Abstract
Pesticides in urban runoff are a major source of pollutants in aquatic ecosystems. Fipronil, a phenylpyrazole insecticide, found in structural pest control products, turf grass control, and home pet flea medication, has recently increased in use and is commonly detected in urban runoff. However, little is known about the effects of fipronil on aquatic organisms at early developmental stages. Here, we evaluated toxicity of fipronil to embryos of Japanese Medaka (Oryzias latipes, Qurt stain) using a high-throughput 96-well plate toxicity test. Male and female embryos (< 6 h post fertilization) were exposed to concentrations of fipronil ranging from 0.1–910 μg L−1 for 14 days or until hatching. Embryos were subjected to gross and microscopic examinations of developmental adverse effects as well as transcriptome analysis using RNA-seq. Results indicated a positive dose-response in reduced hatching success, increased gross deformity (tail curvature) at a lowest-observed-effect concentration (LOEC) of 200 μg L−1 and delayed hatching (~1 day at the highest concentration, LOEC = 600 μg L−1). The transcriptome analysis indicated that fipronil exposure enhanced expression of titin and telethonin, which are responsible for muscle development. It is therefore possible that the formation of a tail curvature is due to asymmetrical overgrowth of muscle. Our results indicate that sub-lethal effects occur in embryonic stages of an aquatic vertebrate following exposure to high concentrations of fipronil, although no adverse effects at the highest published environmentally relevant concentration (6.3 μg L−1) were observed.
Keywords: developmental deformity, multi-model inference, ELISA, RNA-seq
Graphical abstract
Adverse effects:
Decreased hatching success
Delayed hatching
Formation of tail curvature, likely due to overgrowth of muscle (up-regulation of titin and telethonin genes was detected by RNA-seq)
Impaired swimming ability

1. Introduction
Aquatic organisms are exposed to various types of pollutants including pesticides, which originate from agricultural and urbanized areas (Gan et al., 2012; Weston et al., 2005). Pesticides from urban areas are an important pollutant in waterways due to their toxicity to non-target resident aquatic organisms (e.g., Amweg et al., 2006; Bloomquist, 1996; Tingle et al., 2003; Weston et al., 2005, 2009). Fipronil, a phenylpyrazole insecticide found in structural pest control products, turf grass control, and home pet flea medication, has become prominent among the many pesticides detected in surface waters (Jiang et al., 2010; Simon-Delso et al., 2015; Sprague and Nowell, 2008). Fipronil has gained in use as a replacement for the older pesticide classes of pyrethroids and organophosphates in urban areas, causing its environmental concentrations to increase (Gan et al., 2012; Weston et al., 2009). Simon-Delso et al. (2015) estimated that neonicotinoids and fipronil account for roughly 33% of the global insecticide market. A recent study reports environmental concentrations of fipronil in the range of 0.13–12.6 μg L−1 in California (Gan et al., 2012). It is not registered for agricultural use in California, however, it is applied for such use in other states and detections in environmental samples range from 0.01–6.4 μg L−1 (Mize et al., 2008).
Fipronil causes high toxicity to invertebrates through inhibition of gamma-amino butyric acid (GABA)-gated chloride and glutamate-gated chloride channels (Cole et al., 1993; Hainzl et al., 1998; Zhao et al., 2004). Fipronil can affect non-targeted organisms, including vertebrates, by inhibiting repolarization of nerve cells through blocking the influx of chloride anions, leading to hyper-excitation and death (Stehr et al., 2006). Beggel et al (2012) reported up-regulation of the vitellogenin gene, a marker for oestrogenic endocrine disruption, in larval stage of fathead minnow by fipronil exposure, suggesting fipronil acts as an endocrine disruptor and exerts sex-specific effects. The USEPA (1996) reported that fipronil affects growth of Rainbow Trout larvae at a no-observed-effect concentration (NOEC) of 0.0066 ppm and a lowest-observed-effect concentration (LOEC) of 0.015 ppm, and that the metabolite, MB 46136, is more toxic than the parent compound to Rainbow Trout and Bluegill Sunfish. Wirth et al. (2004) reported no effect of fipronil on the fish Cyprinidon variegatus at concentrations up to 5 000 ng L−1 in an estuarine mesocosm experiment, but reduced survival of Grass Shrimp at 355 ng L−1, suggesting potential food web effects for fishes. Thus, the adverse effects of fipronil on fish at larval, juvenile, and adult stages are well documented by previous laboratory studies (Beggel et al., 2012; Bencic et al., 2013; Nillos et al., 2009; USEPA, 1996; Wirth et al., 2004). However, little is known about the developmental toxicity, endocrine disruption, and sex-specific effects of fipronil on the embryonic stage of teleosts.
Developmental toxicity studies offer a unique perspective on ecological and organismal health. Organisms at the embryonic stage are the most vulnerable to chemical toxicity effects due to a lack of protective mechanisms (Hood, 1996; McKim, 1977; Spitsbergen et al., 1991). Moreover, tissue and vital biological systems are differentiating and developing, thus any disruption or impact can exert lifelong consequences such as formation of deformities. The cumulative effect of these factors makes developing organisms ideal for toxicity studies (McKim, 1977).
Japanese Medaka as used in this study provides several advantages over other commonly used fish models. The primary advantage is that Medaka is sexually dimorphic. At 2 days post fertilization, the sex of a Medaka Qurt strain embryo can be identified based on visible spots on the dorsal side of the head. Males are leucophore positive, a genetically-tied trait (lf= leucophore free, female: Xlf/Xlf and male Xlf/Y+) which causes brown pigmentation to become apparent on the back of the head (Kinoshita, 2009; Wada et al., 1998). Additionally, Medaka is a hardy species that is easy to culture with individual fish providing egg clutches of > 20 eggs per day. Embryos typically hatch within 7–10 days of fertilization (Kinoshita, 2009). This extended development time is ideal for investigating developmental effects given that more time is allowed for toxic action to occur compared to other fish model species such as Zebrafish.
Here, we report the developmental toxicity of fipronil across multiple levels of biological organization (hatching success, phenotypic observation, and transcriptome analysis using RNA-seq) in Japanese Medaka.
2. Materials and methods
2.1. Fipronil solutions
The powder form of fipronil (purity 98%) was purchased from ChemService (West Chester, PA) and was used to prepare a series of stock solutions. All stock solutions were prepared with HPLC grade methanol (Thermo Fisher Scientific, Waltham, MA). First, a 10 000000 μg L−1 stock solution was made, and then diluted in a step-wise manner (1:10 serial dilutions).
Two sets of experimental solutions were prepared; the first set of solutions covers wide range of concentrations at 1:10 serial dilutions (0.1, 1.0, 10.0, 100, and 1 000 μg L−1) and the second set was made to test concentrations between 200 to 1 000 μg L−1 with an increment of 200 (200, 400, 600, 800 and 1 000 μg L−1). The experimental solutions were prepared by spiking stock solutions into reconstituted water: pH 8.0, alkalinity 80 mg L−1 CaCO3, hardness 100 mg L−1 CaCO3 (Horning and Weber, 1985). The final concentration of the methanol in all the experimental solutions was 0.01% (v/v ratio) as suggested by American Society of Testing and Materials (ASTM E1241-92). A 0.01% methanol in reconstituted water (v/v ratio) was used as a vehicle control. A small portion of freshly prepared experimental solution was collected in amber glass vials prior to the initiation of the exposure and stored at −80 °C for measurement of fipronil concentration by enzyme-linked immunosorbent assay (ELISA) as described in the next section (Vasylieva et al., 2015).
2.2. Indirect competitive ELISA
All the buffers were prepared with ultrapure deionized (DI) water and the compositions are listed as follows: phosphate-buffered saline (PBS, 10 mM, pH 7.5), PBST (PBS containing 0.05% Tween 20), coating buffer (14 mM Na2CO3, 35 mM NaHCO3, pH 9.8), blocking buffer (1% BSA in PBST), and substrate buffer (0.1 M sodium citrate/acetate buffer, pH 5.5). Substrate solution contained 0.2 mL of 0.6% 3,3′,5,5′-Tetramethylbenzidine (in dimethyl sulfoxide, w/v), 0.05 mL of 1% H2O2 in 12.5 mL of substrate buffer. Stop solution was 2M H2SO4.
Plates were coated with 1 μg mL−1 antigen (1-CON, Vasylieva et al. 2015) diluted in the coating buffer (100 μL per well). After incubation for 1 h at room temperature (RT), the solution was replaced with the blocking buffer (200 μL per well) and plates were incubated over night at 4 °C or for 1–4 h at room temperature (RT). Plates were then washed with PBST 3 times prior to sample loading. The standards in the assay buffer were prepared in glass vials and loaded on the coated plate in triplicate wells (50 μL per well). The experimental solutions in low concentrations (0.1–10 μg L−1) were directly added to the wells pre-loaded with the assay buffer while samples with high fipronil concentrations (100–1 000 μg L−1) were diluted with the assay buffer before addition to the plate (Vasylieva et al. 2015). An equal volume of anti-fipronil serum (#2268 diluted in PBS, 50 μL per well) was added at 1:4 000 dilution, giving final 1: 8 000 dilution in the plate. The plates were incubated for 1 h at RT and then washed 5 times with PBST. Goat anti-rabbit IgG-HRP conjugate was added at 100 μL per well in a 1:20,000 dilution as instructed by manufacturer (Abcam, Cambridge, MA). The plates were incubated for 1 h at RT and washed 5 times with PBST. Substrate solution was added (100 μL per well) and was left to develop color for 10 min. The reaction was stopped by addition of stop solution (50 μL per well) and absorbance was read at 450 nm. SigmaPlot ver. 11.0 software was used for curve fitting and data analysis to find IC50 values and the equation of the standard curve. The assay was performed four separate times.
2.3. Embryo exposures
Medaka embryos were exposed to nine concentrations of fipronil over the entire embryonic period (the nominal concentrations: 0.1, 1.0, 10.0, 100, 200, 400, 600, 800, and 1 000 μg L−1). A total of five fipronil exposures were run; three of the exposures occurred at concentrations of 0.1, 1.0, 10.0, 100, and 1 000 μg L−1 while the other two were at 200, 400, 600, 800 and 1 000 μg L−1. All the exposures were performed with a newly developed 96-well plate method (Truong et al. 2011). Traditional beaker or petri dish exposure methods generate a large volume of waste, accumulate costly disposal fees, require large and complex temperature control mechanisms (i.e. water baths), and expose embryos in large groups, making tracking of individual responses difficult. Exposure using the 96-well plate method eliminates all of these problems; temperature control is managed via a commonly-used environmental chamber, one well holds only 250 μL of experimental solution, and each developing embryo can be observed without disruption and tracked separately under a dissecting microscope. Medaka embryos (< 6 h post fertilization) were collected from the culture facility at the Aquatic Health Program, University of California, Davis. Immediately after collection, embryos were cleaned with DI water and sorted for health and viability, then placed in 15 mL of experimental solutions in 50 mL Pyrex beakers for batch exposure (50 eggs per beaker, one beaker per concentration). Medaka embryos were kept in the beakers until male and female embryos could be identified (~4 d post-fertilization). At 2 days post-exposure (dpe), a 50% (7.5 mL) water change was performed. At 4 dpe, embryos were separated by sex and moved into a 96-well plate (Costar 9016, Non-sterile, Polystyrene, Flat Bottom, Medium Binding). Embryos were not placed initially in 96-well plates both to allow time to determine sex of the embryos, and to remove unfertilized eggs. This also allowed us to ensure that equal numbers of each sex were exposed and to conclusively attribute hatching failures to experimental conditions. Each well contained 250 μL of fresh experimental solution and one embryo was placed in each well. A total of 32 or more eggs were used for the exposure per treatment. More than 50% of total well volume was changed every other day during the remainder of the experiment. The fish embryos in the 96-well plates were kept in an environmental chamber (Percival Scientific, Perry, IA, Model 136LLVL) at 25 °C on a 16:8 h light:dark cycle with light intensity of 808 Lux during the experiment. All embryos were observed daily, and signs of abnormal development, mortality, and hatching success were recorded. Hatched larvae were kept in the 96-well plates up to 24 h after hatch for assessing abnormal development as well as swimming ability, and then euthanized using tricaine methane sulfonate (Tricaine-S, Western Chemical, Inc., Ferndale, WA). Larval fish lying on their side at the bottom of the wells at 24 h post hatch were considered as “impaired swimming”. Hatchlings were not fed during the exposure. The fish embryos that failed to hatch were terminated at 14 dpe.
2.4. Statistical analysis
We analyzed the data for a sex specific effect of fipronil on hatching success, hatching time, and tail curvature (measured at hatch) using multi-model inference (Anderson et al., 2000; Burnham and Anderson, 2002). For statistical analysis, individual fish in each well were treated as replicates. For both the hatching success and tail curvature data, each model had binomial distributions of error because the response variable is a success/failure (i.e., the fish either hatched or it did not). Using the error distribution appropriate for the type of data (e.g., binomial distribution for success/failure data) provides accurate estimates of confidence intervals (Bolker et al. 2009). Models fit to the hatching success data included H~ (an intercept only model), H~C and H~C+S, where H is hatching success (1 is success, 0 is failure), C is the nominal concentration (μg L−1), and S is a dummy variable for sex (1 is male, 0 is female). Models used to analyze the tail curvature data included TC~ (an intercept only model), TC~C and TC~C+S, where TC is a variable where ‘1’ is a curved tail and ‘0’ is no tail curvature, C is the nominal concentration of fipronil in μg L−1, and S is a dummy variable for sex (1 is male, 0 is female). For the hatching time analysis we built models with Gaussian error distributions. Models included D~ (an intercept only model), D~C, D~C+S, D~C+S+C×S, where D is days to hatching, C is concentration (μg L−1), and S is sex. We included the interaction model to determine whether fipronil influences hatching time differently for each sex, potentially indicating endocrine disrupting effects. Because exposures were run five times over the course of several months, we also tested for an effect of time for each of the responses by including a variable for ‘temporal block’ in the best model and comparing it to a model without the temporal block variable. In each case time was insignificant, so we did not include ‘time’ in the models and present the data from the five time points together. For the tail curvature analysis, more than half the larvae had the deformity at the highest concentration, allowing us to calculate an ED50. We used the methodology described in Hammock et al. (2015) for the calculation.
In addition to the above analyses, we performed three one-way analysis of variances (ANOVAs) to determine the lowest-observed-effect concentration (LOEC) for hatching success, tail curvature, and hatching time. A logistic transformation was used on the hatching success and tail curvature data because the response variables are binary. Concentration was the only independent variable used for each ANOVA. Dunnett’s tests were conducted following all three ANOVAs to find the LOECs.
2.5. RNA-seq
An additional exposure was performed to characterize gene expression patterns of embryos at the lowest concentration of fipronil. For RNA-seq, the lowest-observed effect concentration at which development of the tail curvature was observed (200 μg L−1) was used since non-specific responses due to overdose fipronil exposure (e.g. cell necrosis) were our concern. The exposure was conducted as follows: Medaka embryos were collected as described above (2.3. Embryo exposures), followed by batch exposure in glass beakers (210 embryos in 50 mL of fipronil solution, 200 μg L−1). The same number of embryos were also exposed to 0.01% methanol as a vehicle control. On 4 dpe, the embryos were separated by sex and then placed into a 96-well plate as described above (one embryo per well). The experimental solution (> 50%) was changed every other day. At the end of the exposure (7 dpe, Stage 37: Pericardial cavity formation stage, Kinoshita et al. 2009), all the embryos in each treatment for each sex were pooled together and then randomly divided into 3 groups (n = 10 per group) to obtain a sufficient amount of total RNA for library preparation and to minimize bias from individual variation. The sampling time was chosen to ensure that 1) all the fish were still at embryonic stage and 2) embryos were exposed to fipronil long enough to induce changes in gene expression patterns.
Only male embryos were used for the gene expression analysis in order to standardize test results. Total RNA was isolated using TRIzol Reagent by following the manufacturer’s instructions (Thermo Fisher Scientific). The quality of the total RNA was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) available at the DNA Technologies Core Facility at UC Davis (http://dnatech.genomecenter.ucdavis.edu) and ensured that the quality and integrity of the total RNA samples were above the required criteria for library preparation (RNA Integrity Number > 8.0). No genomic DNA contamination was detected.
The library preparation and sequencing reaction for RNA-seq was performed by the DNA Technologies Core Facility at University of California, Davis. Transcripts with poly(A) tail were enriched from the total RNA samples with Bio-Poly(A) beads (Bioo Scientific, Austin, Texas). Strand-specific RNA-seq libraries were generated with barcoded adapters using the NEXTflex Rapid Directional kit (Bioo Scientific) according to the instructions of the manufacturer. Uniform fragment size distribution for all libraries was verified with a Bioanalyzer. The concentration of the libraries were quantified with Qubit Fluorometer (Thermo Fisher Scientific), pooled equimolarly, and sequenced on a HiSeq 2500 instrument (Illumina, San Diego, CA) with single-end 50 bp reads.
The sequencing data were processed using a series of bioinformatics programs. Prior to the data analysis, the sequence data in FASTQ format were subjected to quality check as well as trimming poor bases using programs in the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html). The Medaka reference genome sequence was obtained from the Ensembl database (downloaded on December 18th 2014, http://www.ensembl.org/index.html) and indexed using Bowtie2-build available within the package of Bowtie2 ver. 2.2.4 (Langmead and Salzberg, 2012). The FASTQ sequence data were used for mapping to the reference genome sequence using TopHat ver. 2.0.13 (Kim et al., 2013). The output files generated for each library were then used for transcript assembly, merging, and statistical analysis using Cufflinks, Cuffmerge, and Cuffdiff within Cufflinks ver. 2.2.1 package, respectively (Trapnell et al., 2010, 2012). All the analyses were performed with the default settings. The gene expression level was expressed as “mapped fragments per kilobase of exon model per million mapped reads (FPKM)” and genes with a false discovery rate (FDR) lower than 0.05 were considered as differentially expressed by the fipronil exposure. A two-tailed t-test was used to test for significance against a null hypothesis that there was no change while a one–tailed t-test using the estimated posterior distribution for that gene was used in cases where there were zero fragments mapped in one condition (Trapnell et al., 2013). The genes for which expression was significantly altered by fipronil exposure were subsequently annotated by BLASTX against protein database (nr) obtained from the NCBI website (downloaded on April 8th 2015, approximately 64 million sequences were available, http://www.ncbi.nlm.nih.gov/). All the data processing with bioinformatics programs was performed using custom workstation built with 2X Xeon E5-2630 6 core CPU with 128GB ECC RAM, 4× HDD in RAID 10 configuration for Data Storage, 64bit Linux system.
Data interpretation of RNA-seq datasets based on a limited number of selected individual genes has a risk of misinterpretation and likely produces inconsistent results when analyzed by researchers with different research backgrounds and expertise. To overcome this issue, a Gene Set Enrichment Analysis (GSEA) was performed to investigate biological functions altered by fipronil exposure using gene ontology terms rather than individual genes. A GSEA was performed using GOstat, a Bioconductor package written in R (Falcon and Gentleman, 2006; R Core Team, 2016). A setting for Cuffdiff (FDR < 0.075) was used to obtain a larger number of differentially expressed genes to perform the analysis, and a total of 2,021 genes were identified as differentially expressed by the fipronil exposure with the setting. The DNA sequences for the differentially expressed Medaka genes by fipronil exposure were retrieved using a custom shell script and further used to obtain Gene Product IDs by running a BLASTX search against the annotated Medaka protein sequences downloaded from the QuickGO database available at the EBI website (http://www.ebi.ac.uk/QuickGO/GAnnotation). For BLASTX, a cutoff value of 95% identity was used to remove sequences showing low similarities, and only non-redundant Gene Product IDs were used for the GSEA. All the Medaka genes expressed in the samples as well as successfully annotated by BLASTX were utilized as “universe” or “background” (Falcon and Gentleman, 2006). The hyperGTest function, which implements the hypergeometric calculation, was used for the analysis (P value cutoff = 0.05).
2.6. Reverse transcriptase quantitative PCR (RT-qPCR)
RNA-seq results for the selected number of genes were validated by RT-qPCR as described in our previous publications (Rochman et al. 2014; Ramírez-Duarte et al. 2016). Briefly, total RNA (1 μg) used for RNA-seq was subjected to DNase treatment (AMPD1, Sigma-Aldroch), followed by cDNA synthesis using Superscript II Reverse Transcriptase following to the manufacturers’ instructions (Life Technologies). The primers for RT-qPCR were designed by PrimerExpress ver. 3.0 (Thermo Fisher Scientific, Waltham, MA, USA) and targets and corresponding primer sequences are listed in Table S1 (Supplementary Data). The reaction cocktail was prepared with Maxima SYBR Green qPCR Master Mix (Thermo Fisher Scientific) and the reactions were performed using an AB7900 HT FAST Thermocycler available at UC Davis Real-time PCR Research and Diagnostics Core Facility (http://www.vetmed.ucdavis.edu/vme/taqmanservice/). Each sample was analyzed in triplicate. The cycle threshold value (Ct) for each target gene was determined and relative expression levels in fold change was calculated by the 29−ΔΔCt method (Schmittgen and Livak, 2008). The geometric means of three house keeping genes, glyceraldehyde-3-phosphate dehydrogenase (GADPH), ribosomal protein L7 (RPL7), and 18S ribosomal RNA were used for data normalization (Zhang and Hu, 2007). A Pearson’s correlation coefficient test was performed to assess the linear relationship of the gene expression data from RNA-seq and RT-qPCR using the package “Hmisc” written in R (R Core Team, 2016). The gene expression data were log2 transformed prior to the statistical test.
3. Results
3.1. Measured fipronil concentrations
The measured fipronil concentrations for the selected experimental solutions are listed in Table S2 (Supplementary Data). The 0.1 μg L−1 concentration was below the limit of detection (LOD) of the ELISA method. Assay #1 found no results for the 1.0 μg L−1 sample (Table S2, Supplementary Data), therefore, only three replications were utilized for measuring the concentration for the 1.0 μg L−1 sample, which produced a mean (± standard deviation) of 3.0 ± 1.14 μg L−1. Four replicate measurements for the 10.0 μg L−1 sample had a mean of 6.3 ± 0.94 μg L−1. The 100 μg L−1 sample had a mean of 86 ± 8.20 μg L−1. A mean of 910 ± 113.52 μg L−1 was found for the 1 000 μg L−1 sample. The experimental solutions collected for ELISA (200, 400, 600, and 800 μg L−1) were accidentally lost during the analysis. Throughout the rest of the paper, we report measured concentrations for the experimental solutions for which the ELISA data are available, otherwise nominal concentrations are reported (0.1, 3.0, 6.3, 86, 200, 400, 600, 800, and 910 μg L−1).
3.2. Embryo exposures
3.2.1. Mortality
Mortality was not observed due to the fipronil exposure even at the highest concentration tested in this study (910 μg L−1).
3.2.2. Hatching success
Embryos exposed to fipronil exhibited a decrease in hatching success in a dose-dependent manner (Fig. 1, Tables 1 and 2). The embryos in the vehicle control group hatched at a success rate of 94.6% whereas 82.8% of embryos hatched successfully at 910 μg L−1 fipronil exposure (Table 1). The influence of fipronil on hatching success was unequivocal, as the two models with a parameter for fipronil concentration received an AICc weight proportion of 0.999 (Table 2), and the fipronil parameter estimate did not overlap zero for the top-ranked model (slope = − 0.00239598, 95% CI = −0.00327, −0.00152). While the top-ranked model did not include sex, the second-ranked model, which had a parameter for sex, received substantial AICc support (AICc weight = 0.45, Table 2). Males had slightly higher hatching success than females, however, the results are inconclusive regarding the influence of sex on hatching success (Tables 1 and 2). The ANOVA mirrored the results of the model comparison, yielding a P value of < 0.0001 (F[9,459] = 5.3433). The Dunnett’s test showed that the LOEC was 910 μg L−1 (P < 0.0001 and P = 0.2091 for control versus 910 μg L−1 and 800 μg L−1, respectively).
Figure 1.
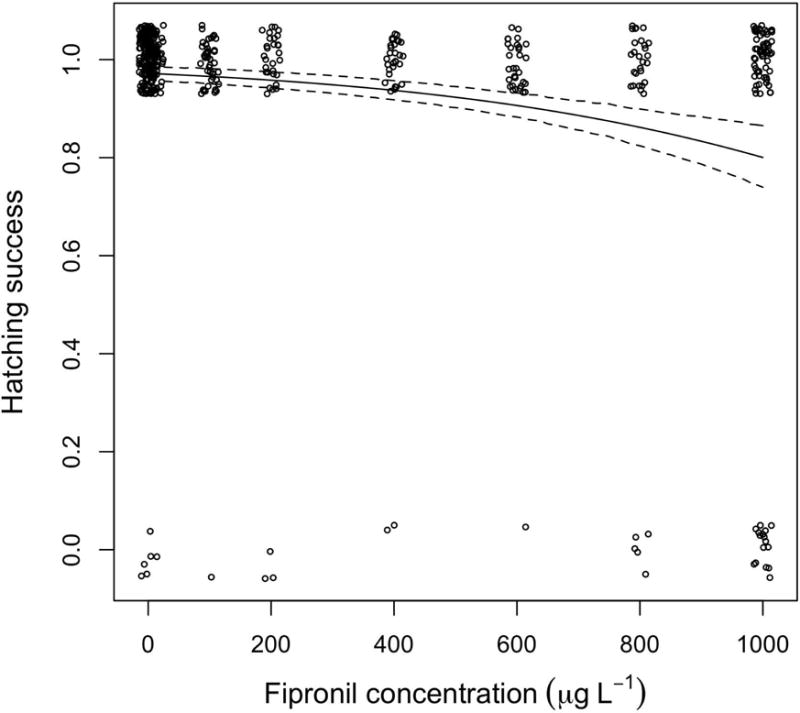
Results and modeling of embryo hatching success following exposure to fipronil (nominal concentrations). The solid line is the top-ranked model (Table 2) and the broken lines represent the 95% confidence interval of the model. The data and predictions encompass all fipronil concentrations to which embryos were exposed. A small random number was added to each point to prevent the points from overlapping (i.e., the ‘jitter’ function in R).
Table 1.
Hatching success of embryos and swimming obstruction and development of tail curvature of hatched larvae following exposure to fipronil. Numbers in parentheses in the columns, “Hatching success”, “Swimming obstruction”, and “Tail curvature”, indicate the percentage of embryos successfully hatched, hatched larvae showing impaired swimming ability, and hatched larvae showing tail curvature, respectively. In this table, data from 4 out of 5 fipronil exposures are presented since tail curvature was not recorded during the first exposure.
| Fipronil (μg L−1) |
Total eggsa |
Hatching success
|
Swimming obstruction
|
Tail curvature
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Total | Male | Female | Totald | Male | Female | Totald | |||||||||||
|
|
|
|
|
||||||||||||||||
| 0.1b | 32 | 16 | (100) | 15 | 93.8 | 31 | (96.9) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) |
| 3.0 | 32 | 15 | (93.8) | 16 | 100 | 31 | (96.9) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) |
| 6.3 | 32 | 16 | (100) | 16 | 100 | 32 | (100) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) |
| 86 | 31 | 15 | (93.8) | 16 | 100 | 31 | (100) | 4 | (26.7) | 4 | (25.0) | 8 | (25.8) | 2 | (13.3) | 1 | (6.7) | 3 | (9.7) |
| 200b | 32 | 16 | (100) | 13 | 81.3 | 29 | (90.6) | 14 | (87.5) | 13 | (100) | 27 | (93.1) | 2 | (12.5) | 5 | (38.5) | 7 | (21.9) |
| 400b | 32 | 15 | (93.8) | 15 | 93.8 | 30 | (93.8) | 14 | (93.3) | 15 | (100) | 29 | (96.7) | 4 | (26.7) | 6 | (40.0) | 10 | (33.3) |
| 600b | 32 | 15 | (93.8) | 16 | 100 | 31 | (96.9) | 15 | (100) | 16 | (100) | 31 | (100) | 7 | (46.7) | 10 | (62.5) | 17 | (54.8) |
| 800b | 32 | 12 | (75.0) | 13 | 81.3 | 25 | (78.1) | 12 | (100) | 13 | (100) | 25 | (100) | 5 | (41.7) | 9 | (69.2) | 14 | (56.0) |
| 910b | 64 | 27 | (84.4) | 26 | 81.3 | 53 | (82.8) | 23 | (85.2) | 25 | (96.2) | 48 | (90.6) | 20 | (74.1) | 21 | (80.8) | 41 | (77.4) |
| VCc | 64 | 31 | (96.9) | 30 | 93.8 | 61 | (94.6) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) |
Equal number of male and female embryos were used for the exposures except for 86 μg L−1 (15 females and 16 males were used for the treatment).
Nominal concentration
Vehicle control
The percentages of larvae showing swimming obstruction and tail curvature were calculated based on the number of hatched fish as a total for each treatment.
Table 2.
Relative support for hatching success, tail curvature, and hatching day models. H refers to proportion hatching success, C to fipronil concentration, and S to sex of the embryo. TC~ refers to proportion tail curvature, C to concentration of fipronil, and S to sex of the embryo. D refers to days to hatching, C to fipronil concentration, S to sex of the embryo, and C×S to the interaction between these parameters.
| Endpoint | Experiment model | df | ΔAICc | AICc wt |
|---|---|---|---|---|
| H~C | 2 | 0 | 0.55 | |
| Hatching success | H~C+S | 3 | 0.4 | 0.45 |
| H~ (Intercept only) | 1 | 30.4 | <0.001 | |
|
| ||||
| TC~C+S | 3 | 0 | 0.59 | |
| Tail curvature | TC~C | 2 | 0.7 | 0.41 |
| TC~ (Intercept only) | 1 | 148.3 | <0.001 | |
|
| ||||
| D~C+S | 4 | 0 | 0.48 | |
| Hatching day | D~C | 3 | 1.2 | 0.26 |
| D~C+S+C×S | 5 | 1.3 | 0.25 | |
| D~ (Intercept only) | 2 | 52.1 | <0.001 | |
df is the degrees of freedom, ΔAICc is the difference in AICc between the model of interest and the top-ranked model, and AICc wt is the proportion of AICc weight.
3.2.3. Tail curvature
Fipronil exposure caused development of tail curvature (Fig. 2, Table 2, slope = 0.00486541, 95% CI = 0.00385, 0.00534) and the frequency of fish with the deformity increased in a dose-dependent manner (Fig. 3). Females showed a slightly higher rate of tail curvature (Table 2, parameter estimate = −0.66, 95% CI = −1.21, −0.11). Based on the model without sex, the ED50 was 668.1 μg L−1 (95% CI = 592.0, 747.9 μg L−1). In Fig. 3, the model represents the probability of an individual acquiring the discrete effect. Approximately 77% of embryos exposed to 910 μg L−1 fipronil developed tail curvature while the embryos in the control group did not exhibited the deformity. The ANOVA also indicated a difference in tail curvature between concentrations (ANOVA, F[9,420] = 41.3618, P =< 0.0001). The Dunnett’s test revealed that the LOEC is 200 μg L−1 (P = 0.0031), while the lowest concentration at which curved tails were observed (though statistically equal to the control) was 86 μg L−1.
Figure 2.
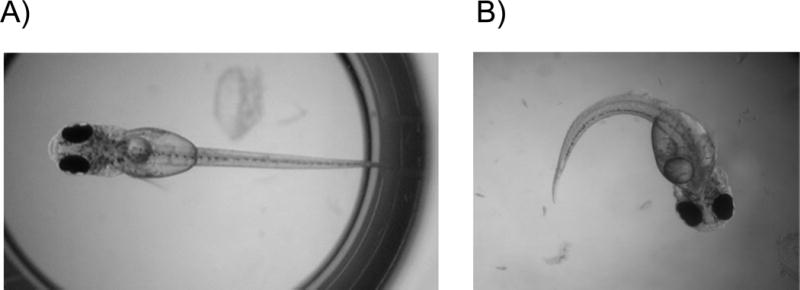
Healthy, swimming larva in vehicle control group (Panel A) versus larva with impaired swimming ability with tail curvature at 910 μg L−1 fipronil exposure group (Panel B).
Figure 3.
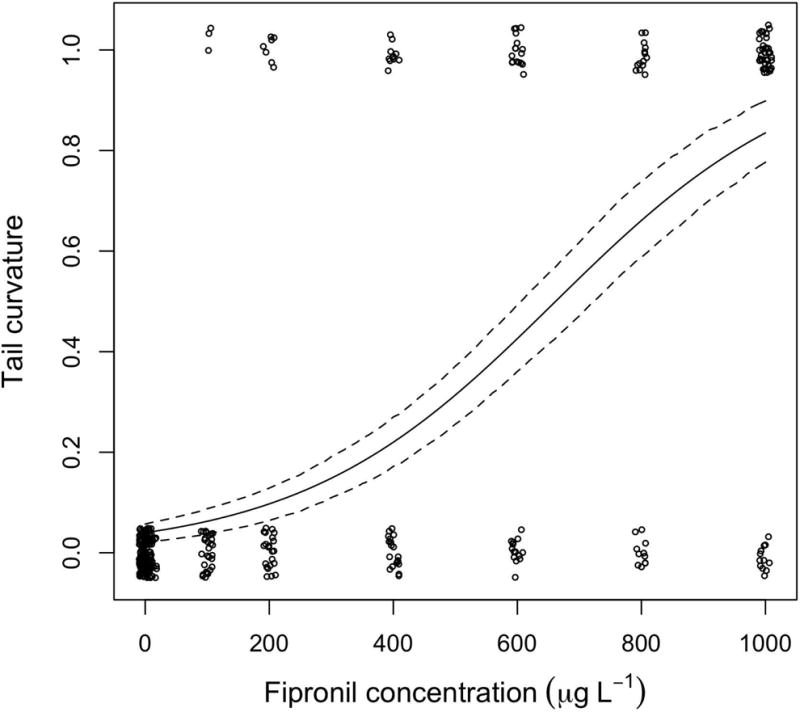
Results and modeling of the tail curvature data. The solid line represents the top-ranked model and the broken lines are the 95% confidence intervals of the model (Table 2). A small random number was added to each point to prevent the points from overlapping (i.e., ‘jitter’ function in R).
3.2.4. Swimming behavior
Beginning at 86 μg L−1, embryos exposed to fipronil exhibited difficulty swimming and were lying on their side (detected by gross observation of concentration versus effect, Table 1). At the concentrations of 200 μg L−1 and above, nearly all hatched larvae exhibited impaired swimming ability (Table 1). This impaired swimming ability was observed in all larvae with tail curvature and in some larvae without the visible physical deformity as well. While such fish were not swimming, they were still alive, with heartbeats and movement of pectoral fins. Counts of heartbeat and movement of pectoral fins were not recorded in this study.
3.2.5. Delayed hatching
Hatching time was delayed by fipronil, as the three top-ranked hatching time models all had a parameter for concentration of fipronil, receiving a combined AICc weight of 0.999 (Table 2). The top-ranked model included parameters for fipronil concentration and sex (Table 2). Females appeared to have slightly longer development time (delayed hatching) than males, but the effect was equivocal as the model without a sex parameter received some AICc weight (0.26; Table 2). This effect was independent of fipronil, as the model with an interaction between sex and concentration did not receive strong AICc support (Table 2, Fig. 4), and the confidence interval for the interaction parameter overlapped zero (0.0015, 95% CI = −0.0045, 0.0075). Fipronil exposure delayed hatching time by ~1 day at 910 μg L−1 in both males and females. An ANOVA (F[9,419] = 6.1892, P =< 0.0001) followed by a Dunnett’s test revealed that the LOEC is 600 μg L−1 (P = 0.0161).
Figure 4.
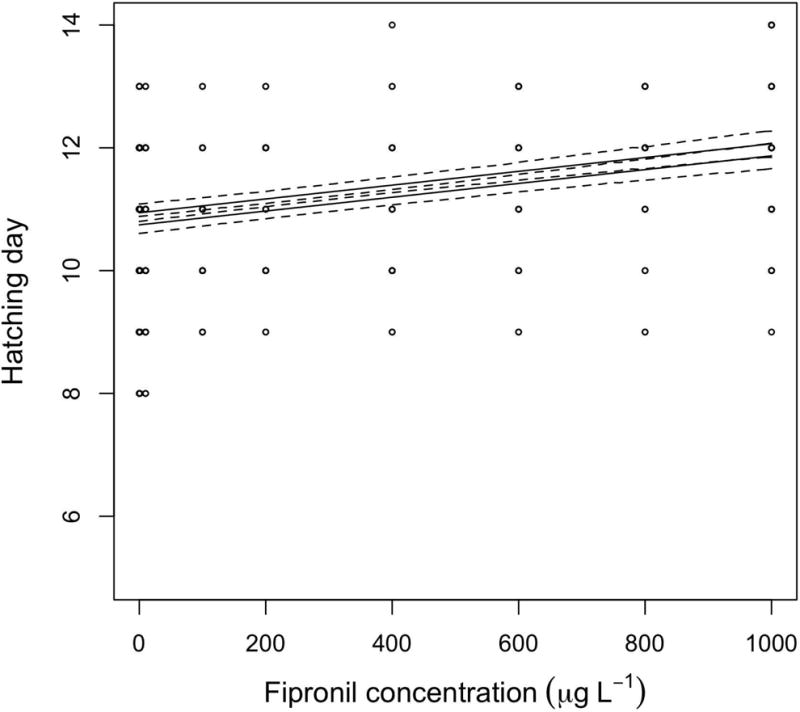
Results and modeling of the time (in days) until hatching following exposure to fipronil (nominal concentrations). The solid lines represent the top-ranked model (Table 2) and the broken lines are the 95% confidence intervals of the model. The lower solid line is the mean prediction for males and the upper solid line is the mean prediction for females.
3.3. RNA-seq
The sequencing reaction generated approximately 61 million (M) reads for fipronil exposed groups and 58 M reads for the control groups (Table 3). Of them, 83 to 86% of the reads were successfully aligned and mapped to the Medaka reference genome sequence, providing over 15 M reads per sample for the analysis (Table 3). In total, 36,102 unique genes, including protein coding and non-coding RNA, were detected in at least one of the six groups. This includes novel genes, which were not predicted in a previous publication (Kasahara et al., 2007). With a false discovery rate (FDR) of 0.05, the expression of 174 genes was altered by the fipronil exposure with 69 and 105 genes significantly up- and down-regulated, respectively. This includes genes belonging to various categories, such as muscle growth and cardiac function (Supplemental Data Table S3).
Table 3.
Summary of sequencing reads for the transcriptome analysis by RNA-seq
| Treatment | Group | No. of reads generated | Read length after trimming (bp) | No. of reads alignged and mapped to the reference genome | Percentage of aligned and mapped reads |
|---|---|---|---|---|---|
| Fipronil | Fipronil 1 | 2,02,14,620 | 41 | 1,73,34,337 | 85.8 |
| Fipronil 2 | 2,04,40,716 | 41 | 1,75,16,671 | 85.7 | |
| Fipronil 3 | 2,04,84,183 | 41 | 1,75,95,651 | 85.9 | |
| Control | Control 1 | 2,01,62,259 | 41 | 1,73,60,820 | 86.1 |
| Control 2 | 1,93,96,458 | 41 | 1,63,31,321 | 84.2 | |
| Control 3 | 1,89,56,560 | 41 | 1,58,86,034 | 83.8 |
The fipronil exposure altered a number of molecular functions as shown in Table 4. This includes biological functions involved in homeostasis of nerve cells in the central nervous system, such as voltage-gated calcium channel activity, calcium channel activity, and calcium ion transmembrane transporter activity. In addition, molecular functions of actin binding and cytoskeletal protein binding were also affected by the fipronil exposure (Table 4). However, alternation of molecular functions that are involved in endocrine system (e.g. thyroid hormone signaling pathway) were not detected.
Table 4.
List of molecular functions altered by fipronil exposure from gene set enrichment analysis. Medaka genes differentially expressed by fipronil exposure (n = 2,021, FDR < 0.075) were used for the analysis.
| GOMFID | Pvalue | OddsRatio | ExpCount | Count | Size | MFTerm |
|---|---|---|---|---|---|---|
| GO:0030414 | < 0.0001 | 5.12 | 1.86 | 8 | 47 | Peptidase inhibitor activity |
| GO:0061134 | < 0.0001 | 4.86 | 1.94 | 8 | 49 | Peptidase regulator activity |
| GO:0061135 | 0.0011 | 4.97 | 1.66 | 7 | 42 | Endopeptidase regulator activity |
| GO:0004866 | 0.0011 | 4.97 | 1.66 | 7 | 42 | Endopeptidase inhibitor activity |
| GO:0004857 | 0.0023 | 3.83 | 2.38 | 8 | 60 | Enzyme inhibitor activity |
| GO:0003964 | 0.0045 | 48.9 | 0.12 | 2 | 3 | RNA-directed DNA polymerase activity |
| GO:0004867 | 0.0047 | 7.03 | 0.71 | 4 | 18 | Serine-type endopeptidase inhibitor activity |
| GO:0005245 | 0.0059 | 10.51 | 0.4 | 3 | 10 | Voltage-gated calcium channel activity |
| GO:0003779 | 0.0064 | 2.59 | 4.63 | 11 | 117 | Actin binding |
| GO:0004842 | 0.0072 | 3.9 | 1.74 | 6 | 44 | Ubiquitin-protein transferase activity |
| GO:0004222 | 0.0078 | 3.06 | 2.89 | 8 | 73 | Metalloendopeptidase activity |
| GO:0005262 | 0.0117 | 5.18 | 0.91 | 4 | 23 | Calcium channel activity |
| GO:0004175 | 0.0161 | 2.08 | 6.7 | 13 | 169 | Endopeptidase activity |
| GO:0016746 | 0.0187 | 2.57 | 3.37 | 8 | 85 | Transferase activity, transferring acyl groups |
| GO:0008134 | 0.0196 | 6.13 | 0.59 | 3 | 15 | Transcription factor binding |
| GO:0008374 | 0.021 | 12.22 | 0.24 | 2 | 6 | O-acyltransferase activity |
| GO:0052689 | 0.0322 | 4.9 | 0.71 | 3 | 18 | Carboxylic ester hydrolase activity |
| GO:0008092 | 0.0325 | 1.87 | 7.37 | 13 | 186 | Cytoskeletal protein binding |
| GO:0015085 | 0.0438 | 3.27 | 1.35 | 4 | 34 | Calcium ion transmembrane transporter activity |
Abbreviations- GOMFID: gene ontology (GO) molecular function identification number, Pvalue: statistical confidence in P value obtained by hypergeometric test, OddsRatio: odds ratio of enrichment of GO ID, ExpCount: the expected count of genes with the given GO term, Count: the count of genes that are annotated to GOID in the set of differently expressed genes, Size: the count of genes that are annotated to GOID in the background, MFTerm: description of molecular function (MF) for GOID.
3.4. RT-qPCR
Pearson’s correlation coefficient test indicated that there was a significant positive correlation between the RNA-seq and RT-qPCR results (r = 0.61, n = 15, P = 0.025, Fig. 5).
Figure 5.
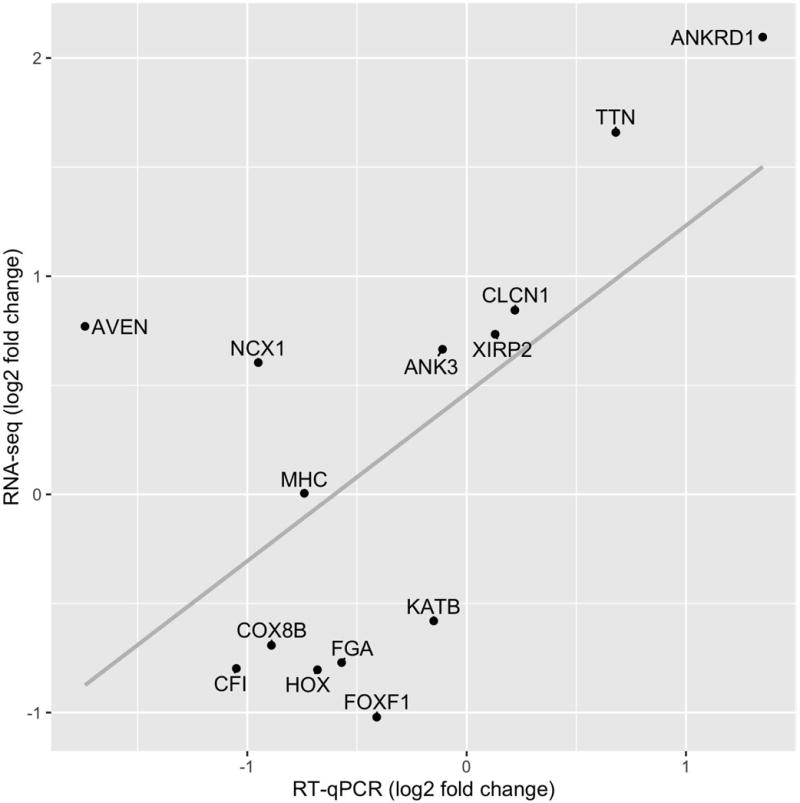
Comparison of RNA-seq and RT-qPCR results for the selected genes. The grey line indicates a linear regression (slope = 0.7690, intercept = 0.4633, R2 = 0.3633). See Table S1 (Supplementary Data) for the gene abbreviations.
4. Discussion
The intent of this study was to investigate developmental toxicity, endocrine disruption, and sex specific effects of fipronil using endpoints across multiple levels of biological organization. Previous studies have applied a similar approach using larval Zebrafish and Fathead Minnow (Beggel et al., 2012; Stehr et al., 2006), however to our knowledge this is the first study to report global transcriptome changes in embryos exposed to fipronil at such an early stage. Results here confirm that previously reported findings on the toxicity of fipronil to larval fish can be reproduced in the embryo (Stehr et al., 2006). Although acute mortality, endocrine disruption, and a sex-specific response were not observed in this study, our results suggest that fipronil exposure during embryonic developmental stages causes various adverse effects such as a decrease in hatching success, induction of tail curvature, impairment of swimming ability, and a delay in hatching time. The transcriptome analysis using RNA-seq indicates that fipronil exposure alters expression of genes which are involved in muscle development among other functions. All the results in this study demonstrate that fipronil disrupts embryonic development and causes various adverse effects to Medaka at high concentrations.
Development of tail curvature was a prominent effect found in this study. The frequency of fish exhibiting the deformity increased in a dose-dependent manner, demonstrating that the effect is due to fipronil. The tails of embryos in fipronil exposed groups appeared thicker and wider compared to the vehicle control group, although this effect was not quantified. Although speculative, it could be due to abnormal growth of the muscle, where one lateral side of the developing tail grows faster than the other, causing the more densely packed, higher growth muscle to contract and lead to the observed tail curvature. For example, Li et al. (2009) observed a tail curvature in Zebrafish embryos exposed to arsenic and suggested that the effect could be due to abnormal cell proliferation and apoptosis in the tail. Beggel et al. (2010) found increased weight and scoliosis in larval Fathead Minnow following fipronil exposure. In addition, Stehr et al., 2006 reported that Zebrafish embryos exposed to fipronil showed disrupted muscle morphology such as shortened myotomes. Similarly, our previous study also reported skeletal deformity in Sacramento Splittail embryos exposed to selenium and splittail larvae exposure to esfenvalerate and diazinon (Teh et al., 2002, 2004). As suggested by other researchers, tail curvature could be a biomarker for adverse developmental effects in fish embryos (Cheng et al., 2000; Li et al., 2009).
The disruption of muscle development due to fipronil exposure is supported by transcriptome analysis by RNA-seq, followed by GSEA. Individual genes involved in muscle development such as titin, telethonin, and myozenin-2 were altered by fipronil exposure (Labeit and Kolmerer, 1995; Valle et al., 1997; Takada et al., 2001). Titin, the titanic protein associated with sarcomere assembly, was up-regulated in embryos exposed to fipronil. Titin is a massive protein involved in the assembly and function of vertebrate striated muscles and plays a central role in muscle contraction and elasticity (Labeit and Kolmerer, 1995). Telethonin, encoded by the titin-cap (TCAP) gene, is involved in the assembly of titin and the anchoring of the Z disk of the sarcomere (Mues et al., 1998; Valle et al., 1997; Zou et al., 2006). Both titin and telethonin were significantly up-regulated in embryos exposed to fipronil compared to the vehicle control. Supporting the result, alternation of the molecular functions of actin binding and cytoskeletal protein binding (muscle-related processes) were detected by GSEA. This is in accord with the hypothesis proposed earlier explaining the tail curvature observed in so many of the larvae exposed to the high concentrations of fipronil. Abnormal muscle development was not directly lethal to the fish in our experiments, however it can dramatically decrease survival of larval fish as it negatively affects their swimming ability (Teh et al., 2004).
The RNA-seq also revealed that the fipronil exposure altered expression of genes belonging to other categories, such as cardiac function. One gene for which expression was enhanced by fipronil was ANKRD1, a marker for early differentiation of cardiac myogenesis, involved in cardiac muscle function (Arimura et al., 2009). ANKRD1 encodes cardiac ankyrin-repeat domain containing protein (CARP), a transcription factor which translocates to the nucleus in response to mechanical stress (Moulik et al., 2009). Aihara et al. (2000) identified CARP as a marker of cardiac hypertrophy. Given the link between cardiac effects and CARP reported in the literature, it is possible that cardiac function was compromised by fipronil exposure. In this study hypertrophy of cardiac muscle or other dysfunction was not observed by gross examination. However, further analyses such as visualization of cardiac muscle by whole mount in situ hybridization or development of transgenic fish with GFP expressing in the heart may provide more insight into effects of fipronil on cardiac function.
Our results regarding the concentrations which cause impairment of swimming ability are congruent with the previous findings reported by Beggel et al. (2010); swimming ability was disrupted in larval Fathead Minnow following exposure to fipronil at concentrations of 142 μg L−1 and higher. Thus, at similar concentrations, fipronil caused similar effects to both embryonic and larval fish. Findings in both studies suggest that the fipronil mechanism of impairing swimming ability is not species- or life stage-specific.
A delay in hatching, an outcome of fipronil exposure as shown in this study, has several potential consequences. The nutritional condition of larvae can be worsened following delayed hatching time (Semmens and Swearer, 2011), possibly leading to starvation after hatching. This is a possibility due to yolk reserve depletion following prolonged incubation. During this extended incubation period, metabolism, growth, and development continue in the embryo, all of which are fueled by energy reserves in the yolk sac. When hatching does occur, the yolk sac is smaller and the larvae are in poor nutritional condition, and each adversely affects the odds of survival (Semmens and Swearer, 2011). The effects of delayed hatching are not well known physiologically or ecologically, highlighting the need for more studies to better understand the consequences of this effect.
5. Conclusion
In this study, we found that embryos exposed to fipronil at high concentrations exhibited impaired embryonic development by causing various adverse effects. Although adverse effects to Medaka embryos was not observed at the current environmentally relevant concentrations of fipronil, aquatic organisms may be exposed to high doses of fipronil by accidental spills and illegal use. In addition, data from the high dose exposures allows us to establish signatures that are related to mechanisms of action of fipronil. This has partially been achieved with RNA-seq in this study, however further investigation on mechanisms of action will pinpoint how exactly the physiological effects surface. Such results will facilitate the hazard identification and will guide regulators as to which concentrations are the threshold for acceptable embryo exposure.
Supplementary Material
Highlights.
Fish embryos exposed to fipronil exhibited impaired embryonic development.
The adverse effects include decreased hatching success and tail curvature.
Fipronil exposure enhanced expression of genes involved in muscle development.
Acknowledgments
Funding for the study was provided by the Aquatic Health Program at UC Davis. Partial support was provided by the Department of Pesticide Regulation #13-C0029. For quantification of fipronil by ELISA, we received funding support from National Institute of Environmental Health Sciences, Superfund Research Program, P42 ES04699 and the National Institute for Occupational Safety and Health Western Regional Center for Agricultural Health Science U50 OH07550. The authors thank Dr. Andrew Whitehead at University of California, Davis, for his constructive suggestions on RNA-seq and GSEA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors’ contributions
SDW performed fish embryo exposure test, participated in data analyses, and wrote the manuscript. BGH guided the frequentist analyses and the model comparisons. NV and SJG performed ELISA with funding support from BDH. CL performed RT-qPCR with guidance with TK. GW carried out the transcriptome analysis (RNA-seq and GSEA) with intellectual support from TK. BGH, TK, and SJT provided suggestions on experimental design and revisions on the manuscript. SJT supervised the research and provided funding support for fish embryo exposure test, statistical analyses, and gene expression analysis. All the co-authors contributed to revision of the manuscript. All the authors read and approve the final manuscript.
Supplemental data
Supplemental data associated with this article are available at the end of the manuscript.
References
- Aihara Y, Kurabayashi M, Saito Y, Ohyama Y, Tanaka T, Takeda S, Tomaru K, Sekiguchi K, Arai M, Nakamura T, Nagai R. Cardiac ankyrin repeat protein is a novel marker of cardiac hypertrophy. Hypertension. 2000;36:48–53. doi: 10.1161/01.hyp.36.1.48. [DOI] [PubMed] [Google Scholar]
- Amweg EL, Weston DP, You J, Lydy MJ. Pyrethroid insecticides and sediment toxicity in urban creeks from California and Tennessee. Environ Sci Tech. 2006;40:1700–1706. doi: 10.1021/es051407c. [DOI] [PubMed] [Google Scholar]
- Anderson DR, Burnham KP, Thompson WL. Null hypothesis testing: problems, prevalence, and an alternative. J Wildlife Manage. 2000;64:912–923. [Google Scholar]
- Arimura T, Bos JM, Sato A, Kubo T, Okamoto H, Nishi H, Harada H, Koga Y, Moulik M, Doi YL, Towbin JA, Ackerman MJ, Kimura A. Cardiac ankyrin repeat protein gene (ANKRD1) mutations in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2009;54:334–342. doi: 10.1016/j.jacc.2008.12.082. [DOI] [PubMed] [Google Scholar]
- ASTM Standard E1241-05, 2013. Standard guide for conducting early life-stage toxicity tests with fishes. ASTM International; West Conshohocken, PA: 2013. www.astm.org. [Google Scholar]
- Beggel S, Werner I, Connon RE, Geist JP. Sublethal toxicity of commercial insecticide formulations and their active ingredients to larval fathead minnow (Pimephales promelas) Sci Total Environ. 2010;408:3169–3175. doi: 10.1016/j.scitotenv.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Beggel S, Werner I, Connon RE, Geist JP. Impacts of the phenylpyrazole insecticide fipronil on larval fish: Time-series gene transcription responses in fathead minnow (Pimephales promelas) following short-term exposure. Sci Total Environ. 2012;426:160–165. doi: 10.1016/j.scitotenv.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Bencic DC, Villeneuve DL, Biales AD, Blake L, Durhan EJ, Jensen KM, Kahl MD, Makynen EA, Martinovic-Weigelt D, Ankley GT. Effects of the insecticide fipronil on reproductive endocrinology in the fathead minnow. Environ Toxicol Chem. 2013;32:1828–1834. doi: 10.1002/etc.2254. [DOI] [PubMed] [Google Scholar]
- Bloomquist JR. Ion channels as targets for insecticides. Annu Rev Entomol. 1996;41:163–190. doi: 10.1146/annurev.en.41.010196.001115. [DOI] [PubMed] [Google Scholar]
- Bolker BM, Brooks ME, Clark CJ, Geange SW, Poulsen JR, Stevens MHH, White JSS. Generalized linear mixed models: a practical guide for ecology and evolution. Trends Ecol Evol. 2009;24:127–135. doi: 10.1016/j.tree.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. Model selection and multimodel inference: a practical information-theoretic approach. second. Springer; New York: 2002. [Google Scholar]
- Cheng SK, Kong-Wai AW, Hung-So C, Sun-Wu RS. Cellular and molecular basis of cadmium-induced deformities in zebrafish embryos. Environ Toxicol Chem. 2000;19:3024–3031. doi: 10.1002/etc.5620191223. [DOI] [Google Scholar]
- Cole LM, Nicholson RA, Casida JE. Action of phenylpyrazole insecticides at the GABA-gated chloride channel. Pestic Biochem Phys. 1993;46:47–54. [Google Scholar]
- Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2006;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- Gan J, Bondarenko S, Oki L, Haver D, Li JK. Occurrence of fipronil and its biologically active derivatives in urban residential runoff. Envir Sci Tech. 2012;46:1489–1495. doi: 10.1021/es202904x. [DOI] [PubMed] [Google Scholar]
- Hainzl D, Cole LM, Casida JE. Mechanisms for selective toxicity of fipronil insecticide and its sulfone metabolite and desulfinyl photoproduct. Chem Res Toxicol. 1998;11:1529–1535. doi: 10.1021/tx980157t. [DOI] [PubMed] [Google Scholar]
- Hammock BG, Lesmeister S, Flores I, Bradburd GS, Hammock FH, Teh SJ. Low food availability narrows the tolerance of the copepod Eurytemora affinis to salinity but not to temperature. Estuar Coast. 2015:1–12. doi: 10.1007/s12237-015-9988-5. [DOI] [Google Scholar]
- Hood RD. Handbook of Developmental Toxicology. first. CRC Press; Florida: 1996. [Google Scholar]
- Horning WB, Weber CI. Short-term methods for estimating the chronic toxicity of effluents and receiving waters to freshwater organisms. EPA/600/4-85/014. 1985:58–75. [Google Scholar]
- Jiang W, Lin K, Haver D, Qin S, Ayre G, Spurlock F, Gan J. Wash-off potential of urban use insecticides on concrete surfaces. Environ Toxicol Chem. 2010;29:1203–1208. doi: 10.1002/etc.184. [DOI] [PubMed] [Google Scholar]
- Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, Tamada T, Nagayasu Y, Doi K, Kasai Y, Jindo T, Kobayashi D, Shimada A, Toyoda A, Kuroki Y, Fujiyama A, Sasaki T, Shimizu A, Asakawa S, Shimizu N, Hashimoto S, Yang J, Lee Y, Matsushima K, Sugano S, Sakaizumi M, Narita T, Ohishi K, Haga S, Ohta F, Nomoto H, Nogata K, Morishita T, Endo T, Shin-I T, Takeda H, Morishita S, Kohara Y. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M, Murata K, Naruse K, Tanaka M. Medaka: biology, management, and experimental protocols. first. Wiley-Blackwell; Iowa: 2009. [Google Scholar]
- Labeit S, Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Lu C, Wang J, Hu W, Cao Z, Sun D, Xia H, Ma X. Developmental mechanisms of arsenite toxicity in zebrafish (Danio rerio) embryos. Aquat Toxicol. 2009;91:229–237. doi: 10.1016/j.aquatox.2008.11.007. [DOI] [PubMed] [Google Scholar]
- McKim J. Evaluation of tests with early life stages of fish for predicting long-term toxicity. J Fish Res Board Can. 1977;34:1148–1154. [Google Scholar]
- Mize SV, Porter SD, Demcheck DK. Influence of fipronil compounds and rice-cultivation land-use intensity on macroinvertebrate communities in streams of southwestern Louisiana, USA. Environ Pollut. 2008;152:491–503. doi: 10.1016/j.envpol.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Moulik M, Vatta M, Witt SH, Arola AM, Murphy RT, McKenna WJ, Boriek AM, Oka K, Labeit S, Bowles NE, Arimura T, Kimura A, Towbin JA. ANKRD1, the gene encoding cardiac ankyrin repeat protein, is a novel dilated cardiomyopathy gene. J Am Coll Cardiol. 2009;54:325–333. doi: 10.1016/j.jacc.2009.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mues A, van der Ven PFM, Young P, Furst DO, Gautel M. Two immunoglobulinlike domains of the Z-disc portion of titin interact in a conformation-dependent way with telethonin. FEBS Lett. 1998;428:111–114. doi: 10.1016/s0014-5793(98)00501-8. [DOI] [PubMed] [Google Scholar]
- Nillos MG, Kunde L, Gan J, Bondarenko S, Schlenk D. Enantioselectivity in fipronil aquatic toxicity and degradation. Environ Toxicol Chem. 2009;28:1825–1833. doi: 10.1897/08-658.1. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. https://www.R-project.org/ [Google Scholar]
- Ramírez-Duarte WF, Jin J, Kurobe T, Teh SJ. Effects of prolonged exposure to low pH on enzymatic and non-enzymatic antioxidants in Japanese Medaka (Oryzias latipes) Sci Total Environ. 2016;568:26–32. doi: 10.1016/j.scitotenv.2016.05.179. [DOI] [PubMed] [Google Scholar]
- Rochman CM, Kurobe T, Flores I, Teh SJ. Early warning signs of endocrine disruption in adult fish from the ingestion of polyethylene with and without sorbed chemical pollutants from the marine environment. Sci Total Environ. 2014;493:656–661. doi: 10.1016/j.scitotenv.2014.06.051. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Semmens D, Swearer SE. Extended incubation affects larval morphology, hatching success and starvation resistance in a terrestrially spawning fish, Galaxias maculatus. J Fish Biol. 2011;79:980–990. doi: 10.1111/j.1095-8649.2011.03074.x. [DOI] [PubMed] [Google Scholar]
- Simon-Delso N, Amaral-Rogers V, Belzunces LP, Bonmatin JM, Chagnon M, Downs C, Furlan L, Gibbons DW, Giorio C, Girolami V, Goulson D, Kreutzweiser DP, Krupke CH, Liess M, Long E, McField M, Mineau P, Mitchell EA, Morrissey CA, Noome DA, Pisa L, Settele J, Stark JD, Tapparo A, Van Dyck H, Van Praagh J, Van der Sluijs JP, Whitehorn PR, Wiemers M. Systemic insecticides (neonicotinoids and fipronil): trends, uses, mode of action and metabolites. Environ Sci Pollut R. 2015;22:5–34. doi: 10.1007/s11356-014-3470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitsbergen JM, Walker MK, Olson JR, Peterson RE. Pathologic alterations in early life stages of lake trout, Salvelinus namaycush, exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin as fertilized eggs. Aquat Toxicol. 1991;19:41–72. [Google Scholar]
- Sprague LA, Nowell LH. Comparison of pesticide concentrations in streams at low flow in six metropolitan areas of the United States. Environ Toxicol Chem. 2008;27:288–298. doi: 10.1897/07-276R.1. [DOI] [PubMed] [Google Scholar]
- Stehr CM, Linbo TL, Incardona JP, Scholz NL. The developmental neurotoxicity of fipronil: notochord degeneration and locomotor defects in zebrafish embryos and larvae. Toxicol Sci. 2006;92:270–278. doi: 10.1093/toxsci/kfj185. [DOI] [PubMed] [Google Scholar]
- Teh SJ, Deng X, Teh FC, Hung SO. Selenium-induced teratogenicity in Sacramento Splittail (Pogonichthys macrolepidotus) Mar Environ Res. 2002;54:605–608. doi: 10.1016/s0141-1136(02)00152-6. [DOI] [PubMed] [Google Scholar]
- Teh SJ, Zhang GH, Kimball T, Teh FC. Lethal and sublethal effects of esfenvalerate and diazinon on Sacramento Splittail larvae. In: Feyrer F, Brown L, Orsi J, Brown R, editors. Early life history of fishes in the San Francisco Estuary and watershed. Am. Fish. S. S. Bethesda; Maryland: 2004. [Google Scholar]
- Tingle C, Rother JA, Dewhurst CF, Lauer S, King WJ. Fipronil: Environmental Fate, Ecotoxicology and Human Health Concerns. Rev Environ Contam Toxicol. 2003;176:1–66. doi: 10.1007/978-1-4899-7283-5_1. [DOI] [PubMed] [Google Scholar]
- Takada F, Woude DLV, Tong HQ, Thompson TG, Watkins SC, Kunkel LM, Beggs AH. Myozenin: an α-actinin- and γ-filamin-binding protein of skeletal muscle Z lines. P Natl Acad Sci USA. 2001;98:1595–1600. doi: 10.1073/pnas.041609698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-Seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong L, Harper SL, Tanguay RL. Evaluation of embryotoxicity using the zebrafish model. Method Mol Biol. 2011;691:271–279. doi: 10.1007/978-1-60761-849-2_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US EPA. Pesticide fact sheet: fipronil. Office of Prevention, Pesticides, and Toxic Substances; Washington, DC: 1996. p. 8. EPA 737-F-96-005. [Google Scholar]
- Valle G, Faulkner G, DeAntoni A, Pacchioni B, Pallavicini A, Pandolfo D, Tiso N, Toppo S, Trevisan S, Lanfranchi G. Telethonin, a novel sarcomeric protein of heart and skeletal muscle. FEBS Lett. 1997;415:163–168. doi: 10.1016/s0014-5793(97)01108-3. [DOI] [PubMed] [Google Scholar]
- Vasylieva N, Ahn KC, Barnych B, Gee SJ, Hammock BD. Development of an immunoassay for the detection of the phenylpyrazole insecticide fipronil. Envir Sci Tech. 2015;49:10038–10047. doi: 10.1021/acs.est.5b01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Shimada A, Fukamachi S, Naruse K, Shima A. Sex-linked inheritance of the lf locus in the Medaka fish (Oryzias latipes) Zool Sci. 1998;15:123–126. doi: 10.2108/zsj.15.123. [DOI] [PubMed] [Google Scholar]
- Weston DP, Holmes RW, You J, Lydy MJ. Aquatic toxicity due to residential use of pyrethroid insecticides. Envir Sci Tech. 2005;39:9778–9784. doi: 10.1021/es0506354. [DOI] [PubMed] [Google Scholar]
- Weston DP, Holmes RW, Lydy MJ. Residential runoff as a source of pyrethroid pesticides to urban creeks. Environ Pollut. 2009;157:287–294. doi: 10.1016/j.envpol.2008.06.037. [DOI] [PubMed] [Google Scholar]
- Wirth EF, Pennington PL, Lawton JC, DeLorenzo ME, Bearden D, Shaddrix B, Sivertsen S, Fulton MH. The effects of the contemporary-use insecticide (fipronil) in an estuarine mesocosm. Environ Pollut. 2004;131:365–371. doi: 10.1016/j.envpol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Hu J. Development and validation of endogenous reference genes for expression profiling of Medaka (Oryzias latipes) exposed to endocrine disrupting chemicals by quantitative Real-Time RT-PCR. Toxicol Sci. 2007;95:356–368. doi: 10.1093/toxsci/kfl161. [DOI] [PubMed] [Google Scholar]
- Zhao X, Yeh JZ, Salgado VL, Narahashi T. Fipronil is a potent open channel blocker of glutamate-activated chloride channels in cockroach neurons. J Pharmacol Exp Ther. 2004;310:192–201. doi: 10.1124/jpet.104.065516. [DOI] [PubMed] [Google Scholar]
- Zou P, Pinotsis N, Lange S, Song YH, Popov A, Mavridis I, Mayans OM, Gautel M, Wilmanns M. Palindromic assembly of the giant muscle protein titin in the sarcomeric Z-disk. Nature. 2006;439:229–233. doi: 10.1038/nature04343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


