Abstract
About 64% of the total aboveground biomass in sugarcane production is from the culm, of which ~90% is present in fiber and sugars. Understanding the transcriptome in the sugarcane culm, and the transcripts that are associated with the accumulation of the sugar and fiber components would facilitate the modification of biomass composition for enhanced biofuel and biomaterial production. The Sugarcane Iso-Seq Transcriptome (SUGIT) database was used as a reference for RNA-Seq analysis of variation in gene expression between young and mature tissues, and between 10 genotypes with varying fiber content. Global expression analysis suggests that each genotype displayed a unique expression pattern, possibly due to different chromosome combinations and maturation amongst these genotypes. Apart from direct sugar- and fiber-related transcripts, the differentially expressed (DE) transcripts in this study belonged to various supporting pathways that are not obviously involved in the accumulation of these major biomass components. The analysis revealed 1,649 DE transcripts between the young and mature tissues, while 555 DE transcripts were found between the low and high fiber genotypes. Of these, 151 and 23 transcripts respectively, were directly involved in sugar and fiber accumulation. Most of the transcripts identified were up-regulated in the young tissues (2 to 22-fold, FDR adjusted p-value <0.05), which could be explained by the more active metabolism in the young tissues compared to the mature tissues in the sugarcane culm. The results of analysis of the contrasting genotypes suggests that due to the large number of genes contributing to these traits, some of the critical DE transcripts could display less than 2-fold differences in expression and might not be easily identified. However, this transcript profiling analysis identified full-length candidate transcripts and pathways that were likely to determine the differences in sugar and fiber accumulation between tissue types and contrasting genotypes.
Introduction
Sugarcane biomass could play a very important role in supporting second generation biofuel production. On average, about 64% of the total aboveground dry biomass in sugarcane production is from the sugarcane culm, while the rest (~36%) is from the trash (leaves) (as reviewed in [1]). In the sugarcane culm-derived biomass, the major components are sugars (mostly sucrose) and fiber (cellulose, hemicellulose and lignin), as reviewed in [2, 3]. In our recent assessment on a diverse sugarcane population [4], these two components, together with other insoluble matters (all known as total solids) make up about 22–39% of the fresh weight, while on a dry biomass basis, the sugar content ranges from 29 to 64% and fiber content from 29 to 61%. Sugarcane sugars (as a food source) have long been used for biofuel production, and in recent years, sugarcane fiber (also referred to in the broader term, lignocellulosic biomass) has been emerging as an alternative option for biofuel production.
Biomass accumulation in sugarcane culms has been shown to be a very highly regulated and tightly connected process in which photosynthetic carbon is partitioned into sugar production or fiber deposition [5–10]. This means that if more carbon is diverted to fiber accumulation, cell-wall synthesis and internode elongation; less would be available for sucrose accumulation and vice versa [11]. The sugarcane culm acts as a carbon sink with both sucrose and fiber (cellulose/hemicellulose) syntheses, requiring a nucleotide sugar, UDP-glucose as a precursor, for a review, see [10]. Cellulose and hemicellulose, once deposited, rarely re-enter the cell’s metabolic process [8], whereas, sucrose has a complex and dynamic accumulation pathway in which it is rapidly synthesized, re-partitioned (into cell-wall polysaccharides) and turned over between the vacuole, the cellular metabolic and apoplastic compartments, depending upon developmental stages [5, 7, 8, 12–14]. The cellulose, hemicellulose and lignin of the fiber fraction are synthesized in different metabolic pathways but are physically linked to each other to form the plant cell-walls [15], and are therefore accumulated in the same target organs [16]. These fiber components are primarily deposited over time during internode elongation (~150 days), and during internode expansion in diameter (for at least 350 days). In an immature internode, around 50% of the incoming carbon goes to the fiber fraction (the rest is mostly partitioned into supporting protein production), whereas, in a more mature internode, only around 8% is diverted to fiber, while most of the remainder is partitioned into storage sucrose (i.e. 66% of that in internode 9) [11].
As the accumulation of sugar and fiber is tightly connected, understanding the regulation (i.e. at the transcriptional level) of their accumulation requires analysis of both fractions. In recent years, efforts have been made to gain more insight into the sugarcane culm transcriptome, particularly, the transcripts that are associated with the sugar and fiber accumulation. These have helped to define the regulation of the carbon flow in the sugarcane plant, as a whole, starting from the leaf (source tissue) and continuing along the culm (sink tissue), as well as various complex metabolic and physiological networks involved in this process [6, 8, 12, 17]. These transcriptome studies have included, for instance, genes expressed in maturing internodes [7], sugar transporters in maturing internodes [18], transcript differential expression in maturing culms [19], transcripts involving in cell-wall metabolism and development [20], transcripts related to cellulose synthase (CesA) and sucrose transporter gene families [14] and the culm transcriptome analysis of contrasting genotypes for lignin content [21–23]. Despite the lack of a reference genome or a complete transcriptome, these studies have provided valuable information about the transcriptome in the sugarcane culms and the expression patterns/regulation of sugar- and fiber-related transcripts.
The aims of this study were: (i) to conduct transcript differential expression analysis between the young and mature internodal tissues of the sugarcane plant, as well as between the contrasting low and high fiber sugarcane genotypes; and (ii) to identify candidate transcripts associated with the carbon partitioning between sugar and fiber components in sugarcane. The analysis was done by using a newly constructed transcriptome reference database which contains transcripts defined as different isoforms, to determine the important genes/transcripts involved in the accumulation of the major biomass components in corresponding samples. The transcriptome database was sequenced by PacBio isoform sequencing (Iso-Seq), as described in Hoang et al. (2017) [24], can be accessed under the NCBI TSA accession number GFHJ00000000.1; and hereafter is referred to as the Sugarcane Iso-Seq Transcriptome (SUGIT) database. Identifying the differential expressed transcripts between immature and mature internodes could suggest potential transcripts that may be involved in sugar and fiber accumulation along the sugarcane culm [25], while those differing between the low and high fiber (or sugar) content would highlight the differences in expression patterns between these two groups. The study set out to increase understanding of the regulation of carbon flux into the major components in the biomass and the genetic basis of these traits at the transcriptional level and support genetic improvement of sugarcane for fiber and/or sugar production.
Materials and methods
Sample selection and preparation
Analysis was performed on 20 internodal samples, belonging to 10 sugarcane genotypes, which were classified into low and high fiber groups (Table 1). These samples were derived from a population previously described in [4]. The RNA extracted from these samples was also used in the construction of the SUGIT database (mentioned earlier), which was employed in the transcript expression analysis in this study. In brief, 5 genotypes were chosen for each of the low and high fiber groups, and for each of the 10 genotypes, 1 top (young) and 1 bottom (mature) internodal tissue samples were collected. The young tissues were defined as the fourth internodes from the top, while the mature tissues as the third internodes from the bottom. For each internodal sample, 4 representative culms from the same genotype were pooled to form 1 biological replicate. Internodes from the pooled culms were harvested, immediately cut into 0.5 cm-thick slices, followed by the removal of the rind and diagonal separation of the remaining pith into small 0.5 cm cubes, using a pair of secateurs. The whole excision process took about 1 min before snap-freezing in liquid nitrogen, and then samples were stored at -80°C until RNA extraction. To avoid changes in the transcriptome due to the different collection times, the excision was conducted between 10 am to 2 pm on the same day. Prior to RNA extraction, frozen samples were pulverized, while kept frozen, into a fine powder in cryo-jars using a Retsch TissueLyser (Retsch, Haan, Germany), as described in [26]. The frequency of 25/S was used and the time was 1 min 30 s. Samples and the cryo-jars were kept in liquid nitrogen, except for when being ground in the TissueLyser. About 1 g of pulverized sample powder was used for RNA extraction.
Table 1. Sugarcane genotypes used in this study, including 5 low and 5 high fiber genotypes.
| Code | Genotypes | %Fibera | Type | Femaleb | Maleb |
|---|---|---|---|---|---|
| 6 | QS99-2014 | 31.2 | Commercial | QA88-1178 | Q205 |
| 1 | QC02-402 | 31.4 | Commercial | QN91-295 | Q200 |
| 12 | KQB08-32953 | 32.5 | Introgression | QBYN04-26042 | QC91-580 |
| 9 | Q200 | 37.6 | Commercial | QN63-1700 | QN66-2008 |
| 8 | Q241 | 39.6 | Commercial | Q138 | SP72-4728 |
| 17 | QBYN04-26041 | 45.0 | Introgression | ROC25 | YN2002-356 |
| 11 | KQB07-23863 | 45.7 | Introgression | KQ228A | MQB89-12554 |
| 10 | QN05-803 | 47.7 | Commercial | QN86-1659 | Q142 |
| 16 | KQB07-24739 | 48.2 | Introgression | QBYC04-10559 | N29 |
| 20 | KQB09-20432 | 49.8 | Introgression | KQ228A | QBYN05-10390 |
a %Fiber on the total dry mass (total solids).
b Parental genotypes
RNA extraction
RNA extraction was conducted using a two-step protocol as described in [26] employing a Trizol kit (Invitrogen), followed by a Qiagen RNeasy Plant minikit (#74134, Qiagen, Valencia, CA, United States). The RNA quality, integrity and quantity were determined by a NanoDrop8000 spectrophotometer (ThermoFisher Scientific, Wilmington, DE, USA), and on a 2100 Agilent Bioanalyser with a plant RNA NanoChip assay (Agilent Technologies, Santa Clara, CA, USA).
RNA-Seq and read data processing
About 3 μg of each of the 20 internodal RNA samples was used for indexed-library preparation (average insert size of 200 bp), employing the TruSeq stranded with Ribo-Zero Plant Library Prep Kit for total RNA library (Illumina Inc.). Each sample was sequenced in two lanes using an Illumina HiSeq4000 instrument to obtain 150 bp paired-end read data, at the Translational Research Institute, The University of Queensland, Australia. This led to more than 80% of the paired-end reads found to be overlapping. The read data was assessed by FastQC [27] for quality and adapter sequences. Read adapters and quality trimming was done in CLC Genomics Workbench v9.0 (CLC-GWB, CLC Bio-Qiagen, Aarhus, Denmark) with a quality score limit <0.01 (Phred Q score ≥20, equivalent to the accuracy of the base calling of 99%), allowing a maximum two ambiguous nucleotides, and removing reads below 75 bp. Only paired-end reads from each lane were kept for each sample and then concatenated into one data file prior to analysis. Since the sequencing resulted in un-balanced read data between the top and bottom internodal samples of each genotype, to reduce the sample size bias in the downstream analysis, the larger samples were down-sized using the seqtk toolkit [28], to obtain approximately uniform sample sizes across each genotype. Further quality control was conducted on the samples in each group and for all samples in the experiment. This was done by first performing counts-per-million (CPM) and then log2 data transformation of the of raw count matrix for each sample, using the script Perl_to_R from the Trinity package. This was used to assess the samples before analysis, and to remove any outliers or potential confounders within the replicates, which could cause batch effects [29], based on transcript expression level, the transcript pairwise comparisons, Pearson correlations and principal component analysis.
Transcript profiling and differential expression analysis
The pipeline was adapted from the Trinity v2.2.0 package [30, 31], designed for transcript profiling and differential expression analysis without reference genome sequences at the transcript isoform level, employing the RNA-Seq by Expectation Maximization (RSEM) software v1.2.31 [32]. To estimate the abundance of each transcript isoform, the RNA-Seq data of each of the samples was aligned against the SUGIT database using the Perl script align_and_estimate_abundance.pl. The Bowtie v2.2.7 program [33] was used with options "—no-mixed—no-discordant—gbar 1000—end-to-end -k 200". The sam alignment output file was converted to a bam file and sorted by samtools-1.3.1 [34]. The sorted bam file was subjected to the program RSEM for quantification of transcript abundance at the isoform level by fractional correcting of read alignment based on the probabilities of the transcript isoforms the reads originally came from, using its iterative process [32]. The transcript abundance estimation result was used to build the matrix of raw read counts and normalized expression values, by the perl script abundance_estimates_to_matrix.pl. The normalized expression measures included fragments per kilobase of feature sequence per million fragments mapped (FPKM) [35] and transcripts per million transcripts (TPM) [36]. The read count matrix for each sample combination was parsed using the script run_DE_analysis.pl for transcript differential expression analysis. To identify the differentially expressed (DE) transcripts, a negative binomial model was used to determine the relationship of the mean and the variance for the dispersion estimation in the DESeq2 package [37]. This was run in a pipeline using script analyze_diff_expr.pl. This DESeq2 package was suitable for general, data driven parameter estimation [38], allowing the selection of differentially expressed transcripts through a dynamic range of data and considering the unbalanced sequencing depth of the different samples. This differential expression analysis pipeline employed R program v3.2.0 [39], with the Bioconductor v3.4 [40] and the following packages: limma [41], ctc [42], and Biobase [43]. Finally, transcripts with a false discovery rate (FDR) adjusted p-value ≤ 0.05 and mean fold change ≥2 were marked as being significantly differentially expressed between the two groups compared. The DE transcripts were clustered by the package cluster 2.0.4 [44] and ape [45], then graphed using the function heatmap.2 in gplots [46]. Lists of up-regulated and down-regulated transcripts were generated for each of comparison.
Functional annotation of identified differentially expressed transcripts
For general function comparison, the DE transcripts were annotated against the Gene Ontology (GO) database, using Blast2GO v4.0.2 [47] with default settings. This used a separate BLASTX homology search result (BLAST+ v2.3.0) with maximum blast hits of 100 against the NCBI non-redundant protein database and an e-value of 1e-10. The GO terms were assigned to each of the DE transcripts, and then the GO terms for each up-regulated and down-regulated transcript sets were extracted, enriched and compared by WEGO [48]. Only enriched GO terms with a p-value cutoff of 0.05 (considered being significant from the Pearson Chi-square test) were used in assessing the over-represented GO terms between the up- and down-regulated transcript sets. For further functional analysis, all DE transcripts were subjected to the program Mercator for automated sequence annotation [49, 50] and functional classifications (bins) were assigned to the DE transcripts. This functional bin annotations were based on: (1) the BLASTX homology searches with a cutoff of 80% against the Arabidopsis TAIR Release 10, PPAP SwissProt/UniProt Plant Proteins, TIGR5 rice proteins and Uniref90; (2) the reverse PSI-BLAST (RPS-blast) searches against the Clusters of orthologous eukaryotic genes database (KOG), conserved domain database (CDD); and (3) InterProScan search against the protein domain databases. The functional bins were visualized and analysed by the ImageAnnotator module, MapMan v3.5.1R2 [51]. This annotation pipeline using Mercator and MapMan (termed as MapMan annotation) assigned the transcripts into the most appropriate bins and reduced the multiple times the transcripts were represented in many bins, which differed from the GO term annotations [51]. Arabidopsis and rice were used as the main sources of information, since these two are still amongst the best annotated plant genomes.
Transcripts specifically involved in accumulation of sugar and fiber
DE transcripts that were potentially involved in the accumulation of the major sugar and fiber components, including cell-wall metabolism, carbohydrate metabolism, photosynthesis, and phenylpropanoid pathway were investigated by using the MapMan annotation bins.
Validation of DE genes using quantitative real-time PCR (qPCR)
To validate the RNA-Seq differential expression, a reverse transcription followed by qPCR was conducted on 4 μg total RNA from each of 8 selected samples including 4 young and 4 mature tissues from 2 low fiber (QC02-401, Q200) and 2 high fiber (QBYN04-26041, QN05-803) genotypes. Three reference genes including glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ubiquitin (UBQ) and clathrin adaptor complex (CAC) were selected for internal normalization, based on published results in literature [52–55]. These genes were elucidated, from the RNA-Seq data, by their stable ubiquitous expression across all the samples. Multiple reference gene normalisation method [56, 57] was applied to the qPCR quantification cycle (Cq) values, and analysed with qbase+ software (VIB, Flanders, Belgium). A total of 8 putative DE genes were chosen for qPCR experiments. Alignment to the putative gene with least symmetry to the multitude of sequence isoforms was ascertained with Clone Manager 9 (Sci-Ed Software, Denver, US). Primer design was accomplished using Primer3 software in NCBI/Primer-BLAST [58]. List of genes and primer sequences are disclosed in S1 Table. Reverse transcription was performed according to the Tetro cDNA Synthesis Kit protocol (Bioline Reagents, London, UK). The cDNA was quantified with the NanoDrop 8000 spectrophotometer (Thermo Scientific, Waltham, US), for equal loading of 100ng cDNA per reaction of each sample run in triplicate. SensiFAST SYBR Lo-ROX Kit (Bioline Reagents, London, UK) was performed as per manufacturer’s protocol on the ViiA7 System (Applied Biosystems, Waltham, US). Accuracy and reliability of results were validated using triplicate biological and technical controls, melt curve analysis and the MIQE guidelines [59]. RNA-Seq TMM-normalized FPKM expression data were correlated against the comparative-Cq qPCR normalised gene expression data, using Microsoft Excel 2013.
Data analysis
All Venn diagrams were generated by the online tool [60]. All Linux-based analyses were performed at the High Performance Computing clusters (Flashlite and Tinaroo), hosted by the Research Computing Center, The University of Queensland, Australia [61]. Analyses using CLC-GWB were run on a CLC Genomics Server, nodes and CLC-clients at QAAFI, the University of Queensland, Australia. All other analyses, unless otherwise stated, were performed using the Data Analysis ToolPak in Microsoft Excel 2013 and RStudio ver.0.9.8/R ver.3.1.2 [39].
Results
RNA-Seq summary
To investigate the differential expression of transcripts between the young and mature tissues of the sugarcane culm, and between the low and high fiber genotypes, an RNA-Seq experiment was conducted. Two groups of 5 low fiber and 5 high fiber genotypes were used, in which, for each genotype, 1 top internodal tissue sample and 1 bottom internodal tissue sample were collected by pooling respective tissues from 4 different sugarcane culms. The total number of trimmed reads obtained for each sample ranged from 6 to 53 million (Table 2). The total number of RNA-Seq reads for all the samples from the low fiber group was 293 million, while that of the high fiber group was 148 million. This made the total read data used in this analysis 441 million reads. The percentage of reads mapped to the SUGIT database ranged from 70 to 82%. In most cases (except for genotype 6), the bottom internodal samples had a higher percentage of reads mapped to the references compared to the top internodal samples (mean of 79.5% and 74.2%, respectively). Amongst 107,598 transcriptome reference sequences, the proportion of the transcripts that had reads mapped to (FPKM>0) ranged from 57% to 76%. This result indicates the proportion of total active transcripts originating from the culm, as the SUGIT reference database was derived from leaf, internode and root tissues.
Table 2. Summary statistics of samples.
| Low fiber genotypes | High fiber genotypes | ||||
|---|---|---|---|---|---|
| Samples | Trimmed reads | %Reads mapped | Samples | Trimmed reads | %Reads mapped |
| T1 | 18,547,122 | 70.76 | T10 | 14,370,404 | 72.10 |
| B1 | 20,000,000 | 78.06 | B10 | 16,000,000 | 81.09 |
| T6 | 12,471,756 | 74.20 | T11 | 8,153,320 | 72.94 |
| B6 | 13,000,000 | 71.51 | B11 | 9,000,000 | 80.71 |
| T8 | 27,639,164 | 76.69 | T16 | 27,263,742 | 75.47 |
| B8 | 29,000,000 | 80.78 | B16 | 29,000,000 | 80.31 |
| T9 | 34,796,280 | 73.64 | T17 | 14,979,572 | 76.92 |
| B9 | 34,920,646 | 80.44 | B17 | 16,000,000 | 80.47 |
| T12 | 49,728,150 | 76.07 | T20 | 6,275,176 | 73.44 |
| B12 | 53,000,000 | 81.64 | B20 | 7,000,000 | 80.15 |
| Total | 293,103,118 | Total | 148,042,214 | ||
T denotes the top internodal samples while B denotes the bottom internodal samples. The number represents the genotype code as listed in Table 1.
Global expression analysis of sugarcane RNA-Seq data
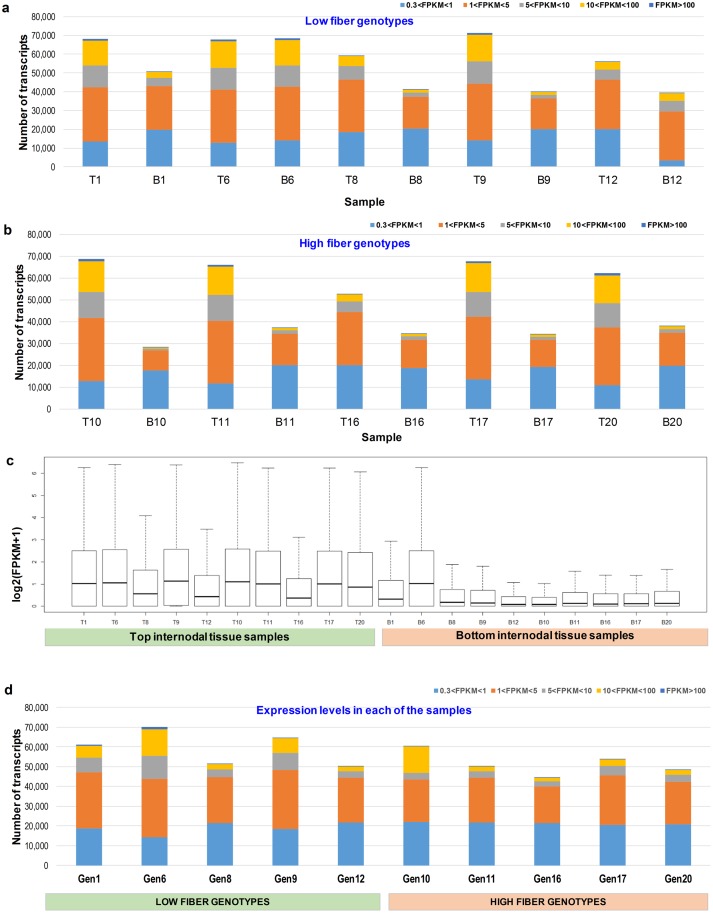
In this analysis, all transcript isoforms were considered, but only transcripts with FPKM greater than 0.3 were counted as significantly expressed as suggested in [62] for RNA-Seq data. Overall, amongst the 20 samples studied, the number of transcript isoforms having FPKM ≥0.3 ranged from 29,339 to 71,156. The total profile of all transcripts expressed at FPKM of different ranges of 0.3–1, 1–5, 5–10, 10–100 and above 100, is presented in Fig 1a and 1b. As expected, there were more transcripts expressed in the top internodal samples (representing the young tissues) compared to that in the bottom internodal samples (representing the mature tissues). There was one exception in the case of genotypes 6 where a similar expression level in the top and bottom internodal samples was observed. This may indicate that the transcriptome expression was still similarly active along the culm in this genotype. In 8/10 genotypes, it was observed that the top internodal tissue samples had a lower proportion of transcript with 0.3<FPKM<1 (representing less active transcripts) and higher proportion of transcripts with FPKM>1 (representing more active transcripts) compared to the bottom internodal tissue samples. It can be seen from the distribution of log2(FPKM+1) in Fig 1c that even for the same tissue type, samples exhibited different expression patterns. Fig 1d shows the global expression of pooled data where the top and bottom internodal samples of each genotype were pooled into a single sample (representing a genotype) prior to mapping to the reference transcriptome. There was not a clear pattern in the total transcript expression level between two groups of low and high fiber genotypes. The total transcripts expressed at FPKM ≥0.3 ranged from 44,698 to 70,064.
Fig 1. Global expression of samples used in this study.
(a) Transcript expression of top and bottom internodal samples of low fiber genotypes. (b) Transcript expression of top and bottom internodal samples of high fiber genotypes. The x-axis indicates the samples while the y-axis represents the number of transcripts expressed. (c) Boxplot of log2(FPKM+1) distribution amongst the top and bottom internodal samples form low and high fiber genotypes. The x-axis indicates the samples while the y-axis represents the log2(FPKM+1). The transcripts with log2(FPKM+1)>6.5 were not shown here. T denotes the top internode while B denotes the bottom internode. (d) Global expression of 10 pooled genotypes. Gen denotes genotype.
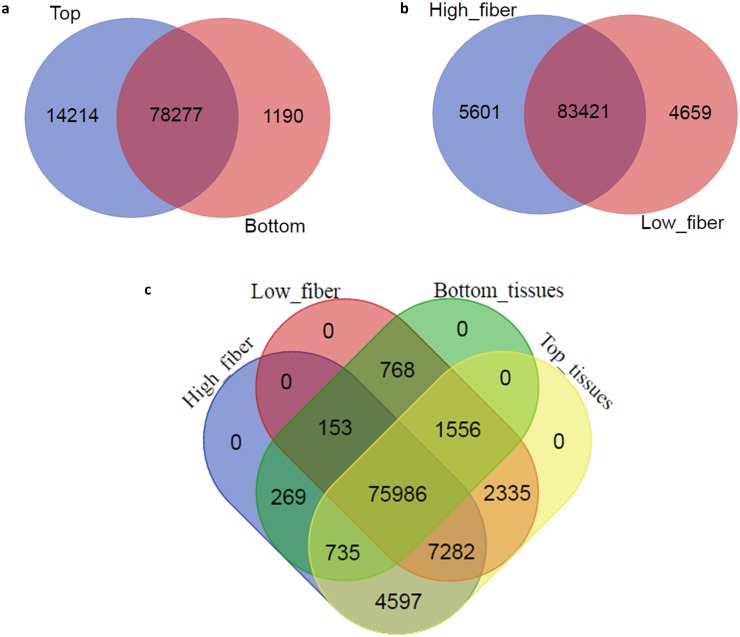
Fig 2a shows that, of the total 93,681 unique transcripts found expressed in all top and bottom internodal samples, 78,277 were common between the two, while 14,214 and 1,190 were uniquely expressed in the top and bottom tissues, respectively. When the total unique expressed transcripts were separated according to low and high fiber genotypes (Fig 2b), the majority of transcripts (83,421) were found to be common between the two groups, only 4,659 and 5,601 transcripts were unique to low fiber and high fiber groups, respectively. In general, when all the samples were considered, there was a similar number of transcripts expressed in the two groups of low and high fiber genotypes in this study. Fig 2c reveals that 75,986 expressed transcript isoforms were common between the 4 fiber content-based and tissue-based comparisons. Taken together these results and data presented in Fig 1, it is suggested that each individual sample had a fraction of unique transcripts that was not detected in the others. Since these samples were derived from the same tissue type, at the global expression level, this could mean that, if one transcript was expressed in sample 1 but not detected in sample 2, it could be that it was not expressed at all in sample 2, or it could mean that it was down-regulated in sample 2 to a low expression level that was not detected (this could also be affected by a low sequencing depth).
Fig 2. Comparison between the number of expressed transcripts (FPKM>0.3).
(a) Between all top and bottom internodal samples, (b) All low and high fiber genotypes and (c) Between the four sample groupings.
Transcript differential expression analysis
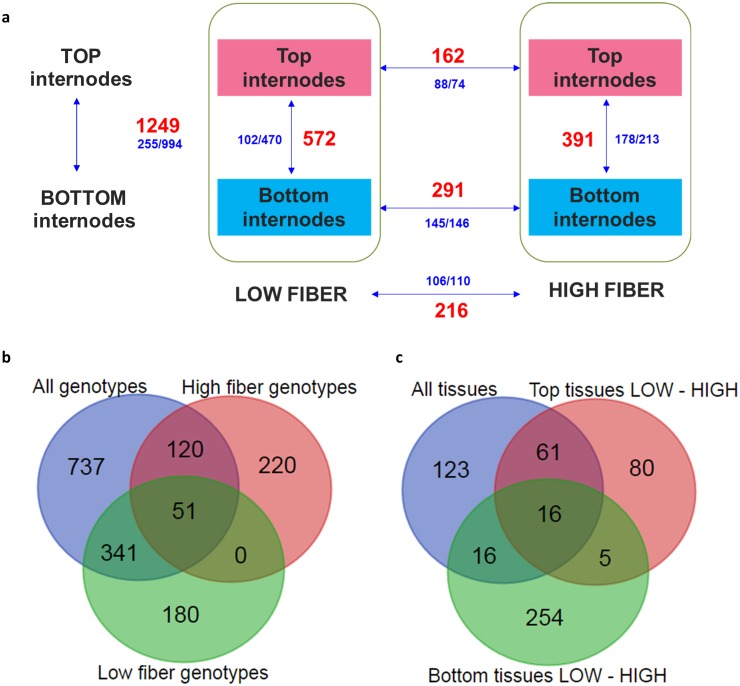
The differentially expressed (DE) transcripts identified by the package DEseq2 with a fold change ≥2 and FDR adjusted p-value ≤0.05 were summarized in Fig 3a. In total, 1,249 DE transcripts were found between all 10 top internodal samples and all 10 bottom internodal samples (referred to as all-genotypes T-B); 572 DE transcripts between the 5 top internodal samples and the 5 bottom internodal samples of the low fiber genotype group (low-fiber T-B); and 391 DE transcripts between the 5 top internodal samples and the 5 bottom internodal samples of the high fiber genotype group (high-fiber T-B), respectively. When compared among these 3 sets of identified transcripts, 51 were found to be common among the 3 comparisons, 341 common only between all-genotypes T-B and the low-fiber T-B, and 120 common only between all-genotypes T-B and the high-fiber T-B (Fig 3b). Amongst the low and high fiber groups, a total of 216, 291 and 162 transcripts were identified as DE transcripts between the pooled tissues of low and high fiber genotypes (referred to as all-tissues L-H), between the bottom internodal samples of the low and high fiber genotypes (bottom-tissues L-H), and between the top internodal samples of the low and high fiber genotypes (top-tissues L-H), respectively (Fig 3c). Of all identified DE transcripts among the 3 comparisons of low and high fiber genotypes, 16 were found to be common among the three, 16 were common between only all-tissues L-H and bottom-tissues L-H, while 61 were common between only all-tissues L-H and top-tissues L-H.
Fig 3. The summary of differentially expressed transcripts.
(a) Differentially expressed transcripts. The red numbers denote the total DE transcripts, while the corresponding blue numbers denote the up-regulated/down-regulated transcripts. (b) Comparison among DE transcripts from all genotypes top vs. bottom tissues, low fiber genotypes top vs. bottom tissues, and high fiber genotypes top vs. bottom tissues. (c) Comparison of DE transcripts from pooled all tissues low vs. high fiber genotypes, bottom tissues low vs. high fiber genotypes, and top tissues low vs. high fiber genotypes.
Transcript differential expression between the young and mature tissues in the sugarcane culm
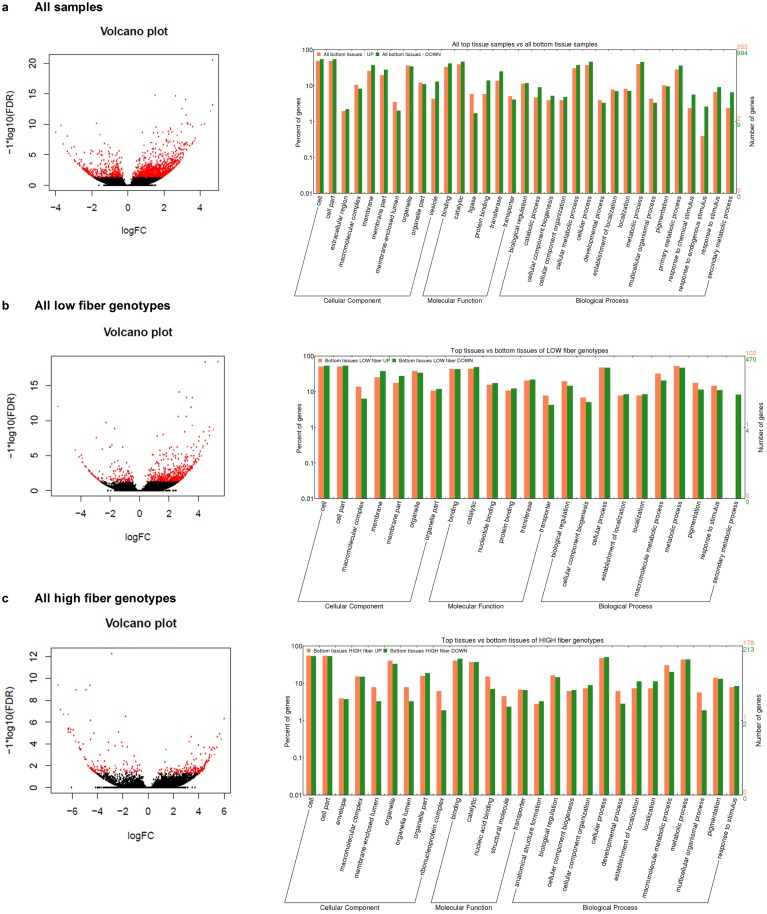
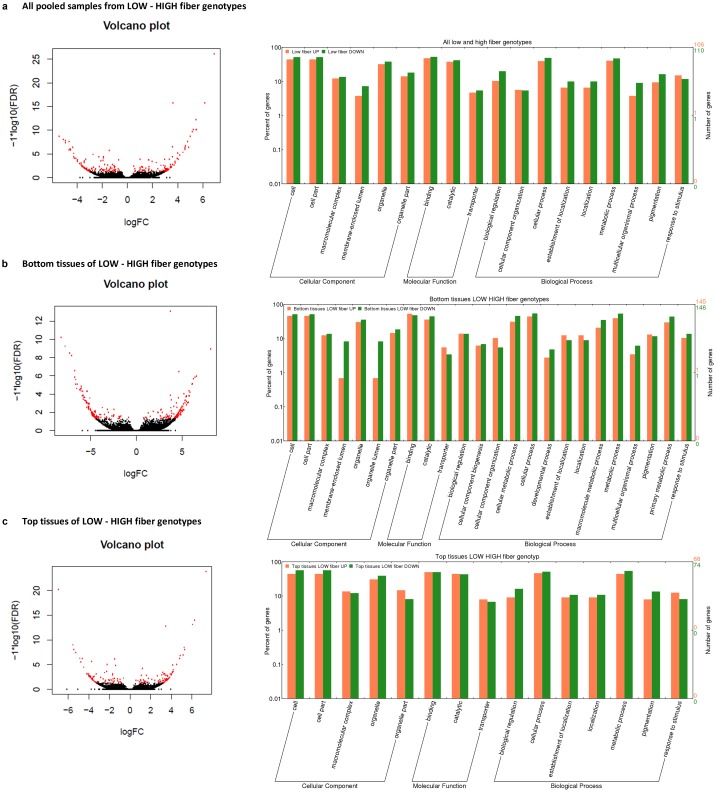
To provide an insight into the difference between the young and mature internodal tissues in the sugarcane culm, the identified DE transcripts derived from 3 tissue-based comparisons were investigated. The bottom internodal tissue samples were considered as the baseline (reference group) when compared to the top internodal tissue samples. Of the total 1,249 transcripts identified in the all-genotypes T-B comparison, 255 transcripts were up-regulated in the bottom tissues while 994 of those were down-regulated in the bottom tissues. Of 572 DE transcripts in the low-fiber T-B comparison, 102 and 470 transcripts were identified as up-regulated and down-regulated in the bottom internodal tissue samples, respectively. Of the 391 DE transcripts high-fiber T-B comparison, 178 and 213 transcripts were up-regulated and down regulated in the bottom internodal tissue samples, respectively. For all the 3 tissue-based comparisons, there were more down-regulated transcripts compared to up-regulated transcripts in the bottom tissues, which could be equivalent to the transcriptome in the top tissues being more active compared to that of the bottom tissues. Amongst all of the DE transcripts identified, there were transcripts with significant log2FC >4 (>16 fold) (Fig 4). In this analysis, since biomass accumulation is a highly regulated process that involves many quantitative trait loci (QTLs) [7, 14, 19, 22], and some of the transcripts were not well annotated, all GO terms that were associated with the up-regulation and down-regulation in the 3 comparisons were considered, enriched and highlighted the over representative functions in each comparison. This analysis revealed that the GO terms that were involved in the up-/ down-regulation between the top and bottom internodal tissues included those in various cellular components, molecular functions, and biological processes. The most abundant GO terms in the 3 comparisons were the cell and organelle part (in the cellular component category—CC), binding and catalytic (in molecular function category—MF), and cellular process and metabolic process (in biological process category—BP), shown in the right panel in Fig 4a–4c. When only significant GO terms that had p-value <0.05 were considered, in all-genotypes T-B comparison, the significant GO terms of the CC included those in vesicle GO:0031982 (down-regulated in the bottom tissues, hereafter referred to as “down”, otherwise, “up” for up-regulated), membrane GO:0016020 and membrane part GO:0044425 (down). Of the MF category, it was catalytic activity GO:0003824 (down), transferase activity GO:0016740 (down), ligase activity GO:0016874 (up), binding GO:0005488 (down), and protein binding GO:0005515 (down). Of the BP category, it was catabolic process GO:0009056 (down), secondary metabolic process GO:0019748 (down), cellular metabolic process GO:0044237 (down), primary metabolic process GO:0044238 (down), cellular process GO:0009987 (down), response to endogenous stimulus GO:0009719 (down), and response to chemical stimulus GO:0042221 (down). In the low-fiber T-B comparison, the significant GO terms were macromolecular complex GO:0032991 (down), membrane GO:0016020 (down), membrane part GO:0044425 (down) (in the CC); secondary metabolic process GO:0019748 (down) and macromolecule metabolic process GO:0043170 (down) (in the BP). In the high-fiber T-B comparison, the significant GO terms were membrane-enclosed lumen GO:0031974 (up), organelle lumen GO:0043233 (up), ribonucleoprotein complex GO:0030529 (up), organelle lumen GO:0043233 (up) (in the CC); nucleic acid binding GO:0003676 (up) (in the MF); macromolecule metabolic process GO:0043170 (up) and multicellular organismal process GO:0032501 (up) (in the BP).
Fig 4. The volcano plots and GO term over-representation analysis between the top and bottom internodal tissues.
(a) All genotypes, (b) Low fiber genotypes and (c) High fiber genotypes. In the left panel, the red dots denote the significant DE transcripts at FDR adjusted p-value <0.05, while the black dots denote the non-significant transcripts between the two cases. In the bar chart (right panel), the right y-axis indicates the number of transcripts for each GO term, while the left y-axis indicates the percentage of transcripts in each GO main category.
The GO term analysis was consistent with the results in Mapman annotation (S2 Table and S1 Fig). Various classifications were found to be involved in the up- and down-regulated DE transcripts. Most function-assigned transcripts were down-regulated in the bottom internodal tissues or up-regulated in the top internodal tissues (blue color), apart from a proportion of DE transcripts were not assigned with a function (red color, bin 35, accounting for 27–32% of total DE transcripts). The most significant classifications included bins 29 (protein), 16 (secondary metabolism), 26 (miscellaneous enzyme families), 30 (signaling), 27 (RNA, including transcription factors), 34 (transport), 20 (stress), 10 (cell-wall related), 13 (amino acid metabolism), 11 (lipid metabolism), 31 (cell), and 17 (hormone metabolism). The important transcripts that were involved in sugar and fiber accumulation included those that were classified in bins of photosynthesis (bin 1), carbohydrates (bin 2 and 3), cell-wall metabolism (cellulose, hemicellulose and sugars—bin 10), and secondary metabolism (including lignin pathway—bin 16).
There were 151 transcripts (within the total 1,649 unique DE transcripts from 3 comparisons) that were directly involved in the accumulation of sugars and fiber, as summarized in Table 3, details in S3 Table. Amongst the identified DE transcripts, there were transcripts associated with carbohydrate metabolism (9 transcripts), photosynthesis (25), cell-wall proteins (8), cellulose synthesis (29), hemicellulose synthesis (6), cell-wall modification (3), cell-wall precursors (6), lignin biosynthesis (58) and dirigent proteins (7). Notably, for cellulose-related transcripts, 24 transcript isoforms were annotated as CesA and CesA-like proteins and 4 as COBRA-like protein precursor. Hemicellulose-related transcripts were those associated with endo-1,4-beta glucanase, plant glycogenin-like starch initiation protein 1 (PGSIP1), GT43 family glycosyltransferases, IRX14 and IRX9 genes. The transcripts for cell-wall precursors included those of UDP-glucose 6-dehydrogenase (EC: 1.1.1.22), UDP-glucose/GDP-mannose dehydrogenase and UDP-XYL synthase 6 (UXS6). Notably, there were a number of transcript isoforms involved in the monolignol metabolism, including 18 transcripts for phenylalanine ammonia lyases—PAL (EC: 4.3.1.24) and PAL (EC: 4.3.1.25), 7 transcripts for 4-coumarate CoA ligase (4CL, EC: 6.2.1.12), 12 transcripts for cinnamoyl CoA reductase (CCR, EC: 1.2.1.44), 3 transcripts for p-coumaroyl shikimate/quinate 3-hydroxylase (C3H, EC: 1.14.13.36), 3 transcripts for caffeic acid/5-hydroxyferulic acid O-methyltransferase—(COMT, EC: 2.1.1.68), 2 for cinnamyl alcohol dehydrogenase (CAD, EC: 1.1.1.195), 10 for caffeoyl CoA O-methyltransferase (CCoAOMT, EC:2.1.1.104) and 1 for ferulate-5-hydroxylase (F5H, EC 1.14.13.-).
Table 3. Differentially expressed transcripts involved in the accumulation of sugars and fiber between the top and bottom tissues in the sugarcane culm, based on MapMan annotation.
| Function classification | Number of DE transcripts |
|---|---|
| Lignin pathway | 58 |
| Cellulose synthesis | 29 |
| Photosynthesis | 25 |
| Carbohydrate metabolism | 9 |
| Cell-wall proteins | 8 |
| Dirigent proteins | 7 |
| Cell-wall precursors | 6 |
| Hemicellulose synthesis | 6 |
| Cell-wall modification | 3 |
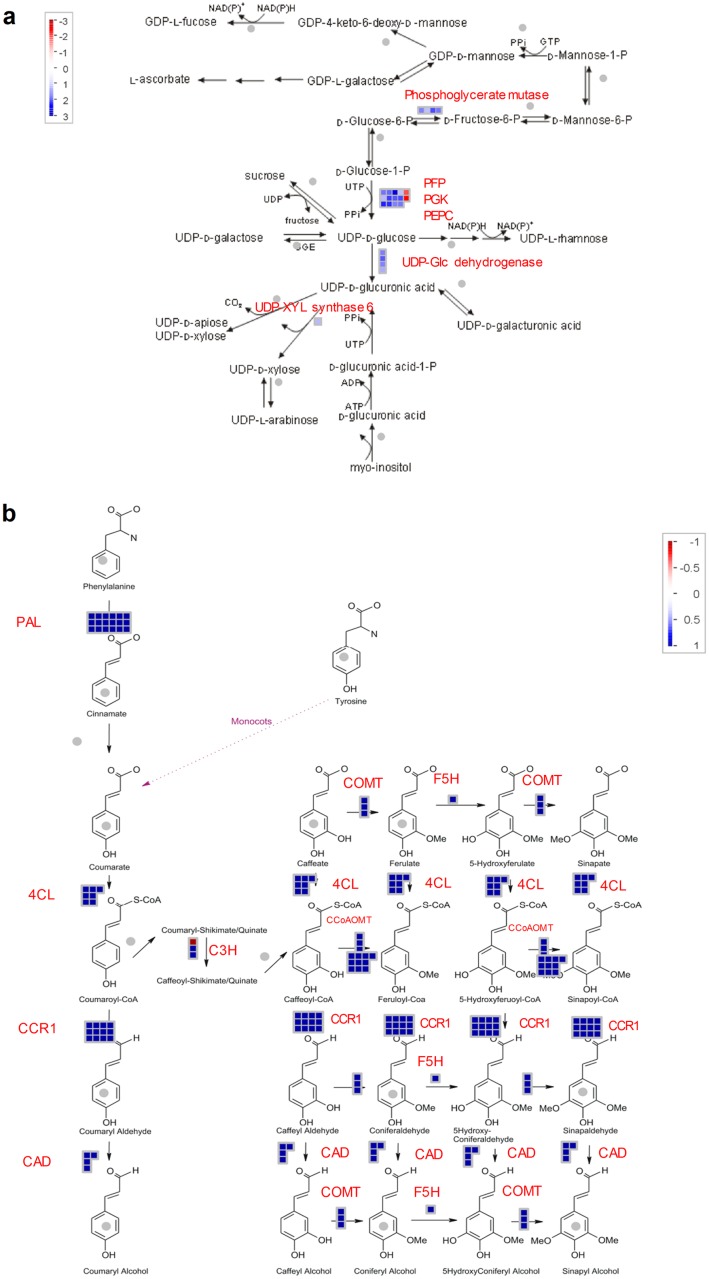
Of the 151 identified transcripts related to sugar and fiber accumulation, all except 5 transcripts, were down-regulated in the bottom internodal tissue. There were 9 DE transcripts that were common among the 3 comparisons, including CesA, IRX9 gene, 4CL-1, two CCR isoforms, and four PAL isoforms. There were more DE transcripts that were common between all-genotypes T-B and only the low-fiber T-B comparisons (40 transcripts), than between all-genotypes T-B and only the high-fiber T-B comparisons (6 transcripts). Fig 5 shows the DE transcripts from the all-genotype T-B comparison, which involved 2 important pathways related to cell-wall precursor metabolism and the lignin pathway; while S2 Fig shows DE transcripts that were annotated as being involved in photosynthesis. The majority of the transcripts in these 3 pathways were down-regulated in the bottom internodal tissues / or up-regulated in the top internodal tissues samples.
Fig 5. Differentially expressed transcripts between the top and bottom tissues in the sugarcane plant.
(a) Cell-wall precursor metabolism. (b) Lignin pathway. Log2FC was used, red color denotes the up-regulated in the bottom internodal tissues while blue color denotes the down-regulated in the bottom internodal tissue. Colored boxes indicate the fold change. Each heatmap is representative of one of the identified differentially expressed transcripts.
Transcript differential expression between low and high fiber sugarcane genotypes
In general, there were fewer DE transcripts identified in this fiber content-based comparison compared to that of the tissue-based comparison. Of 216 total identified DE transcripts from the pooled all-tissues L-H comparison, 106 transcripts were up-regulated and 110 were down-regulated in the bottom tissues of the low fiber group (or up-regulated in the high fiber group). Of the 291 DE transcripts from the bottom-tissues L-H comparison, 145 and 146 transcripts were up- and down regulated in the low fiber group. Of 162 DE transcripts from the top-tissues L-H comparison, 88 and 74 transcripts were up- and down-regulated, respectively, in the top internodal tissue samples of the low fiber group. Fig 6 summarizes the identified DE transcripts between the three comparisons, including those transcripts with significant log2FC >4. The GO analysis also indicated that identified DE transcripts were involved many cellular components, molecular functions and biological processes. This also suggests that the up/down-regulation also involved the same molecular functions and biological processes (Fig 6a–6c).
Fig 6. The volcano plots and GO term over-representation analysis between the low and high fiber genotypes.
(a) All tissues from all genotypes, (b) Bottom tissues from low and high fiber genotypes, and (c) Top tissues from low and high fiber genotypes. The red dots denote the significant DE transcripts, while the black dots denote the non-significant transcripts between the two cases. In the bar chart (right panel), the right y-axis indicates the number of transcripts for each GO term, while the left y-axis indicates the percentage of transcripts in each GO main category.
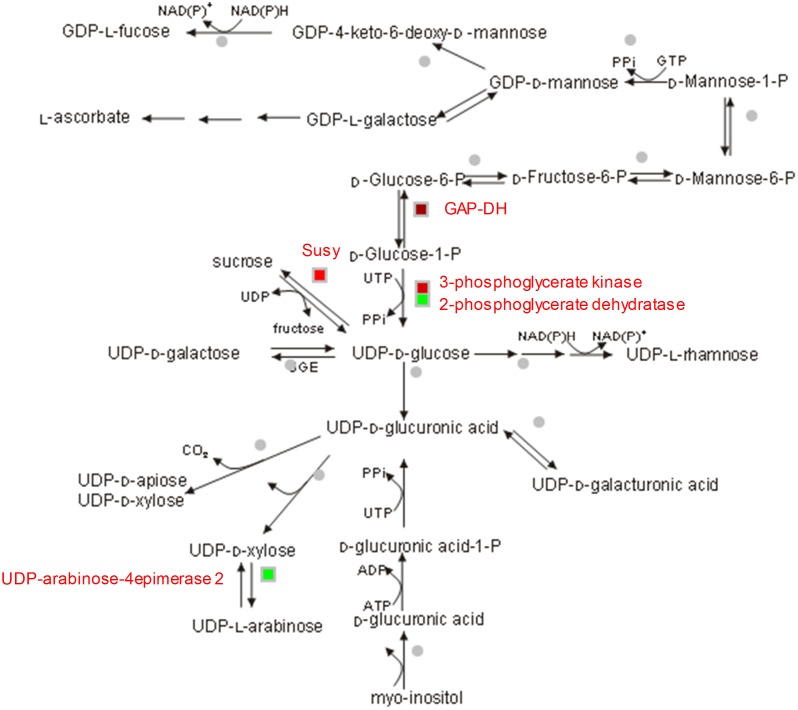
The significant GO term that had a p-value <0.05 in all-tissues L-H comparison was catabolic process GO:0009056, which belonged to the metabolic process in the BP category (up in low fiber genotypes). In the bottom-tissues L-H comparison, the significant GO terms were membrane-enclosed lumen GO:0031974 (down in the bottom tissues of the low fiber genotypes), organelle lumen GO:0043233 (down) (in the CC); metabolic process GO:0008152 (down), macromolecule metabolic process GO:0043170 (down), cellular metabolic process GO:0044237 (down), and primary metabolic process GO:0044238 (down) (in the BP). In the top-tissues L-H comparison, none were significant at p-value <0.05, due to a low number of DE transcripts. The Mapman annotation (S2 Table and S3 Fig) suggested that large proportions of the DE transcripts were attributed to bins 29 (protein), 27 (RNA), 20 (stress), 34 (transport), 16 (secondary metabolism), 31 (cell), 33 (development), 26 (miscellaneous enzyme families), 21 (reduction-oxidation regulation), 30 (signaling), 17 (hormone metabolism) and 13 (amino acid metabolism). A summary of the 17 transcripts that were involved in the sugar and fiber metabolism from MapMan annotation is presented in Table 4. These were derived from the total of 555 unique DE transcripts from the 3 fiber content-based comparisons. The up-regulated transcripts (in the low fiber genotypes) were callose synthase (GSL12) (log2FC = -3.031), alpha-amylase precursor (EC 3.2.1.1) (isozyme 1B) (log2FC = -3.223), photosynthesis helix protein (log2FC = -3.567), fructose-bisphosphate aldolase (log2FC = -4.497), UDP-arabinose 4-epimerase 2 (log2FC = -4.606), 2-phosphoglycerate dehydratase 1 (log2FC = -4.331), and CCoAOMT-5 (log2FC = -4.294). The down-regulated transcripts were glucose-1-phosphate adenylyltransferase large subunit 1 (log2FC = 1.050), granule-bound starch synthase 1b (log2FC = 2.525), sucrose synthase 1 (Susy, EC 2.4.1.13) (log2FC = 4.507), proteasome maturation factor UMP1 (log2FC = 3.575), serine hydroxymethyltransferase (logFC = 1.028), 3-phosphoglycerate kinase (PGK) (log2FC = 2.533), CesA6 (log2FC = 4.515), glyceraldehyde 3-phosphate dehydrogenase (GAP-DH) log2FC = 1.874, CAD (log2FC = 2.294), and COMT (log2FC = 6.619). In addition, 6 dirigent proteins were up-regulated in the high fiber genotypes compared to the low fiber genotypes, having been detected in both all-tissues L-H and bottom-tissues L-H comparisons. The transcripts associated with these proteins had a length ranging from 573 bp to 1,013 bp and with a log2FC ranging from 2.8 to 5.5 (8 to 30-fold). Fig 7 shows the identified DE transcripts involved in cell-wall metabolism.
Table 4. Differentially expressed transcripts involved in sugar and fiber accumulation between the low and high fiber genotypes.
| Function classification | TSA accession | Length (bp) | Description | All tissues (1) | Bottom tissues (3) | Top tissues (2) | |||
|---|---|---|---|---|---|---|---|---|---|
| log2FC | padj | log2FC | padj | log2FC | padj | ||||
| Carbohydrate metabolism | |||||||||
| CHO metabolism | GFHJ01020794.1 | 5097 | Similar to callose synthase; glucan synthase-like 12 (GSL12) | -3.031 | 4.55E-02 | - | - | -3.281 | 4.51E-02 |
| CHO metabolism | GFHJ01023689.1 | 5963 | Alpha-amylase precursor (EC 3.2.1.1) (Isozyme 1B) | - | - | - | - | -3.223 | 4.90E-02 |
| CHO metabolism | GFHJ01037300.1 | 4596 | Glucose-1-phosphate adenylyltransferase large subunit 1 | 1.05 | 3.79E-02 | - | - | - | - |
| CHO metabolism | GFHJ01012375.1 | 1318 | Granule-bound starch synthase 1b, chloroplast precursor (EC 2.4.1.242) | 2.525 | 1.21E-02 | 2.676 | 2.45E-02 | - | - |
| CHO metabolism | GFHJ01015836.1 | 2545 | Sucrose synthase 1 (EC 2.4.1.13) (Sucrose-UDP glucosyltransferase 1) | - | - | 4.507 | 3.77E-02 | - | - |
| Photosynthesis | |||||||||
| Photosynthesis | GFHJ01093755.1 | 727 | Encodes a one helix protein | - | - | - | - | -3.567 | 8.16E-03 |
| Photosynthesis | GFHJ01031690.1 | 1385 | Fructose-bisphosphate aldolase (ALDP) (EC 4.1.2.13) | - | - | -4.497 | 2.47E-02 | - | - |
| Photosynthesis | GFHJ01079929.1 | 562 | Proteasome maturation factor UMP1 | 3.575 | 4.31E-03 | - | - | - | - |
| Photosynthesis | GFHJ01014544.1 | 1813 | Serine hydroxymethyltransferase, mitochondrial precursor (EC 2.1.2.1) | 1.028 | 5.19E-03 | - | - | - | - |
| Photosynthesis | GFHJ01011598.1 | 1658 | Phosphoglycerate kinase, cytosolic (EC 2.7.2.3) | - | - | 2.533 | 2.29E-02 | - | - |
| Cell-wall metabolism | |||||||||
| Hemicellulose synthesis | GFHJ01016359.1 | 1924 | UDP-arabinose 4-epimerase 2 (EC 5.1.3.5) | - | - | -4.606 | 2.29E-02 | - | - |
| Glycolysis | GFHJ01067367.1 | 835 | 2-phosphoglycerate dehydratase 1 (EC 4.2.1.11) | - | - | -4.331 | 3.77E-02 | - | - |
| Cellulose synthesis | GFHJ01021631.1 | 1495 | Encodes a cellulose synthase isomer, related to CESA6 | 4.515 | 1.57E-06 | - | - | 4.462 | 3.29E-05 |
| Glycolysis | GFHJ01029222.1 | 1644 | Chloroplast/plastid localized GAPDH isoforms | - | - | 1.874 | 4.19E-02 | - | - |
| Lignin pathway | |||||||||
| Lignin biosynthesis | GFHJ01052729.1 | 1079 | Caffeoyl-CoA O-methyltransferase 5 CCoAOMT | - | - | -4.294 | 4.32E-02 | - | - |
| Lignin biosynthesis | GFHJ01085183.1 | 589 | Cinnamyl alcohol dehydrogenase (CAD) | 2.294 | 3.60E-05 | - | - | 2.48 | 4.29E-04 |
| Lignin biosynthesis | GFHJ01052435.1 | 1358 | O-methyltransferase ZRP4 (EC 2.1.1.104) (COMT) | - | - | 6.619 | 2.47E-06 | - | - |
| Dirigent proteins | |||||||||
| GFHJ01087645.1 | 573 | Dirigent protein | 3.665 | 3.24E-03 | 8.254 | 5.90E-11 | - | - | |
| GFHJ01086725.1 | 741 | Dirigent protein | 5.454 | 1.96E-09 | 7.791 | 5.48E-10 | - | - | |
| GFHJ01076697.1 | 754 | Dirigent protein | 3.431 | 9.29E-03 | 4.389 | 3.66E-02 | - | - | |
| GFHJ01071693.1 | 872 | Dirigent protein | 2.885 | 1.31E-02 | 3.235 | 4.19E-02 | - | - | |
| GFHJ01078847.1 | 931 | Dirigent protein | 3.223 | 2.27E-02 | 5.211 | 4.92E-03 | - | - | |
| GFHJ01086396.1 | 1013 | Dirigent protein | 3.368 | 1.21E-02 | 5.412 | 2.97E-03 | - | - | |
Negative log2FC values indicate that up-regulated in the low fiber group, while positive log2FC values indicate up-regulated in the high fiber group. padj: adjusted p-value.
(1) Between all pooled samples from low fiber genotypes and high fiber genotypes.
(2) Between top tissues of low and high fiber genotypes.
(3) Between bottom tissues of low and high fiber genotypes. Grey colour indicates the transcripts that were only detected in the pooled sample comparison or only in one comparison.
Fig 7. Differentially expressed transcripts involved in cell-wall precursor metabolism between the low and high fiber genotypes.
Log2FC was used, green color denotes the up-regulated transcript in the low fiber group while red color denotes the down-regulated transcript in the low fiber group. Each heatmap is representative of one of the identified differentially expressed transcripts.
Differentially expressed gene validation by qPCR
To confirm the gene expression measured by RNA-Seq analysis, 8 putative genes differentially expressed between the young and mature tissues and between the low and high fiber genotypes were validated by qPCR. These include genes encoding COBRA-like 5 protein precursor, CAD, CCR, CESA, CESA-like C5, dirigent protein, a putative family 43 glycosyl transferase (IRX9 gene) and sucrose synthase 1 (selected from Table 4 and S3 Table). The reliability of the RNA-Seq data was confirmed by qPCR of 8 selected DE genes, in which each selected gene was shown as a single band at their respective size (data not shown). A significant correlation (r = 0.54, p<0.001, n = 64, df = 62) was found between the two data sets (S4 Table).
Discussion
This study represents an effort to identify the transcripts involved in sugar and fiber accumulation by conducting differential expression analysis between the young and mature internodal tissues in the sugarcane plant, and between two groups of low and high fiber genotypes. The comparison between the young and mature internodes could potentially pinpoint the transcripts that are associated with carbon partitioning to the major biomass components in the sugarcane culm over time. The comparison between the low and high fiber genotypes could reveal the transcripts associated with sugar and fiber accumulation between the two groups. It is important to note that the two groups of low and high fiber genotypes used in this study were respectively equivalent to two groups of high and low sugar genotypes, since on a dry biomass basis, these two components were negatively correlated [4]. Overall, 4 out of 5 genotypes in the low fiber group were commercial sugarcane genotypes, while 4 out of 5 genotypes in the high fiber group were high fiber genotypes derived by introgression of genes from the wild Saccharum spontaneum relatives. The fiber content in these genotypes ranged from 31% to 50% of total dry mass (total solids which include sugars, fiber and others in the sugarcane culm biomass).
Transcript differential expression between the young and mature tissues
According to the literature, sugar and fiber (cellulose, hemicellulose and lignin) in sugarcane are largely developmentally regulated, and consequently, it is expected that genes/transcripts involved in their biosynthesis are also developmentally regulated. In the sugarcane culm, the cell elongation and primary cell-wall deposition commences in internode 1, followed by the deposition of secondary cell-wall in internode 2, and then suberisation and sucrose accumulation [20]. Lignification has also been shown to start early in internode 1 [63], and continues to increase until the fifth or sixth internode. After that, the lignin content appears to be similar between tissues [20, 22]. At the early developmental stage, the internode acts as strong sink for sucrose, supporting the cell-wall synthesis and cell expansion, without an increase in sucrose concentration [14]. The accumulation of sucrose happens later, once the elongation ceases in maturing internodes and reaches a maximum in mature internodes [63]. After this, a major increase in cell-wall thickening (internode elongation) and lignification in the maturing tissues has been reported [6]. It is, therefore, the transcript expression associated with these changes in the culm development that is expected to be regulated, accordingly.
Early studies on Arabidopsis thaliana showed that CesA1, CesA2, CesA3, CesA5, CesA6 and CesA9 function in the biosynthesis of the primary cell-wall [64, 65], while CesA4, CesA 7 and CesA8 take part in the biosynthesis of the secondary cell-wall [66, 67]. Most CesA transcripts are down-regulated in the mature internodes compared to the young internodes, which reflects their roles in this type of tissues of the plant [68], however, CesA3 and CesA5 were found to be up-regulated, which is thought to be necessary for cell-wall maintenance [14, 20] or could be for radial growth in the mature tissue. The pattern of expression of the CesA-like transcripts, on the other hand, being highly abundant in the immature tissues, have been reported to not follow the pattern observed for the CesA in the culm [20]. A recent study [14] suggests that primary cell-wall synthesis in the sugarcane culm could occur throughout the sugarcane culm but is particularly important in the storage parenchyma of the maturing culm, where the internode is fully expanded and actively storing sucrose.
In this study, most of the 151 identified DE transcripts directly involved in the sugar and fiber metabolism, were up-regulated in the top internodal tissues compared to the bottom internodal tissues. These transcripts included those associated with the major carbohydrates, photosynthesis, several CesA and CesA-like transcripts, hemicellulose, cell-wall proteins, cell-wall precursors, major enzymes in the lignin pathway and dirigent proteins. The up-regulation in this tissue-based comparison represents the active growth and metabolism in the young internodal tissues where all the transcripts involved were highly expressed, compared to the less active mature internodal tissues of sugarcane. The top internodal samples were derived from the fourth internodes from the top, while the bottom internodal samples were from the third internode from the bottom of 12-month old sugarcane plants. Considering that each sample was pooled from four different plants and that there was plant-to-plant transcript expression variation, this could mean that the top internodal samples represented both the immature and maturing tissues, while the bottom internodal samples could represent mature tissues. Therefore, there would be highly expressed transcripts that regulate the cell-wall synthesis, lignification and sugar accumulation in the top internodal samples, whereas, these would be less active in the bottom internodal samples where most processes would be stopped or slowed down. That would also explain the up-regulation of the dirigent proteins and down-regulation of only 5 transcripts in the top internodal tissues compared to the bottom tissues. The dirigent proteins are hypothesized to play roles in scaffolding of lignin and biosynthesis of lignan [19, 69, 70] and dirigent domain-containing proteins are involved in the patterning of lignin-based Casparian strip in the root of A. thaliana [71]. However, some studies argued that lignin biosynthesis may not be handled by dirigent proteins, since lignin is not optically active and lignin biosynthesis is chemically controlled [72–74]. These proteins and a group of lignin-related enzymes (PAL, COMT and CCoAOMT) were found to be up-regulated in the maturing tissues when compared to the immature [19].
Differential expression between the low and high fiber genotypes
This study identified 23 transcripts (including 6 dirigent proteins), out of 555 unique DE transcripts between low and high fiber genotypes, which were directly involved in sugar- and fiber-related pathways. The fewer DE transcripts in the fiber content-based analysis compared to the tissue-based analysis could reflect that there were many DE transcripts between these 2 groups whose fold change was at a relatively low level and which was not detected as differential expression. The cutoff for differential expression in this study was set at a minimum of two-fold, while the increase/decrease in fiber content between the two groups of low and high fiber genotypes could be from a combination of many up- or down-regulations involving transcript isoforms at a fold change less two-fold cutoff, as in Fig 6. In the previous tissue-based analysis, the difference between the samples was attributed to the developmental stages in which some of the pathways were significantly active/inactive between the stages, while in this fiber content-based comparison, it could be that all pathways were comparably active between the two groups being compared, except those were identified as DE. These identified DE transcripts could reflect the most important enzymes and pathways that played key roles in the processes making the difference in the fiber content. These included a number of transcripts that involved in carbohydrate metabolism, photosynthesis, cell-wall metabolism, monolignol metabolism and dirigent proteins, as presented in Table 4. The other possibility could be that the selected genotypes were not very contrasting in terms of fiber content, resulting in a narrow difference between the expression of the low and high fiber genotypes. These factors, together with the multiple genes/transcripts controlling the accumulation of sugars and fiber as discussed earlier, could explain the low number of DE detected in this analysis.
In relation to carbohydrate metabolism, callose synthase and alpha-amylase were up-regulated in the top tissues of the low fiber compared to that of the high fiber genotypes, while granule-bound starch synthase 1b and sucrose synthase 1 (Susy) were down-regulated in the bottom tissues of low fiber genotypes. Callose synthases are known to regulate the biosynthesis of callose, a cell-wall polysaccharide found in many higher plant species. Callose is a β-1,3-glucan, and has important roles in many developmental processes, including cell division and growth, tissue differentiation, cell plate formation, pollen development, plasmodesmata and response to stress [75–77]. Alpha-amylase on the other hand, plays roles in the starch degradation in the plant, breaking down the starch for other enzymes to act [78]. It is active when the stored carbohydrates are diverted back to the metabolism when it is required for plant development. Granule-bound starch synthase 1b is responsible for synthesis of starch (amylose) and the final structure of amylopectin [79], while Susy is the major enzyme of sucrose metabolism in sugarcane. The results indicate that, while all metabolic processes were probably happening in tissues of both groups of low and high fiber genotypes, there could be more active processes related to hemicellulose synthesis and starch degradation in the top tissues of low fiber, and more of starch synthesis and sucrose metabolism-related processes in the bottom tissues of high fiber genotypes.
With respect to photosynthesis, there was one up-regulated transcript identified in the top internode of low fiber which associated with one helix protein homologous to cyanobacterial high-light inducible protein. Between the bottom tissues of low and high fiber genotypes, ALDP fructose-bisphosphate aldolase was up-regulated while phosphoglycerate kinase (PGK) was down-regulated in the bottom tissues of low fiber genotypes. ALDP fructose-bisphosphate aldolase is one of the enzymes of the Calvin cycle and is predicted to have the potential to control photosynthetic carbon flux and biomass yields [80], while PGK catalyzes 1,3-biphosphogrycerate and ADP to form 3-phosphoglycerate and ATP [81]. Taking this together with 25 transcripts that were annotated as being associated with photosynthesis in tissue-based analysis (see S1 Fig), it was surprising to detect transcripts belonging to photosynthesis pathway in these samples, since they were derived from the sugarcane culms, a mostly non-photosynthetic tissue, and the rind of the culms was removed during sample excision. None of the data in the literature implies that significant photosynthesis occurs in this type of tissue. Photosynthesis requires a functional photosystem (PS) II and PSI system, a carbon fixation and the reductive pentose phosphate (RPP) pathway [82, 83]. It is noteworthy that the RPP pathway is present in all plastids, which are found in sugarcane culms, as it is crucial for many vital reactions. Some of the enzymes detected are part of the RPP pathway and this could be explained in terms of their roles in converting imported triose-P to starch, lipids and other cellular constituents in the sugarcane culm [9]. However, there were some up-regulated photosynthetic enzymes detected in the DE analysis which belonged to the PSI and PSII, which cannot be explained without further research. It could also be that the photosynthesis was located in the green rind of the sugarcane culms which were not completely removed from the samples.
In the cell-wall metabolism, there were two transcripts (UDP-arabinose 4-epimerase and 2-phosphoglycerate dehydratase) up-regulated in the bottom tissues of low fiber genotypes. UDP-xylose 4-epimerase is the enzyme that catalyzes a reversible reaction between UDP-arabinose and UDP-xylose [84], while 2-phosphoglycerate dehydratase catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate [85]. CesA6 was found down-regulated in the top tissues of low fiber genotypes, and glyceraldehyde-3-phosphate dehydrogenase (GAP-DH) was down-regulated in bottom tissues of low fiber genotypes. CesA6 is critical for cell elongation [86]; and GAP-DH, which can be induced or repressed in sugarcane under abiotic stress, i.e. salt stress tolerance [87], was previously reported to be highly abundant in maturing culms [18]. This result indicates that, apart from the similarly active processes between the two groups, processes related to internode elongation and cellulose synthesis were more active in the top tissues of the high fiber genotypes compared to the top tissues in the low fiber genotypes, while those related to hemicellulose synthesis were more active in the bottom tissues of the low fiber genotypes compared to that of the high fiber genotypes.
The down-regulation of the dirigent proteins and two enzymes in the lignin pathway (CAD and COMT), together with the up-regulation of the CCoAOMT-5 in the low fiber genotypes, reflect their functions in lignin biosynthesis may not the same at a given developmental stage. CAD and COMT are involved in the final steps in the monolignol biosynthesis, while, COMT and CCoAOMT also take part in the production of phenolic compounds [21]. Therefore, while they were present in the lignin pathway, the up- or down-regulation of these enzymes could also be interlinked to other pathways. In comparison of the low and high lignin sugarcane genotypes, PAL-3 and C4H-1 were up-regulated in the low lignin genotype while COMT-1 was up-regulated in high lignin genotype [21]. Only two transcripts related to lignin, C4H and CCoAOMT, were found up-regulated in a study on 11 high-lignin guineagrass genotypes [88]. As discussed in [21], after gathering the results of differential expression of several lignin genes, it was suggested that lignin deposition might be regulated at the transcriptional level, but also the post-transcriptional levels and by enzyme catalytic activities. RNA-Seq data might not reflect all events in the lignin pathway, and therefore the studies at post-transcriptional levels (i.e. proteome and metabolome) would be required to understand further lignin metabolism in the sugarcane plant.
Notes on variation amongst diverse genotype collection and validation of RNA-Seq results
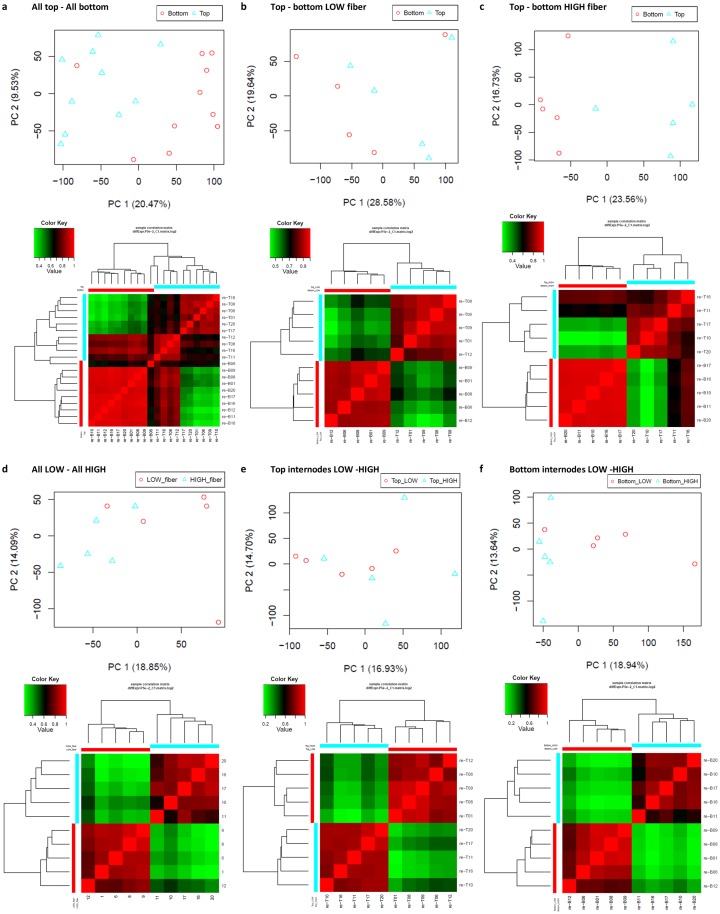
Initially, a diverse collection of 40 samples (from 20 genotypes) were included in the design of this experiment, and it was hypothesized that even though there were dynamic expression patterns and noises within the sample groups of low and high fiber genotypes, the transcripts that play key roles in this difference would always be detected as up- or down-regulated consistently and distinct from the random noises. However, this hypothesis might only work with the traits of high heritability that are controlled by single genes or a few key genes, and with a detectable differential expression level, which can be distinguishable from sample noises. The DE transcript detection soft-wares or models, which are based on the FDR or p-values, normally favor the high fold change due to the un-certainty in reporting the DE transcripts of a low fold change, which could also be sample noises. Biomass traits such as those of sugar and fiber, as mentioned earlier, are controlled by a number of QTLs and the regulation is shown not to be straightforward. This could be reflected in the principal component analysis (PCA) of the transcript expression profiles of the samples. In this PCA, it is suggested that separating samples according to their tissues types (top–bottom tissues) or fiber content (low–high fiber) did not explain the major variation between the respective groups, and there were other factors/dimensions that contribute to the separation of the sample groups used in this study. It was shown in the scatter-plots (Fig 8), that the samples were not tightly clustered according to the tissue types nor fiber groups they belonged to. Overall, the separation according to their tissue types represented more the total variation in the data in comparison to that between the fiber groups, which agreed with the results where more DE transcripts were observed for the tissue-based comparisons than that of the fiber content-based comparisons. The first component (PC1) explained 20–29% of the total variation between the top and bottom internodal tissue groups, while the second component (PC2) accounted for 9–20% of the total variation. Between the low and high fiber groups, the PC1 explained 17–19% of total variation and the PC2 explained 13–15%. Notably, two groups of top and bottom tissue samples were separated into two clusters, except for samples from genotype 6 whose bottom internodal sample was situated in the same cluster as its top internodal sample (top panels Fig 8a and 8b). This was consistent with the global expression analysis which showed these two samples had similar expression patterns, which differed from the other genotypes whose bottom internodal samples had lower expression levels compared to the counterpart top internodal samples. There were less clear clusters between the two groups of low and high fiber genotypes. Overall, the two components explained about 30–48% total variation between the top and bottom internodal tissue samples, and ~32% of total variation between the low and high fiber genotypes. This agrees not only with the assumption that there were multiple factors responsible for the variation between the groups studied in this analysis, but also with the patterns in the dendrograms of Pearson’s pairwise divergence between the groups. The PCA and Pearson analysis could also suggest that there was variation amongst the samples in each group of tissue types or fiber content (bottom panels Fig 8a–8c and 8d–8f). This could be due to the fact that the genotypes were derived from different parental backgrounds, hence each of the genotypes had a unique expression pattern, possibly reflecting their chromosome number and the allelic contribution. The expression of each allele in the homologous group could vary in different environment, genotype or genotype-specific ripening time, which even when the samples were harvested at the same point of time since planting, might not have corresponded to developmental stage, transcriptome-wise. It would be also that the same fiber content in sugarcane results from the up- or down-regulation of different sets of transcripts, which could account for the same contribution towards the increasing/decreasing in fiber content. Using the correlation analysis, the samples whose expression patterns appeared to be very different from the rest in each group were removed, retaining the final collection of 10 genotypes for which the results in this study was based on.
Fig 8. Principal component analysis (PCA) and correlation of expression patterns of the samples.
This was based on all transcripts in the transcriptome dataset. (a) Between all top and all bottom internodal samples. (b) Between top and bottom internodal samples of low fiber genotypes. c, Between top and bottom internodal samples of high fiber genotypes. (d) All pooled low and high fiber genotypes. (e) Between the top internodal samples of low and high fiber genotypes. (f) Between the bottom internodes of low and high fiber genotypes. The value of correlation score was used.
Differentially expressed genes identified by microarrays or RNA-Seq experiments have often been confirmed by qPCR analysis [89, 90]. However, this has been shown to be difficult and un-reliable, especially in polyploid genomes, such as the allohexaploid wheat genome [91]. This is due to the fact that polyploids contain homo(eo)logous and paralogous gene copies, resulting in expression of different but related transcripts (i.e., different isoforms sharing the same exons/introns), which are not discriminated by the technique. It is also likely to be the case for the complex polyploid sugarcane genome, in which multiple homo(eo)logous gene copies (predicted up to 12 [92]), multiple promoter variants and transcript isoforms are likely to exist [14]. Our results showed a significant correlation (r = 0.54, p <0.001) between the RNA-Seq and qPCR results. This is the expected result as a very close correlation would not be expected due to the large number of different closely related genes and transcript isoforms in the highly polyploid sugarcane genome. This could result in amplification of multiple unknown copies of similar genes to the target in the PCR. Poor confirmation rates have been also reported in various studies on polyploid species with inter-platform discrepancies [91]. Designing a homo(eo)logous- and paralogous-specific validation array is complicated [14], RNA-Seq provides a more reliable way to analyse gene expression in a complex polyploid genome like sugarcane, due to the ability to distinguish the many closely related gene transcripts. The RNA-Seq analysis in this study involves a high degree of replication and sample pooling within each treatment providing strong statistical support for the differentially expressed genes identified.
Conclusion
In this study, the newly constructed reference transcriptome, the SUGIT database, which was generated by the PacBio full-length isoform sequencing [24] was used for differential expression analysis. The results showed that 70 to 82% of the total RNA-Seq data to be aligned against this database, and 57–76% of the transcriptome had reads mapped to it, suggesting that the SUGIT database could be used as the reference database for transcript profiling and differential expression analysis. The proportion of RNA-Seq reads mapped is consistent with figures found in the literature, while the percentage of transcripts that had reads mapped indicated the proportion of transcripts that were expressed in the culm tissues out of the total transcriptome database derived from leaf, culm and root tissues.
The accumulation of sugar and fiber in sugarcane is a highly regulated process, and likely involves many genetic factors and QTLs. The differential expression analysis showed that there were many DE transcripts that were directly related to sugar and fiber metabolism, while others were not directly linked but could belong to supporting pathways. In this study, a total of 1,649 differentially expressed transcripts were identified, within which only 151 transcripts were directly related to sugar and fiber metabolism, when comparing between the young and mature internodal samples of the sugarcane culm. Most of these transcripts were up-regulated in the young internodes compared to the mature internodes, highlighting the growth phase of the two tissues being compared. When comparing the two groups of low and high fiber genotypes, there were a total of 555 differentially expressed transcripts identified, of which only 23 transcripts were related to the accumulation of sugars and fiber. This could indicate that the differential expression level between the two groups might be less than two-fold, and due to multi-factors contributing to fiber content and biomass accumulation.
Supporting information
(a) All genotypes, (b) Low fiber genotypes and (c) High fiber genotypes. Classification based on the MapMan annotation. PS denotes photosynthesis while CHO denotes carbohydrates. Red color indicates up-regulation while blue color indicates down-regulation in the bottom internodal tissues.
(XLSX)
Log2FC was used, red color denotes the up-regulated in the bottom internodal tissues while blue color denotes the down-regulated in the bottom internodal tissue. Each heatmap is representative of one of the identified differentially expressed transcripts.
(XLSX)
(a) All tissues from all genotypes, (b) Bottom tissues from low and high fiber genotypes, (c) Top tissues from low and high fiber genotypes. The classification was based on the MapMan annotation. PS denotes photosynthesis while CHO denotes carbohydrates. Blue color indicates up-regulation while red color indicates down-regulation in low fiber.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
The authors are grateful to SRA staff in Brandon station, Burdekin, Queensland, Australia for helping with the sample collecting and processing; to Mr. Ravi Nirmal for helping us in sample collection and transport; and to Dr. Prathima P. Thirugnanasambandam for editorial suggestions.
Data Availability
All RNA-Seq read data are available from the NCBI SRA database under the BioProject PRJNA356226, 40 accession numbers: SRR5258946, SRR5258947, SRR5258948, SRR5258949, SRR5258952, SRR5258953, SRR5258960, SRR5258961, SRR5258968, SRR5258969, SRR5258970, SRR5258971, SRR5258978, SRR5258979, SRR5258980, SRR5258981, SRR5258982, SRR5258983, SRR5258984, SRR5258985, SRR5258986, SRR5258987, SRR5258988, SRR5258989, SRR5258992, SRR5258993, SRR5259000, SRR5259001, SRR5259008, SRR5259009, SRR5259010, SRR5259011, SRR5259018, SRR5259019, SRR5259020, SRR5259021, SRR5259022, SRR5259023, SRR5259024 and SRR5259025. Other relevant data are within the paper.
Funding Statement
This research was carried out as part of NVH's PhD. NVH was awarded an Australian Awards Scholarship, funded by the Australian Agency for International Development (AusAID). Sugar Research Australia provided support in the form of salaries for author FCB, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Singh BP. Industrial crops and uses: Cabi; 2010. [Google Scholar]
- 2.Moore PH, Paterson AH, Tew T. Sugarcane: The Crop, the Plant, and Domestication Sugarcane: Physiology, Biochemistry, and Functional Biology: John Wiley & Sons Ltd; 2013. p. 1–17. [Google Scholar]
- 3.Canilha L, Kumar Chandel A, dos Santos Milessi TS, Fernandes Antunes FA, da Costa Freitas WL, das Gracas Almeida Felipe M, et al. Bioconversion of sugarcane biomass into ethanol: an overview about composition, pretreatment methods, detoxification of hydrolysates, enzymatic saccharification, and ethanol fermentation. Journal of biomedicine & biotechnology. 2012;2012:989572 10.1155/2012/989572 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoang NV, Furtado A, Donnan L, Keeffe EC, Botha FC, Henry RJ. High-Throughput Profiling of the Fiber and Sugar Composition of Sugarcane Biomass. Bioenerg Res. 2016;10(2):400–16. 10.1007/s12155-016-9801-8 [DOI] [Google Scholar]
- 5.Botha F, Whittaker A, Vorster D, Black K. Sucrose accumulation rate, carbon partitioning and expression of key enzyme activities in sugarcane stem tissue. Sugarcane: research towards efficient and sustainable production. 1996:98–101. [Google Scholar]
- 6.Botha FC, Black KG. Sucrose phosphate synthase and sucrose synthase activity during maturation of internodal tissue in sugarcane. Functional Plant Biology. 2000;27(1):81–5. [Google Scholar]
- 7.Carson DL, Botha FC. Genes expressed in sugarcane maturing internodal tissue. Plant Cell Rep. 2002;20 10.1007/s00299-002-0444-1 [DOI] [Google Scholar]
- 8.Whittaker A, Botha FC. Carbon Partitioning during Sucrose Accumulation in Sugarcane Internodal Tissue. Plant Physiol. 1997;115(4):1651–9. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patrick JW, Botha FC, Birch RG. Metabolic engineering of sugars and simple sugar derivatives in plants. Plant biotechnology journal. 2013;11(2):142–56. 10.1111/pbi.12002 [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Nayak S, Koch K, Ming R. Carbon partitioning in sugarcane (Saccharum species). Frontiers in plant science. 2013;4 10.3389/fpls.2013.00201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bindon KA, Botha FC. Carbon allocation to the insoluble fraction, respiration and triose-phosphate cycling in the sugarcane culm. Physiol Plant. 2002;116(1):12–9. Epub 2002/09/05. . [DOI] [PubMed] [Google Scholar]
- 12.Moore P. Temporal and Spatial Regulation of Sucrose Accumulation in the Sugarcane Stem. Functional Plant Biology. 1995;22(4):661–79. 10.1071/PP9950661. [DOI] [Google Scholar]
- 13.Sachdeva M, Bhatia S, Batta SK. Sucrose accumulation in sugarcane: a potential target for crop improvement. Acta Physiologiae Plantarum. 2011;33(5):1571–83. 10.1007/s11738-011-0741-9 [DOI] [Google Scholar]
- 14.Casu RE, Rae AL, Nielsen JM, Perroux JM, Bonnett GD, Manners JM. Tissue-specific transcriptome analysis within the maturing sugarcane stalk reveals spatial regulation in the expression of cellulose synthase and sucrose transporter gene families. Plant Mol Biol. 2015. Epub 2015/10/13. 10.1007/s11103-015-0388-9 . [DOI] [PubMed] [Google Scholar]
- 15.Carpita NC. Structure and biogenesis of the cell walls of grasses. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:445–76. 10.1146/annurev.arplant.47.1.445 [DOI] [PubMed] [Google Scholar]
- 16.Lin F, Waters CL, Mallinson RG, Lobban LL, Bartley LE. Relationships between biomass composition and liquid products formed via pyrolysis. Frontiers in Energy Research. 2015;3 10.3389/fenrg.2015.00045 [DOI] [Google Scholar]
- 17.Walsh KB, Sky RC, Brown SM. The anatomy of the pathway of sucrose unloading within the sugarcane stalk. Functional Plant Biology. 2005;32(4):367–74. 10.1071/FP04102. [DOI] [PubMed] [Google Scholar]
- 18.Casu R, Grof CL, Rae A, McIntyre CL, Dimmock C, Manners J. Identification of a novel sugar transporter homologue strongly expressed in maturing stem vascular tissues of sugarcane by expressed sequence tag and microarray analysis. Plant Mol Biol. 2003;52(2):371–86. 10.1023/A:1023957214644 [DOI] [PubMed] [Google Scholar]
- 19.Casu R, Dimmock C, Chapman S, Grof CL, McIntyre CL, Bonnett G, et al. Identification of Differentially Expressed Transcripts from Maturing Stem of Sugarcane by in silico Analysis of Stem Expressed Sequence Tags and Gene Expression Profiling. Plant Mol Biol. 2004;54(4):503–17. 10.1023/B:PLAN.0000038255.96128.41 [DOI] [PubMed] [Google Scholar]
- 20.Casu RE, Jarmey JM, Bonnett GD, Manners JM. Identification of transcripts associated with cell wall metabolism and development in the stem of sugarcane by Affymetrix GeneChip Sugarcane Genome Array expression profiling. Functional & Integrative Genomics. 2007;7(2):153–67. 10.1007/s10142-006-0038-z [DOI] [PubMed] [Google Scholar]
- 21.Vicentini R, Bottcher A, Brito MdS, dos Santos AB, Creste S, Landell MGdA, et al. Large-Scale Transcriptome Analysis of Two Sugarcane Genotypes Contrasting for Lignin Content. PLoS ONE. 2015;10(8):e0134909 10.1371/journal.pone.0134909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bottcher A, Cesarino I, Santos ABd, Vicentini R, Mayer JLS, Vanholme R, et al. Lignification in sugarcane: biochemical characterization, gene discovery, and expression analysis in two genotypes contrasting for lignin content. Plant Physiol. 2013;163(4):1539–57. 10.1104/pp.113.225250 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santos Brito M, Nobile P, Bottcher A, dos Santos A, Creste S, de Landell M, et al. Expression Profile of Sugarcane Transcription Factor Genes Involved in Lignin Biosynthesis. Tropical Plant Biology. 2015;8(1–2):19–30. 10.1007/s12042-015-9147-y [DOI] [Google Scholar]
- 24.Hoang NV, Furtado A, Mason PJ, Marquardt A, Kasirajan L, Thirugnanasambandam PP, et al. A survey of the complex transcriptome from the highly polyploid sugarcane genome using full-length isoform sequencing and de novo assembly from short read sequencing. BMC Genomics. 2017;18(1):395 10.1186/s12864-017-3757-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watt DA, McCormick AJ, Govender C, Carson DL, Cramer MD, Huckett BI, et al. Increasing the utility of genomics in unravelling sucrose accumulation. Field Crops Research. 2005;92(2–3):149–58. 10.1016/j.fcr.2005.01.012. [DOI] [Google Scholar]
- 26.Furtado A. RNA extraction from developing or mature wheat seeds. Methods in molecular biology. 2014;1099:23–8. 10.1007/978-1-62703-715-0_4 . [DOI] [PubMed] [Google Scholar]
- 27.Andrews S. FastQC A Quality Control tool for High Throughput Sequence Data http://www.bioinformatics.babraham.ac.uk/projects/fastqc2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 28.SEQTK toolkit. https://github.com/lh3/seqtk. Accessed on November 2016.
- 29.Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nature reviews Genetics. 2010;11(10):733–9. http://www.nature.com/nrg/journal/v11/n10/suppinfo/nrg2825_S1.html. 10.1038/nrg2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotech. 2011;29(7):644–52. http://www.nature.com/nbt/journal/v29/n7/abs/nbt.1883.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–512. 10.1038/nprot.2013.084 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12(1):1–16. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Meth. 2012;9(4):357–9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trapnell C, Williams B, Pertea G, Mortazavi A, Kwan G, van Baren M, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory in biosciences = Theorie in den Biowissenschaften. 2012;131(4):281–5. Epub 2012/08/09. 10.1007/s12064-012-0162-3 . [DOI] [PubMed] [Google Scholar]
- 37.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15(12):550 Epub 2014/12/18. 10.1186/s13059-014-0550-8 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seyednasrollah F, Laiho A, Elo LL. Comparison of software packages for detecting differential expression in RNA-seq studies. Briefings in bioinformatics. 2015;16(1):59–70. 10.1093/bib/bbt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Team R Core. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: 2013. ISBN 3-900051-07-0; 2014. [Google Scholar]
- 40.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47 Epub 2015/01/22. 10.1093/nar/gkv007 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucas A, Jasson S. Using amap and ctc Packages for Huge Clustering. The Newsletter of the R Project Volume 6/5, December 2006. 1985;1(3):58. [Google Scholar]
- 43.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12(2):115–21. Epub 2015/01/31. 10.1038/nmeth.3252 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K. Cluster: cluster analysis basics and extensions. R package version. 2012;1(2):56. [Google Scholar]
- 45.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20(2):289–90. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- 46.Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, et al. gplots: Various R programming tools for plotting data. R package version. 2009;2(4). [Google Scholar]
- 47.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–6. Epub 2005/08/06. 10.1093/bioinformatics/bti610 . [DOI] [PubMed] [Google Scholar]
- 48.Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Research. 2006;34(suppl 2):W293–W7. 10.1093/nar/gkl031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mercator Annotator. http://www.plabipd.de/portal/mercator-sequence-annotation. Accessed on 01 September 2016. 2016.
- 50.Lohse M, Nagel A, Herter T, May P, Schroda M, Zrenner R, et al. Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant, cell & environment. 2014;37(5):1250–8. Epub 2013/11/19. 10.1111/pce.12231 . [DOI] [PubMed] [Google Scholar]
- 51.Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M. A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, Maize. Plant, cell & environment. 2009;32(9):1211–29. Epub 2009/04/25. 10.1111/j.1365-3040.2009.01978.x . [DOI] [PubMed] [Google Scholar]
- 52.Iskandar HM, Simpson RS, Casu RE, Bonnett GD, Maclean DJ, Manners JM. Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene expression in sugarcane. Plant Molecular Biology Reporter. 2004;22(4):325–37. 10.1007/BF02772676 [DOI] [Google Scholar]
- 53.Guo J, Ling H, Wu Q, Xu L, Que Y. The choice of reference genes for assessing gene expression in sugarcane under salinity and drought stresses. Scientific reports. 2014;4:7042 Epub 2014/11/14. 10.1038/srep07042 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jain M, Nijhawan A, Tyagi AK, Khurana JP. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochemical and biophysical research communications. 2006;345(2):646–51. 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- 55.Chandna R, Augustine R, Bisht NC. Evaluation of Candidate Reference Genes for Gene Expression Normalization in Brassica juncea Using Real Time Quantitative RT-PCR. PLOS ONE. 2012;7(5):e36918 10.1371/journal.pone.0036918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8(2):R19 Epub 2007/02/13. 10.1186/gb-2007-8-2-r19 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002;3(7):Research0034 Epub 2002/08/20. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134 Epub 2012/06/20. 10.1186/1471-2105-13-134 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical chemistry. 2009;55(4):611–22. Epub 2009/02/28. 10.1373/clinchem.2008.112797 . [DOI] [PubMed] [Google Scholar]
- 60.Draw Venn Diagram. http://bioinformatics.psb.ugent.be/webtools/Venn/. Accessed 20 May 2016. 2016.
- 61.Research Computing Centre. http://www.rcc.uq.edu.au. Accessed 2016. 2016.
- 62.Kang C, Darwish O, Geretz A, Shahan R, Alkharouf N, Liu Z. Genome-Scale Transcriptomic Insights into Early-Stage Fruit Development in Woodland Strawberry Fragaria vesca. The Plant Cell. 2013;25(6):1960–78. 10.1105/tpc.113.111732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jacobsen KR, Fisher D, Maretzki A, Moore P. Developmental changes in the anatomy of the sugarcane stem in relation to phloem unloading and sucrose storage. Botanica Acta. 1992;105(1):70–80. [Google Scholar]
- 64.Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, et al. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2007;104(39):15572–7. Epub 2007/09/20. 10.1073/pnas.0706569104 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, et al. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104(39):15566–71. Epub 2007/09/20. 10.1073/pnas.0706592104 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor NG. Identification of cellulose synthase AtCesA7 (IRX3) in vivo phosphorylation sites—a potential role in regulating protein degradation. Plant Mol Biol. 2007;64(1–2):161–71. Epub 2007/04/12. 10.1007/s11103-007-9142-2 . [DOI] [PubMed] [Google Scholar]
- 67.Taylor NG, Laurie S, Turner SR. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell. 2000;12(12):2529–40. Epub 2001/01/10. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hall H, Ellis B. Transcriptional programming during cell wall maturation in the expanding Arabidopsis stem. BMC Plant Biology. 2013;13(1):14 10.1186/1471-2229-13-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burlat V, Kwon M, Davin LB, Lewis NG. Dirigent proteins and dirigent sites in lignifying tissues. Phytochemistry. 2001;57(6):883–97. Epub 2001/06/26. . [DOI] [PubMed] [Google Scholar]
- 70.Davin LB, Wang HB, Crowell AL, Bedgar DL, Martin DM, Sarkanen S, et al. Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science. 1997;275(5298):362–6. Epub 1997/01/17. . [DOI] [PubMed] [Google Scholar]
- 71.Hosmani PS, Kamiya T, Danku J, Naseer S, Geldner N, Guerinot ML, et al. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(35):14498–503. 10.1073/pnas.1308412110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ralph J, Peng J, Lu F, Hatfield RD, Helm RF. Are Lignins Optically Active? Journal of Agricultural and Food Chemistry. 1999;47(8):2991–6. 10.1021/jf9901136 [DOI] [PubMed] [Google Scholar]
- 73.Wagner A, Donaldson L, Ralph J. Lignification and lignin manipulations in conifers Advances in Botanical Research. 2012. [Google Scholar]
- 74.Hatfield R, Vermerris W. Lignin Formation in Plants. The Dilemma of Linkage Specificity. Plant Physiology. 2001;126(4):1351–7. 10.1104/pp.126.4.1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen X-Y, Kim J-Y. Callose synthesis in higher plants. Plant Signaling & Behavior. 2009;4(6):489–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nedukha OM. Callose: Localization, functions, and synthesis in plant cells. Cytology and Genetics. 2015;49(1):49–57. 10.3103/s0095452715010090 [DOI] [Google Scholar]
- 77.Piršelová B, Matušíková I. Callose: the plant cell wall polysaccharide with multiple biological functions. Acta Physiologiae Plantarum. 2013;35(3):635–44. 10.1007/s11738-012-1103-y [DOI] [Google Scholar]
- 78.Stanley D, Farnden KJ, Macrae EA. Plant α-amylases: functions and roles in carbohydrate metabolism. Biologia. 2005;60(16):65–71. [Google Scholar]
- 79.Nelson O, Pan D. Starch synthesis in maize endosperms. Annual review of plant biology. 1995;46(1):475–96. [Google Scholar]
- 80.Uematsu K, Suzuki N, Iwamae T, Inui M, Yukawa H. Increased fructose 1,6-bisphosphate aldolase in plastids enhances growth and photosynthesis of tobacco plants. J Exp Bot. 2012;63(8):3001–9. Epub 2012/02/11. 10.1093/jxb/ers004 . [DOI] [PubMed] [Google Scholar]
- 81.Buchanan BB, Gruissem W, Jones RL. Biochemistry and molecular biology of plants: John Wiley & Sons; 2015. [Google Scholar]
- 82.Sage RF. The evolution of C4 photosynthesis. New Phytologist. 2004;161(2):341–70. 10.1111/j.1469-8137.2004.00974.x [DOI] [PubMed] [Google Scholar]
- 83.Sage RF, Peixoto MM, Sage TL. Photosynthesis in Sugarcane Sugarcane: Physiology, Biochemistry, and Functional Biology: John Wiley & Sons Ltd; 2013. p. 121–54. [Google Scholar]
- 84.Burget EG, Reiter WD. The mur4 mutant of arabidopsis is partially defective in the de novo synthesis of uridine diphospho L-arabinose. Plant Physiol. 1999;121(2):383–9. Epub 1999/10/12. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Forsthoefel NR, Cushman MA, Cushman JC. Posttranscriptional and posttranslational control of enolase expression in the facultative Crassulacean acid metabolism plant Mesembryanthemum Crystallinum L. Plant Physiol. 1995;108(3):1185–95. Epub 1995/07/01. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, et al. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell. 2000;12(12):2409–24. Epub 2001/01/10. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gupta AK, Kaur N. Sugar signalling and gene expression in relation to carbohydrate metabolism under abiotic stresses in plants. Journal of biosciences. 2005;30(5):761–76. Epub 2006/01/03. . [DOI] [PubMed] [Google Scholar]
- 88.Stabile S, Bodini A, Jank L, Renno F, Santos M, Silva L. Expression of genes from the lignin synthesis pathway in guineagrass genotypes differing in cell-wall digestibility. Grass and Forage Science. 2012;67(1):43–54. [Google Scholar]
- 89.Chakraborty N, Sharma P, Kanyuka K, Pathak RR, Choudhury D, Hooley RA, et al. Transcriptome Analysis of Arabidopsis GCR1 Mutant Reveals Its Roles in Stress, Hormones, Secondary Metabolism and Phosphate Starvation. PLOS ONE. 2015;10(2):e0117819 10.1371/journal.pone.0117819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen J, Hou K, Qin P, Liu H, Yi B, Yang W, et al. RNA-Seq for gene identification and transcript profiling of three Stevia rebaudiana genotypes. BMC Genomics. 2014;15:571 Epub 2014/07/09. 10.1186/1471-2164-15-571 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Poole R, Barker G, Wilson ID, Coghill JA, Edwards KJ. Measuring global gene expression in polyploidy; a cautionary note from allohexaploid wheat. Functional & integrative genomics. 2007;7(3):207–19. [DOI] [PubMed] [Google Scholar]
- 92.Souza GM, Berges H, Bocs S, Casu R, D’Hont A, Ferreira JE, et al. The Sugarcane Genome Challenge: Strategies for Sequencing a Highly Complex Genome. Tropical Plant Biology. 2011;4(3–4):145–56. 10.1007/s12042-011-9079-0 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) All genotypes, (b) Low fiber genotypes and (c) High fiber genotypes. Classification based on the MapMan annotation. PS denotes photosynthesis while CHO denotes carbohydrates. Red color indicates up-regulation while blue color indicates down-regulation in the bottom internodal tissues.
(XLSX)
Log2FC was used, red color denotes the up-regulated in the bottom internodal tissues while blue color denotes the down-regulated in the bottom internodal tissue. Each heatmap is representative of one of the identified differentially expressed transcripts.
(XLSX)
(a) All tissues from all genotypes, (b) Bottom tissues from low and high fiber genotypes, (c) Top tissues from low and high fiber genotypes. The classification was based on the MapMan annotation. PS denotes photosynthesis while CHO denotes carbohydrates. Blue color indicates up-regulation while red color indicates down-regulation in low fiber.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All RNA-Seq read data are available from the NCBI SRA database under the BioProject PRJNA356226, 40 accession numbers: SRR5258946, SRR5258947, SRR5258948, SRR5258949, SRR5258952, SRR5258953, SRR5258960, SRR5258961, SRR5258968, SRR5258969, SRR5258970, SRR5258971, SRR5258978, SRR5258979, SRR5258980, SRR5258981, SRR5258982, SRR5258983, SRR5258984, SRR5258985, SRR5258986, SRR5258987, SRR5258988, SRR5258989, SRR5258992, SRR5258993, SRR5259000, SRR5259001, SRR5259008, SRR5259009, SRR5259010, SRR5259011, SRR5259018, SRR5259019, SRR5259020, SRR5259021, SRR5259022, SRR5259023, SRR5259024 and SRR5259025. Other relevant data are within the paper.