Abstract
Diffusion tensor imaging (DTI) is used extensively in neuroscience to noninvasively estimate white matter (WM) microarchitecture. However, the diffusion signal is inherently ambiguous because it infers WM structure from the orientation of water diffusion and cannot identify the biological sources of diffusion changes. To compare inferred WM estimates to directly labeled axonal elements, we performed a novel within-subjects combination of high-resolution ex vivo DTI with two-photon laser microscopy of intact mouse brains rendered optically transparent by Clear Lipid-exchanged, Anatomically Rigid, Imaging/immunostaining compatible, Tissue hYdrogel (CLARITY). We found that myelin basic protein (MBP) immunofluorescence significantly correlated with fractional anisotropy (FA), especially in WM regions with coherent fiber orientations and low fiber dispersion. Our results provide evidence that FA is particularly sensitive to myelination in WM regions with these characteristics. Furthermore, we found that radial diffusivity (RD) was only sensitive to myelination in a subset of WM tracts, suggesting that the association of RD with myelin should be used cautiously. This combined DTI-CLARITY approach illustrates, for the first time, a framework for using brain-wide immuno-labeling of WM targets to elucidate the relationship between the diffusion signal and its biological underpinnings. This study also demonstrates the feasibility of a within-subject combination of noninvasive neuroimaging and tissue clearing techniques that has broader implications for neuroscience research.
Keywords: Fractional anisotropy, Tissue clearing, Whole brain immunolabeling, Myelin basic protein, Neuroimaging, Multimodal imaging
1. Introduction
Diffusion-weighted MRI is the only noninvasive method available for mapping fiber architecture of tissue in vivo (Le Bihan et al., 1986; Basser et al., 1994) and it is used frequently in both research and clinical settings to assess brain white matter (WM). Diffusion tensor imaging (DTI) is the most commonly employed diffusion model and utilizes the magnitude and orientation of anisotropic water diffusion to estimate the WM structure of brain tissue, thereby producing maps of macroscopic axonal organization in the brain (Basser and Pierpaoli, 1996; Le Bihan et al., 2001; Mori and Zhang, 2006). While DTI has provided extraordinary insights into the WM structure of the brain in both health and disease, the interpretation of diffusion data is often hampered by several important limitations and challenges (Beaulieu, 2002; Jones and Cercignani, 2010). Primary among these limitations is a lack of understanding of the actual biological sources underlying widely used DTI-derived measures.
DTI produces three scalar indices, or diffusivities (λ1, λ2, λ3) that describe the orientation-dependence of water diffusion within individual imaging voxels. These are used to calculate diffusion along the long axis of fibers, termed axial diffusivity (AD or λ ║), diffusion perpendicular to the long axis termed radial diffusivity (RD or λ⊥), and mean diffusivity (MD) or the apparent diffusion coefficient (ADC), which is an average of the three diffusivities. A summary measure that reflects the orientation-dependence of diffusion is called fractional anisotropy (FA), which is significantly higher in brain WM due to the highly restrictive nature of coherent axonal bundles for water diffusion. FA is the most widely used invariant measure of anisotropy and is often used to assess so-called WM “integrity” (Jones et al., 2013). While FA is highly sensitive to microstructural changes, it is not specific to the types of change (e.g. AD or RD changes) and neither FA nor the diffusion tensor can unambiguously identify the underlying cause of anisotropy differences. This has led to considerable debate, since the inception of DTI, over the biological significance of these DTI-derived measures since WM integrity can be affected by many different parameters including axon packing density, axon caliber, myelination, microglia, inflammation, and tissue architecture (Mori and Zhang, 2006; Le Bihan and Johansen-Berg, 2012; Jones et al., 2013). The contribution of each of these parameters cannot be resolved using DTI alone, thus further investigation using other complementary multimodal imaging techniques is imperative.
In the field of light microscopy, there have been several recent technical advances in tissue clearing that enable the molecular examination of entirely intact mouse brains without the need for physical tissue sectioning (Kim et al., 2013; Richardson and Lichtman, 2015). These intact-brain analyses are a vast improvement over traditional slide-based immunohistochemistry that involves sectioning and reconstructing tissue, which is time consuming, error-prone, and does not maintain the structural integrity of neural circuits (Jennings and Stuber, 2014). Clear Lipid-exchanged, Anatomically Rigid, Imaging/immunostaining compatible, Tissue hYdrogel (CLARITY; Chung et al., 2013) is a clearing technique that transforms intact brain tissue into nanoporous, lipid-free, hydrogel-tissue hydrids that are optically transparent but macromolecule permeable. CLARITY achieves tissue transparency to enable deep laser penetration in normally highly light-scattering mouse brains, while preserving proteins in situ and the native 3D structure of brain tissue. When coupled with two-photon or light-sheet laser microscopy, whole mouse brains can be imaged entirely intact with sub-cellular resolution (Tomer et al., 2014). This intact analysis approach can be especially beneficial for studying WM tracts within the brain, which have complex 3D architectures spanning multiple biological scales.
Therefore, to directly examine the contribution of myelination in DTI-derived metrics, we used a within-sample design combining ex vivo DTI at ultra high field (11.7 T) with CLARITY whole-brain immunolabeling of myelin basic protein (MBP; Baumann and Pham-Dinh, 2001; Boggs, 2006). We found that CLARITY MBP immunolabeling produced 3D whole-brain maps of myelinated WM structures and that MBP expression within these structures significantly correlated with DTI-derived FA measures. Moreover, when examining a subset of WM tracts, we found that the FA-MBP correlation was most robust specifically in WM regions known to have higher myelination and high fiber coherence, as measured by geometric diffusion tensor indices. Therefore, a brain-wide analysis of MBP indicates that FA is sensitive to axon myelination and that this multimodal approach incorporating 3D CLARITY analysis can be potentially used to resolve other molecular factors that contribute to the diffusion signal.
2. Materials and methods
2.1. Subjects
For initial pilot experiments to test the compatibility of performing the CLARITY protocol following ex vivo DTI in the same brains, we used three male thy1-eYFP-H mice (Jackson Laboratories, Bar Harbor ME), 3–6 months old (Chang et al., in press). For the main experiments, we used four male C57BL/6 J mice (Jackson Laboratories), 3–6 months old. Mice were housed under 12 h light/dark cycle with ad libitum access to food and water. All animal procedures were approved by the Feinstein Institute Medical Research Institutional Animal Care and Use Committee and maintained according to National Institutes of Health guidelines.
2.2. ex vivo DTI
Mice were transcardially perfused with a customized CLARITY liquid hydrogel (see Sec 2.3 for details) that also included 0.1 mM gadopentetate dimeglumine (Gd-DTPA; Magnevist). Brains were then removed and incubated in 4% PFA-liquid hydrogel for 3 days at 4 °C on a laboratory rocker. Prior to DTI scanning, the brains were immersed in a PBS-Magnevist solution for 48 h. Magnevist was used as a T1-shortening contrast agent for MRI to achieve shorter repetition times (TR), thereby allowing faster DTI acquisitions while maintaining a good signal-to-noise ratio (Aggarwal et al., 2010; Jiang and Johnson, 2011). For MRI, the brains were placed in 15 mm NMR glass tubes, which were filled with Fomblin® oil (Solvay Solexis, Thorofare, NJ), an MR-invisible liquid for susceptibility matching and limiting tissue dehydration. Diffusion-weighted images were acquired on an 11.7 T NMR spectrometer (Bruker BioSpin, Billerica, MA) using a three-dimensional gradient-and-spin-echo (3D DW-GRASE) sequence with twin navigator-echo phase correction (Aggarwal et al., 2010), along 15 independent directions and a b-value of 1500 s/mm2. Total scanning time for each specimen was approximately 16.5 h. The temperature of the specimens was maintained at 28 °C during imaging in order to avoid polymerization of the CLARITY hydrogel.
DTI parameters were as follows: effective echo time (TE) of 31 ms, pulse repetition time (TR) of 800 ms, 4 signal averages with phase cycling, diffusion gradient duration/separation of 3/12 ms, and receiver bandwidth of 100 kHz. Typical imaging field of view and matrix size were 12.8×8.0×16.3 mm and 128×80×162, respectively. The native resolution was 100 μm isotropic. Images were reconstructed using IDL6.4 (ITT Visual Information Solutions, Boulder, CO) with zero-padding of the k-space data by a factor of 2. From the diffusion-weighted images, maps of FA, AD, RD, MD, Cl, and Cp were calculated using FSL (http://www.fmrib.ox.ac.uk/fsl) software as follows: AD=λ1, RD=(λ2+λ3)/2, MD=(λ1+λ2+λ3)/3, Cl=(λ1 - λ2)/ λ1, Cp = (λ2 - λ3)/λ1, and . TrackVis (http://www.trackvis.org) was used for post hoc calculations and visualization. DTI regions of interest (ROIs) were created in TrackVis with manually drawn ROIs using the Free Hand tool by E.H.C.
2.3. CLARITY whole-brain clearing and imaging
We performed the CLARITY procedure as previously described (Chung et al., 2013; Tomer et al., 2014; Yang et al., 2014) with custom modifications in order to maximize antibody penetration and minimize tissue expansion . We used a hydrogel containing 4% paraformaldehyde (PFA), 1.75% acrylamide, 0.01875% bis-acrylamide, and 0.25% VA-044 initiator. Prior to DTI, we perfused brains with this custom hydrogel solution, then performed the ex vivo DTI scanning. Following DTI, brains were then polymerized at 37 °C for 3 h to form the brain-hydrogel hybrid. There was only one perfusion and one CLARITY hydrogel used in this study (Fig. 1A). Tissue clearing was achieved with passive CLARITY (PACT; Tomer et al., 2014; Yang et al., 2014) until brains were optically transparent (30–40 days). While this passive technique is markedly slower, we found that it achieved excellent tissue transparency and structural preservation without tissue inflation. Transmittance through cleared whole brains was measured at three laser wavelengths (Chang et al., in press) using a fluorescence spectrometer (Public Lab, Cambridge, MA). Once transparent, the brains were washed in 1× PBS+0.1% Triton X for 2 days, changing the solution every 24 h. Brains were then incubated in primary antibody solution, anti-myelin basic protein (1:50, EMD Millipore), 0.5 M sodium borate solution, (pH 8.5) and 0.1% Triton-X (wt/vol) for 14 days at 37 °C on an orbital shaker. Brains were then washed for 7 days in 0.5 M sodium borate with 0.1% Triton-X on a shaker at 37 °C, followed by secondary antibody labeling with Alexa Fluor 633 (1:50, Life technologies) in 0.5 M sodium borate + 0.1% Triton-X for another 14 days at 37 °C on an orbital shaker. The brains were placed in a final wash for 7 days following secondary labeling. Prior to two-photon laser scanning microscopy (2P-LSM) imaging, brains were immersed in refractive index matching solution (RIMS; Yang et al., 2014) for 3 days at RT and sealed with Molykote 111 compound (Dow Corning) in a custom imaging chamber (iSpacer, Jun Lab) protected from light.
Fig. 1.
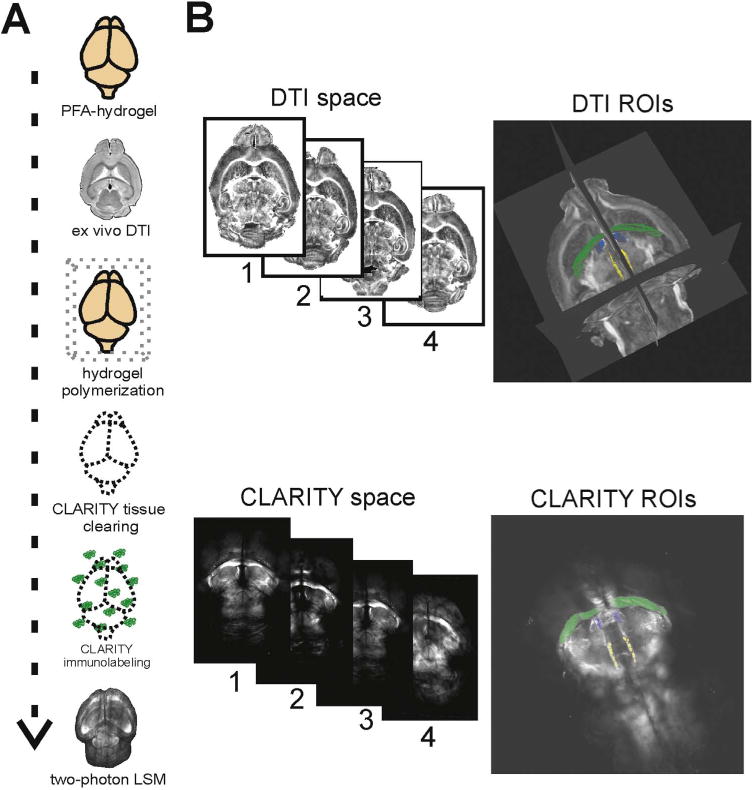
Brain-wide analyses combining ex vivo DTI and CLARITY. (A) Schematic of experimental design for DTI-CLARITY multimodal approach. (B) DTI images were registered to a single reference brain within DTI space. CLARITY MBP images were registered to the same reference brain within CLARITY space. WM ROIs were manually delineated in DTI FA space (TrackVis) and CLARITY MBP space (Imaris). Our procedure yielded corresponding WM ROIs that correlated in a near linear relationship between volume comparisons (Fig. 3B). Displayed ROI volumes are: hippocampal commissure (green), stria medullaris (blue), and fasciculus retroflexus (yellow). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
For 2P-LSM, we used an Olympus FV1000-MPE with Mai Tai DeepSee Ti:Sapphire laser (SpectraPhysics) and an Olympus XLFLUOR 340 objective with a magnification of 4× and 0.28 NA. A subset of high-resolution images was acquired using an Olympus XLPLN10XSVMP objective (10×, 0.6 NA). Whole brain images were acquired using FluoView Mosiac stage control and tiling software to construct stitched z-stacks at a resolution of 3.98×3.98×50 μm. The dorsal half of the brain was imaged first in the iSpacer chamber, then inverted to image the ventral half. Image z-stacks were translated and stitched using ImageJ and Imaris 8.0 (Bitplane; Zurich, Switzerland) to produce whole brain volumes. A subset of image z-stacks were imaged at a higher resolution of 1.99×1.99×25 μm in order to better resolve fiber fascicles. This higher resolution data is shown in Fig. 5. Multitiled (15% overlap) whole brain images were visualized and analyzed in 3D using Imaris 8.0. WM ROIs were traced (by E.H.C.) using Imaris in Surpass mode and automatic Surface creation with absolute intensity thresholding, followed by manual refinements. MBP fluorescence intensity values ranged from 0–4095 and mean intensity values within ROIs were used for comparisons.
2.4. DTI-CLARITY analyses
Two separate coordinate systems were used for measurement and ROI definitions: DTI space and CLARITY space. These spaces were defined via registration to reference images from one selected reference brain (mouse #3) in the DTI and CLARITY space separately. This particular mouse brain was acquired in very similar orientation in both DTI and CLARITY space, therefore finding anatomical landmarks on the corresponding images was straightforward in these pairs of images. Similar anatomical structures were then identified and traced in each brain using the Surfaces module (semi-automatic based on intensity thresholding) in Imaris for CLARITY images and manual ROI tracing using TrackVis for DTI images. This resulted in 14 discrete 3D ROIs for each mouse brain, one set of 14 in the DTI space and another in the CLARITY space (Fig. 1B). The corresponding structures were visually checked based on anatomical landmarks, and subsequently their volumes showed excellent correlations between the two image spaces (Fig. 3B). WM ROIs were identified by tracing the MBP immunofluorescence signal in Imaris and the WM boundaries on FA maps in TrackVis). The identified ROI structures (Fig. 2B, C) were the anterior commissure (posterior aspect), corpus callosum (genu, body, splenium), fimbria (left, right), fornix (left, right), stria medullaris (left, right), fasciculus retroflexus (left, right) and the mammillothalamic tract (left, right). Once 3D ROIs were identified in the corresponding space, we registered FA images by applying a twelve-parameter affine linear transformation with FMRIB Linear Image Registration Tool (FLIRT; Jenkinson et al., 2002) and then applying the transform to the other scalar images. For CLARITY images, we down-sampled (1/10) and transformed the microscopy images into a 3D Nifti format with an in-house R (http://www.R-project.org/) script. Downsampling was necessary because at the original resolution, the required memory was over the capacity of our workstation (> 400GB of RAM). We then applied a twelve-parameter affine registration (with FLIRT) to individual MBP maps to the corresponding reference brain in the MBP space. Transformation matrices for the original resolution were calculated by concatenating the transformations (down-sampling, registration, up-sampling) and applied to the native-resolution microscopy images. We further refined registrations manually using 3D Slicer (http://www.slicer.org; Fedorov et al., 2012).
The mean MBP immunofluorescence intensities (from Imaris) and mean FA values (from TrackVis) within WM ROIs were measured and used for statistical comparisons. Statistical tests were performed using IBM SPSS version 22.0 and R (version 3.0) with mixed effect model analyses. For mixed models, we used anatomical region (ROI) as a categorical variable (fixed effect) and the DTI-derived metric (FA, AD, or RD) as a continuous variable to predict MBP, controlling for subject variability (random effect). Comparisons were performed using ANOVA or Spearman's correlation, with a P value of <0.05 considered statistically significant.
3. Results
3.1. CLARITY polymerization and tissue clearing are normal following ex vivo DTI
In order to test the feasibility of CLARITY following DTI, we first tested three thy1-eYFP-H mice that underwent ∼18 h of ex vivo DTI in a high-field scanner. These brains were perfused with a modified liquid hydrogel solution containing 0.1 mM Gd-DTPA, a MRI contrast agent. Brains were then immersed in PBS for 48 h prior to scanning and then in Fomblin during DTI. Following DTI, brains were then polymerized and cleared passively according to the CLARITY protocol. We found that polymerization, passive clearing, fluorescence intensity, and structural integrity were well-preserved in these brains (Fig. 1 in Chang et al., in press). This demonstrated that CLARITY could be successfully performed despite the presence of a contrast agent in the hydrogel and minor deviations from the original published protocol (Chung et al., 2013).
3.2. Whole brain maps of MBP-labeled fiber tracts
For CLARITY MBP-labeled mouse brains, we followed the same experimental design with the additional steps of MBP immunolabeling (Fig. 1A). The selection of a protein marker for myelination was particularly important as lipids, of which myelin is primarily composed, are removed during CLARITY tissue clearing. CLARITY whole brains achieved excellent optical transparency and were imaged entirely intact using a 2P-LSM to produce a 3D brain-wide map of MBP-positive WM structures (Fig. 2A, B).
Fig. 2.
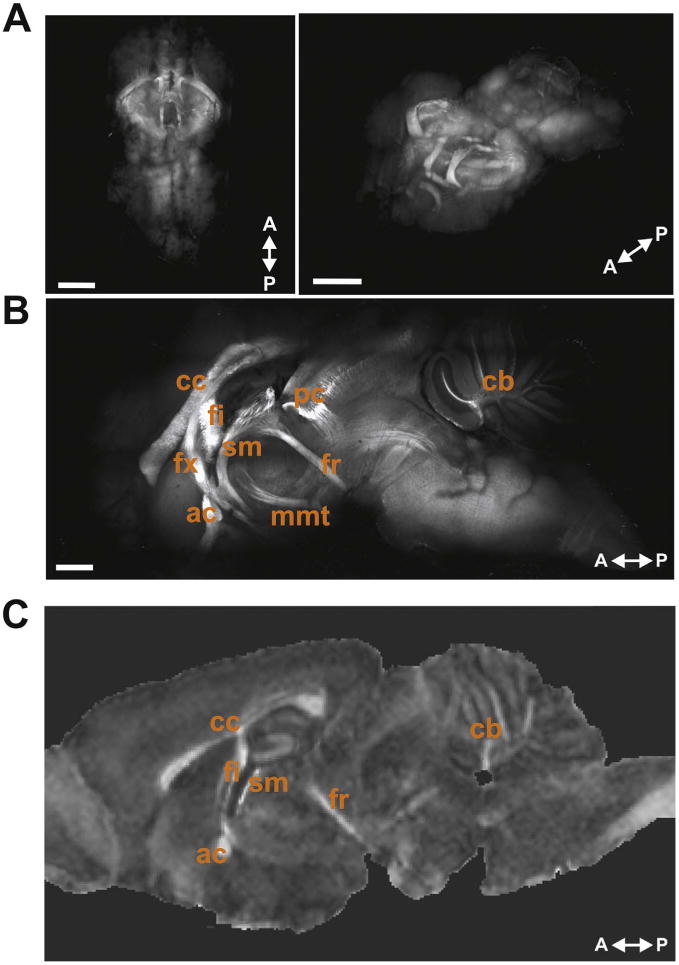
Whole mouse brain CLARITY MBP immunolabeling. (A) Images of CLARITY MBP whole brain in 3D showing a top-down view (left) and rotated horizontal view (right). Scale bars, 2 mm. (B) 1500 µm sagittal optical section near the midline of a CLARITY MBP brain shows many of the major myelinated WM structures within the mouse brain. Labeled WM tracts are: corpus callosum (cc), fimbria (fi), fornix (fx), anterior commissure (ac), stria medullaris (sm), posterior commissure (pc), fasciculus retroflexus (fr), mammillothalamic tract (mmt), and cerebellar WM (cb). Scale bar, 1.0 mm. (C) Representative sagittal image slice from FA map showing some of the same WM structures.
3.3. CLARITY MBP correlates with DTI-derived FA measures
We compared MBP labeling of WM to DTI scalar metrics, namely FA, RD, AD, and MD across multiple ROIs in the same brain samples (Fig. 3). There was a highly significant positive correlation between the volume of CLARITY MBP-labeled structures and volumes for these same ROIs in DTI (Fig. 3B; Spearman's correlation, ρ=0.876, P < 0.00001), indicating excellent structural correspondence. When analyzing FA, the most widely used metric for WM “integrity” (Jones et al., 2013), we found, using a mixed effect model, that FA significantly correlated with MBP immunofluorescence across the full set of ROIs (Fig. 3C; Χ2(1)=7.74, P<0.01). Next, we examined the MBP relationships with the directional diffusivities AD and RD. Using mixed effect models, we found a lack of significant correlations between MBP immunofluorescence and AD (Fig. 3D; Χ2(1)=0.27, P=0.61) and between MBP immunofluorescence and RD (Fig. 3E; Χ2(1)=0.05, P=0.83). This was unexpected for the scalar metric RD, which is considered to be sensitive to the degree of myelination and thus was expected to provide a significant negative correlation (Song et al., 2002; Sun et al., 2006a, 2006b).
Fig. 3.
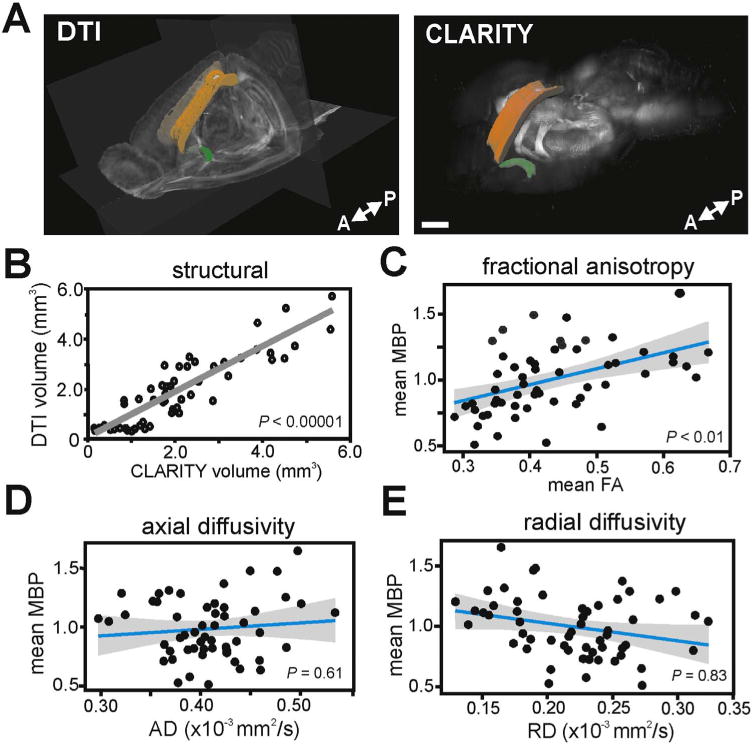
FA is correlated with MBP immunofluorescence. (A) Images show ROI-based comparisons of WM ROIs in DTI space (left) and MBP-positive WM structures in CLARITY space (right). Displayed ROIs are corpus collosum (genu, body, splenium) in orange and anterior commissure (posterior aspect) in green. Scale bar, 1000 µm. (B) DTI volumes significantly correlated with CLARITY volumes across all quantified WM structures (P<0.00001). (C) Mean MBP immunofluorescence correlated significantly with mean FA (P< 0.01). However, MBP did not correlate with measures of directional diffusivity, mean AD (D) or mean RD (E) across all WM ROIs measured. Linear regressions are shown with 95% confidence intervals. Each data point is a mean MBP immunofluorescence and mean FA measure from a single WM ROI, in one mouse brain. MBP immunofluorescence values were normalized to a global average. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. FA and RD correlations with MBP vary across different WM tract subsets
To further examine the relationship between FA and RD with MBP immunofluorescence, we parsed out a subset of WM structures that exhibited the expected inverse relationship between MBP and RD (Song et al., 2002). This subset of WM structures included the corpus collosum (genu, body, splenium), fimbria, and the anterior commissure. We found that MBP immunofluorescence of these structures was positively correlated with FA (Fig. 4A; Χ2(1)=12.56, P < 0.0005) and negatively correlated with RD (Χ2(1)=10.54, P < 0.005). In the remaining WM structures (fornix, stria medullaris, fasciculus retroflexus, and mammillothalamic tract), MBP immunofluorescence correlated with FA (Fig. 4B; Χ2(1)=10.62, P < 0.005) but not with RD (Χ2(1)=0.804, P=0.37). This highlights that the contribution of myelination to the variance of DTI-derived measures appears to vary depending on which WM tracts are being examined.
Fig. 4.
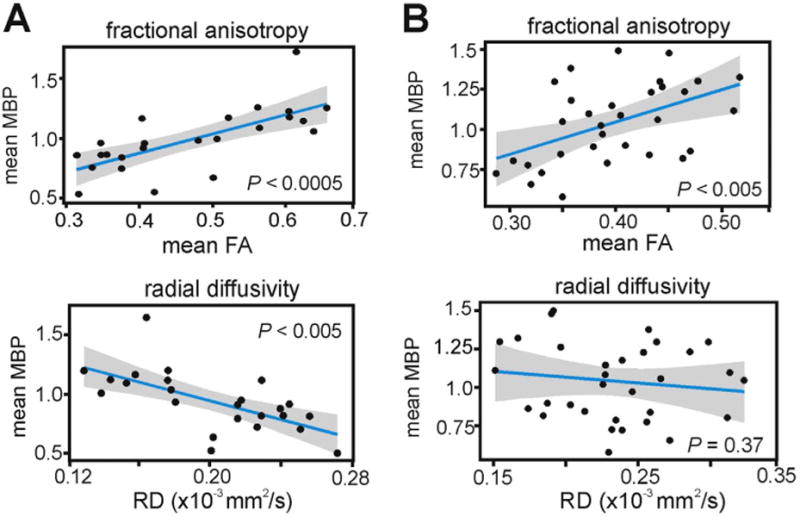
RD does not correlate with myelination in a subset of WM tracts. (A) Within a subset of mainly commissural WM tracts, FA and RD were significantly correlated with MBP immunofluorescence. (B) RD did not correlate with MBP in the remaining WM tracts, indicating that other factors such as fiber architecture, axon density, or axon caliber may be more significant contributors to diffusion anisotropy and directional diffusivity in these WM regions. Linear regressions are shown with 95% confidence intervals. Each data point is a mean MBP immunofluorescence and mean FA measure from a single WM ROI, in one mouse brain. MBP immunofluorescence values were normalized to a global average.
3.5. Within-tract analysis of stria medullaris and corpus callosum
It has been estimated that up to 90% of WM voxels in a DTI map contain complex fiber architecture and cannot be modeled accurately using a single tensor model (Jeurissen et al., 2013). This is evident when comparing the coarse DTI voxel to microscope images of the same location in the CLARITY sample. Moving from macroscopic to microscopic scales further highlights the discrepancy between the principal eigenvectors and the physical fiber bundles that are actually present at those locations in the brain (Fig. 5A and B).
In order to examine the FA-MBP relationship in further detail, we performed additional within-tract analysis on two WM ROIs, the stria medullaris and corpus callosum. The stria medullaris is a WM tract within the thalamus that carries axons from the septum to the habenula (Watson et al., 2012). We segmented the stria medullaris tract into 10 individual ROIs to perform within-tract correlations of CLARITY MBP with DTI-derived scalar metrics and found that FA-MBP correlations were highest in ROIs of the stria medullaris where fascicles were compact and coherent, with mean FA values greater than 0.5 (Fig. 5C; ρ=0.666, P < 0.00001, Spearman's correlation, n=44 regions). Conversely, across ROIs with mean FA values below 0.5, the FA-MBP correlation was not significant (P=0.118). To quantify the fiber coherence in these WM regions, we used the DTI-derived geometric indices, linear case (Cl) and planar case (Cp) anisotropy, which characterize the shape of the apparent diffusion tensor to estimate intravoxel fiber architecture (Westin et al., 2002; O'Donnell and Westin, 2011). High FA ROIs of the stria medullaris had high linear anisotropy (Cl=0.51 ± 0.021, mean ± SEM), indicative of highly coherent fiber organization. In contrast, regions where clearly diverging fascicles could be observed in the CLARITY sample had lower linear anisotropy (Cl=0.35 ± 0.024) and also lower FA-MBP correlations (ρ=0.185, P=0.388, n=24 regions).
The corpus callosum is the largest WM structure in the brain, connecting both hemispheres of the brain. We segmented the corpus callosum into 16 discrete regions (4 genu, 8 body, 4 splenium) and found that the pattern of MBP immunofluorescence (Fig. 5D) was consistent with known regional differences of myelin densities in the mouse brain (Sepehrband et al., 2015; West et al., 2016). Specifically, mean MBP in the genu and splenium were both higher than in the body, indicating that MBP immunofluorescence could be used to index the density of myelinated axons within a given WM region. This is consistent with the understanding that the amount of compact myelin is known to correlate with MBP, at least in genetic models of hypomyelination (Shine et al., 1992). When examining FA-MBP correlations within each of the callosal subregions, we found that there were significant positive correlations for the genu (P < 0.0005) and splenium (P < 0.0005), but not for the body (P=0.138; Fig. 5D). Geometric anisotropy indices estimated that the body of the corpus collosum contained more crossing or mixing fiber architecture as reflected by its lower linear anisotropy (Cl=0.36 ± 0.030) and higher planar anisotropy (Cp=0.29 ± 0.031), compared to the genu and splenium (genu: Cl=0.53 ± 0.033, Cp=0.17 ±0.019; splenium, Cl=0.48±0.049, Cp=0.19 ±0.011).
Fig. 5.
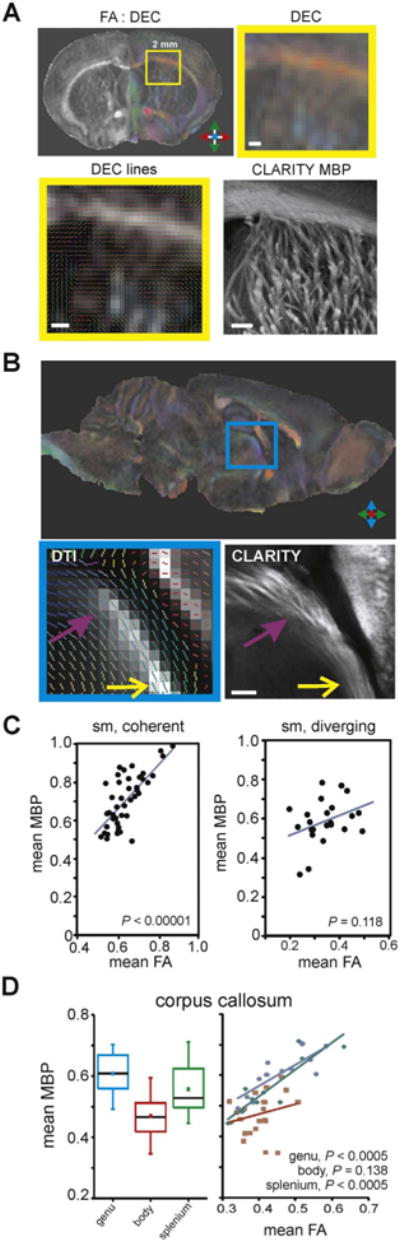
Spatial resolution differences and within-tract analyses. (A) Comparison of coronal FA and direction-encoded color (DEC) FA maps to CLARITY MBP microscopy. The low spatial resolution of the DTI images, in comparison to microscopy, becomes evident when moving from macroscopic to microscopic scales. Large, coherent WM structures (e.g. corpus callosum) are well represented by DTI, but more complex fiber
4. Discussion
By utilizing intact brain-wide analysis combining ex vivo DTI and CLARITY MBP immunolabeling in the same samples, we have demonstrated that FA measures significantly correlate with a direct label of axon myelination. Within-tract analyses of the corpus callosum and stria medullaris showed that the FA-MBP correlation was most robust in regions that are known to be densely myelinated and exhibit coherent fiber orientation, suggesting that FA measures in these types of WM regions may be more sensitive to myelination than in regions containing crossing or dispersing fiber architectures. The contribution of myelination to the variance of FA and specific directional diffusivities, such as RD, varied widely across WM tracts, and therefore it's important to keep in mind the other factors that contribute to these measures (Wheeler-Kingshott and Cercignani, 2009). Given the widespread usage of DTI for assessing WM, we believe that similar multimodal imaging studies, possibly using additional molecular markers to quantify additional factors, are worth pursuing to better understand the biological sources of diffusion changes.
WM in the brain has a very complex structure that cannot be accurately estimated using a single diffusion tensor model, such as DTI. Nonetheless, DTI remains a powerful method for noninvasively estimating WM structure in the brain with high sensitivity, but low specificity. A number of DTI validation studies have examined the potential anatomical underpinnings of the diffusion signal in animal and human post-mortem tissue (Budde et al., 2007; Leergaard et al., 2010; Seehaus et al., 2015), however, these studies have relied on traditional 2D slide-based immunohistochemistry which have major limitations for examining complex 3D structures. Using 3D-immunohistochemistry (3D-IHC), we show that WM regions with highly coherent fiber orientations, such as subregions of the mouse stria medullaris and the corpus collosum (Fig. 5C and D), exhibit strong positive correlations between FA and myelination. In our data, MBP immunofluorescence in these WM subregions accounted for up to nearly half of the variance in the FA measure (FA-MBP correlation in stria medullaris, high FA; r2=0.487), providing evidence that diffusion anisotropy measures in these subregions are highly sensitive to myelination. However, additional factors such as fiber architecture, axonal diameter, glial cells, and inflammation also contribute to diffusion anisotropy.
In this study, we have attempted to minimize the contribution of distortions and artifacts in our neuroimaging signals. Common sources of noise for in vivo neuroimaging include head motion, cardiac pulses, and arousal, which we have excluded by imaging ex vivo. A significant challenge in understanding WM is the vast difference in scale between the biology and the tools used to measure it. The spatial resolution of diffusion MRI is low, typically on the order of 1–2 mm in the human brain in vivo, while myelinated axons are characterized on a micron scale. This large gap makes it difficult to make assessments of the microbiology that is leading to measured mesoscopic and macroscopic changes (e.g. altered FA or RD). While diffusion anisotropy does carry microscopic anatomical information, this information is averaged over the large volume of an imaging voxel. In an effort to obtain a high spatial resolution (100 μm isotropic), we used an ultra high field 11.7 T scanner with a 3D DW-GRASE sequence that minimized distortion while allowing for a 12-fold acceleration compared to spin echo imaging (Aggarwal et al., 2010). However, even at this 100 μm resolution and ∼17 h scan times, each imaging voxel can contain anywhere from 1,000 to 13,000 axons, over 80 oligodendrocytes, and more than 20 astrocytes (Walhovd et al., 2014). This is likely a small subset of the entire biological repertoire within a single imaging voxel. Also, our scanning was performed at a relatively low temperature of 28 °C, which was necessary to prevent polymerization of the hydrogel. While temperature and fixation affect diffusivity values, and ADC in fixed tissue is significantly lower than in vivo (Richardson et al., 2014), anisotropy values are comparable between in vivo and ex vivo samples (Sun et al., 2001, 2003; Zhang et al., 2012).
The relationship between FA and myelin is complex and cannot be resolved using this correlational approach alone. In this study, we chose to label MBP as a putative biological index for myelination because it is the predominant protein within CNS myelin, representing as much as 30% of the protein in myelin (Baumann and Pham-Dinh, 2001) and is essential to myelin formation (Boggs, 2006). Many variables related to myelin may contribute to diffusion anisotropy, including the variable thickness and lengths of myelin sheaths that can affect inter-axonal space (Vorísek and Syková, 1997). Also, the percentage of myelinated axons and myelin sheath thickness are not necessarily indexed by MBP expression (Shine et al., 1992). Indeed, diffusion anisotropy is present even when compact myelin is completely absent, although it is significantly reduced (Roach et al., 1985; Watanabe et al., 2010). Nonetheless, the lack of significant correlation between RD and MBP across the brain-wide group of WM ROIs (Fig. 3E) was unexpected, which is why we decided to examine this association further in specific WM subsets (Fig. 4). This additional analysis indicated that RD is sensitive to myelination within specific tracts, specifically the corpus collosum, fimbria, and anterior commissure. Compared to the other WM ROIs, this subset tended to have larger volumes, which may minimize a potential partial volume effect that may have confounded the RD-MBP association in smaller WM tracts. The association of RD to myelination has been previously studied in some of these same WM structures, such as the corpus collosum and anterior commissure (Song et al., 2002), however, many other WM structures analyzed here have not been examined previously. Additionally, some of the RD-myelination association evidence is based on studies involving myelin damage to the mouse optic nerve (Song et al., 2003, 2006), which may differ from WM tracts within brain tissue. None of the prior studies utilized a 3D-IHC approach for making comparisons to diffusion measures.
We have used CLARITY in this study for its superior immunolabeling capability but there are several other tissue clearing techniques available to researchers that were developed to preserve fluorescence, such as Scale (Hama et al., 2011), 3DISCO (Ertürk et al., 2012), SeeDB (Ke et al., 2013), and CUBIC (Susaki et al., 2014). Many of these clearing techniques result in structural alterations or are incompatible with immunolabeling, however, new tissue clearing protocols are evolving rapidly and include further improvements on the CLARITY approach (Yang et al., 2014; Epp et al., 2015). Techniques such as PACT/PARS (Yang et al., 2014) and iDISCO (Renier et al., 2014) extend clearing, immunolabeling, and imaging of large intact volumes to a wide variety of organs other than the brain. In addition to emerging tissue clearing techniques, there are also other noninvasive imaging modalities that complement DTI information, such as free water imaging (Pasternak et al., 2009) and magnetization transfer (MacKay et al., 1994). These other imaging modalities provide additional experimental avenues for comparisons of 3D-IHC to WM-related targets. In this study, we have only examined MBP but there are many additional axon and myelin-related proteins (e.g. myelin oligodendrocyte glycoprotein, myelin-associated glycoprotein, neuregulin-1, neurofilament) that can be immunolabeled and compared using a similar multimodal study design.
Our findings should be considered in the context of several limitations. First, while it appears that we have labeled the major myelinated WM tracts of the mouse brain, there may be areas that are not fully captured due to incomplete antibody penetration or uneven labeling, which have been reported by other groups using intact brain approaches (Epp et al., 2015; Li et al., 2015). Fixation is known to cause reductions in diffusivity, however, imaging of live and fixed brains has been shown to provide similar diffusion anisotropy results (Sun et al., 2001, 2003; Zhang et al., 2012). Also, because we compared both DTI and CLARITY in fixed tissue ex vivo, the effects of fixation are not expected to affect the orientation-dependencies of diffusivity or their comparison across WM tracts. Another point of consideration is that the liquid hydrogel may have changed diffusivity during DTI in some unpredictable way. While we have not seen evidence for uneven labeling or altered diffusivity in our samples, we cannot rule these out as possibilities. Lastly, we have used MBP immunofluorescence as a marker of myelination, however, multiple variables can influence the quantitation of fluorophores in fixed tissue samples (Straight, 2007; Waters, 2009) and this interpretation must be considered cautiously. Additional studies, ideally including electron microscopy to directly quantify axonal myelination, are needed for further validation of our results.
The experimental framework used here is an important step towards resolving the biological contributors to the diffusion signal and provides an opportunity for more detailed investigation of brain WM in both animal models and in human tissue. For example, it is possible to generate structural tensors from histological sections (Budde and Frank, 2012; Seehaus et al., 2015), and structure tensor informed fiber tractography (STIFT) has already been reported using high field gradient echo MRI (Kleinnijenhuis et al., 2012). A similar structural tensor approach could be applied to CLARITY-cleared tissue volumes (Ye et al., 2016), including blocks of human brain, for direct within-sample comparison to diffusion tractography. 3D-IHC structural tensors can also potentially be used to corroborate a growing collection of connectivity data in the mouse brain that includes a probabilistic tractography connectome (Calabrese et al., 2015). This type of data could be very useful for addressing specific inherent limitations of DTI tractography that have been recently highlighted by others (Thomas et al., 2014). Additional multimodal possibilities include overlaying directional information (which DTI does not provide) of fiber pathways onto DTI-derived scalar maps by employing viral-mediated anterograde and retrograde tracing combined with CLARITY (Menegas et al., 2015). This would provide valuable information on the functional direction of neural communication along WM tracts and could significantly increase the utility of diffusion-weighted MRI based maps of the brain, as it would indicate the directionality of WM tracts and possibly identify new patterns of functional connectivity that have not been revealed using traditional slide-based histology. Perhaps most importantly, our results demonstrate that this 3D-IHC approach can be used to produce a brain-wide map of a molecular target. The true potential of CLARITY, and other immunolabeling-compatible whole-brain imaging techniques, is in the flexibility of being able to generate 3D maps of potentially any target for which an antibody exists. As full genome (Lein et al., 2007), proteome (Sharma et al., 2015), and connectivity databases (Mitra, 2014; Oh et al., 2014) become widely available, we can begin to integrate 3D brain-wide molecular analyses with these large-scale efforts.
5. Conclusions
This is the first study to combine DTI with CLARITY within-subject and demonstrates that the clearing technique can be successfully performed despite modifications in the protocol made due to DTI scanning. It is also the first time, to our knowledge, that whole-brain labeling of MBP has been performed and examined in 3D within intact mouse brains. By using MBP as a proxy for myelination, we showed that it correlated with DTI-derived FA across intact WM structures, particularly in coherent and highly organized fiber regions. RD only correlated with MBP in a smaller subset of WM structures. We believe that this 3D-IHC approach can be combined with other noninvasive neuroimaging modalities in an effort to better understand the molecular underpinnings of several commonly used neuroimaging measures.
Acknowledgments
The authors thank Dr. Amanda Chan for technical assistance and Drs. Aristotle Voineskos, Majnu John, and Todd Lencz for useful feedback on the manuscript. This work was supported by the Center for Intervention Development and Applied Research (P50MH080173). Funding agencies had no role in the study design, acquisition or interpretation of data, or in writing the manuscript.
References
- Aggarwal M, Mori S, Shimogori T, Blackshaw S, Zhang J. Three-dimensional diffusion tensor microimaging for anatomical characterization of the mouse brain. Magn Reson Med. 2010;64:249–261. doi: 10.1002/mrm.22426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J Magn Reson B. 1996;111:209–211. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, Le Bihan D. MR diffusion tensor spectroscopy and imaging. Biophys J. 1994;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann N, Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervouse system. Physiol Rev. 2001;81:871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- Beaulieu C. The basis of anisotropic water diffusion in the nervous system – a technical review. NMR Biomed. 2002;15:434–455. doi: 10.1002/nbm.782. [DOI] [PubMed] [Google Scholar]
- Boggs JM. Myelin basic protein: a multifunctional protein. Cell Mol Life Sci. 2006;63:1945–1961. doi: 10.1007/s00018-006-6094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde MD, Frank JA. Examining brain microstructure using structure tensor analysis of histological sections. NeuroImage. 2012;63:1–10. doi: 10.1016/j.neuroimage.2012.06.042. [DOI] [PubMed] [Google Scholar]
- Budde MD, Kim JH, Liang HF, Schmidt RE, Russell JH, Cross AH, et al. Toward accurate diagnosis of white matter pathology using diffusion tensor imaging. Magn Reson Med. 2007;57:688–695. doi: 10.1002/mrm.21200. [DOI] [PubMed] [Google Scholar]
- Calabrese E, Badea A, Cofer G, Qi Y, Johnson A. A diffusion MRI tractography connectome of the mouse brain and comparison with neuronal tracer data. Cereb Cortex. 2015;25:4628–4637. doi: 10.1093/cercor/bhv121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EH, Argyelan M, Aggarwal M, Chandon TS, Karlsgodt KH, Mori S, Malhotra AK. Diffusion tensor imaging measures of white matter compared to myelin basic protein immunofluorescence in tissue cleared intact brains. Data in Brief. doi: 10.1016/j.dib.2016.12.018. http://dx.doi.org/10.1016/j.dib.2016.12.018. [DOI] [PMC free article] [PubMed]
- Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp JR, Niibori Y, Hsiang HL, Mercaldo V, Deisseroth K, Josselyn SA, et al. Optimization of CLARITY for clearing whole-brain and other intact organs. eNeuro. 2015;2 doi: 10.1523/ENEURO.0022-15.2015. (ENEURO. 0022-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertürk A, Becker K, Jährling N, Mauch CP, Hojer CD, Egen JG, et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc. 1983;7(11):95. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, et al. 3D slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging. 2012;30:1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Bannister PR, Brady JM, Smith SM. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Jennings JH, Stuber GD. Tools for resolving functional activity and connectivity within intact neural circuits. Curr Biol. 2014;24:R41–R50. doi: 10.1016/j.cub.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeurissen B, Leemans A, Tournier JD, Jones DK, Sijbers J. Investigating the prevalence of complex fiber configurations in white matter tissue with diffusion magnetic resonance imaging. Hum Brain Mapp. 2013;34:2747–2766. doi: 10.1002/hbm.22099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Johnson GA. Microscopic diffusion tensor atlas of the mouse brain. NeuroImage. 2011;56(3):1235–1243. doi: 10.1016/j.neuroimage.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DK, Cercignani M. Twenty-five pitfalls in the analysis of diffusion MRI data. NMR Biomed. 2010;23:803–820. doi: 10.1002/nbm.1543. [DOI] [PubMed] [Google Scholar]
- Jones DK, Knösche TR, Turner R. White matter integrity, fiber count, and other fallacies: The do's and don'ts of diffusion MRI. NeuroImage. 2013;73:239–254. doi: 10.1016/j.neuroimage.2012.06.081. [DOI] [PubMed] [Google Scholar]
- Ke MT, Fujimoto S, Imai T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci. 2013;16:1154–1161. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- Kim SY, Chung K, Deisseroth K. Light microscopy mapping of connections in the intact brain. Trends Cogn Sci. 2013;17:596–599. doi: 10.1016/j.tics.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Kleinnijenhuis M, Barth M, Alexander DC, van Cappellen van Walsum AM, Norris DG. Structure Tensor Informed Fiber Tractography (STIFT) by combining gradient echo MRI and diffusion weighted imaging. NeuroImage. 2012;59:3941–3954. doi: 10.1016/j.neuroimage.2011.10.078. [DOI] [PubMed] [Google Scholar]
- Le Bihan D, Johansen-Berg H. Diffusion MRI at 25: exploring brain tissue structure and function. NeuroImage. 2012;61:324–341. doi: 10.1016/j.neuroimage.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bihan D, Breton E, Lallemand D, Grenier P, Cabanis E, Laval Jeantet M. MR Imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology. 1986;161:401–407. doi: 10.1148/radiology.161.2.3763909. [DOI] [PubMed] [Google Scholar]
- Le Bihan D, Mangin JF, Poupon C, Clark CA, Pappata S, Molko N, et al. Diffusion tensor imaging: concepts and appplications. J Magn Reson Imaging. 2001;13:534–546. doi: 10.1002/jmri.1076. [DOI] [PubMed] [Google Scholar]
- Leergaard TB, White NS, de Crespigny A, Bolstad I, D'Arceuil H, Bjaalie JG, et al. Quantitative histological validation of diffusion MRI fiber orientation distributions in the rat brain. PLoS One. 2010;5:e8595. doi: 10.1371/journal.pone.0008595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Li J, Czajkowsky DM, Li X, Shao Z. Fast immuno-labeling by electrophoretically driven infiltration for intact tissue imaging. Sci Rep. 2015;5:10640. doi: 10.1038/srep10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay A, Whittall K, Adler J, Li D, Paty D, Graeb D. In vivo visualization of myelin water in brain by magnetic resonance. Magn Reson Med. 1994;31(6):673–677. doi: 10.1002/mrm.1910310614. [DOI] [PubMed] [Google Scholar]
- Menegas W, Bergan JF, Ogawa SK, Isogai Y, Umadevi Venkataraju K, et al. Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass. Elife. 2015;4:e10032. doi: 10.7554/eLife.10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra P. The circuit architecture of whole brains at the mesoscopic scale. Neuron. 2014;83:1273–1283. doi: 10.1016/j.neuron.2014.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Zhang J. Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron. 2006;51:527–539. doi: 10.1016/j.neuron.2006.08.012. [DOI] [PubMed] [Google Scholar]
- O'Donnell LJ, Westin CF. An introduction to diffusion tensor image analysis. Neurosurg Clin N Am. 2011;22:185–196. doi: 10.1016/j.nec.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak O, Sochen N, Gur Y, Intrator N, Assaf Y. Free water elimination and mapping from diffustion MRI. Magn Reson Med. 2009;62(3):717–730. doi: 10.1002/mrm.22055. [DOI] [PubMed] [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Richardson DS, Lichtman JW. Clarifying tissue clearing. Cell. 2015;162:246–257. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S, Siow B, Panagiotaki E, Schneider T, Lythgoe MF, Alexander DC. Viable and fixed white matter: diffusion magnetic resonance comparisons and contrasts at physiological temperature. Magn Reson Med. 2014;72:1151–1161. doi: 10.1002/mrm.25012. [DOI] [PubMed] [Google Scholar]
- Roach A, Takahashi N, Pravtcheva D, Ruddle F, Hood L. Chromosomal mapping of mouse myelin basic protein gene and structure and transcription of the partially deleted gene in shiverer mutant mice. Cell. 1985;42:149–155. doi: 10.1016/s0092-8674(85)80110-0. [DOI] [PubMed] [Google Scholar]
- Seehaus A, Roebroeck A, Bastiani M, Fonseca L, Bratzke H, Lori N, et al. Histological validation of high-resolution DTI in human post-mortem tissue. Front Neuroanat. 2015;9:98. doi: 10.3389/fnana.2015.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepehrband F, Clark KA, Ullmann JF, Kurniawan ND, Leanage G, Reutens DC, et al. Brain tissue compartment density estimated using diffusion-weighted MRI yields tissue parameters consistent with histology. Hum Brain Mapp. 2015;36:3687–3702. doi: 10.1002/hbm.22872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique-Hoyos N, et al. Cell type- and brain region-resolved mouse brain proteome. Nat Neurosci. 2015;18:1819–1831. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine HD, Readhead C, Popko B, Hood L, Sidman RL. Morphometric analysis of normal, mutant, and transgenic CNS: correlation of myelin basic protein expression to myelinogenesis. J Neurochem. 1992;58:342–349. doi: 10.1111/j.1471-4159.1992.tb09316.x. [DOI] [PubMed] [Google Scholar]
- Song SK, Sun SW, Ramsbottom MJ, Chang C, Russell J, Cross AH. Dysmyelination revealed through MRI as increased radial (but unchanged axial) diffusion of water. NeuroImage. 2002;17:1429–1436. doi: 10.1006/nimg.2002.1267. [DOI] [PubMed] [Google Scholar]
- Song SK, Sun SW, Ju WK, Lin SJ, Cross AH, Neufeld AH. Diffusion tensor imaging detects and differentiates axon and myelin degeneration in mouse optic nerve after retinal ischemia. NeuroImage. 2003;20:1714–1722. doi: 10.1016/j.neuroimage.2003.07.005. [DOI] [PubMed] [Google Scholar]
- Straight AF. Fluorescence protein applications in microscopy. Methods Cell Biol. 2007;81:93–113. doi: 10.1016/S0091-679X(06)81006-X. [DOI] [PubMed] [Google Scholar]
- Sun SW, Neil JJ, Liang HF, He YY, Schmidt RE, Hsu CY, et al. Formalin fixation alters water diffusion coefficient magnitude but not anisotropy in infarcted brain. Magn Reson Med. 2001;53:1447–1451. doi: 10.1002/mrm.20488. [DOI] [PubMed] [Google Scholar]
- Sun SW, Neil JJ, Song SK. Relative indices of water diffusion anisotropy are equivalent in live and formalin-fixed mouse brains. Magn Reson Med. 2003;50:743–748. doi: 10.1002/mrm.10605. [DOI] [PubMed] [Google Scholar]
- Sun SW, Liang HF, Trinkaus K, Cross AH, Armstrong RC, Song SK. Noninvasive detection of cuprizone induced axonal damage and demyelination in the mouse corpus callosum. Magn Reson Med. 2006;55:302–308. doi: 10.1002/mrm.20774. [DOI] [PubMed] [Google Scholar]
- Sun SW, Liang HF, Le TQ, Armstrong RC, Cross AH, Song SK. Differential sensitivity of in vivo and ex vivo diffusion tenor imaging to evolving optic nerve injury in mice with retinal ischemia. NeuroImage. 2006;32:1195–1204. doi: 10.1016/j.neuroimage.2006.04.212. [DOI] [PubMed] [Google Scholar]
- Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, et al. Whole brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- Thomas C, Ye FQ, Irfanoglu MO, Modi P, Saleem KS, Leopold DA, et al. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc Natl Acad Sci USA. 2014;111:16574–16579. doi: 10.1073/pnas.1405672111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid andhigh-resolution imaging of intact tissues. Nat Protoc. 2014;9(7):1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorísek I, Syková E. Evolution of anisotropic diffusion in the developing rat corpus callosum. J Neurophysiol. 1997;78(2):912–919. doi: 10.1152/jn.1997.78.2.912. [DOI] [PubMed] [Google Scholar]
- Walhovd KB, Johansen-Berg H, Káradóttir RT. Unraveling the secrets of white matter–bridging the gap between cellular, animal and human imaging studies. Neuroscience. 2014;276:2–13. doi: 10.1016/j.neuroscience.2014.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Frahm J, Michaelis T. Myelin mapping in the living mouse brain using manganese-enhanced magnetization transfer MRI. NeuroImage. 2010;49(2):1200–1204. doi: 10.1016/j.neuroimage.2009.09.050. [DOI] [PubMed] [Google Scholar]
- Waters JC. Accuracy and precision in quantitative fluorescence microscopy. J Cell Biol. 2009;185:1135–1148. doi: 10.1083/jcb.200903097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson C, Paxinos G, Puelles L. The Mouse Nervous System. Academic Press; London, U.K: 2012. [Google Scholar]
- West KL, Kelm ND, Carson RP, Does MD. A revised model for estimating g-ratio from MRI. NeuroImage. 2016;125:1155–1158. doi: 10.1016/j.neuroimage.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin CF, Maier SE, Mamata H, Nabavi A, Jolesz FA, Kikinis R. Processing and visualization for diffusion tensor MRI. Med Image Anal. 2002;6:93–108. doi: 10.1016/s1361-8415(02)00053-1. [DOI] [PubMed] [Google Scholar]
- Wheeler-Kingshott CA, Cercignani M. About “axial” and “radial” diffusivities. Magn Reson Med. 2009;61(5):1255–1260. doi: 10.1002/mrm.21965. http://dx.doi.org/10.1002/mrm.21965. [DOI] [PubMed] [Google Scholar]
- Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E, et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Allen WE, Thompson KR, Tian Q, Hsueh B, Ramakrishnan C, Wang AC, Jennings JH, Adhikari A, Halpern CH, Witten IB, Barth AL, Luo L, McNab JA, Deisseroth K. Wiring and molecular features of prefrontal ensembles representing distinct experiences. Cell. 2016;165:1776–1788. doi: 10.1016/j.cell.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jones MV, McMahon MT, Mori S, Calabresi PA. In vivo and ex vivo diffusion tensor imaging of cuprizone induced demyelination in the mouse corpus callosum. Magn Reson Med Mar. 2012;67(3):750–759. doi: 10.1002/mrm.23032. [DOI] [PMC free article] [PubMed] [Google Scholar]


