Graphical Abstract
Decellularized cartilage microparticles, and all associated native signals, are delivered to hMSC populations in a dense, type I collagen matrix. Hybrid usage of native tissue signals and the engineering control of collagen matrices show the ability to induce local infiltration and differentiation of hMSCs. Additionally, the solid cartilage microparticles inhibit bulk cell-mediated contraction of the composite.
Keywords: 3D microenvironment, differentiation, mesenchymal stem cells, reduction and reconstitution, guanidine hydrochloride
Current use of decellularized articular cartilage as a regenerative platform suffers from limited implant diffusion characteristics and cellular infiltration. Attempts to address this concern using decellularized cartilage microparticles allows for customized implant shape, tailored porosity, and improved cell infiltration. However, these developments utilize severe crosslinking agents that adversely affect cell differentiation, and fail to attain clinically relevant mechanical properties required for implant survival. We have overcome these issues through the formation of a composite approach, combining the advantages of mature, decellularized tissue with tunable features of a reconstituted collagen hydrogel system. Through the application of a plastic compression regime, we formed cellularized composite structures that exceeded the percolation threshold of the cartilage microparticles and exhibited clinically relevant mechanical properties. We also utilized chemical reduction and mechanical reconstitution methods to investigate the contributions of GAG and collagenous components to chondrogenic induction and matrix properties. With the inclusion of human mesenchymal stem cells into the composite system, we showed microenvironment dependent cell morphology and phenotype when in contact with cartilage microparticles. Our work demonstrates a cartilage microparticle composite matrix with clinically relevant mechanical properties, and chondrogenic differentiation of hMSCs that infiltrate both native and chemically reduced cartilage microparticles.
1. Introduction
Decellularization has been advanced as a viable strategy for tissue regeneration in the face of traumatic injury and degenerative diseases [1, 2]. Decellularized tissues contain the mature, healthy extracellular matrix (ECM) of the target tissue, where the native decellularized ECM is known to direct host cell infiltration, migration, and phenotype induction, and ultimately promote integration with the host tissue [3–6]. The biological makeup of ECM components of various tissues is highly conserved and largely non-immunogenic, allowing for autologous, allogenic, and xenogenic transplantation of decellularized tissues with relatively minimized risk to the host [1, 7]. Decellularization has the potential to be useful for the treatment of articular cartilage defects, which are resilient to natural repair and subject to high mechanical forces, promoting further tissue degeneration [1, 8, 9].
Recent studies have focused on the in vitro and in vivo regeneration potential of whole chondral or osteochondral tissue [5, 7, 10, 11]. These studies have noted that the relatively dense ECM morphology of articular cartilage limits complete decellularization due to the lack of penetration of decellularization agents [7, 12]. Additionally, once the tissue has been decellularized, the high tissue density prevents cellular infiltration and remodeling in the long term, often requiring additional modification of the ECM (e.g. guanidine-hydrochloride reduction of glycosaminoglycans) to increase tissue porosity [1]. However, additional modifications are often not successful in promoting bulk cellular infiltration, which can lead to global degeneration and limited utility of the implant in vivo [1].
Recently, the field has developed methods that help to retain the local mechanical, structural, and biochemical microenvironment of native articular cartilage, but allow for easier penetration of decellularization agents as well as means to increase cellular infiltration into the dense native tissue [6, 11–13]. These methods are largely focused on the use of atomized (a.k.a. pulverized, microparticulated) cartilage tissue that is size controlled to create a powder of fragments of cartilage. Fragments are then either crosslinked to each other or suspended in a polymerizable medium to create three-dimensional constructs of tunable size and/or cartilage microparticle density, which can promote cell attachment and upregulation of cartilage specific genes (e.g. type II collagen, SOX9, aggrecan) to form neocartilage [2]. These studies have been performed with primary chondrocytes and a variety of stem cell sources, typically showing local induction when in the presence of decellularized cartilage microparticles. Additionally, in vivo rabbit models of these techniques have shown significant cartilage regeneration as compared to an empty defect [2]. However, current development of these constructs is largely dependent on hostile exogenous crosslinking conditions, requiring cell-seeding post-crosslinking such that cells are not globally distributed in the 3D construct. Further, the cartilage microparticle density is largely dependent on the packing efficiency of the particles but otherwise uncontrolled. Finally, it remains unknown what specific aspects of the cartilage structural or biochemical nature that contributes to the positive results seen in previous research.
There remains a need to (1) create a more tunable, controlled system of cartilage microparticle construct formation without the need for exogenous crosslinkers and (2) further examine the contribution of the complex native cartilage microenvironment to the induction of chondrogenic differentiation. Consequently, we developed a tunable system of decellularized cartilage microparticles and a polymerizable collagen matrix for the formation of a 3D composite material. We further applied a densification regime, recently described by our laboratory [14], to increase the strength of the collagen matrix and increase the packing and spacing of the cartilage microparticles to a controlled level. Finally, we added a series of guanidine reduction and mechanical reconstitution mixes of cartilage microparticles to investigate the contribution of covalently bound GAGs and the structural, collagenous ECM. We specifically investigated the following objectives, i.e. to: (1) create cartilage microparticle powders of various stages of reduction and reconstitution using guanidine-hydrochloride (Figure 1), (2) fabricate tunable microparticle-collagen composites of controlled microparticle density, (3) measure the mechanical and ultrastructural properties of microparticle-collagen composites with the various stages of reduction and reconstitution, and (4) investigate the cellular induction qualities of unmodified, reduced, and reconstituted cartilage microparticle-collagen composites on a human mesenchymal stem cells over a short term culture period.
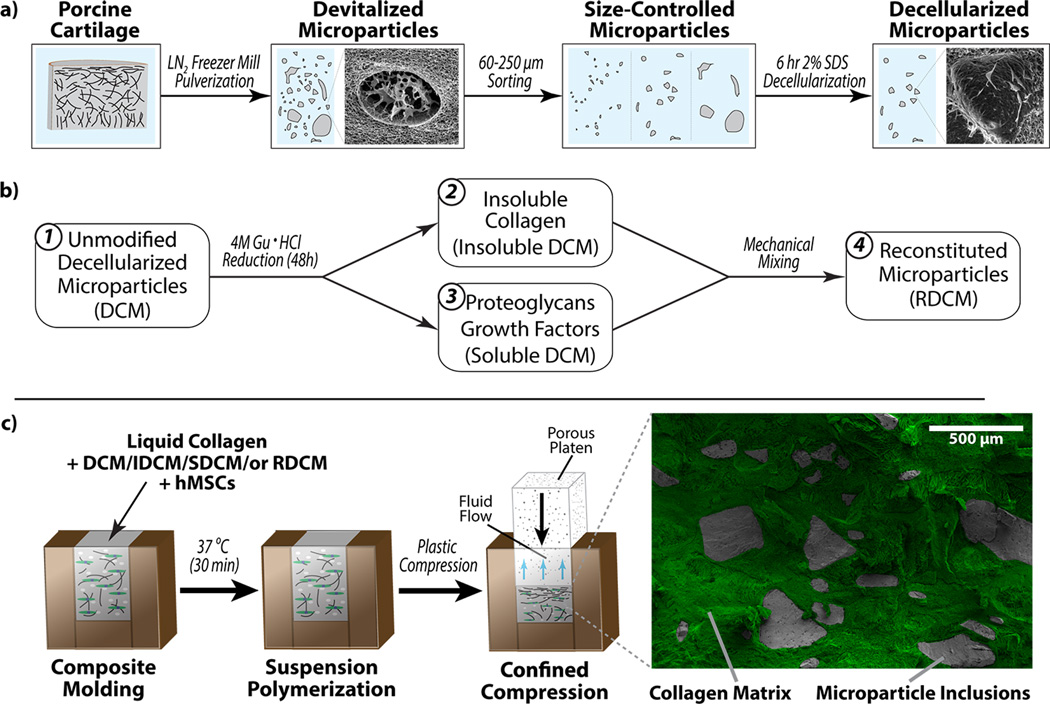
Figure 1.
Decellularized cartilage microparticles, or their constituents, suspended in a densified collagen composite matrix. a) Porcine cartilage tissue was harvested, devitalized and pulverized in a liquid nitrogen freezer mill and size controlled for the formation of standard decellularized cartilage microparticles. SEM images show fragmented cell bodies representing cell devitalization and empty lacunae representing the result of tissue decellularization. b) Four cartilage microparticles groups were developed including (1) unmodified decellularized cartilage microparticles (DCM), (2) insoluble (mostly collagenous) fraction of cartilage microparticles from Gu·HCl reduction (IDCM), (3) soluble (proteoglycan and glycoprotein) fraction from Gu·HCl reduction (SDCM), and (4) mechanical mixing of SDCM and IDCM to form reconstituted cartilage microparticles (RDCM). c) Various microparticle groups (false colored grey), and viable cells, were polymerized in a type I collagen matrix (false colored green) and compressed to create high strength, high-density microparticle-collagen composite matrices.
2. Results and Discussion
2.1. Microparticles Increase Composite Mechanical Properties Via Percolation
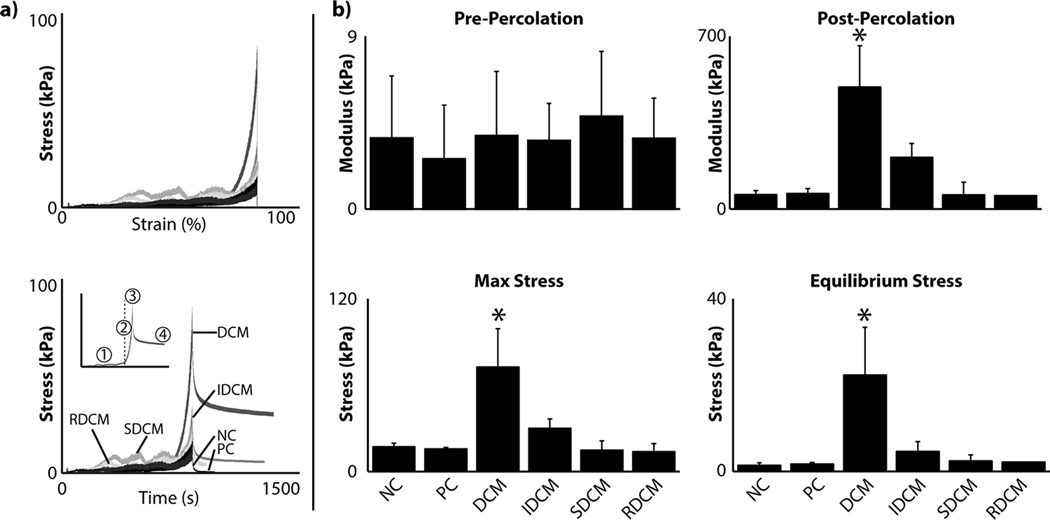
The mechanical properties of the composite matrices were found to be dependent on the cartilage microparticle groups (Figure 2). These groups include otherwise unmodified decellularized cartilage microparticles (DCM), the insoluble product of the guanidine treatment to cartilage microparticles (IDCM), the soluble product of the guanidine treatment to cartilage microparticles (SDCM), and the reconstitution of the two guanidine products (IDCM+SDCM=RDCM). Additionally, positive and negative control groups (PC and NC, respectively) were included for cellular analysis. Both PC (with TGF-β3 supplement) and NC (without supplement) samples were composed of polymerized type I collagen matrices without the inclusion of any native cartilage component. The linear compression modulus at low strain (pre-percolation) was not statistically dependent on the cartilage microparticle treatment (p=0.820). At high strain, however, near the end of the densification regime, the linear modulus of compression was highly dependent on cartilage microparticle treatment (p<0.001), where samples with the unreduced microparticles (DCM group) had a significantly higher linear modulus compared to other groups. It is likely that the percolation threshold was met for these samples, where the previously suspended particles reached continuous contact throughout the composite matrix, allowing the mechanical properties of microparticles to contribute on a bulk scale. Similarly, max stress and equilibrium stress were found to be dependent on cartilage microparticle groups (p<0.001 for both), where the unreduced DCM group exhibited higher max stress and equilibrium stress.
Figure 2.
Cartilage microparticles influenced composite matrix mechanical properties. a) Representative stress plots show differences in cartilage microparticle groups plotted against both strain and time. Inset shows different regions of interest for mechanical analysis: (1) low strain linear modulus, (2) high strain, post-percolation threshold modulus, (3) maximum stress, and (4) equilibrium stress, where the dashed line shows the percolation threshold. b) At low strain, where the collagen matrix dominates the mechanical properties, there was no significant difference among samples (p=0.820). Post-percolation threshold and for the duration of the mechanical compression, the response was significantly different for percolation linear modulus, maximum stress, and equilibrium stress, where the DCM sample was higher in all categories (*=p<0.001).
Notably, composite matrices using the reduced soluble component (SDCM and RDCM groups) were much more difficult and fragile to handle. Post-compression, we observed splitting and fraying of both composites, such that the geometric integrity of the matrices were not intact, where all other samples were easily handled and exhibited geometric integrity throughout compression and transfer processes. As a result, we measured the maintenance of the soluble portion through dimethylmethylene blue (DMMB) analysis in the composite matrices to ensure leaching did not occur in the short term. Our one-week study showed readings in the surrounding culture media below the noise level for DMMB colorimetric and 280 nm absorbance analysis at 4 and 7 days, indicating that, by our systems of measurement, no soluble cartilage microparticle (SDCM) leaching was occurring.
Through the densification of the composite matrices, we were able to tune the final concentration of cartilage microparticles. Interestingly, all samples reached the percolation threshold, wherein the macro-scale structure of the composite matrix was compacted to a point where continuous contact between particles occurred throughout the bulk of the matrix [15], as apparent by the transition in mechanical properties to the much higher properties of the cartilage microparticles via stress shielding to the collagen network. Since this effect was not seen in the control samples or previous densification work [14], it is likely that the percolation occurred between cartilage microparticles, and not as an effect of percolation in the surrounding collagen matrix. Composite matrices were able to be formed at a range of 1–4% of extracellular matrix components (wet weight), at the range, and exceeding, that of native cartilage [8]. However, as noted, higher densities of microparticles led to fragmenting and a lack of structural integrity of the bulk matrix, likely due to failure of the interstitial collagen matrix due to loading or shearing between cartilage microparticles.
The mechanical response of the composite matrices was found to be highly dependent on microparticle group, where the highest mechanical response was seen in the unmodified DCM group, with all collagen-GAG interactions inherent in the native cartilage present. The IDCM group, in absence of the covalently bound GAGs, showed a statistically similar mechanical response to collagen-only NC and PC samples, consistent with previous literature where GAGs are implicated as the primary contributor to cartilage compressive properties [1, 16]. However, all differences in mechanical response were seen at high strain in the compression regime, past the percolation threshold (Figure 2) where cartilage microparticle mechanics dominated. At lower strain levels, linear moduli between samples were not statistically different, as the strain was likely localized to the surrounding collagen matrix. Further, samples which include the reduced, soluble GAG component (SDCM and RDCM) were found to have much lower mechanical properties and additionally had little structural integrity and were difficult to handle, suggesting that the soluble GAG component interfered with the fibril structure of the collagen matrix, similar to what has been reported previously [17].
2.2. Isolated Proteoglycan Components Alter Collagen Fibril Morphology
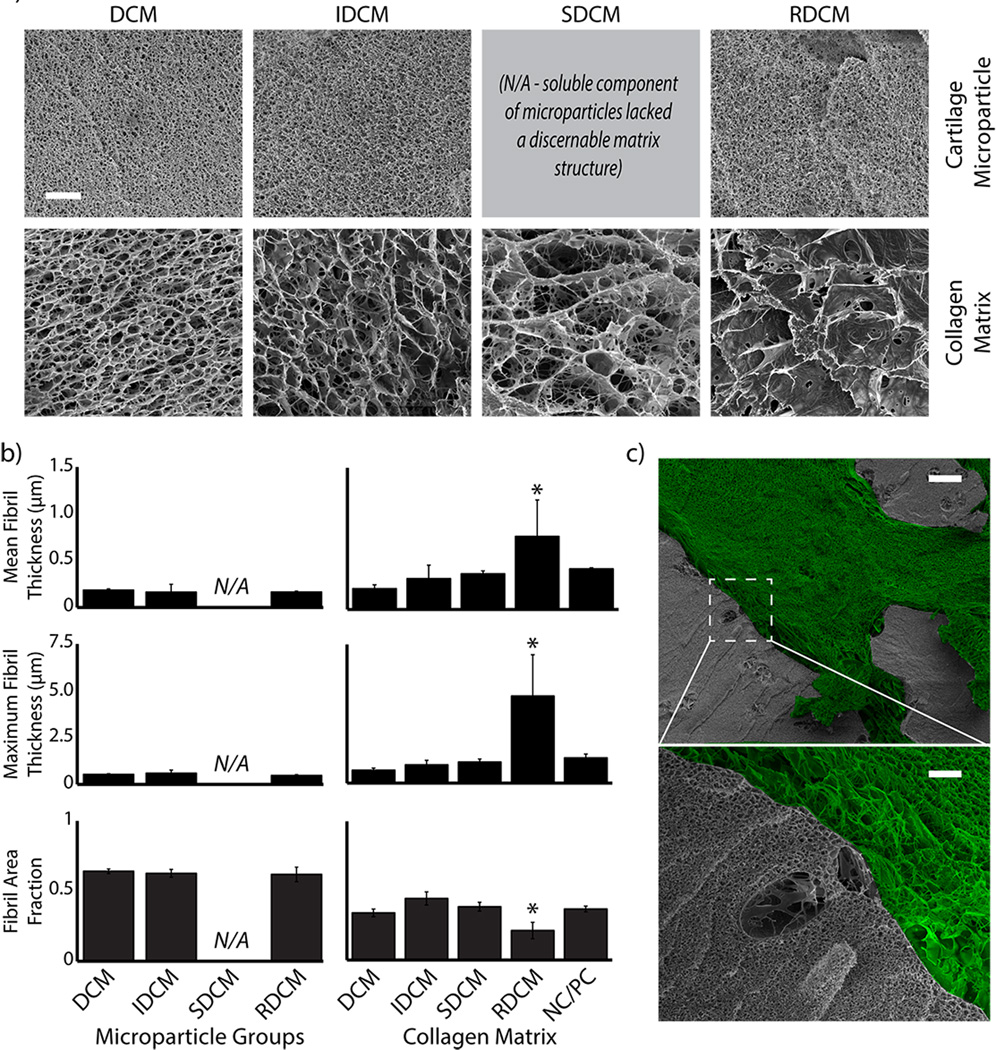
Cryo-SEM analysis revealed differences in matrix fibril morphology as a result of compression across various cartilage microparticle treatments (Figure 3). Measurements of cartilage microparticles showed no significant difference between DCM, IDCM, and RDCM groups in mean fibril thickness (p=0.850), maximum fibril thickness (p=0.347), and fibril area fraction (p=0.750) (Figure 3b). The cartilage matrix, however, was impacted by the various microparticles. Mean fibril thickness and maximum fibril thickness was found to be significantly higher in RDCM samples (p=0.028 and p=0.002, respectively). As a likely result, fibril area fraction was found to be significantly lower in RDCM samples (p=0.001).
Figure 3.
Cartilage microparticle inclusion altered the collagen matrix morphology. a) Representative cryo-SEM images showed similar fibril morphology across cartilage microparticles (top), but varied fibril morphology in the surrounding collagen matrix (bottom) (20,000×, scale bar = 5 µm). SDCM was the addition of the soluble component of the tissue after Gu·HCl reduction. As a result, this material solubilized back into the fluid prior to polymerization and there were no discernable solid microparticles. b) Quantitative analysis of the SEM images confirm no statistical difference between cartilage microparticles regardless of Gu·HCl reduction or mechanical reconstitution. SDCM samples were found to have larger collagen fibril bundles and much larger pores than all other samples as well as control. c) Examination of the cartilage-collagen interface shows the differences in fibril morphology, as well as the maintenance of cell lacunae in cartilage microparticles (scale bar: top = 250 µm, bottom = 5 µm). False coloration was used to show the difference between cartilage microparticles (greyscale) and the densified collagen matrix (green). (*=p<0.05 compared to the NC/PC group.)
SEM analysis qualitatively showed porosity and density differences between the cartilage microparticles and the surrounding cartilage matrix, as well as the interface between them (Figure 3a,c). Qualitative examination of the DCM, IDCM, and RDCM groups showed cartilage microparticles characterized by high-density fibril regions and empty lacunae of the former chondron complex. The interface between the microparticles and the collagen matrix showed greater porosity and aligned fibril regions, suggesting shear effects between the cartilage microparticles and collagen matrix during compression.
Ultrastructural analysis revealed conservation of porosity and fibril characteristics within cartilage microparticles, including chondron lacunae, which was unaffected by Gu·HCl reduction of GAGs [1]. Interestingly, this is converse to previous reports utilizing Gu·HCl, which were shown to increase the porosity of cartilage tissue [1]. Despite the changes in mechanical properties and structural integrity, SDCM samples did not show significant changes in fibril morphology. The collagen matrix only exhibited changes in fibril morphology in the RDCM group. This was likely due to the decreased integrity of the collagen matrix induced by the soluble GAG extract coupled with shearing due to cartilage microparticle interaction beyond the post-percolation threshold. The reduced integrity of the collagen matrix was likely caused by the interaction between the soluble proteoglycan component and the collagen fibrils, as mentioned previously[17].
2.3. Microparticles Inhibit Bulk Cell-Mediated Contraction
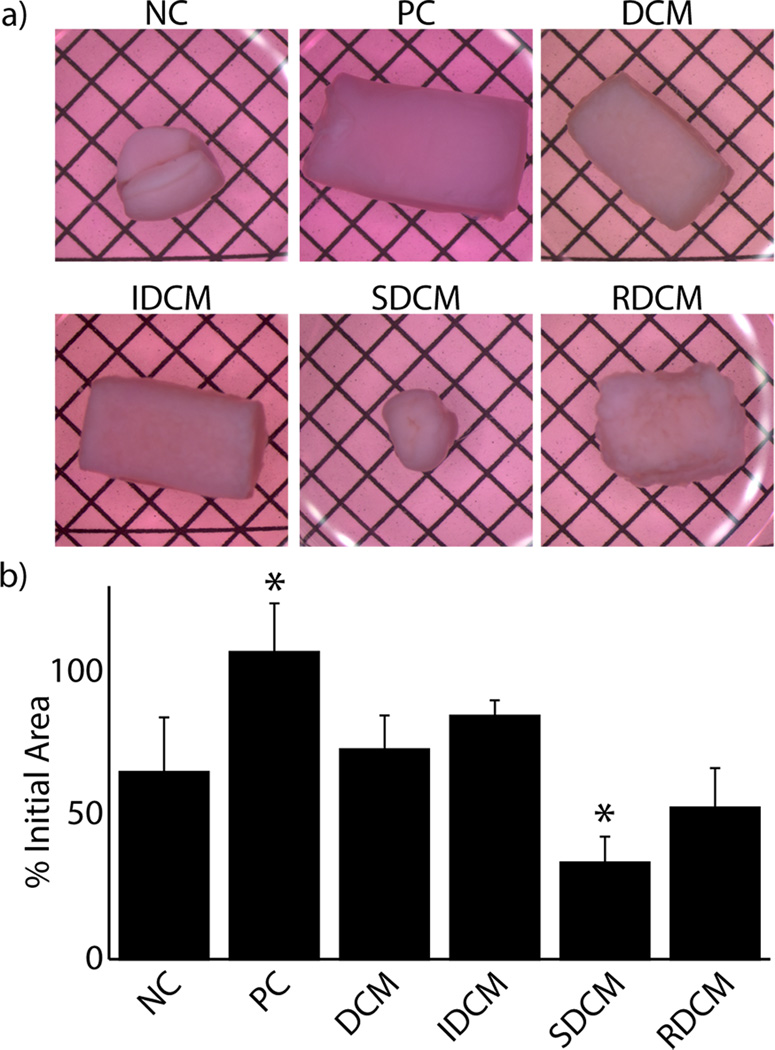
Six sample groups were included for investigation of cellularized matrices. As in the mechanical analysis, DCM, IDCM, SDCM, and RDCM groups were produced as explained in the methods. Further, positive control (PC) and negative control samples (NC) were included for analysis and were composed of cell encapsulated type I collagen matrices cultured in chondrogenic inductive (PC) and non-inductive (NC) media. Qualitative and quantitative analysis of the cellularized composite matrices at two weeks showed significant differences in morphology due to cell-mediated contraction. PC, DCM, and IDCM groups were shown to have the lowest cell-mediated contraction (p<0.001, Figure 4). In most of the groups with lower equilibrium mechanical properties (NC, SDCM, RDCM), significant deformation was observed both qualitatively and quantitatively (Figure 4b).
Figure 4.
Treatment groups exhibited varied cell-mediated contraction through the two week cell study. a) Representative images of cellularized samples at the end of the two week culture period show maintained bulk morphology in positive control (PC, i.e. a TGF-β3 supplemented culture media in the absence of any microparticles), DCM, and IDCM samples (grid spacing = 2.1 mm). b) Quantitative analysis of these data suggest significantly increased cell-mediated contraction in SDCM and RDCM samples. (*=p<0.001 compared to the NC group.)
Cell-mediated contraction was clearly mitigated in PC, DCM, and IDCM samples, where all other samples had large deformations in bulk morphology. As the culture conditions for the PC group were pro-chondrogenic, it is likely that slowed metabolic rate of the cells account for the lack of cell-mediated contraction. Further, it is possible that the pro-chondrogenic media inhibited fibroblast differentiation of the cells, further preventing cell-mediated contraction. For DCM and IDCM samples, the only difference in culture conditions versus negative controls, which did show large deformations, was the presence of the solid cartilage microparticles. This suggests that the composite mechanical structure of the DCM and IDCM particles either mechanically inhibited cell mediated contraction, induced a phenotype in the stem cells that mitigated contractile forces, or a combination thereof. This result is also consistent with previous work showing increased mechanics leading to mitigated cell-mediated contraction [18]. However, when the isolated soluble extract was presented to the construct (SDCM and RDCM groups), which has already been shown to alter mechanical properties and fibril morphology, cell-mediated contraction and construct deformation occurred on a level consistent with the negative control samples, even in the presence of physical microparticles (the IDCM component of the RDCM group). It is likely that, even in the presence of solely fibroblast-like cells, the solid microparticles of the DCM and IDCM groups contributed to the bulk of the resistance to cell-mediated contraction. Further, the maintenance of geometry suggests minimized disruption of the composite structure, such that the type I collagen matrix maintained a complete network surrounding the compressed microparticles. SDCM and RDCM groups, however, exhibited collagen matrix fraying, suggesting a disruption of the complete network structure, further supported by the diminished mechanical properties in these groups.
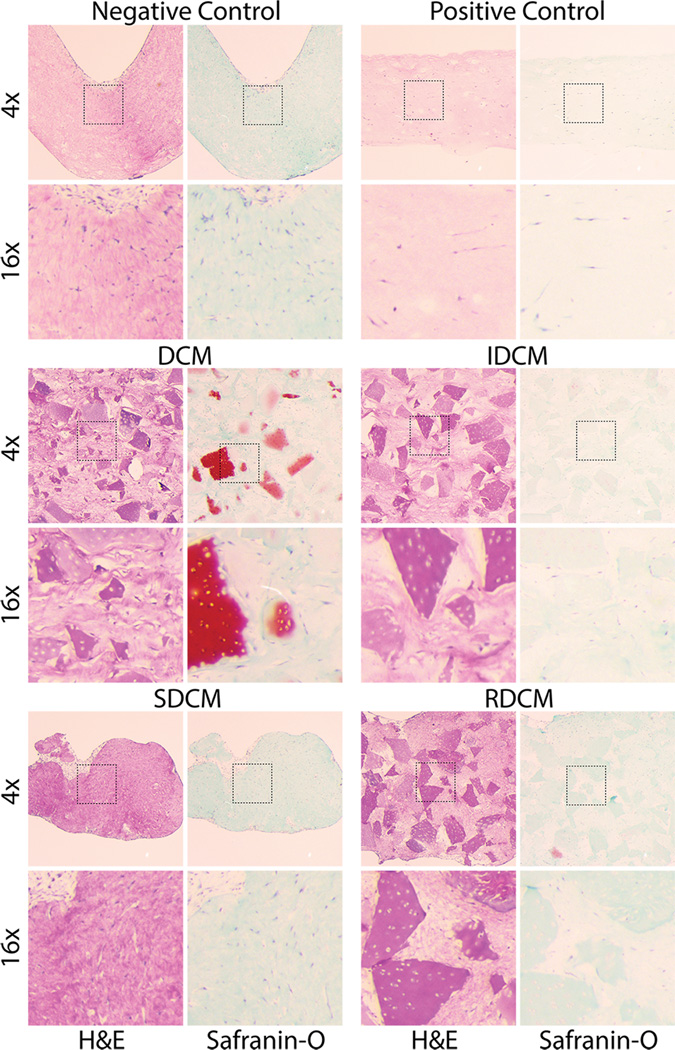
2.4. Unmodified and Insoluble Cartilage Microparticles Showed Cellular Infiltration and Proteoglycan Production
Histological analysis revealed the morphological and biochemical differences between each of the treatment groups. Negative control samples showed distorted morphology of the matrix, but an intact collagen structure via H&E stain. Safranin-O stain showed no presence of proteoglycans or the formation of articular cartilage tissue. Both stains showed elongated cell bodies homogenously distributed throughout the sample. Positive control samples, alternatively, showed via H&E stain no distortion in morphology, but similar cell body elongation and no presence of articular cartilage formation, despite the presence of chondro-inductive growth factors (TGF-β3). DCM samples showed an even distribution of cartilage microparticles throughout the collagen matrix and clear safranin-o staining of characteristic cartilage tissue (Figure 5). Interestingly, cartilage microparticles that maintained red safranin-o staining also showed cellular infiltration of cartilage lacunae, where those that showed little cell infiltration consistently showed little cartilage staining (Figure 5). As expected, IDCM groups, depleted of GAGs via Gu·HCl reduction, showed no red safranin-o staining of the cartilage microparticles, while showing similar distribution and size within the composite matrix to the DCM group. Interestingly, those cartilage microparticles that showed cellular infiltration additionally showed mild red safranin-o staining near the lacunae edges. Cells not within the microparticles showed cellular elongation. SDCM groups, as noted previously, did not contain any solid cartilage pieces but contained the soluble reduction from the Gu·HCl treatment. As a result, no cartilage microparticles are present and similar elongated cell bodies were seen. Similar to as previously described, SDCM samples were not structurally intact and did not exhibit any cartilage specific staining. RDCM samples, similar to both IDCM and SDCM, did not exhibit high levels of cartilage specific safranin-o staining, but did maintain the cartilage microparticle structures similar to DCM and IDCM. However, as noted above, RDCM samples showed low structural integrity and similarly show cell body elongation in the interstitial spaces between cartilage microparticles.
Figure 5.
Histology revealed structural and biochemical differences between samples, as well as overall cell morphology and cellular infiltration of cartilage particles. Major differences in the biochemical makeup of DCM vs reduced IDCM samples can be seen in Safranin-O (proteoglycan) staining, where DCM samples stain positive for cartilage whereas IDCM samples stain very weakly for cartilage. Interestingly, we noted a distribution of red stained and unstained particles in the DCM samples, where the samples that retain positive stain showed cellular infiltration and presence in the native lacunae. This potentially suggests infiltration of the native chondron structure and maintenance of the cartilage tissue where non-cellularized particles show low red staining, suggesting degradation of the biochemical makeup. Interestingly, no samples show positive safranin-o staining between cartilage microparticles, or in samples without particles, including the positive control (PC). Additionally, histological analysis at higher magnification (16×) showed cellular infiltration in cartilage microparticles. Cells were seen to infiltrate microparticles in DCM, IDCM, and RDCM samples, that is, all samples with solid cartilage microparticles. In particles with cell infiltration, positive Safranin-O staining is seen, suggesting a chondrogenic phenotype for those cells that have infiltrated native lacunae. Additionally, these images showed the elongated cell body morphology for the cell population localized to the collagen matrix in between cartilage particles, suggesting non-chondrogenic differentiation and no influence of the cartilage particles except to those cells directly in contact.
The histological analysis showed cellular infiltration in chondron lacunae and maintenance of positive cartilage safranin-o staining for DCM samples, similar to previous research [6]. Additionally, cellular infiltration into chondron lacunae was apparent in IDCM samples as well. Otherwise, all cells in the collagen matrix and in other samples exhibited elongated cell bodies. As expected, unmodified DCM samples exhibit positive safranin-o staining where IDCM samples did not due to the Gu·HCl extraction of GAGs. Interestingly, within DCM samples, particles that exhibit less infiltration also show less overall safranin-o staining, suggesting that infiltrated cells contribute to the maintenance of the cartilage microenvironment. Additionally, IDCM particles with infiltrated cells show light safranin-o staining, further supporting possible chondrogenic induction very localized to specific cellular interactions with cartilage microparticles. Further, this may suggest a heightened role of the mechanical and biochemical properties of the collagenous cartilage matrix, and merits further investigation into the manipulation of type II collagen structures to higher physiological relevance to investigate their potential in cartilage regeneration. All other samples, including positive control samples, show little evidence of positive safranin-o staining or chondrogenic cell morphology. As a result, the large majority of the cell population, even in DCM and IDCM samples, do not have contact with the cartilage microparticles and may not receive the signals necessary for chondrogenic induction. This is similar with previous research in this field, where the majority of cells reside in the interstitium between particles with little cartilage positive staining [6]. The small number of cells that do interact with cartilage microparticles show chondrogenic morphology and mitigation of local GAG degradation (Figure 5–6).
Figure 6.
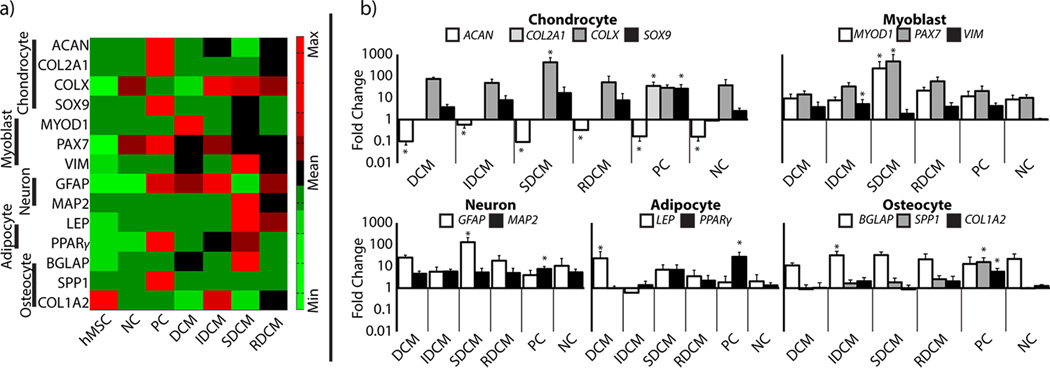
Quantitative PCR analysis shows that microparticle group cell populations were dominated by higher concentrations of cells located in the inter-particle type I collagen matrix, and largely exhibit gene response to the physicochemical properties of the type I collagen. a) Heat map of gene array shows qualitative differences between sample groups, as compared to the undifferentiated hMSCs. Gene expressions have been standardized across the sample groups to show comparable heat map between gene expression values. It is clear that positive control samples exhibit the highest expression of chondrocyte markers, although they also exhibit upregulation of markers for all other differentiation pathways as compared to hMSCs. DCM, IDCM, and RDCM samples all showed upregulation of myoblast markers. SDCM and RDCM additionally showed upregulation of adipocyte markers. b) Quantitative analysis shows statistical comparisons of differentiation as compared to the undifferentiated hMSCs. Here, it can be seen that only positive control samples exhibit significant chondrocyte markers, all samples are shown to downregulate ACAN and only positive controls were shown to upregulate COL2A1 and SOX9 as a population. IDCM samples were found to upregulate myoblast and osteocyte markers. SDCM samples were also shown to upregulate myoblast markers as well as neuron markers. Interestingly, positive control samples also show upregulation of neuron, adipocyte, and osteocyte markers in addition to chondrocyte markers. Due to the high number of cells not in contact with the cartilage microparticles, these data showed no cartilage specific differentiation on average in any experimental groups.
2.5. Local MSC Differentiation Occurred in Cells in Contact with Microparticles
Gene expression analysis showed varying stem cell differentiation as a result of treatment group (Figure 6). Across groups, significant gene differences were seen for: cartilage markers-ACAN (p<0.001), COL2A1 (p<0.001), COLX (p<0.001), and SOX9 (p=0.002); myoblast markers-MYOD1 (p<0.001), PAX7 (p=0.027), and VIM (p=0.040); neuron markers-GFAP (p<0.001) and MAP2 (p=0.057); adipocyte markers-LEP (p=0.035) and PPARγ (p<0.001); and osteocyte markers-SPP1 (p<0.001), COL1A2 (p<0.001) and BGLAP (p=0.057). When compared to undifferentiated hMSCs as a control, all samples downregulated ACAN (p<0.05) while COL2A1 and SOX9 were only significantly upregulated in positive controls (p<0.05). COLX, a cartilage hypertrophy indicator, was significantly upregulated in SDCM samples. Myoblast markers were significantly upregulated in IDCM (VIM, p<0.05) and SDCM (MYOD1 and PAX7, p<0.05 for both). Neuron markers were upregulated in SDCM (GFAP, p<0.05) and PC (MAP2, p<0.05) samples. Adipocyte markers were upregulated in DCM (LEP, p<0.05) and PC (PPARγ, p<0.05) samples. Osteocyte markers were upregulated in IDCM (BGLAP, p<0.05) and PC (SPP1 and COL1A2, p<0.05 for both) samples. Additionally, positive control samples exhibited upregulation in MAP2, PPARγ, SPP1, and COL1A2 (p<0.05 for all).
A high percentage of the cell population was found to be suspended in the collagen matrix without interaction with cartilage microparticles. We did not observe a rounded, chondrogenic morphology in these cells and expected a large number of cells to show differentiation in a non-chondrogenic pathway (Figure 5). Real time quantitative PCR supports this hypothesis, showing no significant upregulation of cartilage markers in any sample except the positive control sample (Figure 6). This is converse to previous reports, although this study did not look at markers other than chondrogenic genes [6]. Our data suggests that the undifferentiated cells respond to direct interaction with the critical signals (in the case of positive controls, the presence of diffused TGF-β3), and not just the presence of the microparticles. However, we might expect samples containing the soluble extract (SDCM and RDCM) to contain enough diffused GAGs to interact globally with encapsulated cells. It is possible that the signal provided by the soluble extract is not sufficient to induce differentiation in the undifferentiated stem cell line. Additionally, it is likely that significant GAG degradation is occurring over time, as seen in DCM samples, as opposed to the positive control, which is consistently supplemented with TGF-β3. Interestingly, the positive control samples also exhibit upregulation in neuron, adipocyte, and osteocyte markers, though not to the level and consistency as chondrocyte markers. SDCM, in the absence of interaction with the solid cartilage microparticles structure, showed hypertrophic chondrocyte markers (COLX) and showed significant myoblast differentiation. It appears as though, while not globally inducing chondrocyte differentiation, the presence of microparticles in DCM and IDCM samples mitigated differentiation down non-chondrogenic pathways. Previous studies similar to these suggest that, given more time, global chondrogenic differentiation would occur in DCM and potentially IDCM samples [6]. This suggests an importance of the physicochemical structure of native cartilage tissue in the differentiation and maintenance of the chondrocyte phenotype. Further investigation of this cell-matrix interaction with the unmodified (DCM) and reduced (IDCM) and the potential for engineering a similar system with an adequate type II collagen system.
3. Conclusion
In summary, our study demonstrated the ability to create unique, tunable cartilage microparticle-collagen composite matrices containing significant mechanical, ultrastructural, and cell-matrix interaction properties. Additionally, we were able to investigate the short term differentiation potential of these systems in a human stem cell line, showing the potential for xenogenic microparticle systems. Further, we showed the importance of the structural nature of the native cartilage structure and biochemistry in bulk cell-matrix interaction and local cell differentiation. We believe further investigation of these xenogenic systems, as well as alternate suspension matrices, such as type II collagen, have the potential to create tunable composite systems with the ability to induce chondrogenic differentiation and regeneration for osteoarthritis defect repair.
4. Experimental Section
4.1. Decellularized Cartilage Microparticles Isolation and Reduction
All cartilage tissue for this study was sourced from market weight porcine tissue (at least 200 separate animals) within 48 hours of slaughter. Cartilage tissue was harvested by exposure of knee joint space and scalpel removal of cartilage tissue with care to not include any calcified tissue. Harvested tissue was immediately frozen at −80 °C until further processing. Tissue was devitalized via pulverization in a liquid nitrogen magnetic freezer mill as previously described [19]. Crushed tissue particles were subsequently sorted via a microsieve stack (Electron Microscopy Sciences, Hatfield PA) and collected to have a distributed particle diameter of 67–500 µm (35–230 standard mesh size) similar to previously described [2]. Sorted cartilage microparticles were decellularized in 2% sodium dodecyl sulfate for 6 hours to remove cellular and genetic material to specifications as previously described (Figure 1a) [3, 7]. Cartilage particles that underwent no further modification were then 3× rinsed in ddH2O, flash frozen in LN2, and lyophilized.
To examine the potential contribution of the many physicochemical components of native tissue, the cartilage microparticles were reduced using previously established Gu·HCl-based procedures [20]. Briefly, lyophilized decellularized cartilage microparticles were subject to reduction via treatment in 4M Gu·HCl with 1% protease inhibitor and 25mM EDTA under rotation for 48 hours at 4 °C [21]. After Gu·HCl treatment, samples were centrifuged at 1600 ×g for 15 min. The supernatant was then collected in a separate container for soluble protein extraction. The insoluble pellet was washed in ddH2O, flash frozen in liquid nitrogen, and lyophilized for the extraction of insoluble decellularized cartilage microparticles [20]. The supernatant was precipitated in 100% EtOH overnight at −20 °C, washed with 100% EtOH, and subsequently dialyzed against ddH2O at 25 °C to completion (MW cutoff ≈ 3500 Da, Spectrapor, Rancho Dominguez CA) [20, 22]. Dialyzed solution was flash frozen in liquid nitrogen and lyophilized forming a soluble reduction component of the decellularized cartilage microparticles. Gu·HCl reduction was verified via SDS-PAGE analysis [20, 23].
4.2. Formation of Dense Collagen-Cartilage Microparticle Composite Matrices
Collagen-Cartilage Microparticle composites were formed via encapsulation and densification within polymerized collagen matrices. Cartilage microparticles were used separately or mechanically mixed to form four groups of collagen-cartilage microparticle composites (Figure 1b): unmodified decellularized cartilage microparticles (DCM), Gu·HCl extracted insoluble component of decellularized cartilage microparticles (IDCM), Gu·HCl extracted soluble component of decellularized cartilage microparticles (SDCM), and reconstituted components of decellularized cartilage microparticles (RDCM). DCM microparticles were mixed into the collagen at 1% w/v (10mg/mL) initial concentration and then subject to matrix densification via plastic compression to a strain of 90% to effectively increase the DCM concentration to approximately 10% w/v, approaching physiological levels [14]. These levels were chosen as a result of a series of pilot studies investigating both initial microparticle density and size. Initial densitites of 2% w/v and greater led to significant shearing within the construct when compressed, compromising the structural integrity of the whole composite (data not shown). Similarly, particles sized larger than 500µm were found to reach percolation and begin shearing the composite at lower strain than particles sized < 500µm (data not shown). Similarly, IDCM and SDCM microparticles were mixed with the collagen at appropriate ratios as measure from the reduction (0.8% w/v for IDCM, 0.2% w/v for SDCM) and similarly compressed. RDCM composites were formed by mechanically mixing the soluble and insoluble reduction components to at the same previously described concentrations (0.8% w/v IDCM + 0.2% w/v SDCM→1% w/v RDCM) and similarly compressed.
Microparticle components were mixed with neutralized type I collagen oligomers (5mg/mL), placed in a rectangular mold (10×5×14 mm, w×t×h) and polymerized at 37 °C for 30 min [24]. Samples (n=5) were then placed in a servoelectric mechanical testing system (TestResources, Shakopee MN) and compressed via confined compression with a porous platen (porosity=15%, pore size=35µm, Small Parts Inc., Miami Lakes, FL) to a final strain magnitude of 90% at a strain rate of 0.1% s−1 [14]. Mechanical response and properties of the composites were measured during compression including pre-percolation modulus, post-percolation modulus, and maximum stress (Figure 2). The percolation threshold was defined as the strain level where the linear modulus of compression suddenly increases, suggesting bulk contact of cartilage microparticles. Compression was additionally held after the test until equilibrium where equilibrium/residual stress was measured.
It is important to note that the soluble components, although lyophilized, was soluble again once introduced to the unpolymerized collagen solution. Therefore, unlike other groups, the SDCM group contained no solid cartilage particulate or microparticles of any sort. To examine the potential leaching of the SDCM component into the media, we placed SDCM composites into isotonic PBS for 1 week, sampling at 3 and 7 days with fluid change at each sampling point. Leached material was measured via both DMMB assay and A280 protein spectroscopy (data not shown, NanoDrop ND-1000, Thermo Fischer Scientific, Waltham MA).
4.3. Ultrastructural Analysis
The ultrastructure of cartilage and collagen components was assessed via cryo-SEM (Nova NanoSEM, FEI, Hillsboro OR) using an Everhart-Thornley (<10,000× magnification) or immersion lens (>10,000× magnification) detector. Samples were frozen in critical state liquid nitrogen, fractured to the imaging plane, and sublimated for 13 minutes to reveal collagen ultrastructure. Images were acquired at magnifications between 300× and 40,000× to visualize gross structure and specific fibril areas within cartilage microparticles and the collagen matrix. Images at 10,000× were obtained at three separate locations to determine the ultrastructural differences between sample groups. Mean fibril thickness, maximum fibril thickness, and fibril area fraction were quantified and analyzed at three separate locations in both cartilage and collagen regions using the Thickness and Volume Fraction functions in BoneJ, developed to measure these properties in trabecular bone, as previously described [25]. SEM images were thresholded and binarized prior to analysis. Prior to analysis, BoneJ software was validated through the development of a custom MATLAB script, outputting artificial fibril networks of specified mean size and distribution.
4.4. Human Mesenchymal Stem Cell Differentiation Study
To examine the signaling potential of the cartilage microparticle composites, low passage human MSCs were additionally encapsulated prior to polymerization and compression. Low passage hMSCs (≤ passage 5, Lonza, Basel, Switzerland) were seeded at 1E6 cells mL−1 initial concentration, with an average post-compression concentration of 10E6 cells mL−1. A total of 6 groups (n=6 for each) were initiated for a 2 week differentiation study examining the chondrogenic induction potential of cartilage microparticles within a collagen matrix. The negative control (NC) group was composed of densified collagen in the absence of any microparticles and cultured in non-differentiating media. Positive controls (PC) were also composed of densified collagen in the absence of microparticles but cultured in chondrogenic induction media (Lonza, Basel, Switzerland) containing TGF-β3, the current gold standard for chondrogenic differentiation induction [26, 27]. Four experimental groups were additionally cultured using cartilage microparticles as previously described (DCM, IDCM, SDCM, and RDCM) in non-inducting media. Media was exchanged every two days. At the conclusion of the study, four samples from each group of six were utilized for qPCR assessment while two samples were immediately fixed in 4% paraformaldehyde at 4 °C overnight and sent for histological processing (hematoxylin and eosin and safranin-o). Prior to separation, samples were measured for cell-mediated contraction in the length×width dimension utilizing a dissection microscope (Leica M80, Leica Microsystems, Buffalo Grove IL, USA) backed by a grid of known spacing (2.1 mm).
4.5. Real-Time Quantitative Polymerase Chain Reaction
Total RNA isolation was performed using the Aurum Total RNA Mini Kit (Bio-Rad, Hercules CA, USA). Collagen matrices were homogenized (TissueRuptor, QIAzol Lysis Reagent, Qiagen, The Netherlands) and cleaned from protein using two rounds of chloroform precipitation. Total RNA was reverse transcripted into cDNA (iScript Reverse Transcription Supermix, Bio-Rad) using a thermocycler and Quantitative Real-Time PCR (CFX96 Touch, Bio-Rad) was performed using SsoAdvanced SYBR Green Supermix and the CFX96 Touch adthermocycler (Bio-Rad, Hercules CA, USA). Amplification curves were analyzed via sigmoidal curve fitting analysis[32].
A variety of genes were investigated for hMSC differentiation. For all samples, GAPDH was utilized as the housekeeping gene. Known chondrocyte (ACAN, COL2A1, COLX, and SOX9), myoblast (MYOD1, PAX7, VIM), neuron (GFAP, MAP2), adipocyte (LEP, PPARγ), and osteocyte (BGLAP, SPP1, COL1A2) differentiation genes were measure for all samples, including undifferentiated hMSCs (Table S1). All samples were normalized to the housekeeping gene and further normalized to the identical cell line gene expression prior to the start of the study, where the cells expressed surface markers for undifferentiated hMSCs, to investigate cell differentiation as a result of the different microenvironments presented (cartilage microparticle groups). All primers were specific for all known isotypes and are separated by at least one intron or span an exon-exon junction, if splicing information was available.
4.6. Statistical Analysis
Mechanical, ultrastructure, and cell-mediated contraction data were analyzed via one-way ANOVA with cartilage microparticle treatment (PC, NC, DCM, IDCM, SDCM, and RDCM) as the main effect. Post-hoc analyses were performed using Tukey’s pairwise analysis with 95% confidence. qPCR data was analyzed via one-way ANOVA with cartilage microparticle treatment as the main effect, and measurements from the undifferentiated hMSCs were included in the analysis. Post-hoc analyses for qPCR data was analyzed via Dunnett’s test and compared to a control level of the gene expression values for undifferentiated hMSCs.
Supplementary Material
Acknowledgments
The authors would like to acknowledge funding from NIH R01 AR063712, NIH R21 AR064178, NIH R21 AR066230, and NSF Graduate Research Fellowship Program under Grant No. DGE-0833366 (TN). The authors would like to thank Purdue Life Science Microscopy Facilities for SEM core facility use and Cook Research, Inc. for histology processing and staining.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Tyler Novak, Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN 47907, USA.
Benjamin Seelbinder, Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN 47907, USA; Department of Mechanical Engineering, University of Colorado Boulder, Boulder, CO 80304, USA.
Celina M. Twitchell, Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN 47907, USA
Sherry L. Voytik-Harbin, Prof., Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN 47907, USA Department of Basic Medical Sciences, Purdue University, West Lafayette, IN 47907, USA.
Corey P. Neu, Prof., Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN 47907, USA; Department of Mechanical Engineering, University of Colorado Boulder, Boulder, CO 80304, USA.
References
- 1.Schwarz S, Koerber L, Elsaesser AF, Goldberg-Bockhorn E, Seitz AM, Dürselen L, Ignatius A, Walther P, Breiter R, Rotter N. Tissue Eng Part A. 2012;18:2195. doi: 10.1089/ten.TEA.2011.0705. [DOI] [PubMed] [Google Scholar]
- 2.Kang H, Peng J, Lu S, Liu S, Zhang L, Huang J, Sui X, Zhao B, Wang A, Xu W, Luo Z, Guo Q. J Tissue Eng Regen Med. 2014;8:442. doi: 10.1002/term.1538. [DOI] [PubMed] [Google Scholar]
- 3.Crapo PM, Gilbert TW, Badylak SF. Biomaterials. 2011;32:3233. doi: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xue JX, Gong YY, Zhou GD, Liu W, Cao Y, Zhang WJ. Biomaterials. 2012;33:5832. doi: 10.1016/j.biomaterials.2012.04.054. [DOI] [PubMed] [Google Scholar]
- 5.Yang Q, Peng J, Guo Q, Huang J, Zhang L, Yao J, Yang F, Wang S, Xu W, Wang A, Lu S. Biomaterials. 2008;29:2378. doi: 10.1016/j.biomaterials.2008.01.037. [DOI] [PubMed] [Google Scholar]
- 6.Chang CH, Chen CC, Liao CH, Lin FH, Hsu YM, Fang HW. J Biomed Mater Res A. 2014;102:2248. doi: 10.1002/jbm.a.34897. [DOI] [PubMed] [Google Scholar]
- 7.Elder BD, Eleswarapu SV, Athanasiou KA. Biomaterials. 2009;30:3749. doi: 10.1016/j.biomaterials.2009.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mow VC, Ratcliffe A, Poole AR. Biomaterials. 1992;13:67. doi: 10.1016/0142-9612(92)90001-5. [DOI] [PubMed] [Google Scholar]
- 9.Martel-Pelletier J, Boileau C, Pelletier JP, Roughley PJ. Best Pract Res Clin Rheumatol. 2008;22:351. doi: 10.1016/j.berh.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Peretti GM, Randolph MA, Caruso EM, Rossetti F, Zaleske DJ. J Orthop Res. 1998;16:89. doi: 10.1002/jor.1100160115. [DOI] [PubMed] [Google Scholar]; Cheng NC, Estes BT, Awad HA, Guilak F. Tissue Eng Part A. 2009;15:231. doi: 10.1089/ten.tea.2008.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]; Diekman BO, Rowland CR, Lennon DP, Caplan AI, Guilak F. Tissue Eng Part A. 2010;16:523. doi: 10.1089/ten.tea.2009.0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peretti GM, Randolph MA, Villa MT, Buragas MS, Yaremchuk MJ. Tissue Eng. 2000;6:567. doi: 10.1089/107632700750022206. [DOI] [PubMed] [Google Scholar]
- 12.Benders KE, van Weeren PR, Badylak SF, Saris DB, Dhert WJ, Malda J. Trends Biotechnol. 2013;31:169. doi: 10.1016/j.tibtech.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Chen CC, Liao CH, Wang YH, Hsu YM, Huang SH, Chang CH, Fang HW. J Orthop Res. 2012;30:393. doi: 10.1002/jor.21541. [DOI] [PubMed] [Google Scholar]
- 14.Novak T, Seelbinder B, Twitchell CM, van Donkelaar CC, Voytik-Harbin SL, Neu CP. Advanced Functional Materials. 2016 doi: 10.1002/adfm.201503971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genin GM, Kent A, Birman V, Wopenka B, Pasteris JD, Marquez PJ, Thomopoulos S. Biophys J. 2009;97:976. doi: 10.1016/j.bpj.2009.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu XL, Wan LQ, Guo XE, Mow VC. J Biomech. 2010;43:673. doi: 10.1016/j.jbiomech.2009.10.026. [DOI] [PubMed] [Google Scholar]
- 17.Yang YL, Sun C, Wilhelm ME, Fox LJ, Zhu J, Kaufman LJ. Biomaterials. 2011;32:7932. doi: 10.1016/j.biomaterials.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keogh MB, O'Brien FJ, Daly JS. Acta Biomater. 2010;6:4305. doi: 10.1016/j.actbio.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Bailey JL, Critser PJ, Whittington C, Kuske JL, Yoder MC, Voytik-Harbin SL. Biopolymers. 2011;95:77. doi: 10.1002/bip.21537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sampath TK, Reddi AH. Proc Natl Acad Sci U S A. 1981;78:7599. doi: 10.1073/pnas.78.12.7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Didangelos A, Yin X, Mandal K, Saje A, Smith A, Xu Q, Jahangiri M, Mayr M. Mol Cell Proteomics. 2011;10:M111.008128. doi: 10.1074/mcp.M111.008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pepinsky RB. Anal Biochem. 1991;195:177. doi: 10.1016/0003-2697(91)90315-k. [DOI] [PubMed] [Google Scholar]
- 23.Kreger ST, Bell BJ, Bailey J, Stites E, Kuske J, Waisner B, Voytik-Harbin SL. Biopolymers. 2010;93:690. doi: 10.1002/bip.21431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novak T, Voytik-Harbin SL, Neu CP. Acta Biomater. 2015;11:274. doi: 10.1016/j.actbio.2014.09.031. [DOI] [PubMed] [Google Scholar]
- 25.Doube M, Kłosowski MM, Arganda-Carreras I, Cordelières FP, Dougherty RP, Jackson JS, Schmid B, Hutchinson JR, Shefelbine SJ. Bone. 2010;47:1076. doi: 10.1016/j.bone.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joyce ME, Roberts AB, Sporn MB, Bolander ME. J Cell Biol. 1990;110:2195. doi: 10.1083/jcb.110.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorpe SD, Buckley CT, Vinardell T, O'Brien FJ, Campbell VA, Kelly DJ. Ann Biomed Eng. 2010;38:2896. doi: 10.1007/s10439-010-0059-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.