Abstract
The structures of ribosomes in complex with inhibitors of translation have not only shed light on the interactions of antibiotics with the ribosome but also on the underlying mechanisms by which they interfere with the ribosome function. Several recent papers [1–4] have correlated the available ribosome structures with the wealth of biochemical data [5]. In this review we shall focus on the lessons learned for drug specificity rather than presenting a comprehensive survey of the known structures of ribosome complexes with antibiotics.
INTRODUCTION
In order for translational inhibitors to be useful in a clinical setting they must be highly effective against eubacterial target, yet not inhibit the eukaryotic ribosome. Since protein translation is essential and highly conserved across all kingdoms of life, both demands are quite often at odds with each other, giving rise to side effects during treatment [6].
Specificity of antibiotics that bind at the decoding site of the ribosome
One of the most common side effects of aminoglycoside and tuberactinomycin treatment is irreversible loss of hearing [7]. The toxicity of these drugs is correlated with their limited selectivity between binding to bacterial and mitochondrial ribosomes [8]. After administration, aminoglycosides are cleared from the blood stream within days, but persist for weeks, even months, in the tissues and fluids of the inner ear [9]. In the hair cells of the inner ear the aminoglycosides promote biochemical events that elicit apoptotic response leading to irreversible loss of hearing [10].
The binding of either aminoglycosides or tuberactinomycins to the ribosome increases the misincorporation of aminoacids in vitro, due to a mismatching between the tRNA anticodon and the mRNA codon in the A-site of the ribosome [11–14]. An amino-acylated tRNA progresses through multiple steps before its amino acid is incorporated into a nascent polypeptide chain. First, the amino-acylated tRNA is delivered to the ribosome in a ternary complex with EF-Tu and GTP (Fig. 1C). Initially, the ternary complex binds to the ribosome in a rapid codon-independent and reversible process. After this initial binding, the anticodon region of the tRNA pairs with the codon region of the mRNA in the A-site of the small ribosomal subunit [15], which consists of parts of the head, shoulder and the top of h44a) domains (Fig. 1A, B). The two universally conserved nucleotides, A1492 and A1493b), flip out from an internal loop at the top of h44 (Fig. 1B) to monitor the Watson-Crick base-pair geometry of the first two codon-anticodon base pairs (Fig. 1B, D–F). Together with ribosomal protein S12 and G530, whose base flips from syn to anti conformation (Fig. 1F), A1492 and A1493 form a network of interactions across the minor groove of the codon-anticodon helix that induces the head and the shoulder domains of the small subunit to close on each other. This domain closure constrains the anticodon loop of the decoded amino-acylated tRNA (Fig. 1C) [16,17].
Figure 1. Decoding center of eubacterial 70S ribosome.
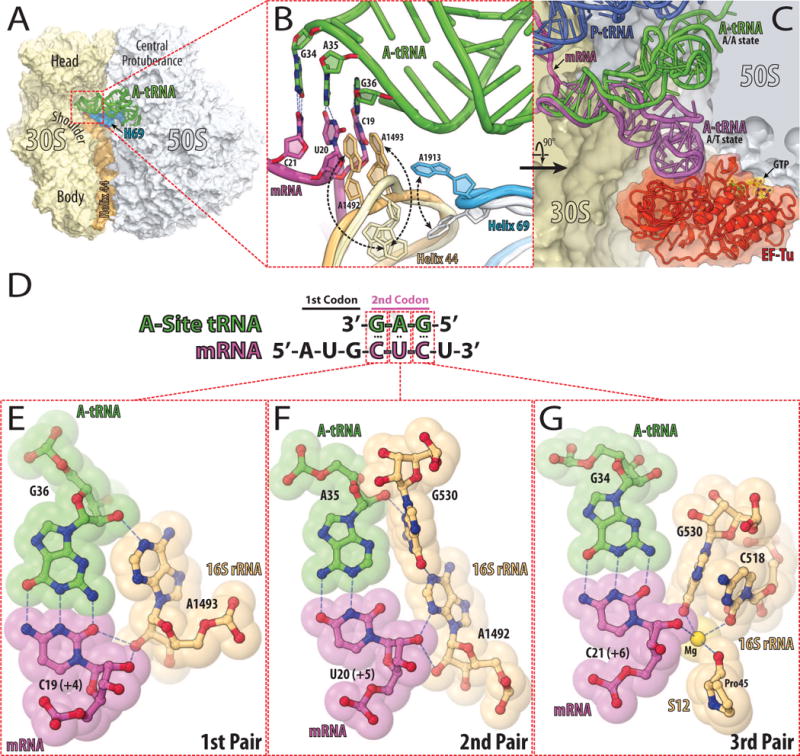
(A) The structure of 70S ribosome from eubacteria Thermus thermophilus with bound tRNAs and mRNA [57]. The 30S subunit is shown in light yellow with h44 of the 16S rRNA in light orange. The 50S subunit is in light blue with H69 of the 23S rRNA in marine. The tRNA bound in the A-site is in green. (B) Close up view of the decoding center of the 70S ribosome in the unliganded [58] and tRNA bound states [57]. The subunits and A-site bound tRNA are colored as in (A), the mRNA is in magenta. The conformational change of each nucleotide upon tRNA binding is indicated with black dashed arrows. Nucleotides involved in the codon-anticodon interactions (hydrogen bonds in blue dashed lines) are represented as sticks with their nitrogen and oxygen atoms in dark blue and red, respectively. (C) Comparison of the tRNA in the A/T state (purple) [59] with the accommodated A/A state (green) [57]. EF-Tu is shown in red, GTP in yellow, the 30S subunit in light yellow, 50S subunit in light blue, the P-site bound tRNA in dark blue, and the mRNA in magenta. (D) Schematic diagram of codon-anticodon interactions between mRNA (magenta) and cognate tRNA (green). (E, F, G) Codon-anticodon recognition by nucleotides of the 16S rRNA [60]. Cognate A-site bound tRNA is displayed in green, mRNA in magenta, nucleotides of 16S rRNA and portion of protein S12 are in light orange. Nitrogen, oxygen, and magnesium atoms are colored in blue, red, and yellow, respectively.
The binding of both tuberactinomycins, such as viomycin or capreomycin, as well as amnioglycosides, such as paromomycin, stabilizes the flipped out conformations of A1492 and A1493, enabling them to make contacts with the minor groove of the codon-anticodon helix even if they do not form a perfect Watson-Crick base-pair geometry (Fig. 2D, E).
Figure 2. Antibiotics bound to the decoding center of the eubacterial 70S ribosome.
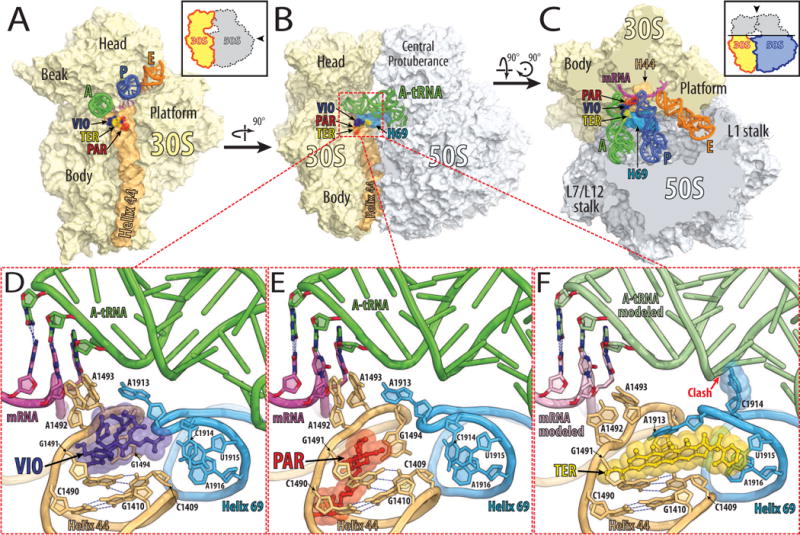
(A, B, C) Overview of the superimposed binding sites of viomycin (purple) [18], paromomycin (red) [57], and thermorubin (yellow) [26] on the Thermus thermophilus 70S ribosome viewed from three different perspectives. Shown in light yellow is the 30S subunit with h44 in light orange and in light blue is the 50S subunit with H69 in marine. The tRNAs are displayed in green for the A-site, in dark blue for the P-site, and in orange for the E-site bound tRNA. The mRNA is shown in magenta. In (A), the 30S subunit is viewed from the 50S subunit, as indicated by the inset. The view in (B) is from the cytoplasm onto the A-site. The view in (C) is from the top after removing the head of the 30S subunit and protuberances of the 50S subunit, as indicated by the inset. Close up views of the binding sites of viomycin (VIO, purple), paromomycin (PAR, red), and thermorubin (TER, yellow) are shown in panels (D), (E), and (F), respectively.
The binding site for tuberactinomycins lies at the interface between h44 of the small ribosomal subunit and H69 of the large ribosomal subunit (Fig. 2A–C). Their binding site is formed by nucleotides A1493 and G1494 of h44 and nucleotides A1913 and C1914 at the tip of H69 (Fig. 2D). The stacking of the macrocyclic scaffold of the drug against G1491 and G1494, positions the guanidinium moiety of the capreomycidine side chain close enough to the phosphate group of A1493 of the 16S rRNA to form a salt bridge. This salt bridge effectively locks the tuberacitnomycins over A1492 and A1493 in their flipped-out conformations, preventing their return into the internal loop [18].
Crystal structures of the 30S subunit and the 70S ribosome, in complex with paromomycin, show that it binds only in the major groove of h44 [19,20], where it forms hydrogen bonds with the bases and the backbone of both rRNA strands of the helix (Fig. 2E). One of its cyclic sugar moieties inserts into the internal loop of h44 where it stacks against G1491 and forms hydrogen bonds to the base of A1408 and the phosphate of A1493. In this position, the cyclic sugar moiety not only displaces A1492 from the internal loop of h44, but also stabilizes A1493 in its flipped-out position [19]. In addition to priming A1492 and A1493 to make contact within the minor groove of the codon-anticodon helix, paromomycin also induces a partial closing movement of protein S12 toward h44, thereby not only stabilizing the binding of near-cognate tRNAs but also promoting their accommodation on the ribosome [21].
Currently, the risk of drug-induced hearing loss can be minimized by proper dosage with constant monitoring of the drug levels in the blood [22]. Despite these measures, aminoglycosides can still induce hearing loss in some patients. Among those predisposed to hearing loss are patients carrying A1490G [23] or C1410U [24] mutations in the decoding center of the mitochondrial ribosome. The equivalent positions C1490 and G1410 in eubacterial 16S rRNA form a perfect Watson-Crick base-pair (Fig. 2E), which is disrupted in wild type mitochondrial ribosomes by an A•C mismatch. Either of the aforementioned mutations restores the eubacterial type of base-pairing which increases the binding affinity for aminoglycosides and, thereby, the sensitivity of these patients to aminoglycoside antibiotics [25].
Cytosolic ribosomes have perfect base-pairing between 1490- and 1410-equivalent positions, but nevertheless are resistant to aminoglycosides. However, an adenine to guanine transition in position 1408 from eubacterial/mitochondrial to eukaryotic ribosomes bestows the innate resistance upon eukaryotic ribosomes. Interestingly, A1408G is the dominant drug-resistant mutation in clinical strains of eubacteria [8].
However, the innate resistance of mitochondrial and cytoplasmic ribosomes to tuberactinomycins results from a mismatch between G1491 and C1409, which form a base-pair in eubacteria (Fig. 2D). Any disruption of this eubacteria-specific base-pair confers resistance to tuberactinomycins [14]. The same base-pair can explain the specificity of thermorubin for eubacterial ribosomes [26]. The tetracyclic moiety of thermorubin stacks against the bases of G1491and C1409 (Fig. 2F), allowing A1913 of the 23S rRNA to extend along the conjugated aromatic tetracyclic moiety. This ties h44 of the 16S rRNA and H69 of the 23S rRNA together, preventing the motion of these helices relative to each other, that is essential for the translocation during protein synthesis. The correlation between the low inhibitory effect of the drug and the absence of the equivalent G1491:C1409 base-pair in mitochondrial and eukaryotic ribosomes suggests that the observed extensive stacking of thermorubin is crucial for its ribosome selectivity.
Specificity of antibiotics that bind in the ribosome exit tunnel
While eubacterial ribosomes are sensitive to aminoglycosides, macrolides and lincosamides [27], mammalian mitochondrial ribosomes are only sensitive to aminoglycosides but not to macrolides or lincosamides [24,28,29]. Also, cytoplasmic ribosomes are not sensitive to any of these classes of antibiotics [27].
Both macrolides and lincosamides bind downstream of the peptidyl transferase center (PTC) in the peptide exit channel, through which nascent polypeptide exits the large ribosomal subunit during protein synthesis (Fig. 3A, B). When macrolides are bound to the ribosomes, the hydrophobic side of their lactone ring rests on the surface that is formed by the bases of U2611, A2058, and A2059 (Fig. 3D) [30–34]. Since they obstruct the exit tunnel, macrolides allow the synthesis of only short oligopeptides [35].
Figure 3. Antibiotics bound in the exit tunnel and the peptidyl transferase center of eubacterial 70S ribosome.
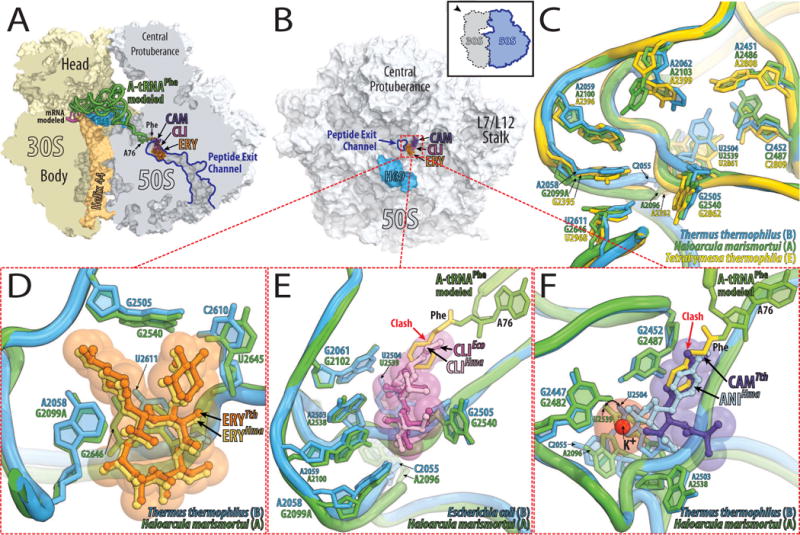
(A, B) Overview of the superimposed binding sites of erythromycin (ERY, orange), clindamycin (CLI, pink), and chloramphenicol (CAM, purple) bound to the Thermus thermophilus 70S ribosome viewed from two different perspectives. (A) 70S ribosome is cut open along the exit tunnel. Shown in light yellow is the 30S subunit with h44 in light orange and in light blue is the 50S subunit with H69 in marine. The mRNA is shown in magenta. Bound to the A-site is an amino-acylated tRNAPhe shown in green with its 3′-terminal phenylalanine residue in yellow. (B) The 50S subunit viewed from the 30S subunit as indicated by the inset and with the same color-coding as in (A). (C) A close up view of the superimposed antibiotic binding sites in ribosomes from the three different kingdoms of life. The drugs are omitted for clarity. As an example of the eubacterial binding site (B) the 23S rRNA from Thermus thermophilus (blue) is shown, for arecheal (A) the 23S rRNA from Haloarcula marismortui (green), and for eukaryotic (E) the 28S rRNA from Tetrahymena thermophila (yellow). (D, E, F) Close up views of the comparisons between eubacterial (blue) and archaeal (green) antibiotic binding sites. (D) Comparison of the structures of erythromycin (ERY) bound to Thermus thermophilus 70S ribosome (orange) and bound to Haloarcula marismortui 50S subunit carrying a G2099A mutation (yellow). (E) Clindamycin bound to Escherichia coli 70S ribosome is displayed in dark pink and in light pink when bound to Haloarcula marismortui 50S subunit carrying a G2099A mutation. (F) Chloramphenicol bound to Thermus thermophilus 70S ribosome is displayed in purple and in light blue is anisomycin when bound to Haloarcula marismortui 50S subunit. Potassium ion is shown in red.
Lincosamides bind in the same pocket as macrolides, forming extensive hydrogen bonds with A2058, A2059, A2503, and G2505 in E. coli (Fig. 3E) [31]. At the same time, they extend from this binding pocket into the PTC, where they interfere with the accommodation of the incoming aminoacyl-tRNA into the A-site [31,33].
Adenine in position 2058 is well conserved throughout all eubacteria [8], while eukaryotes, archaea and mitochondria have guanine in this position, which correlates with their observed insensitivity to macrolides and lincosamides (Fig. 3C, Fig. 4). In general, organisms with guanine in position 2058 are much more resistant to macrolides or lincosamides compared to those with adenine in this position [36]. The presence of guanine in position 2058 not only prevents essential hydrogen bonding between macrolides/lincosamides and the 23S rRNA, but its solvated amino-group hinders the stacking between the hydrophobic surface of the lactone ring of the antibiotic and the base of 2058. The G2058A mutation in the large ribosomal subunit of the halophilic archaeon Haloarcula marismortui increases the affinity for these antibiotics by four orders of magnitude [33].
Figure 4. Secondary structure and phylogenetic variations of the eubacterial 23S rRNA that forms the peptidyl transferase ring.
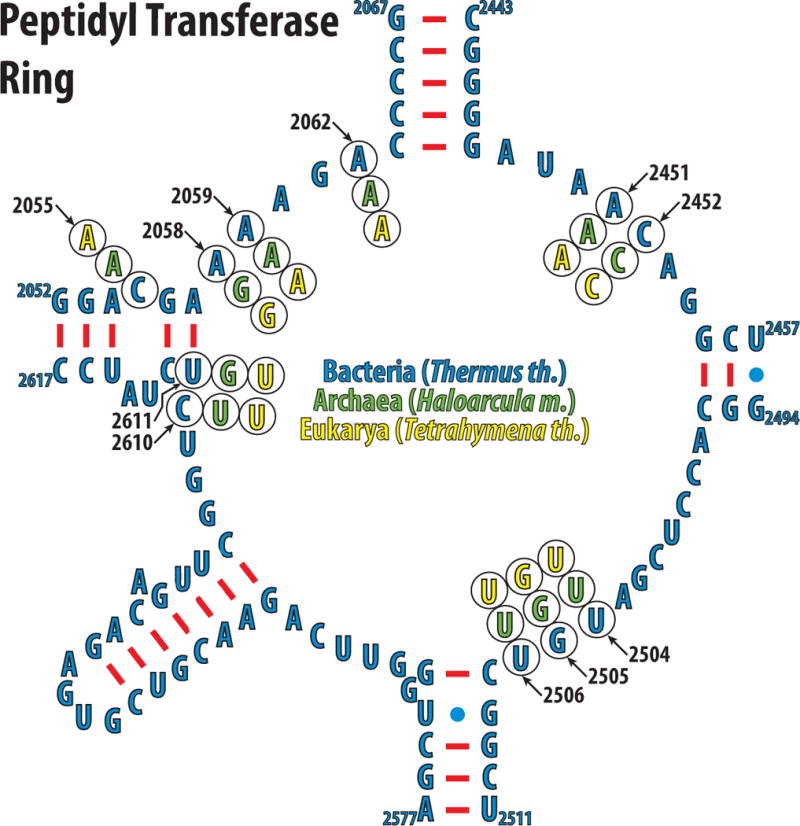
Nucleotides of the peptidyl transferase ring of the eubacteria Thermus thermophilus are shown in blue. The phylogenetic variations of the nucleotides discussed in the text are shown in green for archeon Haloarcula marismortui, and yellow for eukaryote Tetrahymena thermophila.
Despite the appeal of this explanation, variations in the structures of different macrolides as well as phylogenetic differences outside nucleotide 2058 can influence the specificity of each drug idiosyncratically. Although wild type H. marismortui has a guanine in position 2058 of 23S rRNA, it is still sensitive to some macrolides such as carbomycin and tylosin [37]. Ribosomes from Saccharomyces cerevisiae that carry a G2058A mutation are nevertheless resistant to the macrolide erythromycin [38], confirming that nucleotides outside the antibiotic-binding pocket can modulate ribosome’s response to a particular drug.
A comparison of the known structures of the lincosamide clindamycin bound to the eubacterial and archaeal ribosomes reveals that the positions of nucleotides 2504–2507 vary depending on the system studied (Fig. 3E) [31,33,39]. All the observed differences emanate from U2504 and its interactions with the nucleotide in position 2055. In the G2058A mutant of H. marismortui, the flexibility of U2504 is limited by its stacking onto A2055. In E. coli, a cytosine in position 2055 allows U2504, along with G2505, U2506, and C2507 to adopt a conformation that not only allows an additional hydrogen bond between G2505 and the antibiotic, but also increases the van der Waals contacts between the drug and the ribosome. Thus, the phylogenetic differences in position 2055 lead to conformational changes of the conserved nucleotides U2504, G2505, U2506, and C2507. A C2055A mutation in eubacteria not only increases the resistance to clindamycin, as expected, but also to chloramphenicol [40].
Specificity of antibiotics binding to the A-site crevice
Chloramphenicol inhibits protein synthesis by both bacterial and mitochondrial, but not cytoplasmic ribosomes [41]. Inhibition of mitochondrial protein synthesis during chloramphenicol treatment is generally accepted to be the underlying mechanism of dose-dependent reversible bone marrow suppression [42]. Currently, chloramphenicol is rarely used, due to its severe side effects and the availability of newer antibiotics with safer clinical profiles [22].
Chloramphenicol binds in the so-called A-site crevice that is formed by the bases of A2451 and C2452 of the 23S rRNA and prevents the amino-acid side chain of an incoming aminoacylated tRNA from binding (Fig. 3F). Chloramphenicol’s nitrobenzyl ring stacks onto the base of C2452 of the 23S rRNA, and its methylene-hydroxyl group interacts with an adjacent potassium ion that is coordinated by G2061, G2447, and C2501 [30].
The binding of chloramphenicol mirrors the binding of another drug, anisomycin, which is specific for archaeal and eukaryotic, but not eubacterial ribosomes (Fig. 3F). Both antibiotics are coordinated by an adjacent potassium ion, and both of their aromatic rings stack on the C2452 base. However, the different positions of U2504 in eubacteria vs. both archaea and eukaryotes, determine the specificity of chloramphenicol for eubacteria, and of anisomycin for archaea/eukaryotes [30].
The nucleotide in position 2055, which is either a cytosine in eubacteria or adenine in archaea and eukaryotes (Fig. 3C, Fig. 4) [43], determines the specificity not only for chloramphenicol and anisomycin but also for a whole spectrum of A-site inhibitors [44,45], including linezolid [46,47]. Similar to chloramphenicol, linezolid causes bone marrow suppression and lactic acidosis, which is directly linked to the inhibition of mitochondrial protein synthesis [48].
Phylogenetic variations in the rRNA nucleotide sequences must have similar effects on drug affinity and specificity as spontaneous resistance mutations. For instance, structural studies of anisomycin-resistant mutant ribosomes from H. marismortui elucidated that the most common mutations conferring resistance either destabilize the antibiotic bound state or sterically block the antibiotic binding pocket. A less common trend in acquiring drug resistance is the increase of the energetic barrier for antibiotic binding, that can be achieved either by stabilization of the unbound state or by introducing an additional state with a lower energy than the unbound state. At the same time, all the mutations leading to antibiotic resistance cannot exceed the energetic barriers that would prevent the substrate from binding [49].
The affinity and selectivity of antibiotics can be determined not only by the phylogenetic variations in ribosomal RNA, but also by variations in ribosomal proteins, as observed in the case of E-site specific antibiotics.
Specificity of E-site specific antibiotics
Haloarcula marismortui ribosomal protein L44e (homologous to L36A in Tetrahymena thermophila and to RPL41 in Saccharomyces cerevisiae) significantly contributes to the binding of the deacylated tRNA into the E-site, however, it is only present in archaea and eukaryotes and has no counterpart in eubacteria. This protein provides the binding platform for 13-deoxytedanolide to the E-site enabling it to compete with the 3′-terminal adenine of the deacylated tRNA (Fig. 5A, B) [50,51]. In eubacteria, L44e is replaced by the structurally unrelated ribosomal protein L28, which partially occupies the space required for the binding of 13-deoxytedanolide (Fig. 5B), therefore conferring natural resistance to eubacterial ribosomes against this antibiotic. In yeast, a single amino acid substitution in the unstructured loop of RPL41 (L44e homolog) that is necessary for 13-deoxytedanolide binding confers resistance to eukaryotic-specific cycloheximide [52]. A recent X-ray crystal structure of the Tetrahymena thermophila 60S ribosomal subunit confirmed the cycloheximide binding pocket in the E-site of the large ribosomal subunit (Fig. 5C, D). However, the detailed interactions of cycloheximide with the ribosome are still elusive, as the resolution of the electron density map was not sufficient to place the drug unambiguously [53].
Figure 5. Antibiotics that compete with the binding of the deacylated tRNA to the E-site.
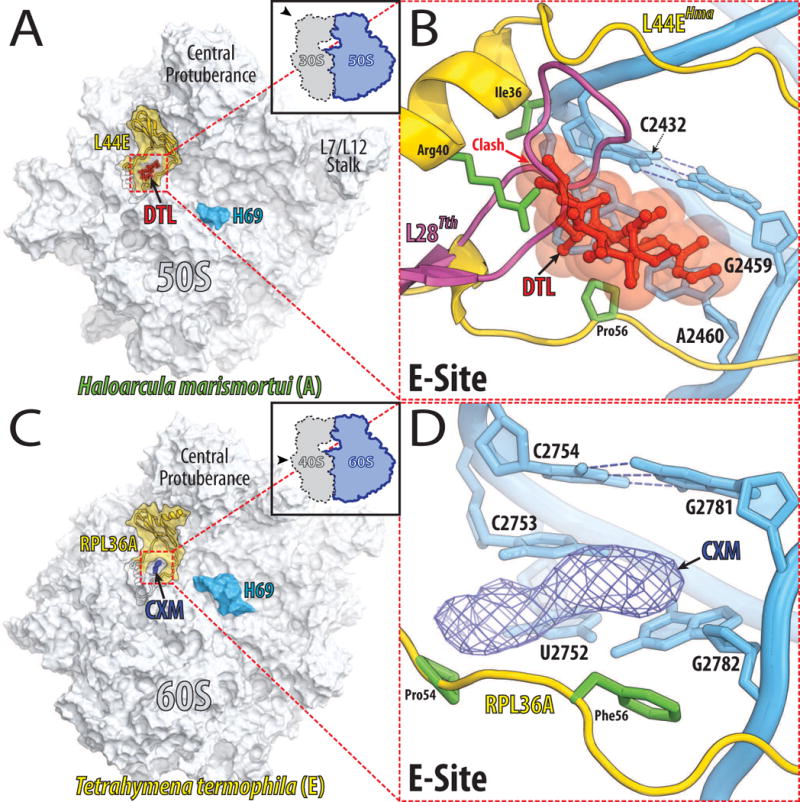
(A) Overview of the binding sites of 13-deoxytedanolide (DTL, red) on the large ribosomal subunit from archaeon Haloarcula marismortui as viewed from the 30S subunit [50]. Black contour outlines L44e protein, which surrounds the E-site and whose visible portion is highlighted in yellow. (B) A close up view of the 13-deoxytedanolide binding site. The 23S rRNA is colored in blue and L44e is in yellow with its Ile36, Arg40, and Pro56 side-chains in green. A model of the L28 ribosomal protein from Thermus thermophilus is shown in pink. Note, that L28 partially occupies the space required for 13-deoxytedanolide binding to ribosome. (C) Overview of the cycloheximide binding site on the 60S ribosomal subunit from eukaryote Tetrahymena thermophila [53]. The 60S subunit (light blue) is viewed from the 40S ribosomal subunit. Colored in yellow is the visible portion of RPL36A, which encircles the E-site. The electron density map for cycloheximide (CXM) contoured at 3σ is represented as a blue mesh. (D) A close up view of the cycloheximide binding site. The 26S rRNA is colored in blue and RPL36A is in yellow with the Pro54 and Phe56 side-chains in green. The electron density map for cycloheximide (CXM) is represented as a blue mesh.
CONCLUSION
At the beginning of the last century Paul Ehrlich recognized the problem of drug specificity and proposed the concept of “selective toxicity” [54]. Today, the development of new drugs with minimal toxicity is still a challenge [6]. Numerous side effects of antimicrobial inhibitors targeting the ribosomes are linked to inadvertent inhibition of protein synthesis in mitochondria [55]. Despite similar responses to many antibiotics by eubacterial and mitochondrial ribosomes, the ribosomes are distinctively different from each other. Ribosomes isolated from mitochondria of different species range in size from 55S to 80S. Mammalian mitochondrial ribosomes are close in size and weight to E. coli ribosomes. However, they contain less than half of the rRNA, even missing a 5S rRNA homolog, and nearly twice the number of ribosomal proteins compared to E. coli ribosomes [56]. Despite all the insights gained from the comparison of the structures of ribosomes isolated from organisms belonging to different kingdoms of life, inadvertent inhibition of mitochondrial protein synthesis remains unpredictable. Presumably, an atomic structure of mitochondrial ribosome could significantly facilitate rational design of ribosome targeting antibiotics with improved clinical profiles, effective against pathogenic bacteria yet with minimal side effects.
HIGHLIGHTS.
-
➢
Correlation between antibiotic specificity and observed structural differences
-
➢
Mitochondrial and eubacterial ribosomes respond differently to the same antibiotics
-
➢
Selectively targeting eubacterial over mitochondrial ribosomes can minimize drug side effects
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Helices of the 23S and 16S rRNA are indicated with upper-case H and lower-case h, respectively
E. coli numbering for nucleotides of ribosomal RNA is used throughout the text
References
- 1•.Wilson DN. The A–Z of bacterial translation inhibitors. Critical Reviews in Biochemistry and Molecular Biology. 2009;44:393–433. doi: 10.3109/10409230903307311. An extensive review of the recent structures of inhibitors bound to the ribosome. [DOI] [PubMed] [Google Scholar]
- 2••.Sohmen D, Harms JM, Schlunzen F, Wilson DN. Enhanced SnapShot: Antibiotic inhibition of protein synthesis II. Cell. 2009;139:212–212 e211. doi: 10.1016/j.cell.2009.08.009. An appealing graphical representation of the effects of many different classes of inhibitors on protein translation. [DOI] [PubMed] [Google Scholar]
- 3.Poehlsgaard J, Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat Rev Microbiol. 2005;3:870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- 4.Kannan K, Mankin AS. Macrolide antibiotics in the ribosome exit tunnel: species-specific binding and action. Antimicrobial Therapeutics Reviews: Antibiotics That Target the Ribosome. 2011;1241:33–47. doi: 10.1111/j.1749-6632.2011.06315.x. [DOI] [PubMed] [Google Scholar]
- 5•.Spahn CM, Prescott CD. Throwing a spanner in the works: antibiotics and the translation apparatus. J Mol Med (Berl) 1996;74:423–439. doi: 10.1007/BF00217518. in combination with [1] and [41] represent most of the knowledge accumulated on inhibitors of protein translation over the past decades. [DOI] [PubMed] [Google Scholar]
- 6.Bottger EC. Antimicrobial agents targeting the ribosome: the issue of selectivity and toxicity - lessons to be learned. Cell Mol Life Sci. 2007;64:791–795. doi: 10.1007/s00018-007-6431-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryskier A. Antimicrobial agents: antibacterials and antifungals. Washington, D.C.: ASM Press; 2005. [Google Scholar]
- 8.Bottger EC, Springer B, Prammananan T, Kidan Y, Sander P. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Rep. 2001;2:318–323. doi: 10.1093/embo-reports/kve062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forge A, Schacht J. Aminoglycoside antibiotics. Audiol Neurootol. 2000;5:3–22. doi: 10.1159/000013861. [DOI] [PubMed] [Google Scholar]
- 10.Hutchin T, Cortopassi G. Proposed molecular and cellular mechanism for aminoglycoside ototoxicity. Antimicrob Agents Chemother. 1994;38:2517–2520. doi: 10.1128/aac.38.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies J, Davis BD. Misreading of Ribonucleic Acid Code Words Induced by Aminoglycoside Antibiotics - Effect of Drug Concentration. Journal of Biological Chemistry. 1968;243:3312. [PubMed] [Google Scholar]
- 12.Marrero P, Cabanas MJ, Modolell J. Induction of translational errors (misreading) by tuberactinomycins and capreomycins. Biochem Biophys Res Commun. 1980;97:1042–1047. doi: 10.1016/0006-291x(80)91481-3. [DOI] [PubMed] [Google Scholar]
- 13.Hobbie SN, Akshay S, Kalapala SK, Bruell CM, Shcherbakov D, Bottger EC. Genetic analysis of interactions with eukaryotic rRNA identify the mitoribosome as target in aminoglycoside ototoxicity. Proc Natl Acad Sci U S A. 2008;105:20888–20893. doi: 10.1073/pnas.0811258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akbergenov R, Shcherbakov D, Matt T, Duscha S, Meyer M, Wilson DN, Bottger EC. Molecular basis for the selectivity of antituberculosis compounds capreomycin and viomycin. Antimicrob Agents Chemother. 2011;55:4712–4717. doi: 10.1128/AAC.00628-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 17••.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. A comprehensive review on A-site decoding. By combining structural, biochemical and genetic evidence the author put forward how the structurally observed induced fit of tRNA binding can explain the observed biochemical and genetic data. [DOI] [PubMed] [Google Scholar]
- 18.Stanley RE, Blaha G, Grodzicki RL, Strickler MD, Steitz TA. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat Struct Mol Biol. 2010;17:289–293. doi: 10.1038/nsmb.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 20.Selmer M, Dunham CM, Murphy FVT, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 21.Pape T, Wintermeyer W, Rodnina MV. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat Struct Biol. 2000;7:104–107. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- 22••.Cohen BH. Pharmacologic Effects on Mitochondrial Function. Developmental Disabilities Research Reviews. 2010;16:189–199. doi: 10.1002/ddrr.106. A concise introduction to the side effects of antibiotics as a result of mitochondrial dysfunction. [DOI] [PubMed] [Google Scholar]
- 23.Prezant TR, Agapian JV, Bohlman MC, Bu X, Oztas S, Qiu WQ, Arnos KS, Cortopassi GA, Jaber L, Rotter JI, et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 24.Zhao H, Li R, Wang Q, Yan Q, Deng JH, Han D, Bai Y, Young WY, Guan MX. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am J Hum Genet. 2004;74:139–152. doi: 10.1086/381133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hobbie SN, Bruell CM, Akshay S, Kalapala SK, Shcherbakov D, Bottger EC. Mitochondrial deafness alleles confer misreading of the genetic code. Proc Natl Acad Sci U S A. 2008;105:3244–3249. doi: 10.1073/pnas.0707265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bulkley D, Johnson F, Steitz TA. The Antibiotic Thermorubin Inhibits Protein Synthesis by Binding to Inter-Subunit Bridge B2a of the Ribosome. J Mol Biol. 2012 doi: 10.1016/j.jmb.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cammarano P, Teichner A, Londei P, Acca M, Nicolaus B, Sanz JL, Amils R. Insensitivity of archaebacterial ribosomes to protein synthesis inhibitors. Evolutionary implications. EMBO J. 1985;4:811–816. doi: 10.1002/j.1460-2075.1985.tb03702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denslow ND, O’Brien TW. Antibiotic susceptibility of the peptidyl transferase locus of bovine mitochondrial ribosomes. Eur J Biochem. 1978;91:441–448. doi: 10.1111/j.1432-1033.1978.tb12696.x. [DOI] [PubMed] [Google Scholar]
- 29.Ibrahim NG, Burke JP, Beattie DS. The sensitivity of rat liver and yeast mitochondrial ribosomes to inhibitors of protein synthesis. J Biol Chem. 1974;249:6806–6811. [PubMed] [Google Scholar]
- 30.Bulkley D, Innis CA, Blaha G, Steitz TA. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc Natl Acad Sci U S A. 2010;107:17158–17163. doi: 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunkle JA, Xiong L, Mankin AS, Cate JH. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci U S A. 2010;107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson DN, Harms JM, Nierhaus KH, Schlunzen F, Fucini P. Species-specific antibiotic-ribosome interactions: implications for drug development. Biol Chem. 2005;386:1239–1252. doi: 10.1515/BC.2005.141. [DOI] [PubMed] [Google Scholar]
- 33.Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 34•.Hansen JL, Ippolito JA, Ban N, Nissen P, Moore PB, Steitz TA. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol Cell. 2002;10:117–128. doi: 10.1016/s1097-2765(02)00570-1. Based on the presented structure the authors proposed that a G2099A mutation should enable the binding of erythromycin to Haloarcula marismortui ribosomes. This hypothesis was proven to be right later in [33] [DOI] [PubMed] [Google Scholar]
- 35.Tenson T, Lovmar M, Ehrenberg M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J Mol Biol. 2003;330:1005–1014. doi: 10.1016/s0022-2836(03)00662-4. [DOI] [PubMed] [Google Scholar]
- 36.Vester B, Douthwaite S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob Agents Chemother. 2001;45:1–12. doi: 10.1128/AAC.45.1.1-12.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanz JL, Marin I, Urena D, Amils R. Functional-Analysis of 7 Ribosomal Systems from Extremely Halophilic Archaea. Canadian Journal of Microbiology. 1993;39:311–317. [Google Scholar]
- 38.Bommakanti AS, Lindahl L, Zengel JM. Mutation from guanine to adenine in 25S rRNA at the position equivalent to E. coli A2058 does not confer erythromycin sensitivity in Sacchromyces cerevisae. RNA. 2008;14:460–464. doi: 10.1261/rna.786408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 40.Long KS, Poehlsgaard J, Hansen LH, Hobbie SN, Bottger EC, Vester B. Single 23S rRNA mutations at the ribosomal peptidyl transferase centre confer resistance to valnemulin and other antibiotics in Mycobacterium smegmatis by perturbation of the drug binding pocket. Mol Microbiol. 2009;71:1218–1227. doi: 10.1111/j.1365-2958.2009.06596.x. [DOI] [PubMed] [Google Scholar]
- 41.Vázquez DI. Inhibitors of Protein Biosynthesis. Berlin; New York: Springer-Verlag; 1979. [Google Scholar]
- 42.Yunis AA. Chloramphenicol: relation of structure to activity and toxicity. Annu Rev Pharmacol Toxicol. 1988;28:83–100. doi: 10.1146/annurev.pa.28.040188.000503. [DOI] [PubMed] [Google Scholar]
- 43.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davidovich C, Bashan A, Auerbach-Nevo T, Yaggie RD, Gontarek RR, Yonath A. Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity. Proc Natl Acad Sci U S A. 2007;104:4291–4296. doi: 10.1073/pnas.0700041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gurel G, Blaha G, Moore PB, Steitz TA. U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome. J Mol Biol. 2009;389:146–156. doi: 10.1016/j.jmb.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ippolito JA, Kanyo ZF, Wang D, Franceschi FJ, Moore PB, Steitz TA, Duffy EM. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J Med Chem. 2008;51:3353–3356. doi: 10.1021/jm800379d. [DOI] [PubMed] [Google Scholar]
- 47.Wilson DN, Schluenzen F, Harms JM, Starosta AL, Connell SR, Fucini P. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc Natl Acad Sci US A. 2008;105:13339–13344. doi: 10.1073/pnas.0804276105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garrabou G, Soriano A, Lopez S, Guallar JP, Giralt M, Villarroya F, Martinez JA, Casademont J, Cardellach F, Mensa J, et al. Reversible inhibition of mitochondrial protein synthesis during linezolid-related hyperlactatemia. Antimicrob Agents Chemother. 2007;51:962–967. doi: 10.1128/AAC.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blaha G, Gurel G, Schroeder SJ, Moore PB, Steitz TA. Mutations outside the anisomycin-binding site can make ribosomes drug-resistant. J Mol Biol. 2008;379:505–519. doi: 10.1016/j.jmb.2008.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schroeder SJ, Blaha G, Tirado-Rives J, Steitz TA, Moore PB. The structures of antibiotics bound to the E site region of the 50 S ribosomal subunit of Haloarcula marismortui: 13-deoxytedanolide and girodazole. J Mol Biol. 2007;367:1471–1479. doi: 10.1016/j.jmb.2007.01.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51•.Schmeing TM, Moore PB, Steitz TA. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA. 2003;9:1345–1352. doi: 10.1261/rna.5120503. First structural definition of the E-site on the large ribosomal subunit. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawai S, Murao S, Mochizuki M, Shibuya I, Yano K, Takagi M. Drastic alteration of cycloheximide sensitivity by substitution of one amino acid in the L41 ribosomal protein of yeasts. J Bacteriol. 1992;174:254–262. doi: 10.1128/jb.174.1.254-262.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53••.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–948. doi: 10.1126/science.1211204. One of the first high resolution structures of the eukaryotic large ribosomal subunit. [DOI] [PubMed] [Google Scholar]
- 54.Parascandola J. The Theoretical Basis of Ehrlich,Paul Chemotherapy. Journal of the History of Medicine and Allied Sciences. 1981;36:19–43. doi: 10.1093/jhmas/xxxvi.1.19. [DOI] [PubMed] [Google Scholar]
- 55.Jones CN, Miller C, Tenenbaum A, Spremulli LL, Saada A. Antibiotic effects on mitochondrial translation and in patients with mitochondrial translational defects. Mitochondrion. 2009;9:429–437. doi: 10.1016/j.mito.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 56.O’Brien TW, Denslow ND, Faunce WH, Anders JC, Liu J, O’Brien BJ. Structure and function of mammalian mitochondrial ribosomes. In: Nierhaus KH, Franceschi F, Subramanian AR, Erdmann VA, Wittmann-Liebold B, editors. The Translational apparatus: structure, function, regulation, evolution. Plenum Press; 1993. pp. 575–586. [Google Scholar]
- 57.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 59.Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FVT, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484:256–259. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- 61•.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. First X-ray crystal structure addressing the ribosomal peptidyl transferase reaction. [DOI] [PubMed] [Google Scholar]


