Abstract
The aryl hydrocarbon receptor (AHR) is a basic helix-loop-helix transcription factor conserved across phyla from flies to humans. Activated by a number of endogenous ligands and environmental toxins, studies on AHR function and gene regulation have largely focused on a toxicological perspective relating to aromatic hydrocarbons generated by human activities and the often-deleterious effects of exposure on vertebrates mediated by AHR activation. A growing body of work has highlighted the importance of AHR in physiologic processes, including immune cell differentiation and vascular patterning. Here we dissect the contribution of the 3 zebrafish AHRs, ahr1a, ahr1b and ahr2, to endothelial cyp1a1/b1 gene regulation under physiologic conditions and upon exposure to the AHR ligand Beta-naphthoflavone. We show that in fish multiple AHRs are functional in the vasculature, with vessel-specific differences in the ability of ahr1b to compensate for the loss of ahr2 to maintain AHR signaling. We further provide evidence that AHR can regulate the expression of the chemokine receptor cxcr4a in endothelial cells, a regulatory mechanism that may provide insight into AHR function in the endothelium.
Introduction
The aryl hydrocarbon receptor (AHR) was originally identified for its role in mediating toxicity of environmental compounds [1]. Dubbed the “dioxin receptor,” AHR is a ligand-activated transcription factor that binds a number of halogenated aromatic hydrocarbons (HAHs) and polycyclic aromatic hydrocarbons (PAHs) [2]. Many of these ligands are environmental toxins like dioxin (see below), but others are derived from compounds in food, gut microbiota, and circulating lipids/hormones. In its unliganded form, AHR is confined to the cytoplasm in a complex with two HSP90 proteins, a p23 and X-associated protein (XAP) chaperones [3]. When ligand bound, conformational changes allow AHR to dissociate from the cytoplasmic complex and bind to a cofactor, aryl hydrocarbon nuclear translocator (ARNT, also called HIF-1B), and together these proteins translocate to the nucleus and bind AHR-response elements in the promoters of target genes [4, 5]. While the AHR has many transcriptional targets, the two best characterized are the cytochrome P450 1A1 and 1B1 (CYP1A1 and CYP1B1) genes [6]. These enzymes belong to the CYP1 family of monoxygenases, and are responsible for the metabolism of a variety of endogenous and xenobiotic molecules into bioactive compounds and toxic derivatives, respectively.
The “dioxin receptor” was so-called because of its role in mediating the carcinogenic effects of the environmental contaminant 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) [7]. The affinity of AHR for TCDD is extremely high, and the dissociation constant (Kd) for binding is in the order of picomolar concentrations of TCDD [8]. In comparison, the PAH Beta-naphthoflavone (BNF) is about 100x less potent, and the tryptophan metabolite kynurenine (an AHR agonist in vivo) binds AHR with a Kd of ~4 uM [9, 10]. The Drosophila homologue cannot bind dioxin [11], which -combined with the fact that common environmental toxins were not found before widespread human development- suggests that AHR has physiological roles besides regulating xenobiotic metabolism.
AHR-deficient mice indicate physiological functions of AHR signaling within the vasculature. Notably, AHR -/- mice have smaller livers, displaying portosystemic shunting that prevents blood from circulating through the organ [12]. This phenotype is the result of an intact ductus venosus, a fetal vascular connection between the portal vein and inferior vena cava that must be closed at birth. This event requires AHR function in ECs, as EC-specific Tie2:Cre-mediated deletion reproduces the shunt phenotype, while Albumin:Cre-mdiated deletion in hepatocytes does not [13]. AHR -/- mice also have vascular patterning defects in the eye (ectopic loops) and kidney (decreased capillary density), but these are relatively uncharacterized [12].
Interestingly, shear stress plays a role in the AHR/CYP1A/1B signaling axis. The expression of AHR and cyp1a1/1b1 are positively regulated by shear stress, and nuclear translocation of AHR is enhanced in areas of high shear [14, 15]. Han et al. showed that CYP induction by flow required AHR binding to its promoter, and that flow-induced cell cycle inhibition required AHR. CYP enzymes can metabolize lipids like arachnidonic acid into vasoactive HETEs and EETs that modulate vessel tone [16]. Accordingly, AHR -/- and CYP1B1 -/- mice have low blood pressure, while CYP1A1 -/- animals are hypertensive [17, 18]. Kynurenine, a known endogenous AHR ligand, exhibits vasodilatory properties [19]. Still, the role of AHR in developmental vascular patterning is unknown.
The zebrafish model has been used primarily to investigate AHR-mediated toxicity of HAHs and PAHs during development. In contrast to mammals, which contain a single AHR gene, the zebrafish genome has 3 AHR co-orthologues. Phylogenetic analysis supports a model where a single ancestral AHR gene in invertebrates underwent a duplication to yield an AHR1 and AHRR, and these are present in all chordate lineages. A subsequent duplication of AHR1 led to two AHR genes (AHR1/AHR2) [20]. Mammals are thought to have lost the AHR2 gene. Fish retained the AHR1/AHR2 gene pair, and this was expanded by the whole genome duplication that occurred in the lineage of ray-finned fish after the separation of bony and cartilaginous fish [21], yielding 4 co-orthologues: AHR1a/AHR2a and AHR1b/AHR2b. In the zebrafish Danio rerio, one of these co-orthologues has been lost, yielding 3 AHR genes that have been identified: ahr1a, ahr1b and ahr2 [22]. TCDD administration causes pericardial edema, growth failure, circulation loss, craniofacial malformations and death in zebrafish embryos [23]. AHR -/- mice are protected from these deleterious effects of dioxin [24]. Of the zebrafish AHRs, ahr2 is responsible for most TCDD-induced toxicity and knocking it down dramatically improves survival [25]. However, data on vascular patterning in ahr2-deficient zebrafish sans exposure to toxic chemicals is scarce. A flow defect has been reported in a subset of cranial vessels in embryos exposed to AHR agonists, along with some subtle patterning variations [26–28]. Recently, it has been shown that ahr1b can functionally compensate for the loss of ahr2 in the endothelium to upregulate cyp1a1 in response to exogenous AHR ligands [29]. However, no analysis was done in these embryos without exposure to chemicals, and there is no report of vascular patterning defects in zebrafish lacking all AHR function.
Materials and methods
Fish lines
Zebrafish were housed in an aquaculture facility maintained at 28.5°C with a 14 hrs day/night cycle, as per standard fish husbandry practice [30]. Fish carrying the vascular transgene Tg(kdrl:EGFP)s843 [31] were used to examine vessel morphologies in live embryos. ahr2hu3335 mutants used in this study were described previously [29]. All animal experiments were performed in compliance with the relevant laws and institutional guidelines of the Max Planck Institute for Molecular Biomedicine and were approved by local animal ethics committees of the Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen.
Live imaging and drug treatments
Embryos were maintained at 28.5°C in E3 medium, supplemented with 0.003% phenylthiourea to prevent pigment formation. For live imaging experiments, embryos were anaesthetized in 1x tricaine (168 mg/L). Imaging was done on an inverted Sp5 (Leica Microsystems) confocal microscope, with embryos mounted in 1% low melting point agarose (Life Technologies) in glass bottom dishes (Wilco Wells). Embryos from ISH experiments were stored in 75% glycerol/PBST and imaged on standard stereo dissection microscopes, with high magnification images taken on a Zeiss AxioImager microscope. For drug treatments, nifedipine (Sigma-Aldrich) and Beta-naphthoflavone (Sigma-Aldrich) were dissolved in DMSO to 10 mM stocks, and diluted in E3 medium to experimental concentrations listed. For nifedipine treatments, 2x tricaine was also included to anaesthetize embryos. Treatments were administered to embryos in 24-well plates, with 25 embryos/well in 1mL of E3.
TALEN mutagenesis
All of the mutants generated in the course of this study were made by TALEN mutagenesis. TALEN pairs were designed using the online TAL Effector Nucleotide Targeter (https://tale-nt.cac.cornell.edu/). Criteria for optimal TALEN pairs were followed as described in Cermak et al., 2011 [32]. The following TALEN pairs were selected for optimal length and presence of a restriction site in the spacer.
ahr1a 5’ TALEN arm
DNA sequence: 5’-gaagttctggccagcttggc-3’
RVD sequence: NH NI NI NH NG NG HD NG NH NH HD HD NI NH HD NG NG NH NH HD
ahr1a 3’ TALEN arm
DNA sequence: 5’-tccacaggagatcgtatcc-3’
RVD sequence: NH NH NI NG NI HD NH NI NG HD NG HD HD NG NH NG NH NH NI
ahr1b 5’ TALEN arm
DNA sequence: 5’-cggttaaactctgag-3’
RVD sequence: HD NN NN NG NG NI NI NI HD NG HD NG NN NI NN
ahr1b 3’ TALEN arm
DNA sequence: 5’-tcttcttccatttcc-3’
RVD sequence: NN NN NI NI NI NG NN NN NI NI NN NI NI NN NI
The Golden Gate TALEN kit v2.0 used for generation of TALENs was a gift from Daniel Voytas and Adam Bogdanove (Addgene kit # 1000000024). ahr1a and ahr1b TALENs were assembled using NH RVD module plasmids instead of NN RVDs, and cloned directly into the GoldyTALEN vector [33]. TALENs were linearized with SacI and mRNA produced with T3 mMessage Machine kit (Ambion). mRNA for each TALEN arm was co-injected (1:1 mix of 5’:3’ TALEN arm) into 1-cell stage WT embryos. TALEN efficacy was checked by DNA extraction from 20 pooled embryos from each injection (see Genotyping section for details). PCR was performed with standard TAQ polymerases with no proof-reading ability, and undigested bands were gel extracted for subsequent TOPO® cloning by TA overhangs and sequencing to identify founder fish and establish stable lines. Total TALEN mRNA injected was 50 pg (ahr1a) and 20 pg (ahr1b), and ~100 fish raised for the founder (F0) generation.
Genotyping
Standard PCR genotyping protocol was followed with HotMaster TAQ polymerase (PeqLab, 01–8210) using the following primers and conditions:
ahr1a: forward primer GGGCGATGTCTAAATCTATTCT and reverse primer GAGAGATTAATGCATAACCAATTT, digested with HinfI. ahr1b: forward primer TGTATATTTTCATGCCATCC and reverse primer GTGTTTCGTCTGACCCAG, digested with MspI, ahr2: forward primer GCTCAATGTCCCTGGGAACACCTGG and reverse primer CTGAATTCCTGGTTAGCAGCCAAGTTATTCTG.
The annealing temperature for all primer pairs was 60°C. PCR for ahr1a yields a 499 bp product, cleaving to 327 bp and 172 bp fragments in WT. PCR for ahr1b yields a 447 bp product, cleaving to 255 bp and 192 bp fragments in WT. PCR amplicons for ahr2 were sequenced to identify the WT and hu3335 allele.
In situ hybridization
In situ hybridization to detect transcripts in whole embryos was performed as previously described [34]. The cxcr4a probe was previously described [35]. To generate all other probes, RNA was extracted from pooled 48 hpf WT embryos using the Qiagen Micro RNeasy kit (Qiagen, 74004). cDNA was produced with the iScript cDNA Synthesis Kit from Bio-Rad (Bio-Rad, 1708890), and the following primers were used to amplify gene-specific cDNA using standard TAQ polymerase. ahr1a: forward primer CGGCATGAGTTTCAGAGACA and reverse primer AAGAGGCAGGATCAGAAGAT, ahr1b: forward primer CCAGAAAGGAGCAGGTACGGATGAAGTTAC and reverse primer GTTGACGGCTGTCTGCGAGAGGG, ahr2: forward primer GATGGAGTCAACTTCTCAGAAGGGGAGC and reverse primer ACTACTAGTATCCATTCCCTCTTGGATGTTCATTC, cyp1a1: forward primer CGGAAACAACCCACATTTGAGTCTGAC and reverse primer CGGTGAACTTTAACCTTTGCAGCAGGAT, cyp1b1: forward primer CGAATGGCTCAGAAATACGGCGAC and reverse primer TGGACCAGCACAGACGTGAAGAGGAA, glut1: forward primer GCAGGAGGAACTCAATGCTC and reverse primer TGGACCAGCACAGACGTGAAGAGGAA. These PCR amplicons were TOPO cloned into the pCRII-TOPO TA vector from Invitrogen (Invitrogen, 450640), and digested/sequenced to determine orientation for antisense probe generation following linearization of the plasmid.
Quantification and statistical analysis
All experiments were performed 2–3 times, with the exception of confocal imaging of the vasculature in live embryos. This was performed once to document the lack of gross vascular defects in ahr1a -/- and triple AHR mutants. To analyze different AHR mutants, embryos for ISH experiments were obtained from incrosses of ahr2hu3335 +/- or ahr1amu153 +/-; ahr1bmu145 +/-; ahr2hu3335 +/- adult fish. Embryos from WT incrosses were used for experiments only comparing DMSO and BNF treatments. For analysis of ISH experiments, all embyros were imaged and genotyped. Numbers are reported as “Number of embyros of a particular genotype with staining pattern as in image/total number of embryos of that genotype analyzed”. Statisical analysis for counts of cxcr4a-positive cells in the hindbrain was performed using GraphPad Prism 6.0. For live imaging experiments, fish were imaged blindly and genotyped aferwards. All data available from the authors upon request.
Results
Zebrafish AHR genes are differentially expressed during development
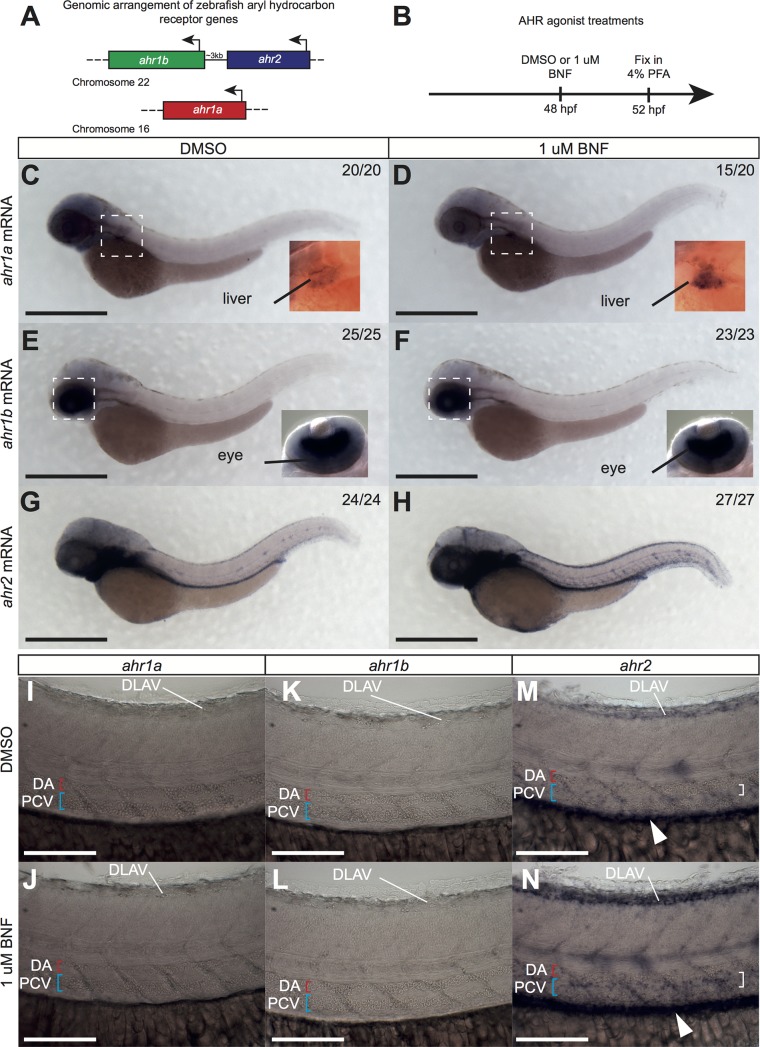
Whereas mammals possess a single AHR gene, the zebrafish Danio rerio contains 3 distinct AHR genes in its genome [22]: linkage of ahr1b and ahr2 on chromosome 22, and a single ahr1a on chromosome 16 (Fig 1A). To address the role of AHR in embryonic vascular development, we first determined the expression patterns of all 3 AHR genes at 52 hpf by in situ hybridization. In addition to endogenous transcripts, we also investigated AHR expression in embryos exposed to the AHR agonist Beta-naphthoflavone (BNF) (Fig 1B). We found differential expression of AHR genes in the embryo, with partially overlapping domains and responsiveness to BNF treatment. ahr1a mRNA was only weakly detected in the liver, and was upregulated by BNF (Fig 1C and 1D). In contrast, ahr1b was highly expressed only in the developing eye. This expression was not influenced by BNF (Fig 1E and 1F). Finally, ahr2 was the most broadly expressed of the 3 genes and was strongly upregulated by BNF (Fig 1G and 1H). Higher magnification images of the trunk indicate expression of ahr2, but not ahr1a or ahr1b, in the tissue between the dorsal aorta (DA) and posterior cardinal vein (PCV) and around the dorsal longitudinal anastomotic vessel (DLAV) (Fig 1I–1N). These results show that different AHR genes are expressed in distinct tissues and respond differently to agonist treatment.
Fig 1. Zebrafish AHR genes are differentially expressed during development.
(A) Schematic of genomic arrangement of zebrafish AHR genes. Note linkage of ahr1b and ahr2 (~3 kb intergenic distance). (B) Experimental design. Embryos were exposed to DMSO or 1 uM BNF for 4 hrs starting at 48 hpf, then fixed in 4% PFA for ISH. (C-H) Stereomicroscope images of whole mount ISH in 52 hpf embryos exposed to DMSO or 1 uM BNF showing expression of ahr1a (C, D), ahr1b (E, F) and ahr2 (G, H). Boxes indicate region of high magnification image in inset. Numbers indicate embryos with indicated expression pattern/total embryos analyzed. Scale bar is 500 um. (I-N) High magnification images of trunk in 52 hpf embryos exposed to DMSO or 1 uM BNF showing expression of ahr1a (I, J), ahr1b (K, L) and ahr2 (M, N). No staining evident for ahr1a or ahr1b in either condition. ahr2 expression is detected in DLAV, gut (white arrowhead) and region between DA and PCV (white bracket), and is increased by BNF-treatment. Scale bar is 100 um. Abbreviations—AHR: aryl hydrocarbon receptor, BNF: beta-naphthoflavone, DA: dorsal aorta, DLAV: dorsal longitudinal anastomotic vessel, DMSO: dimethylsulfoxide, hpf: hours post fertilization, ISH: in situ hybridization, kb: kilobase pair PCV: posterior cardinal vein, PFA: paraformaldehyde.
Differential vascular expression of cytochrome p450 1 genes under basal and BNF-induced conditions
AHR signaling activates expression of CYP1 genes. We therefore analyzed the endogenous and BNF-induced expression of cyp1a1/b1 in order to understand where AHR signaling might be activated, paying particular attention to vascular-specific domains. Both cyp1a1 and cyp1b1 were strongly upregulated in embryos exposed to BNF (Fig 2A and 2B). In the trunk neither enzyme showed strong endogenous expression in the epidermis, and cyp1a1 was the only gene detected in the vasculature (S1A and S1B Fig). Specifically, we observed strong expression in the PCV, intersegmental vessels (ISVs), and DLAV, with limited expression in a few cells of the DA. BNF led to high induction of cyp1a1 expression in the epidermis, precluding imaging of blood vessels located more deeply within the tissue (S1C Fig). cyp1b1 expression was induced in the epidermis to a lower extent by BNF, permitting the visualization of internal structures. We detected induced cyp1b1 expression in the DA, DLAV and ISVs of the trunk (S1D Fig). Thus, cyp1a1 is the predominate CYP gene expressed in the trunk vasculature and is highly enriched in the vein and small vessels with less expression in the primary artery.
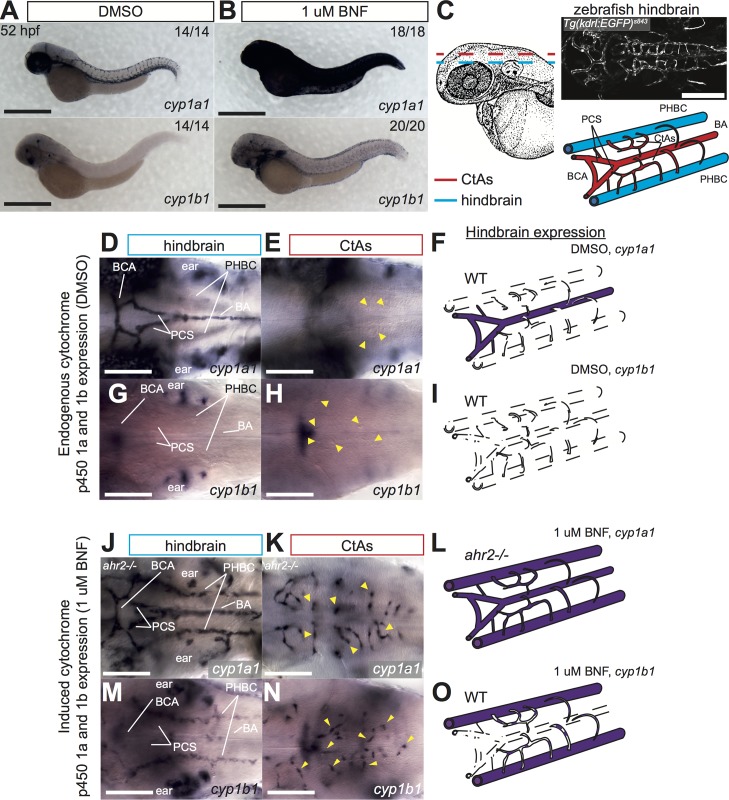
Fig 2. Differential vascular expression of CYP genes under basal and BNF-induced conditions.
(A) Stereomicroscope images of whole mount ISH for cyp1a1 and cyp1b1 in 52 hpf WT embryos after 4 hrs exposure to DMSO (A) or 1 uM BNF (B). Strong upregulation of cyp1a1 in skin can be detected. cyp1b1 is also induced in the skin, but to a lesser extent. Numbers indicate embryos with indicated expression pattern/total embryos analyzed. Scale bar is 500 um. (C). Camera lucida drawing of 48 hpf zebrafish embryo from Kimmel, et al 1995 [36]. Dashed lines indicate plane of images to visualize central arteries (CtAs, red) and hindbrain vessels (blue). Confocal image shows dorsal view of hindbrain vasculature of live embryo at 48 hpf (scale bar is 200 um). Cartoon schematic depicts arrangement of hindbrain vessels. (D-I) High magnification images of basal cyp1a1/b1 expression at 52 hpf in the hindbrain (D, G) and CtAs (E, H) of the head vasculature in embryos treated with DMSO. Schematics in F,I summarize hindbrain expression patterns of both genes. (J-O) High magnification images of BNF-induced cyp1a1/b1 expression at 52 hpf in the hindbrain (J, M) and CtAs (K, N) of the head vasculature in embryos treated with 1 uM BNF. Schematics in L,O summarize hindbrain expression patterns of both genes. Analysis of cyp1a1 expression was performed in ahr2 mutant embryos to permit visualization of hindbrain vasculature. Yellow arrowheads mark CtAs. Scale bar is 100 um in all high magnification images. Abbreviations–AHR: aryl hydrocarbon receptor, BA: basilar artery, BCA: basal communicating artery, BNF: beta-naphthoflavone, CtA: central artery, CYP: cytochrome p450, DMSO: dimethylsulfoxide, hpf: hours post fertilization, ISH: in situ hybridization, PCS: posterior communicating segments, PHBC: primordial hindbrain channel, WT: wildtype.
We next asked if endogenous CYP1 expression might be similarly distributed between arteries, veins and capillaries in other vascular beds and turned our attention to the zebrafish cranial vasculature (Fig 2C). At this stage, the hindbrain vasculature is composed of two bilaterally paired veins, the primordial hindbrain channels (PHBCs). Between them lies the primary arterial input into the brain, the basilar artery (BA), branching via the paired posterior communicating segments (PCS) to form the basal communicating artery (BCA). The central arteries (CtAs) extend dorsally and medially to connect the PHBCs to the BA and irrigate the neuronal tissue. In the plane below these brain vessels the bifurcated lateral dorsal aortae (LDA) collect blood from the heart to be delivered posteriorly toward the trunk via the DA (S1E Fig).
We found cyp1a1 to be restricted to cranial arteries, including the BA, BCA, and PCS (Fig 2D). While high expression was detected in the liver and the LDA (asterisk in S1F Fig), cyp1a1 was notably excluded from the CtAs and PHBCs (Fig 2D–2F). In the anterior head, we also detected cyp1a1 in the paired ventral primitive internal carotid arteries (PICA) between the eyes, and in the optic artery (OA) (S1G and S1H Fig). Additionally, cyp1a1 was highly expressed in the outer eye epithelium but not in the optic furrow (S1I Fig, arrow)
In comparison to cyp1a1, cyp1b1 displayed a more narrow expression pattern and was detected in the developing ear and a region in the middle of the brain, but was not detectable in any vascular structures (Fig 2G–2I). This finding extended to the anterior head as well, where cyp1b1 expression was absent from the liver as well as the LDA, PICA and OA vessels (S1J–S1L Fig). Therefore, as in the trunk, the vascular structures of the head are normally devoid of cyp1b1 expression. Interestingly, in the eye, cyp1b1 was primarily expressed near the base of and in a swath along the optic furrow (S1M Fig, arrow). Therefore, these two CYP genes display a complementary pattern of expression in the embryonic eye, with cyp1b1 expression covering those optic regions lacking cyp1a1.
Lastly, we analyzed BNF-induced changes in CYP1 expression. As already shown, BNF upregulates cyp1a1 to such an extent as to make imaging of interior structures impossible with conventional light microscopy (Fig 2B). We therefore leveraged a previously published ahr2 mutant [29] to examine BNF-induced cyp1a1 expression in the cranial vasculature. This mutant fails to induce cyp1a1 in the skin, enabling the visualization of interior vessels. cyp1a1 expression was preserved in the BA, PCS and BCA, but BNF treatment expanded its expression into the PHBC and CtAs as well, rendering all brain vessels cyp1a1-positive (Fig 2J–2L). In the anterior head, ahr2 mutants had cyp1a1 expression in the liver, LDA and PICA vessels, and strong upregulation in the eye (S1N–S1Q Fig). In wildtype embryos, we found that BNF induced the expression of cyp1b1 in the LDA, but did not cause an upregulation in the liver (S1R Fig). Strikingly, in the hindbrain BNF treatment induced cyp1b1 expression only in a subset of vessels. Expression was readily detected in the PHBC, but not the BCA, BA or PCS (Fig 2M). cyp1b1 expression was induced in the CtAs as well, but in a somewhat more punctate fashion than cyp1a1 (compare Fig 2K to 2N). Therefore, in the hindbrain cyp1b1 induction is excluded in those vessels that are highly cyp1a1-positive under normal conditions (Fig 2O). In the anterior head, cyp1b1 induction was observed in the PICA vessels, but not the OA (S1S and S1T Fig). In the eye, the induction of epidermal cyp1b1 expression by BNF in addition to the basal domain in the optic furrow was also observed (S1U Fig, compare to S1M). Thus, while cyp1a1 showed basal levels of expression within endothelial cells, we detected cyp1b1 expression within blood vessels only after AHR agonist treatment. The marked induction of both cyp1a1/b1 in the CtAs, which are normally negative for either CYP gene, prompted us to examine if the CtAs had an otherwise normal gene expression profile, and we therefore analyzed glut1 expression in BNF treated embryos. This gene encodes a glucose transporter integral for blood-brain-barrier (BBB) formation known to be enriched in the zebrafish brain [37]. We found that vascular glut1 expression was restricted to brain capillaries including the CtAs, and this expression was reduced by BNF treatment (S2 Fig), indicating that BNF-induced AHR activation might affect BBB maturation.
These findings illustrate that, similarly to aryl hydrocarbon receptors, CYP function in the embryo appears partitioned to different genes with different expression domains that complement one another. This is tightly regulated such that the brain arteries (but not capillaries or veins) are cyp1a1-positive in contrast to the trunk vasculature, which has high cyp1a1 expression in the vein and capillaries but less in the primary artery. Interestingly, hyper-activation of the AHR pathway by BNF induces cyp1b1 in blood vessels that ordinarily do not express high levels of either CYP gene (e.g. the PHBCs and CtAs).
Generation of zebrafish ahr1a and ahr1b mutants
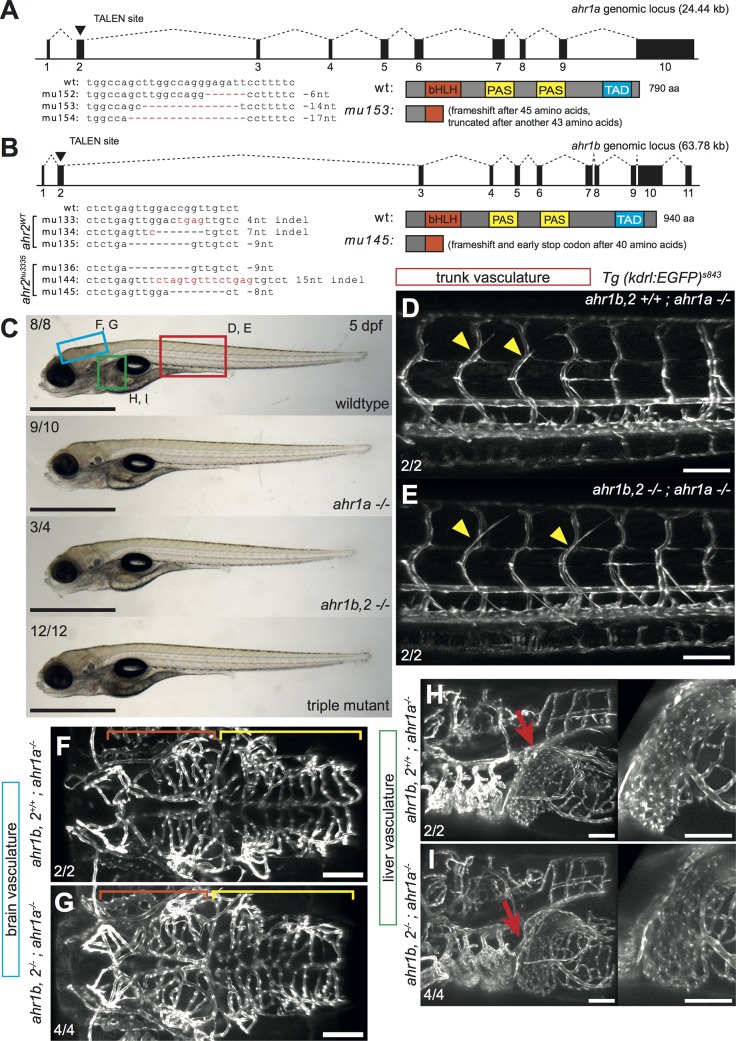
To study the function of AHR in vascular development, we generated zebrafish mutants that lack all AHR function. While a TILLING allele (hu3335) in ahr2 [29] was readily available, we used TALEN pairs targeted to a region in the bHLH domain of exon 2 in both ahr1b and ahr1a to generate further mutants (Fig 3A and 3B). Importantly, since ahr1b and ahr2 are tightly linked, we obtained ahr1b mutants in WT (mu133-mu135) and ahr2hu3335/hu3335(mu136, mu144, mu145) genetic backgrounds to ensure that we could study the function of ahr1b alone in single-mutants or in ahr1b/ahr2hu3335 double mutants (hereafter written as ahr1b,2 -/-) (Fig 3B). We chose the ahr1amu153 and ahr1bmu145 alleles for subsequent analysis because both consist of even-numbered nucleotide deletions that lead to predicted frameshifts and early stop codons, and together allow for the study of triple AHR mutant embryos. To our surprise, ahr1a -/- and triple AHR-deficient embryos were viable and morphologically indistinguishable from their WT siblings, forming normal swim bladders and feeding by 5dpf (Fig 3C). We analyzed a number of vascular beds at different developmental stages, but did not observe obvious patterning defects. Angiogenesis in the trunk proceeded normally, and triple mutants developed the full complement of ISVs and had circulation (Fig 3D and 3E). The brain had no overt vascular defects, and both midbrain and hindbrain compartments were well vascularized (Fig 3F and 3G). Due to the reported liver vascular defects in AHR -/- mice [12], we examined the liver vasculature in our mutants at 5 dpf (Fig 3H and 3I). However, there were no gross differences in size and complexity and no obvious evidence of shunting, suggesting that AHR is dispensable for embryonic vascular patterning in zebrafish.
Fig 3. Generation of zebrafish ahr1a and ahr1b mutants.
(A, B) Depiction of genomic locus for ahr1a (A) and ahr1b (B). TALEN pairs were designed targeting the bHLH domain in exon 2 of both genes. Genomic lesions in the TALEN spacer indicated in alignment to WT sequence, and cartoons show predicted protein products. For ahr1b, alleles mu133-mu135 were generated in a WT genetic background, while mu136-mu145 were made in an ahr2hu3335 homozygous mutant background. (C) Brightfield images of WT, ahr1a -/-, ahr1b,2 -/- and triple AHR mutant embryos at 5 dpf. No gross morphological defects in AHR mutants are apparent. Boxes indicate region of imaging for different vascular beds in D-I. Numbers correspond to “number of embryos with inflated swim bladders/total number of embryos of that genotype”. Scale bar is 1000 um. (D-I) Maximum intensity projections of confocal z-stacks comparing the trunk (D, E), brain (F, G) and liver (H, I) vascular beds in ahr1a -/- and triple AHR mutant zebrafish embryos at 5 dpf. Vascular patterning appears normal in both mutants compared to WT. Yellow arrowheads point to forming intercostal vessels in the trunk. Orange brackets mark midbrain vessels, yellow brackets mark hindbrain vessels. Red arrows show the liver. Numbers indicate the “number of embryos of a particular genotype with vascular characteristics as in image/total numbers of that genotype analyzed.” Scale bar is 100 um. Abbreviations—AHR: aryl hydrocarbon receptor, bHLH: basic helix-loop-helix, hpf: hours post fertilization, TALEN: transcription activator-like effector nuclease, WT: wildtype.
Dissection of AHR-dependent and independent control of CYP1 expression
We considered the following 3 possibilities to explain the lack of detectable vascular phenotypes in triple AHR mutant embryos:
no requirement for AHR during vascular development in zebrafish
as yet unnoticed subtle defect(s) in specific vascular beds or specific developmental stages
AHR alleles generated by TALEN mutagenesis are not loss of function
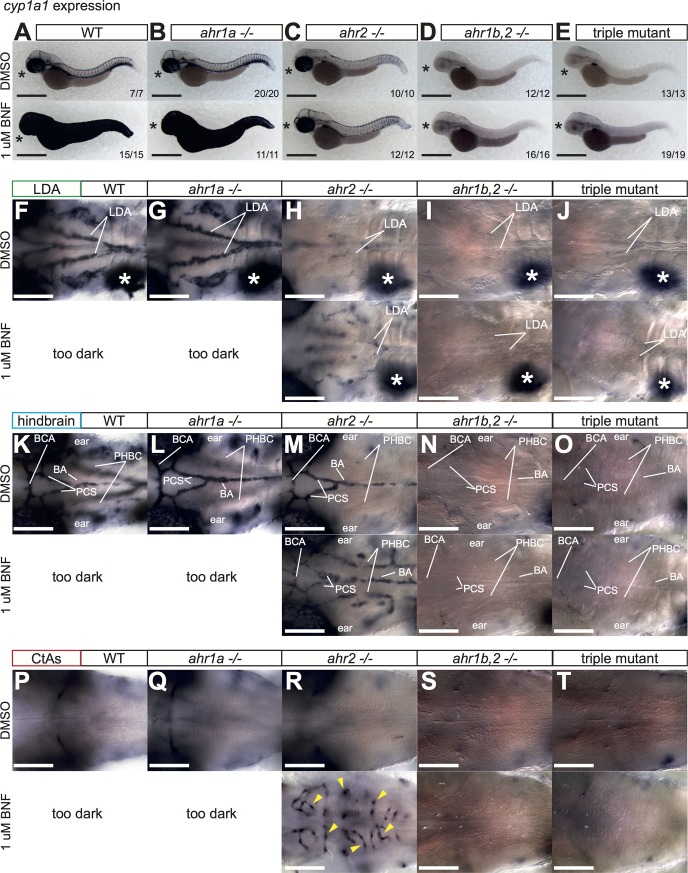
As CYP1 expression is routinely used as a proxy for active AHR signaling, we employed the previous BNF/CYP1 assay with our newly generated mutants to determine if our alleles were indeed nonfunctional, and unravel the contribution of individual AHRs to endogenous and pharmacologically-induced CYP1 expression, starting with cyp1a1. Initial experiments showed no differences in cyp1a1 expression in ahr1b -/- embryos compared to WT, and we therefore focused our analysis on embryos obtained from in-crosses of ahr1amu153 +/-; ahr1bmu145 +/-; ahr2hu3335 +/- adults, yielding clutches containing WT, ahr1a -/-, ahr1b,2 -/- and triple AHR mutant embryos. We found no difference between WT and single ahr1a mutants (Fig 4A and 4B) in terms of endogenous or BNF-induced cyp1a1 expression. ahr2 mutants maintained high levels of cyp1a1 in the eye, but had clearly reduced endogenous expression of cyp1a1 in the trunk vasculature (Fig 4C, upper panel). Closer examination of the trunk revealed that in ahr2 mutants, endogenous cyp1a1 expression was almost completely lost in the DA and PCV vessels compared to WT and ahr1a -/- mutants, while somewhat reduced cyp1a1 levels persisted in the ISVs and DLAV (S3A–S3C Fig, upper panels). This remaining vascular expression could be augmented by BNF treatment, observable owing to the aforementioned loss of cyp1a1 induction in the skin of ahr2 mutants (Fig 4C and S3C Fig, lower panels). Finally, analysis of ahr1b,2 and triple AHR mutants showed loss of all cyp1a1 expression in the eye and in the trunk blood vessels, while gut and liver expression were unaffected (Fig 4D and 4E, and S3D and S3E Fig, lower panels). These results demonstrate firstly that ahr1bmu145 is very likely a loss of function allele, and that ahr1b can functionally compensate for loss of ahr2 in the eye and in some, but not all, zebrafish blood vessels. Secondly, they also indicate that not all cyp1a1 expression is dependent on AHR.
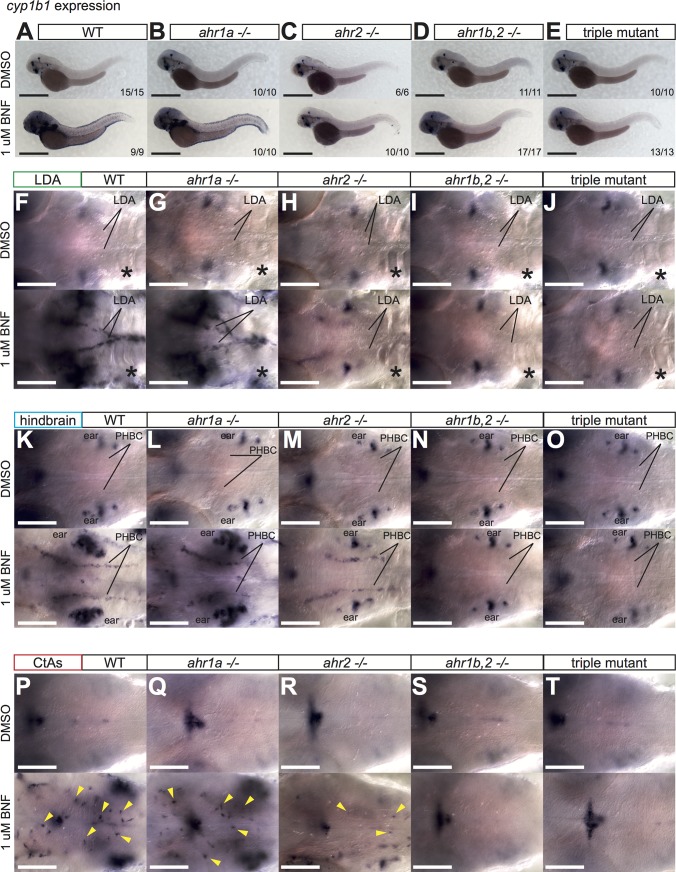
Fig 4. Regulation of cyp1a1 by AHR.
(A) Overview pictures of cyp1a1 expression at 52 hpf in DMSO or BNF-treated WT (A), ahr1a -/- (B), ahr2 -/- (C), ahr1b,2 -/- (D) and triple AHR mutants (E). ahr1a -/- are indistinguishable from WT. (*) indicates expression in the eye in DMSO and BNF-treated embryos, which is lost only in ahr1b,2 -/- and triple AHR mutants. Note reduction of endogenous vascular cyp1a1 expression in ahr2 mutants (C, upper panels) that is lost in ahr1b,2 -/- and triple AHR mutants (D, E upper panels). ahr2 -/- embryos treated with BNF do not induce cyp1a1 expression in the skin, revealing staining in blood vessels underneath (C, lower panel), which is abolished in ahr1b,2 -/- and triple AHR mutants (D, E, lower panels). Scale bar is 500 um. (F-T) High magnification images of blood vessels of 3 different planes of the head vasculature: the LDA (F-J), the hindbrain blood vessels (K-O) and the hindbrain capillaries (P-T) in all genetic combinations with DMSO and BNF treatment. Note strong dependence of LDA (but not hindbrain arteries, PHBC or CtAs) on ahr2 for full cyp1a1 expression. White asterisk denotes liver. Yellow arrowheads label individual CtAs. Numbers indicate embryos with indicated expression pattern/total embryos of that genotype analyzed. Scale bar is 100 um. Abbreviations–AHR: aryl hydrocarbon receptor, BNF: beta-naphthoflavone, CtA: central artery, CYP: cytochrome p450, DMSO: dimethylsulfoxide, hpf: hours post fertilization, LDA: lateral dorsal aortae, PHBC: primordial hindbrain channel, WT: wildtype.
To address whether functional overlap exists between ahr1b and ahr2 in other vessels, we extended our analysis of cyp1a1 expression to the cranial vasculature in different AHR mutants. In the LDA, cyp1a1 expression was normal in ahr1a mutants, almost completely abrogated in ahr2 mutants and lost in ahr1b,2 and triple AHR-deficient embryos (Fig 4F–4J, upper panels). cyp1a1 could be detected in the liver for all genetic combinations of AHR alleles. BNF could weakly stimulate cyp1a1 in the LDA of ahr2 mutants, but not double ahr1b,2 and triple AHR mutants (Fig 4H–4J, lower panels). Analysis of the hindbrain revealed that the endogenous cyp1a1 expression in the BA, BCA and PCS vessels was refractory to loss of ahr1a and ahr2, and was only eliminated in ahr1b,2 and triple AHR mutants (Fig 4K–4O, upper panels). The induction of cyp1a1 in the PHBC vessels by BNF previously observed in ahr2 mutants did not occur in double- and triple AHR fish (Fig 4M–4O, lower panels). Similarly, cyp1a1 induction by BNF in the normally cyp1a1-negative CtAs persisted in ahr2 mutants, and was abolished in ahr1b,2 and triple AHR mutants (Fig 4P–4T).
Together, these results highlight a differential capacity for ahr1b to compensate for ahr2 in controlling cyp1a1 expression in embryonic vessels: strong ahr1b activity in the ISVs, DLAV, BA, PCS, PHBCs and CtAs, with minimal contribution to cyp1a1 expression in the LDA, DA and PCV.
Analysis of cyp1b1 expression showed no effect of loss of any or all AHR gene function on the endogenous levels of cyp1b1 (Fig 5A–5E, upper panels). Both WT and ahr1a mutants displayed high levels of cyp1b1 in the skin in response to BNF, which did not occur in ahr2, ahr1b,2 and triple AHR mutants (Fig 5A–5E, lower panels). In the trunk, the vascular cyp1b1 expression induced by BNF weakly persisted in the ISVs and DLAV of ahr2 mutants and was absent from ahr1b,2 and triple AHR mutants, mirroring the results we obtained for cyp1a1, namely that ahr1b is functional in the DLAV and ISVs and less in the DA and PCV (S3F–S3J Fig). Similarly, analysis of the cranial vessels showed no induction of cyp1b1 in the LDA of ahr2 mutants, while induction in the CtAs and PHBC could occur in the absence of ahr2, but not in embryos lacking ahr1b,2 or all 3 AHRs (Fig 5F–5T). These results highlight the importance of both ahr1b and ahr2 in zebrafish endothelium, and emphasize the nuanced spatial requirements for AHR function across the embryo in physiologic and toxicological responses.
Fig 5. Regulation of cyp1b1 by AHR.
(A-E) Overview pictures of cyp1b1 expression at 52 hpf in DMSO or BNF-treated WT (A), ahr1a -/- (B), ahr2 -/- (C), ahr1b,2 -/- (D) and triple AHR mutants (E). No changes in expression pattern were evident in DMSO-treated embryos that lacked any AHR genes. Induction in the skin by BNF was dramatically reduced in ahr2 -/- (compare A to C, lower panels). Scale bar is 500 um. F-T) High magnification images of blood vessels in 3 different planes of the head vasculature: the LDA (F-J), the hindbrain blood vessels (K-O) and the hindbrain capillaries (P-T) in all genetic combinations with DMSO and BNF treatment. Black asterisks denote approximate location of liver. Yellow arrowheads label individual CtAs. Note maintenance of endogenous cyp1b1 expression in the ear (K-O, upper panels) and head (P-T, upper panels) of all mutants. Expression of cyp1b1 was not detected in the liver for either treatment in any genetic combination (F-J). Note lack of BNF-induced staining in LDA of ahr2 mutants, but strong staining in PHBCs (H and M, lower panels). Numbers indicate embryos with indicated expression pattern/total embryos of that genotype analyzed. Scale bar is 100 um. Abbreviations–AHR: aryl hydrocarbon receptor, BNF: beta-naphthoflavone, CtA: central artery, CYP: cytochrome p450, DMSO: dimethylsulfoxide, hpf: hours post fertilization, LDA: lateral dorsal aortae, PHBC: primordial hindbrain channel, WT: wildtype.
Transcriptional response of the chemokine receptor cxcr4a to BNF is AHR-dependent
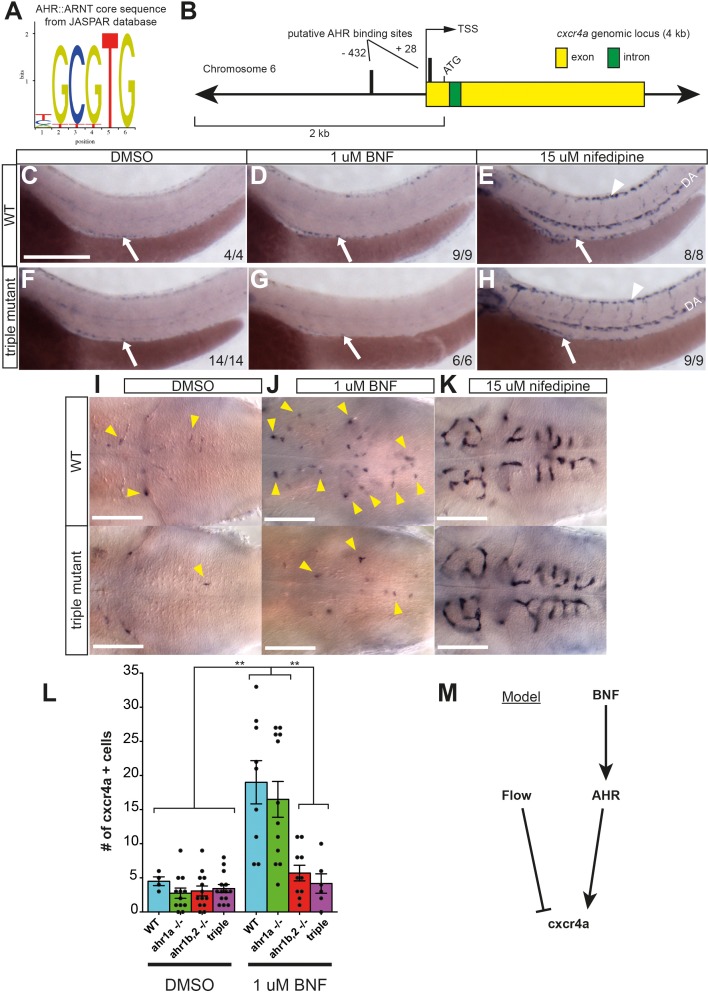
The chemokine receptor cxcr4a has been reported to be differentially expressed in zebrafish embryos exposed to AHR ligands [38]. However, this study was performed by microarray analysis of whole embryo lysate, and therefore no spatial information on the nature of this regulation is known. The AHR::ARNT heterodimer binds a canonical AHR response element, (TNGCGTG) (Fig 6A) [39]. Using the 2 kb promoter sequence 5’ of the ATG start codon in cxcr4a as a template, a search of the JASPAR online database of transcription factor binding motifs revealed 2 putative AHR binding sites (Fig 6B). To test the hypothesis that AHR may be a transcription factor capable of regulating cxcr4a expression, we treated WT embryos from 48–52 hpf with DMSO or 1 uM BNF and performed an ISH for cxcr4a. At this stage, vascular cxcr4a expression was essentially limited to the aortic arch vessels (not shown), the forming subintestinal vasculature (Fig 6C, arrow) and a few hindbrain CtAs (Fig 6I, arrowheads). Interestingly, BNF treatment led to an increased expression of cxcr4a specifically in endothelial cells, such as the subintestinal vasculature (Fig 6D, arrow), and particularly in the hindbrain (Fig 6J, arrowheads). We next asked if this induction was similar to the upregulation of cxcr4a known to occur in embryos without blood flow [40]. Nifedipine is a calcium channel blocker, and its administration to embryos stops the heartbeat, causing blood flow to cease. This leads to dramatic upregulation of cxcr4a in the DA (Fig 6E) and DLAV (Fig 6E, arrowhead) of the trunk and in the cranial vessels (Fig 6K). This response was stronger than that induced by BNF, and seemed to occur in more ECs of the blood vessels. To determine if AHR was responsible for any endogenous or pharmacologically-induced cxcr4a expression, we analyzed triple AHR mutant embryos. Basal expression in the subintestinal vasculature was unaltered in triple AHR mutants treated with DMSO (asterisk in Fig 6F, arrow). However, the BNF-induced increase in cxcr4a expression in the hindbrain was attenuated (Fig 6J, arrowheads). The response to flow block appeared unchanged, and we observed strong induction of cxcr4a in the trunk and head vessels to the same extent as WT (Fig 6H and 6K). Due to the rather robust upregulation of cxcr4a in the CtAs under BNF treatment, we turned to this vascular bed to analyze cxcr4a regulation by BNF in AHR mutants in more detail. We counted the number of cxcr4a-positive cells in WT, ahr1a -/-, ahr1b,2 -/- and triple AHR mutant embryos treated with DMSO and BNF. The endogenous expression of cxcr4a appeared unaffected by loss of any AHR genes, whereas the BNF-induced increase in expression was abrogated in ahr1b,2 -/- and triple AHR mutants compared to WT and ahr1a -/- (Fig 6L). Our results therefore lead to a model where flow and AHR have opposing and independent effects on cxcr4a transcription. Ectopic activation of AHR by BNF enhances the expression of cxcr4a in endothelial cells, whereas blood flow serves to negatively regulate this chemokine receptor, and AHR is not responsible for the increase in cxcr4a expression when blood flow is compromised (Fig 6M).
Fig 6. Transcriptional control of cxcr4a by AHR.
(A) Core DNA binding sequence of AHR::ARNT complex from JASPAR database. (B) Schematic of the zebrafish cxcr4a gene. Using strict search criteria (a relative profile score threshold of 90%), the promoter sequence spanning 2 kb from the start codon was analyzed for AHR binding sites in the JASPAR database. Two putative AHR binding sites lie close to the transcription start site (TSS). (C-H) Whole mount ISH for cxcr4a in WT and triple AHR mutant embryos treated from 48 hpf to 52 hpf with DMSO (C, F), 1 uM BNF (D, G) or 15 uM nifedipine (E, H). Arrows indicate subintestinal vasculature, while arrowheads point to DLAV. Scale bar is 250 um. (I-K) High magnification images of hindbrain capillaries in WTs and triple AHR mutants. Note similar levels of cxcr4a-positive CtAs (yellow arrowheads) in WTs and triple AHR mutants treated with DMSO (I), and markedly fewer cxcr4a-positive cells in triple mutants treated with BNF compared to WT (J). Both WTs and triple AHR mutants display marked increase in cxcr4a expression in all CtA endothelial cells after nifedipine treatment (K). Scale bar is 100 um. (L) Quantification of cxcr4a-positive endothelial cells in high magnification images of the hindbrain of WT, ahr1a -/-, ahr1b,2 -/- and triple AHR mutants treated from 48–52 hpf with DMSO or nifedipine. All genetic combinations of AHR mutations have similar numbers of cxcr4a-positive cells at basal levels. The BNF-induced increase of cxcr4a expression in WT still occurs in ahr1a -/- embryos, but does not show in ahr1b,2 -/- or triple AHR mutants. Analyzed by One-way ANOVA (DMSO treatment N = 4 WT, 12 ahr1a -/-, 13 ahr1b,2 -/- and 14 triple AHR fish. 1 uM BNF treatment N = 9 WT, 12 ahr1a -/-, 10 ahr1b,2 -/- and 6 triple AHR fish.) (M) Summary of transcriptional control of cxcr4a expression by flow (negative regulator) and BNF (positive regulator). Induction by BNF is in an AHR-dependent manner. **P<0.01, error bars indicate s.e.m. Abbreviations–AHR: aryl hydrocarbon receptor, ARNT: aryl hydrocarbon nuclear translocator, BNF: beta-naphthoflavone, CtA: central artery, CYP: cytochrome p450, DLAV: dorsal longitudinal anastomotic vessel, DMSO: dimethylsulfoxide, hpf: hours post fertilization, ISH: in situ hybridization, LDA: lateral dorsal aortae, PHBC: primordial hindbrain channel, TSS: transcription start site, WT: wildtype.
Discussion
Generation of AHR-deficient zebrafish
Studying AHR function in fish is challenging based simply on the number of AHR family genes in their genomes. This work is the first to examine the effect of genetic deletion of all AHR gene products in the zebrafish, and to correlate their respective contributions to AHR signaling in different tissues. In line with published expression data, we observed broad embryonic expression of ahr2, including the endoderm, skin and close to the trunk axial vessels [23]. We show that the other two zebrafish AHRs, ahr1a and ahr1b, display more restricted expression patterns, and we only detected them in the liver and eye, respectively. We could demonstrate functional redundancy of both ahr1b and ahr2 in the eye, where their overlapping expression ensures cyp1a1 transcription in ahr1b- or ahr2-single but not double mutants. The transcription of zebrafish AHRs responds differentially to the agonist BNF, which induces ahr2 and ahr1a but not ahr1b expression.
Our gain of function experiments with BNF confirmed previous findings that ahr2 is responsible for most of the ligand-induced cyp1a1/cyp1b1 expression in zebrafish [25, 29]. However, these studies reported only qPCR data on CYP transcripts in whole embryo lysate (no spatial information) or only focused on the effect of ahr2 knockdown on induced CYP levels (little attention to endogenous CYP transcript with knockdown or knockout of ahr2). Our analysis of different vascular beds revealed distinct requirements for ahr1b and ahr2 in specific vessels. ahr1b is required to maintain basal cyp1a1 expression in the arteries of the hindbrain and the ISVs of the trunk in the absence of ahr2. Surprisingly, this compensation does not occur in all vessels, and we observed that the DA, PCV and LDA were highly dependent on ahr2 for full cyp1a1 expression. By amplifying AHR activity with BNF, we could demonstrate overlapping function of ahr1b and ahr2 in inducing cyp1a1 and cyp1b1 in the PHBCs and CtAs of the hindbrain, vessels that are normally negative for both CYP1 genes. Our analysis of triple AHR mutants also demonstrates that endogenous cyp1b1 expression and a baseline level of cyp1a1 expression in the liver are AHR independent.
The spatial differences in vascular CYP1 expression and differential reliance on ahr1b and ahr2 are striking. Why, for example, is the primary axial vein highly cyp1a1-positive while the hindbrain veins do not express either CYP gene? Why does ahr1b maintain cyp1a1 expression in the hindbrain arteries but not the LDA in ahr2 mutants? Why does BNF fail to induce cyp1b1 expression in the cyp1a1-positive hindbrain arteries? Possible explanations include differential expression of AHRs themselves. ISH may not be sensitive enough to detect low levels of AHR transcript, and it may be that ahr1b is lowly expressed in the BA, PCS, BCA, CtAs and ISVs but not in the LDA, DA and PCV, thereby determining the vessels in which compensation is possible. We also did not examine changes in the expression patterns of all the AHR genes in our genetic mutants, and cannot rule out the possibility that loss of one may induce compensatory upregulation of the others.
The combined effects of AHR and hemodynamics could further explain the unique CYP expression profile in distinct vessels. cyp1a1 and cyp1b1 are the two most strongly upregulated genes in HUVECs exposed to shear stress in vitro [14, 41]. Regulation of cyp1a1 by shear stress was dependent on AHR binding sites in the rat cyp1a1 promoter in vitro [15]. In that study, fluid shear stress also led to an increase in expression and nuclear translocation of AHR. We recently showed that the PCV in the trunk experiences considerable hemodynamic forces at this stage of development [42]. A differential hemodynamic profile could potentially explain the lack of cyp1a1 expression in the hindbrain veins. It is worth noting that the loss of endogenous cyp1a1 expression we observed in our triple AHR mutants is a striking finding, as these embryos have no obvious flow defect. It remains to be seen if any zebrafish AHRs are flow-regulated themselves, or if they control the flow-responsiveness of cyp1a1 in vivo. Interestingly, we did not detect any endothelial cyp1b1 expression in baseline conditions. cyp1b1 was shown to be one of the most highly expressed CYP enzymes in human brain microvessels from patient biopsies [43]. More recently it was shown that B-catenin positively regulates cyp1b1, but not cyp1a1, in endothelioma cell lines [44], and the authors concluded that the ability of cyp1b1 to synthesize retinoic acid (from retinol) and 20-HETE (from arachnidonic acid) were essential in modulating BBB function. Furthermore, this study demonstrated that B-catenin was required for the AHR-dependent induction of cyp1b1 expression by TCDD. This could explain our observations that BNF treatment upregulated cyp1b1 in a restricted set of blood vessels in brain, specifically those that are cyp1a1-negative in endogenous situations. In support of this, Ziegler et al. found an inverse relationship between the levels of cyp1a1 and cyp1b1 expression with and without B-catenin function, suggesting their expression was mutually exclusive. Regarding BBB integrity, our findings that BNF induces the downregulation of glut1 expression in brain capillaries may be an additional mode of AHR ligand-induced toxicity. Dioxin has been shown to reduce the amount of tight junction proteins Claudin and ZO-1 in endothelial cells in an in vitro BBB system [45], and it also causes a reduction in Glut1 expression when applied to mouse embryonic carcinoma cells [46]. Our data are consistent with these findings and provide further in vivo evidence that impaired BBB function may result from AHR hyper-activation.
AHR and vascular patterning
Our failure to detect a vascular phenotype in triple AHR mutants is somewhat perplexing, given the observed vascular defects in liver, eye and kidney of AHR-deficient mice [12]. Triple AHR mutant zebrafish are viable, and we observed no obvious abnormalities in vascular patterning during embryonic development. This does not exclude subtle defects in distinct vessels, phenotypes relating to altered cell cycle progression or vascular tone for example, that might require careful quantification to resolve. We could show that the ahr1bmu145 allele is likely non-functional, and it is very probable that ahr1amu153 is also a null allele, as it is targeted to the same bHLH region of the open reading frame. It has been reported that zebrafish ahr1a does not bind well to the AHR agonist TCDD or transcriptionally activate AHR luciferase reporters in in vitro experiments [22], and this led to the suggestion that ahr1a was an inactive pseudogene. However, more recent work suggested that induction of cyp1a1 in the zebrafish liver in response to the compound leflunomide was ahr1a-dependent [29], and could indicate simply that ahr1a has higher affinity for different ligands. Application of leflunomide to the triple AHR mutants described here could provide evidence that the ahr1amu153 allele is a functional null.
An interesting finding is the potential for AHR to transcriptionally regulate the expression of the chemokine receptor cxcr4a in response to exogenous ligands. While such regulation was reported in a microarray study [38], our study shows that this regulation occurs in endothelial cells. cxcr4a has been shown to be dramatically upregulated in zebrafish embryos lacking blood flow [40], and this hemodynamic regulation occurs normally in AHR mutants. Thus, our results show that flow and AHR have opposite effects on cxcr4a regulation, in contrast to their mutual positive effect on cyp1a1 levels. At the time point of our analysis, we did not observe a change in endogenous cxcr4a expression in triple AHR mutants, nor did AHR-deficient embryos recapitulate the published vascular phenotypes seen in cxcr4aum20 mutants in LDA or hindbrain patterning [35, 40]. This indicates that AHR is dispensable for normal cxcr4a function during zebrafish embryonic development. Nevertheless, it is tempting to speculate that a link between AHR and cxcr4 may be functionally relevant, as AHR -/- and CXCR4 -/- mice have remarkably similar vascular phenotypes in the kidney [12, 47]. Analysis of vascular remodeling at later stages of fish development may provide evidence of such a link.
Supporting information
A-D) High magnification images of cyp1a1/b1 expression in trunk vasculature of DMSO (A, B) and BNF (C, D) treated embryos. Arrowheads point to ISVs. cyp1a1 is expressed highly in PCV, ISVs and DLAV, and in isolated DA cells under normal conditions (A), and possible induction in blood vessels by BNF is obscured by high skin staining (C). No endogenous vascular cyp1b1 can be detected (B), but it is induced in DA, DLAV and ISVs by BNF (D). E) Camera lucida drawing of 48 hpf zebrafish embryo from Kimmel, et al 1995 [36]. Dashed lines indicate plane of images in this figure to visualize lateral dorsal aortae (LDA, green). Confocal image shows dorsal view of LDA in live embryo at 48 hpf (scale bar is 200 um). Cartoon schematic depicts arrangement of LDA and DA. F-M) Cranial expression of cyp1a1/b1 in DMSO-treated embryos. Expression of cyp1a1 is mostly vascular-specific and restricted to arteries (LDA, PICA and OA). (*) marks the liver. No vascular or liver expression of cyp1b1 is detected, and staining is only evident in the ear, middle of the brain and parts of the ventral eye. Arrow marks optic furrow. N-U) Cranial expression of cyp1a1/b1 in BNF-treated embryos. In addition to the basal expression domains, specific blood vessels upregulate cyp1b1 (the LDA and PICA). Note lack of cyp1b1 expression in the liver even under BNF stimulation (* in R). Embryos in N-Q are ahr2 mutants to enable imaging of interior vessels. Numbers of embryos analyzed are the same as in Fig 2. All scale bars are 100 um. Abbreviations–AHR: aryl hydrocarbon receptor, BNF: beta-naphthoflavone, CYP: cytochrome p450, DA: dorsal aorta, DLAV: dorsal longitudinal anastomotic vessel, DMSO: dimethylsulfoxide, hpf: hours post fertilization, ISV: intersegmental vessel, LDA: lateral dorsal aortae, OA: optic artery, PCV: posterior cardinal vein, PICA: primitive internal carotid artery.
(TIF)
A-C) Overview and high magnification images of whole mount ISH for glut1 in WT embryos at 52 hpf. Vascular expression is limited to the brain capillaries (yellow arrowheads indicate individual CtAs). D-F) Glut1 expression in WT embryos treated with 1 uM BNF. Staining intensity is notably decreased in brain vessels. Scale bar in overview is 500 um, and 100 um in high magnification images. Abbreviations–BNF: beta-naphthoflavone, CtA: central artery, DMSO: dimethylsulfoxide, hpf: hours post fertilization, ISH: in situ hybridization.
(TIF)
A-J) High magnification images of cyp1a1/b1 expression in the trunk vasculature at 52 hpf in DMSO or BNF-treated WT (A, F), ahr1a -/- (B, G), ahr2 -/- (C, H), ahr1b,2 -/- (D, I) and triple AHR mutants (E, J). Expression patterns of both genes in ahr1a -/- are indistinguishable from WT in either condition. In ahr2 mutants a dramatic loss of endogenous cyp1a1 expression from the PCV and DA is observed, together with a weaker but persistent expression in DLAV and ISVs that is enhanced by BNF treatment (C). This remaining vascular expression is lost in ahr1b,2 -/- and triple AHR mutants (D, E). Note the AHR-independent expression of cyp1a1 in the gut (arrows in E) Similar results are seen in the BNF-induced cyp1b1 expression, which is weakly maintained in ISVs and DLAV of ahr2 mutants and lost in ahr1b,2 -/- and triple AHR mutant embryos (F-J). Numbers of embryos analyzed are the same as in Fig 4 (cyp1a1) and Fig 5 (cyp1b1). All scale bars are 100 um. Abbreviations–AHR: aryl hydrocarbon receptor, BNF: beta-naphthoflavone, CYP: cytochrome p450, DA: dorsal aorta, DLAV: dorsal longitudinal anastomotic vessel, DMSO: dimethylsulfoxide, hpf: hours post fertilization, ISV: intersegmental vessel, PCV: posterior cardinal vein, WT: wildtype.
(TIF)
Acknowledgments
The authors would like to thank Reinhild Bussmann for excellent fish care, Inna Maier for technical assistance and Martin Lange for generating the glut1 ISH probe.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by the Max Planck Society (A.F.S.), the Deutsche Forschungsgemeinschaft (DFG SI-1374/3-2; DFG SI-1374/4-1; DFG SI-1374/5-1; A.F.S.), and a European Research Council (ERC) starting grant (260794-ZebrafishAngio; A.F.S.). This work was supported by the Deutsche Forschungsgemeinschaft (DFG) Cells-in-Motion Cluster of Excellence (EXC 1003-CIM), University of Münster, Germany. This study was partially supported from the Instituto de Salud Carlos III (Ministerio de Economía y Competitividad and Fondos Feder, Spain) grant no. CB16-10-00238, and from the Consejería de Economía y Conocimiento (Junta de Andalucía, Spain) grant no. P10-CTS-5784 (D.A.-C.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–40. doi: 10.1146/annurev.pa.35.040195.001515 . [DOI] [PubMed] [Google Scholar]
- 2.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–34. doi: 10.1146/annurev.pharmtox.43.100901.135828 . [DOI] [PubMed] [Google Scholar]
- 3.Zhang N. The role of endogenous aryl hydrocarbon receptor signaling in cardiovascular physiology. J Cardiovasc Dis Res. 2011;2(2):91–5. doi: 10.4103/0975-3583.83033 ; PubMed Central PMCID: PMCPMC3144625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffman EC, Reyes H, Chu FF, Sander F, Conley LH, Brooks BA, et al. Cloning of a factor required for activity of the Ah (dioxin) receptor. Science. 1991;252(5008):954–8. . [DOI] [PubMed] [Google Scholar]
- 5.Lindsey S, Papoutsakis ET. The evolving role of the aryl hydrocarbon receptor (AHR) in the normophysiology of hematopoiesis. Stem Cell Rev. 2012;8(4):1223–35. doi: 10.1007/s12015-012-9384-5 ; PubMed Central PMCID: PMCPMC3463730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin J, Sheng B, Qiu Y, Yang K, Xiao W, Yang H. Role of AhR in positive regulation of cell proliferation and survival. Cell Prolif. 2016;49(5):554–60. doi: 10.1111/cpr.12282 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fracchiolla NS, Annaloro C, Guidotti F, Fattizzo B, Cortelezzi A. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) role in hematopoiesis and in hematologic diseases: A critical review. Toxicology. 2016;374:60–8. doi: 10.1016/j.tox.2016.10.007 . [DOI] [PubMed] [Google Scholar]
- 8.Bradfield CA, Kende AS, Poland A. Kinetic and equilibrium studies of Ah receptor-ligand binding: use of [125I]2-iodo-7,8-dibromodibenzo-p-dioxin. Mol Pharmacol. 1988;34(2):229–37. . [PubMed] [Google Scholar]
- 9.Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478(7368):197–203. doi: 10.1038/nature10491 . [DOI] [PubMed] [Google Scholar]
- 10.Zordoky BN, El-Kadi AO. 2,3,7,8-Tetrachlorodibenzo-p-dioxin and beta-naphthoflavone induce cellular hypertrophy in H9c2 cells by an aryl hydrocarbon receptor-dependant mechanism. Toxicol In Vitro. 2010;24(3):863–71. doi: 10.1016/j.tiv.2009.12.002 . [DOI] [PubMed] [Google Scholar]
- 11.Butler RA, Kelley ML, Powell WH, Hahn ME, Van Beneden RJ. An aryl hydrocarbon receptor (AHR) homologue from the soft-shell clam, Mya arenaria: evidence that invertebrate AHR homologues lack 2,3,7,8-tetrachlorodibenzo-p-dioxin and beta-naphthoflavone binding. Gene. 2001;278(1–2):223–34. . [DOI] [PubMed] [Google Scholar]
- 12.Lahvis GP, Lindell SL, Thomas RS, McCuskey RS, Murphy C, Glover E, et al. Portosystemic shunting and persistent fetal vascular structures in aryl hydrocarbon receptor-deficient mice. Proc Natl Acad Sci U S A. 2000;97(19):10442–7. doi: 10.1073/pnas.190256997 ; PubMed Central PMCID: PMCPMC27043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walisser JA, Glover E, Pande K, Liss AL, Bradfield CA. Aryl hydrocarbon receptor-dependent liver development and hepatotoxicity are mediated by different cell types. Proc Natl Acad Sci U S A. 2005;102(49):17858–63. doi: 10.1073/pnas.0504757102 ; PubMed Central PMCID: PMCPMC1308889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conway DE, Sakurai Y, Weiss D, Vega JD, Taylor WR, Jo H, et al. Expression of CYP1A1 and CYP1B1 in human endothelial cells: regulation by fluid shear stress. Cardiovasc Res. 2009;81(4):669–77. doi: 10.1093/cvr/cvn360 ; PubMed Central PMCID: PMCPMC2642602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han Z, Miwa Y, Obikane H, Mitsumata M, Takahashi-Yanaga F, Morimoto S, et al. Aryl hydrocarbon receptor mediates laminar fluid shear stress-induced CYP1A1 activation and cell cycle arrest in vascular endothelial cells. Cardiovasc Res. 2008;77(4):809–18. doi: 10.1093/cvr/cvm095 . [DOI] [PubMed] [Google Scholar]
- 16.Fleming I. The cytochrome P450 pathway in angiogenesis and endothelial cell biology. Cancer Metastasis Rev. 2011;30(3–4):541–55. doi: 10.1007/s10555-011-9302-3 . [DOI] [PubMed] [Google Scholar]
- 17.Agbor LN, Elased KM, Walker MK. Endothelial cell-specific aryl hydrocarbon receptor knockout mice exhibit hypotension mediated, in part, by an attenuated angiotensin II responsiveness. Biochem Pharmacol. 2011;82(5):514–23. doi: 10.1016/j.bcp.2011.06.011 ; PubMed Central PMCID: PMCPMC3148414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agbor LN, Walsh MT, Boberg JR, Walker MK. Elevated blood pressure in cytochrome P4501A1 knockout mice is associated with reduced vasodilation to omega-3 polyunsaturated fatty acids. Toxicol Appl Pharmacol. 2012;264(3):351–60. doi: 10.1016/j.taap.2012.09.007 ; PubMed Central PMCID: PMCPMC3494483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Liu H, McKenzie G, Witting PK, Stasch JP, Hahn M, et al. Kynurenine is an endothelium-derived relaxing factor produced during inflammation. Nat Med. 2010;16(3):279–85. doi: 10.1038/nm.2092 ; PubMed Central PMCID: PMCPMC3556275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn ME, Karchner SI, Shapiro MA, Perera SA. Molecular evolution of two vertebrate aryl hydrocarbon (dioxin) receptors (AHR1 and AHR2) and the PAS family. Proc Natl Acad Sci U S A. 1997;94(25):13743–8. ; PubMed Central PMCID: PMCPMC28377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. doi: 10.1038/nature12111 ; PubMed Central PMCID: PMCPMC3703927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karchner SI, Franks DG, Hahn ME. AHR1B, a new functional aryl hydrocarbon receptor in zebrafish: tandem arrangement of ahr1b and ahr2 genes. Biochem J. 2005;392(Pt 1):153–61. doi: 10.1042/BJ20050713 ; PubMed Central PMCID: PMCPMC1317674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreasen EA, Spitsbergen JM, Tanguay RL, Stegeman JJ, Heideman W, Peterson RE. Tissue-specific expression of AHR2, ARNT2, and CYP1A in zebrafish embryos and larvae: effects of developmental stage and 2,3,7,8-tetrachlorodibenzo-p-dioxin exposure. Toxicol Sci. 2002;68(2):403–19. . [DOI] [PubMed] [Google Scholar]
- 24.Fernandez-Salguero PM, Hilbert DM, Rudikoff S, Ward JM, Gonzalez FJ. Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol Appl Pharmacol. 1996;140(1):173–9. doi: 10.1006/taap.1996.0210 . [DOI] [PubMed] [Google Scholar]
- 25.Prasch AL, Teraoka H, Carney SA, Dong W, Hiraga T, Stegeman JJ, et al. Aryl hydrocarbon receptor 2 mediates 2,3,7,8-tetrachlorodibenzo-p-dioxin developmental toxicity in zebrafish. Toxicol Sci. 2003;76(1):138–50. doi: 10.1093/toxsci/kfg202 . [DOI] [PubMed] [Google Scholar]
- 26.Dong W, Teraoka H, Tsujimoto Y, Stegeman JJ, Hiraga T. Role of aryl hydrocarbon receptor in mesencephalic circulation failure and apoptosis in zebrafish embryos exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Sci. 2004;77(1):109–16. doi: 10.1093/toxsci/kfh023 . [DOI] [PubMed] [Google Scholar]
- 27.Teraoka H, Ogawa A, Kubota A, Stegeman JJ, Peterson RE, Hiraga T. Malformation of certain brain blood vessels caused by TCDD activation of Ahr2/Arnt1 signaling in developing zebrafish. Aquat Toxicol. 2010;99(2):241–7. doi: 10.1016/j.aquatox.2010.05.003 ; PubMed Central PMCID: PMCPMC3040289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kubota A, Stegeman JJ, Woodin BR, Iwanaga T, Harano R, Peterson RE, et al. Role of zebrafish cytochrome P450 CYP1C genes in the reduced mesencephalic vein blood flow caused by activation of AHR2. Toxicol Appl Pharmacol. 2011;253(3):244–52. doi: 10.1016/j.taap.2011.03.025 ; PubMed Central PMCID: PMCPMC3143178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodale BC, La Du JK, Bisson WH, Janszen DB, Waters KM, Tanguay RL. AHR2 mutant reveals functional diversity of aryl hydrocarbon receptors in zebrafish. PLoS One. 2012;7(1):e29346 doi: 10.1371/journal.pone.0029346 ; PubMed Central PMCID: PMCPMC3252317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westerfield M. The zebrafish book A guide for the laboratory use of zebrafish (Danio rerio). 4th edition ed. Eugene: Univ. of Oregon Press; 2000. [Google Scholar]
- 31.Beis D, Bartman T, Jin SW, Scott IC, D'Amico LA, Ober EA, et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132(18):4193–204. doi: 10.1242/dev.01970 . [DOI] [PubMed] [Google Scholar]
- 32.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39(12):e82 doi: 10.1093/nar/gkr218 ; PubMed Central PMCID: PMCPMC3130291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG 2nd, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491(7422):114–8. Epub 2012/09/25. doi: 10.1038/nature11537 ; PubMed Central PMCID: PMC3491146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3(1):59–69. Epub 2008/01/15. doi: 10.1038/nprot.2007.514 . [DOI] [PubMed] [Google Scholar]
- 35.Siekmann AF, Standley C, Fogarty KE, Wolfe SA, Lawson ND. Chemokine signaling guides regional patterning of the first embryonic artery. Genes Dev. 2009;23(19):2272–7. doi: 10.1101/gad.1813509 ; PubMed Central PMCID: PMCPMC2758748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. Epub 1995/07/01. doi: 10.1002/aja.1002030302 . [DOI] [PubMed] [Google Scholar]
- 37.Umans RA, Henson HE, Mu F, Parupalli C, Ju B, Peters JL, et al. CNS angiogenesis and barriergenesis occur simultaneously. Dev Biol. 2017;425(2):101–8. doi: 10.1016/j.ydbio.2017.03.017 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goodale BC, Tilton SC, Corvi MM, Wilson GR, Janszen DB, Anderson KA, et al. Structurally distinct polycyclic aromatic hydrocarbons induce differential transcriptional responses in developing zebrafish. Toxicol Appl Pharmacol. 2013;272(3):656–70. doi: 10.1016/j.taap.2013.04.024 ; PubMed Central PMCID: PMCPMC4098828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swanson HI, Chan WK, Bradfield CA. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J Biol Chem. 1995;270(44):26292–302. . [DOI] [PubMed] [Google Scholar]
- 40.Bussmann J, Wolfe SA, Siekmann AF. Arterial-venous network formation during brain vascularization involves hemodynamic regulation of chemokine signaling. Development. 2011;138(9):1717–26. Epub 2011/03/25. doi: 10.1242/dev.059881 ; PubMed Central PMCID: PMC3074448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCormick SM, Eskin SG, McIntire LV, Teng CL, Lu CM, Russell CG, et al. DNA microarray reveals changes in gene expression of shear stressed human umbilical vein endothelial cells. Proc Natl Acad Sci U S A. 2001;98(16):8955–60. Epub 2001/08/02. doi: 10.1073/pnas.171259298 [pii]. ; PubMed Central PMCID: PMC55355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugden WW, Meissner R, Aegerter-Wilmsen T, Tsaryk R, Leonard EV, Bussmann J, et al. Endoglin controls blood vessel diameter through endothelial cell shape changes in response to haemodynamic cues. Nature cell biology. 2017;19(6):653–65. doi: 10.1038/ncb3528 ; PubMed Central PMCID: PMCPMC5455977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dauchy S, Dutheil F, Weaver RJ, Chassoux F, Daumas-Duport C, Couraud PO, et al. ABC transporters, cytochromes P450 and their main transcription factors: expression at the human blood-brain barrier. J Neurochem. 2008;107(6):1518–28. doi: 10.1111/j.1471-4159.2008.05720.x . [DOI] [PubMed] [Google Scholar]
- 44.Ziegler N, Awwad K, Fisslthaler B, Reis M, Devraj K, Corada M, et al. beta-Catenin Is Required for Endothelial Cyp1b1 Regulation Influencing Metabolic Barrier Function. J Neurosci. 2016;36(34):8921–35. doi: 10.1523/JNEUROSCI.0148-16.2016 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyazaki W, Fujiwara Y, Katoh T. The effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin on the development and function of the blood-brain barrier. Neurotoxicology. 2016;52:64–71. doi: 10.1016/j.neuro.2015.11.003 . [DOI] [PubMed] [Google Scholar]
- 46.Tonack S, Kind K, Thompson JG, Wobus AM, Fischer B, Santos AN. Dioxin affects glucose transport via the arylhydrocarbon receptor signal cascade in pluripotent embryonic carcinoma cells. Endocrinology. 2007;148(12):5902–12. doi: 10.1210/en.2007-0254 . [DOI] [PubMed] [Google Scholar]
- 47.Takabatake Y, Sugiyama T, Kohara H, Matsusaka T, Kurihara H, Koni PA, et al. The CXCL12 (SDF-1)/CXCR4 axis is essential for the development of renal vasculature. J Am Soc Nephrol. 2009;20(8):1714–23. Epub 2009/05/16. ASN.2008060640 [pii] doi: 10.1681/ASN.2008060640 ; PubMed Central PMCID: PMC2723985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A-D) High magnification images of cyp1a1/b1 expression in trunk vasculature of DMSO (A, B) and BNF (C, D) treated embryos. Arrowheads point to ISVs. cyp1a1 is expressed highly in PCV, ISVs and DLAV, and in isolated DA cells under normal conditions (A), and possible induction in blood vessels by BNF is obscured by high skin staining (C). No endogenous vascular cyp1b1 can be detected (B), but it is induced in DA, DLAV and ISVs by BNF (D). E) Camera lucida drawing of 48 hpf zebrafish embryo from Kimmel, et al 1995 [36]. Dashed lines indicate plane of images in this figure to visualize lateral dorsal aortae (LDA, green). Confocal image shows dorsal view of LDA in live embryo at 48 hpf (scale bar is 200 um). Cartoon schematic depicts arrangement of LDA and DA. F-M) Cranial expression of cyp1a1/b1 in DMSO-treated embryos. Expression of cyp1a1 is mostly vascular-specific and restricted to arteries (LDA, PICA and OA). (*) marks the liver. No vascular or liver expression of cyp1b1 is detected, and staining is only evident in the ear, middle of the brain and parts of the ventral eye. Arrow marks optic furrow. N-U) Cranial expression of cyp1a1/b1 in BNF-treated embryos. In addition to the basal expression domains, specific blood vessels upregulate cyp1b1 (the LDA and PICA). Note lack of cyp1b1 expression in the liver even under BNF stimulation (* in R). Embryos in N-Q are ahr2 mutants to enable imaging of interior vessels. Numbers of embryos analyzed are the same as in Fig 2. All scale bars are 100 um. Abbreviations–AHR: aryl hydrocarbon receptor, BNF: beta-naphthoflavone, CYP: cytochrome p450, DA: dorsal aorta, DLAV: dorsal longitudinal anastomotic vessel, DMSO: dimethylsulfoxide, hpf: hours post fertilization, ISV: intersegmental vessel, LDA: lateral dorsal aortae, OA: optic artery, PCV: posterior cardinal vein, PICA: primitive internal carotid artery.
(TIF)
A-C) Overview and high magnification images of whole mount ISH for glut1 in WT embryos at 52 hpf. Vascular expression is limited to the brain capillaries (yellow arrowheads indicate individual CtAs). D-F) Glut1 expression in WT embryos treated with 1 uM BNF. Staining intensity is notably decreased in brain vessels. Scale bar in overview is 500 um, and 100 um in high magnification images. Abbreviations–BNF: beta-naphthoflavone, CtA: central artery, DMSO: dimethylsulfoxide, hpf: hours post fertilization, ISH: in situ hybridization.
(TIF)
A-J) High magnification images of cyp1a1/b1 expression in the trunk vasculature at 52 hpf in DMSO or BNF-treated WT (A, F), ahr1a -/- (B, G), ahr2 -/- (C, H), ahr1b,2 -/- (D, I) and triple AHR mutants (E, J). Expression patterns of both genes in ahr1a -/- are indistinguishable from WT in either condition. In ahr2 mutants a dramatic loss of endogenous cyp1a1 expression from the PCV and DA is observed, together with a weaker but persistent expression in DLAV and ISVs that is enhanced by BNF treatment (C). This remaining vascular expression is lost in ahr1b,2 -/- and triple AHR mutants (D, E). Note the AHR-independent expression of cyp1a1 in the gut (arrows in E) Similar results are seen in the BNF-induced cyp1b1 expression, which is weakly maintained in ISVs and DLAV of ahr2 mutants and lost in ahr1b,2 -/- and triple AHR mutant embryos (F-J). Numbers of embryos analyzed are the same as in Fig 4 (cyp1a1) and Fig 5 (cyp1b1). All scale bars are 100 um. Abbreviations–AHR: aryl hydrocarbon receptor, BNF: beta-naphthoflavone, CYP: cytochrome p450, DA: dorsal aorta, DLAV: dorsal longitudinal anastomotic vessel, DMSO: dimethylsulfoxide, hpf: hours post fertilization, ISV: intersegmental vessel, PCV: posterior cardinal vein, WT: wildtype.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.