in WT but lost in KO shown as red line. (f) Contact insulation map for 3Mb window centered on cPcdh locus. (Top) Heatmaps from WT and KO cortical neurons for 9 bands, from 0–80kb to 920–1040kb distance. KO shows loss of superTADcPcdh insulation. (Bottom) Two representative insulation bands reveal Setb1-sensitive insulation zones in KO neurons aligned with excess CTCF peaks, as indicated.
Setdb1 shields neuronal genomes from excess CTCF occupancy
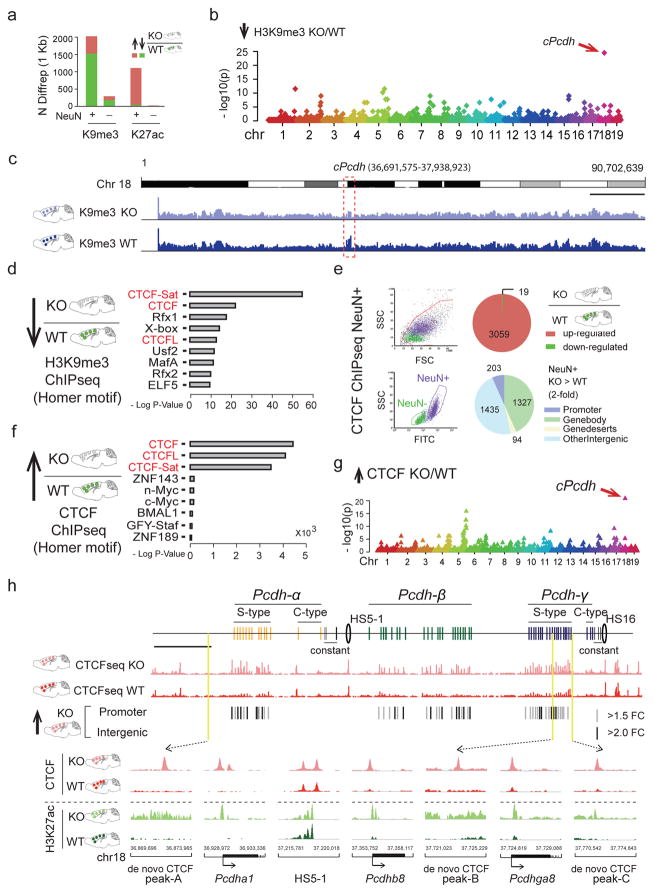
How could neuronal Setdb1 ablation trigger such highly localized alterations in chromosomal conformations? To explore the role of Setdb1-regulated repressive histone methylation, we charted the Setdb1 product, trimethyl-histone H3-lysine 9 (H3K9me3), in NeuN+ and, for comparison, NeuN− nuclei sorted from adult Cre+Setdb12lox/2lox and CK-Cre− Setdb12lox/2lox cortex. DiffRep-based analysis with 1kbsw revealed that 75% of 2,021 differentially H3K9me3-tagged sequences were hypomethylated in mutant neurons. These deficits were specific, because ChiP-seq profiling for open chromatin-associated acetyl-H3-lysine 27 (H3K27ac) showed that 96.4% of 1,112 differentially tagged sequences were hyperacetylated in Setdb1-deficient neurons (Figure 2a, Supplementary Tables 1,2). Furthermore, cPcdh emerged genome-wide as top scoring locus for H3K9me3 hypomethylation (1Mbsw: H3K9me3, 35-fold enrichment, observed/expected 21/0.61, Poisson test P=3.19e−25) (Figure 2b), with the densely concentrated H3K9me3 deficit readily visible in whole chromosome 18 browser views (Figure 2c). In contrast, NeuN− nuclei sorted from the same cortical specimens were only minimally affected (Figure 2a, Supplementary Figure 3, Supplementary Tables 3,4).
Figure 2. Histone modification and CTCF landscapes in Setdb1-deficient neuronal nuclei.
(a) DiffRep counts (1kbsw) for H3K9me3 methylation and H3K27ac acetylation (ChIP-seq) from KO compared to WT adult cortex, for NeuN+ and NeuN−. H3K9me3 >1.5-fold; H3K27ac >2-fold; FDR P<0.05. (b) Manhattan plot with linear representation of autosomes, showing localized enrichments (1MBsw) for H3K9me3 hypomethylation in KO. Top-scoring chromosome 18 cPcdh locus corresponds to site affected by loss of long-range loop bundles (Figure 1d). (c) Mouse total chromosome 18 (mm10; merged fastQ N=3 animals) H3K9me3 landscape for NeuN+ KO and WT, with ~1.2 Mb (chr18:36,691,575-37,938,923) cPcdh locus flagged. Scale bar, 10 Mb. (d) CTCF motif (red) enrichment in sequences H3K9me3 hypomethylated in KO. (e) (Left) FACS plots showing separation of crosslinked NeuN+ from NeuN− nuclei (adult cortex) for cell-type specific CTCF ChIP-seq. (Right) Mutant NeuN+ showed 3059 CTCF up- and only 19 CTCF down-regulated sequences, affecting primarily inter- and intragenic DNA (>2-fold KO/WT NeuN+ nuclei, N=4 animals/group, FDR P <0.05. (f) Dramatic CTCF motif (red) enrichment among the 3059 CTCF-up sequences. (g) Autosomal genome Manhattan plot showing localized clustering of CTCF up sequences in KO neurons, with cPcdh as top ranking locus (1Mbsw). (H) (Top) cPcdh CTCF landscapes in KO and WT NeuN+ as indicated. Significantly up-regulated (KO>WT) promoter-bound and intergenic CTCF sequences marked separately. Scale bar, 100 kb. (Bottom) Coordinate increase of (red) CTCF and (green) H3K27ac at selected cPcdh promoters and intergenic DNA in KO neurons. In contrast, robust peaks at baseline, independent of genotype, at DNaseI hypersensitive HS5-1(HS5-1a+HS5-1b).
We then analyzed motifs in H3K9me3 hypomethylated sequences in Setdb1-deficient neurons. Strikingly, 3/5 top scoring motifs matched to the transcriptional regulator and key 3D genome organizer CTCF, including the CTCFL/BORIS paralog (HOMER enrichment17, P<10−60) (Figure 2d, Supplementary Table 5). Furthermore, we uncovered in published Setdb1 ChIP-seq data from stem cells and CD19+ B lymphocytes significant CTCF motif enrichment (Supplementary Figure 4a, Supplementary Tables 6,7). Of note, other types of H3K9 methyltransferase, including G9a/Glp, completely lacked CTCF motif enrichment (Supplementary Figure 4b). We therefore predicted altered CTCF occupancy in the Setdb1-deficient neuronal genome. Strikingly, ChIP-seq on NeuN+ from adult CK-Cre+Setdb12lox/2lox and CK-Cre− Setdb12lox/2lox cortex showed that 99.4% (3059/3078) of sequences with altered CTCF binding represented up-regulated and de novo peaks (Figure 2e, Supplementary Table 8), including many promoters and enhancers (Supplementary Figure 5). There was extreme over-representation for CTCF motifs (HOMER enrichment P<10−1000) (Figure 2f, Supplementary Table 9) independent of filter conditions (Supplementary Figure 6, Supplementary Table 10), affecting cis-regulatory elements and H3K9me3 hypomethylated sequences (Supplementary Table 11). Therefore, Setdb1 shields mature neuronal genomes from excess CTCF occupancy at cryptic binding sites. Of note, cPcdh again emerged as top scoring locus genome-wide (1Mbsw, CTCF NeuN+ up-peaks: 18.7-fold enrichment, Poisson test P=1.32e−21, Figure 2g,h). Additional localized enrichments of excess CTCF occupancies matched to loci on chromosomes 5 and 7 affected by loss of long-range chromosomal contacts and H3K9 hypomethylation (Figure 2g).
Given that CTCF—a key regulator of higher order chromatin including domain insulation18 —is upregulated at thousands of positions in the Setdb1-deficient neuronal genome, we assessed genome-wide domain insulation in the in situ Hi-C datasets from our mutant and wildtype cortical neurons. We first focused on CTCF de novo peaks, filtered for (i) proximity to TAD boundary (20% of total TAD length) and (ii) vicinity (±100kb) of sequences with altered H3K9me3 after Setdb1 ablation. Of these, 52–57% of de novo CTCF showing stronger insulation scores in mutants across 8/9 insulation bands, covering 80–1040kb contact distance (Supplementary Figure 7). At sites with conserved CTCF peaks, insulation scores showed very minimal differences between mutant and control neurons (Supplementary Figure 8). Therefore, excess of CTCF on a genome-wide scale conveys a subtle shift towards increased insulation strength in mutant neurons, with the notable exception of Setdb1-sensitive superTADs affected by structural collapse and loss of insulation. Our findings, in conjunction with recent genome-scale studies reporting loss of domain insulation in glioma cells due to decreased CTCF binding19, suggest that spatial architectures of chromosomes are highly sensitive to bidirectional changes in CTCF occupancies. Next, we explored alterations in A/B compartments, defined as multi-Mb chromosomal segments representing ‘A’ open/’B’ condensed chromatin tending to interact with other loci sharing similar levels of chromatin accessibility2. Because A/B compartments are defined on a continuum2 (as opposed to a biphasic signal), we quantified ‘compartment-ness’ from the intrachromosomal contact matrices generated by HiC-Pro at 100kb bin resolution (see Online Methods). Of note, the total number of A/B-specific compartment bins was only minimally different between genotypes. Strikingly, however, 6032/11048 or 54% of ‘A’ and 8468/12977 or 65% of ‘B’ bins had higher ‘compartment-ness’ scores in mutant compared to wildtype neurons (Fisher’s exact P <10−210) (Supplementary Figure 9a). However, Setdb1-sensitive superTADs did not follow this genome-wide trend, as exemplified by the weakened ‘B’ signal at cPcdh in mutant neurons (Supplementary Figure 9b).
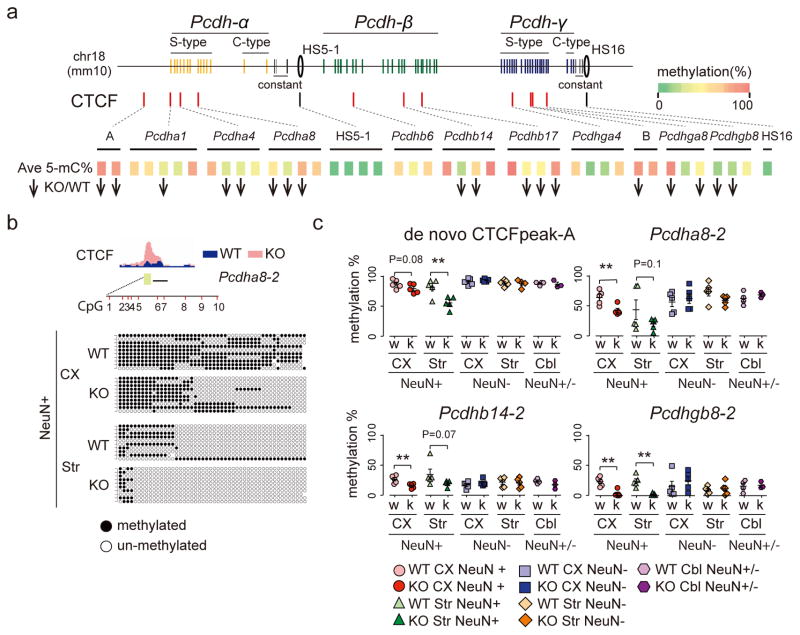
Which molecular mechanisms contribute to the CTCF excess at the H3K9me3 hypomethylated sites? Of note, the majority of cPcdh sequences affected show coordinate increases in CTCF binding and histone hyperacetylation (Figure 2h), suggesting a shift towards open/permissive chromatin states. To this end, alterations in DNA cytosine methylation—reducing CTCF’s DNA affinity20, 21 via interaction with the seventh of CTCF’s 11 zinc fingers22—could play a key role, because Setdb1 functions as upstream regulator for DNA methylation23. To explore, we quantified by bisulfite sequencing (bis-seq)24 levels of cytosine mC5 methylation with 43 PCR amplicons targeting 13 cPcdh sites including cortical and striatal NeuN+ and NeuN− nuclei from adult CK-Cre+Setdb12lox/2lox and CK-Cre− Setdb12lox/2lox brain, plus cerebellar tissue as additional control, comprising 46 individual samples altogether (Supplementary Table 12). As expected, mC5 in non-neuronal nuclei and cerebellum remained unaltered. However, in Setdb1-deficient neurons, intergenic and promoter sequences affected by excess/de novo CTCF showed significant mC5 deficits. In contrast, mC5 levels were extremely low while CTCF peaks were very robust, independent of genotype, at sites harboring strong cPcdh enhancers elements including HS5-1 and HS1625–29 (Figure 3, Supplementary Tables 12,13). Therefore, excess CTCF in Setdb1-deficient neuronal genomes is associated with ‘open’ chromatin state conversion at cryptic CTCF binding sites. This includes reduced DNA methylation levels, and weakening of regulatory mechanism designed to prevent excess CTCF binding.
Figure 3. DNA methylation profiling at the cPcdh locus.
(a) (Top) red tick marks for 11 cPcdh promoters and 2 intergenic sequences (A,B) with excess/de novo CTCF occupancy in KO neurons; black tick marks for HS5-1 and HS16 with robust CTCF peaks in both KO and WT (see also Figure 2h). (Bottom) Averaged 5mC DNA methylation levels (green-red=0–100%) of 43 amplicons representing the set of 13 regulatory sequences show in Top panel. Bis-seq data were averaged across 47 DNA samples from cortical and striatal NeuN+ and NeuN−, and cerebellar homogenate. Downward arrows: 18/43 bis-seq amplicons show mC5 deficit in Setb1-deficient neurons. 0/43 show increase (P<0.5–0.1/amplicon) (see Supplementary Table 13 for details on quantification). (b) Representative bis-seq example from Pcdha8 amplicon no.2 capturing 10 CpG sites. Score cards from 50 randomly selected DNA molecules: circles black/white methylated/not methylated. (c) Quantification of bis-seq amplicons expressed as %methylated. *,**P<0.05(0.01) unpaired one-tailed t-test. Each symbol represents 1 sample from 1 animal. Note methylation deficits specifically for cortical (CX) and striatal (Str) NeuN+ from Setdb1-deficient (k) neurons, compared to wildtype (w).
TAD-specific regulation of gene expression
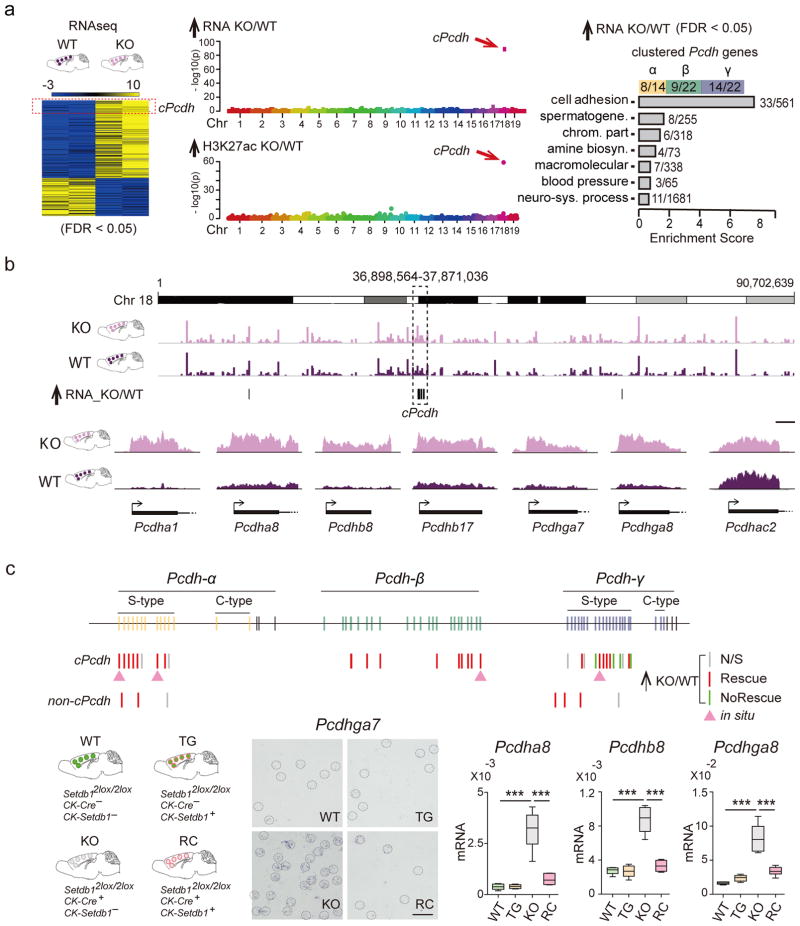
To explore whether disintegration of superTADcPcdh affects gene expression, we mapped transcriptomes and neuronal H3K27ac in Cre+Setdb12lox/2lox and CK-Cre− Setdb12lox/2lox cortex. Consistent with Setdb1’s repressor function8, the majority of transcripts altered in mutant were up-regulated (208/321) (Supplementary Table 14). Importantly, 20% of the entire pool of Setdb1-sensitive genes located to cPcdh, affected Protocadherins and non-Protocadherins, resulting in a unique, 543-fold enrichment on a genome-wide scale (Poisson test P=2.32e−89; 1Mbsw sliding windows applied to N=208 transcripts) (Figure 4a,b). Similarly, among the 1070 sequences with histone hyperacetylation in mutant neurons (DiffRep 1kbsw, adj. P<0.05, Supplementary Table 2), the cPcdh locus was uniquely affected with 96-fold enrichment on a genome-wide scale (1MBsw observed/expected 38/0.42, Poisson test P=6.06e−60) (Figure 4a).
Figure 4. Transcriptional dysregulation at the cPcdh locus.
(a) (Left) genome-wide RNAseq heatmap, blue-yellow range show average levels of expression (log 2), for transcripts with significant (FDR P<0.05) difference in expression of KO compared to WT PFC. (Right) Gene Ontology of differentially expressed genes (FDR P<0.05) highlight Setdb1-dependent regulation of cPcdh cell adhesion genes. (Middle) Manhattan plots for autosomal genome (mouse chromosomes 1–19), showing singular enrichment (1MBsw) for (Top) upregulated transcripts and (Bottom) histone hyperacetylated chromatin at chromosome 18 cPcdh locus, as indicated. (b) (Top) Whole chromosome 18 view on cortex RNA-seq, merged FastQ N=2KO (light purple) and N=2WT (dark purple). (Bottom) representative RNAseq tracks for first exon of specific Pcdhα, Pcdhβ, and Pcdhγ genes. Scale bar, 500bp. Notice increased expression primarily from non-C type Pcdh genes with stochastic expression pattern (S-type), while C-type Pcdh genes remain unaffected. (c) Top: Pcdh and non-Pcdh transcripts tested in adult cortex from mutant and transgenic rescue mice and their respective controls by qRT-PCR and in situ hybridization as indicated. Bottom: Transgenic rescue for representative S-type Pcdh, shown by ISH from middle layers of lateral cerebral cortex (scale bar, 50 μm). Box plots (1st/3rd quartile, median, whiskersmin,max) summarizing qRT-PCR in PFC of WT, TG (CK-Setdb1+ transgenic line), KO and RC (CK-Setdb1+ transgenic rescue of conditional CK-Cre+, Setdb12flox/2lox mutants). N=6/group, ***P<0.001. Pcdha8, t (WT/KO) = 9.59, t (KO/RC)=8.33; Pcdhb8, t (WT/KO)=11.07, t (KO/RC)=10.03; Pcdhga8, t (WT/KO)=10.07, t (KO/RC)=7.39. One-way ANOVA, Bonferroni corrected. Additional ISH gene expression data are shown in Supplementary Figures 12–15.
To confirm that such extremely locus-specific accumulation of up-regulated transcripts and H3K27ac hyperacetylated sequences is indeed driven by neuronal ablation of Setdb1, we reintroduced full length Setdb1, via a CK-Setdb1 transgene30, into the conditional mutant line (Supplementary Figure 10). Parallel testing of four genotypes, or CK-Cre+ Setdb12lox/2lox and CK-Cre− Setdb12lox/2lox(2lox/wt) each with and without CK-Setdb1 (N=6 mice/genotype) confirmed complete rescue with return to baseline expression for 33/43, or 76% of α, β and γ Protocadherins and of additional (non-Protocadherin) genes in cPcdh (Figure 4c, Supplementary Table 15). To further test whether cPcdh’s unique vulnerability is specific for Setdb1 deletions in postnatal neurons, we profiled adult cortical and striatal transcriptomes after CK-Cre ablation of neuronal G9a/Glp, encoding a H3K9 methyltransferase complex essential for normal brain function31, 32. In addition, we profiled transcriptomes from embryonic day E15.5 Nestin-Cre+,Setdb12lox/2lox cortex. None of these various transcriptome sets showed local enrichment at cPcdh (Supplementary Figure 11). Therefore, Setdb1 exerts unique transcriptional control across the cPcdh domain specifically in mature neurons. However, this regulatory layer is not representative for other types of H3K9 methyltransferase, or for the prenatal Setdb1-deficient brain.
Of note, 31/53 S-type (single-neuron stochastically expressed) Protocadherin α, β and γ genes, critically important for neuronal diversity and connectivity33, 34, showed increased expression after neuronal Setdb1 ablation (Figure 4b,c). We studied S-type expression patterns by in situ hybridization providing single cell resolution, using probes specific for individual S-type Pcdha1, Pcdha8, Pcdhb22 and Pcdhga7. Strikingly, brain sections from Setdb1 conditional mutants but none of the three control genotypes including transgenic rescue showed massively increased numbers of robustly stained neurons diffusely distributed across cortical layers II-VI and hippocampus (Figure 4C). Cerebellar cortex, which in contrast to forebrain is lacking CK-Cre expression, remained unaffected in conditional mutant brain (Figure 4c, Supplementary Figures 12–15). Therefore, S-type single neuron stochastic constraint is severely compromised in Setdb1-deficient neurons, contributing to up-regulated expression at the cPcdh locus. Given the critical importance of orderly cPcdh expression—including single-cell stochastic constraint of S-type Pcdh genes—for neuronal morphology and connectivity13, 33, 34, 49, we quantified spine densities and diameters from layer III apical dendrites from Setdb1wt/wt and Setdb12lox/2lox mice, crossed into a conditional line expressing membrane-bound GFP (GFP-F)50 for Golgi-like labeling after low-titer AAV8hSYN1-CreGFP delivered to adult PFC. Indeed, spines from Setdb1-deficient neurons showed 40–50% increased density, and overall decreased size (Supplementary Figure 1g), providing a morphological correlate for dysregulated cPcdh expression.
Balanced facilitative and repressive conformations at cPcdh
Next, we wanted to gain deeper mechanistic insight into the molecular mechanisms mediating the unique position of the cPcdh locus within the Setdb1-sensitive transcriptome and epigenome space. Because CTCF associates with RNA polymerase subunits35 and transcriptional activators36, 37, CTCF-upregulation at H3K27ac-hyperacetylated S-type Pcdh α/β/γ promoters in Setdb1-deficient neurons could facilitate expression, including loss of single cell-stochastic constraint. However, promoter-bound CTCF alone is not sufficient to up-regulate transcription because from genome-wide 63 genes with excess CTCF around the transcription start site, only transcripts within the cPcdh locus were increased. Of note, promoter-enhancer loopings furnished by the CTCF-cohesin scaffolding complex contribute to transcriptional regulation of cPcdh genes20 and therefore, excess CTCF occupancy in cPcdh sequences from Setdb1-deficient neurons could trigger alterations in higher order chromatin. Indeed, excessive CTCF binding at the cPcdh locus of Setdb1 mutant neurons was not limited to promoters, because multiple CTCF peaks emerged de novo in intergenic DNA upstream from α, and within the γ cluster (peaks A-C in Figure 2h). Importantly, these de novo peaks were surrounded by broad >100–200kb stretches of H3K9me3-tagged chromatin that underwent significant ‘shrinkage’ after neuronal Setdb1 ablation (labeled ‘R1’ and ‘R2’ in Figure 5a). Importantly, ‘R1’ and ‘R2’ marked the anchor regions of massive bundles of long-range chromosomal conformations in wildtype neurons. Thus, densely spaced H3K9me3-tagged ‘R1’ loopings, emanating from 100–200kb wide blocks of repressive chromatin upstream of Pcdhα genes, radiated towards many sites within cPcdh, even reaching the distal-most Pcdhγ sequences. However, these long-range loopings became completely dissolved upon structural disintegration of the superTADcPcdh (Figure 1e). Among H3K9me3-tagged conformations lost after neuronal Setdb1 deletion were multiple loopings interconnecting R1 and R2 with two DNAse I hypersensitive enhancer elements, HS16- and HS5-1, previously shown to broadly facilitate cPcdh expression25–29. These defects in HS16/HS5-1 bound long-range contacts were highly specific, because mutant neurons fully maintained shorter-range loopings from Protocadherin gene promoters to HS16/HS5-1 enhancers within the subTADs (Figure 1e). We confirmed these Hi-C findings, including specific weakening of long-range R1—HS16, R1—R2 and R2—HS5-1 and preservation of shorter-range contacts, in neuron-specific chromosome conformation capture (3C) PCR assays from adult mutant and control cortex (Figure 5b, Supplementary Figure 16a). These studies, taken together, would suggest that in wildtype, HS16 and HS5-1 enhancer sequences are ‘locked’ into H3K9me3-tagged repressive chromatin. Upon Setdb1 deletion, loss of R1/R2 repressive loop formations could release the ‘epigenomic brake’, thereby shifting the balance from repressive towards facilitative contacts furnished by HS16 and HS5-1-bound promoter-enhancer loopings, thereby triggering increased expression across the cPcdh locus (Figure 5c). To test this hypothesis, we transfected NG108 neuroblastoma cells with small RNA guided (sgRNA) Cas9-SunTag protein scaffolds38 designed to load ten copies of the potent transcriptional activator, VP64, onto single HS16 sites (Figure 5d). Therefore, such type of HS16Cas9-SunTag(10xVP64) ‘epigenomic superactivation’ could, like the loss of R1—HS16 and R2—HS5-1 repressive loopings after Setdb1 ablation, increase transcription at multiple positions across the entire 1Mb cPcdh locus via promoter-enhancer contacts and other mechanisms. Dual labeled cells (sgRNA-dCas9-10xGCN4’SunTag’-blue fluorescent protein and scFv(recognizing GCN4 epitope)-superfold GFP-VP6438) were compared to controls expressing exactly the same types of vectors but without the sgRNA. Indeed, HS16 epigenomic superactivation was associated with increased expression of 3/6 cPcdh transcripts (pre-selected for consistent baseline expression in neuroblastoma cells), closely mimicking the transcriptional phenotype in Setdb1 mutant cortex (Figure 5e).
Figure 5. Epigenomic editing at the cPcdh locus.
(a) cPcdh locus and surrounding sequences ~2Mb of mouse chr. 18, including TADs called (TADtree) and H3K9me3 tracks for KO and WT. Notice ‘shrinkage’ of broadly (>100–200kb) stretched ‘R1’ and ‘R2’ blocks of H3K9me3-tagged chromatin in KO neurons. (b) Overview on cell-type specific 3C-PCR, cropped agarose gels showing specific loop products for cPcdh and B2m control. No lig=3C without DNA ligase, L=100bp DNA ladder. Dot graphs summarizing 3C-PCR (mean±S.E.M.; 1dot=1animal) cPcdh loop1,2,3 as indicated. All data normalized to B2m 3C, N(Loop1)=3/group, *P(Loop1)=0.05, Mann Whitney, one-tailed; N(Loop2)=4/group, *P(Loop2)=0.014,Mann Whitney, two-tailed. Loop defects in KO include A/R1(de novo CTCF peak A in R1)-HS16 and A/R1-B/R2 (de novo CTCF peak B in R2). In contrast, shorter-range Pcdha8 promoter-HS5 enhancer loop is maintained in KO neurons. Complete gels shown in Supplementary Figure 16c. (c) Summary presentation of 3C-PCR. (d) dCas9-SunTag superactivation of HS16 cPcdh enhancer with U6-sgRNA cassette upstream of CK-dCas9-10xGCN4epitope-BFP cassette, and CK-svFv-sGFP-VP64 cassette on separate vector. Representative FACS sort shows dually labeled BFP+GFP+ NG108 cells. NC=negative control (e) RT-PCR quantification (mean±S.E.M.; 1dot=1cell culture or animal) of Pcdha3, Pcdha8, Pcdhb16, Pcdhgb2 and Pcdhgb8 transcripts (black arrows in panel d mark genomic positions), normalized to Gapdh RNA. (Top) BFP+GFP+ NG108 cells with (HS16/VP64) and without (VP64) sgRNAHS16 cassette. (Bottom) adult KO and WT PFC. N=4 VP64/3 hs16/vp64 (NG108 cells), *P(Pcdha8)=0.0268, *P(Pcdha11)=0.0437, *P(Pcdhgb8)=0.0126, unpaired t test, one-tailed; N=6/group (mice), **P(Pcdha3)=0.002, **P(Pcdha8)=0.002, *P(Pcdha11)=0.026, **P(Pcdhgb8)=0.0022, Mann Whitney, two-tailed. See also Supplementary Figure 16A for additional 3C-PCR loop quantifications.
Conserved regulation of human and mouse superTADcPCDH
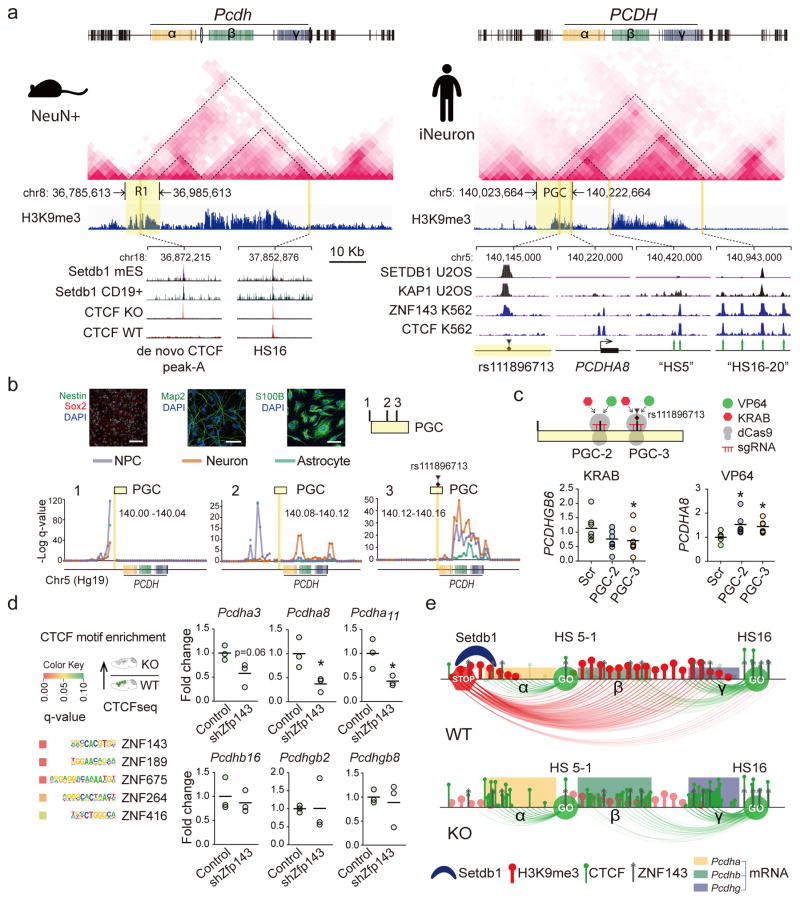
The linear arrangement of α, β and γ clusters with S- and C-type Protocadherin genes is highly conserved across vertebrate genomes13. We showed that higher order chromatin, including broad >100–200kb stretches of intergenic Setdb1-regulated H3K9me3-tagged sequence associated with repressive loop bundles, critically regulates transcription across the cPcdh locus. We asked whether such types of 3D genome conformations, just like the linear genome, could be conserved across mammalian lineages. To examine, we generated in situ Hi-C interaction matrices in human glutamatergic neurons differentiated from induced pluripotent stem cell-derived neural precursors by controlled expression of Neurogenin 2, and compared the 3D genome map to wildtype mouse cortex NeuN+ nuclei (of which >80% are from excitatory neurons). Indeed, TAD landscapes surrounding the cPcdh/PCDH locus (Mm10 chr. 18; Hg19 chr. 5) showed startling similarities between mouse cortex NeuN+ nuclei and human neurons, including complete preservation of cluster-specific subTADs nested into a large Mb-scale superTADcPCDH. In addition, human and mouse neuronal chromatin exhibited highly similar-shaped H3K9me3 landscapes, including the broadly stretched aforementioned Setdb1-sensitive ‘R1’ at the superTAD’s 5’ end and ‘R2’ around the 5’end of the γ cluster (Figure 6a). We were surprised to discover that ‘R1’ near-perfectly matched a risk haplotype (chr5:140,023,664-140,222,664) of the Psychiatric Genomics Consortium39. This haplotype (no. 108 in reference39, referred to as ‘PGC’ hereafter) significantly contributes, independently from another 107 loci genome-wide, to schizophrenia heritability39, with a small INDEL as the lead polymorphism (rs111896713 chr5:140,143,66439). This risk polymorphism matched to robust Setdb1 peaks conserved in human and mouse cells including brain, but ‘replaced’ by de novo CTCF peaks upon Setdb1 ablation (Figure 6a, Supplementary Figure 16b,d). Therefore, we predicted that higher order chromatin organization at these positions will be highly conserved in human brain cells, and specifically in neurons, with long-range loopings radiating from ~200kb R1 towards cPCDH promoter and enhancers primarily anchored in chromatin at and around the Setdb1 peak. To explore, we surveyed with 40kb resolution the cPCDH-bound chromosomal contacts in our in situ Hi-C datasets generated from human neurons and their isogenic neural precursors cells (NPC) and NPC-differentiated astrocytes. Strikingly, 40kb bins within PGC showed a step-wise progression in contact intensities with cPCDH sequences, culminating in massively increased contact frequencies at the bin harboring a robust Setdb1 peak (‘PGC-3’ in Figure 6b). This effect was pronounced in neurons and NPC, while corresponding loopings were much weaker or missing altogether in our contact maps from astrocytes, indicating strong cell type-specific regulation of local 3D genome architectures (Figure 6b). Next, we asked whether these within-PGC haplotype differences in cPCDH interaction frequencies translate into differential repressive potential. To this end, we introduced small guide RNAs (sgRNA) into two stable NPC lines, expressing 1. dCas9-KRAB fusion protein tethering KAP1 (KRAB-associated protein 1)-Setdb1 repressor complex8, 40, or 2. dCas9-VP64 to dock the VP64 activator at different positions within the schizophrenia risk haplotype. We then measured expression levels for S-type γPcdh genes expressed in NPC at comparatively high levels at baseline (data not shown). Interestingly, KRAB recruited to sequences close to the Setdb1 peak at the risk haplotype’s lead polymorphism (‘PGC-3’ in Figure 6c), was consistently associated in 3/3 experiments with a robust multifold decrease in expression of PCDHGB6 (but not PCDHGA3). In contrast, dCas9-KRAB docked to a non-Setdb1 binding site (‘PGC-2’ in Figure 6c) or to a scrambled control sequence (‘Scr’ in Figure 6c) remained ineffective and did not suppress PCDHGB6 and PCDHGA3 expression (Figure 6c). Of note, VP64 epigenomic editing at ‘PGC-3’ and neighboring ‘PGC-2’ was associated with increased expression of a subset of cPCDH genes (Figure 6c). These findings, taken together, suggest that repressive effects on Protocadherin gene expression are specific for loop-bound KRAB positioned at intergenic ‘PGC-3’ sequences upstream of the cPCDH gene clusters.
Figure 6. Regulatory mechanisms at human and mouse TADcPCDH.
(a) (Top) Neuronal in situ Hi-C interaction matrices, and H3K9me3 landscapes, for ~2Mb of mouse and human cPCDH, including superTAD spanning across α,β,γ clusters. (Bottom left) Setdb1 peaks in mouse embryonic stem cell and lymphocytes match to de novo CTCF peak in Setdb1 KO neurons. (Bottom right) ‘PGC’, Psychiatric Genomics Consortium risk haplotype chr5:140,023,664-140,222,664 with lead polymorphism rs111896713 matching to Setdb1, KAP1 and ZNF143 peaks. Note epigenomic similarities of ‘R1’ (mouse) and ‘PGC’ (human). (b) Neural progenitor cell (NPC) differentiation into neurons and astrocytes, with phenotypic markers as indicated. Scale bar, NPC (neuron/astrocyte) 100 (50) μm. Conformations for three representative 40kb bins from 200kb ‘PGC’ haplotype, with bin harboring the index polymorphism (‘PGC-3’) showing dramatically increased cPCDH contact. (c) Dot graphs show cPCDH gene expression in epigenomically edited NPC, with PCDHGB6 (but not PCDHGA8) transcript decreased by sgRNA-guided dCas9-KRAB in 3/3 experiments. dCas9-VP64 elicits increased expression of a subset of Protocadherin transcripts. Scr, scrambled control. All data normalized to 18srRNA, shown as fold change. N=6–9/group, P < 0.05, Mann Whitney, two-tailed. (d) ZNF-specific motif enrichments in CTCF-up sequences. Dot graphs (1dot/cell culture) summarize expression of specific Pcdhα, β and γ genes after shRNA-induced Zfp143 knock-down in NG108 neuroblastoma cells. Unpaired t test, two-tailed, N=3 per treatment. *P<0.05 (e) Schematic summary of TADcPcdh epigenomic architectures in WT and KO neurons. Loss of repressive long-range contacts in KO shifts the balance towards facilitative shorter range promoter-enhancer loopings.
It is remarkable that KRAB—a critical module in KRAB-zinc finger proteins (KRAB-ZNF/Zfp) important for sequence-specific docking of the KAP1-Setdb1 repressor complex8, 40—inhibits cPCDH expression via long-range loopings, bypassing 644kb linear genome in case of PCDHGB6 (Figure 6c). Therefore, intergenic R1/risk haplotype-bound Setdb1 is likely to function as key transcriptional regulator at the cPCDH locus. These intergenic sequences harbor in the ENCODE database matching peaks for Setdb1, KAP1 and multiple KRAB-ZNF proteins including ZNF 27441 and ZFP143 (Figure 6a). Importantly, ZFP143 recognition sequences emerged as top scoring zinc finger motifs enriched at sites with excessive CTCF binding in Setdb1-deficient mouse neurons (Figure 6d, Supplementary Tables 9,10). ZNF143, like CTCF and cohesion considered a key organizer for the 3D genome42, 43, co-assembles with positive and negative regulators of transcription depending on local chromatin context44. Unsurprisingly then, ZNF143 at the cPCDH locus occupies in addition to R1 repressive chromatin also promoters and HS16/HS5-1 enhancers (Figure 6a). Therefore, alterations in ZNF143 supply, affecting facilitative and repressive chromatin, could destabilize cPcdh expression. Indeed, small RNA-mediated Zfp143 knock-down in mouse neuroblastoma cells was associated with decreased expression of multiple cPcdh genes (Figure 6d). Our studies, taken together, suggest that 1. regulatory 3D genome architectures at the cPCDH locus are highly conserved between mouse and human, 2. include SETDB1-KRAB-ZNF143 and CTCF as key organizers of local repressive and facilitative chromosomal conformations, 3. ‘bundles’ or ‘aggregates’ of Setdb1-dependent long-range repressive loopings radiating from intergenic DNA (‘R1’, ‘R2’) function as ‘epigenomic brakes’ for transcriptional control, counterbalancing facilitative shorter range promoter-enhancer contacts (Figure 6e).
Discussion
Neuronal Setdb1 ablation triggers structural disintegration of megabase-scale TADs, including the Protocadherin α/β/γ locus as the only Setdb1-sensitive TAD harboring a gene cluster. TADs affected in mutant neurons showed shrinkage of broadly stretched H3K9me3-tagged chromatin, in conjunction with localized hotspots of excess and de novo CTCF binding. Setdb1-regulated long-range repressive cPcdh loopings were highly enriched in neurons as compared to their isogenic precursors, and carried DNA polymorphisms conferring liability for schizophrenia. 3D genome conformations at cPCDH could have even broader relevance for neuropsychiatric disease, given that SETDB1 microdeletions and structural variants are associated with neurodevelopmental delay51, 52, with CpG hypermethylation reported for orthologous CTCF binding sites within the PCDH gene cluster in Down syndrome (trisomy 21) including the mouse model53, and cPcdh DNA promoter methylation linked to depression and anxiety54–56. Furthermore, mice exposed to chronic variable stress, a preclinical paradigm frequently implied in psychiatric disease57, show hyperexpression of β Protocadherin genes (Supplementary Figure 17).
We show that Setdb1, maintaining high levels of DNA methylation and low levels of histone acetylation at sequences in close vicinity or partial overlap with potential CTCF binding sites, critically shields neuronal genomes from uncontrolled CTCF docking at thousands of cryptic binding sites genome-wide. However, upon neuronal Setdb1 ablation, the shield becomes defunct, triggering collapse of vulnerable TADs. Remarkably, our findings on excessive cPcdh CTCF occupancies and increased cPcdh expression and the resulting increase in spine densities in Setdb1-deficient neurons are perfect opposites to the previously reported decreases in cPcdh expression and spine densities after neuronal Ctcf ablation58. Likewise, switching ‘reverse-forward’ strand orientations in CTCF binding sequences25 could disrupt promoter-enhancer loopings and broadly dampen transcription across α/β/γ clusters (figure 2d in reference25). These findings strongly point to delicate regulatory mechanisms governing chromosomal conformations, with genomes excessively populated by CTCF, including the Setdb1-deficient neurons of the present study, showing disintegration of higher order chromatin in locus-specific manner. The structural collapse of the Setdb1-sensitive superTADs was highly specific, given that the genome-wide excess of CTCF binding in mutant neurons triggered a genome-wide increase in insulation strength and ‘compartment-ness’.
Future work will clarify whether neuronal Setdb1 overexpression30, 59, or loss of other proteins assigned with regulation of cPcdh expression, including DNMT3b cytosine methyltransferase34 and SMCHD160 and WIZ61 repressors, could trigger TAD-specific 3D genome changes in neurons. We note that Setdb1 is primarily located towards the 5’ and 3’ ends of superTADcPcdh (Figure 6a,e, Supplementary Figure 18). In any case, the TAD-specific phenotypes in Setdb1 mutant neurons point towards unexplored modular complexities in the regulatory mechanisms governing the 3D genome. Thus, rewiring or disintegration of specific TAD units may not be exclusive to chromosomal microdeletion and –duplication events62, 63, because as shown here, loss of Setdb1 function triggers the disintegration of highly select subset of neuronal TADs. With each chromosome furnishing hundreds of TAD-like structures, it will be an exciting and challenging task to dissect ‘TAD-by-TAD’ and in cell-type specific manner, the multilayered mechanisms governing locus-specific higher order chromatin in highly differentiated brain cells.
ONLINE METHODS
Human Stem Cell Lines
All work with human induced pluripotent stem cell lines has been approved by the Institutional Review Board of the Mount Sinai School of Medicine, in accordance with Mount Sinai’s Federal Wide Assurances (FWA#00005656, FWA#00005651) to the Department of Health and Human Services. No new stem cell lines had been generated for the work presented here. Informed consent had been obtained from all participating subjects. See Supplementary Methods for differentiation into neural pregenitors, glutamatergic neurons and astrocytes.
Animal studies
All animal work was approved by the Institutional Animal Care and Use Committee of the Icahn School of Medicine at Mount Sinai. Mice were held under specific pathogen-free conditions with food and water being supplied ad libitum in an animal facility with a reversed 12 h light/dark cycle (light off at 7:00 am) under constant conditions (21 ± 1°C; 60% humidity). All animals were group housed (2–5/cage).
Generation of Setdb1 conditional mutant and rescue mice
Setdb12lox/2lox mice were generated by Ozgene, Australia. In brief, two loxP sites were inserted to endogenous Setdb1 locus (Setdb1 ENSMUSG00000015697; SET domain, bifurcated 1, MGI:1934229) flanking exon 3. To generate conditional Setdb1 knockout mice, Setdb12lox/2lox mice were first crossed with CK-Cre+/− transgenic mice to generate Setdb12lox/+, CK-Cre+/0 heterozygous, which were further crossed with Setdb12lox/2lox mice to generate Setdb12lox/2lox, CK-Cre+/0 homozygous. Cre recombinase mediated excision of Setdb1 exon 3 causes frame shift and generates a stop codon at the new junction of exon 2 and exon 4, and results in early termination of Setdb1 translation. Gender and age matched littermates with genotype Setdb12lox/2lox, CK-Cre0/0 were used as controls with wildtype SETDB1 levels. CK-Setdb1 transgenic mice, described previously64, express full-length mouse Setdb1 cDNA driven by CK promoter in postnatal and adult mouse forebrain. To generate Setdb1 rescue mice, the CK-Setdb1 transgene was introduced into Setdb12lox/2lox conditional knockout background and CK-Setdb1+/0, Setdb12lox/2lox mice were crossed with Setdb12lox/+, CK-Cre+/0 to generate Setdb12lox/2lox, CK-Cre+/0, CK-Setdb1+/0 rescue mice. Gender and age matched littermates with genotype Setdb12lox/2lox, CK-Cre0/0, CK-Setdb10/0 were used as wildtype controls. All genetically engineered lines were backcrossed to the C57BL6/J line for at least 10 generations.
See Supplementary Methods for information on Nestin-Cre conditional mutagenesis, Mendelian survival ratios, Histology, Comet-assay and RNA quantifications including RNA-seq.
Chromatin assays
Chromatin assays (ChIP-seq, in situ Hi-C, 3C-PCR) and RNA-seq were conducted in young adult mice, at 3 months of age (± 2 weeks). Supplemental Tables 1–4, 8 and 12 provide additional information for each chromatin assay, including number of animals and sex ratios.
Nuclei preparation, immunotagging and fluorescence-activated sorting
For fluorescence-activated nuclei sorting, nuclei were extracted from mouse cerebral cortex or human prefrontal cerebral cortex (control, PFC, male, PMI 17) and anterior cingulate cortex (control, ACC, female, PMI 27) as described before65. In brief, brain tissue was homogenized in hypotonic lysis solution, purified by ultra-centrifugation, and then re-suspended in 1 ml DPBS containing 0.1% BSA, 1:1000 Anti-NeuN antibody, clone A60, Alexa Fluor®488 conjugated (EMD MILLIPORE CORP MAB377X). Samples were incubated for at least 45 min by rotating in the dark at 4°C. DAPI was added before FACS to label all the nuclei. Sorting was done on Flow Cytometry Center at Mount Sinai. Nuclei were separated into NeuN+ and NeuN- population and then pelleted for following applications. For XChIP, 3C and in situ HiC experiments, 10 minutes of 1% formaline fixation at room temperature was incorporated right after brain homogenization. Cross-linking was quenched by incubating with 125 mM glycine. Nuclei were then purified, stained and sorted as described above.
ChIP-seq
Native immunoprecipitation (NChIP) was performed as described65. In brief, NeuN+ (neuronal) and NeuN− (non-neuronal) nuclei were pelleted post-FACS and then resuspended in 300 ul of MNase digestion buffer (10 mm Tris, pH 7.5; 4 mm MgCl2; and 1 mm Ca2+), digested with 3ul of MNase (0.2 U/μl) for 5 min at 28°C to obtain mono-nucleosomes. Reaction was stopped with 50mM EDTA, pH 8. Nuclei were swollen to release chromatin after addition of hypotonization buffer (0.2 mm EDTA, pH 8, containing PMSF, DTT, and benzamidine). Chromatin was incubated with anti-H3K9me3 (Abcam AB8898) and anti-H3K27ac (Active Motif, #39133) antibodies overnight at 4°C. The DNA-protein-antibody complexes were captured by Protein AG Magnetic Beads (Thermo Scientific™ 88803) by incubating at 4°C for 2 hours. Magnetic beads were then washed with low-salt buffer, high-salt buffer, and TE buffer. DNA was eluted from the beads, and treated with RNase A followed by proteinase K digestion. DNA was purified by phenol-chloroform extraction and ethanol precipitation.
For XChIP on crosslinked preparations, formaldehyde-fixed NeuN+ nuclei after FACS were resuspended in lysis buffer containing 0.1%SDS, sonicated (Bioruptor® Plus sonication device, Diagenode) at the ‘high’ setting for 30 minutes on ice. The size of DNA fragmentation was between 100 bp to 500 bp with an average size of 300bp. Chromatin was then incubated with anti-CTCF (EMD Millipore, # 07729) or anti-SETDB1 (Santa Cruz H-300X ##sc-66884 X) or anti-SETDB1 (Thermofisher 5H6A12 #MA5-15722) and captured with Protein AG Magnetic Beads. After washing and elution, DNA was incubated at 65°C overnight for reverse cross-linking, followed by RNase A, proteinase K treatment and DNA precipitation. Setdb1 occupancies were measured by conventional ChIP-PCR. For CTCF ChIP-seq library preparation, ChIP DNA was end repaired (End-it DNA Repair kit; Epicentre) and A tailed (Klenow Exo-minus; Epicentre). Adaptors (Illumina) were ligated to the ChIP-DNA (Fast-Link kit; Epicentre) and then PCR amplified using Illumina TruSeq ChIP Library Prep Kit. Library DNA with expected size (NChIP, ~275bp; XChIP, 350bp to 500bp) was selected by Pippin and submitted to New York Genomic Center and sequenced with Illumina HiSeq 2000, 75bp, paired end. The ChIP-seq data was first checked for quality using the various metrics generated by FastQC (v0.11.2). Raw sequencing reads were then aligned to the mouse mm10 genome (or Hg19 for human) using default settings of Bowtie (v2.2.0). Uniquely mapped reads were retained and the alignments were subsequently filtered using the SAMtools package (v0.1.19) to remove duplicate reads. Differential analysis between mutant and control samples was performed using diffReps with window size 1000 bp and moving step size 500 bp, and FDR<5% as significance cutoff66 and data visualized on the genome using the Integrative Genomics Viewer (IGV) program67. For H3K27ac and CTCF ChIPseq, peak-calling was performed using MACS (v2.1.1) with a FDR cutoff of 0.05. Gene Ontology enrichment of annotated genes, with significant hits from diffReps within gene bodies or within 3Kb around transcriptional starting sites, was further analyzed using DAVID Functional Annotation Bioinformatics tools (Resources 6.7, National Institute of Allergy and Infectious Diseases, NIH). Significant hits from diffReps for decrease in H3K9me3 and increase in CTCF ChIPseq were subjected to motif analysis, using the Homer package (v4.8.3) at default settings68. Manhattan plots for genome-wide differential epigenetic profiling of conditional mutants and controls were constructed after the genome was divided into non-overlapping 1Mb bins, including the 1Mb bin spanning the clustered Pcdh genes chr18:36,870,001-37,870,000, mm10. The number of occurrences of each signal was tabulated within each bin. The probability of the number of occurrence of each signal per 1Mb bin was then modeled using a Poisson distribution with the maximum likelihood estimator for the lambda parameter given by the calculated mean number of occurrences. The Poisson models for each signal were used to calculate the probability of occurrence of the signal observed in every 1Mb bin (including the Pcdh bin).
Chromosome Conformation Capture (3C)
3C was performed using standard protocols with minor modifications69. In brief, nuclei were fixed and extracted from mouse cerebral cortex and FACS sorted as described above. NeuN positive (neuronal) nuclei were then pelleted and digested with Hind III restriction enzyme (NEB) at 37°C overnight, washed, and treated with T4 DNA ligase at room temperature for 4 hr. 3C DNA was then incubated at 65°C overnight for reverse crosslinking followed by DNA purification and precipitation. 3C primers were listed Primers are listed in Supplementary Table 16. Sequence-verified PCR products were measured semi-quantitatively with UVP Bioimaging system/Labworks 4.5 software. Neighboring primers at B2m gene locus was used for normalization.
in situ Hi-C including bioinformatical analyses
Nuclei were fixed and extracted from mouse cerebral cortex and human postmortem anterior cingulate cortex and sorted into NeuN+ (neuronal) and NeuN− (non-neuronal) populations, which were then processed using an in situ Hi-C protocol70, with minor modifications. Briefly, the protocol involves a restriction digest of the cross-linked chromatin within intact nuclei, followed by biotinylation of the strand ends, re-ligation, sonication and size selection for 300–500bp fragments, followed by standard library preparation for Illumina sequencing. The resulting data were mapped, filtered, and normalized using HiC-Pro71 (v2.7.8) and visualized on the Washington University Epigenome Browser. To explore localized enrichments in the in situ Hi-C datasets, we tabulated for each 40kb bin along chromosome 18 the number of long-range interactions greater than 200kb that were disrupted (ie. significantly decreased in conditional CK-Cre mutants versus control, as detected using DESeq2 at P<0.05). The probability of observing the number of disrupted interactions at each bin was then modeled using a Poisson distribution with maximum likelihood of mean (0.165) calculated from the data. Topological associating domains (TAD) were predicted using TADtree70 using the 20kb Hi-Cpro data as input with the following parameter settings: maximum size of TAD in bins (S) = 60; maximum number of TADs in each tad-tree (M) = 10; boundary index parameter (p) = 6; boundary index parameter (q) = 24; balance between boundary index and squared error in score function (gamma) = 500; number of TADs to use (N) = 400 (chr18) or 700 (chr5).
In addition, initial processing of the raw 2×125bp read pair FASTQ files was performed using the HiC-Pro analysis pipeline. In brief, HiC-Pro performs four major tasks: aligning short reads, filtering for valid pairs, binning, and normalizing contact matrices. HiC-Pro implements the truncation-based alignment strategy using Bowtie v2.2.372, mapping full reads end-to-end or the 5’ portion of reads preceding a GATCGATC ligation site that results from restriction enzyme digestion with MboI followed by end ligation. Invalid interactions such as same-strand, dangling-end, self-cycle, and single-end pairs are not retained. Binning was performed in 40kb and 100kb non-overlapping, adjacent windows across the genome and resulting contact matrices were normalized using iterative correction and eigenvector decomposition (ICE) as previously described73.
Starting with the 20 kb resolution intra-chromosomal contact matrices generated by HiC-Pro, we first generated 100 kb resolution contact matrices by summing the interaction frequencies of the 20 kb bins within each 100 kb bin. We next generated the corresponding log2(observed/expected) matrices, where the observed/expected values are the ratio of the contact values of each interaction bin to the average contact values of all interaction bins the same distance apart. The Pearson’s correlation matrices were then calculated from the log2(observed/expected) matrices, and PCA was performed on them. The first principal component (PC1) was then used to differentiate the compartments. When the first principal component value was positively correlated to gene density and gene expression (we found that the first principal component always correlates with both gene density and gene expression in the same direction), bins with positive PC1 values are assigned as compartment A while bins with negative PC1 values are assigned as compartment B. Conversely, when PC1 is negatively correlated to gene density and gene expression, bins with negative PC1 values are assigned as compartment A while bins with positive PC1 values are assigned as compartment B.
Higher-resolution topologically associated domain (TAD) calls were made following the procedure described by Dixon, et al.74 using the directionality index (DI) metric. DI was calculated using raw interaction counts between 40kb or 100kb bins and respective window sizes of 2Mb or 5Mb to capture observed upstream or downstream interaction bias of genomic regions. A Hidden Markov model (HMM) was then trained to infer true bias states. TADs were defined by pairing adjacent regions of inferred downstream or upstream bias states.
To identify significantly enriched interactions involving a bin of interest, the expected interaction counts for each interaction distance were estimated by calculating the mean of all intrachromosomal bin-bin interactions of the same separation distance throughout the entire ICE-normalized contact matrix. We estimated the probability of observing an interaction between a bin-of-interest and some other bin by calculating the expected interaction between those two bins divided by the sum of all expected interactions between the bin-of-interest and all other intrachromosomal bins. We then calculated the p-value of observing the observed number of interaction counts or more between the bin-of-interest and some other bin using a binomial test where the number of successes was defined as the observed interaction count, the number of tries as the total number of observed interactions between the bin-of-interest and all other intrachromosomal bins, and the success probability as the probability of observing the bin-bin interaction estimated from the expected mean interaction counts. To control false discovery rate, the R package, qvalue, was used to estimate q-values from the calculated binomial p-values.
The Insulation Analysis was performed with reference to Crane et al75 and Vietri Rudan et al76. Briefly, using the 20kb resolution HiC matrix, we calculated (at each 20kb bin) the average interaction frequency of the chromosomal bins within a certain distance band. The normalized “insulation” score along the chromosome for each band was then calculated as the log2 ratio of average interaction frequency at each 20kb bin to the average of all 20kb bins in that band. Regions along the chromosome that display a dip/valley/minima of normalized insulation values represent regions of reduced interactions, and can be interpreted as TAD boundaries or regions of high local insulation.
DNA methylation
Targeted bis-seq was utilized for fine-mapping of methylation patterns in cPcdh candidate sequences. Genomic DNA (1 μg) was bisulfite-converted using the EpiTect Bisulfite Kit (Qiagen). Primers were designed in MethPrimer77 and bisulfite-converted DNA was amplified and multiplexed by high throughput PCR using a Fluidigm AccessArray instrument. PCRs were performed in duplicate and the duplicates pooled. Primers are listed in Supplementary Table 16. The library was diluted to a final concentration of 10pM with 35% of PhiX library. Paired-end reads (250 bp) were generated with an Illumina MiSeq sequencer. Fastq files were generated by the MiSeq sequencer. After trimming for low-quality bases (Phred score<30), Illumina and Fluidigm adaptors and reads with a length <40 bp with TrimGalore, the reads were aligned to the mouse genome (mm10) using Bismark78 using the following settings: -D 50 -R 10 --score_min L,0,-0.6. Since the sequences are PCR-based, reads were not deduplicated. Methylation calling was performed using Bismark extractor78. Net methylation was assessed when the coverage was at least 100X and reported by CpG and averaged across amplicon. Graphical representations of random samples of 50 sequenced DNA fragments were generated using R. Briefly, using CpG context output files generated by Bismark methylation extractor78 which reports CpG methylation status for each individual sequenced DNA fragments (taking into account paired-end reads), methylation patterns for each DNA fragment were reconstructed based on the coordinates of the covered CpGs and their methylation status. Off-target reads mapping outside the amplicon coordinates were discarded. After random sampling of 50 sequences using the R sample function, their methylation patterns were plotted using the R plot function. Only CpGs present in the reference genome were represented and sequences were represented on positive strand. Circles represent consecutive CpGs, with each line being a unique DNA fragment. White circles are unmethylated CpGs and black circles are methylated CpGs.
To ensure sufficient library complexity, we pooled two PCRs for each amplicon. The median coverage per library ranged from 1000X to 2600X (Supplementary Figure 19A). Since we used a PCR based targeted bis-seq approach, the sequence start points are constrained for each amplicon. Therefore the constrained start sites generated duplication levels which should not be treated as technical duplicates nor removed by bioinformatics deduplication. These duplication levels will be reflected in the distribution of specific sequences that are identified using start sites and therefore not informative to assess the library complexity. Therefore, since some level of randomness of DNA methylation patterns in a cell population is present, to estimate the library complexity, we assessed the number of distinct methylation patterns (i.e. specific sequences based on C>T conversion) observed for each amplicon in a given library. Because of biological duplicates (genuinely distinct DNA molecules with the same methylation pattern), this metric provides highly stringent coverage information. In addition, to take into account sequencing errors, only methylation patterns representing more than 1% of the total reads covering a given amplicon were counted. Overall, the median of methylation patterns per amplicon was 5 (range:1–12.5), with 81.2% of the amplicons with less than 5 methylation patterns covering fully methylated (>80% of methylation) or fully unmethylated (<20%) regions, which are expected to show lower randomness level (Supplementary Figure 19B).
Lenti-shRNA knockdown of Zfp143
Mouse shRNA lentiviral particles targeting Zfp143 (4 unique 29mer target-specific shRNA, 1 scramble control) were purchased from Origene (TL502149V), with sequences in the shRNA expression cassettes are verified by the manufacturer to correspond to the target gene Zfp143 (Gene ID 20841) with 100% identity, to produce 70% or more gene expression knock-down provided a minimum transfection efficiency of 80%. NG108 cells (NG108-15 #108CC15, Vendor: ATCC, Organism: Mus musculus (neuroblastoma); Rattus norvegicus (glioma)) were seeded in 12-well plates 12 hours before viral transduction. 72 hours after transduction, cells were lysed with 500ul of Trizol, total RNA was extracted, reverse transcribed and Pcdh gene expression quantified by real-time PCR. Primers are listed in Supplementary Table 16.
dCas9-SunTag(10xVP64) epigenomic editing
The CRISPR/dCas9_Suntag_VP64 two-plasmid system79 was used for genomic editing on Pcdh locus in the NG108 neuronal cell line. For pLV-U6-sgRNA-CK-dCas9-10xGCN4-BFP construct, lentiviral backbone and dCas9 cassette were cloned from plasmid Lenti-dCAS-VP64_Blast (Addgene, #61425), CamK-II promoter (1293 bp) was cloned from plasmid pAAV-CamKII-hChR2(T159C)-p2A-ETFP-WPR (a gift from Javier Maeso), and 10XGCN4-P2A-BFP cassette was cloned from pHRdSV40-dCas9-10xGCN4_V4-P2A-BFP(Addgene, #60903). The U6-sgRNA cassette was inserted upstream of CamK-II promoter. For the pLV-CK-scFv-GCN4-sfGFP-VP64-GB1-NLS construct, the lentiviral backbone and CK promoter cloned as described above. The scFV-GCN4-sfGFP-RsrII-GB1-NLS (1875 bp) sequence was cloned from plasmid pHR-scFv-GCN4-sfGFP-GB1-NLS-dWPRE (Addgene, #60906), and the VP64 cassette was cloned from plasmid pHRdSV40-scFv-GCN4-sfGFP-VP64-GB1-NLS (Addgene, #60904). All plasmid construction was done by the VectorBuilder team from Cyagen Biosciences. Three sgRNAs (Supplementary Table 16) were designed to target mouse HS16 site at the Pcdh locus. For transfection, control cells received a two-vector system pLV-CK-dCas9-10xGCN4-BFP lacking the sgRNA cassettes and pLV-CK-scFv-GCN4-sfGFP-VP64-GB1-NLS; Cell cultures for HS16 superactivation were transfected with pLV-U6-sgRNA-CK-dCas9-10xGCN4-BFP plasmid that included the sgRNA cassettes targeting the HS16 site, and pLV-CK-scFv-GCN4-sfGFP-VP64-GB1-NLS. 72 hours after transfection, cells were harvested and sorted by FACS to collect BFP and GFP double positive cells. Total RNA was then extracted, reverse transcribed and Pcdh gene expression quantified by real-time PCR. Primers are listed. Primers are listed in Supplementary Table 16.
dCas9-KRAB epigenomic editing
Human neural precursor cells (NPCs) were maintained at high density, grown on growth factor reduced Matrigel (BD Biosciences) coated plates in NPC media (Dulbecco’s Modified Eagle Medium/Ham’s F12 Nutrient Mixture (ThermoFisher Scientific), 1x N2, 1x B27-RA (ThermoFisher Scientific) and 20 ng ml−1 FGF2 (R & D Systems, 233-FB-10) and split 1:3 every week with Accutase (Millipore, Billerica, MA, USA). The hiPSC-NPC line 553-S1-1, as previously described and validated80,81 was used in all NPC editing experiments. Generation of stable dCas9-KRAB NPCs: 3.5×106 NPCs per well were seeded onto growth factor reduced Matrigel coated 6-well plates in NPC media. The following day lentiviruses generated as above using either the lentiviral vectors dCas9:VP64-T2A-puro and dCas9:KRAB-T2A-puro were added and cultures were spinfected (1 hour, 1000xg, 25°C). Following spinfection, plates were transferred to a cell culture incubator for 3 hours. Media was then removed and replaced with fresh NPC media. The following day, fresh NPC medium containing 1 μg/ml puromycin (Sigma, #P7255) was added and cells were maintained in NPC medium containing 1 μg/ml puromycin for the remainder of the experiment. Stable NPC lines were validated via FACS using Cas9-AF488 antibody (Cell Signaling Technologies, 5uL/1×106 cells #34963S). sgRNA design and cloning: The sgRNAs were designed using the Optimized CRISPR Design tool (See URLs) at the genomic regions of interest. Guide RNAs were selected based on their specific locations at decreasing distances from region of interest as well as strand specificity and lack of predicted off targets. Synthesic oligonucleotides (Supplementary Table 16) were annealed (95°C for 5 min, ramp down to 25°C at 5°C per minute), diluted 1:100 and then ligated into BsmB1 digested lentiGuide-dTomato. NPC lentiviral transduction and FACS: 100,000 dCas9-KRAB NPCs per well were seeded onto growth factor reduced Matrigel coated 24-well plates in NPC medium containing 1 μg/ml puromycin. The following day lentiviruses with scrambleD sgRNAs and pooled sgRNAs targeting PGC-1, PGC-2 and PGC-3 region were added to cultures in the presence of polybrene. 48 hours after transduction, NPC cells were FACS sorted and live cell population with dTomato signal were collected directly into Trizaol LS (Thermo Fisher, 10296028). Total RNA was extracted for RT-PCR.
Data Availability
All next generation sequencing data for genome-scale analysis in this publication have been deposited in NCBI’s Gene Expression Omnibus82 and are accessible through GEO Series accession number GSE99363 (See URLs). All other data discussed are included in the publication and available from the Authors upon request.
URLs
Optimized CRISPR Design tool, http://crispr.mit.edu/
GSE99363, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE99363
Supplementary Material
Acknowledgments
Supported by National Institutes of Health, U.S.A. (NIH) grants R01MH106056 (S.A.), P50MH096890 (E.N.), R01MH101454, NIA U01P50AG005138-30-1 and U01AG046170 (K.B.), R01AG050986, R01MH109677, (P.Rou.), R01NS091574 (A.S.), and NIH training and fellowship awards 1F30MH113330 (P.Ra.) and T32-AG049688 (S.C.). Additional support was provided by a Grant-in-Aid for AMED-CREST, AMED, Japan and the Japan-U.S. Brain Research Cooperation Program to T.Y., the Veterans Affairs Merit grant BX002395 (P.Rou.), the Brain & Behavior Research Foundation (Y.J., P.Rou.), the Alzheimer’s Association (P.Rou.), the New York Stem Cell Foundation (K.B.) and the Brain Research Foundation (S.A.).
Footnotes
Competing Financial Interest: The Authors declare no conflicts of interest.
Author Contributions: Performed experiments: Y.J., P.Raj., T.H., B.K., B.H., S-M.H., B.J., L.K., R.P., S.C., C.D., C.P., J.TCW, B.S. Conceived and designed experiments: Y.J., S.A. Performed statistical analyses: Y.J.. Bioinformatics and genomic analyses: E.L., P.Raj., W.L., P.Rou., L.S.. Contributed materials: A.S., B.R. (G9a/Glp transcriptome data); B.L., E.N. (mouse stress model/transcriptome data). Supervised the DNA methylation analysis: B.T.. Supervised research: H.M., K.B., T.Y., L.S., S.A.. Wrote the paper: Y.J., Y-H. E.L., B.T., L.S., S.A. with contributions from co-authors.
References for main text
- 1.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merkenschlager M, Nora EP. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu Rev Genomics Hum Genet. 2016;17:17–43. doi: 10.1146/annurev-genom-083115-022339. [DOI] [PubMed] [Google Scholar]
- 4.Cubenas-Potts C, Corces VG. Topologically Associating Domains: An invariant framework or a dynamic scaffold? Nucleus. 2015;6:430–434. doi: 10.1080/19491034.2015.1096467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Won H, et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan SL, et al. Essential roles of the histone methyltransferase ESET in the epigenetic control of neural progenitor cells during development. Development. 2012;139:3806–3816. doi: 10.1242/dev.082198. [DOI] [PubMed] [Google Scholar]
- 10.Rowe HM, et al. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development. 2013;140:519–529. doi: 10.1242/dev.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leung D, et al. Regulation of DNA methylation turnover at LTR retrotransposons and imprinted loci by the histone methyltransferase Setdb1. Proc Natl Acad Sci U S A. 2014;111:6690–6695. doi: 10.1073/pnas.1322273111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elsasser SJ, Noh KM, Diaz N, Allis CD, Banaszynski LA. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015;522:240–244. doi: 10.1038/nature14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen WV, Maniatis T. Clustered protocadherins. Development. 2013;140:3297–3302. doi: 10.1242/dev.090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yagi T. Molecular codes for neuronal individuality and cell assembly in the brain. Front Mol Neurosci. 2012;5:45. doi: 10.3389/fnmol.2012.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhan Y, et al. Reciprocal insulation analysis of Hi-C data shows that TADs represent a functionally but not structurally privileged scale in the hierarchical folding of chromosomes. Genome Res. 2017;27:479–490. doi: 10.1101/gr.212803.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sofueva S, et al. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vietri Rudan M, et al. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flavahan WA, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Y, et al. CTCF/cohesin-mediated DNA looping is required for protocadherin alpha promoter choice. Proc Natl Acad Sci U S A. 2012;109:21081–21086. doi: 10.1073/pnas.1219280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renda M, et al. Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J Biol Chem. 2007;282:33336–33345. doi: 10.1074/jbc.M706213200. [DOI] [PubMed] [Google Scholar]
- 23.Du J, Johnson LM, Jacobsen SE, Patel DJ. DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol. 2015;16:519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paliwal A, et al. Comparative anatomy of chromosomal domains with imprinted and non-imprinted allele-specific DNA methylation. PLoS Genet. 2013;9:e1003622. doi: 10.1371/journal.pgen.1003622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maze I, et al. Critical Role of Histone Turnover in Neuronal Transcription and Plasticity. Neuron. 2015;87:77–94. doi: 10.1016/j.neuron.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kehayova P, Monahan K, Chen W, Maniatis T. Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proc Natl Acad Sci U S A. 2011;108:17195–17200. doi: 10.1073/pnas.1114357108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monahan K, et al. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-alpha gene expression. Proc Natl Acad Sci U S A. 2012;109:9125–9130. doi: 10.1073/pnas.1205074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribich S, Tasic B, Maniatis T. Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proc Natl Acad Sci U S A. 2006;103:19719–19724. doi: 10.1073/pnas.0609445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yokota S, et al. Identification of the cluster control region for the protocadherin-beta genes located beyond the protocadherin-gamma cluster. J Biol Chem. 2011;286:31885–31895. doi: 10.1074/jbc.M111.245605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y, et al. Setdb1 histone methyltransferase regulates mood-related behaviors and expression of the NMDA receptor subunit NR2B. J Neurosci. 2010;30:7152–7167. doi: 10.1523/JNEUROSCI.1314-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maze I, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaefer A, et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 2009;64:678–691. doi: 10.1016/j.neuron.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thu CA, et al. Single-cell identity generated by combinatorial homophilic interactions between alpha, beta, and gamma protocadherins. Cell. 2014;158:1045–1059. doi: 10.1016/j.cell.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toyoda S, et al. Developmental epigenetic modification regulates stochastic expression of clustered protocadherin genes, generating single neuron diversity. Neuron. 2014;82:94–108. doi: 10.1016/j.neuron.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 35.Chernukhin I, et al. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol. 2007;27:1631–1648. doi: 10.1128/MCB.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holwerda SJ, de Laat W. CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120369. doi: 10.1098/rstb.2012.0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Golan-Mashiach M, et al. Identification of CTCF as a master regulator of the clustered protocadherin genes. Nucleic Acids Res. 2012;40:3378–3391. doi: 10.1093/nar/gkr1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iyengar S, Ivanov AV, Jin VX, Rauscher FJ, 3rd, Farnham PJ. Functional analysis of KAP1 genomic recruitment. Mol Cell Biol. 2011;31:1833–1847. doi: 10.1128/MCB.01331-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frietze S, O’Geen H, Blahnik KR, Jin VX, Farnham PJ. ZNF274 recruits the histone methyltransferase SETDB1 to the 3’ ends of ZNF genes. PLoS One. 2010;5:e15082. doi: 10.1371/journal.pone.0015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heidari N, et al. Genome-wide map of regulatory interactions in the human genome. Genome Res. 2014;24:1905–1917. doi: 10.1101/gr.176586.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailey SD, et al. ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters. Nat Commun. 2015;2:6186. doi: 10.1038/ncomms7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie D, et al. Dynamic trans-acting factor colocalization in human cells. Cell. 2013;155:713–724. doi: 10.1016/j.cell.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ibrahim HM, Tamminga CA. Schizophrenia: treatment targets beyond monoamine systems. Annu Rev Pharmacol Toxicol. 2011;51:189–209. doi: 10.1146/annurev.pharmtox.010909.105851. [DOI] [PubMed] [Google Scholar]
- 46.Owen MJ, Sawa A, Mortensen PB. Schizophrenia. Lancet. 2016;388:86–97. doi: 10.1016/S0140-6736(15)01121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnsten AF, Girgis RR, Gray DL, Mailman RB. Novel Dopamine Therapeutics for Cognitive Deficits in Schizophrenia. Biol Psychiatry. 2016 doi: 10.1016/j.biopsych.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nee DE, D’Esposito M. The Representational Basis of Working Memory. Curr Top Behav Neurosci. 2016 doi: 10.1007/7854_2016_456. [DOI] [PubMed] [Google Scholar]
- 49.Keeler AB, Molumby MJ, Weiner JA. Protocadherins branch out: Multiple roles in dendrite development. Cell Adh Migr. 2015;9:214–226. doi: 10.1080/19336918.2014.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chakravarthy S, et al. Cre-dependent expression of multiple transgenes in isolated neurons of the adult forebrain. PLoS One. 2008;3:e3059. doi: 10.1371/journal.pone.0003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Q, et al. Chromosomal microarray analysis in clinical evaluation of neurodevelopmental disorders-reporting a novel deletion of SETDB1 and illustration of counseling challenge. Pediatr Res. 2016;80:371–381. doi: 10.1038/pr.2016.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cukier HN, et al. The expanding role of MBD genes in autism: identification of a MECP2 duplication and novel alterations in MBD5, MBD6, and SETDB1. Autism Res. 2012;5:385–397. doi: 10.1002/aur.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mendioroz M, et al. Trans effects of chromosome aneuploidies on DNA methylation patterns in human Down syndrome and mouse models. Genome Biol. 2015;16:263. doi: 10.1186/s13059-015-0827-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garafola CS, Henn FA. A change in hippocampal protocadherin gamma expression in a learned helpless rat. Brain Res. 2014;1593:55–64. doi: 10.1016/j.brainres.2014.08.071. [DOI] [PubMed] [Google Scholar]
- 55.Suderman M, et al. Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(Suppl 2):17266–17272. doi: 10.1073/pnas.1121260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGowan PO, et al. Broad epigenetic signature of maternal care in the brain of adult rats. PLoS One. 2011;6:e14739. doi: 10.1371/journal.pone.0014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.LaPlant Q, et al. Role of nuclear factor kappaB in ovarian hormone-mediated stress hypersensitivity in female mice. Biol Psychiatry. 2009;65:874–880. doi: 10.1016/j.biopsych.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep. 2012;2:345–357. doi: 10.1016/j.celrep.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 59.Bharadwaj R, et al. Conserved higher-order chromatin regulates NMDA receptor gene expression and cognition. Neuron. 2014;84:997–1008. doi: 10.1016/j.neuron.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen K, et al. Genome-wide binding and mechanistic analyses of Smchd1-mediated epigenetic regulation. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E3535–3544. doi: 10.1073/pnas.1504232112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Isbel L, et al. Wiz binds active promoters and CTCF-binding sites and is required for normal behaviour in the mouse. Elife. 2016;5 doi: 10.7554/eLife.15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Franke M, et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature. 2016;538:265–269. doi: 10.1038/nature19800. [DOI] [PubMed] [Google Scholar]
- 63.Redin C, et al. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat Genet. 2017;49:36–45. doi: 10.1038/ng.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang Y, et al. Setdb1 histone methyltransferase regulates mood-related behaviors and expression of the NMDA receptor subunit NR2B. J Neurosci. 2010;30:7152–67. doi: 10.1523/JNEUROSCI.1314-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kundakovic M, et al. Practical Guidelines for High-Resolution Epigenomic Profiling of Nucleosomal Histones in Postmortem Human Brain Tissue. Biol Psychiatry. 2017;81:162–170. doi: 10.1016/j.biopsych.2016.03.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen L, et al. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One. 2013;8:e65598. doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bharadwaj R, et al. Conserved higher-order chromatin regulates NMDA receptor gene expression and cognition. Neuron. 2014;84:997–1008. doi: 10.1016/j.neuron.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Servant N, et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Imakaev M, et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–4. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–31. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 78.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–2. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–46. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu J, et al. Inhibition of STEP61 ameliorates deficits in mouse and hiPSC-based schizophrenia models. Mol Psychiatry. 2016 doi: 10.1038/mp.2016.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Topol A, et al. Dysregulation of miRNA-9 in a Subset of Schizophrenia Patient-Derived Neural Progenitor Cells. Cell Rep. 2016;15:1024–36. doi: 10.1016/j.celrep.2016.03.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All next generation sequencing data for genome-scale analysis in this publication have been deposited in NCBI’s Gene Expression Omnibus82 and are accessible through GEO Series accession number GSE99363 (See URLs). All other data discussed are included in the publication and available from the Authors upon request.