Abstract
In order to effectively control and monitor schistosomiasis, new diagnostic methods are essential. Taking advantage of computational approaches provided by immunoinformatics and considering the availability of Schistosoma mansoni predicted proteome information, candidate antigens of schistosomiasis were selected and used in immunodiagnosis tests based on Enzime-linked Immunosorbent Assay (ELISA). The computational selection strategy was based on signal peptide prediction; low similarity to human proteins; B- and T-cell epitope prediction; location and expression in different parasite life stages within definitive host. Results of the above-mentioned analysis were parsed to extract meaningful biological information and loaded into a relational database developed to integrate them. In the end, seven proteins were selected and one B-cell linear epitope from each one of them was selected using B-cell epitope score and the presence of intrinsically disordered regions (IDRs). These predicted epitopes generated synthetic peptides that were used in ELISA assays to validate the rational strategy of in silico selection. ELISA was performed using sera from residents of areas of low endemicity for S. mansoni infection and also from healthy donors (HD), not living in an endemic area for schistosomiasis. Discrimination of negative (NEG) and positive (INF) individuals from endemic areas was performed using parasitological and molecular methods. All infected individuals were treated with praziquantel, and serum samples were obtained from them 30 and 180 days post-treatment (30DPT and 180DPT). Results revealed higher IgG levels in INF group than in HD and NEG groups when peptides 1, 3, 4, 5 and 7 were used. Moreover, using peptide 5, ELISA achieved the best performance, since it could discriminate between individuals living in an endemic area that were actively infected from those that were not (NEG, 30DPT, 180DPT groups). Our experimental results also indicate that the computational prediction approach developed is feasible for identifying promising candidates for the diagnosis of schistosomiasis and other diseases.
Introduction
Schistosomiasis remains one of the most prevalent parasitic diseases in the world with more than 240 million people infected in 78 countries [1]. Control strategies have been based on chemotherapy, but these attempts have failed to interrupt transmission. Part of this failure could be attributed to the absence of an accurate method of diagnosis that is able to determine the real prevalence of the disease in populations, and that could monitor the success of interventions and assess healing after therapeutic intervention [2–4]. Parasitological tests are still the most widely used diagnostic methods of schistosomiasis control programs [5,6], and of these, the Kato-Katz technique is the most used due to its low cost, abillity to detect different helminths infection and greater sensibility in áreas high intensity infections [7–9]. However, low parasite burdens require examination of more slides or association of the parasitological tests with serological and molecular techniques to have an accurate diagnosis of the disease. In fact, molecular and immunological techniques have proven to be more sensitive and promising in identifying infection in individuals with negative coproscopic results [10–15].
An example of an immunological technique that has provided satisfactory results in the diagnosis of schistosomiasis is the urine-based point-of-care (POC-CCA) assay for detecting circulating cathodic antigen in urine samples (Rapid Medical Diagnostics, Pretoria, South Africa). This test has demonstrated promise for use in epidemiological studies, in clinical laboratories and in endemic areas, with higher sensitivity than the Kato-Katz technique [16–19]. However, more studies are necessary to assess the efficacy of this technique in the field, especially in areas of low endemicity [20,21].
Immunodiagnostic methods based on serology have been widely used and have greater sensitivity than parasitological methods [6,22–24], particularly in areas of low endemicity [25,26]. Among serological tests, ELISA assay is widely used for the diagnosis of schistosomiasis, but one of the difficulties in using this method is the choice of the parasite`s antigen. Crude antigens may exhibit cross-reactivity with other helminthes, as well as possess low sensitivity [27]. To overcome this obstacle, purified and recombinant antigens [28–35] have been used.
Immunoinformatics emerged as a new multidisciplinary approach for identifying diagnostic targets at the beginning of 21st century, with the accumulation of genomic data in public domain databases. Indeed, some bioinformatic tools have already been used to identify Schistosoma antigens. In silico analyses were used to search for protein tandem repeats in the genome of S. mansoni, and seven diagnostic candidates were identified as a result of this study [36]. Guo et al. (2012) used a bioinformatics analysis to find target sequences for molecular diagnosis from S. japonicum retrotransposons [37]. Also working with S. japonicum, Zhang et al. (2007) searched for B-cell epitopes in three pre-selected proteins in order to design two multi-epitope chimeric proteins to be used as diagnostic targets, and showed that these proteins were able to react with sera of S. japonicum infected patients [38]. Immunoinformatics has also been used to identify diagnostic targets of other eukaryotic pathogens, such as the use of proteomics combined with B-cell epitope search to find potential antigens from Cryptococcus gattii proteome [39].
In this context, in a previous work, our research group has scanned the predicted proteome of S. mansoni using tools from SchistoDB 2.0 [40] and online bioinformatic`s tools. From these previous studies we were able to identify six potential diagnostic candidates [41], among which was Sm200, a 200 kDa tegumental protein. Although this protein was highly sensitive in identifying infected patients from endemic areas, it was not able to differentiate between non-infected and infected individuals from an area of low endemicity [42].
Besides the classical immunoinformatics tools, such as epitope prediction and subcellular location prediction, another approach that has emerged for identifying immunogenic proteins is the search for B-cell epitopes associated with structurally disordered regions. In fact, it has already been demonstrated that immunogenic peptides may be closely associated with flexibility [43], which could promote the exposure of these immunogenic regions to the immune system. This strategy also takes into account that intrinsically disordered regions (IDRs) were present in approximately 60–70% of the proteins from the eukaryotics’ predicted proteomes analyzed, and are responsible for important biological functions [44].
Based on the above, the present work was developed due to the necessity to find a target to be used in a test for the diagnosis of schistosomiasis with higher specificity and sensitivity than the tests currently used, and also able to identify cured individuals after chemotherapy. The developed and implemented strategy involves epitope prediction, associated with subcellular location and expression in different stages of S. mansoni, to find proteins from the whole predicted proteome of the parasite that could be used as diagnostic targets. In addition, a relational database was created to integrate all of the data generated. From the proteins selected, peptides were submitted to manual curation and selected based on their score in B-cell epitopes prediction and on the presence of IDRs in the neighborhood of the epitope location in the protein. These peptides were synthesized and evaluated experimentally by ELISA in order to validate the computational prediction.
Methods
Genomic data
13,273 transcripts from S. mansoni were obtained from SchistoDB 2.0 [40]. Data from protein expression linked with different parasite life cycle stages in human host, including egg, schistosomula, lung schistosomula, and adult worm, were also downloaded for computational downstream analysis.
Epitope prediction
All proteins from the parasite S. mansoni were screened for B-cell and TCD4+ epitopes. For B-cell epitope prediction, we used BepiPred 1.0 [45], AAP12 [46] and BCPred12 [47]; and for TCD4+ epitope prediction, we used NetMHCII 2.2 [48], which predicts the affinity of epitopes to 17 different alleles.
Prediction of intrinsic disordered proteins (IDPs)
Prediction of protein disorder was based on the consensus prediction obtained by the following algorithms: DisEMBL [49], GlobPlot [50], IUPred [51], and VSL2B [52]. A region of a protein was considered disordered when 40 or more consecutive aminoacids were predicted as disordered, since prediction accuracy was shown to be higher in longer disordered regions [53].
Prediction of subcellular location
It was also important to investigate the subcellular location of the proteins, since this information is of great significance to the functional analysis of the proteins themselves and also of specific regions. For this, the following algorithms were selected: Sigcleave [54], TargetP 1.1 [55]and SignalP 4.0 [56] to verify the presence of signal peptides; TMHMM 2.0 [57,58] to verify transmembrane domains; and SherLoc2 [59] to verify subcellular location.
Sequence similarity searches
The Standalone BLAST (Basic Local Alignment Search Tool) algorithm [60] was used to perform sequence similarity searches of pre-selected diagnostic targets from S. mansoni against predicted human proteins. This analysis is necessary in order to avoid cross-reaction with proteins from host organisms.
Workflow of analysis for selection of target diagnostic proteins
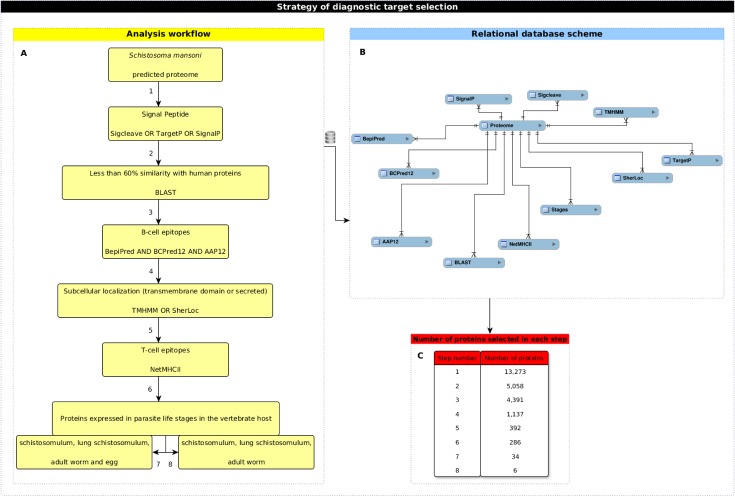
All predictions described above were performed on local servers and loaded into the computational analytical workflow developed in order to select potential diagnostic antigens (Fig 1A). The first step of our approach involved selection of proteins with signal peptides as predicted by Sigcleave, TargetP 1.1 or SignalP. Next, we used the BLAST algorithm to exclude proteins with more than 60% of similarity with human predicted proteome. After these analyses, we selected proteins with at least one linear B-cell epitope predicted by BepiPred 1.0, BCPreds, and AAP12. Epitopes with prediction scores above one on these three predictors were chosen. Proteins harboring the predicted epitopes were subsequently submitted to subcellular location prediction with SherLoc2 and scanned for transmembrane domains with TMHMM 2.0. Target proteins predicted at the extracellular compartment or at plasmatic membrane were selected. These target proteins were analyzed for the presence of TCD4+ epitopes with affinity to 17, 16, 15, 14 or 13MHCII alleles predicted by NetMHCII 2.2. Finally, in order to use the information regarding the parasite life cycle stages on the definitive host, two different strategies were applied. In the first one, proteins predicted as simultaneously expressed on schistosomulum, lung schistosomulum, adult worm and egg were selected. In the second one, proteins simultaneously expressed in all the above stages except egg were chosen.
Fig 1. Strategy of diagnostic target selection.
(A) Each step of the workflow analysis was numbered according to the strategy used for target selection. (B) Schematic representation of the relational database developed to integrate data obtained after computational predictions. (C) Number of proteins selected after each step from (A).
Data integration
Data from S. mansoni predicted proteome, from parasite life cycle stages, and from all predictions performed were automatically integrated in a MySQL relational database (Fig 1B). This database was developed in order to make it possible to extract biologically relevant information related to diagnostic targets using the previously described workflow analysis.
Selection of peptides
Custom peptide synthesis was performed using the sequence information from the predicted epitopes from the proteins selected by the workflow described above.
The strategy of peptides’ selection was based on the following steps: a) sequences predicted as linear B-cell epitopes by AAP12 and BCPreds with a score of at least 1, and by BepiPred 1.0 with a score of at least 1.5; b) if the analyses failed to find a peptide, a second query was performed considering just AAP12 and BepiPred 1.0, and the same scores described above; and c) if no epitope has been found yet, the cutoff scores were changed to at least 0.9 for AAP12 and at least 1 for BepiPred 1.0. After the selection was finished, an additional filtering step was performed removing epitopes with the following characteristics: a) located at predicted signal peptides regions; b) located at predicted transmembrane domains regions; and c) predicted as identical to human epitopes.
In the final step, we preferentially selected epitopes containing IDRs in their neighborhood and predicted by BCPreds, even with a lower score. After selection of the epitopes, we performed another BLAST step against host`s genome and all the proteins from Ancylostoma ceylanicum; A. duodenale; Ascaris lumbricoides; Enterobius vermicularis; Hymenolepis nana; Necator americanus; Taenia solium; T. saginata; Trichuris trichiura available in NCBI database to avoid similar peptides.
Synthesis of peptides
Peptides were identified with numbers from 1 to 7 in order to facilitate the description of results. Peptide 1 was synthesized using PS3TM synthesizer (Protein Technologies, Inc) by chemical solid phase synthesis using the Fmoc strategy. Cleavage of peptides was carried out manually as described by Chan & White (2000)[61], and purification was performed by high performance liquid chromatography, with using a Shimadzu Prominence chromatograph. The other peptides were synthesized by the company Genescript with more than 90% purity. All peptides were diluted in DMSO (dimethyl sulfoxide) at a concentration of 1 μg/μL and stored at -70°C.
Human serum samples
In this study, sera from 58 individuals (female/male: 24/34) from an area of low endemicity for S. mansoni infection (Pedra Preta, Minas Gerais, Brazil) and 13 sera from healthy donors (HD group) (female/male: 06/07) not living in an endemic area for schistosomiasis were used. Pedra Preta is a little village in a schistosome endemic area next do Montes Claros, MG. The sera from de individuals evaluated in our study was collected in a previous study performed by our group in a rural zone of Pedra Preta with approximately 230 residents. The previous study evaluated 201 individuals. The prevalence determined by examination of two Kato-Katz slides, which is recommended by the Brazilian Ministry of Heath to determine infection, was 8% with a mean parasite load of 40.6 opg. However a 35.8% of positivity were found when feces samples were analyzed using Kato-Katz (18 slides) and TF-Test® techniques [62]. Individuals co-infected with Ascaris lumbricoides, Thricuris trichiura, hookworm, Taenia sp, Hymenolepis nana and Enterobius vermiculares were observed in this endemic area, but in our study only sera of S. mansoni monoinfected individuals were used before and after treatment.
The discrimination of negative (NEG group) and positive (INF group) serum samples of individuals from endemic areas was performed according to parasitological and molecular methodology described by Siqueira et al (2015)[62]. Briefly, each participant provided four separate stool samples on each of four consecutive days for use in the Kato-Katz [7], TF-Test® [63,64] and PCR-ELISA [65] techniques. Individual serum samples were obtained after centrifugation of blood samples at 3000g for 5 minutes. These samples were kept at -20°C until moment of use. Sera samples from twenty six individuals infected with S. mansoni and from 25 individuals from endemic area with negative stool examination were evaluated.
All participants with positive stool examination for schistosomiasis determined by parasitological methods were treated with praziquantel in a single dose of 60mg/Kg for children and 50mg/Kg for adults, as recommended by the Brazilian Ministry of Health. Sera were obtained 30 and 180 days post-treatment (groups 30DPT and 180DPT), at these time points feces were also collected. Cure rate was determined by examination of 18 Kato-Katz slides and TF-Test®. Seventeen and nine sera samples were evaluated 30 and 180 days post-treatment, respectively. Some sera samples were from individuals previously evaluated in the infected group.
Ethics
The use of both healthy donor sera and sera from individuals living in the Pedra Preta endemic area for schistosomiasis were approved by the Ethical Research Committee of the René Rachou Research Institute -Fiocruz (CEPSH/CPqRR 03/2008: 105/2004-OF.215-TEC) and the National Ethical Board (784/2008, CONEP14886). All participants and parents/legal guardians were informed about the purpose and objectives of the study before providing written informed consent to participate of this study.
ELISA against human sera
The ELISA assay was first standardized using 1 or 2μg/ml of synthetic peptide incubated with pool of sera from individuals belonging to the healthy donor group; negative group or infected group in serial dilution in PBST beginning at 1:20 and ending at 1:1,280. The conjugate was evaluated in a dilution of 1:60,000 (anti-human IgG-HRP). After standardization, the assay was performed in flat bottom plates (Maxisorp NUNC®) sensitized with 100μl/well of carbonate bicarbonate buffer 0.05M, pH 9.6, containing peptide at a concentration of 1μg/mL (peptides 1,5,6 and 7) or 2μg/mL (peptides 2 and 3), and incubated for approximately 16 hours at 4°C. To remove the unbound proteins, the plates were washed three times with 150mM PBS containing 0.05% Tween 20 (PBST) and blocked with PBST plus 10% fetal bovine serum for 2 hours at 37°C. The sera from patients were diluted 1:40 (peptides 4 and 7), 1:80 (peptides 1 and 2), 1:160 (peptides 3 and 6) and 1:320 (peptide 5) in PBST. These sera were added (100μl/well) to the plates and incubated for 2 hours at 37°C. After washing the plates (3X), an anti-human IgG secondary antibody conjugated to peroxidase (Sigma Aldrich) was used at a dilution of 1: 60,000 in PBST for one hour at 37°C. After three washes, the reaction was revealed for 10 minutes with 100μl of liquid substrate 3, 3', 5, 5'-tetramethylbenzidine (TMB/Sigma). The reaction was stopped with 50μL of sulfuric acid and the optical density determined by an automatic ELISA reader (Multiskan, Thermo Scientific), using a filter of 450nm.
Statistical analysis
The software package GraphPad Prism 4.0 (GraphPad Software, San Diego, CA, USA) was used for statistical analysis. Kruskal-Wallis test followed by Dunn post-test were performed on samples without a normal distribution. Receptor Operating Characteristic curves (ROC curves) were used to calculate sensitivity, specificity and the cutoff points between infected and healthy donor groups or between infected and uninfeced individuals from endemic area. Positive predictive values (PPV) and Negative Predictive Values (NPV) was determined by the following formula: PPV = number of true positives/(number of true positives + number of false positives); NPV = number of true negatives/(number of true negatives + number of false negatives)
Results
Selected proteins
In this study, target proteins to be used in schistosomiasis diagnosis were selected using the computational workflow described in methods section (Fig 1C). The analysis started with the predicted proteome of S. mansoni, comprised of 13,273 proteins. Only 5,058 proteins had a signal peptide predicted in its sequence and remained in the workflow. 4,391 of the proteins that remained in the workflow presented less than 60% of similarity with human proteins. Prediction of B cell epitope was the third filtering step, which resulted in a reduction of the number of proteins to 1,137. From these, 392 proteins, predicted to be located on the plasmatic membrane or to be secreted, were selected. For T CD4+ cell epitope prediction, 17 MHC alleles were analyzed (14 HLA-DR; H2-IAb, H2-IAd e H2-IAs). The adopted criterion took into account the predicted epitopes located along the complete extension of the protein with affinity to bind the maximum number of alleles. As result, none of the 392 proteins possessed epitopes with affinity to the 17 MHC alleles, five were found to have epitopes with affinity to 16 MHC alleles, 72 presented epitopes predicted to bind 15 MHC alleles, 186 presented predicted epitopes with affinity to 14 MHC alleles and 286 proteins presented epitopes with affinity to the 13 MHC alleles. Finally, in each set of proteins, we analyzed predicted profile of expression in different parasite life cycle stages within the vertebrate host. Two different approaches were used. First, proteins predicted to be expressed simultaneously on schistosomulum, lung schistosomulum, adult worm and egg stages were selected. The combination of this criterion with the criteria described above resulted in the selection of two proteins with epitopes predicted to bind 16 MHC alleles, eight proteins containing epitopes predicted to bind 15 MHC alleles, fourteen proteins presenting epitopes with affinity to 14 MHC alleles and ten proteins with epitopes predicted to bind 13 MHC alleles (S1 Table). Second, proteins predicted to be expressed simultaneously on schistosomulum, lung schistosomulum and adult worm were selected, resulting in the selection of five proteins with epitopes predicted to bind 14 MHC alleles and one protein presenting epitopes with affinity to 13 MHC alleles (S1 Table). The seven proteins with predicted epitope with affinity to bind the highest number of MHC alleles in each approach were selected. In the group of proteins predicted to be expressed simultaneously on schistosomulum, lung schistosomulum, adult worm and egg, two proteins that had predicted epitopes with affinity to 16 MHC alleles were selected. In the group of proteins predicted to be simultaneously expressed in all of the above-mentioned stages except egg, five proteins with epitopes predicted to bind 14 MHC alleles were selected (Table 1).
Table 1. Schistosoma mansoni proteins selected after in silico analysis, according to the strategy outlined in the workflow.
| ID | Function Predicted |
Amino acids Length |
Life stages a | Number of alleles |
|---|---|---|---|---|
| Smp_136560 | Expressed protein | 1995 | 1 | 16 |
| Smp_141860 | Heat containing protein, putative | 4619 | 1 | 16 |
| Smp_093840 | Trispanning orphan receptor; TORE, putative | 239 | 2 | 14 |
| Smp_126160 | Poly (p) /ATP NAD kinase, putative | 1077 | 2 | 14 |
| Smp_150390.1 | Expressed protein | 668 | 2 | 14 |
| Smp_167240 | Expressed protein | 776 | 2 | 14 |
| Smp_180240 | F-spondin, putative | 941 | 2 | 14 |
a Parasite life stages in definitive host
1—Schistosomulum, lung chistosomulum, adult worm and egg
2—Schistosomulum, lung schistosomulum and adult worm
Serological tests
Seven peptides (one for each selected protein), with no similarity host or other heelminths’ proteins, were chosen to be used as antigens in the serological tests. Peptide number, protein ID (systematic name), life stages in which they are expressed, peptide sequence, degree of purity, prediction of IDRs and disorder predictors for each selected peptide are described in Table 2. Three of the peptides having predicted IDRs in their neighborhood were selected.
Table 2. Epitopes selected to be used as synthetic peptides in ELISA test.
|
Peptide Number |
ID (Initial and final coordinates) |
Life stagesa |
Sequence of peptide |
Degree of purity (%) |
Prediction of IDRs (Initial and final coordinates) |
Disorder predictors |
|---|---|---|---|---|---|---|
| 1 | Smp_136560 (1564–1578) | 1 | ITEGNNSREGNSEKV | 60.1 | 1501–1658 | REM465, GlobPipe e IUPred |
| 2 | Smp_141860 (1694–1709) | 1 | NHSMDKDDDDFSDIDK | 95,31 | 1693–1793 | REM465, GlobPipe, IUPred e VSL2B |
| 3 | Smp_093840(219–233) | 2 | TTTNKDDTQINSAPS | 96,69 | 182–239 | REM465, GlobPipe, IUPred e VSL2B |
| 4 | Smp_126160(438–452) | 2 | LVTPESKYYSSLPGN | 95,97 | - | - |
| 5 | Smp_150390.1(216–230) | 2 | SLPSNAHNNDNNSSD | 95,55 | - | - |
| 6 | Smp_167240(213–228) | 2 | QCDLDTQWNPAGTEYS | 97,74 | - | - |
| 7 | Smp_180240(339–353) | 2 | RDWPTTLTGAGGSTT | 97,67 | - | - |
a Life stages of parasite in definitive host
1 –Schistosomulum, lung chistosomulum, adult worm and egg
2—Schistosomulum, lung schistosomulum and adult worm.
These synthetically produced peptides were employed in ELISA tests against human sera from the following experimental groups: HD, NEG, INF, 30DPT and 180DPT, as described in the Materials and Methods section. Significant IgG reativity against peptides 1, 3, 4, 5 and 7 were observed in the group of S. mansoni infected individuals in comparison with healthy donor. Significant IgG levels against peptides 1, 3, 4, and 5 were also observed in infected group when compared to individuals from the endemic area with negative stool examination (Fig 2). A significant reduction in serum reactivity to peptide 5 was observed in the groups 30DPT and 180DPT (Fig 2). Reactivity to peptide 7 was still observed in sera from the 180 days post-treatment group. Paired analysis before and after treatment were performed for some individuals. Data demonstrate that a significant descrease in IgG levels against peptides 1, 3 and 5 is observed 30 and 180 days post-treatment. Decreased IgG levels against peptides 2 and 4 were also observed 30 and 180 days post-treatment, respectivelly (S1 Fig).
Fig 2. Human IgG-specific response against synthetic peptides from S. mansoni targeted for use in diagnosis.
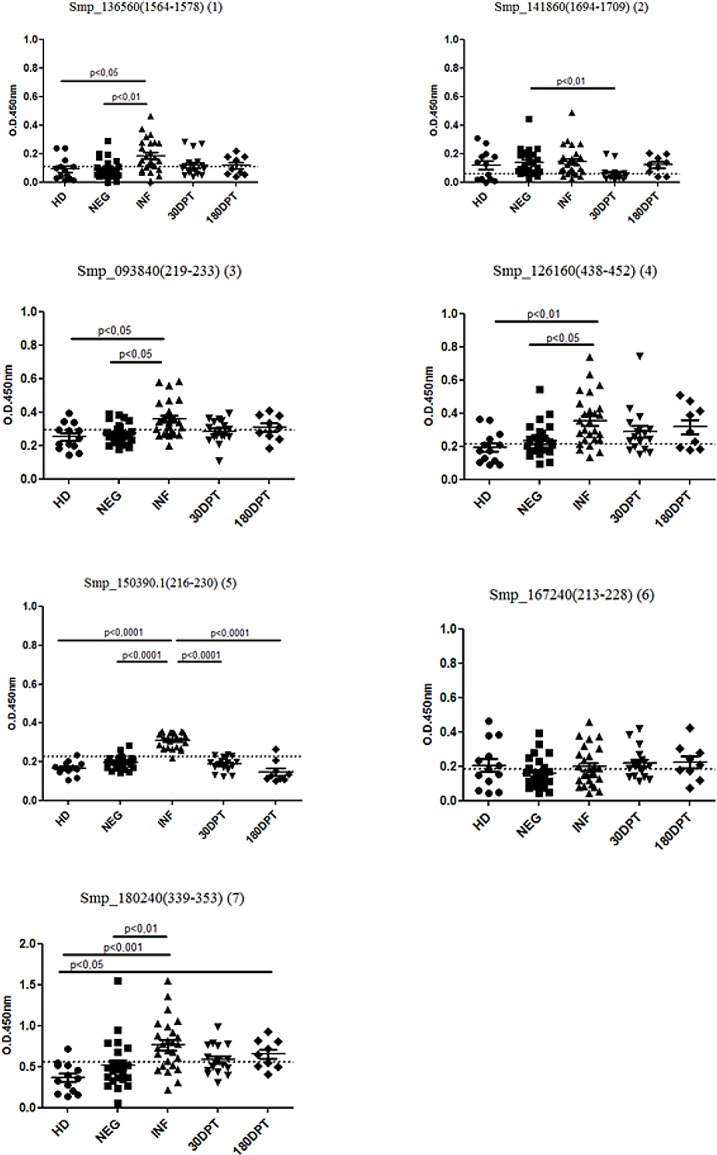
Serum samples were collected from 51 individuals living in an area of low endemicity for S. mansoni infection. Samples from 25 individuals with negative stool examination (NEG) and from 26 individuals with positive stool examination were evaluated (INF). Sera from patients treated 30 and 180 days after treatment were also evaluated. Additionally, sera from 13 healthy donors (HD) not living in an endemic area for schistosomiasis were used as a negative control. Significant differences between groups are indicated in the graphic. Dashed lines represent the cutoff in the absorbance level which determines specificity and sensitivity.
Specificities for the ELISA assay were 76.92%, 46.15%, 76.92%, 69.23%, 100%, 53,85% and 92,31% while sensitivities were 73.08%, 84.62%, 69.23%, 84.62%, 96.15%, 53.85% and 73.08% for peptides 1, 2, 3, 4, 5, 6 and 7, respectively when the cutoff point was determined using the data from healthy donor and infected groups (S2 Table). Similar results were observed when cutoff point was determined using the data from uninfected individuals of endemic area (S3 Table) The performance of each peptide used as an antigen in the ELISA can be observed in the ROC curve (S2 and S3 Figs) and in S2 and S3 Tables, which summarizes important performance values.
Discussion
Currently, schistosomiasis transmission in Brazil is maintained by individuals with low levels of infection, and so there is a need to develop effective approaches for prevention, control and elimination of this disease [66]. A diagnostic test with high specificity and sensitivity would be an important tool for limiting the transmission of this parasite, potentially resulting in its control and elimination. Diagnostic tests with low sensitivity may not detect individuals with low-levels of infection, who then remain infected and contribute substantially to schistosomiasis transmission.
The strategy that we used to select potential proteins for immunodiagnostic assays was based on criteria linked to important characteristics of recognition by the host immune system. These criteria included the presence of signal peptides, low similarity with human proteins, liner B and T CD4+ epitopes and favorable subcellular location (plasma membrane or secreted). We also analyzed the predicted parasite life-cycle stages in the definitive host, considering two approaches: a) selection of proteins for schistosoma diagnostic, covering all the stages in definitive host (schistosomulum, lung schistosomulum, adult worm and egg); and b) selection of proteins for post-treatment diagnosis of cure, excluding the proteins expressed in the egg stage, since after treatment the adult worm is eliminated, but the egg remains for a longer period of time, and its antigens could stimulate antibody production. Because the rational applyed for selecting antigens to be used in post-treatment diagnosis of cure, is based on the use of antigens expressed in schistsosmula and adult worm, reactivity in case of reinfection might occur. Thus making it difficult for the test to descriminate between treatment failure and reinfection. Paired analysis from data of reativicty to peptides before and after treatment suggest that cases of reinfection might have occurred in some individuals (S1 Fig).
After performing the workflow, we observed that some of the selected proteins were annotated as expressed proteins without predicted molecular function (hypothetical proteins) and consequently have been poorly studied. These proteins may be good targets for schistosomiasis serological diagnosis, since they matched all criteria of selection. Other proteins were annotated as putative and with a functional annotation that suggests their function, sometimes describing a conserved domain. These findings suggest the low quality of genome annotation linked with the parasite proteins.
Recently, recombinant antigens [41,67–69] and synthetic peptides [70] have been used in order to increase the specificity of serological tests. One recombinant antigen that has been widely used for identifying patients with low parasite burdens is CCA (circulating cathodic antigen) [69]. In the present work, we verified that this protein matched all the criteria established in our workflow, suggesting our methodology reflects a promising rationale for diagnostic. However, this protein was not selected in our study, because we decided to synthetize peptides from proteins containing epitopes predicted to bind to at least 14 different MHC alleles, and CCA contains epitopes predicted to bind only 13 different MHC alleles.
To validate our strategy of targets selection for use in schistosmiasis diagnosis, linear B cell epitopes were identified in the target proteins and synthetically produced to be tested in ELISA. The approach for epitope selection was based on the benchmark of prediction algorithms described by Resende et al. (2012), which showed that the best combination of algorithms for linear B cell epitope prediction in parasites is BepiPred and AAP12 [71]. Besides, we selected epitopes harboring IDRs in their neighborhood, since the importance of this relationship had already been shown in an immunogenic B cell epitope from Plasmodium vivax [43]. However, we could not confirm this for S. mansoni in our analysis since the peptide with the best performance in ELISA tests was not associated with IDRs.
For the ELISA assays, sera from individuals of an area of low endemicity for S. mansoni infection and sera from healthy donors (HD group) not living in an endemic area for schistosomiasis were used. The synthetic peptides showed a diversity of sensitivities and specificities. Among the seven peptides tested, three (peptides 1, 5 and 7) performed better, with sensitivities and specificities higher than 73%. It is important to highlight that peptide 5 had the best results, with a specificity of 100% and a sensitivity of 96.15%. Furthermore, this peptide was able to differentiate sera from individuals of an area of low endemicity before and after treatment.
Although some peptides tested here did not exhibit high sensitivity and specificity, all of them performed better than the Kato-Katz technique (S4 Table). In this study, Kato-Katz exhibited a sensitivity of 36.46% when two slides from two fecal samples were analyzed, and a sensitivity of 46.15% when 18 slides from four fecal samples were evaluated.
Other studies have also focused on searching for new antigens to schistosomiasis serological diagnosis using different approaches. Xu and collaborators (2014), using computational tools, selected and expressed a recombinant protein known as SjSP, which was tested in ELISA against human sera and had a sensitivity of 90.4% and a specificity of 98.9% [72] in the diagnosis of S. japonicum infection. Zhong and collaborators (2010) also reported good performance in ELISA testing using the S. japonicum recombinant proteins SjLAP and SjFBPA, with sensitivity ranging from 87.8% to 98.1% and 84.7% to 100%, respectively [35].
Taking into account studies based on S. mansoni recombinant proteins or peptides, our group has recently reported a recombinant protein, Sm200, with a sensitivity of 90% and a specificity of 93% in ELISA tests [42]. Another recombinant protein, SmRP26, was also used in ELISA tests for the diagnosis of S. mansoni infection in acute and chronic phases of the disease, with sensitivities of 83% and 32%, respectively, and a specificity of 97% [73]. Grenfell and collaborators used a recombinant circulating cathodic antigen (CCA) protein and two peptides derived from CCA as S. mansoni antigens in ELISA tests for the diagnosis of the disease. The results demonstrated 96% sensitivity for recombinant CCA protein and 80% and 74% for peptides 1 and 2, respectively [69]. Moreover, a pool of synthetic peptides was also tested as antigens for the diagnosis of S. mansoni infection. This ELISA test showed a sensitivity of 86.8% and a specificity of 94.2% [33]. Similar sensitivities were observed in our study using the peptides identified, but in contrast to the studies described above, we also analyzed peptide recognition by sera from individuals living in an endemic area of schistosomiasis, but characterized as non-infected by parasitological and molecular tests. In this regard, ELISA using peptide 5 achieved the best performance, since it could discriminate individuals living in endemic area who were actively infected from those who were not. Therefore an ELISA test using this peptide has the potential to be used in epidemiological surveys, as well as to monitor treatment efficiency.
In summary, we demonstrated that a bioinformatics approach was successful in selecting good candidates for use in the diagnosis of schistosomiasis, with peptide 5 being the most promising.
Supporting information
Significant diferences are pointed in the graphs.
(TIF)
(TIF)
(TIF)
aLife stages of parasite in definitive host.
1—schistosomulum, lung chistosomulum, adult worm and egg.
2—schistosomulum, lung schistosomulum and adult worm.
(DOCX)
aCI—confidence interval; bPPV = positive predictive value; PPV = number of true positives/(number of true positives + number of false positives); cNPV—negative predictive value; NPV = number of true negatives/(number of true negatives + number of false negatives).
(DOCX)
aCI—confidence interval; bPPV = positive predictive value; PPV = number of true positives/(number of true positives + number of false positives); cNPV—negative predictive value; NPV = number of true negatives/(number of true negatives + number of false negatives).
(DOCX)
Nd—not determined.
O.D.—optical density at 560nm.
*—serum from patient evalueted before treatament in the infected group (INF).
nd—not determined.
r—antigen-reactive sera.
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Fellowships were provided by Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG) to G.B.F.C., by Coordenação de Aperfeiçoamento de Nível Superior (CAPES) to D.M.R. and L.M.V.S. and by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) to J.C.R., P.Z.C., A.T. and C.T.F. This study received financial support from the Fundação de Amparo à Pesquisa do Estado de Minas Gerais (PPM - 00377-15 – C.T.F.; APQ-01661-13 – J.C.R.; PPM-00710-15 – J.C.R. and APQ-03535-13 – C.T.F.), Conselho Nacional de Desenvolvimento Científico e Tecnológico (303711/2015-9 – C.T.F.; 407702/2012-1 – C.T.F.; 301526/2015-0 – J.C.R. and 486618/2013-7 – J.C.R.), Programa de Pós-Graduação em Ciências da Saúde do CPqRR and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
References
- 1.WHO.2014. Shistosomiasis fact sheet 115. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs115/en/. [Google Scholar]
- 2.Berhe N, Medhin G, Erko B, Smith T, Gedamu S, Bereded D, et al. Variations in helminth faecal egg counts in Kato–Katz thick smears and their implications in assessing infection status with Schistosoma mansoni. Acta Trop. 2004;92: 205–212. doi: 10.1016/j.actatropica.2004.06.011 [DOI] [PubMed] [Google Scholar]
- 3.Utzinger J, N’Goran EK, Caffrey CR, Keiser J. From innovation to application: Social–ecological context, diagnostics, drugs and integrated control of schistosomiasis. Acta Trop. 2011;120, Supplement 1: S121–S137. [DOI] [PubMed] [Google Scholar]
- 4.Weerakoon KGAD, Gobert GN, Cai P, McManus DP. Advances in the diagnosis of human schistosomiasis. Clin Micro biol Rev. 2015;28: 939–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu YC. Immunodiagnosis and its role in schistosomiasis control in China: a review. Acta Trop. 2005;96: 130–136. doi: 10.1016/j.actatropica.2005.07.007 [DOI] [PubMed] [Google Scholar]
- 6.Doenhoff MJ, Chiodini PL, Hamilton JV. Specific and sensitive diagnosis of schistosome infection: can it be done with antibodies? Trends Parasitol. 2004;20: 35–39. [DOI] [PubMed] [Google Scholar]
- 7.Katz N, Chaves A, Pellegrino J. A simple device for quantitative stool thick-smear technique in Schistosomiasis mansoni. Rev Inst Med Trop Sao Paulo. 1972;14: 397–400. [PubMed] [Google Scholar]
- 8.The control of schistosomiasis. Second report of the WHO Expert Committee. World Health Organ Tech Rep Ser. 1993;830: 1–86. [PubMed] [Google Scholar]
- 9.Hawkins KR, Cantera JL, Storey HL, Leader BT, de Los Santos T. Diagnostic Tests to Support Late-Stage Control Programs for Schistosomiasis and Soil-Transmitted Helminthiases. PLoS Negl Trop Dis. 2016;22: 10 doi: 10.1371/journal.pntd.0004985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enk MJ, Lima ACL, Massara CL, Coelho PMZ, Schall VT. A combined strategy to improve the control of Schistosoma mansoni in areas of low prevalence in Brazil. Am J Trop Med Hyg. 2008;78: 140–146. [PubMed] [Google Scholar]
- 11.Gonçalves MML, Barreto MGM, Peralta RHS, Gargioni C, Gonçalves T, Igreja RP, et al. Immunoassays as an auxiliary tool for the serodiagnosis of Schistosoma mansoni infection in individuals with low intensity of egg elimination. Acta Trop. 2006;100: 24–30. doi: 10.1016/j.actatropica.2006.09.004 [DOI] [PubMed] [Google Scholar]
- 12.Sandoval N, Siles-Lucas M, Pérez-Arellano JL, Carranza C, Puente S, López-Abán J, et al. A new PCR-based approach for the specific amplification of DNA from different Schistosoma species applicable to human urine samples. Parasitology. 2006;133: 581–587. doi: 10.1017/S0031182006000898 [DOI] [PubMed] [Google Scholar]
- 13.Alarcón de Noya B, Ruiz R, Losada S, Colmenares C, Contreras R, Cesari IM, et al. Detection of schistosomiasis cases in low-transmission areas based on coprologic and serologic criteria: The Venezuelan experience. Acta Trop. 2007;103: 41–49. doi: 10.1016/j.actatropica.2007.04.018 [DOI] [PubMed] [Google Scholar]
- 14.Kinkel HF, Dittrich S, Bäumer B, Weitzel T. Evaluation of eight serological tests for diagnosis of imported schistosomiasis. Clin Vaccine Immunol CVI. 2012;19: 948–953. doi: 10.1128/CVI.05680-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavalcanti MG, Silva LF, Peralta RHS, Barreto MGM, Peralta JM. Schistosomiasis in areas of low endemicity: a new era in diagnosis. Trends Parasitol. 2013;29: 75–82. doi: 10.1016/j.pt.2012.11.003 [DOI] [PubMed] [Google Scholar]
- 16.Colley DG, Binder S, Campbell C, King CH, TchuemTchuenté L-A, N’Goran EK, et al. A five-country evaluation of a point-of-care circulating cathodic antigen urine assay for the prevalence of Schistosoma mansoni. Am J Trop Med Hyg. 2013;88: 426–432. doi: 10.4269/ajtmh.12-0639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamberton PHL, Kabatereine NB, Oguttu DW, Fenwick A, Webster JP. Sensitivity and specificity of multiple Kato-Katz thick smears and a circulating cathodic antigen test for Schistosoma mansoni diagnosis pre- and post- repeated- praziquantel Treatment. PLoS Negl Trop Dis. 2014;8 doi: 10.1371/journal.pntd.0003139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adriko M, Standley CJ, Tinkitina B, Tukahebwa EM, Fenwick A, Fleming FM, et al. Evaluation of circulating cathodic antigen (CCA) urine-cassette assay as a survey tool for Schistosoma mansoni in different transmission settings within Bugiri District, Uganda. Acta Trop. 2014;136: 50–57. doi: 10.1016/j.actatropica.2014.04.001 [DOI] [PubMed] [Google Scholar]
- 19.Greter H, Krauth SJ, Ngandolo BNR, Alfaroukh IO, Zinsstag J, Utzinger J. Validation of a Point-of-Care Circulating Cathodic Antigen Urine Cassette Test for Schistosoma mansoni Diagnosis in the Sahel, and Potential Cross-Reaction in Pregnancy. Am J Trop Med Hyg. 2016;94: 361–364. doi: 10.4269/ajtmh.15-0577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lodh N, Mwansa JCL, Mutengo MM, Shiff CJ. Diagnosis of Schistosoma mansoni without the stool: comparison of three diagnostic tests to detect Schistosoma mansoni infection from filtered urine in Zambia. Am J Trop Med Hyg. 2013;89: 46–50. doi: 10.4269/ajtmh.13-0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legesse M, Erko B. Field-based evaluation of a reagent strip test for diagnosis of schistosomiasis mansoni by detecting circulating cathodic antigen (CCA) in urine in low endemic area in Ethiopia. Parasite. 2008;15: 151–155. doi: 10.1051/parasite/2008152151 [DOI] [PubMed] [Google Scholar]
- 22.Ruppel A, Shi YE, Wei DX, Diesfeld HJ. Sera of Schistosoma japonicum-infected patients cross-react with diagnostic 31/32 kD proteins of S. mansoni. Clin Exp Immunol. 1987;69: 291–298. [PMC free article] [PubMed] [Google Scholar]
- 23.Tsang VC, Wilkins PP. Immunodiagnosis of schistosomiasis. Screen with FAST-ELISA and confirm with immunoblot. Clin Lab Med. 1991;11: 1029–1039. [PubMed] [Google Scholar]
- 24.Silva RM, Kanamura HY, Camargo ED, Chiodelli SG, Nakamura PM, Gargioni C, et al. A comparative study on IgG-ELISA, IgM-IFT and Kato-Katz methods for epidemiological purposes in a low endemic area for schistosomiasis. Mem Inst Oswaldo Cruz. 1998;93: 279–282. [DOI] [PubMed] [Google Scholar]
- 25.Alarcón de Noya B, Cesari IM, Losada S, Colmenares C, Balzán C, Hoebeke J, et al. Evaluation of alkaline phosphatase immunoassay and comparison with other diagnostic methods in areas of low transmission of schistosomiasis. Acta Trop. 1997;66: 69–78. [DOI] [PubMed] [Google Scholar]
- 26.Hamilton JV, Klinkert M, Doenhoff MJ. Diagnosis of schistosomiasis: antibody detection, with notes on parasitological and antigen detection methods. Parasitology. 1998;117 Suppl: S41–57. [DOI] [PubMed] [Google Scholar]
- 27.Alarcón de Noya B, Colmenares C, Lanz H, Caracciolo MA, Losada S, Noya O. Schistosoma mansoni: immunodiagnosis is improved by sodium metaperiodate which reduces cross-reactivity due to glycosylated epitopes of soluble egg antigen. Exp Parasitol. 2000;95: 106–112. doi: 10.1006/expr.2000.4515 [DOI] [PubMed] [Google Scholar]
- 28.Dunne DW, Bain J, Lillywhite J, Doenhoff MJ. The stage-, strain- and species-specificity of a Schistosoma mansoni egg antigen fraction (CEF6) with serodiagnostic potential.Trans R Soc Trop Med Hyg. 1984;78: 460–470. [DOI] [PubMed] [Google Scholar]
- 29.Tsang VC, Tsang KR, Hancock K, Kelly MA, Wilson BC, Maddison SE. Schistosoma mansoni adult microsomal antigens, a serologic reagent. I. Systematic fractionation, quantitation, and characterization of antigenic components. J Immunol. 1983;130: 1359–1365. [PubMed] [Google Scholar]
- 30.Ruppel A, Diesfeld HJ, Rother U. Immunoblot analysis of Schistosoma mansoni antigens with sera of schistosomiasis patients: diagnostic potential of an adult schistosome polypeptide. Clin Exp Immunol. 1985;62: 499–506. [PMC free article] [PubMed] [Google Scholar]
- 31.Doenhoff MJ, Butterworth AE, Hayes RJ, Sturrock RF, Ouma JH, Koech D, et al. Seroepidemiology and serodiagnosis of schistosomiasis in Kenya using crude and purified egg antigens of Schistosoma mansoni in ELISA. Trans R Soc Trop Med Hyg. 1993;87: 42–48. [DOI] [PubMed] [Google Scholar]
- 32.Bligh J, Schramm G, Chiodini PL, Doenhoff MJ. Serological analysis of the outcome of treatment of Schistosoma mansoni infections with praziquantel. Ann Trop Med Parasitol. 2010;104: 511–520. doi: 10.1179/136485910X12786389891245 [DOI] [PubMed] [Google Scholar]
- 33.de Oliveira EJ, Kanamura HY, Takei K, Hirata RDC, Valli LCP, et al. Synthetic peptides as an antigenic base in an ELISA for laboratory diagnosis of schistosomiasis mansoni. Trans R Soc Trop Med Hyg. 2008;102: 360–366. doi: 10.1016/j.trstmh.2007.11.008 [DOI] [PubMed] [Google Scholar]
- 34.Sulbarán GS, Ballen DE, Bermúdez H, Lorenzo M, Noya O, Cesari IM. Detection of the Sm31 antigen in sera of Schistosoma mansoni-infected patients from a low endemic area. Parasite Immunol. 2010;32: 20–28. doi: 10.1111/j.1365-3024.2009.01152.x [DOI] [PubMed] [Google Scholar]
- 35.Zhong Z, Zhou H, Li X, Luo Q, Song X, Wang W, et al. Serological proteome-oriented screening and application of antigens for the diagnosis of Schistosomiasis japonica. Acta Trop. 2010;116: 1–8. doi: 10.1016/j.actatropica.2010.04.014 [DOI] [PubMed] [Google Scholar]
- 36.Kalenda YDJ, Kato K, Goto Y, Fujii Y, Hamano S. Tandem repeat recombinant proteins as potential antigens for the sero-diagnosis of Schistosoma mansoni infection.Parasitol Int. 2015;64: 503–512. doi: 10.1016/j.parint.2015.06.012 [DOI] [PubMed] [Google Scholar]
- 37.Guo JJ, Zheng HJ, Xu J, Zhu XQ, Wang SY, Xia CM. Sensitive and specific target sequences selected from retrotransposons of Schistosoma japonicum for the diagnosis of schistosomiasis. PLoS Negl Trop Dis. 2012;6 doi: 10.1371/journal.pntd.0001579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H, Zhu YC, Si J, Zhao S, Wang XT, Yin XR, Cao LM, Cao GQ, Hua WQ, Xu M, Liang YS. Development and identification of the multiple B cell epitope antigens of Schistosoma japonicum. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2007;4: 285–289. [PubMed] [Google Scholar]
- 39.Martins LMS, Andrade HM de, Vainstein MH, Wanke B, Schrank A, Balaguez CB, et al. Immunoproteomics and immunoinformatics analysis of Cryptococcus gattii: novel candidate antigens for diagnosis. Future Microbiol. 2013;8: 549–563. doi: 10.2217/fmb.13.22 [DOI] [PubMed] [Google Scholar]
- 40.Zerlotini A, Heiges M, Wang H, Moraes RLV, Dominitini AJ, Ruiz JC, et al. SchistoDB: a Schistosoma mansoni genome resource. Nucleic Acids Res. 2009;37: 579–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carvalho GBF, Silva-Pereira RA da, Pacífico LGG, Fonseca CT. Identification of Schistosoma mansoni candidate antigens for diagnosis of schistosomiasis. Mem Inst Oswaldo Cruz. 2011;106: 837–843. [DOI] [PubMed] [Google Scholar]
- 42.Carvalho GBF de, Pacífico LGG, Pimenta DLF, Siqueira LMV, Teixeira-Carvalho A, Coelho PMZ, et al. Evaluation of the use of C-terminal part of the Schistosoma mansoni 200kDa tegumental protein in schistosomiasis diagnosis and vaccine formulation. Exp Parasitol. 2014;139: 24–32. doi: 10.1016/j.exppara.2014.02.003 [DOI] [PubMed] [Google Scholar]
- 43.Bueno LL, Lobo FP, Morais CG, Mourão LC, de Ávila RAM, Soares IS, et al. Identification of a highly antigenic linear B cell epitope within Plasmodium vivax apical membrane antigen 1 (AMA-1). PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0021289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruy P de C, Torrieri R, Toledo JS, Alves V de S, Cruz AK, Ruiz JC. Intrinsically disordered proteins (IDPs) in trypanosomatids. BMC Genomics. 2014;15 doi: 10.1186/1471-2164-15-1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larsen JEP, Lund O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2: 2 doi: 10.1186/1745-7580-2-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Liu H, Yang J, Chou K-C. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids. 2007;33: 423–428. doi: 10.1007/s00726-006-0485-9 [DOI] [PubMed] [Google Scholar]
- 47.EL-Manzalawy Y, Dobbs D, Honavar V. Predicting linear B-cell epitopes using string kernels. J Mol Recognit JMR. 2008;21: 243–255. doi: 10.1002/jmr.893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nielsen M, Lund O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinformatics. 2009;10: 296 doi: 10.1186/1471-2105-10-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11: 1453–1459. [DOI] [PubMed] [Google Scholar]
- 50.Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31: 3701–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dosztányi Z, Csizmók V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347: 827–839. doi: 10.1016/j.jmb.2005.01.071 [DOI] [PubMed] [Google Scholar]
- 52.Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006;7: 208 doi: 10.1186/1471-2105-7-208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han Pengfei, Zhang Xiuzhen, and Feng Zhi-Ping. Predicting disordered regions in proteins using the profiles of amino acid indices. BMC Bioinformatics. 2009;10: (Suppl 1) S42 doi: 10.1186/1471-2105-10-S1-S42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14: 4683–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting Subcellular Localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300: 1005–1016. doi: 10.1006/jmbi.2000.3903 [DOI] [PubMed] [Google Scholar]
- 56.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8: 785–786. doi: 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 57.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 2001;305: 567–580. doi: 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 58.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6: 175–182. [PubMed] [Google Scholar]
- 59.Briesemeister S, Blum T, Brady S, Lam Y, Kohlbacher O, Shatkay H. SherLoc2: A high-accuracy hybrid method for predicting subcellular localization of proteins. J Proteome Res. 2009;8: 5363–5366. doi: 10.1021/pr900665y [DOI] [PubMed] [Google Scholar]
- 60.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215: 403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 61.Chan W. C.; White P. D.—Fmoc Solid Phase Peptide Synthesis–A Practical Approach. Editora Oxford University Press, 2000. [Google Scholar]
- 62.Siqueira LMV, Gomes LI, Oliveira E, de Oliveira ER, de Oliveira ÁA, Enk MJ, et al. Evaluation of parasitological and molecular techniques for the diagnosis and assessment of cure of schistosomiasis mansoni in a low transmission area. Mem Inst Oswaldo Cruz. 2015;110: 209–214. doi: 10.1590/0074-02760140375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gomes JF, Hoshino-Shimizu S, S. Dias LC, Araujo AJSA, Castilho VLP, Neves FAMA. Evaluation of a novel kit (TF-Test) for the diagnosis of intestinal parasitic infections. J Clin Lab Anal. 2004;18: 132–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Siqueira LMV, Coelho PMZ, Oliveira ÁA de, Massara CL, Carneiro NF de F, Lima ACL, et al. Evaluation of two coproscopic techniques for the diagnosis of schistosomiasis in a low-transmission area in the state of Minas Gerais, Brazil. Mem Inst Oswaldo Cruz. 2011;106: 844–850. [DOI] [PubMed] [Google Scholar]
- 65.Gomes LI, dos Santos Marques LH, Enk MJ, de Oliveira MC, Coelho PMZ, Rabello A. Development and evaluation of a sensitive PCR-ELISA system for detection of Schistosoma infection in feces. PLoS Negl Trop Dis. 2010;4 doi: 10.1371/journal.pntd.0000664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lustigman S, Prichard RK, Gazzinelli A, Grant WN, Boatin BA, McCarthy JS, et al. A research agenda for helminth diseases of humans: the problem of helminthiases. PLoS Negl Trop Dis. 2012;6 doi: 10.1371/journal.pntd.0001582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schramm G, Gronow A, Knobloch J, Wippersteg V, Grevelding CG, Galle J, et al. IPSE/alpha-1: A major immunogenic component secreted from Schistosoma mansoni eggs. Mol Biochem Parasitol. 2006;147: 9–19. doi: 10.1016/j.molbiopara.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 68.Doenhoff MJ, Wheeler JG, Tricker K, Hamilton JV, Sturrock RF, Butterworth AE, et al. The detection of antibodies against Schistosoma mansoni soluble egg antigens (SEA) and CEF6 in ELISA, before and after chemotherapy.Ann Trop Med Parasitol. 2003;97: 697–709. doi: 10.1179/000349803225002354 [DOI] [PubMed] [Google Scholar]
- 69.Grenfell R, Harn DA, Tundup S, Da’dara A, Siqueira L, Coelho PMZ. New approaches with different types of circulating cathodic antigen for the diagnosis of patients with low Schistosoma mansoni load. PLoS Negl Trop Dis. 2013;7 doi: 10.1371/journal.pntd.0002054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oliveira EJ de, Kanamura HY, Takei K, Hirata RDC, Nguyen NY, Hirata MH. Application of synthetic peptides in development of a serologic method for laboratory diagnosis of schistosomiasis mansoni. Mem Inst Oswaldo Cruz. 2006;101: 355–357. [DOI] [PubMed] [Google Scholar]
- 71.Resende DM, Rezende AM, Oliveira NJ, Batista IC, Corrêa-Oliveira R, Reis AB, et al. An assessment on epitope prediction methods for protozoa genomes. BMC Bioinformatics. 2012;13: 309 doi: 10.1186/1471-2105-13-309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu X, Zhang Y, Lin D, Zhang J, Xu J, Liu Y, et al. Serodiagnosis of Schistosoma japonicum infection: genome-wide identification of a protein marker, and assessment of its diagnostic validity in a field study in China. Lancet Infect Dis. 2014;14: 489–497. doi: 10.1016/S1473-3099(14)70067-2 [DOI] [PubMed] [Google Scholar]
- 73.Makarova E, Goes TS, Leite MF, Goes AM. Detection of IgG binding to Schistosoma mansoni recombinant protein RP26 is a sensitive and specific method for acute schistosomiasis diagnosis. Parasitol Int. 2005;54: 69–74. doi: 10.1016/j.parint.2004.12.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Significant diferences are pointed in the graphs.
(TIF)
(TIF)
(TIF)
aLife stages of parasite in definitive host.
1—schistosomulum, lung chistosomulum, adult worm and egg.
2—schistosomulum, lung schistosomulum and adult worm.
(DOCX)
aCI—confidence interval; bPPV = positive predictive value; PPV = number of true positives/(number of true positives + number of false positives); cNPV—negative predictive value; NPV = number of true negatives/(number of true negatives + number of false negatives).
(DOCX)
aCI—confidence interval; bPPV = positive predictive value; PPV = number of true positives/(number of true positives + number of false positives); cNPV—negative predictive value; NPV = number of true negatives/(number of true negatives + number of false negatives).
(DOCX)
Nd—not determined.
O.D.—optical density at 560nm.
*—serum from patient evalueted before treatament in the infected group (INF).
nd—not determined.
r—antigen-reactive sera.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.