Abstract
Burkholderia multivorans is a significant health threat to persons with cystic fibrosis (CF). Infections are difficult to treat as this pathogen is inherently resistant to multiple antibiotics. Susceptibility testing of isolates obtained from CF respiratory cultures revealed that single agents selected from different antibiotic classes were unable to inhibit growth. However, all isolates were found to be susceptible to ceftazidime when combined with the novel non-β-lactam β-lactamase inhibitor, avibactam (all minimum inhibitor concentrations (MICs) were ≤8 mg/L of ceftazidime and 4 mg/L of avibactam). Furthermore, a major β-lactam resistance determinant expressed in B. multivorans, the class A carbapenemase, PenA was readily inhibited by avibactam with a high k2/K of (2 ± 1) × 106 μM−1 s−1 and a slow koff of (2 ± 1) × 10−3 s−1. Mass spectrometry revealed that avibactam formed a stable complex with PenA for up to 24 h and that avibactam recyclized off of PenA, re-forming the active compound. Crystallographic analysis of PenA–avibactam revealed several interactions that stabilized the acyl–enzyme complex. The deacylation water molecule possessed decreased nucleophilicity, preventing decarbamylation. In addition, the hydrogen-bonding interactions with Lys-73 were suggestive of a protonated state. Thus, Lys-73 was unlikely to abstract a proton from Ser-130 to initiate recyclization. Using Galleria mellonella larvae as a model for infection, ceftazidime–avibactam was shown to significantly (p < 0.001) improve survival of larvae infected with B. multivorans. To further support the translational impact, the ceftazidime–avibactam combination was evaluated using susceptibility testing against other strains of Burkholderia spp. that commonly infect individuals with CF, and 90% of the isolates were susceptible to the combination. In summary, ceftazidime–avibactam may serve as a preferred therapy for people that have CF and develop Burkholderia spp. infections and should be considered for clinical trials.
Keywords: β-lactamase, β-lactamase inhibitor, cystic fibrosis, ceftazidime, avibactam, Burkholderia
Graphical Abstract
According to the Cystic Fibrosis Foundation, approximately 1000 people are diagnosed with cystic fibrosis (CF) each year in the United States.1 CF is a life-threatening disease caused by mutations in the CF transmembrane conductance regulator (CFTR) protein that leads to secretory malfunctions in the lungs as well as in the digestive system. Thick mucus builds up in the lungs of persons with CF, leading to ineffective clearance and increased susceptibility to infection. The hallmarks of CF include chronic microbial infections and inflammation in the lungs. Antibiotic therapy to target bacterial pathogens can significantly increase the life expectancy of individuals with CF. However, highly antibiotic resistant strains of bacteria have emerged, resulting in limited treatment options for people with CF.
A significantly problematic group of bacterial pathogens that infect individuals with CF is the Burkholderia cepacia complex (Bcc), which consists of 20 unique species.2 Within this group of species, Burkholderia multivorans is among the most commonly identified species isolated from CF respiratory specimens. This species has been associated with “cepacia syndrome”, a rapidly progressive necrotizing pneumonia.3 B. multivorans possesses a large genome (three chromosomes of ~7.0 Mbp) that carries a multitude of antibiotic resistance mechanisms. One of the major determinants of resistance is the class A β-lactamase, PenA. PenA possesses a very broad substrate profile that includes carbapenems and β-lactamase inhibitors.4
Here, we analyze a panel of B. multivorans clinical isolates that carry the blapenA gene and identify a novel β-lactam–β-lactamase inhibitor combination, ceftazidime–avibactam, which is effective against these highly drug resistant strains. Furthermore, we biochemically characterize PenA with avibactam using steady-state kinetics, mass spectrometry, and crystallography and determine the mechanism of inactivation of PenA. Moreover, in vivo testing reveals that ceftazidime–avibactam improves the survival of Galleria mellonella infected with B. multivorans. In addition, the combination is effective in vitro against other Bcc species that infect people with CF. Our findings lead us to propose that ceftazidime–avibactam should be considered as an agent for clinical trials or for salvage therapy in severe necrotizing pneumonia.
RESULTS
Clinical Isolates of B. multivorans Are Diverse and Highly Drug Resistant
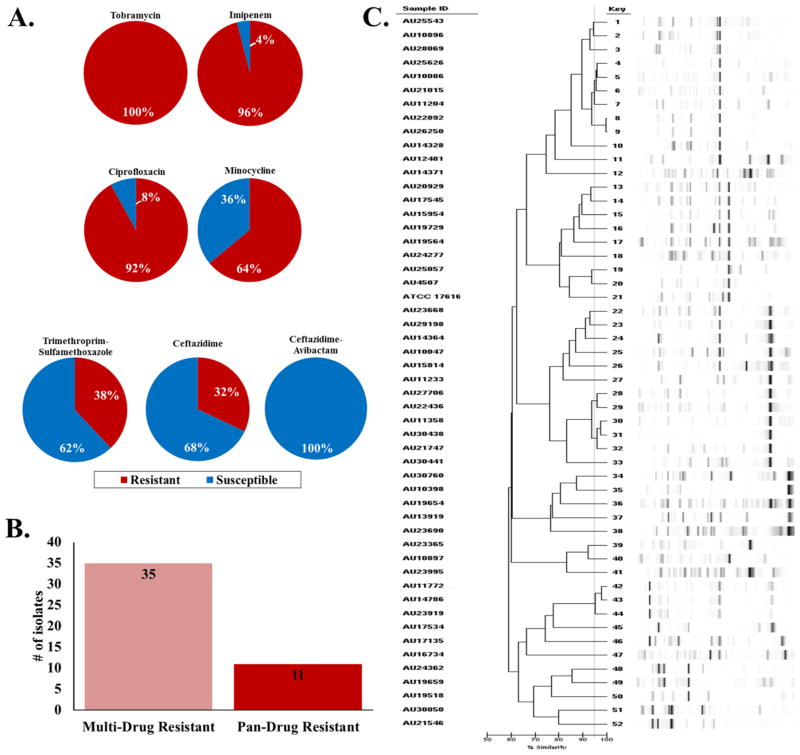
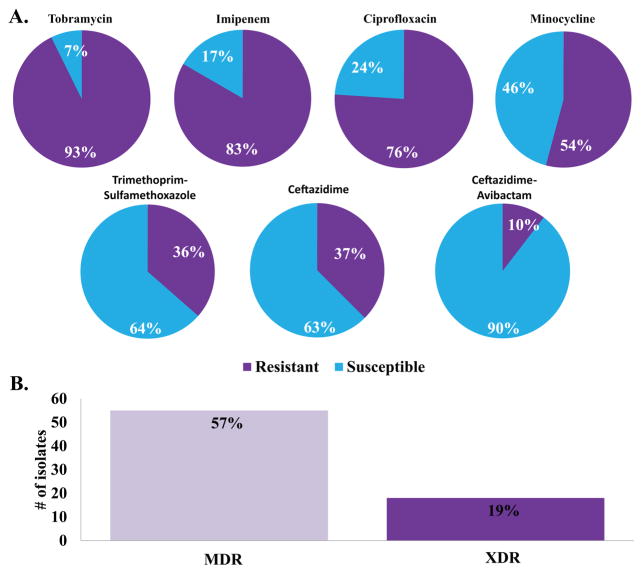
A collection of 50 B. multivorans strains isolated from CF patients was tested against a selected panel of antibiotics (tobramycin, imipenem, ciprofloxacin, minocycline, trimethroprim–sulfamethoxazole, ceftazidime, and ceftazidime–avibactam) using the agar dilution minimum inhibitory concentration (MIC) method. Greater than 90 percent of the isolates were resistant to tobramycin, imipenem, and ciprofloxacin (Figure 1A and Supplemental Table 1). Minocycline possessed some activity with 36% of the isolates testing susceptible to this agent. The two “first-line” agents for the treatment of Bcc infections, ceftazidime and trimethoprim–sulfamethoxazole, demonstrated only 68 and 62% susceptibility, respectively, limiting their choice as empiric therapy. Overall, 70% of the strains were multidrug resistant (MDR) or resistant to at least two major classes of antibiotics (Figure 1B). Additionally, 22% of the strains were extremely drug resistant (XDR) or resistant to all of the major classes of antibiotics. Using repetitive sequence-based PCR (rep-PCR), the group of 50 isolates was shown to represent a genetically diverse set of strains (Figure 1C). Rep-PCR generates a strain-specific bacterial genome fingerprint based on highly conserved repetitive sequence elements amplified via PCR.5
Figure 1.
Characteristics of the 50 clinical B. multivorans isolates: (A) summary pie charts of the susceptibility testing results (susceptible (blue) and resistant (red)) conducted with tobramycin, imipenem, ciprofloxacin, minocycline, trimethroprim–sulfamethoxazole, ceftazidime, and ceftazidime–avibactam; (B) bar graph representing the number of isolates that are MDR and XDR; (C) dendrogram of the rep-PCR results.
Avibactam Restores the Activity of Ceftazidime against B. multivorans
Ceftazidime–avibactam was approved by the U.S. Food and Drug Administration in 2015 and the European Medicines Agency in 2016 for the treatment of serious infections (e.g., urinary tract infections) caused by MDR Enterobacteriaceae. The benefit of this combination, compared to other β-lactams and β-lactam–β-lactamase inhibitor combinations, resides in the ability of avibactam to inhibit class A and C β-lactamases, including class A carbapenemases (e.g., KPC-2).6 Given (i) the high level of resistance observed in the clinical isolates and (ii) that B. multivorans possesses a class A carbapenemase, blapenA, and a class C β-lactamase, blaBmampC, the novel β-lactam–β-lactamase inhibitor combination, ceftazidime– avibactam, was tested against these isolates. Remarkably, the addition of avibactam to ceftazidime restored susceptibility to ceftazidime for all strains tested (Figure 1A and Supplemental Table 1).
Avibactam Is a Potent Inactivator of the PenA β-Lactamase
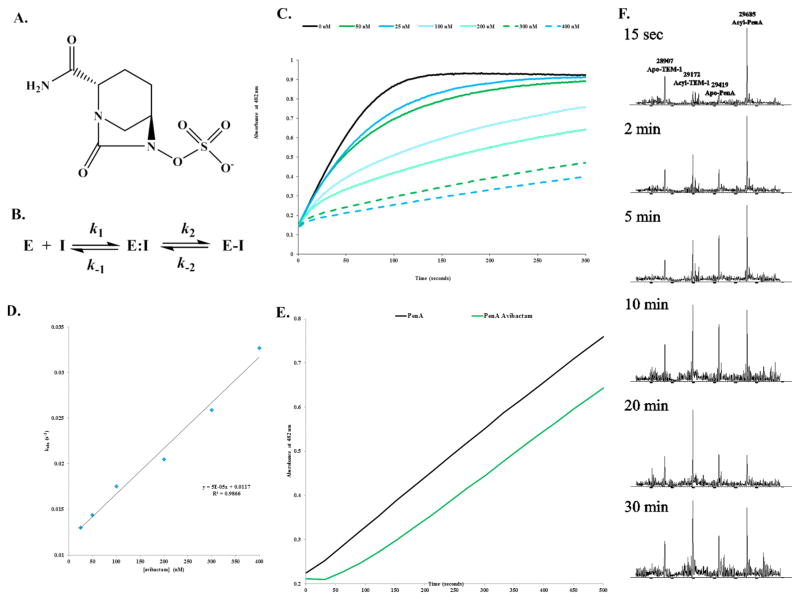
Biochemical analysis using steady-state kinetics and mass spectrometry supports the potent activity of ceftazidime–avibactam against B. multivorans expressing the PenA β-lactamase. Avibactam was previously shown to form a reversible acyl–enzyme complex with class A and C β-lactamases (Figure 2A,B).6 On the basis of this kinetic mechanism, the on- (k2/K) and off-rates (koff) of avibactam were assessed with PenA. Avibactam acylated PenA with a rapid a k2/K value of (2 ± 1) × 106 μM−1 s−1 (Table 1 and Figure 2C,D). Avibactam was slow to come off PenA (Figure 2E and Table 1) and stayed bound for up to 24 h with no fragmentation or hydrolysis (Table 1). In addition, carbamylation by avibactam was reversible as avibactam can be transferred to another β-lactamase, TEM-1, from PenA and back to PenA during time (Figure 2F). Thus, avibactam remained active after recyclization.
Figure 2.
The avibactam inhibition mechanism of PenA. (A) Chemical structure of avibactam. (B) Scheme representing the interactions of PenA with avibactam.6a In this model, formation of the noncovalent complex, E:I is represented by the dissociation constant, Kd, which is equivalent to k−1/k1. k2 is the first-order rate constant for the acylation step, or formation of E-I. k−2 is the first-order rate constant for the recyclization step or re-formation of E:I. (C) Inhibition of nitrocefin hydrolysis by PenA using increasing concentrations of avibactam measured in absorbance at λ482 nm (absorbance units (a.u.)). (D) Data from panel C were fit to obtain kobs values, and here the kobs values were plotted versus [avibactam]. (E) Recovery of nitrocefin hydrolysis activity by PenA after inhibition by avibactam (green line); PenA alone (black line) without inhibition. (F) Acyl transfer of avibactam from PenA to TEM-1 during a time course started at 15 s and up to 30 min. Molecular weights of apo-PenA, acyl-PenA, apo-TEM-1, and acyl-TEM-1 are 29419 ± 3, 29685 ± 3, 28907 ± 3, and 29172 ± 3 Da, respectively. By 20 min, most of the avibactam had transferred to TEM-1, and by 30 min, some of the avibactam had transferred back to PenA.
Table 1.
Steady-State Kinetic Parameters and ESI-MS Results for PenA with Nitrocefin and Avibactam
| parameter | value |
|---|---|
| NCF Km (μM) | 147 ± 15 |
| NCF kcat/Km (μM−1 s−1) | 3.2 ± 0.3 |
| AVI Ki app (μM) | 0.5 ± 0.1 |
| AVI k2/K (μM−1 s−1) | (2 ± 1) × 106 |
| AVI koff (s−1) | (2 ± 1) × 10−3 |
| AVI ton 5 min | 1 |
| PenA alone (amu) | 29417 ± 3 |
| PenA–avibactam 5 min (amu) | 29682 ± 3 |
| PenA–avibactam 5 h (amu) | 29682 ± 3 |
| PenA–avibactam 24 h (amu) | 29682 ± 3 |
Crystal Structure of PenA with Avibactam
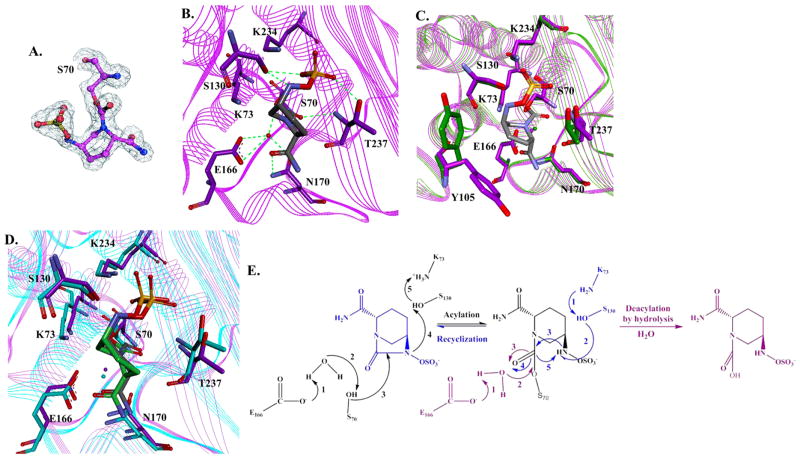
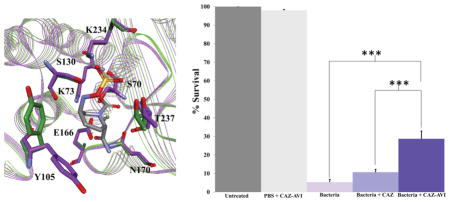
The PenA β-lactamase was crystallized in space group C2 with three molecules in the asymmetric unit (Table 2). The three-dimensional structure of PenA acylated by avibactam was resolved to a resolution of 1.6 Å with an Rwork of 0.160 and an Rfree of 0.230 (Table 2 and Figure 3A,B). An overlay of the apo-PenA crystal structure with the PenA–avibactam acyl structure revealed a major movement in Tyr-105 (Figure 3C). The avibactam-bound active site of PenA was similar to the previously reported avibactam complex with KPC-2 (Figure 3D).7 However, we demonstrate that important novel findings into the inactivation mechanism of avibactam with class A β-lactamases can be observed from the PenA–avibactam structure.
Table 2.
X-ray Data Collection and Results from Phenix Refinement
| parameter | value |
|---|---|
| source | Photon Factor AR-NW12A |
| detector | ADSC Quantum 210r |
| exposure time (s) | 3 |
| oscillation angle (deg) | 1.0 |
| no. of frames | 130 |
| temp (K) | 100 |
| wavelength (Å) | 1.00 |
| space group | C2 |
| cell dimensions | |
| a (Å) | 120.12 |
| b (Å) | 69.43 |
| c (Å) | 84.63 |
| α = γ (deg) | 90.000 |
| β (deg) | 90.049 |
| dmin (highestres.shell) (Å) | 1.60 (1.63–1.60) |
| observations | 380200 (18572) |
| unique reflections | 89867 (4422) |
| completeness (%) | 98.0 (96.6) |
| redundancy | 4.2 (4.2) |
| Iav/σ(I) | 14.4 (2.3) |
| Rsym(I) | 0.089 (0.701) |
| resolution range (Å) | 20–1.6 |
| no. of reflections used [F > 0σ(F)] | 89812 |
| Rwork/Rfreea (%) | 0.164/0.199 |
| Rtotal (%) | 0.165 |
| residues in Ramachandran zones | |
| favored/allowed/disallowed (%) | 99.0/1.0/0 |
| rmsd values from ideality | |
| bond lengths (Å) | 0.0006 |
| bond angles (deg) | 1.10 |
| mean B factors (no. of atoms) | |
| protein (no. of atoms) | 15.1(5852) |
| avibactam (no. of atoms) | 17.4 (51) |
| water molecules (no. of atoms) | 29.8 (746) |
| all atoms (no. of atoms) | 16.0 (6649) |
Rfree was calculated from 4512 reflections (5%).
Figure 3.
Active site structure of carbamylated PenA β-lactamase. (A) Electron density map of acylated Ser-70 and avibactam with omit map. (B) Snapshot of the active site of PenA crystal structure (PDB 3WZR) with avibactam (gray). Potential hydrogen-bonding interactions are indicated by dashed green lines. (C) Overlay of the crystal structures of apo-PenA (PDB 3W4Q) (green) and PenA–avibactam (purple-gray). (D) Overlay of the crystal structures of PenA–avibactam (purple-gray) and KPC-2–avibactam (PDB 4ZBE) (cyan-green). (E) Proposed mechanistic schemes of avibactam carbamylation, decarbamylation, and recyclization with PenA based on crystallographic and biochemical analyses.
Interactions of Avibactam with Serine β-Lactamases
Serine β-lactamases that are acylated by avibactam were shown to undergo two pathways toward the regeneration of free enzyme (Figure 3E).6,7 First, decarbamylation proceeds through a hydrolytic pathway by nucleophilic attack of the carbamoyl bond using a catalytic water. Second, a recyclization pathway occurs in which there is a nucleophilic attack by the nitrogen atom of the secondary amine of avibactam, resulting in the re-formation of active avibactam; active site residues Lys-73 and Ser-130 are predicted to initiate this process via a proton shuttle.
Decarbamylation Is Disfavored in the PenA–Avibactam Complex
Focusing on the first pathway, in the acylated PenA structure, the deacylation water could be detected at the bottom of the active site cavity as was observed with KPC-2 (Figure 3B,D).7 The most important observation in the carbamylated PenA structure was that the deacylation water was within hydrogen bonding distance of three adjacent atoms (Glu-166, Asn-170, and the O of the carboxamide side chain of avibactam) (Figure 3B). This observation suggests that an oxonium water molecule may form in the PenA–avibactam structure; thus, the nucleophilicity of the deacylation water is lowered.
Recyclization Occurs Slowly in the PenA–Avibactam Complex
In the second pathway, the sulfate group of avibactam was bound in a similar position as in the KPC-2–avibactam complex (Figure 3D). The nitrogen atom of the secondary amine of avibactam of the acylated PenA was in the same space as the acylated KPC-2–avibactam structure. In addition, the secondary amine of avibactam was close to Ser-130:Oγ and within hydrogen-bonding distance in both structures. In the acylated PenA structure, the Lys-73 amino group was within hydrogen-bonding distance of Ser-70:Oγ, Asn-132:Oδ, and Ser-130:O, suggesting that Lys-73 was in a protonated state and less likely to join the proton relay toward recyclization.
In Vivo Efficacy of Ceftazidime–Avibactam
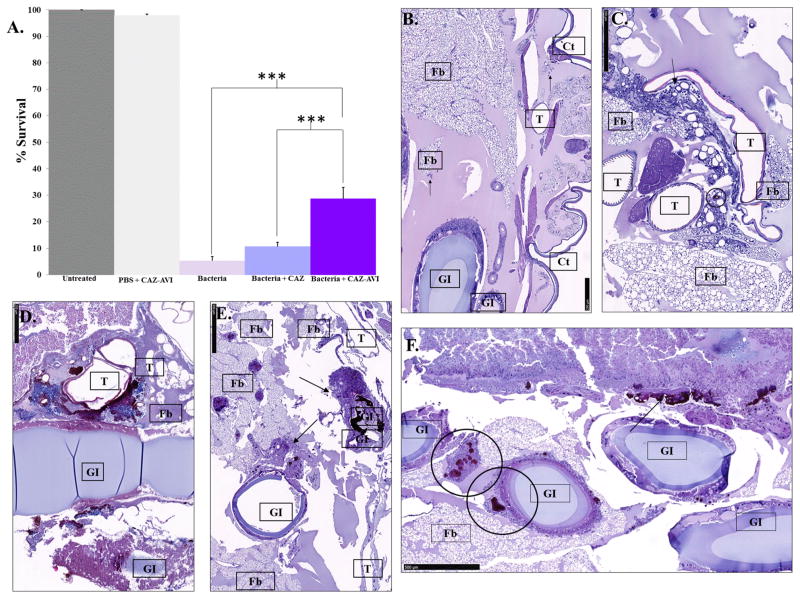
As clinical trials and animal models are difficult to perform with Bcc, we assessed the ability of the ceftazidime–avibactam combination to affect the survival in vivo of larvae of G. mellonella infected with B. multivorans AU14786 (ceftazidime MIC = 32 mg/L; ceftazidime–avibactam MIC = 4 mg/L) and treated with ceftazidime or ceftazidime–avibactam. We found that the combination of ceftazidime–avibactam significantly improved the survival of G. mellonella compared to the untreated controls (p < 0.001) and those treated with ceftazidime alone (p < 0.001) (Figure 4A).
Figure 4.
G. mellonella survival assays. (A) Percent survival of G. mellonella after infection by B. multivorans AU14786 (bacteria) treated with ceftazidime (CAZ) or ceftazidime–avibactam (CAZ–AVI) or mock-infected (PBS + CAZ–AVI). (***) =p value <0.001. Histological sections of G. mellonella. Tissue labels: trachea (T), gastrointestinal tract (GI), fat body (FB), subcuticular region (Ct). (B) Uninfected. (C) Mock-infected. (D) Bacteria alone. (E) Bacteria + CAZ. (F) Bacteria + CAZ–AVI.
Histological analysis of the infected larvae revealed hallmarks of infection that are alleviated by the ceftazidime–avibactam combination. Normal histology of an uninfected larva revealed a few hemocytes (arrows) circulating in the hemolymph (Figure 4B). In a mock-infected larva, histological sections displayed a small melanized nodule (circle) and some recruited hemocytes in peritracheal areas near the fat body (arrow) (Figure 4C). In an infected untreated larva, heavily damaged larval tissues with large melanized nodules all around with necrotic tracheal and intestinal walls were observed (Figure 4D). In an infected larva treated with ceftazidime alone, the larval tissues were less damaged than those observed in Figure 4D, with melanized nodules of medium and large size (arrows) mainly localized within intestinal walls (Figure 4E). The tracheae appeared uninjured. In an infected larva treated with the ceftazidime–avibactam combination, only small- and medium-sized nodules (circles) were observed inside and near the gastrointestinal tract, the fat body, and subcuticolar areas (Figure 4F).
Activity of Ceftazidime–Avibactam against Other Common Bcc and Burkholderia spp. Isolated from CF Respiratory Specimens
To further confirm the utility of the ceftazidime–avibactam combination for the treatment of Burkholderia spp. infections in people with CF, we conducted susceptibility testing using 96 non-B. multivorans clinical isolates from CF respiratory specimens. Greater than 50% of the isolates were resistant to tobramycin, imipenem, ciprofloxacin, and minocycline (Figure 5A and Supplemental Table 2). Similarly to B. multivorans, 36 and 37% of the Burkholderia spp. isolated from CF respiratory specimens were resistant to first-line agents trimethoprim–sulfamethoxazole and ceftazidime, respectively. Overall, 57% of the strains were MDR, and 19% of the strains were XDR (Figure 5B). Avibactam combined with ceftazidime was the most effective against these strains, with 90% of isolates being susceptible to the drug combination.
Figure 5.
Susceptibility testing of the 96 clinical non-B. multivorans Burkholderia spp. isolates from CF patients. (A) Summary pie charts of the susceptibility testing results (susceptible (blue) and resistant (purple)) conducted with tobramycin, imipenem, ciprofloxacin, minocycline, trimethroprim–sulfamethoxazole, ceftazidime, and ceftazidime–avibactam. (B) Bar graph representing the number of isolates that are MDR and XDR.
DISCUSSION
Infections of the respiratory tract in individuals with CF are a significant contributor to morbidity and mortality. Highly drug resistant pathogens, such as Bcc, severely limit treatment options. Here, we found that when avibactam is combined with ceftazidime, susceptibility to ceftazidime in MDR and XDR clinical strains of Burkholderia spp. isolated from CF respiratory specimens is restored. The panel of isolates analyzed here was genetically diverse, thus showing the breadth and potential utility of this combination against Burkholderia spp. isolates. Others have previously tested the ceftazidime–avibactam combination against Bcc, but the number of strains and variety of species in these studies were limited.8
Avibactam was also shown to be a potent inhibitor of a major drug resistance determinant, PenA, which is expressed by B. multivorans. Unexpectedly, we found that PenA’s acylation rate (k2/K) was 100-fold higher than that of KPC-2.6a,7b In addition, the “off-rate” of avibactam was also 100-fold higher for PenA than for KPC-2. Simply put, avibactam gets onto PenA more quickly than KPC-2, but recyclizes off more rapidly.
The crystallographic analyses presented herein suggest a novel mechanism of inactivation of PenA by avibactam (Figure 3E). For carbamylated PenA, deacylation or hydrolysis of avibactam was unlikely. In most class A β-lactamases, the deacylation water was stabilized by two strong hydrogen bonds, to Glu-166:Oε1 and Asn-170:Oε. However, the deacylation water in the PenA–avibactam structure possessed one more hydrogen bond to the carbonyl oxygen of the carboxamide group of the avibactam molecule, thus reducing its nucleophilicity. This observation is similar to the meropenem:SHV-1 β-lactamase crystal structure in which a third hydrogen bond to the O of the hydroxyl group of meropenem reduced the nucleophilicity of the deacylation water. In the meropenem:SHV-1 complex, a protonated Glu-166 was observed.9 Thus, despite PenA’s being a carbapenemase, this class A enzyme was inhibited by avibactam in a similar manner as a non-carbapenemase (SHV-1). We suspect that the deacylation water of PenA does not form a third hydrogen bond with carbapenems as seen with avibactam; thus, unlike SHV-1, PenA is able to hydrolyze carbapenems. Studies are in progress to assess this hypothesis.
Intramolecular decarbamylation of PenA to regenerate avibactam appeared unlikely (Figure 3E). On the basis of our observations, for recyclization of avibactam to occur, the process needs a proton to be abstracted from the secondary amine of avibactam by Ser-130:Oγ and deprotonated by Lys-73. In the carbamylated PenA, we observed that the Lys-73 amino group was hydrogen bonded to Ser-70:Oγ, Asn-132:Oδ, and Ser-130:O, suggesting Lys-73 was in a protonated state and was unable to participate in the proton shuttle. An alternative pathway may exist in which the OAG atom of avibactam sulfonate, which resides 2.79 Å from Ser130:Oγ, is the candidate proton acceptor for recyclization (Figure 3E). We note that this is less possible because the sulfonate is a weaker base. Overall, the active site structure of the PenA–avibactam complex was in an unfavorable configuration to promote recyclization of avibactam. Moreover, these observations help explain why the PenA– avibactam structure is stable. This unique biochemical property combined with ready cell entry into a difficult-to-treat Gram-negative pathogen provides a rationale for the efficacy of this β-lactam–β-lactamase inhibitor combination against Bcc.
Finally, we show in a nonmammalian model of infection that the combination of ceftazidime–avibactam significantly attenuated the impact of Bcc infection. In addition, histological analysis of the larval tissues revealed that damage to the larva was much less upon treatment with ceftazidime–avibactam than with ceftazidime alone. Ceftazidime–avibactam may serve as an alternative therapy for individuals with CF that develop Burkholderia spp. infections. Ceftazidime–avibactam was shown to possess favorable pharmacokinetic and pharmacodynamic properties in the lung.10 Taken together, our observations support further testing of ceftazidime–avibactam in clinical trials of CF patients challenged by this deadly pathogen or as a salvage therapy for patients with severe necrotizing pneumonia when other agents are ineffective.
MATERIALS AND METHODS
Bacterial Strains and Plasmids
The methods for cloning of blapenA into the pBC SK(+) and pGEX-6P2 vectors were described previously.4 The blapenA gene was expressed on pBC SK(+) in Escherichia coli DH10B for susceptibility testing and on pGEX-6P2 in E. coli Origami 2 (DE3) for protein expression. blaTEM-1 was expressed from the pET24a(+) vector in E. coli BL21(DE3) RP+ cells for protein expression.11 The Bcc clinical isolates used in this study came from the collection of the Cystic Fibrosis Foundation B. cepacia Research Laboratory and Repository at the University of Michigan. Each Bcc isolate was identified to the species level by using species-specific polymerase chain reaction, recA RFLP, and/or DNA sequencing of the recA gene.12
Repetitive Sequence-Based PCR (Rep-PCR)
DNA was isolated from the 50 B. multivorans clinical isolates using the Mo Bio UltraClean Microbial DNA Isolation Kit (Mo Bio Laboratories, Carlsbad, CA, USA). Subsequently, the DNA was subjected to rep-PCR amplification using the DiversiLab fingerprinting kit (BioMérieux, USA) for Pseudomonas spp., according to the manufacturer’s instructions. The Diversilab system is based on rep-PCR amplification of noncoding repetitive sequences interspersed throughout the bacterial genome. Rep-PCR was performed using the following parameters: initial denaturation (94 °C) for 2 min, and then 35 cycles of 30 s of denaturation (94 °C), 30 s of annealing (50 °C), and 90 s of extension (70 °C), followed by 3 min of final extension (70 °C) and ending at (4 °C). The amplification products were separated with an Agilent B2100 Bioanalyzer. Five microliters of DNA standard and 1 μL of the rep-PCR product were loaded. The data were entered into the DiversiLab software system and analyzed to generate a dendrogram.
Expression and Purification of PenA and TEM-1
The PenA β-lactamase was purified as previously described;4 purification of TEM-1 is described herein. Briefly, E. coli BL21 DE3 producing TEM-1 was grown in superoptimal broth to an optical density at 600 nm (ODλ600 nm) of 0.6, 1 mM isopropyl β-D-1-thiogalactopyranoside was added, and the cells were grown for 3 h under induction. The cells were pelleted, frozen, and subjected to stringent periplasmic fractionation and preparative isoelectric focusing, as previously described.13 Fast protein liquid chromatography on an ÄKTA purifier 10 using a Sephadex 16/60 gel filtration chromatography column (GE Life Sciences) was used for further polishing steps. The purities of PenA and TEM-1 were assessed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Protein concentrations were determined by measuring the absorbance at λ280 nm and using the protein’s extinction coefficients, which were obtained using the ProtParam tool at the ExPASy SIB Bioinformatics Resource Portal.
Compounds
Tobramycin, ciprofloxacin, minocycline, trimethoprim, sulfamethoxazole, ceftazidime, and aztreonam were purchased from Sigma-Aldrich. Imipenem–cilastatin was obtained from its commercial source. Avibactam, batches AFCH005151 and C565/5, were kind gifts from AstraZeneca; the chemical structure is presented in Figure 2A. Nitrocefin was a kind gift from Dr. ShahriarMobashery at the University of Notre Dame in South Bend, IN, USA.
In Vitro Susceptibility Test Methods
MICs for the clinical Bcc isolates were determined by the MH agar dilution method according to Clinical and Laboratory Standards Institute (CLSI) guidelines.14 Overnight cultures were grown in MH broth at 37 °C in a shaking incubator and then read in a Spectronic Genesys 5 spectrophotometer at ODλ600 nm. The amount of cells added to the Steers replicator was adjusted according to each isolate’s ODλ600 nm reading to ensure that equal amounts of cells were added to each well. The Steers replicator delivered 10 μL of each diluted culture containing approximately 104 colony-forming units to each MH plate. The MIC plates were read after 24 h, and CLSI guidelines were used for interpretations.15 Any variability (>1 doubling-dilution difference between experiments) in triplicate results was resolved by the disk diffusion (Becton-Dickinson) method according to CLSI guidelines. Avibactam was tested at 4 mg/L in combination with increasing antibiotic concentrations of ceftazidime or aztreonam.
Steady-State Kinetic Analysis
Steady-state kinetic parameters were determined using an Agilent 8453 diode array spectrophotometer. Briefly, each assay was performed in 10 mM phosphate-buffered saline (PBS) at pH 7.4 at room temperature.
For determination of Vmax and Km, PenA was maintained at 14 nM with nitrocefin in excess molar concentration to establish pseudo-first-order kinetics, as previously described.16
The proposed interactions between PenA and avibactam are depicted in Figure 2B based on previous studies with class A β-lactamases and avibactam.6b,17 Determination of kinetic values Ki apparent (app), k2/K, kcat/kinact, and koff were previously described, and full methodology and equations used were defined in Papp-Wallace et al.16 Briefly, for the determination of the Ki app value, PenA was maintained at 14 nM, and using a direct competition assay with the reporter substrate, nitrocefin at 100 μM was mixed with various concentrations of avibactam (100–500 nM). For the k2/K assessment, progress curves were obtained by incubating 14 nM PenA with increasing concentrations of avibactam (50–800 nM) using nitrocefin (50 μM) as a reporter substrate. The partition ratio (kcat/kinact) at 5 min for PenA with avibactam was determined by incubating 1 μM PenA with 1 μM avibactam, at which point >90% inhibition of 100 μM nitrocefin hydrolysis was obtained. The koff value was determined by incubating 10 μM PenA with an avibactam concentration of 50 μM for 30 min. Samples were serially diluted to a final enzyme concentration of 2.0 nM, and hydrolysis of 100 μM nitrocefin was measured. The progress curves were fit to a single-exponential decay equation.
Electrospray Ionization (ESI) Mass Spectrometry (MS)
To discern the nature of the intermediates of inactivation by avibactam in the reaction pathway with the PenA β-lactamase, ESI-MS was performed on a Waters SynaptG2-Si quadrupole time-of-flight mass spectrometer equipped with a LockSpray dual electrospray ion source, as previously described.18 For the experiments, 10 μM PenA was incubated with 10 μM avibactam for set times (i.e., 5 min, 5 h, and 24 h) at room temperature in 10m MPBS (pH 7.4). Reactions were terminated by the addition of 0.1% formic acid and 1% acetonitrile.
Acyl-Transfer Experiment
To assess if avibactam recyclizes to re-form active compound, an acyl-tranfer ESI-MS experiment was conducted using PenA as the donor and TEM-1 as the recipient.6b Ten micromolar PenA was fully acylated with <10 μM avibactam such that no free avibactam was present. Ten micromolar TEM-1 β-lactamase was added, and at time points of 15 s and 2, 5, 10, 20, and 30 min, samples were terminated and prepared for ESI-MS as described above.
Crystallization and Structure Refinement
The PenA β-lactamase was crystallized by the vapor diffusion method using a 250 μL reservoir with a 4 μL hanging drop (2 μL of reservoir solution + 2 μL of protein solution). For PenA, the well solution contained 25% polyethylene glycol of 8 kDa (PEG8K), 0.2 M sodium chloride, 0.1 M HEPES at pH 7.0, and 15 mg/mL PenA in 2 mM HEPES (pH 7.5). Crystals appeared in 1–2 weeks, reaching sizes of 0.4–0.6 mm. Before data collection, a pregrown crystal was soaked at room temperature for 30 min in the 30% PEG holding solution (pH 7.5) containing 1mM avibactam. The crystals were cryoprotected by dipping them into a reservoir solution containing 20% glycerol, flush-cooled, and kept at 100 K with a nitrogen gas stream.
The 0.5° oscillation images were collected on an ADSC quantum Q210r CCD detector with synchrotron radiation (λ = 1.00 Å at beamline NW12E of the Advanced Ring of the Photon Factory, Tsukuba, Japan). The HKL2000 program was used to index and scale X-ray intensities (Table 2).
The 27.5 kDa PenA β-lactamase crystallized in space group C2 with three molecules in the asymmetric unit and the following cell dimensions: a = 120.1 Å, b = 69.4 Å, and c = 84.6 Å, α = 90.000°, β = 90.049°, and γ = 90.000° (100 K). Using the apo-PenA structure (PDB 3W4Q) as a model, initial rigid body refinement was done with PHENIX. Model building and further refinement were performed with the programs COOT and PHENIX, respectively. Table 2 lists the final refinement statistics. The coordinates were deposited in the Protein Data Bank (PDB 3WZR).
G. mellonella Survival Assays
To assess the activity of ceftazidime–avibactam against B. multivorans in vivo, the G. mellonella insect model of infection was used.19 Larvae weighing between 200 and 400 mg were maintained on wood chips in the dark at 4 °C. Briefly, B. multivorans AU14786 was harvested from an 18 h culture by resuspending 1 × 109 cells in 500 μL of cold PBS. Ceftazidime and avibactam were also suspended in cold PBS.
A Hamilton syringe was used to inject 5 μL of the diluted bacterial suspension via the left proleg of each larva. After 30 min, the infected larvae were then treated with either ceftazidime (4 μg) or ceftazidime–avibactam (4 μg/4 μg) by injecting 5 μL into the right proleg. Two control groups of larvae were used. The first group was untreated. The second group, to test the impact of trauma, was injected with 5 μL of PBS and, after 30 min, 5 μL of ceftazidime/avibactam (4 μg/4 μg). One hundred G. mellonella larvae were used in each condition and incubated at (37 °C) in a sterile Petri dish for 24 h intervals for 48 h total. After 24 h of incubation, infected and control larvae were subsequently treated with ceftazidime (4 μg), ceftazidime/avibactam (4 μg/4 μg), or PBS + ceftazidime/avibactam (4 μg/4 μg) respectively, by injecting 5 μL in the right proleg. Larvae were considered dead when they displayed no movement in response to gentle prodding with a glass rod. Three replicates with 100 larvae per Petri dish were performed for each condition. The proportion that survived under each of the five treatments was compared using a test of proportions. Pairwise treatment comparisons were then performed, using a Bonferroni p value adjustment for the multiple tests. All data analysis was performed using R version 3.2.2.
Larval Histology
The Histology Core facility at Case Western Reserve University embedded a larva from each treatment condition in paraffin, sectioned the larva, and stained slides using hematoxylin–eosin staining. Scientists at the Division of Human Pathology, University of Milan, analyzed the stained slides.
Supplementary Material
Acknowledgments
These studies were partly supported by the Grant-in-Aid for Scientific Research (C), No. 26460534, Japan Society for the Promotion of Science. Research reported in this publication was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, the Veterans Affairs Merit Review Program BX002872 (K.M.P.-W.) and BX001974 (R.A.B.) from the U.S. Department of Veterans Affairs Biomedical Laboratory Research and Development Service and the Geriatric Research Education and Clinical Center VISN 10 to R.A.B. The contents do not represent the views of the U.S. Department of Veterans Affairs or the U.S. government. Funding was also provided by The National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Awards R21AI114508, R01AI100560, R01AI063517, and R01AI072219 to R.A.B. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was also supported by funding (to J.J.L.) from the Cystic Fibrosis Foundation. We thank David Kuo and Dr. Menachem Shoham in the Department of Biochemistry at Case Western Reserve University for providing guidance on working with G. mellonella. We thank AstraZeneca for providing the avibactam powder used in this study.
Footnotes
ORCID
Robert A. Bonomo: 0000-0002-3299-894X
Author Contributions
Conceived and designed the experiments: K.M.P.-W. and R.A.B. Performed the experiments: K.M.P.-W., M.N., N.O., M.L.W., E.T.Z., S.A.B., J.A.G., and M.F.M. Analyzed the data: K.M.P.-W., M.N., M.F., D.T., E.B., B.M.W., and R.A.B. Contributed reagents/materials/analysis tools: K.M.P.-W., M.N., J.J.L., and R.A.B. Wrote the paper: K.M.P.-W., S.A.B., M.N., and R.A.B.
Notes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.7b00020.
Supplemental Table 1: MICs in mg/L of selected antibiotics against clinical isolates of B. multivorans.
Supplemental Table 2: MICs in mg/L against clinical isolates of Burkholderia spp. (PDF)
References
- 1.(a) Cystic Fibrosis Foundation. https://www.cff.org/; (b) Elborn JS. Cystic fibrosis. Lancet. 2016;388(10059):2519–2531. doi: 10.1016/S0140-6736(16)00576-6. [DOI] [PubMed] [Google Scholar]
- 2.(a) Horsley A, Jones AM, Lord R. Antibiotic treatment for Burkholderia cepacia complex in people with cystic fibrosis experiencing a pulmonary exacerbation. Cochrane Database Syst Rev No. 2016;1:CD009529. doi: 10.1002/14651858.CD009529.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Regan KH, Bhatt J. Eradication therapy for Burkholderia cepacia complex in people with cystic fibrosis. Cochrane Database Syst Rev. 2016;11:CD009876. doi: 10.1002/14651858.CD009876.pub3. [DOI] [PubMed] [Google Scholar]
- 3.Shafiq I, Carroll MP, Nightingale JA, Daniels TV. Cepacia syndrome in a cystic fibrosis patient colonised with Burkholderia multivorans. BMJ Case Rep. 2011;2011 doi: 10.1136/bcr.08.2010.3296. bcr0820103296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papp-Wallace KM, Taracila MA, Gatta JA, Ohuchi N, Bonomo RA, Nukaga M. Insights into β-lactamases from Burkholderia species, two phylogenetically related yet distinct resistance determinants. J Biol Chem. 2013;288(26):19090–19102. doi: 10.1074/jbc.M113.458315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olive DM, Bean P. Principles and applications of methods for DNA-based typing of microbial organisms. J Clin Microbiol. 1999;37(6):1661–1669. doi: 10.1128/jcm.37.6.1661-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Durand-Reville TF, Lahiri S, Thresher J, Livchak S, Gao N, Palmer T, Walkup GK, Fisher SL. Kinetics of avibactam inhibition against class A, C, and D β-lactamases. J Biol Chem. 2013;288(39):27960–27971. doi: 10.1074/jbc.M113.485979. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Kern G, Walkup GK, Fisher SL. Avibactam is a covalent, reversible, non-β-lactam β-lactamase inhibitor. Proc Natl Acad Sci U S A. 2012;109(29):11663–11668. doi: 10.1073/pnas.1205073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Lahiri SD, Mangani S, Durand-Reville T, Benvenuti M, De Luca F, Sanyal G, Docquier JD. Structural insight into potent broad-spectrum inhibition with reversible recyclization mechanism: avibactam in complex with CTX-M-15 and Pseudomonas aeruginosa AmpC β-lactamases. Antimicrob Agents Chemother. 2013;57(6):2496–2505. doi: 10.1128/AAC.02247-12. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Xu H, Hazra S, Blanchard JS. NXL104 irreversibly inhibits the β-lactamase from Mycobacterium tuberculosis. Biochemistry. 2012;51(22):4551–4557. doi: 10.1021/bi300508r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Mushtaq S, Warner M, Livermore DM. In vitro activity of ceftazidime+NXL104 against Pseudomonas aeruginosa and other non-fermenters. J Antimicrob Chemother. 2010;65(11):2376–2381. doi: 10.1093/jac/dkq306. [DOI] [PubMed] [Google Scholar]; (b) Everaert A, Coenye T. Effect of β-Lactamase inhibitors on in vitro activity of β-lactam antibiotics against Burkholderia cepacia complex species. Antimicrob Resist Infect Control. 2016;5:44. doi: 10.1186/s13756-016-0142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nukaga M, Bethel CR, Thomson JM, Hujer AM, Distler A, Anderson VE, Knox JR, Bonomo RA. Inhibition of class A β-lactamases by carbapenems: crystallographic observation of two conformations of meropenem in SHV-1. J Am Chem Soc. 2008;130(38):12656–12662. doi: 10.1021/ja7111146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Berkhout J, Melchers MJ, van Mil AC, Seyedmousavi S, Lagarde CM, Schuck VJ, Nichols WW, Mouton JW. Pharmacodynamics of ceftazidime and avibactam in neutropenic mice with thigh or lung infection. Antimicrob Agents Chemother. 2016;60(1):368–375. doi: 10.1128/AAC.01269-15. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Berkhout J, Melchers MJ, van Mil AC, Seyedmousavi S, Lagarde CM, Nichols WW, Mouton JW. Pharmacokinetics and penetration of ceftazidime and avibactam into epithelial lining fluid in thigh- and lung-infected mice. Antimicrob Agents Chemother. 2015;59(4):2299–2304. doi: 10.1128/AAC.04627-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drawz SM, Bethel CR, Doppalapudi VR, Sheri A, Pagadala SR, Hujer AM, Skalweit MJ, Anderson VE, Chen SG, Buynak JD, Bonomo RA. Penicillin sulfone inhibitors of class D β-lactamases. Antimicrob Agents Chemother. 2010;54(4):1414–1424. doi: 10.1128/AAC.00743-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Mahenthiralingam E, Bischof J, Byrne SK, Radomski C, Davies JE, Av-Gay Y, Vandamme P. DNA-Based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J Clin Microbiol. 2000;38(9):3165–3173. doi: 10.1128/jcm.38.9.3165-3173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Payne GW, Vandamme P, Morgan SH, Lipuma JJ, Coenye T, Weightman AJ, Jones TH, Mahenthiralingam E. Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl Environ Microbiol. 2005;71(7):3917–3927. doi: 10.1128/AEM.71.7.3917-3927.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papp-Wallace KM, Bethel CR, Distler AM, Kasuboski C, Taracila M, Bonomo RA. Inhibitor resistance in the KPC-2 β-lactamase, a preeminent property of this class A β-lactamase. Antimicrob Agents Chemother. 2010;54(2):890–897. doi: 10.1128/AAC.00693-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. Approved Standard, CLSI document M7-A7. 7. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2006. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. 25th Informational Supplement M100-S27. Wayne, PA, USA: 2017. Performance Standards for Antimicrobial Susceptibility Testing. [Google Scholar]
- 16.Papp-Wallace KM, Winkler ML, Gatta JA, Taracila MA, Chilakala S, Xu Y, Johnson JK, Bonomo RA. Reclaiming the efficacy of β-lactam-β-lactamase inhibitor combinations: avibactam restores the susceptibility of ceftazidime against CMY-2-producing Escherichia coli. Antimicrob Agents Chemother. 2014;58:4290–4297. doi: 10.1128/AAC.02625-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrison JF, Walsh CT. The behavior and significance of slow-binding enzyme inhibitors. Adv Enzymol Relat Areas Mol Biol. 2006;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 18.Papp-Wallace KM, Becka SA, Taracila MA, Winkler ML, Gatta JA, Rholl DA, Schweizer HP, Bonomo RA. Exposing a β-lactamase “twist”: the mechanistic basis for the high level of ceftazidime resistance in the C69F variant of the Burkholderia pseudomallei PenI β-lactamase. Antimicrob Agents Chemother. 2016;60(2):777–788. doi: 10.1128/AAC.02073-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(a) Mil-Homens D, Ferreira-Dias S, Fialho AM. Fish oils against Burkholderia and Pseudomonas aeruginosa: in vitro efficacy and their therapeutic and prophylactic effects on infected Galleria mellonella larvae. J Appl Microbiol. 2016;120(6):1509–1519. doi: 10.1111/jam.13145. [DOI] [PubMed] [Google Scholar]; (b) Kamal F, Dennis JJ. Burkholderia cepacia complex phage-antibiotic synergy (PAS): antibiotics stimulate lytic phage activity. Appl Environ Microbiol. 2015;81(3):1132–1138. doi: 10.1128/AEM.02850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Thomas RJ, Hamblin KA, Armstrong SJ, Muller CM, Bokori-Brown M, Goldman S, Atkins HS, Titball RW. Galleria mellonella as a model system to test the pharmacokinetics and efficacy of antibiotics against Burkholderia pseudomallei. Int J Antimicrob Agents. 2013;41(4):330–336. doi: 10.1016/j.ijantimicag.2012.12.009. [DOI] [PubMed] [Google Scholar]; (d) Mil-Homens D, Bernardes N, Fialho AM. The antibacterial properties of docosahexaenoic omega-3 fatty acid against the cystic fibrosis multiresistant pathogen Burkholderia cenocepacia. FEMS Microbiol Lett. 2012;328(1):61–69. doi: 10.1111/j.1574-6968.2011.02476.x. [DOI] [PubMed] [Google Scholar]; (e) Brackman G, Cos P, Maes L, Nelis HJ, Coenye T. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob Agents Chemother. 2011;55(6):2655–2661. doi: 10.1128/AAC.00045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Seed KD, Dennis JJ. Development of Galleria mellonella as an alternative infection model for the Burkholderia cepacia complex. Infect Immun. 2008;76(3):1267–1275. doi: 10.1128/IAI.01249-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.