Abstract
Increases in fruit weight of cultivated vegetables and fruits accompanied the domestication of these crops. Here we report on the positional cloning of a quantitative trait locus (QTL) controlling fruit weight in tomato. The derived allele of Cell Size Regulator (CSR-D) increases fruit weight predominantly through enlargement of the pericarp areas. The expanded pericarp tissues result from increased mesocarp cell size and not from increased number of cell layers. The effect of CSR on fruit weight and cell size is found across different genetic backgrounds implying a consistent impact of the locus on the trait. In fruits, CSR expression is undetectable early in development from floral meristems to the rapid cell proliferation stage after anthesis. Expression is low but detectable in growing fruit tissues and in or around vascular bundles coinciding with the cell enlargement stage of the fruit maturation process. CSR encodes an uncharacterized protein whose clade has expanded in the Solanaceae family. The mutant allele is predicted to encode a shorter protein due to a 1.4 kb deletion resulting in a 194 amino-acid truncation. Co-expression analyses and GO term enrichment analyses suggest association of CSR with cell differentiation in fruit tissues and vascular bundles. The derived allele arose in Solanum lycopersicum var cerasiforme and appears completely fixed in many cultivated tomato’s market classes. This finding suggests that the selection of this allele was critical to the full domestication of tomato from its intermediate ancestors.
Author summary
Starting about 10,000 years ago, during the Neolithic period, human societies began the transformation from a hunting and gathering-dependent lifestyle to an agrarian lifestyle. This transformation was accompanied by plant and animal domestication. Tomato shows a huge increase in fruit weight that has arisen as a consequence of its domestication. We identified a gene that encodes a poorly characterized protein that controls fruit weight in tomato. The mutation that led to the increase in fruit weight arose early during the cultivation of tomato and is now incorporated in all large tomato varieties. The gene regulates cell size in the fruit and is called Cell Size Regulator. The increases in cell size are proposed to relate to cellular maturation that accompanies fruit growth.
Introduction
Rapid morphological diversification among closely related organisms often arise in response to strong selection pressures such as those imposed by domestication. Starting approximately 10,000 years ago, during the Neolithic period, human societies began the transformation from a hunting and gathering-dependent lifestyle to an agrarian lifestyle which was accompanied by plant and animal domestication [1, 2]. The process of domestication is associated with the taming of wild relatives and the selections of types that benefit human use in terms of food production and clothing as well as shelter and companionship. Specifically for vegetable and fruit crops, the domesticated plants feature much larger produce, reduced plant branching and often a determinate growth habit [1].
The fully wild ancestor of tomato (Solanum pimpinellifolium) is indigenous largely to the coastal regions of Ecuador and Peru [3, 4]. Selections to the intermediate type, S. lycopersicum var cerasiforme, took place in the Andean mountain region of Ecuador and Northern Peru and further selections to the earliest domesticate type, S. lycopersicum var lycopersicum, took place in Central America [4]. Continued selections after the initial domestication events led to a huge diversity of size and shape with fruit weight increasing as much as a 1,000 fold [5].
Even though the fruit is a terminal structure, the parameters that determine its final shape and weight are rooted throughout the plant’s lifespan. The development of the fruit starts as early as the formation of the inflorescence and floral meristem 3 weeks after sowing [6, 7]. In tomato, floral development from inflorescence meristem to anthesis-stage flowers takes place in approximately 3 weeks whereas fruit development from opened flower to ripe fruit lasts between 4.5 and 7 weeks depending on the genotype [7, 8]. Final fruit morphology is the result of a combination of processes that regulate meristem organization, overall cell division rates and duration, cell shape and cell expansion. Meristem organization processes that lead to larger meristems support the formation of a bulkier fruit. For example, natural mutations in the tomato orthologs of the meristem architecture genes WUSCHEL (WUS) and CLAVATA3 (CLV3) lead to fruit with more locules associated with large and flat fruits [9]. Induced mutations in genes in the same WUS-CLV3 pathway also support the notion that enlarging meristems lead to bulkier fruit [10]. Distinct from meristem organization are the cell division, cell shape and cell expansion processes which can be controlled at multiple time points throughout the ontogeny of the ovary and fruit. These cellular processes are typically defined to take place before or after anthesis [11]. Whereas details of cell division, cell shape and cell expansion are not well described for tomato ovary development, processes following pollination and fertilization of the ovules are much better understood. The initial stages of fruit development following successful pollination and fertilization are marked by a rapid increase in cell proliferation through reinitiating cell divisions [7, 8]. In most fruit tissues such as the pericarp, cell division ceases 5 to 10 days after anthesis with the exception of the epidermal cell layer. Following this rapid cell division stage, fruit growth continues by extensive cell enlargements that last for three to five weeks until the fruit ripening stage [7, 8]. Thus far, two natural mutations in tomato impact cell number by increasing the number of cell layers in the pericarp of the fruit. This increase in cell layers is achieved by changing cell division rates and/or duration in the developing ovary or fruit [12, 13]. CNR/FW2.2 changes cell layers in the ovary walls and these organs are already larger at anthesis [13]. SlKLUH/FW3.2 also impacts cell layers mainly in the pericarp but of the developing fruit instead of the ovary [12]. In addition to fruit size, the mutation in SlKLUH is associated with delayed ripening suggesting that the duration of cell division is extended [12]. Derived alleles for CNR/FW2.2 and SlKLUH/FW3.2 are resulting in expression level changes as gene expression is different in the NILs. Moreover, association mapping led to the most significant polymorphisms in the promoter of these two fruit weight genes [12, 14]. Thus far, natural mutations in tomato genes that impact cell size have not yet been identified. However, increases in cell size are associated with tomato domestication and the emergence of larger fruit [15].
The increase in size from a 1 to 2-mm wide ovary to a 5 to 10-cm wide fruit is predominantly the result of the dramatic increase in cell enlargement in the pericarp that follows the cell proliferation stage [7, 8]. In the developing fruit, the transition and maintenance of cell enlargement is not well understood. On the other hand, the transition from proliferation to enlargement is better described in root development [16, 17]. Cells differentiate from the root meristem zone to the transition zone which is controlled by the antagonistic action of auxin and cytokinin [16, 18]. Certain cytokinin signaling regulators, the B-type Arabidopsis Response Regulators (ARRs), appear to play crucial roles in synchronizing the entry into the endoreduplication cycle in roots of Arabidopsis [16, 17, 19]. In roots, the transition zone also marks the initiation of endoreduplication, a process marked by DNA replication without nuclear division. This process is known to occur in maturing cells and results in higher nuclear ploidy levels by bypassing chromatid segregation and cytokinesis [20, 21]. Endoreduplication is proposed to occur in many multicellular organisms as a potential mechanism for cell enlargement and cellular differentiation. In particular for tomato, the proper development of the tomato fruit is intimately associated with endoreduplication [15, 22].
We here report on the cloning and characterization of a novel tomato fruit weight gene Cell Size Regulator (CSR). This quantitative trait locus had been identified in a prior study as a minor QTL explaining only 8% of the phenotypic variance [23]. Despite its small effect on the trait in a segregating F2 population, the heritability in progeny studies was high, facilitating the fine mapping and eventual cloning of the gene. The derived allele CSR-D increases fruit mass through cell enlargement which is accompanied by small increases in nuclear ploidy level of pericarp cells. Expression and co-expression analyses with CSR suggests a role in cell differentiation during the later stages of fruit development, including vascular development. We propose that CSR impacts cellular differentiation leading to a potentially indirect effect on endoreduplication during tomato fruit maturation. The CSR-D mutation likely arose in S. lycopersicum var cerasiforme and became entirely fixed in the large fruited tomato germplasm suggesting that this improvement was an important step towards the domestication of tomato from the cherry type to the cultivated S. lycopersicum var lycopersicum.
Results
Solyc11g071940 underlies fw11.3 and controls weight by increasing cell size
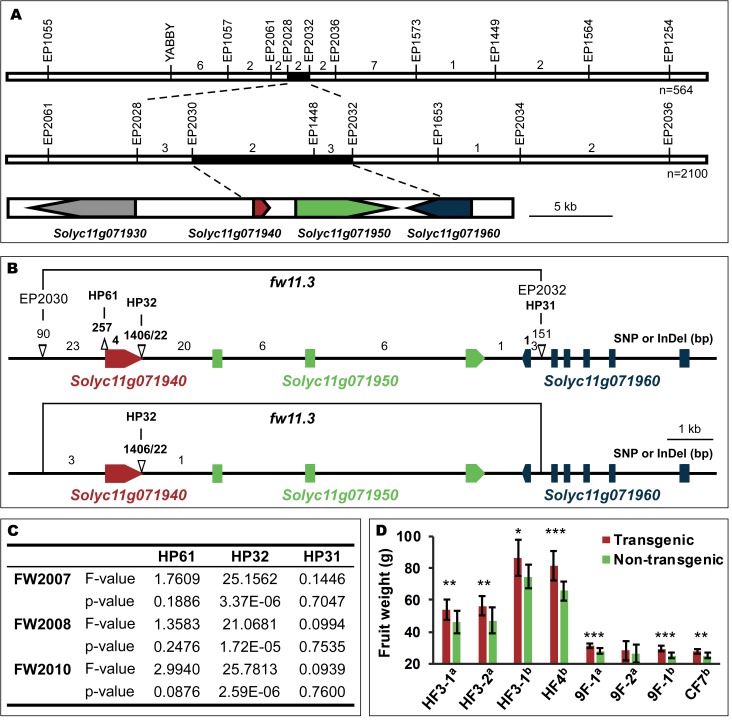
The fw11.3 locus was previously mapped to a 149 kb region on chromosome 11 [24] and further fine-mapped to a 13kb region between markers EP2030 and EP2032 (Fig 1A, S1 Table). This region included two full open reading frames, Solyc11g071940 and Solyc11g071950, and the partial Solyc11g071960. The common nucleotide polymorphisms among the fw11.3 segregating populations were a large deletion at the 3’ end of Solyc11g071940 and four SNPs in the non-coding region surrounding the same gene (Fig 1B; S1 Table). We next conducted an association study to determine whether the partial gene deletion was correlated with fruit weight variation in the tomato germplasm. Using a population that was developed to conduct association studies for fruit weight in tomato [25], the 3’ deletion of Solyc11g071940 was shown to be significantly associated with fruit enlargement and over three years of fruit weight evaluations (Fig 1C). The association of the large deletion in Solyc11g071940 with fruit weight implies that this gene underlies fw11.3.
Fig 1. Cloning of the fw11.3 QTL.
(A) The fw11.3 locus was mapped to the EP2028-EP2032 interval on tomato chromosome 11. The fw11.3 locus was further narrowed down to EP2030-EP2032 interval including two and a half open reading frames. Numbers above the chromosome indicate number of recombinant plants in the respective interval that were progeny tested. The black region delineates the fine mapped locus on the genome. The three candidate genes are shown in red, green and blue. (B) The DNA polymorphisms at the fw11.3 locus among LA1589 and Howard German/Rio Grande (upper) or Yellow Pear and Gold Ball Livingston (lower) segregating populations. Bold numbers above the chromosome indicate polymorphisms in the coding region; non-bold numbers indicate polymorphisms in the non-coding region. EP2030, EP2030, HP61, HP32 and HP31 denote markers. Δ: deletion or insertion. (C) Association mapping results of fruit weight with markers HP61, HP32 and HP31. The years used to collect the data are shown as FW2007, FW2008 and FW2010. (D) Transgenic complementation tests. Average fruit weight of 10 to 13 plants from T1 generation transgenic and non-transgenic sib plants cultivated under the same growing conditions are shown. Error bar: standard deviation. HF, Howard German background and 9F/CF, VIR347 background. Data collected during the 2013 field season is denoted with an “a”. Data collected during the 2014 field season is denoted with a “b”. Significance determined by paired t-tests and transgenic (transgenically carrying the CSR-D allele) plants were compared to their non-transgenic (CSR-WT) sibs. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
Detailed sequence analyses showed that the derived allele carried a 1,406 bp deletion and 22 bp insertion resulting in a 194 amino-acid truncation of the predicted wild type protein. Interestingly, the truncation might not result in a loss-of-function of fw11.3, as the derived allele was partially dominant over the wild type allele [24]. To further corroborate that Solyc11g071940 was the likely candidate FW11.3 gene and that the derived allele constituted a gain-of-function mutation, we constructed transgenic complementation lines by transferring the partially dominant allele of Solyc11g071940 to the lines in the wild type fw11.3 background. Fifteen independent transgenic complementation lines (T0) were obtained of which four progeny families (T1) were evaluated at least once. Fruit weight was significantly increased in nearly all transgenic progeny lines compared to their non-transgenic sibs (Fig 1D, S2 Table). Therefore, the results from the transgenic complementation lines demonstrate that Solyc11g071940 controls tomato fruit size and that the deletion found in the derived allele is indeed acting in a dominant manner. This finding supports the notion that fw11.3-D functions as a gain-of-function mutation.
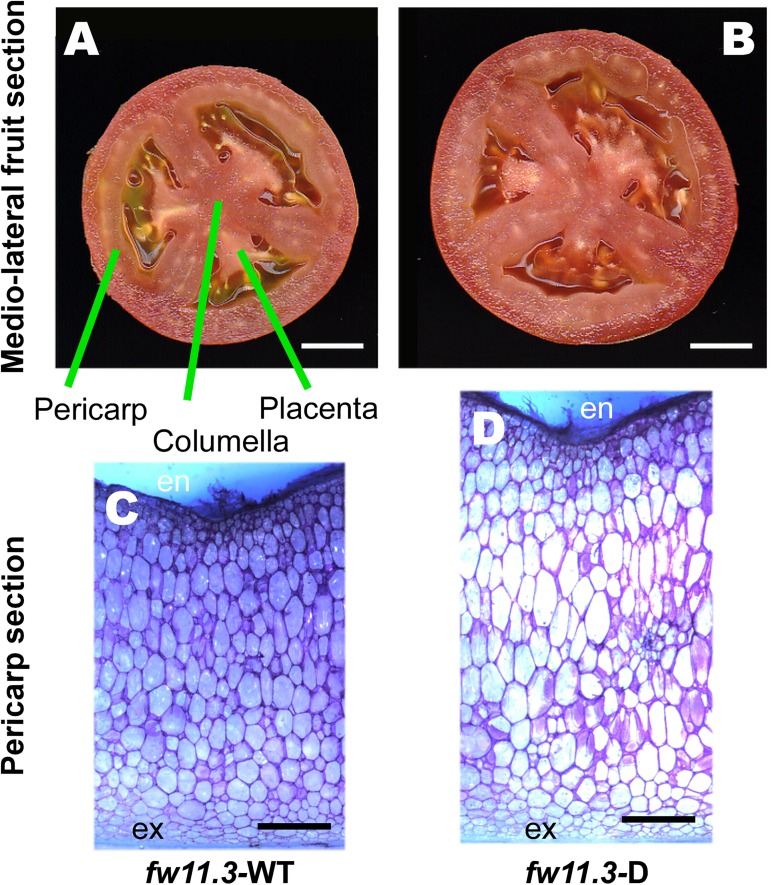
As expected, the fruit weight, fruit area and fruit perimeter were significantly larger in the fw11.3-D nearly isogenic lines (NIL) in the cultivated background (Table 1). To evaluate the effect of the QTL in the wild species background, we developed NILs where fw11.3-D was introgressed in the mostly LA1589 background, an accession of S. pimpinellifolium that is the closest fully wild ancestor species of cultivated tomato. These NILs were either differing for just fw11.3 or were fixed for the derived alleles at fw2.2 and fw3.2 while differing at the fw11.3 locus. Similar to the NILs in the cultivated background, fruit weight, fruit area and fruit perimeter were significantly larger in the fw11.3-D in the wild species background (Table 1). To evaluate which parts of the fruit were enlarged by the derived allele, we analyzed the pericarp, columella and placenta areas in the NILs (Fig 2). The pericarp area was significantly enlarged in the NILs that carry the derived allele in both cultivated and wild tomato species backgrounds. By contrast, the columella and placenta area were not consistently different in the NILs (Table 1). Because the effect of fw11.3 was most pronounced in the pericarp in all the NILs, we evaluated whether increased cell layers (number) in the mesocarp or increased cell size contributed to the larger pericarp area values. Our data showed that the number of cell layers was mostly unaltered in NILs in both cultivated and wild species backgrounds whereas mesocarp cell size was increased in all the lines carrying the derived allele of fw11.3 (Fig 2, Table 2). Moreover, transgenic lines that complemented the large fruit phenotype also showed increased cell sizes compared to non-transgenic control. Thus, the role of FW11.3 is to increase cell size and therefore we named Solyc11g071940 to Cell Size Regulator (CSR).
Table 1. Fruit structure analyses in medio-lateral sections of the fw11.3 NILs.
| Population (background) | 12S114 (RG) | 12S116 (RG) | 12S147 (HG) | 12S148 (HG) | 15S107 (LA1589) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele | fw11.3-D | fw11.3-WT | fw11.3-D | fw11.3-WT | fw11.3-D | fw11.3-WT | fw11.3-D | fw11.3-WT | fw2.2/fw3.2/fw11.3-D | fw2.2/fw3.2/fw11.3-WT | fw11.3-D | fw11.3-WT |
| Trait | ||||||||||||
| Fruit Weight (g) | 88.0 ± 5.3 | 73.2 ± 1.9*** | 66.3 ± 3.8 | 53.0 ± 2.6**** | 91.6 ± 4.5 | 72.6 ± 2.2**** | 88.4 ± 2.2 | 68.2 ± 4.6**** | 2.13 ± 0.13 | 1.49 ± 0.10**** | 1.51 ± 0.12 | 1.26 ± 0.04** |
| Fruit Perimeter (cm) | 17.7 ± 0.64 | 16.6 ± 0.3** | 16.0 ± 0.3 | 14.8 ± 0.5*** | 15.8 ± 0.4 | 14.3 ± 0.3**** | 16.0 ± 0.4 | 14.3 ± 0.4**** | 5.11 ± 0.22 | 4.64 ± 0.10** | 4.61 ± 0.17 | 4.31 ± 0.1* |
| Fruit Area (cm2) | 22.2 ± 1.5 | 19.6 ± 0.7** | 18.2 ± 0.7 | 15.7 ± 1*** | 17.9 ± 1 | 14.7 ± 0.5**** | 18.4 ± 0.9 | 14.7 ± 0.9**** | 1.89 ± 0.16 | 1.56 ± 0.07** | 1.55 ± 0.11 | 1.34 ± 0.06* |
| Pericarp Area (cm2) | 11.08 ± 0.92 | 9.53 ± 0.35** | 9.89 ± 0.30 | 8.16 ± 0.47**** | 8.57 ± 0.42 | 6.87 ± 0.29**** | 8.15 ± 0.36 | 6.59 ± 0.51**** | 0.82 ± 0.08 | 0.70 ± 0.03* | 0.62 ± 0.05 | 0.55 ± 0.02* |
| Pericarp Area/Total Area | 0.50 ± 0.02 | 0.49 ± 0.01 | 0.54 ± 0.02 | 0.52 ± 0.02 | 0.48 ± 0.02 | 0.47 ± 0.01 | 0.44 ± 0.02 | 0.45 ± 0.03 | 0.43 ± 0.01 | 0.45 ± 0.003*a | 0.40 ± 0.01 | 0.40 ± 0.004 |
| Pericarp Thickness (cm) | 0.77 ± 0.05 | 0.72 ± 0.02* | 0.78 ± 0.03 | 0.69 ± 0.03*** | 0.67 ± 0.03 | 0.58 ± 0.02**** | 0.61 ± 0.03 | 0.54 ± 0.02*** | 0.21 ± 0.01 | 0.20 ± 0.003 | 0.27 ± 0.01 | 0.25 ± 0.005* |
| Placenta and Columella Area (cm2) | 3.52 ± 0.28 | 3.00 ± 0.22** | 2.26 ± 0.25 | 2.19 ± 0.29 | 2.32 ± 0.25 | 1.96 ± 0.20* | 2.91 ± 0.4 | 2.30 ± 0.3** | 0.065 ± 0.008b | 0.050 ± 0.009*,b | 0.033 ± 0.005b | 0.033 ± 0.002b |
| Placenta and Columella Area/Total Area | 0.16 ± 0.01 | 0.15 ± 0.01 | 0.12 ± 0.01 | 0.14 ± 0.01*,a | 0.13 ± 0.01 | 0.13 ± 0.01 | 0.16 ± 0.02 | 0.16 ± 0.01 | 0.034 ± 0.002b | 0.032 ± 0.005b | 0.021 ± 0.003b | 0.025 ± 0.002b |
RG, Rio Grande
HG, Howard German
LA1589, an accession of the wild species S. pimpinellifolium. Significance determined by paired t-Test and compared to fw11.3-D in the same NIL.
*, p ≤ 0.05
**, p ≤ 0.01
***, p ≤ 0.001
****, p ≤ 0.0001.
a Note the value is larger for the fw11.3-WT.
b Values reflect the columella area only.
Fig 2. Fruit and cellular structure of fw11.3 NILs in the Howard German background.
(A) Medio-lateral section of fw11.3-WT fruit. Size bar = 1 cm. The pericarp, columella and placenta tissues are shown. (B) Medio-lateral section of fw11.3-D fruit. Size bar = 1 cm. (C) Hand cut section of a fw11.3-WT pericarp from a representative mature green fruit stained with toluidine blue. Size bar = 1 mm. (D) Hand cut section of a fw11.3-D pericarp from a representative mature green fruit stained in toluidine blue. Size bar = 1 mm. The inner epidermis is denoted as “en” whereas the outer epidermis is denoted as “ex”. The mesocarp are represented by the cells between the inner and outer epidermis.
Table 2. Cell size and cell number comparisons in the pericarp of fw11.3 NILs and VIR347 transgenic lines.
| 12S147 Howard German NILs | 14S81 Rio Grande NILs | ||||||
| fw11.3-D | fw11.3-WT | P-value | fw11.3-D | fw11.3-WT | P-value | ||
| n = 5 | n = 5 | n = 11 | n = 9 | ||||
| Cell layers | 18.8 ± 0.6 | 18.6 ± 0.6 | 0.664 | 30.39 ± 2.12 | 29.62 ± 1.34 | 0.360 | |
| Largest cell size (mm2) | 0.21 ± 0.02 | 0.16 ± 0.02 | 0.006 | 0.16 ± 0.02 | 0.14 ± 0.01 | 0.048 | |
| Average cell size (mm2) | 0.10 ± 0.02 | 0.08 ± 0.01 | 0.042 | 0.07 ± 0.012 | 0.057 ± 0.009 | 0.015 | |
| 15S107 LA1589 NILs | |||||||
| fw2.2/fw3.2/fw11.3-D | fw2.2/fw3.2/fw11.3-WT | P-value | fw11.3-D | fw11.3-WT | P-value | ||
| n = 5 | n = 6 | n = 5 | n = 6 | ||||
| Cell layers | 16.34 ± 0.50 | 15.82 ± 0.72 | 0.237 | 13.86 ± 0.70 | 13.02 ± 0.18 | 0.030 | |
| Largest cell size (mm2) | 0.054 ± 0.006 | 0.045 ± 0.005 | 0.034 | 0.050 ± 0.005 | 0.035 ± 0.004 | 6.66E-04 | |
| Average cell size (mm2) | 0.031 ± 0.007 | 0.025 ± 0.003 | 0.118 | 0.027 ± 0.003 | 0.016 ± 0.001 | 0.001 | |
| 14S68 VIR347 transformed lines | 14S70 VIR347 transformed lines | ||||||
| Transgenic | Non-transgenic | P-value | Transgenic | Non-transgenic | P-value | ||
| n = 10 | n = 10 | n = 8 | n = 9 | ||||
| Cell layers | 18.69 ± 0.78 | 18.63 ± 0.73 | 0.855 | 19.67 ± 0.31 | 19.51 ± 0.72 | 0.560 | |
| Largest cell size (mm2) | 0.12 ± 0.02 | 0.09 ± 0.01 | 0.002 | 0.12 ± 0.01 | 0.09 ± 0.01 | 5.55E-05 | |
| Average cell size (mm2) | 0.069 ± 0.014 | 0.052 ± 0.011 | 0.007 | 0.067 ± 0.009 | 0.046 ± 0.007 | 8.42E-05 | |
Cell layers reflect the mesocarp cell number in the pericarp between the epidermis and the endodermis.
Largest cell size is based on the average size of the six largest cells in the pericarp found in the hand cut sections.
Average cell size reflects the average cell size in the mesocarp by dividing the number of cells by a set area. The set area was 6 mm2 for 12S147; 9.466 mm2 for 14S81; 0.543 mm2 and 0.506 mm2 for the LA1589 NILs respectively; and 2.036 mm2 for 14S68 and 14S70.
"n" reflects the number of plants evaluated. For each plant, at least four sections from three fruits were evaluated.
Other phenotypes associated with CSR in the fw11.3 NILs
To determine whether fw11.3-D affected other plant growth characteristics, we evaluated several traits such as yield per plant and leaf size. In addition to fruit weight, fw11.3 controlled fruit number per plant such that fw11.3-WT produced more fruit than fw11.3-D (S3 Table). Overall yield per plant was not altered, which implied that increased fruit weight was compensated by fewer fruit per plant. Higher fruit number could have resulted from increased length of the side shoots which in turn would have led to more inflorescences even though the latter trait was not significantly different in the NILs. It was unlikely that fw11.3 controlled the source-sink relationship because reducing the number of fruit concommittantly led to larger fruits in both fw11.3-WT and fw11.3-D backgrounds (S3 Table). The effect on fruit weight was manifested late in development, namely after anthesis since ovary size was the same in the fw11.3 NILs, and without an effect on ripening time. Other architectural and morphological traits were not consistently altered in the NILs implying that the effect of fw11.3 was nearly exclusively on fruit weight by increasing cell size (S3 Table).
Cells in fruits of many plant species undergo endoreduplication leading to increased nuclear ploidy levels. The increase in ploidy and nuclear size is positively correlated to cell size in tomato [15]. To determine whether CSR might control cell size in the pericarp by increasing the nuclear size, we determined the changes in ploidy levels in mature green fruit collected from the fw11.3 NIL plants. At the mature green stage, the fruit had reached its largest size which occurred between 35 (mature green) and 40 to 43 DPA (ripening) in the Rio Grande background [26]. The flow cytometry results showed the most prevalent ploidy levels of 8C, 16C, and 32C but also some nuclei with levels as high as 512C in the tomato pericarp (P10 in S1 Fig). Over multiple experiments, the Rio Grande fw11.3-D NIL pericarps featured more nuclei at higher ploidy level (64C to 512C) compared to fw11.3-WT for at least one C-value (S4 Table). In one replicate, the increased number of higher ploidy level nuclei was accompanied by a reduced number of lower ploidy nuclei in fw11.3-D compared to fw11.3-WT. The endoreduplication index (EI') in two out of five replicates was significantly increased, suggesting that CSR-D leads to more nuclei at higher ploidy levels compared to WT. We also evaluated changes in nuclei ploidy levels in the pericarp of the NILs in the LA1589 wild species background. The EI' was significantly higher in one of two sets of NILs, which is consistent with the results from the Rio Grande NILs. Nuclear ploidy levels were also evaluated in columella tissues (S4 Table). In the Rio Grande background, nuclei ploidy levels at certain higher C-values were the same or decreased in fw11.3-D, contrary to the ploidy level observations in the pericarp. In all, EI' in columella was never significantly increased in fw11.3-D in either cultivated or wild species background. Combined, these results implied that the effect of fw11.3 on endoreduplication might be indirect and if so, it only affects the cells in the tomato pericarp.
Protein sequence and phylogeny analysis of CSR
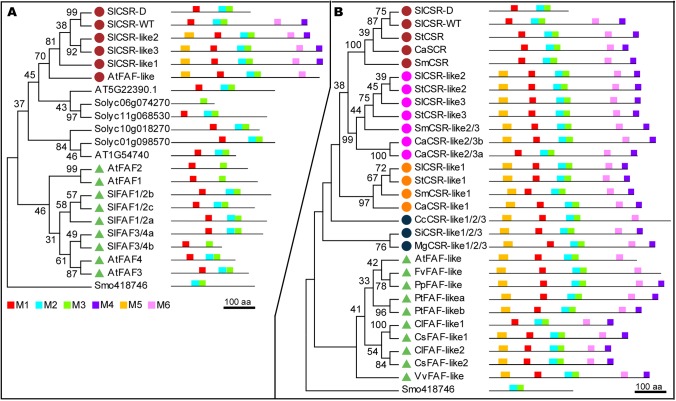
The predicted CSR protein featured a domain that is recognized as the FAF domain (formely known as DUF3049, Pfam accession PF11250). The tomato genome harbored 13 predicted proteins that represented FAF domain family. In Arabidopsis, the FAF domain was found in 10 proteins which included four FANTASTIC FOUR (FAF) and a single FAF-like protein. In tomato, three genes were recognized as CSR paralogs because their proteins were closely related to one another and shared similar motif patterns. We named these three paralogs CSR-like1, CSR-like2 and CSR-like3 (Fig 3). At the protein level, the MEME motifs 2 and 3 represented the FAF domain whereas motif 6 defined the CSR family proteins. Motifs 4 and 5 were also often found in members of the CSR clade. The single copy Arabidopsis FAF-like protein was the most similar to the tomato CSR clade, whereas the four FAFs formed a separate clade (Fig 3A). Likely orthologs of the four Arabidopsis FAFs were identified in tomato, including three SlFAF1/2 and two SlFAF3/4. The FAF proteins have been recognized as controlling shoot meristem size [27].
Fig 3. Phylogenic and motif analysis of FAF domain-containing proteins.
(A) Phylogenic tree of tomato and Arabidopsis proteins, using FAF domain sequences in the tree construction. Motif structure is based on full length protein sequences. (B) Phylogenic tree and motif analysis of CSR-like proteins found in different plant species: Solanum lycopersicum (Sl), Solanum tuberosum (St), Solanum melongena (Sm), Capsicum annuum (Ca), Coffea canephora (Cc), Sesamum indicum (Si), Mimulus guttatus (Mg), Arabidopsis thaliana (At), Fragaria vesca (Fv), Prunus persica (Pp), Populus trichocarpa (Pt), Citrullus lanatus (Cl), Cucumis sativus (Cs), Vitis vinifera (Vv), and Selaginella moellendorffii (Smo). Selaginella moellendorfii (Smo) is used as outgroup. The same colored dots or triangles represent same subclade. Scale bar: 100 amino acids. Renamed proteins: Solyc06g073940 (SlCSR-like1), Solyc01g009260 (SlCSR-like2), Solyc01g009270 (SlCSR-like3), Solyc06g084280 (SlFAF1/2a), Solyc06g008990 (SlFAF1/2b), Solyc09g065140 (SlFAF1/2c), Solyc01g079740 (SlFAF3/4a), Solyc06g054310 (SlFAF3/4b), PGSC0003DMP400005394 (StCSR), CA11g16000 (CaCSR), Sme2.5_00683.1_g00009 (SmCSR), PGSC0003DMP400023251 (StCSR-like2), PGSC0003DMP400023246 (StCSR-like3), Sme2.5_01340.1_g00001 (SmCSR-like2/3), CA01g13730 (CaCSR-like2/3b), CA01g13720 (CaCSR-like2/3a), PGSC0003DMP400010456 (StCSR-like1), Sme2.5_00076.1_g00022 (SmCSR-like1), CA06g22610 (CaCSR-like1), Cc01g07830 (CcCSR-like1/2/3), Sin1010620 (SiCSR-like1/2/3), mgv1a004589m (MgCSR-like1/2/3), mrna23163.1-v1.0-hybrid (FvFAF-like), ppa002898m (PpFAF-like), Potri.009G016600 (PtFAF-like_a), Potri.001G216000.2 (PtFAF-like_b), Cla008617 (ClFAF-like1), Csa6M426380 (CsFAF-like1), Cla007326 (ClFAF-like2), Csa1M635920 (CsFAF-like2), VIT206s0009g003101 (VvFAF-like).
Only one CSR member was found in Arabidopsis whereas four CSR paralogs were found in tomato. To further explore the apparent expansion of the CSR family, we searched databases for protein members that were closely related to CSR and included motifs 4 and/or 6. The most likely orthologs of tomato CSR and CSR-like 1 to 3 were found in other Solanaceous species namely potato, eggplant and chile pepper (Fig 3B). When comparing species with sequenced genomes outside the Solanaceae family but within the Asterids clade, only one copy of CSR was found in coffee (Gentianales), sesame and mimulus (Lamiales) (Fig 3B). However, based on motif structure, this single paralog appeared more similar to the CSR-like1 to 3 encoded proteins than to CSR. For species in the Rosids clade, most of them carried only one copy of a CSR-like except watermelon, cucumber and poplar. These results suggested a single common ancestor of CSR that evolved independently in Rosids and Asterids clades, and only expanded to four in the Solanaceace family.
Expression analysis of CSR and related genes
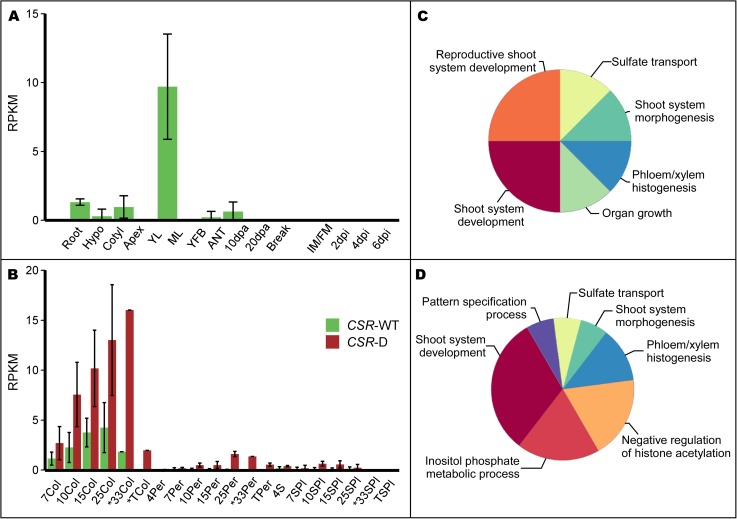
To further understand the role in tomato fruit growth, we evaluated the temporal and spatial expression patterns of CSR. Despite its effect only on fruit weight, CSR-WT expression was not detected in whole developing fruit (Fig 4A). Low expression however, was detected in mature leaf tissues (Fig 4A, S5 Table). Moreover, analyses of other RNA-seq datasets showed that CSR expression was undetectable in tomato meristems (http://tomatolab.cshl.edu/efp/cgi-bin/efpWeb.cgi), as well as ovary and 4 days old fruit tissues collected by laser captured microscopy (http://ted.bti.cornell.edu/cgi-bin/TFGD/digital/experiment.cgi?ID=D009). These results demonstrated that CSR expression was undetectable from meristems, young floral buds, anthesis to the cell proliferation stage of fruit development. Although CSR expression was undetectable when evaluating the whole fruit, we expected gene expression in certain tissues within this organ at later developmental stages. Indeed when sampling the pericarp, columella and seed&placenta tissues separately from maturing fruits, the expression of CSR was found to be low but detectable. CSR was expressed the highest in the columella where its expression increased between 7 to 33 days post anthesis (DPA), and decreased again at the turning stage (Fig 4B). CSR was also expressed in pericarp and seed/placenta tissues albeit at low or very low levels (Fig 4B). Interestingly, CSR-D showed higher expression compared to CSR-WT but with a similar profile in both columella and pericarp. Recently, higher resolution expression information in developing tomato fruits (from J.K.C Rose, Cornell University) also supported the notion that CSR-D was highest expressed in the columella. Moreover, the expression was particularly high in the vascular bundles of the developing tomato pericarp (S2 Fig).
Fig 4. Expression of CSR in tomato plant organs and fruit tissues.
(A) Expression of CSR-WT in tomato LA1589 plant tissues. Root: root; Hypo: hypocotyl; Cotyl: cotyledon; Apex: shoot apex including the SAM and youngest leaves; YL: young leaf; ML: mature leaf; YFB: young floral buds from 10 days after initiation and younger; ANT: whole flower at anthesis; 10 and 20 dpa: 10 and 20 days post anthesis developing fruit, respectively; Break: breaker stage fruit which is before turning color; IM/FM: inflorescence and floral meristem; 2, 4, and 6 dpi: 2, 4, and 6 days post initiation flower buds, respectively. Error bar indicates standard deviation. (B) CSR transcript accumulation in fw11.3 NILs fruit tissues during fruit development. Error bar: standard deviation. *The 33Col, 33Per, 33SPl and TCol samples represent one replicate. Col: columella; Per: pericarp; S: seeds; SPl: seeds and placenta; T: turning stage fruit. Numbers associated with the sample names represent the fruit developmental stage as days post anthesis. Hence, 7Col represents columella tissue collected 7 days post anthesis. (C) ClueGO enrichement in the CSR-D coexpression cluster. (D) ClueGO enrichment in the CSR-WT coexpression cluster. The dimension of the pie chart wedges is proportional to the number of terms included in each category. The most significant term of the group was used for annotation.
Of all the genes that encode the FAF motif, CSR-like1 was the highest expressed in nearly all plant tissues examined including seedling tissues (cotyledons, hypocotyl, shoot apex and roots) and in the columella of developing fruit (S5 Table). In developing fruit, CSR-like1 featured the most similar expression dynamic as CSR, even though CSR-like1 was often 15 fold or higher expressed than CSR-D (S5 Table). Expression of CSR-like2 and CSR-like3 was specifically detected in hypocotyl, cotyledon and shoot apex (Fig 4A, S5 Table). In all, CSR was expressed dynamically in developing fruit and CSR-D allele was higher expressed than CSR-WT allele. On the other hand, tomato FAF1/2a showed the highest expression in floral meristem tissues and early developing floral buds; FAF1/2c was particularly well expressed in emerging leaves; and FAF3/4a was well expressed in seedling tissues as well as floral meristem and early floral buds. In sum, CSR and CSR-like genes show distinct expression in maturing tissues whereas the FAFs are highest expressed in meristems and young tisues.
Co-expression patterns of CSR in developing fruit
DESeq2 and other differential gene expression methods showed few genes that were significantly differentially expressed at different time points and tissue types throughout fruit development when contrasting the fw11.3-D and fw11.3-WT NILs [26]. And very few of those were consistent among more than one time point or tissue type. Instead, we sought to identify genes that were expressed with similar dynamics as CSR. Genes with common expression patterns could share related functions as they may control or be controlled by similar processes in the plant [28]. To identify potential shared developmental and/or metabolic processes with CSR, we conducted co-expression cluster analyses and then performed functional enrichment using the RNA-Seq data from the fw11.3-D and fw11.3-WT NILs separately. Cluster analyses showed that CSR-D had a 99.3% probability to be part of cluster 11 which was comprised of 493 genes (probability of 90% or higher) (S3A Fig, S1 Dataset). Functional enrichment using ClueGO identified six pathways that were significantly enriched. In the CSR-D co-regulated cluster, the pathway “shoot system development” was especially highly enriched with 31 genes, 17 of which encoded putative transcription factors (Fig 4C, S2 Dataset). The enriched pathway “phloem or xylem histogenesis” was also of interest due to the relatively high expression of CSR-D in the vascular tissues (S2 Fig). In this pathway, six genes of which four encoded putative transcription factors were found.
We next identified the CSR-WT co-expressed genes using the RNA-seq data set that was generated with the fw11.3-WT NIL. Because many of the genes were found in both the CSR-D and the CSR-WT co-regulated clusters, we expected similar pathways to be enriched. Of the 558 CSR-WT cluster genes with a probability of 90% or higher, 63% (352) were shared with the CSR-D co-expressed cluster 11 (S1 Dataset, S3B Fig). Indeed, the pathway “shoot system development” was similarly enriched in the CSR-D and CSR-WT clusters. Likewise, the pathway “phloem or xylem histogenesis” was enriched in both clusters (Fig 4D). These findings suggest a role for CSR in regulating cell size during shoot and vascular development in growing tissues.
Genes of interest in the co-regulated clusters that were not part of an enriched GO term were also found. One gene of particular interest was CSR-like1, which showed similar expression patterns in developing fruits as CSR (S5 Table). Also, several putative orthologs in the cytokinin signaling and biosynthesis pathway such as LOG3 (Solyc11g069570), IPT5 (Solyc01g080150), WOL (Solyc04g008110), ARR12 (Solyc07g005140) and ARR1 or ARR2 (Solyc01g0655540) were found in one or both CSR co-expressed gene clusters. Moreover, a likely ortholog of an auxin efflux carrier PILS5 (Solyc03g032080), an auxin signaling gene ARF11 (Solyc05g0560400), and a gene encoding a pleckstrin domain-containing protein (Solyc08g066860) which is thought to be a component of the auxin canalization pathway were found in the CSR co-regulated gene clusters.
Origin of CSR-D during the evolution of tomato
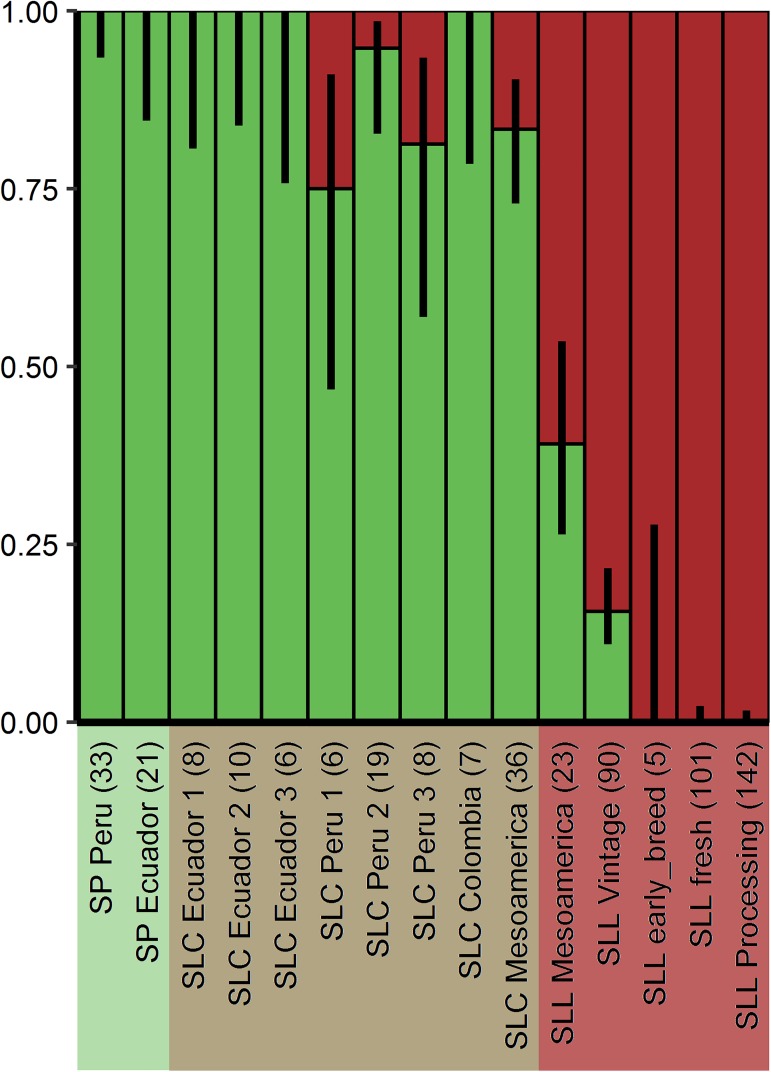
Fruit weight was an important selection criterion that drove the evolution of tomato from a wild relative bearing small fruit to the large tomatoes found in grocery stores today. To determine whether selections for the derived allele of CSR might have taken place, we evaluated the origin and distribution of CSR-D in wild, semi-domesticated, early landraces as well as modern breeding germplasm (Fig 5). The mutation was found in low frequency in S. lycopersicum var cerasiforme from Peru and Mesoamerica. However, the distribution of the derived allele frequency greatly increased in the Mesoamerican domesticated landraces and is now completely fixed in the large fruited contemporary germplasm. The contemporary germplasm included processing and fresh market tomatoes, the latter which were comprised of globe/large used for slicing and plum tomatoes used for soups and stews [4]. These results imply that CSR-D arose late but became rapidly fixed during the selections by early farmers.
Fig 5. Distribution of CSR wild type and derived allele in the tomato germplasm.
Ancestral allele in green, derived allele in burgundy. Black lines show binomial confidence intervals at 95%. Background colors highlight different species: S. pimpinellifolium (light green), S. lycopersicum var. cerasiforme (light brown) and S. lycopersicum var. lycopersicum (pink). Number of accessions in each category is given in parenthesis above the different genetically distinct classes [4].
Discussion
In this study, we demonstrated by fine mapping, association mapping and plant transformation experiments that the tomato fw11.3 QTL is controlled by Solyc11g071940 encoding a largely uncharacterized protein. The 1.4 kb deletion caused a 194 amino acid truncation at the C-terminus of the encoded protein creating a partial dominant allele. The heavier fruit was predominantly caused by increased pericarp area resulting from larger cells in this tissue, hence the name CSR which is short for Cell Size Regulator. Increases in cell size in the pericarp were correlated to increases in nuclear ploidy levels in this same tissue albeit that CSR’s effect on this process appears to be small. Fixation of the derived allele in cultivated tomato supported the notion that increased cell size mediated by CSR was indeed critical for the recent evolution of the crop.
CSR coincides with the cell enlargement stage in developing tomato fruit
In roots, cellular differentiation occurs when cells progress from the meristematic (dividing) zone through the transition zone into the elongation and differentiation zone. Development of the tomato pericarp may follow a similar trajectory as described for the root: immediately following pollination and fertilization and after a short period of cell division, pericarp cells transition to the cell expansion stage. In small fruited tomatoes, the cell layers in the walls of the tomato increase from 10 to 20 in less than five days following pollination [7]. This period is followed by approximately 3.5 weeks of cell enlargement prior to ripening. Expression of CSR coincides with the cell expansion stage and therefore, this gene might be associated with cellular differentiation. A conserved domain FAF with unknown function has been identified in CSR and CSR-like proteins. This domain is named after the Arabidopsis FANTASTIC FOUR (FAF) proteins regulating shoot meristem size [27]. Whereas FAFs are expressed strongly in meristematic cells and young growing tissues, CSR expression was undetectable in these tissues and instead only found in maturing plant organs such as the fruits prior to ripening (S5 Table). Thus, the function of CSR and FAFs is not likely the same at the tissue level even though biochemically they might be involved in similar processes within the cell. Our research efforts focused on the effect of CSR in the pericarp because this tissue contributes greatly to overall fruit weight, and the area was significantly expanded through cell size increases in the fw11.3-D NILs. However, expression of CSR was the highest in the columella and not the pericarp (Fig 4). Recently, we obtained higher resolution expression of CSR-D in developing tomato fruit which showed that expression of this gene is high in and around vascular bundles in the pericarp. Thus, the high expression of CSR in the columella could be due to the relatively high vasculature density in columella tissues. We attempted to evaluate cell size in the columella which ranged from very small (in the center) to very large (towards the periphery). However, coumella structure varied a lot from fruit to fruit and therefore, it was not feasible to evaluate cell size similarly in the columella sections taken from different fruits. Since the entire vascular bundle was captured by laser captioned microdissection, it is not known where in or around the bundles CSR is expressed. To further understand the function of CSR, future studies should be directed to evaluate its role in vascular development and in the columella.
CSR might be involved in the antagonistic action of auxin and cytokinin to regulate cell differentiation
Even though CSR encodes a protein of unknown function, co-expression analyses may have revealed the cellular processes that provide insights into the function of CSR. The antagonistic roles of auxin and cytokinin might possibly be associated with CSR function since several genes associated with these pathways were found in the co-expression clusters. For example in Arabidopsis root development, the B-type response regulators ARR2 and ARR12 play important roles in the mitotic exit by upregulating the expression of CCS52A1, an activator of the APC/C complex, and SHY2/IAA3, leading to inhibition of auxin signaling [17, 19]. Putative orthologs of ARR2 (Solyc01g0655540) and ARR12 (Solyc07g005140) were found in the CSR co-expressed dataset. Additional cytokinin signaling and biosynthetic proteins were identified including LOG3 (Solyc11g069570) of unknown function but thought to play a role in the biosynthesis of cytokinins, a cytokinin synthase protein IPT5 (Solyc01g080150) [29], a cytokinin receptor CRE/AHK4 (Solyc04g008110) [30], and a cytokinin sensitivity protein PRL1 (Solyc01g094480) [31] were found in the CSR gene clusters in the fw11.3 NILs. Auxin signaling and response genes were found in these clusters as well, namely genes encoding auxin transport protein PILS5 (Solyc03g032080) and ABCB15 (Solyc02g087410) [32, 33], an auxin signaling protein ARF11 (Solyc05g0560400), a pleckstrin domain-containing protein (Solyc08g066860) that is involved in vascular patterning and auxin canalization [34], a HB-2 transcription factor (Solyc08g078300) involved in cell expansion and response to auxin [35, 36], IBR5 (Solyc12g005990) a dual specificity phosphatase that promotes auxin responses and acts as a regulator of organ size in Arabidopsis [37], and a KNOTTED-like protein 1 (Solyc04g077210) whose expression is repressed by auxin resulting in the promotion of leaf fate [38, 39]. GO term enrichment for genes in the CSR expression clusters also implied that plant vascular development is one of the processes that may be affected by CSR and CSR-like1 which was consistent with its expression in this tissue. Indeed, deeper searches in the entire list of co-regulated genes led to the identification of additional genes that may be critical in vascular development. These include genes that encode putative orthologs of receptor-like kinases XIP1 (Solyc04g077010) involved in the maintenance of cell files or cell morphology in conductive elements [40] and BRI1-like 2 (Solyc04g008430) associated with the development of provascular/procambium cells [41–43], as well as several phloem expressed lectin genes (Solyc02g069020, Solyc02g069030, Solyc02g069060, Solyc02g031740). In addition, putative orthologs of genes involved in vascular development [44] were found in the dataset including APL (Solyc12g017370), CNA (Solyc12g044410) and BRX-like 4 (Solyc12g044410). With CSR expression particularly high in vascular bundles and the columella (which is enriched for xylem and phloem tissues), these co-regulated genes suggest that CSR and perhaps CSR-like1 play a role in cellular maturation in the vascular bundle leading indirectly to increases in pericarp cell size and nuclear ploidy. Therefore, CSR might be involved in the antagonistic effects of auxin and cytokinin as a mechanism for cellular differentiation and enlargement in different tissue types.
Cell enlargement is also associated with enhanced endoreduplication and larger fruit weights [15]. Ubiquitously found in both higher plants and animals, the function of endoreduplication is not well understood other than its association with increased cell size. The transition from cell proliferation to enlargement in roots coincides with the initiation of endoreduplication [16, 17, 19, 20]. Other studies have suggested a role for endoreduplication in enhanced metabolism [45] or to sustain growth under adverse conditions including pathogen attack [46, 47]. Even though the function is not well understood, the mechanisms regulating the core entry and progression of endoreduplication has been reasonably well established. Distinct stages of cell division are regulated by CYCLINS (CYC), Cyclin-Dependent Kinases (CDK) and CDK inhibitors (CKI). The onset and progression of endoreduplication is mediated by these same core cell cycle proteins through transcription factors regulating gene expression as well as regulators that control the ubiquitination of the proteins which then targets them for proteolytic degradation. Specifically, CYCLINS and CDKs that regulate the M stage are suppressed when endoreduplication is promoted [20, 48]. This suppression is mediated by activation of a specific E3 ubiquitin ligase named the Anaphase Promoting Complex/Cyclosome (APC/C) that leads to ubiquitination of the mitotic Arabidopsis proteins CDKB1;1 and CYCA2;3 which is then followed by proteasome-mediated protein degradation [49]. Activators of APC/C are for example Cell Cycle Switch52 A1 (CCS52A1) encoding a WD-repeat protein and mutations or transcriptional downregulation leads to termination of cell expansion and reduced endoreduplication. It is unlikely, however that CSR plays an important role in endoreduplication because none of the known aforementioned endoreduplication genes such as those encoding CYCLINS, CDK and CKI or any related to APC/C proteasome complex and most of their transcriptional and translational regulators, were present in the CSR gene expression clusters (S1 Dataset) [26]. Moreover, the impact of CSR-D on enhancing endoreduplication was very small.
Evolution of CSR in dicot plants
How did the CSR clade including the FAF-like genes evolve in plants? In our study, we observed that CSR expanded in the Solanaceae family resulting in four paralogs: CSR-like1, CSR-like2, CSR-like3 and CSR. Among them, CSR-like2 and CSR-like3 might have been the result of a recent tandem duplication event because they shared high sequence similarity to one another and were located next to each other on chromosome 1. When compared with selected species in the Solanaceae family, most of the four paralogs corresponded to an ortholog in pepper, eggplant and potato. The clustered CSR-like2 and 3 shared two orthologs in potato and pepper, but only one in eggplant. The eggplant genome sequence is not yet available and thus, it was possible that eggplant also carried two CSR-like2/3 gene copies. As for other species in the Asterids clade, coffee, sesame and mimulus carried only one paralog of the CSR family genes. This result suggest that the duplication resulting in CSR, CSR-like1 and CSR-like2/3 occurred after the Solanales diverged from the Gentianales and Lamiales orders, but before the Solanaceae family split to tomato, potato, pepper and eggplant. A single FAF-like gene is found in monocot and dicot species and is proposed to be the ancestor to FAF [27] as well as the CSR clade based on our findings (Fig 3A). In fact, the predicted protein motif structure of FAF-like in Arabidopsis is more similar to CSR than to FAF. As CSR and CSR-like genes only expanded in the Solanaceae family, they may have evolved specific functions that are specific to the family. Regardless and as mentioned above, the CSR subclade of the FAF proteins may be involved in cellular differentiation and enlargement resulting from the antagonistic action of auxin and cytokinin. And this might be the unifying role of CSR/CSR-like/FAF-like genes in land plants.
Materials & methods
Plant materials
Tomato seeds or DNA were obtained from the TGRC (http://tgrc.ucdavis.edu/; S. pimpinellifolium LA1589); Tomato Growers Supply Co (Howard German, Rio Grande, Yellow Pear); Dr. Mathilde Causse (Institut National de la Recherche Agronomique-Avignon, France; Tomato core collection for association mapping [25]); Drs. Maria José Díez and Jose Blanca (Universitat Politècnica de València, Spain; Instituto de Conservación y Mejora de la Agrodiversidad Valenciana collection [4]); Dr. David Francis (The Ohio State University, United States; SolCAP collection [50]). Plants were grown in field (for fruit weight and plant evaluations) and greenhouse (for population development and evaluation of additional plant phenotypes) at Ohio Agricultural Research and Development Center (Wooster, OH USA) or at the University of Georgia (Athens, GA USA) under standard conditions. For the greenhouse, the plants were grown in 2-gallon pots using 15-9-12 Osmocote slow release supplemented by 20-20-20 fertilization in Fafard 3B growing media with supplemental lighting.
Population development, fine mapping and progeny testing
To fine map fw11.3, a population derived from a cross between S. lycopersicum c.v. Howard German (HG) and the wild species S. pimpinellifolium accession LA1589 was used [51] (S4 Fig). New markers were developed for the fine mapping and were based on the tomato genome sequence and known marker sequences (S6 Table). Eight recombinants were obtained through screening of 1906 seedlings, and two additional recombinants were obtained through screening 732 seedlings. Fruit weight was compared between fw11.3-WT and fw11.3-D plants within each recombinant family using Student’s t-test (S1 Table). Additional mapping was conducted in populations derived from crosses between S. lycopersicum c.v. Rio Grande (RG) × LA1589 and Gold Ball Livingston (GBL) × Yellow Pear (YP). In RG, three recombinants were progeny tested (S1B Table) whereas another three recombinants were progeny tested from an GBL × YP F2 population [52] (S1C Table).
NILs in the cultivated background were developed from the fine mapping populations in the HG and RG populations and only differed at the locus of interest while all other parts of the genome were fixed. Resulting from initial backcrosses to the cultivated parent, 75% and 87.5% of the loci in these NILs were estimated to be fixed for the HG or RG parent, respectively. The HG NILs originated from BC1F7 11S62-2 plant (S4 Fig), with the introgression region of around 36kb. The RG NILs originated from BC2F5 12S114-5 plant in RG mapping population (S4B Fig), with the introgression region around 532kb. A second set of RG NILs were derived from the BC2F6 heterozygous plant 12S255-11, with the introgressed region of around 131kb.
The fw11.3 NILs in LA1589 background were developed by marker-assisted selection after several generations of backcrossing and selfing. Breeding scheme for NIL development is schematically shown in S4C Fig. The entire introgressed region at fw11.3 locus (73 kb) contained 11 annotated genes (ITAG2.4 tomato genome annotation release), including CSR. In all cases, the NILs were grown in a randomized plot design and therefore cultivated under the same conditions.
Association mapping
The accessions used for the association mapping were a Core Collection that included 93 S. pimpinellifolium, S. lycopersicum var cerasiforme and S. lycopersicum var lycopersicum accessions [25] (S7 Table). Association mapping was performed using three InDel markers, HP61, HP32 and HP31 (S6 Table) found in the 13 kb region spanning the locus in a population described by [25]. Association analysis was performed using MLM model of TASSEL2.1 software [53]. Population structure matrix Q and kinship matrix K were generated with STRUCTURE 2.2 [54] and SPAGeDi [55], respectively. Twenty EST-SSR markers distributed throughout the genome were used to generate Q and K matrix [25].
Plant transformation
Fosmid SL_FOS0119H09 (provided by Dr. J.J. Giovannoni, USDA-ARS, Ithaca NY) from tomato cultivar Heinz1706 spanned the fw11.3-D locus. To release the insert for transformation, the clone was digested with AvrII and SgrAI corresponding to the Solyc11g071940 coding region, 7,101-bp upstream region and 3,466-bp downstream region. The resulting 11,314 bp insert was cloned into the Agrobacterium transformation vector pHaoN, modified from pCAMBIA1300 (by adding the following selectable marker: Pnos-KAN-Tnos), linearized by digestion with XbaI and XmaI. The enzymers AvrII and XbaI, SgrAI and XmaI are two pairs of isocaudomers that generate compatible ends after digestion. The complementation construct pHORF2 was transformed into VIR347, carrying the wild type allele of fw11.3 similar to LA1589; and also in the HG NIL containing the LA1589 wild type allele (11S167 carrying fw11.3-WT, S4A Fig). Transformation was conducted at the Plant Transformation Core Research Facility at the University of Nebraska-Lincoln (Dr. T. Clemente) and independent transgenic lines were identified after Southern blot hybridizations of EcoRI and EcoRV-digested genomic DNA following standard procedures. For each transgenic family, 10 to 13 transgenic complementation plants and non-transgenic sib plants were identified from the T1 generation using marker assisted selection, transplanted in a random plot design, and evaluated under the same conditions. A total of four independent transgenic events (HF3, HF4, 9F, CF7) were evaluated in 2013 and 2014, and HF3 and 9F were replicated in both years. For the T1 transgenic lines from HF3-1 and HF4 grown in 2014, only a few fruit had matured at the time of harvest and 8 to 12 fruit per plant were weighted individually. Due to large variations among plants within same genotype, plants that carried the largest and smallest fruit of the same genotype were removed prior to statistical evaluations [26] (S2 Table). Average fruit weight per plant was calculated by divding the fruit number by total weight.
Cytological evaluations of the fw11.3 NILs and VIR347 transgenic complementation plants
Cell layer and mesocarp cell area were measured in pericarp of breaker-stage fruit of the fw11.3 NILs, and the VIR347 transgenic and non-transgenic sib plants. Two to three slices per fruit and two representative fruit per plant were evaluated. Transverse sections of the pericarp (approximately 1 mm thick and 1 cm long) cut from the equatorial region were stained by adding a drop of 0.5% Toluidine Blue in 0.1% Na2CO3 solution for 1 to 2 seconds. The samples were rinsed with water to prevent staining of the internal cell layers. The stained sections were photographed using the attached digital camera (SPOT RT KE, Diagnostic Instruments) on the Leica MZFLIII dissecting microscope (Leica Microsystems, Switzerland). The cell layers were counted four times in each section from the exocarp to endocarp avoiding the vascular bundles. Largest cell size was measured by tracing the six largests cells with ImageJ. The average cell size was measured by counting the cell number in an equal sized rectangle and divided by the rectangular area. Student t-test were used to compare the cell number and cell size difference between the two genotypes.
Ploidy analysis
Representative mature green fruit were used for ploidy analyses. Five slices of fresh pericarp tissue (1 mm thick slice of 0.5–1 cm2 area, avoiding septum tissue) from each fruit were chopped finely under 1.2 ml nuclei extraction buffer (100 mM Tris-Cl pH 7, 85 mM NaCl, 5 mM MgCl2, and 0.1% Triton X100), with a razor blade A 100 μm nylon mesh filter (Sysmex Partec GmbH, Görlitz, Germany) was used to filter the nuclei from the chopped tissue suspension (600 μL). Three μL of DAPI solution (0.2 mg/mL) was added to each sample prior to loading onto the BD LSRII flow cytometer (Biomedical Research Tower Facility, Columbus, OH) or the CyAn ADP flow cytometer (Beckman Coulter, Cytometry Shared Resource Laboratory, Athens GA) for fluorescence-activated cell sorting (FACS) analysis. 10,000 nuclei were counted using 405 nm laser excitation and blue emission filter 450/50, Only 3,000 nuclei were counted if gating became clogged (possibly due to debris interference). A 7 DPA fruit and mature leaves were used as internal control to calibrate nuclei content C-values for the RG NILs and LA1589 NILs, respectively. After manually adjusting the gating to exclude background noise, the histograms of different nuclei level (C-values) events were generated (S1 Fig). The percentage of each ploidy level from all nuclei counts was calculated. The lower C value nuclei were not evaluated as those peaks were not discernable above background noise. The percentage of each ploidy level was compared between fw11.3-D and fw11.3-WT. To calculate the Endoreduplication Index (EI), we modified the established formula by removing 2C and 4C ploidy levels and calculated EI as EI’ = [4C*1+ 8C*2+ 16C*3+ 32C*4+ 64C*5+ 128C*6+ 256C*7+ 512C*8] / [total counts from 4C to 512C] [56].
Ovary, fruit, seed, plant architecture, leaf structure, and source-sink relationship analyses
Ovaries at anthesis were collected for size measurement. The middle part of ovaries were infltrated and fixed with 3% glutaraldehyde, 2% paraformaldehyde in 0.1 M potassium phosphate buffer pH7.4 overnight, dehydrated with a graded ethanol series from 25% to 100%. Ovaries were cut transversely in the middle with sharp blade after critical point drying and before platinum coating. Samples were scanned and imaged recorded with Hitachi S-3500N scanning electron microscope (Hitachi Ltd, Japan) under high vacuum. Ovary sizes were measured with the images using ImageJ software. Total yield of ripe and green fruit were recorded separately by the weight and number of the fruit according to previously established protocols [57]. Fruit ripening was recorded with hand pollinated first two flowers of each inflorescence. Six to ten fruits per plant that set well were evaluated. The dates were recorded as each fruit turned to orange (30%-60% surface color change to orange) and red (>90% surface color change to red). Fruit quality was measured as the total soluble solid content (degree of Brix). A quarter of each 14 representative ripe fruits were blended together per plant. The homogenized juice was filtered with Kimwipes and Brix was measured by a pocket refractometer (ATAGO CO., LTD, Tokyo, Japan). Seeds were extracted from fully ripe fruit and soaked in 12.5% HCl, rinsed and air dried in the laboratory for 6 days on mesh screens with paper towels, weighted and counted. Inflorescence number and flower number per inflorescence per plant were counted with greenhouse grown plants under 20-20-20 with Calcium supplement or 14-7-14 fertilizer condition. Developing inflorescences and aborted flowers were also included. Plant architecture was measure as plant height, node number, side shoot number and total side shoot length at 55 and 66 days after sowing (DAS) in the greenhouse and 87 and 108 DAS or 42 and 63 days after transplanting (DAT) in the field. Leaf structure were measured with the mature leaves at 8th, 9th, and 10th nodes counting from cotyledon. Leaf weight, rachis length, petiole length, intercalary leaflet number, secondary leaflet number, tertiary leaflet number and terminal leaflet size (width and length) were measured. To test source-sink relationship, two fruits per inflorescence of a total of 7 inflorescences were kept and fruit weight from these plants was compared with control plants with no fruit removal. All phenotypic evaluations were performed with fw11.3 NILs under RG background with two replications except source-sink relationship experiment, each with 6 to 13 plants (S3 Table). Student t-tests were performed to compare each trait between fw11.3-D and fw11.3-WT NIL.
CSR protein sequence analysis and phylogeny tree building
To identify conserved domains in the CSR protein, the Conserved Domain Database (CDD, https://www.ncbi.nlm.nih.gov/cdd/) [58] was used. The FAF domain (Pfam accession PF11250) was identified in 14 proteins in tomato genome protein sequence ITAG2.4 release (SGN, http://solgenomics.net/tools/blast/; Solyc01g009260.1.1, Solyc01g009270.1.1, Solyc01g079740.2.1, Solyc01g098570.2.1, Solyc06g008990.1.1, Solyc06g054310.1.1, Solyc06g073940.2.1, Solyc06g074270.1.1, Solyc06g084280.1.1, Solyc09g065140.1.1, Solyc10g018270.1.1, Solyc11g068530.1.1, and Solyc11g071940.1.1 (the latter corresponds to CSR). Thirteen of the tomato proteins were used in this study, except Solyc04g072650.1.1, which appeared to be an outlier in the protein phylogeny. To ensure the accuracy of protein sequences, all encoded proteins were confirmed using the predicted mRNA sequences with the ExPASy translation tool [59]. Additional motif searches were conducted using the full length protein sequence in MEME 4.10.0 [60] using the settings of 6 different motifs with 6–50 motif width. Three predicted proteins (Solyc06g073940.2.1, Solyc01g009260.1.1, and Solyc01g009270.1.1) featured the most similar motif patterns and highest similarity with CSR (E-value ≤ 1e-80), and therefore these were defined as paralogs. The FAF domain was found in 10 Arabidopsis proteins (TAIR, http://www.arabidopsis.org/wublast/index2.jsp) and seven of them were used in this study. Of these, only one was considered the closest paralogs of CSR because of high MEME motif similarities. Four FAF domain-containing Arabidopsis proteins corresponded to the FANTASTIC FOUR (FAF) clade.
Multiple sequence alignments of FAF by CLUSTALW2 [61] with default settings were performed and the results were exported to MEGA6 [62] for phylogenetic analysis. Neighbor-joining tree was constructed for 13 tomato and seven Arabidopsis FAF domain sequences with 1000 replicates for bootstrap validation. A FAF domain from Selaginella moellendorffii (Phytozome ID: 418746) was used as outgroup. Two proteins (Solyc01g079740.2.1 and Solyc06g054310.1.1) were closely related to FAF3 and FAF4 and were renamed SlFAF3/4a and SlFAF3/4b. Three proteins (Solyc06g084280.1.1, Solyc06g008990.1.1 and Solyc09g065140.1.1) were also closely related and presented the FAF1 and FAF2 subclade, and were renamed to SlFAF1/2a, SlFAF1/2b, and SlFAF1/2c, respectively.
To identify potential orthologs in other crop species, the full length protein sequence of CSR-WT and its three paralogs were used as a query using the SGN database (http://solgenomics.net/tools/blast/), Cucurbit Genomics Database (http://www.icugi.org/cgi-bin/ICuGI/tool/blast.cgi), Phytozome version 9.1 (http://www.phytozome.net/) and Genome Database for Rosaceae (GDR; http://www.rosaceae.org/) and paralogs were identified in potato, eggplant, pepper, cocoa, sesame, mimulus, watermelon, cucumber, grape, poplar, peach, and strawberry (E-value ≤ 1e-80). MEME analysis was conducted to identify the most likely orthologs in each species by selecting those with the most similar motif patterning proteins. Multiple sequence alignment and phylogeny tree construction were performed as the same method described above.
Fruit tissue collection for expression analysis
Three to thirty fruit from 10 fw11.3-WT and 10 fw11.3-D NIL plants each were collected between 1:00 pm to 3:00 pm and dissected tissues were immediately frozen in liquid nitrogen. The three tissues dissected were pericarp, columella, and developing seeds with placenta at the following developmental stages: 4, 7, 10, 15, 25, 33 DPA and turning stage fruit. For 4 DPA fruit, the columella, placenta and developing seeds were collected together. Most samples consisted of four replicates, except 7 DPA and 25 DPA (three replicates), 33 DPA (one replicate) and turning stage (two replicates). The low number of replicates for the latter tissues was due to severe incidence of blossom-end rot in the greenhouse.
RNA isolation, RNA-seq library preparation and sequencing
Hot borate RNA extraction method [63] was used for total RNA extraction from 25, 33 DPA and turning stage fruit. The TRIzol (Invitrogen, Carlsbad, CA) RNA extraction method was used for 4, 7, 10 and 15 DPA fruit following the manufacturer’s specifications. RNA quantity and quality were assessed using a Qubit 2.0 fluorometer RNA Assay Kit (Invitrogen Inc. USA) and an Agilent 2100 Bioanalyzer RNA 6000 Nano kit (Agilent, USA). Approximately 5μg total RNA was used to prepare strand-specific libraries of approximately 250bp fragments [64, 65]. Libraries were barcoded and 8 samples were pooled per lane on the flowcell. Fifty-one bp single-end reads were generated on the Illumina HiSeq2000 at Genomics Resources Core Facility at Weill Medical College (New York, NY).
Read alignments and library quality checks
The pre-processing, read alignment and quantification of gene levels were performed using previously established protocols in the lab [64] with minor modifications. Briefly, ribosomal RNA-free reads were mapped to the Solanum lycopersicum reference genome (Build SL2.50) using tomato gene model annotation (ITAG2.4 release) to facilitate mapping reads across exon-exon junctions. The final expression data were shown as reads per kilobase of exon model per million mapped reads (RPKM). For the expression of selected genes at different developmental stages, the average RPKM were used. Summary statistics for each of the RNA-seq libraries are shown in S8 Table.
The correlations among samples were evaluated because the results would address reproducibility among samples (S9 Table). The columella tissue at the turning stage (TCol) showed low correlation between the two replicates (64% for fw11.3-WT and 85% for fw11.3-D, respectively). After ruling out the possibility of fw11.3-D or fw11.3-WT sample switch by evaluating the SNPs at fw11.3, we found that unlike TCol_rep1, TCol_rep2 was more correlated with other turning stage tissue types (pericarp and seed/placenta) than TCol_rep1. This suggested mixed up tissue samples for TCol_rep2. We therefore decided to discard TCol_rep2 and only use TCol_rep1 to represent turning stage columella tissue.
Co-expression clustering and GO term enrichment
The gene expression data used for co-expression cluster analysis was the gene expression RPKM values of three fruit tissue types at seven developmental stages (six stages for columella tissue). The identification of co-expressed genes with CSR was done separately with CSR-D and CSR-WT alleles. The data was first pre-processed by Mfuzz [66] for normalization. The heatmap.2 function in R was then employed to generate a heatmap based on the normalized RPKM. Twelve clusters for CSR-D and 10 clusters for CSR-WT were identified visually based on the heatmap results. Fuzzy C-means clustering in Mfuzz was applied with cluster number as 12 and 10 (CSR-D and CSR-WT, respectively) and default settings. Soft clustering was chosen in “visualization” to generate clusters. Finally, the clustering results with the probabilities of each gene in each cluster were exported to Excel. Genes with the probabilities below 90% were removed from the clusters.
Arabidopsis ortholog genes were obtained for CSR-D and CSR-WT co-expressed overlapping genes by using BLASTP against TAIR10 amino acid sequence (p-value ≤ 7.00E-06). Cytoscape plug-in ClueGO (Version 2.3.2) [67] was used to perform the Gene Ontology (GO) analysis using the GO biological process available on November 17 2016. The ClueGO networks were set to ‘medium’ and their connectivity was based on a kappa score of 0.4. GO Term grouping was selected with an initial group size of 1 and group merging set at 50%. Two-sided hypergeometric tests were applied and p-value correction was carried out using the Bonferroni step-down method. GO terms with adjusted p ≤ 0.05 were considered as significant.
Accession numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession number SRP017242, SRP089936, SRP089970.
Data records
The raw FASTQ files for the RNA-seq libraries were deposited at NCBI Sequence Read Archive (SRA) with SRA study accession SRP089936. Gene expression data (RPKM) are available through a Tomato Functional Genomic Database (TFGD; http://ted.bti.cornell.edu/cgi-bin/TFGD/digital/home.cgi).
Supporting information
The single nuclei gating (upper) and nuclei ploidy histogram (lower) is shown for each genotype. In the upper graphs, the P1 box was manually set prior to counting the single nuclei that showed different ploidy levels (non-red dots). Colors were automatically generated by the flow cytometry BD FACSDIVATM software, and correspond to P2 to P9 nuclei. Red color represent debris. P2 represents 2C nuclei and was difficult to discern among the debris. P3 to P10 represent polyploid nuclei ranging from 4C to 512C shown on the X-axis. The widths of each nuclei level were manually adjusted. The horizontal bars in the histogram showed the area used for the nuclei counts.
(PDF)
(A) Expression of CSR in developing fruit tissues from anthesis to ripe fruit. (B) Expression of CSR in developing pericarp tissues after fruit set.
(PDF)
(A) fw11.3-D NIL gene expression cluster analysis. CSR-D and its co-expressed genes are included in cluster 11. (B) fw11.3-WT NIL gene expresssion cluster analysis. CSR-WT and its co-expressed genes are included in cluster 1. Normalized RPKM were used. Horizontal axis represents the following tissues and stages: 7Col (1), 10Col, 15Col, 25Col (4), 33Col, TCol, 4Per (7), 7Per, 10Per, 15Per, 25Per (11), 33Per, TPer, 4S, 7SPl, 10SPl (16), 15SPl, 25SPl, 33SPl, TSPl. (Col: columella; Per: pericarp; SPl: seeds and placenta. Numbers in parenthesis are show in the figure as x-axis labels).
(PDF)
(A) Pedigree of plants used for progeny test, fine-mapping and genetic transformation in Howard German background. PP: fw11.3-WT NIL. (B) Pedigree of plants used for fine-mapping and phenotypic evaluations in Rio Grande background. (C) Pedigree of LA1589 fruit weight NILs.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
Dr. Tea Meulia at the Molecular and Cellular Imaging Center OSU, Wooster for the microscopy analyses. Dr. Joss Rose from Cornell University for sharing CSR expression data prior to public release. Dr. Christian Chevalier from INRA, Bordeaux in France for sharing insightful discussions about endoreduplication in tomato. Bruce Williams, Horticulture and Crop Science at OSU, for field preparation and care; Meghan Fisher, Shin-Ruei Lee, Jiheun Cho and Kacey Jones for greenhouse care.
Data Availability
The raw FASTQ files for the RNA-seq libraries were deposited at NCBI Sequence Read Archive (SRA) with SRA study accession SRP089936. Gene expression data (RPKM) are available through a Tomato Functional Genomic Database (TFGD; http://ted.bti.cornell.edu/cgi-bin/TFGD/digital/home.cgi).
Funding Statement
This research was funded by the National Science Foundation IOS 0922661 and IOS 1564366 to EvdK. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Paran I. and van der Knaap E. Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. J Exp Bot. 2007; 58:3841–3852. doi: 10.1093/jxb/erm257 [DOI] [PubMed] [Google Scholar]
- 2.Purugganan M.D. and Fuller D.Q. The nature of selection during plant domestication. Nature. 2009; 457:843–848. doi: 10.1038/nature07895 [DOI] [PubMed] [Google Scholar]
- 3.Blanca J., Canizares J., Cordero L., Pascual L., Diez M.J., et al. Variation revealed by SNP genotyping and morphology provides insight into the origin of the tomato. PLoS One. 2012; 7:e48198 doi: 10.1371/journal.pone.0048198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanca J., Montero-Pau J., Sauvage C., Bauchet G., Illa E., et al. Genomic variation in tomato, from wild ancestors to contemporary accessions. BMC Genomics. 2015; 16:257 doi: 10.1186/s12864-015-1444-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanksley S.D. The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell. 2004; 16 Suppl:S181–189. doi: 10.1105/tpc.018119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welty N., Radovich C., Meulia T. and van der Knaap E. Inflorescence development in two tomato species. Can J Bot. 2007; 85:111–118. [Google Scholar]
- 7.Xiao H., Radovich C., Welty N., Hsu J., Li D., et al. Integration of tomato reproductive developmental landmarks and expression profiles, and the effect of SUN on fruit shape. BMC Plant Biol. 2009; 9:49 doi: 10.1186/1471-2229-9-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillaspy G., Ben-David H. and Gruissem W. Fruits: A Developmental Perspective. Plant Cell. 1993; 5:1439–1451. doi: 10.1105/tpc.5.10.1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez G.R., Munos S., Anderson C., Sim S.C., Michel A., et al. Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 2011; 156:275–285. doi: 10.1104/pp.110.167577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu C., Liberatore K.L., MacAlister C.A., Huang Z., Chu Y.H., et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat Genet. 2015; 47:784–792. doi: 10.1038/ng.3309 [DOI] [PubMed] [Google Scholar]
- 11.Van der Knaap E., Chakrabarti M., Chu Y.H., Clevenger J.P., Illa-Berenguer E., et al. What lies beyond the eye: the molecular mechanisms regulating tomato fruit weight and shape. Front Plant Sci 2014; 5:227 doi: 10.3389/fpls.2014.00227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakrabarti M., Zhang N., Sauvage C., Munos S., Blanca J., et al. A cytochrome P450 CYP78A regulates a domestication trait in tomato (Solanum lycopersicum). Proc Natl Acad Sci U S A. 2013; 110:17125–17130. doi: 10.1073/pnas.1307313110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frary A., Nesbitt T.C., Grandillo S., van der Knaap E., Cong B., et al. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science. 2000; 289:85–88. [DOI] [PubMed] [Google Scholar]
- 14.Lin T., Zhu G., Zhang J., Xu X., Yu Q., et al. Genomic analyses provide insights into the history of tomato breeding. Nat Genet. 2014; 46:1220–1226. doi: 10.1038/ng.3117 [DOI] [PubMed] [Google Scholar]
- 15.Cheniclet C., Rong W.Y., Causse M., Frangne N., Bolling L., et al. Cell expansion and endoreduplication show a large genetic variability in pericarp and contribute strongly to tomato fruit growth. Plant Physiol. 2005; 139:1984–1994. doi: 10.1104/pp.105.068767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dello Ioio R., Linhares F.S., Scacchi E., Casamitjana-Martinez E., Heidstra R., et al. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol. 2007; 17:678–682. doi: 10.1016/j.cub.2007.02.047 [DOI] [PubMed] [Google Scholar]
- 17.Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., et al. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008; 322:1380–1384. doi: 10.1126/science.1164147 [DOI] [PubMed] [Google Scholar]
- 18.Ishida T., Adachi S., Yoshimura M., Shimizu K., Umeda M., et al. Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development. 2010; 137:63–71. doi: 10.1242/dev.035840 [DOI] [PubMed] [Google Scholar]
- 19.Takahashi N., Kajihara T., Okamura C., Kim Y., Katagiri Y., et al. Cytokinins control endocycle onset by promoting the expression of an APC/C activator in Arabidopsis roots. Curr Biol. 2013; 23:1812–7. doi: 10.1016/j.cub.2013.07.051 [DOI] [PubMed] [Google Scholar]
- 20.Breuer C., Braidwood L. and Sugimoto K. Endocycling in the path of plant development. Curr Opin Plant Biol. 2014; 17:78–85. doi: 10.1016/j.pbi.2013.11.007 [DOI] [PubMed] [Google Scholar]
- 21.Edgar B.A., Zielke N. and Gutierrez C. Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nat Rev Mol Cell Biol. 2014; 15:197–210. doi: 10.1038/nrm3756 [DOI] [PubMed] [Google Scholar]
- 22.Chevalier C. Cell cycle control and fruit development, in Cell cycle control and plant development. Inze D., Editor. 2007; Blackwell Publishing: Oxford: p. 269–293. [Google Scholar]
- 23.Van der Knaap E. and Tanksley S.D. The making of a bell pepper-shaped tomato fruit: identification of loci controlling fruit morphology in Yellow Stuffer tomato. Theor Appl Genet. 2003; 107:139–147. doi: 10.1007/s00122-003-1224-1 [DOI] [PubMed] [Google Scholar]
- 24.Huang Z. and van der Knaap E. Tomato fruit weight 11.3 maps close to fasciated on the bottom of chromosome 11. Theor Appl Genet. 2011; 123:465–474. doi: 10.1007/s00122-011-1599-3 [DOI] [PubMed] [Google Scholar]
- 25.Ranc N., Munos S., Santoni S. and Causse M. A clarified position for Solanum lycopersicum var. cerasiforme in the evolutionary history of tomatoes (solanaceae). BMC Plant Biol. 2008; 8:130 doi: 10.1186/1471-2229-8-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mu, Q. The cloning and cellular basis of a novel tomato fruit weight gene: cell size regulator (FW11.3/CSR). 2015; Masters Thesis in Horticulture and Crop Science. Ohio State University: Columbus Ohio.
- 27.Wahl V., Brand L.H., Guo Y.L. and Schmid M. The FANTASTIC FOUR proteins influence shoot meristem size in Arabidopsis thaliana. BMC Plant Biol. 2010; 10:285 doi: 10.1186/1471-2229-10-285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allocco D.J., Kohane I.S. and Butte A.J. Quantifying the relationship between co-expression, co-regulation and gene function. BMC Bioinformatics. 2004; 5:18 doi: 10.1186/1471-2105-5-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kakimoto T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP isopentenyltransferases. Plant Cell Physiol. 2001; 42:677–685. [DOI] [PubMed] [Google Scholar]
- 30.Yamada H., Suzuki T., Terada K., Takei K., Ishikawa K., et al. The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 2001; 42:1017–1023. [DOI] [PubMed] [Google Scholar]
- 31.Nemeth K., Salchert K., Putnoky P., Bhalerao R., Koncz-Kalman Z., et al. Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 1998; 12:3059–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbez E., Kubes M., Rolcik J., Beziat C., Pencik A., et al. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature. 2012; 485:119–122. doi: 10.1038/nature11001 [DOI] [PubMed] [Google Scholar]
- 33.Kaneda M., Schuetz M., Lin B.S., Chanis C., Hamberger B., et al. ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. J Exp Bot. 2011; 62:2063–2077. doi: 10.1093/jxb/erq416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou H., Erickson J., Meservy J. and Schultz E.A. FORKED1 encodes a PH domain protein that is required for PIN1 localization in developing leaf veins. Plant J. 2010; 63:960–973. doi: 10.1111/j.1365-313X.2010.04291.x [DOI] [PubMed] [Google Scholar]
- 35.Schena M., Lloyd A.M. and Davis R.W. The HAT4 gene of Arabidopsis encodes a developmental regulator. Genes Dev. 1993; 7:367–379. [DOI] [PubMed] [Google Scholar]
- 36.Steindler C., Matteucci A., Sessa G., Weimar T., Ohgishi M., et al. Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development. 1999; 126:4235–4245. [DOI] [PubMed] [Google Scholar]
- 37.Johnson K.L., Ramm S., Kappel C., Ward S., Leyser O., et al. The Tinkerbell (Tink) Mutation Identifies the Dual-Specificity MAPK Phosphatase INDOLE-3-BUTYRIC ACID-RESPONSE5 (IBR5) as a Novel Regulator of Organ Size in Arabidopsis. PLoS One. 2015; 10:e0131103 doi: 10.1371/journal.pone.0131103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chuck G., Lincoln C. and Hake S. KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell. 1996; 8:1277–1289. doi: 10.1105/tpc.8.8.1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez-Perez J.M., Candela H., Robles P., Lopez-Torrejon G., del Pozo J.C., et al. A role for AUXIN RESISTANT3 in the coordination of leaf growth. Plant Cell Physiol. 2010; 51:1661–1673. doi: 10.1093/pcp/pcq123 [DOI] [PubMed] [Google Scholar]
- 40.Bryan A.C., Obaidi A., Wierzba M. and Tax F.E. XYLEM INTERMIXED WITH PHLOEM1, a leucine-rich repeat receptor-like kinase required for stem growth and vascular development in Arabidopsis thaliana. Planta. 2012; 235:111–122. doi: 10.1007/s00425-011-1489-6 [DOI] [PubMed] [Google Scholar]
- 41.Clay N.K. and Nelson T. VH1, a provascular cell-specific receptor kinase that influences leaf cell patterns in Arabidopsis. Plant Cell. 2002; 14:2707–2722. doi: 10.1105/tpc.005884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cano-Delgado A., Yin Y., Yu C., Vafeados D., Mora-Garcia S., et al. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development. 2004; 131:5341–5351. doi: 10.1242/dev.01403 [DOI] [PubMed] [Google Scholar]
- 43.Ceserani T., Trofka A., Gandotra N. and Nelson T. VH1/BRL2 receptor-like kinase interacts with vascular-specific adaptor proteins VIT and VIK to influence leaf venation. Plant J. 2009; 57:1000–1014. doi: 10.1111/j.1365-313X.2008.03742.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Rybel B., Mahonen A.P., Helariutta Y. and Weijers D. Plant vascular development: from early specification to differentiation. Nat Rev Mol Cell Biol. 2016; 17:30–40. doi: 10.1038/nrm.2015.6 [DOI] [PubMed] [Google Scholar]
- 45.Lee H.O., Davidson J.M. and Duronio R.J. Endoreplication: polyploidy with purpose. Genes Dev. 2009; 23:2461–2477. doi: 10.1101/gad.1829209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Veylder L., Larkin J.C. and Schnittger A. Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 2011; 16:624–634. doi: 10.1016/j.tplants.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 47.Bao Z. and Hua J. Interaction of CPR5 with cell cycle regulators UVI4 and OSD1 in Arabidopsis. PLoS One. 2014; 9:e100347 doi: 10.1371/journal.pone.0100347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inze D. and De Veylder L. Cell cycle regulation in plant development. Annu Rev Genet. 2006; 40:77–105. doi: 10.1146/annurev.genet.40.110405.090431 [DOI] [PubMed] [Google Scholar]
- 49.Marrocco K., Bergdoll M., Achard P., Criqui M.C. and Genschik P. Selective proteolysis sets the tempo of the cell cycle. Curr Opin Plant Biol. 2010; 13:631–639. doi: 10.1016/j.pbi.2010.07.004 [DOI] [PubMed] [Google Scholar]
- 50.Sim S.C., Van Deynze A., Stoffel K., Douches D.S., Zarka D., et al. High-density SNP genotyping of tomato (Solanum lycopersicum L.) reveals patterns of genetic variation due to breeding. PLoS One. 2012; 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonzalo M.J. and van der Knaap E. A comparative analysis into the genetic bases of morphology in tomato varieties exhibiting elongated fruit shape. Theor Appl Genet 2008; 116:647–656. doi: 10.1007/s00122-007-0698-7 [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez G.R., Kim H.J. and van der Knaap E. Mapping of two suppressors of OVATE (sov) loci in tomato. Heredity (Edinb). 2013; 111:256–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bradbury P.J., Zhang Z., Kroon D.E., Casstevens T.M., Ramdoss Y., et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007; 23:2633–2635. doi: 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- 54.Falush D., Stephens M. and Pritchard J.K. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes. 2007; 7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hardy O.J. and Vekemans X. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes. 2002; 2:618–620. [Google Scholar]
- 56.Barow M. and Meister A. Lack of correlation between AT frequency and genome size in higher plants and the effect of nonrandomness of base sequences on dye binding. Cytometry. 2002; 47:1–7. [DOI] [PubMed] [Google Scholar]
- 57.Nesbitt T.C. and Tanksley S.D. fw2.2 directly affects the size of developing tomato fruit, with secondary effects on fruit number and photosynthate distribution. Plant Physiol. 2001; 127:575–583. [PMC free article] [PubMed] [Google Scholar]
- 58.Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., et al. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015; 43:D222–6. doi: 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003; 31:3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bailey T.L., Williams N., Misleh C. and Li W.W. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006; 34:W369–73. doi: 10.1093/nar/gkl198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23:2947–2948. doi: 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 62.Tamura K., Stecher G., Peterson D., Filipski A. and Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013; 30:2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pang M., Stewart J.M. and Zhang J. A mini-scale hot borate method for the isolation of total RNA from a large number of cotton tissue samples African J Biotech. 2011; 10:15430–15437. [Google Scholar]
- 64.Huang Z., Van Houten J., Gonzalez G., Xiao H. and van der Knaap E. Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol Genet Genomics. 2013; 288:111–129. doi: 10.1007/s00438-013-0733-0 [DOI] [PubMed] [Google Scholar]
- 65.Zhong S., Joung J.-G., Zheng Y., Chen Y.-r., Liu B., et al. High-Throughput Illumina Strand-Specific RNA Sequencing Library Preparation. Cold Spring Harbor Protocols. 2011; 2011:940–949. doi: 10.1101/pdb.prot5652 [DOI] [PubMed] [Google Scholar]
- 66.Kumar L. and EM F. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007; 2:5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009; 25:1091–1093. doi: 10.1093/bioinformatics/btp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The single nuclei gating (upper) and nuclei ploidy histogram (lower) is shown for each genotype. In the upper graphs, the P1 box was manually set prior to counting the single nuclei that showed different ploidy levels (non-red dots). Colors were automatically generated by the flow cytometry BD FACSDIVATM software, and correspond to P2 to P9 nuclei. Red color represent debris. P2 represents 2C nuclei and was difficult to discern among the debris. P3 to P10 represent polyploid nuclei ranging from 4C to 512C shown on the X-axis. The widths of each nuclei level were manually adjusted. The horizontal bars in the histogram showed the area used for the nuclei counts.
(PDF)
(A) Expression of CSR in developing fruit tissues from anthesis to ripe fruit. (B) Expression of CSR in developing pericarp tissues after fruit set.
(PDF)
(A) fw11.3-D NIL gene expression cluster analysis. CSR-D and its co-expressed genes are included in cluster 11. (B) fw11.3-WT NIL gene expresssion cluster analysis. CSR-WT and its co-expressed genes are included in cluster 1. Normalized RPKM were used. Horizontal axis represents the following tissues and stages: 7Col (1), 10Col, 15Col, 25Col (4), 33Col, TCol, 4Per (7), 7Per, 10Per, 15Per, 25Per (11), 33Per, TPer, 4S, 7SPl, 10SPl (16), 15SPl, 25SPl, 33SPl, TSPl. (Col: columella; Per: pericarp; SPl: seeds and placenta. Numbers in parenthesis are show in the figure as x-axis labels).
(PDF)
(A) Pedigree of plants used for progeny test, fine-mapping and genetic transformation in Howard German background. PP: fw11.3-WT NIL. (B) Pedigree of plants used for fine-mapping and phenotypic evaluations in Rio Grande background. (C) Pedigree of LA1589 fruit weight NILs.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The raw FASTQ files for the RNA-seq libraries were deposited at NCBI Sequence Read Archive (SRA) with SRA study accession SRP089936. Gene expression data (RPKM) are available through a Tomato Functional Genomic Database (TFGD; http://ted.bti.cornell.edu/cgi-bin/TFGD/digital/home.cgi).