Abstract
Background and aim: Understanding the molecular biological mechanisms underlying laryngeal squamous cell carcinoma (LSCC) invasion and metastasis is crucial for diagnosis, treatment, and prognosis. We aimed to examine the expression of the tumor suppressor microRNA-204-5p (miR-204-5p) and its target gene, forkhead box C1 (FOXC1), in human LSCC and explore their roles in the malignant behaviors of LSCC Hep-2 and TU-177 cells.
Methods: The regulatory effects of miR-204-5p on the 3′ untranslated region of FOXC1 predicted by bioinformatics were tested by dual-luciferase reporter assay. Quantitative RT-PCR was used to detect mRNA expression in 43 fresh samples of LSCC and corresponding adjacent normal mucosa (ANM). FOXC1 protein expression was examined by immunohistochemistry. miR-204-5p mimics and FOXC1 siRNA were transfected into LSCC cell lines Hep-2 and TU-177 to observe malignant behavior. miR-204-5p mimics were injected into Hep-2 or TU-177 xenograft tumors in nude mice to examine tumor growth.
Results: The miR-204-5p mRNA level was lower in all 43 LSCC samples than in the ANM samples, but the FOXC1 level was higher in the LSCC samples than in the ANM samples. The miR-204-5p level was lower for stage III and IV cancer and lymph node N+ status samples than for stage I and II cancer and N0 status samples. FOXC1 mRNA and protein levels were higher for N+ than for N0 LSCC. The miR-204-5p mRNA levels were lower in Hep-2 and TU-177 cells than in ANM tissues, but FOXC1 mRNA levels were higher in Hep-2 and TU-177 cells than in ANM tissues. Dual-luciferase reporter assays demonstrated the targeted regulatory effects of miR-204-5p on the FOXC1 3′ UTR. Cell proliferation and colony formation was facilitated with miR-204-5p mimics and FOXC1 siRNA, with weaker cell migration and invasion than the controls. Moreover, miR-204-5p overexpression or FOXC1 knockdown inhibited the EMT process in LSCC cells. In vivo experiments demonstrated that injection of miR-204-5p into Hep-2 and TU-177 xenograft tumors in nude mice significantly inhibited tumor growth.
Conclusions: miR-204-5p is involved in the invasion and metastasis of LSCC. It has a targeted regulatory effect on FOXC1 expression; malignant LSCC behaviors, including cell proliferation, invasion, and metastasis, are suppressed, and tumor growth in vivo is inhibited. This suggests that miR-204-5p may be a target for molecular therapy of LSCC in the future.
Keywords: hsa-miR-204-5p, FOXC1, laryngeal squamous cell carcinoma, invasion, metastasis
Introduction
Head and neck squamous cell carcinoma (SCC) is the sixth most common cancer worldwide 1. Laryngeal SCC (LSCC) originates from the laryngeal mucosa epithelium and is the second most common head and neck SCC 2. Recently, the worldwide incidence of LSCC has slightly increased, with an upward trend among younger populations 3-6. In northern China, including Shanxi Province, the incidence of LSCC is high 7, 8. Given the anatomical structure of the larynx and its rich submucosal lymphatic structures, LSCC exhibits malignant biological behaviors, including local invasion and cervical lymph node metastasis 9, that are important risk factors for recurrence and poor prognosis 10. Over the past 3 decades, the 5-year survival rate for LSCC has not markedly improved 4-6. Investigating the molecular biological mechanisms underlying LSCC invasion and metastasis is crucial to the diagnosis, treatment, and prognosis of the disease 7.
MicroRNAs (miRNAs) are a class of recently discovered small endogenous non-coding single-stranded RNAs of approximately 22 nt. They exist in eukaryotes and can bind to the 3′ untranslated region (3′ UTR) of target mRNAs by degrading the mRNA or inhibiting translation. miRNAs are closely associated with tumor activity. They act as oncogenes or tumor suppressors and play important roles in tumor cell growth, proliferation, differentiation, apoptosis, the epithelial-mesenchymal transition (EMT), and stem cell differentiation 11-13. For example, Let-7a 14, 15, miR-7 16, miR-2117, 18, miR-31 19, miR-133a 20, miR-137 21, miR-138 22, miR-218 23, miR-148a 24, and miR-489 25, among others, play essential roles in tumor development.
Thousands of miRNAs have been identified, but their molecular biological functions and related mechanisms in LSCC development remain unknown. miRNAs show a tissue-specific distribution 11; miRNA microarray is an important method for screening differential miRNAs. Our pilot experiments with Agilent human miRNA Microarray 19.0 showed that miR-204-5p expression was significantly lower in LSCC compared to adjacent normal mucosa (ANM) tissue 26. These findings were consistent with other gene chip results 27-30. A few studies have shown that miR-204-5p can inhibit tumor cell proliferation, migration, invasion, and metastasis 28-33, suggesting it may be a tumor suppressor in LSCC.
Our bioinformatics analysis revealed that the 3′ UTR of the transcription factor Forkhead box C1 (FOXC1) contains two binding sites for miR-204-5p. Proteins in the FOX family act as nuclear transcription factors and are widely involved in tumor cell proliferation, differentiation, apoptosis, metastasis, metabolism, and other biological processes 34. FOXC1 might be a risk factor for LSCC 35. FOXC1 can promote cancer cell proliferation, migration, and invasion and may play an important role in EMT 36-39. However, the biological role of FOXC1 in LSCC remains unexplored.
We hypothesized that miR-204-5p might play an important role in LSCC invasion and metastasis by regulating FOXC1. To test this hypothesis, we compared transcription levels in 43 LSCC and corresponding ANM samples, examined FOXC1 protein expression in LSCC and analyzed the association of LSCC clinical pathology parameters with miR-204-5p and FOXC1 expression. The regulatory effect of miR-204-5p on FOXC1 was verified by dual-luciferase reporter assay. We examined the proliferation, colony formation, migration, and invasion of LSCC cell lines Hep-2 and TU-177 using gene transfection to restore miR-204-5p expression. Finally, we assessed the effect of miR-204-5p on tumor growth in a nude mouse xenograft model. These findings may provide a theoretical basis for the development of molecular targeted therapy for LSCC.
Materials and Methods
Ethics statement
The study was approved by the medical ethics committee of Shanxi Medical University and complied with the Declaration of Helsinki. Informed consent per institutional guidelines was obtained from all patients who agreed to participate in this study.
Clinical samples and patient population
We obtained 43 frozen LSCC and paired corresponding ANM tissues from patients undergoing surgery at the Department of Otolaryngology Head and Neck Surgery of The First Hospital Affiliated with Shanxi Medical University. LSCC was diagnosed by histology and was untreated by radiotherapy or chemotherapy. ANM tissues were isolated from surgical excision specimens at least 1 cm from the neoplastic edge. The fresh specimens were divided into 2 samples: 1 was placed in liquid nitrogen for quantitative RT-PCR (qRT-PCR), and the other was embedded in paraffin for hematoxylin and eosin staining to confirm the diagnosis of LSCC and ANM. Staging followed the Tumor, Node, Metastasis (TNM) staging system of the Union for International Cancer Control (UICC, 2010). The histological types of LSCC were determined by the World Health Organization (WHO) system. A summary of the study population and clinicopathological variables is shown in Table 1.
Table 1.
Demographics and clinical features of 43 patients with laryngeal squamous cell carcinomas.
| Parameter | No. (%) |
|---|---|
| Age, years (range 45-73) | |
| >59 | 24 (55.8) |
| ≤59 | 19 (44.2) |
| Gender | |
| Male | 39 (90.7) |
| Female | 4 (9.3) |
| Cancer differentiation | |
| Well | 18 (41.9) |
| Moderate or poor | 25 (58.1) |
| Tumor stage | |
| T1+T2 | 25 (58.1) |
| T3+T4 | 18 (41.90) |
| Cervical lymph node status | |
| N0 | 24 (55.8) |
| N+ | 19 (44.2) |
| Clinical stage | |
| I + II | 14 (32.6) |
| III + IV | 29 (67.4) |
| Tumor site | |
| Supraglottis + subglottis | 16 (37.2) |
| Glottis | 27 (62.8) |
Cell culture and transfection
The human laryngeal cancer cell lines Hep-2 (China Center for Type Culture Collection, Wuhan, China) and TU-177 (Bioleaf Biotech Co., Shanghai, China) were maintained in RPMI-1640 or DMEM/F12 (1:1) containing 10% heat-inactivated fetal bovine serum (Biological Industries, CT, USA), 100 U/mL penicillin, and 0.1 mg/mL streptomycin in a humidified atmosphere of 5% CO2 and 95% air at 37°C.
siRNA and miRNA mimics transfection
siRNA targeting FOXC1 (FOXC1-siRNA) and negative control siRNA (NC) were designed and manufactured by Genechem Co. (Shanghai, China). The siRNA sequences are shown in Table S1. Cells were transiently transfected with the hsa-miR-204-5p mimic (UUCCCUUUGUCAUCCUAUGCCU) or negative control mimic (NC) (UUUGUACUACACAAAAGUACUG) (RiboBio Co., Guangzhou, China) or with FOXC1-siRNA or NC siRNA. Briefly, cells were seeded in 6-well plates 24 h before transfection and were transiently transfected at 30% to 40% confluency. The transfection was conducted using Lipofectamine RNAi MAX Transfection Reagent (Invitrogen, Carlsbad, CA, USA) following manufacturer's instructions.
Purification of total RNA and qRT-PCR (qPCR)
Total RNA was purified from frozen tissues or cells using TRIzol (Invitrogen). The miRNA-specific or mRNA reverse transcription was performed using the miRNA first-strand cDNA synthesis kit (BLKW Biotechnology Co., Beijing, China) or the ReverTra Ace qPCR RT Kit (TOYOBO, Shanghai, China) with 1 μg total RNA. PCR amplification was performed using the SYBR Green Realtime PCR Master Mix (TOYOBO) with the ABI PRISM 7500 Sequence Detection System (Applied Biosystems, CA, USA). The cycling conditions included a 5-min initial denaturation at 95°C, followed by 40 cycles at 95°C for 15 s, 60°C for 15 s, and 72°C for 32 s. The stem-loop primer sequence for hsa-miR-204-5p was 5'-ACACTCCAGCTGGGTTCCCTTTGTCATCCTAT-3'; the reverse primer for hsa-miR-204-5p was 5'-CTCAACTGGTGTCGTGGA -3' (72 bp). The following qPCR primers for FOXC1 were used: forward, 5'-CATTTTGGTCTAGGGTGGTTTC-3'; reverse, 5'-TCTGATTGGCAGGGCAGAT -3' (156 bp). The primer sequences for internal controls U6 and 18S RNA were described previously 40, 41. All reactions were performed in triplicate, and the experiment was repeated 3 times. The relative expression of hsa-miR-204-5p and FOXC1 was analyzed using the 2-ΔΔCT method 42.
Antibodies and immunohistochemistry staining
Paraffin sections were deparaffinized and dehydrated in a graded series of ethanol solutions. Samples were processed in an autoclave for 2 min and 10 sec with EDTA, pH 9.0, for FOXC1 antigen retrieval. Endogenous peroxidase activity was blocked by immersing tissue sections in 3% H2O2 in methanol (v/v) at room temperature for 10 min and washed with phosphate buffered saline (PBS). Nonspecific background staining was reduced by incubating sections with normal nonimmune serum (Boster Co., Wuhan, China) for 15 min at room temperature. The sections in a moist chamber were incubated overnight at 4°C with the FOXC1 antibody (ab97742, 1:150) (Abcam, Cambridge, UK) and washed 3 times with PBST for 5 min each. Sections were incubated with the secondary antibody from the Super Vision (SV) HRP-Polymer anti-Goat IHC kit (Boster Co., Wuhan, China) for 15 min at room temperature and washed 3 times for 5 min with PBST. The DAB substrate detection system was used (Boster Co.). Counterstaining was performed using hematoxylin, and sections were dehydrated and mounted with coverslips.
Evaluation and scoring of immunohistochemistry
The FOXC1 expression was semiquantitatively scored using a mean EI (E×I) score, which combined the extent (E) of staining (percentage of positive tumor cells) with the staining intensity (I, unequivocally positive immunohistochemical staining of tumor cells). Extent (E) was graded on a scale from 0 to 3: 0 = negative, 1 = 1-30%, 2 = 31-60%, 3 > 60%. Intensity (I) was graded on a scale of 0-3: 0 = none, 1 = weak staining, 2 = moderate staining, 3 = strong staining. The mean EI score (varying from 0 to 9) was calculated for each LSCC specimen. EI scores of 0-3 were considered low expression, and scores > 3 were considered high expression. Each immunohistochemically stained section was independently evaluated by two pathologists, with agreement reached by consensus. The slides were coded, and evaluators remained unaware of the code to avoid observer bias. To determine the expression pattern, at least 10 high-power fields (40×10) and at least 100 cancer cells per field were randomly selected for microscopy. Non-small-cell lung cancer was selected as an appropriate positive control for FOXC1 38. Negative controls were generated by substituting the primary antibody with PBS.
miRNA target prediction
TargetScan Human Release 7.1 (http://www.targetscan.org), the database miRDB (http://mirdb.org/miRDB/), and microRNA.org (http://www.microrna.org) were used to analyze the potential hsa-miR-204-5p binding site in the FOXC1 3′ UTR.
Luciferase assay
Luciferase assays were performed in 293T cells (ATCC, Manassas, VA, USA). The luciferase reporter plasmids were constructed using the psi-CHECK2 vector (Promega Biotech Co., Ltd, USA) with 3'UTR-FOXC1 or 3'UTR-mut-FOXC1-1/2/3 (Table S2). Cells were separately transfected or co-transfected with 50 nM hsa-miR-204-5p mimics and 0.5 μg plasmid. Cells were harvested and lysed for luciferase assays after transfection for 48 h. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). All experiments were performed in triplicate.
Western blot analysis
Whole cell lysates were prepared using the Mammalian Protein Extraction Kit (Cwbiotech, Beijing, China). Protein concentrations were determined using the Bicinchoninic Acid (BCA) protein assay kit (Cwbiotech). Protein samples with 2× loading buffer (0.25 mol/L Tris-Cl, pH 6.8, 10% SDS, 0.5% bromophenol blue, and 50% glycerol) were boiled for 5 min, loaded into 10% Tris-HCl polyacrylamide gels (80 V, 60 min), and transferred to Immobilon-P Transfer Membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5% nonfat milk at 4°C overnight, incubated with goat polyclonal antibody for FOXC1 (ab97742, Abcam) overnight at 4°C, and incubated with horseradish peroxidase (HRP)-conjugated rabbit anti-goat IgG(H+L) (Southern Biotech, Birmingham, AL, USA) for 2 h at room temperature. Immunoreactive bands were detected and developed using the Immobilon Western Chemiluminescent HRP substrate (Millipore). HRP-conjugated GAPDH monoclonal antibody was used as the loading control (KangChen Bio-tech, Shanghai, China). Grayscale analysis was conducted with Quantity One 4.0 (Bio-Rad, Hercules, CA, USA). All experiments were performed in triplicate.
Cell growth analysis
MTS and colony formation assays were used to evaluate cell growth. Briefly, cells undergoing digestion were seeded in 96-well plates at 1 × 105/ml and 100 μl/well. The CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega Biotech Co., USA) was performed after incubation for 0, 24, 48, and 72 h. After 4 h of incubation at 37°C with 5% CO2, absorption was measured at 490 nm with a Multiscan MK3 automatic ELISA reader (Thermo Fisher Scientific, Hudson, NH, USA). The growth rate was calculated as (OD-0 h OD/0 h OD)×100% and data were normalized by cell group. Each treatment was carried out in triplicate.
Colony formation assay
Cells were seeded at 200 cells/well in 96-well plates, moved to the cell incubator for 7 days and stained with crystal violet. The number of colony-forming cells was calculated using the AID iSpot Reader (AID Diagnostika, GmbH, Germany). The colony formation rate was calculated as the number of colonies/number of seeded cells ×100%, and data were normalized by cell group. Each treatment was carried out in triplicate.
Scratch assay
Lines at intervals of 0.5-1 cm were created in six-well plates using a marker pen. Each well had at least 5 lines. Cells, which were transfected for 12 h, were seeded onto plates at 1×106/well and allowed to adhere up to 60%-70% confluency, incubated with 5-20 μg/mL mitomycin (Sigma, MO, USA) for 1 h at 37°C to inhibit cellular division, and washed with fresh serum-free medium 3 times. A straight line (i.e., “scratch”) was created in the confluent cell monolayer using an aseptic 200-μL pipette tip, and cells were washed 3 times with fresh serum-free medium to clear cell debris and suspension cells. Fresh serum-free medium was added, and cells were allowed to migrate for 48 h to close the wound. Images were obtained at 0, 6, 24 and 48 h in the same position per the reference lines and at the same magnification. Each sample was performed in triplicate.
Cell invasion assay
Cells were transfected for 24 h and suspended in 100 μL serum-free medium. Then, 1×105 cells were seeded into the top chamber of a Matrigel-coated BD Falcon Cell Culture Insert, containing polyethylene terephthalate and 8-μm pores (BD Biosciences, San Jose, CA, USA), in a 24-well BD Falcon Companion TC Plate (BD Biosciences). The bottom chamber was filled with 600 μL medium with 10% FBS as a chemoattractant. After incubation with 5% CO2 at 37°C for 48 h, cells on the top surface of the chamber were carefully removed with a cotton swab, and cells on the bottom surface of the chamber were fixed with 4% paraformaldehyde, stained with crystal violet, and eluted with 33% acetic acid. Invasiveness was quantified by determination of OD570 absorbance, and data were normalized by cell group. Each sample was tested in triplicate.
Animal experiments
Animal experimental procedures followed the Health Guide for the Care and Use of Laboratory Animals approved by the medical ethics committee of Shanxi Medical University. Six SPF-grade male BALB/C nude mice, aged 7 weeks, were purchased from SLRC Laboratory Animal Co. (Changsha, Hunan, China, certificate No. HNASLKJ20120391). SPF-grade mouse feed was purchased from Guangdong Medical Laboratory Animal Center (Certificate No. 0088859). The nude mice were maintained in the laboratory for 1 week and used for further experiments if no abnormal behaviors were observed. Mice were subcutaneously injected with 2×105 Hep-2 (3 mice) or TU-177 (3 mice) cells on the dorsal sides of the left and right thighs (0.1 ml/mouse). The tumor sizes were measured twice a week with calipers, and the volume was determined by long diameter × short diameter2/243. Once tumors were formed, mice were intratumorally injected with drugs as previously described 44-46. Briefly, the left or right site was injected with 10 nmol NC-agomir or miR-204-5p agomir (2'-O-methyl +5' cholesterol modification, GenePharma, Shanghai, China) with 0.1 mL Invivofectamine 2.0 (Invitrogen) once per week per site for 5 weeks; the mice were then killed. Tumor bodies were harvested, weighed, and photographed at the end of treatment.
Statistical analysis
Statistical analysis was performed using SPSS v19.0 (SPSS Inc., Chicago, IL, USA). Pearson's chi-square or Fisher's exact tests were used to assess the association between the clinicopathological variables and immunohistochemical status. The association between the clinical and histopathological variables and FOXC1 immunohistochemical staining was investigated using the Mann-Whitney test. Correlations between the miR-204-5p level and FOXC1 mRNA and protein levels were determined by bivariate correlation. Quantitative data were analyzed by t-test. For all tests, a two-tailed P ≤ 0.05 was considered significant.
Results
Expression of miR-204-5p in LSCC and ANM tissue and association with cervical lymph node metastasis and clinical stage
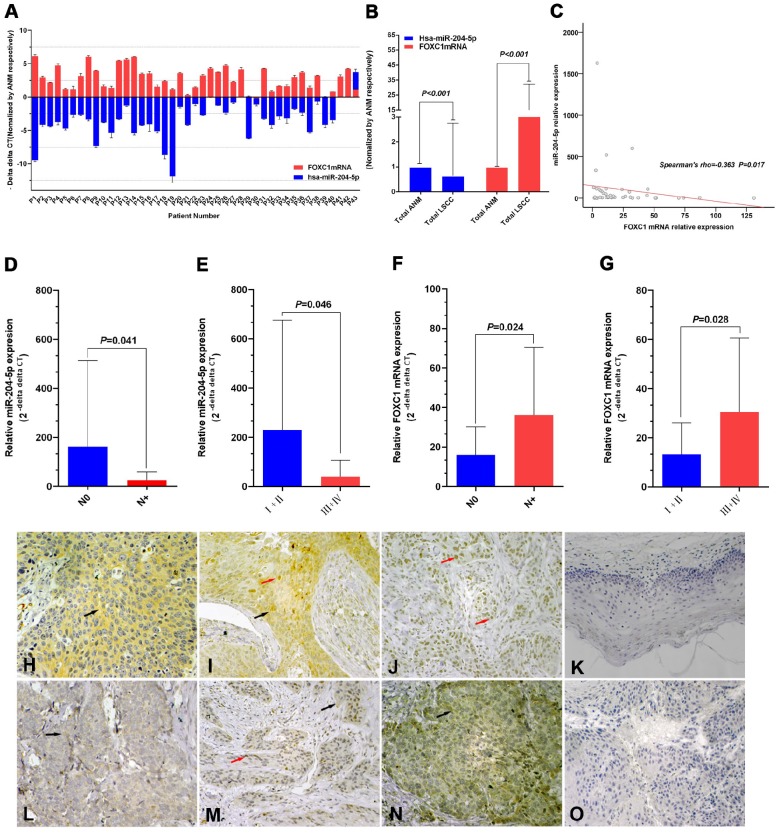
The relative expression of miR-204-5p was lower in 43 LSCC tissues relative to ANM tissues (overall 0.64±2.11-fold, P<0.001; Fig. 1A, 1B). The relative expression of miR-204-5p for stage N+ was 25.00±34.20, which was lower than 161.96±351.17 for the N0 LSCC samples (Mean Rank 17.61 vs. 25.48, P=0.041). For stages III and IV (III+IV), the relative expression of miR-204-5p was 39.54±67.82, which was lower than 229.67±446.03 for stage I and II LSCC (I+II) (Mean Rank 19.34 vs. 27.50, P=0.046) (Fig. 1D, 1E).
Figure 1.
Expression of miR-204-5p and forkhead box C1 (FOXC1) in LSCC tissues. A: Quantitative RT-PCR (qPCR) analysis of relative RNA expression of miR-204-5p and FOXC1 in 43 LSCC tissue samples and corresponding adjacent normal mucosa (ANM). Data are presented as -ΔΔCT (mean±SD). B: qPCR analysis of relative RNA expression of miR-204-5p and FOXC1 in all 43 LSCC samples and corresponding ANM. C: Correlation of expression between miR-204-5p and FOXC1 RNA levels in LSCC tissues (r=-0.363, P=0.017). D-E: Relative RNA expression of miR-204-5p by lymph node and clinical staging in LSCC and ANM samples. F-G: Relative mRNA expression of FOXC1 by lymph node and clinical staging in LSCC and ANM samples. H-J: Immunohistochemical staining of the FOXC1 protein in LSCC tissue sections. Strong cytoplasmic FOXC1 staining in LSCC cells (H). Strong nuclear and cytoplasmic FOXC1 staining in LSCC cells (I). Strong nuclear FOXC1 staining in LSCC cells (J). K: Immunohistochemical staining of FOXC1 protein in adjacent normal mucosa. L: Low FOXC1 expression in tissues with high miR-204-5p expression; weak positive granular deposition in the cytoplasm of LSCC cells. M: Low FOXC1 expression in tissues with high miR-204-5p expression; weak positive granular deposition in the cytoplasm and nuclei of LSCC cells. N: Pronounced FOXC1 expression in tissues with low miR-204-5p expression; notable positive granular deposition in the cytoplasm of LSCC cells. O: FOXC1 negative control section (primary antibody replaced by PBS during the IHC process of LSCC tissue section). Black arrows show positive cytoplasmic staining; red arrows show positive nuclear staining; magnification 40×10. Data are presented as the mean ± SD.
Elevated FOXC1 mRNA and protein levels in LSCC tissue and correlation with cervical lymph node metastasis
The relative FOXC1 mRNA level was higher in the 43 LSCC tissues than in ANM tissues (14.19±18.26-fold, P<0.001; Fig. 1A, 1B). For stage N+, the relative expression of FOXC1 mRNA was 36.09±34.40, which was higher than 16.11±14.14 for the stage N0 LSCC samples (Mean Rank 26.84 vs. 18.17, P=0.024; Fig. 1F). For stages III and IV (III+IV), the relative expression of FOXC1 mRNA was 30.56±30.01, which was higher than 13.30±12.73 for stage I and II LSCC (I+II) (Mean Rank 24.93 vs. 15.93, P=0.028) (Fig. 1G).
FOXC1 protein expression was higher in LSCC tissues than in ANM tissues (Fig. 1H-1K). Consistent with previous reports 47, 48, our results showed that FOXC1 was localized to the cytoplasm and/or nuclei of the cancer cells (Fig. 1H-1J). FOXC1 protein expression was higher in the stage N+ LSCC samples than in the stage N0 LSCC samples (84.2%, 16/19 vs 50.0%, 12/24) (P = 0.019). Rank sum testing showed that FOXC1 protein expression was significantly higher in the stage N+ samples than in the stage N0 LSCC samples (P = 0.021). Moreover, FOXC1 protein expression was higher in LSCC stages III and IV (III+IV) than in stages I and II (I+II) (79.3%, 23/29 vs 35.7%, 5/14) (P = 0.005). Rank sum testing showed that FOXC1 protein expression was significantly higher in stage (III+IV) than (I+II) (P = 0.005). Age, gender, tumor stage, degree of differentiation, and primary cancer site were not associated with the ratio and strength of FOXC1 protein expression (Table 2).
Table 2.
Association of clinical features and FOXC1 protein level in 43 LSCC patients.
| Variables | No. (n=43) |
FOXC1 protein level (n, %) | P | FOXC1 protein level | P ** | |
|---|---|---|---|---|---|---|
| Low | High | Mean rank | ||||
| Age, years (range 45-76) | ||||||
| ≤59 | 19 | 5 (26.3) | 14 (73.7) | 0.294 | 23.84 | 0.300 |
| >59 | 24 | 10 (41.7) | 14 (58.3) | 20.54 | ||
| Gender | ||||||
| Male | 39 | 15 (38.5) | 24 (61.5) | 0.280* | 21.23 | 0.129 |
| Female | 4 | 0 (0.0) | 4 (100.0) | 29.50 | ||
| Differentiation | ||||||
| Well | 18 | 6 (33.3) | 12 (66.7) | 0.856 | 22.33 | 0.858 |
| Moderate or poor | 25 | 9 (36.0) | 16 (64.0) | 21.76 | ||
| Tumor stage | ||||||
| T1+T2 | 25 | 10 (40.0) | 15 (60.0) | 0.407 | 20.90 | 0.412 |
| T3+T4 | 18 | 5 (27.8) | 13 (72.2) | 23.53 | ||
| Cervical lymph node | ||||||
| N0 | 24 | 12 (50.0) | 12 (50.0) | 0.019 | 18.75 | 0.021 |
| N+ | 19 | 3(15.8) | 16 (84.2) | 26.11 | ||
| Clinical stage | ||||||
| I + II | 14 | 9 (64.3) | 5 (35.7) | 0.005 | 15.68 | 0.005 |
| III + IV | 29 | 6 (20.7) | 23 (79.3) | 25.05 | ||
| Tumor site | ||||||
| Supraglottis or subglottis | 16 | 4 (25.0) | 12 (75.0) | 0.295 | 24.13 | 0.301 |
| Glottis | 27 | 11 (40.7) | 16 (59.3) | 20.74 | ||
| miR-204-5p level | ||||||
| Low | 21 | 4 (19.0%) | 17 (81.0%) | 0.033 | 25.40 | 0.035 |
| High | 22 | 11 (50.0%) | 11 (50.0%) | 18.75 | ||
| FOXC1 mRNA level | ||||||
| Low | 21 | 11 (52.4%) | 10 (47.6%) | 0.019 | 18.24 | 0.020 |
| High | 22 | 4 (18.2%) | 18 (81.8%) | 25.59 | ||
* By Fisher's exact test, ** by Mann-Whitney U test.
Negative correlation between miR-204-5p and FOXC1 expression in LSCC tissues
The FOXC1 mRNA level was negatively correlated with the miR-204-5p level in LSCC tissues (Spearman's rho=-0.363, P=0.017; Fig. 1C). The expression level of the miR-204-5p or FOXC1 mRNA was classified by median relative expression, with expression higher than the median considered high expression and lower than the median considered low expression. Both FOXC1 mRNA and protein levels were negatively correlated with miR-204-5p expression, but the FOXC1 mRNA level was positively correlated with the FOXC1 protein level (Table 3). Among 22 cases of high miR-204-5p expression, 11 showed low FOXC1 protein expression (50.0%) (Fig. 1L, M). Among 21 cases of low miR-204-5p expression, 17 showed high FOXC1 protein expression (81.0%) (Fig. 1N).
Table 3.
Correlations among miR-204-5p level and FOXC1 mRNA and protein levels in 43 samples of LSCC.
| FOXC1 protein level | P* | FOXC1 mRNA level | P* | |
|---|---|---|---|---|
| Spearman's rho coefficient | Spearman's rho coefficient | |||
| miR-204-5p level | -0.325 | 0.034 | -0.303 | 0.048 |
| FOXC1 protein level | 0.359 | 0.018 |
* P < 0.05 (2-tailed) considered significant
Direct regulation of FOXC1 by miR-204-5p
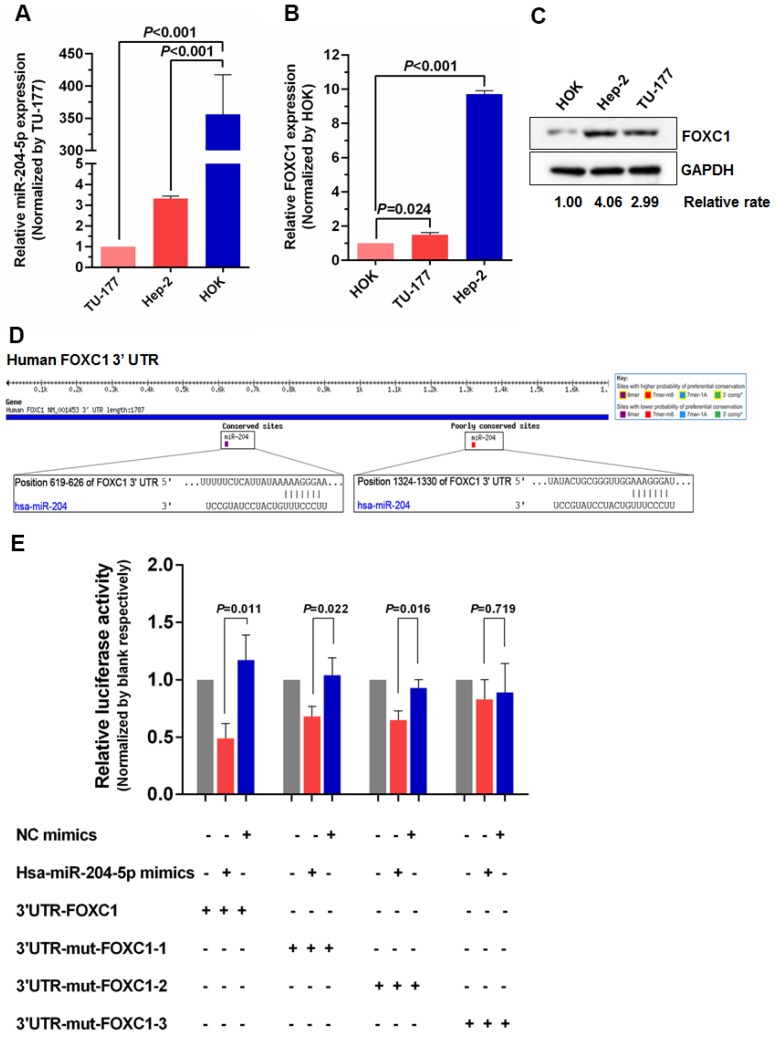
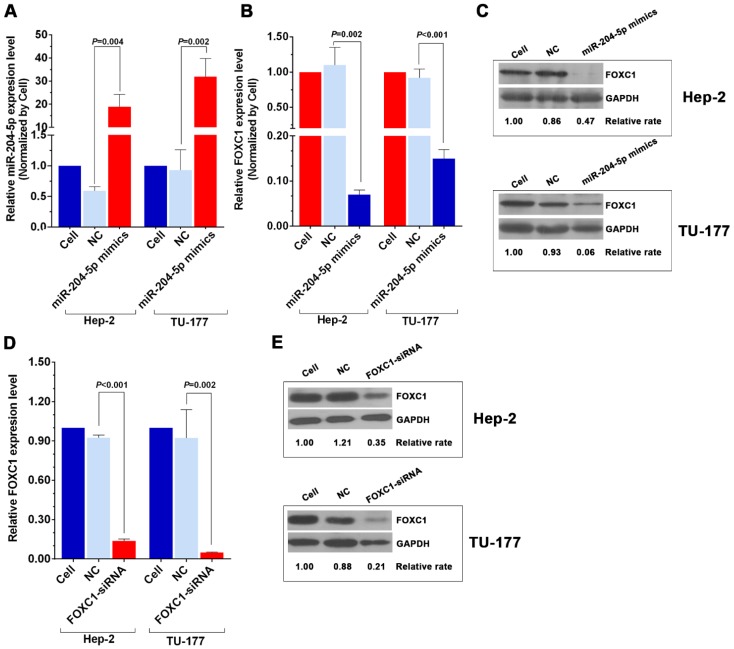
To further investigate the relationship between miR-204-5p and FOXC1, expression of miR-204-5p and FOXC1 in LSCC cell lines Hep-2 and TU-177 and control cell line HOK was determined by qPCR. As shown in Figure 2A and 2B, miR-204-5p was lower in both Hep-2 and TU-177 cells than in HOK cells, while the FOXC1 mRNA level was higher in Hep-2 and TU-177 cells than in HOK cells. Consistently, expression of FOXC1 protein in Hep-2 and TU-177 cells was higher than in HOK cells (Fig. 2C). These results suggest that miR-204-5p might directly regulate FOXC1 expression. Prediction using TargetScan Human 7.1, microrna.org, and miRDB each revealed two miR-204-5p binding sites on the FOXC1 3' UTR (Fig. 2D). The dual-luciferase reporter assay revealed that the luciferase activities of 3′UTR-FOXC1 (wild-type), 3′UTR-mut-FOXC1-1, and 3′UTR-mut-FOXC1-2 were significantly decreased when co-transfected with the miR-204-5p mimics compared with the NC mimics, but 3′ UTR-mut-FOXC1-3 (complete mutations on both sites) showed no significant changes (Fig. 2E). Thus, miR-204-5p may bind to both sites in the FOXC1 3′ UTR and play a regulatory role. We further confirmed that miR-204-5p overexpression inhibits endogenous FOXC1 mRNA and protein expression in LSCC cells (Fig. 3A-3C). These data collectively suggest that miR-204-5p directly inhibits FOXC1 expression in LSCC cells.
Figure 2.
miR-204-5p directly inhibits FOXC1 expression. qPCR analysis of miR-204-5p (A) and FOXC1 (B) expression in LSCC cell lines Hep-2 and TU-177 and in normal control cell line HOK. B: Expression of FOXC1 in LSCC cell lines Hep-2 and TU-177 and in normal control cell line HOK was determined by western blotting. D: miR-204-5p binding sites in the FOXC1 3′ UTR predicted by TargetScan. E: Luciferase activity assay of predicted miR-204-5p binding sites in the FOXC1 3′ UTR. Data are presented as the mean ± SD.
Figure 3.
miR-204-5p mimics and FOXC1 siRNA inhibit FOXC1 expression in LSCC cells. A: qPCR analysis of the miR-204-5p level in miR-204-5p mimic-transfected (50 nM, 48 h) Hep-2 and TU-177 cells. qPCR and western blotting analysis of FOXC1 mRNA (B) and protein levels (C) in miR-204-5p mimics-transfected (50 nM, 48 h) Hep-2 and TU-177 cells. qRT-PCR and western blotting analysis of FOXC1 mRNA (D) and protein levels (E) in FOXC1 siRNA-transfected (50 nM, 48 h) Hep-2 and TU-177 cells. Data are presented as the mean ± SD.
Inhibitory effect of miR-204-5p and FOXC1 siRNA on LSCC cell proliferation and colony formation
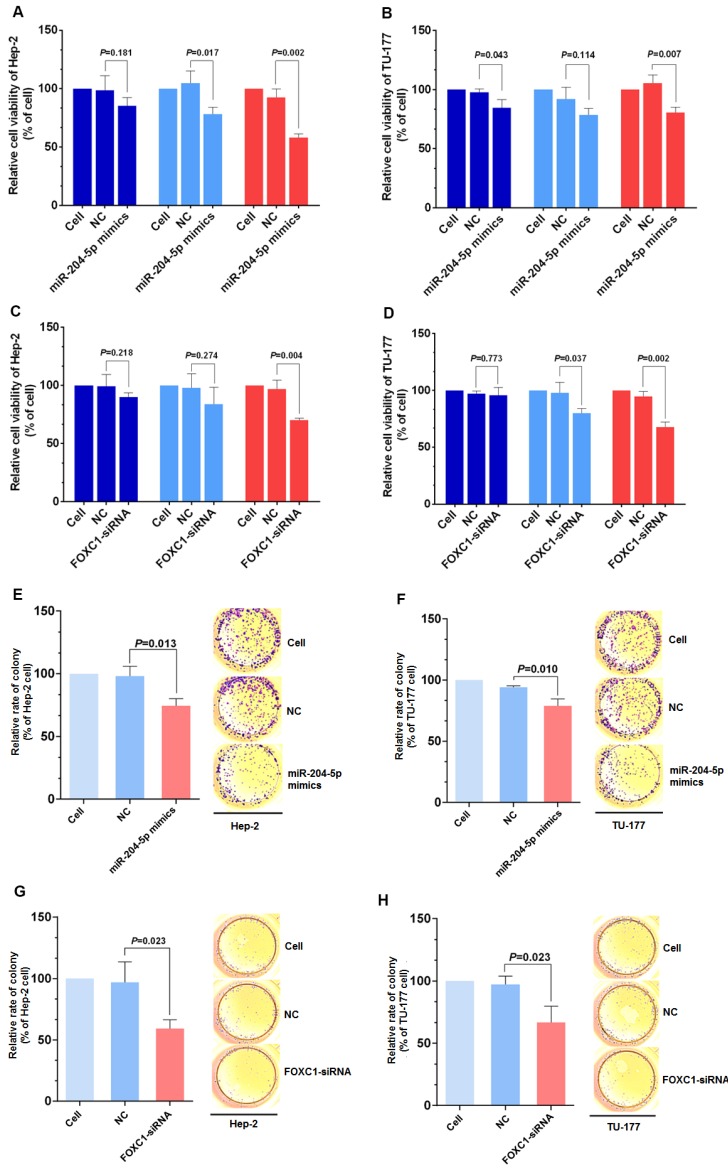
Knockdown of FOXC1 was performed using siRNA that targeted the CDS region of the FOXC1 mRNA, and effective knockdown was obtained with 50 nM si-FOXC1-2 (Fig. S1). Therefore, si-FOXC1-2 was used in subsequent experiments for FOXC1 knockdown (Fig. 3D-3E). To investigate the role of miR-204-5p and FOXC1 in LSCC growth, the MTS assay was performed by measuring optical density values at 490 nm at 0, 24, 48, and 72 h after transfection of Hep-2 and TU-177 cells with miR-204-5p mimics or FOXC1 siRNA. The results showed that cell proliferation was inhibited after the miR-204-5p mimics or FOXC1 siRNA were transfected (Fig. 4A-4D). The colony-formation assay confirmed that the miR-204-5p mimics and FOXC1 siRNA inhibited proliferation of LSCC cells (Fig. 4E-4H).
Figure 4.
miR-204-5p overexpression or FOXC1 knockdown inhibit LSCC cell proliferation. MTS assay of Hep-2 and TU-177 cells transfected with miR-204-5p mimics (50 nM) (A, B) or FOXC1 siRNA (50 nM) (C, D) for 24, 48, and 72 h. Colony formation assay of Hep-2 and TU-177 cells transfected with miR-204-5p mimics (50 nM) (E, F) or FOXC1 siRNA (50 nM) (G, H). Data are presented as the mean ± SD.
miR-204-5p overexpression or FOXC1 knockdown inhibits migration, invasion, and the epithelial-mesenchymal transition in LSCC cells
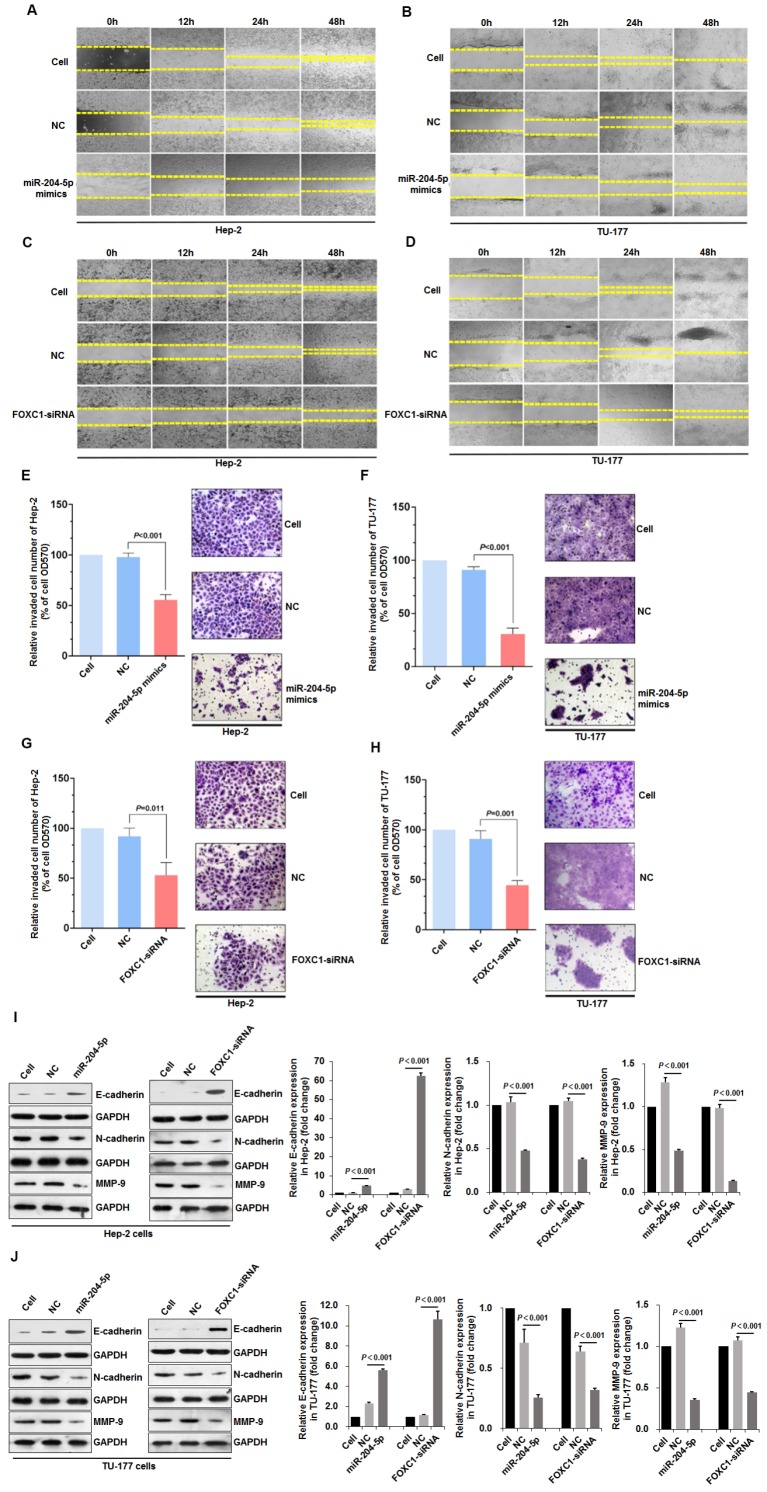
Scratch assays that were performed to assess wound healing by Hep-2 and TU-177 cells at 12, 24, and 48 h after transfection with the miR-204-5p mimics or FOXC1 siRNA revealed reduced cell migration after transfection (Fig. 5A-5D). Furthermore, Transwell assays revealed the reduced invasion abilities of Hep-2 and TU-177 cells with the miR-204-5p mimics or FOXC1 siRNA transfections (Fig. 5E-5H).
Figure 5.
miR-204-5p overexpression or FOXC1 knockdown inhibits LSCC cell migration, invasion, and the EMT process. Scratch assay of cell migration by Hep-2 and TU-177 cells transfected with miR-204-5p mimics (50 nM) (A, B) or FOXC1 siRNA (50 nM) (C, D) for 12, 24, and 48 h. Relative number of invaded cells in a Transwell assay of Hep-2 and TU-177 cells transfected with miR-204-5p mimics (50 nM) (E, F) or FOXC1-siRNA (50 nM) (G, H). Hep-2 and TU-177 cells were transfected with miR-204-5p mimics (50 nM) or FOXC1-siRNA for 48 h, and expression of EMT markers E-cadherin, N-cadherin, and MMP-9 in Hep-2 cells (I) and TU-177 cells (J) was detected by immunoblotting. Data are presented as the mean ± SD.
The EMT process is an important mechanism by which tumor cells become capable of invasion and metastasis 49. The expression of EMT markers E-cadherin, N-cadherin, and MMP-9 in LSCC cells after transfections with the miR-204-5p mimics or FOXC1-siRNA was determined by immunoblotting. miR-204-5p overexpression or FOXC1 knockdown upregulated E-cadherin but downregulated N-cadherin and MMP-9 significantly in Hep-2 and TU-177 cells (Fig. 5I-5J). These results suggest that miR-204-5p overexpression or FOXC1 knockdown inhibits EMT in LSCC cells.
Inhibitory effect of miR-204-5p on growth of xenograft tumors in vivo
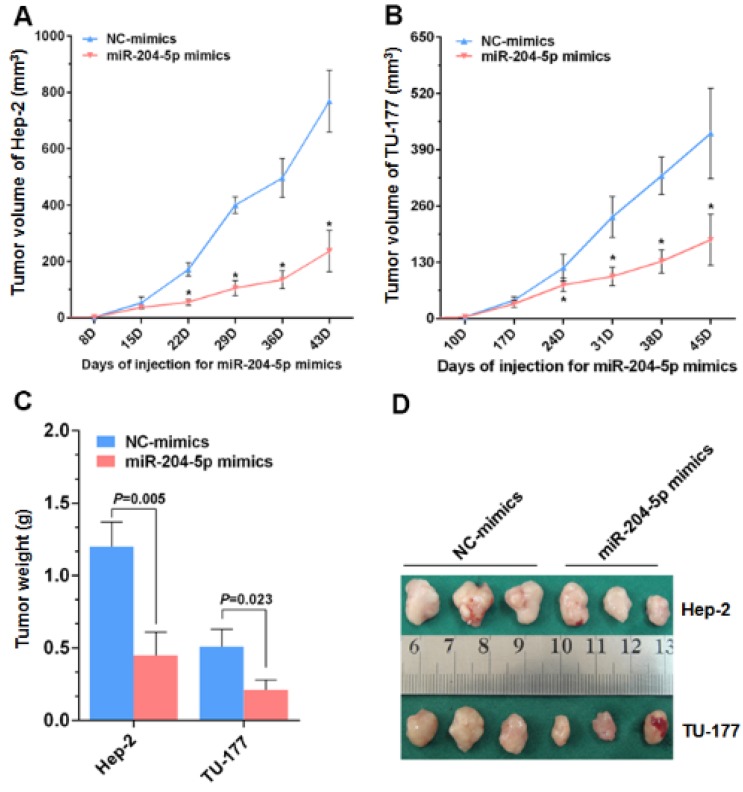
From the nude mice tumor model generated with Hep-2 and TU-177 cells, the tumor volumes and final tumor weights were smaller with the miR-204-5p mimics than with the NC-mimics injections (Fig. 6A-6D). These results suggest that miR-204-5p suppresses the growth of LSCC xenograft tumors in nude mice.
Figure 6.
miR-204-5p inhibits Hep-2 and TU-177 tumor growth in vivo. After injections with the miR-204-5p mimics, the volumes of the Hep-2 (A) or TU-177 (B) tumors were decreased compared with the NC mimics group. Mean final weights of the Hep-2 (C) and TU-177 (D) tumors with the miR-204-5p mimics or NC mimics. Data are presented as the mean ± SD. *, P<0.05.
Discussion
Metastasis is a basic characteristic of malignant tumors and a major cause of recurrent disease and death in many patients 50, 51. The process of tumor cell metastasis is accompanied by disrupted regulation of oncogenes and tumor suppressor genes. miRNAs are small non-coding single-stranded RNA molecules that completely or partially bind to the 3′ UTR regions of target genes, thereby inhibiting the translation of or directly degrading target gene transcripts 11. The discovery of miRNAs has opened new horizons for investigations into regulatory crosstalk between oncogenes and tumor suppressor genes during tumor invasion and metastasis.
Using miRNA microarrays, we revealed that miR-204-5p expression was downregulated 5.02-fold in human LSCC tissue relative to normal tissue (P<0.001, data not shown). This trend is consistent with trends in oral cancer 19, hypopharyngeal cancer 25, squamous cell maxillary sinus cancer 27, squamous cell carcinoma 28, endometrial cancer 29, and bile duct epithelial cancer 30. We found the relative expression of miR-204-5p to be lower in 43 human LSCC samples than in the adjacent normal tissues. Moreover, the miR-204-5p level was lower in stage N+ and clinically advanced (III and IV) LSCC tissues than in stage N0 and early-to-mid-stage (I and II) LSCC. Lee et al. observed that miR-204 localized to a cancer-associated genomic region (CAGR), 9q21.1-q22.3, but no candidate tumor suppressor gene was identified in the CAGR; miR-204 expression was low in the head and neck SCC JSQ3 (nasal cavity) and SQ38 (pyriform sinus) cell lines. After restoration of miR-204 expression in JSQ3 and SQ38 cells, cell adhesion, invasion, and metastasis were reduced. Similar results were observed in studies of endometrial cancer 29, gastric cancer 31, renal cell carcinoma 32, and liver cancer 33. Thus, miR-204-5p might be involved in LSCC invasion and metastasis and be a tumor suppressor gene.
FOXC1 may be a genetic risk factor for LSCC 35. Our bioinformatics analysis predicted FOXC1 as a target gene for miR-204-5p. The relative expression of FOXC1 was higher in 43 LSCC samples than in the adjacent normal tissues and negatively correlated with the miR-204-5p level. miR-204-5p expression was lower, but FOXC1 expression was higher, in Hep-2 and TU-177 cells than in control cells HOK, suggesting that miR-204-5p may have a targeted regulatory effect on FOXC1. Our data further confirmed that miR-204-5p directly bound to the 3' UTR of the FOXC1 mRNA to inhibit expression.
High FOXC1 expression has been observed in breast cancer with lymph node and brain metastases and is associated with a poor prognosis 36. FOXC1 transcription and translation in non-small-cell lung cancer were increased and associated with lymph node metastasis. FOXC1 was found to be an independent prognostic risk factor for non-small-cell lung cancer 38. A study of hepatocellular carcinoma showed significantly increased FOXC1 mRNA and protein levels in hepatocellular carcinoma tissues, especially when microvascular invasion was evident. Similarly, in this study, the ratio of FOXC1 expression and the expression strength were both significantly higher in stage N+ than in stage N0 LSCC. Thus, FOXC1 may play an important role in LSCC invasion and metastasis.
Snail and twist are key signaling molecules in EMT. After knockdown of FOXC1 expression in Bel-7402 liver cancer cells, E-cadherin was upregulated, vimentin and N-cadherin were downregulated, and the EMT phenotype of the tumor cells was weakened 39. Thus, FOXC1 might be involved in tumor invasion and metastasis by regulating EMT or by directly participating in tumor microvessel formation and lymphangiogenesis 37. In this study, our data demonstrated that knockdown of FOXC1 inhibits EMT in LSCC cells and suggested that miR-204-5p may inhibit LSCC cell invasion and migration by suppressing FOXC1 expression and the EMT process.
To determine whether miR-204-5p suppresses LSCC cell proliferation, invasion, and metastasis by inhibiting FOXC1, we transfected miR-204-5p mimics or FOXC1 siRNA into Hep-2 and TU-177 cells. FOXC1 transcription and translation were decreased after transfection, and cell proliferation, colony formation, invasion, and metastasis of Hep-2 and TU-177 were all suppressed after transfections with the miR-204-5p mimics or FOXC1 siRNA. These data suggest that miR-204-5p overexpression suppresses tumor growth by inhibiting its target gene, FOXC1. Studies of endometrial and breast cancer showed similar results 29, 36. miR-204-5p can also inhibit ezrin expression and indirectly inhibit Ras activation, thereby suppressing proliferation and metastasis of cancer cells 31. Additionally, our data revealed that growth of xenograft LSCC tumors in nude mice was significantly inhibited after injections with the miR-204-5p mimics. Therefore, our in vitro and in vivo studies indicate that miR-204-5p has significant potential for molecular targeted therapy of LSCC.
In conclusion, we found that disrupted expression and regulation of FOXC1 by miR-204-5p is a frequent molecular event during LSCC invasion and metastasis. miR-204-5p can suppress malignant LSCC behaviors, including cell proliferation, invasion, and metastasis, by directly inhibiting FOXC1 expression. Moreover, miR-204-5p suppressed LSCC tumor growth in vivo. These findings provide new insights into the molecular mechanism of LSCC invasion and metastasis and offer potential targets and markers for LSCC treatment and prognosis. Further study is required to unravel the mechanism underlying dysregulation of miR-204-5p in LSCC.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 81572670, 81402256, 81602394), China Postdoctoral Science Foundation (2016M591412), Natural Science Foundation of Shanxi Province (2014011039-5, 2015021198, 201601D011087), The Scientific and Technological Achievements Transformation Guidance Foundation of Shanxi Province (201604D131002), The Key Scientific and Technological Innovation Platform Foundation for Head and Neck Cancer Research of Shanxi Province (201605D151003), Scientific and Technological Innovation Programs of Higher Education Institutions in Shanxi (STIP, 2016-92, 2016-93), Research Project of Shanxi Province Health and Family Planning Commission (201301073, 2014028, 201601037, 201601038), Excellent talent science and technology innovation project of Shanxi Province (201605D211029), and Outstanding Youth Development Foundation of The First Hospital Affiliated with Shanxi Medical University (YR1601).
Author Contributions
Conceived and designed the experiments: BQW, SXW, CMZ, WG.
Performed the experiments: WG, YYW, XLH, CMZ, MXZ, BC, QQL, XKQ, WYL.
Analyzed the data: WG, YYW, CMZ, HXL, SXW.
Contributed reagents/materials/analysis tools: BQW, SXW, WG, CMZ.
Wrote the paper: WG, YYW, XLH, CMZ, MXZ.
References
- 1.Black RJ, Bray F, Ferlay J, Parkin DM. Cancer incidence and mortality in the European Union: cancer registry data and estimates of national incidence for 1990. Eur J Cancer. 1997;33(7):1075–107. doi: 10.1016/s0959-8049(96)00492-3. [DOI] [PubMed] [Google Scholar]
- 2.Chu EA, Kim YJ. Laryngeal cancer: diagnosis and preoperative work-up. Otolaryngol Clin North Am. 2008;41(4):673–95. doi: 10.1016/j.otc.2008.01.016. v. [DOI] [PubMed] [Google Scholar]
- 3.Dequanter D, Lothaire P, Zouaoui K, Brohee D. Epidemiology and clinical characteristics of larynx and hypopharynx carcinoma: a comparative study in the Hainaut and review of the literature. Acta Chir Belg. 2012;112(6):423–5. doi: 10.1080/00015458.2012.11680866. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Thomas A, Murray T, Thun M. Cancer statistics, 2002. CA Cancer J Clin. 2002;52(1):23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- 5.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 6.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 7.Gao W, Zhang C, Feng Y, Chen G, Wen S, Huangfu H. et al. Fascin-1, ezrin and paxillin contribute to the malignant progression and are predictors of clinical prognosis in laryngeal squamous cell carcinoma. PLoS One. 2012;7(11):e50710. doi: 10.1371/journal.pone.0050710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji W, Guan C, Pan Z. Analysis of curative effects on laryngeal carcinoma patients in the northeast region of China. Acta Otolaryngol. 2008;128(5):574–7. doi: 10.1080/00016480701596104. [DOI] [PubMed] [Google Scholar]
- 9.Hermans R. Staging of laryngeal and hypopharyngeal cancer: value of imaging studies. Eur Radiol. 2006;16(11):2386–400. doi: 10.1007/s00330-006-0301-7. [DOI] [PubMed] [Google Scholar]
- 10.Yurut-Caloglu V, Caloglu M, Turan FN, Ibis K, Karagol H, Kocak Z. et al. The better performance status, the better outcome: laryngeal carcinoma treated with definitive radiotherapy. J Otolaryngol Head Neck Surg. 2008;37(6):799–806. [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122(1):6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 13.van Rooij E. The art of microRNA research. Circ Res. 2011;108(2):219–34. doi: 10.1161/CIRCRESAHA.110.227496. [DOI] [PubMed] [Google Scholar]
- 14.Wang YY, Ren T, Cai YY, He XY. MicroRNA let-7a Inhibits the Proliferation and Invasion of Nonsmall Cell Lung Cancer Cell Line 95D by Regulating K-Ras and HMGA2 Gene Expression. Cancer Biother Radiopharm; 2012. [DOI] [PubMed] [Google Scholar]
- 15.Yu CC, Chen YW, Chiou GY, Tsai LL, Huang PI, Chang CY. et al. MicroRNA let-7a represses chemoresistance and tumourigenicity in head and neck cancer via stem-like properties ablation. Oral Oncol. 2011;47(3):202–10. doi: 10.1016/j.oraloncology.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Xiong S, Zheng Y, Jiang P, Liu R, Liu X, Chu Y. MicroRNA-7 inhibits the growth of human non-small cell lung cancer A549 cells through targeting BCL-2. Int J Biol Sci. 2011;7(6):805–14. doi: 10.7150/ijbs.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu C, Li B, Cheng Y, Lin J, Hao J, Zhang S. et al. MiR-21 plays an important role in radiation induced carcinogenesis in BALB/c mice by directly targeting the tumor suppressor gene Big-h3. Int J Biol Sci. 2011;7(3):347–63. doi: 10.7150/ijbs.7.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology (Williston Park) 2010;79(3-4):313–20. doi: 10.1159/000323283. [DOI] [PubMed] [Google Scholar]
- 19.Liu CJ, Tsai MM, Hung PS, Kao SY, Liu TY, Wu KJ. et al. miR-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Cancer Res. 2010;70(4):1635–44. doi: 10.1158/0008-5472.CAN-09-2291. [DOI] [PubMed] [Google Scholar]
- 20.Kinoshita T, Nohata N, Fuse M, Hanazawa T, Kikkawa N, Fujimura L. et al. Tumor suppressive microRNA-133a regulates novel targets: moesin contributes to cancer cell proliferation and invasion in head and neck squamous cell carcinoma. Biochem Biophys Res Commun. 2012;418(2):378–83. doi: 10.1016/j.bbrc.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 21.Deng Y, Deng H, Bi F, Liu J, Bemis LT, Norris D. et al. MicroRNA-137 targets carboxyl-terminal binding protein 1 in melanoma cell lines. Int J Biol Sci. 2011;7(1):133–7. doi: 10.7150/ijbs.7.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Wang C, Chen Z, Jin Y, Wang Y, Kolokythas A. et al. MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochem J. 2011;440(1):23–31. doi: 10.1042/BJ20111006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu DW, Cheng YW, Wang J, Chen CY, Lee H. Paxillin predicts survival and relapse in non-small cell lung cancer by microRNA-218 targeting. Cancer Res. 2010;70(24):10392–401. doi: 10.1158/0008-5472.CAN-10-2341. [DOI] [PubMed] [Google Scholar]
- 24.Guo SL, Peng Z, Yang X, Fan KJ, Ye H, Li ZH. et al. miR-148a promoted cell proliferation by targeting p27 in gastric cancer cells. Int J Biol Sci. 2011;7(5):567–74. doi: 10.7150/ijbs.7.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kikkawa N, Hanazawa T, Fujimura L, Nohata N, Suzuki H, Chazono H. et al. miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC) Br J Cancer. 2010;103(6):877–84. doi: 10.1038/sj.bjc.6605811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang C, Gao W, Wen S, Wu Y, Fu R, Zhao D. et al. Potential key molecular correlations in laryngeal squamous cell carcinoma revealed by integrated analysis of mRNA, miRNA and lncRNA microarray profiles. Neoplasma. 2016;63(6):888–900. doi: 10.4149/neo_2016_608. [DOI] [PubMed] [Google Scholar]
- 27.Nohata N, Hanazawa T, Kikkawa N, Sakurai D, Fujimura L, Chiyomaru T. et al. Tumour suppressive microRNA-874 regulates novel cancer networks in maxillary sinus squamous cell carcinoma. Br J Cancer. 2011;105(6):833–41. doi: 10.1038/bjc.2011.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y, Yang X, Huang Y, Fan H, Zhang Q, Wu Y. et al. Network modeling identifies molecular functions targeted by miR-204 to suppress head and neck tumor metastasis. PLoS Comput Biol. 2010;6(4):e1000730. doi: 10.1371/journal.pcbi.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung TK, Lau TS, Cheung TH, Yim SF, Lo KW, Siu NS. et al. Dysregulation of microRNA-204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int J Cancer. 2012;130(5):1036–45. doi: 10.1002/ijc.26060. [DOI] [PubMed] [Google Scholar]
- 30.Chen L, Yan HX, Yang W, Hu L, Yu LX, Liu Q. et al. The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. J Hepatol. 2009;50(2):358–69. doi: 10.1016/j.jhep.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 31.Lam EK, Wang X, Shin VY, Zhang S, Morrison H, Sun J. et al. A microRNA contribution to aberrant Ras activation in gastric cancer. Am J Transl Res. 2011;3(2):209–18. [PMC free article] [PubMed] [Google Scholar]
- 32.Mikhaylova O, Stratton Y, Hall D, Kellner E, Ehmer B, Drew AF. et al. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell. 2012;21(4):532–46. doi: 10.1016/j.ccr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng L, Yu J, Huang T, Jia H, Dong Q, He F. et al. Differential combinatorial regulatory network analysis related to venous metastasis of hepatocellular carcinoma. BMC Genomics. 2012;13(Suppl 8):S14. doi: 10.1186/1471-2164-13-S8-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7(11):847–59. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 35.Mirisola V, Mora R, Esposito AI, Guastini L, Tabacchiera F, Paleari L. et al. A prognostic multigene classifier for squamous cell carcinomas of the larynx. Cancer Lett. 2011;307(1):37–46. doi: 10.1016/j.canlet.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 36.Ray PS, Wang J, Qu Y, Sim MS, Shamonki J, Bagaria SP. et al. FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Res. 2010;70(10):3870–6. doi: 10.1158/0008-5472.CAN-09-4120. [DOI] [PubMed] [Google Scholar]
- 37.Seo S, Fujita H, Nakano A, Kang M, Duarte A, Kume T. The forkhead transcription factors, Foxc1 and Foxc2, are required for arterial specification and lymphatic sprouting during vascular development. Dev Biol. 2006;294(2):458–70. doi: 10.1016/j.ydbio.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 38.Wei LX, Zhou RS, Xu HF, Wang JY, Yuan MH. High expression of FOXC1 is associated with poor clinical outcome in non-small cell lung cancer patients. Tumour Biol; 2012. [DOI] [PubMed] [Google Scholar]
- 39.Xu ZY, Ding SM, Zhou L, Xie HY, Chen KJ, Zhang W. et al. FOXC1 contributes to microvascular invasion in primary hepatocellular carcinoma via regulating epithelial-mesenchymal transition. Int J Biol Sci. 2012;8(8):1130–41. doi: 10.7150/ijbs.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xin Q, Li J, Dang J, Bian X, Shan S, Yuan J. et al. miR-155 Deficiency Ameliorates Autoimmune Inflammation of Systemic Lupus Erythematosus by Targeting S1pr1 in Faslpr/lpr Mice. J Immunol. 2015;194(11):5437–45. doi: 10.4049/jimmunol.1403028. [DOI] [PubMed] [Google Scholar]
- 41.Fabre AL, Colotte M, Luis A, Tuffet S, Bonnet J. An efficient method for long-term room temperature storage of RNA. Eur J Hum Genet. 2014;22(3):379–85. doi: 10.1038/ejhg.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Naito S, von EAC, Giavazzi R, Fidler IJ. Growth and metastasis of tumor cells isolated from a human renal cell carcinoma implanted into different organs of nude mice. Cancer Res. 1986;46(8):4109–15. [PubMed] [Google Scholar]
- 44.Behlke MA. Progress towards in vivo use of siRNAs. Mol Ther. 2006;13(4):644–70. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q. et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19(2):232–43. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Li D, Liu X, Lin L, Hou J, Li N, Wang C. et al. MicroRNA-99a inhibits hepatocellular carcinoma growth and correlates with prognosis of patients with hepatocellular carcinoma. J Biol Chem. 2011;286(42):36677–85. doi: 10.1074/jbc.M111.270561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang LY, Li LS, Yang Z. Correlation of FOXC1 protein with clinicopathological features in serous ovarian tumors. Oncol Lett. 2016;11(2):933–8. doi: 10.3892/ol.2015.3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J, Xu Y, Li L, Wang L, Yao R, Sun Q. et al. FOXC1 is associated with estrogen receptor alpha and affects sensitivity of tamoxifen treatment in breast cancer. Cancer Med. 2017;6(1):275–87. doi: 10.1002/cam4.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED. et al. Epithelial-mesenchymal and mesenchymal-epithelial transitions in carcinoma progression. J Cell Physiol. 2007;213(2):374–83. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- 50.Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–8. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 51.Poste G, Fidler IJ. The pathogenesis of cancer metastasis. Nature. 1980;283(5743):139–46. doi: 10.1038/283139a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.