Abstract
Barro-Soria et al. highlight work that reveals how β1 and β3 subunits modulate NaV1.5 channel function.
The human voltage-gated sodium channel NaV1.5 plays a critical role in the human heart, in which it generates inward sodium currents that underlie cardiomyocyte depolarization. The NaV1.5 protein is composed of more than 2,000 amino acids, organized into four homologous domains (Catterall et al., 2017), which equip the channel with one central pore domain and four peripheral voltage sensor domains. In the human heart, NaV1.5 interacts with several other proteins to form a macromolecular complex. Among important interaction partners are the four β subunits (β1–4), which each have one transmembrane segment, an extracellular N terminus, and an intracellular C terminus (Abriel, 2010). All four β subunits are expressed in the heart and modulate the trafficking and biophysical properties of NaV1.5, although the functional effect of the different β subunits are still debated (Abriel, 2010). Correct voltage dependence and kinetics of NaV1.5 channel activation and inactivation, together with correct NaV1.5 channel density in the plasma membrane, are critical for cardiac function. As a consequence, mutations in the gene encoding NaV1.5 have been linked to cardiac arrhythmias, including Brugada syndrome, Long QT Syndrome type 3, and cardiac conduction disease (Veerman et al., 2015). Moreover, multiple mutations in the genes encoding β1–4 have been associated with altered NaV1.5 function and cardiac arrhythmias (Abriel, 2010). In this issue, Silva and co-workers study the mechanism by which β1 and β3 modulate the activity of NaV1.5. β1 and β3 are noncovalently bound to NaV1.5 (in contrast to β2 and β4, which are covalently bound) and have previously been shown to shift the voltage dependence of channel inactivation. However, the direction and magnitude of these shifts are not conclusive and appear to vary with expression system (Abriel, 2010). Moreover, the molecular understanding of how β1 and β3 interact with NaV1.5 to alter voltage dependence has remained poor. In their work in this issue, Zhu et al. use optical approaches to resolve some of these questions. Molecular insights into how β subunits modulate NaV1.5 channel function are important for our understanding of the physiological relevance of each β subunit and how mutations interfere with NaV1.5–β subunit interactions.
Voltage clamp fluorometry (VCF) to track movement in distinct domains
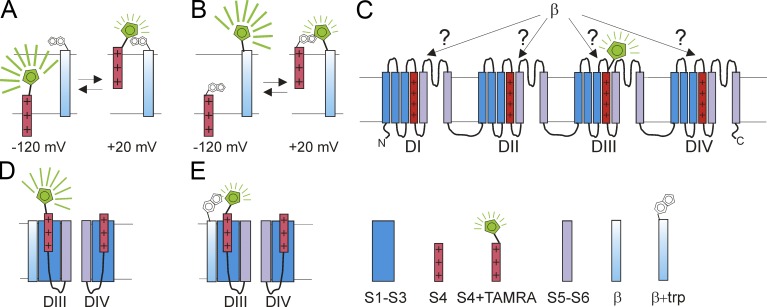
The NaV1.5 channel is opened by membrane depolarization, which is triggered by the outward movement of transmembrane segment S4 in the voltage sensor domains. A few milliseconds after channel opening, the channels inactivate. This fast inactivation is believed to be triggered by the exposure of an intracellular inactivation motif, composed of a few amino acids in the loop between domains III and IV, which interacts with other intracellular motifs in NaV1.5 to block the pore (Catterall et al., 2017). Conformational changes such as these can be detected by the powerful fluorescence technique, VCF (Mannuzzu et al., 1996; Chanda and Bezanilla, 2002). To conduct VCF on NaV1.5, Zhu et al. (2017) introduce a cysteine at the external end of the voltage sensor S4 in each of the four domains, one at a time, and then covalently label the cysteine with the thiol-reactive fluorescent probe TAMRA-maleimide (Fig. 1 A). The rationale behind this is that the voltage sensors will move in response to voltage changes and this movement will cause the voltage sensor–bound TAMRA compound to move from one micro environment to another (Fig. 1 A). Most fluorophores are sensitive to their surrounding environment, such that their fluorescence intensities or fluorescence spectra, will alter when they encounter a different environment (Fig. 1 A). These different microenvironments could differ in hydrophobic/hydrophilic nature or in their proximity to quenching groups such as tryptophans (Mannuzzu et al., 1996; Pantazis and Olcese, 2012). In VCF, this fluorescence change is used as a reporter for monitoring voltage sensor movement (Mannuzzu et al., 1996; Chanda and Bezanilla, 2002). The Silva group has previously shown that, by labeling the different voltage sensors in the four domains of NaV1.5, they can measure the voltage dependence and kinetics of individual voltage sensors in these four domains (Varga et al., 2015). Indeed, VCF has previously been used to show that movement of the voltage sensor in the fourth domain (DIV-S4) correlates with NaV channel inactivation, whereas movements of the voltage sensors in the three first domain (DI-S4, DII-S4, DIII-S4) correlate more with NaV channel activation (Chanda and Bezanilla, 2002; Capes et al., 2013).
Figure 1.
Using VCF to identify conformational changes in NaV1.5. (A) A fluorophore attached to the voltage sensor S4 could experience changes in its microenvironment when S4 moves outward in response to a membrane depolarization. The change in microenvironment alters the fluorescence from the fluorophore, e.g., by changes in the hydrophobic/hydrophilic nature of the environment or by approaching a quenching residue. (B) Similarly, the fluorescence from a fluorophore attached to an immobile protein segment could change when S4 moves outward, if the outward moving S4 changes the microenvironment around the fluorophore. (C) Using VCF, one can label, one at a time, the four different S4s with a fluorophore (here shown the construct with the fluorophore attached to DIII-S4). When each of these constructs, one at a time, are coexpressed with β1 or β3, one can detect whether β1 and/or β3 alter the S4 movement in a specific domain (here DIII). (D and E) By introducing a quenching tryptophan residue in a β subunit (E), one can detect whether the β subunit is close to an S4 in a specific domain. If the β subunit is located close to DIII-S4, then one would expect to see a tryptophan-induced change in the fluorescence signal from the construct with a fluorophore attached to DIII-S4 (compare fluorescence in D and E).
Zhu et al. (2017) use VCF to detect the effects of different β subunits on the voltage dependence and kinetics of voltage sensor movement in the four different domains of NaV1.5 (Fig. 1 C). Voltage sensor movement has traditionally been detected using gating current measurements. However, in gating current measurements, the currents from all four voltage sensor domains are measured simultaneously, which makes it hard to conclude that a certain effect of a β subunit is on a specific voltage sensor domain. In contrast, VCF allows each voltage sensor domain to be labeled separately, one at a time, and can detect the effects on each individual voltage sensor domain. This makes VCF particularly suited to a study of the specific effects of β subunits on individual voltage sensor domains (Fig. 1 C).
When using VCF, one has to remember that it is an indirect technique, in which one infers conformational changes in the protein from fluorescence changes of a fluorophore attached to the protein domain of interest (in this case, one of the four different voltage sensors). In a VCF experiment, it is not always clear why the fluorescence changes in response to changes in membrane voltage. It could be caused by movement of the domain to which the fluorophore is attached (the voltage sensor; Fig. 1 A), or it could be that some other part of the protein moves toward an immobile fluorophore and thereby alters its local environment (Fig. 1 B). In addition, a β subunit could affect a specific voltage sensor domain either directly (by associating with it; Fig. 1, D and E) or indirectly (through an allosteric mechanism). Zhu et al. (2017) use several different experiments to distinguish between all these different possible interpretations of the VCF signals. To test for direct interactions between β subunits and a specific voltage sensor domain, they introduce a tryptophan residue in the β subunits to see whether it can alter (quench) the fluorescence from the labeled voltage sensor (Fig. 1, D and E). To test for allosteric effects versus direct effects, they uncouple the voltage sensor domain from the pore domain by uncoupling mutations in the S4–S5 linker and the S6 domain. If effects of the β subunit on a specific voltage sensor persist even in the presence of the uncoupling mutations, then the effects are most likely caused by direct association of the β subunit and that particular voltage sensor domain.
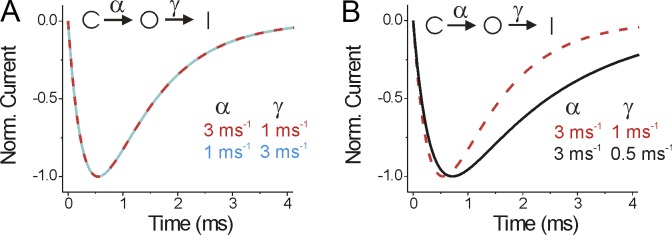
Caution should be exercised in interpreting fluorescence signals (or currents) from voltage-gated ion channels, as these are complex proteins with many different states. This makes it hard to conclude which rates or transitions are altered by different conditions, such as the effects of β subunits in this study. Even in a very simple model, it can be hard to dissect which rates are altered from data obtained by standard voltage protocols. Take a simple three-state model with one closed, one open, and one inactivated state, for example. From a simple voltage step protocol, it is impossible to tell whether activation (the closed to open transition) or inactivation (the open to inactivated transition) is the faster transition (Fig. 2 A). Similarly, it is impossible to tell whether a β subunit affects activation or inactivation (or both), because, for example, changes to just the inactivation rate will alter both the rise and fall of the ionic current time course (Fig. 2 B). This is because of the coupled nature of these transitions. To really demonstrate which transitions are affected, one must either uncouple the different transitions (as in Zhu et al., 2017) or perform experiments with much more complex voltage protocols with several sequential voltage pulses.
Figure 2.
Currents from a simple three-state ion channel model. (A) Simulated currents from a model where the channels transit from a closed state C to an open state O (by the rate α) and then to the inactivated state I (by the rate γ). Both sets of parameters (α = 3 ms−1 and γ = 1 ms−1 or α = 1 ms−1 and γ = 3 ms−1) generated the same normalized current time course. (B) Slowing only the inactivation rate γ (from 1 ms−1 to 0.5 ms−1) changes both the activation time course and the inactivation time course.
β1 and β3 subunits affect the S4 of domains III and IV
By studying macroscopic currents generated by human NaV1.5 overexpressed in Xenopus oocytes, the authors first show that coexpression of either the β1 or β3 subunit shifts steady-state inactivation toward more positive voltages. In contrast, the conductance versus voltage (G(V)) curve is not affected by either β1 or β3. β3 slows down inactivation but does not change the rate of recovery of inactivation. In contrast, β1 speeds up the rate of recovery of inactivation but does not change the rate of inactivation. This β1- and β3-induced modulation of channel inactivation is expected to allow more NaV1.5 channels to be available for activation in a physiological voltage range. β1- and β3-induced shifts in steady-state inactivation toward more positive voltages are consistent with several previous studies (e.g., An et al., 1998; Fahmi et al., 2001) but in contrast to others (Ko et al., 2005).
The authors then turn to gating currents generated by NaV1.5 to provide insights into the molecular mechanism of β1 and β3 modulation. Gating current recordings revealed β1- and β3-induced shifts also of the gating charge versus voltage (Q(V)) curve toward positive voltages, in combination with steeper Q(V) curves. These findings indicate that these β subunits shift the voltage dependence of activation for one or several S4 segments. The authors then take advantage of the VCF technique and their NaV1.5 VCF constructs to provide important insights into which specific voltage sensor domain is affected by β1 and β3. They systematically study the voltage dependence and the kinetics of the fluorescence change for each voltage sensor domain.
Zhu et al. (2017) find that both β1 and β3 shift the voltage dependence of S4 in domain IV (DIV-S4) toward positive voltages. Moreover, β1 speeds up deactivation of the S4s in DIV and DIII. As domain IV is closely linked to NaV1.5 channel inactivation, the shifted voltage dependence and faster deactivation of this particular S4 induced by β1 could well explain the β1 effect on steady-state inactivation. The authors propose that the faster deactivation of DIV-S4 (and maybe that of DIII-S4) underlies the faster recovery of inactivation of NaV1.5. In addition, the faster deactivation of DIV-S4 most likely contributes, at least partly, to the shift in steady-state inactivation.
In addition to shifting the voltage dependence of DIV-S4 toward positive voltages, β3 has a similar shifting effect on DIII-S4. However, in contrast to β1, β3 speeds up deactivation of DIII-S4, but not deactivation of DIV-S4. The fact that β3 only speeds up DIII-S4 deactivation (and not DIV-S4 deactivation) might explain why β3 does not affect NaV1.5 channel recovery from inactivation. Zhu et al. (2017) propose the alternative explanation that β3 might decouple DIII-S4 deactivation from recovery of inactivation and that this might be why a β3-induced faster deactivation of DIII-S4 does not speed up recovery of inactivation.
Curiously, β3 slows down the activation time course but does not shift the G(V) for NaV1.5. Activation of DI–DIII S4s has been proposed to be rate limiting for activation in NaV1.4 channels (Chanda and Bezanilla, 2002). However, Zhu et al. (2017) find that β3 shifts the voltage dependence of DIII-S4 to more positive voltages but does not (at least with the present resolution of the fluorescence signals) change the activation rate of DI–DIII S4s in NaV1.5. How can these contradictory effects be explained? NaV channels have four voltage sensors, each with different voltage dependences and kinetics. It is generally assumed that the first three voltage sensors are mainly responsible for the activation of NaV channels and the fourth voltage sensor is mainly responsible for the inactivation of NaV channels. If activation of all three DI–DIII voltage sensors are necessary for channel opening, however, then the G(V) will be mainly determined by the voltage sensor with the most positive voltage dependence and the kinetics of opening will be mainly determined by the voltage sensor with the slowest kinetics of these three voltage sensors. It is possible that, for NaV1.5 at least, the DIII-S4 is not very important for determining the G(V). This is the explanation that Zhu et al. (2017) give for the lack of effect of DIII-S4 on the G(V), as the DIII-S4 voltage dependence is much more negative than the voltage dependence of the G(V). But what gives rise to the slower channel opening time course in the presence of β3? One possibility is that the slower activation is just an indirect effect of the slowed inactivation (Fig. 2 B). Another possibility is that one of the voltage sensors (whichever is the rate limiting for channel opening in NaV1.5) activates more slowly in the presence of β3 but that the resolution (noisiness) of the fluorescence signal doesn’t allow detection of this change in S4 activation.
The authors then perform a series of experiments to determine whether β1 and β3 interact with DIV-VSD and DIII-VSD directly. Using the N1765A mutation in the DIV-S6 of NaV1.5, which decouples the voltage sensor of domain IV from the pore of domain IV, the authors show that β1 shifts the voltage dependence of DIV-S4 even though the voltage sensor is decoupled from the pore. The authors therefore conclude that β1 directly targets the voltage sensor domain, and not the pore, of domain IV. Likewise, for β3, the authors used the N1765A mutation in DIV-S6 and the A1330W mutation in the DIII S4–S5 linker of the NaV1.5 channel, which are known to uncouple DIV-S4 and DIII-S4, respectively, from the pore domain. Even in the case where the voltage sensors were uncoupled from the pore, β3 still right-shifted the voltage dependence of DIII- and DIV-S4s, suggesting that β3 directly interacts with those two voltage sensor domains. The authors then tested the proximity of β1 and β3 to the four voltage sensor domains by introducing a tryptophan at the top of the transmembrane part of β1 or β3. Because S4 moves outwardly upon depolarization, they reasoned that, for example, a tryptophan at the top of β1 would quench the fluorophore linked to DIV-S4 if β1 was close to DIV-VSD (within van der Waals contact distance, <15 Å, of the fluorophore). However, the authors could not detect β1-induced quenching of the fluorescence intensity from fluorophores linked to any of the four voltage sensors. This could be because a tryptophan at the top of β1 is not close enough to the fluorophores in any voltage sensor. However, the shift in DIV-S4 voltage dependence induced by the β1 subunit with an additional tryptophan was very small, suggesting that maybe the tryptophan somehow interferes with the β1 modulation of NaV1.5. In contrast, they found that a tryptophan introduced at the top of β3 quenches the fluorescence intensity from DIII-S4, which led them to conclude that β3 and the DIII-S4 segment are within van der Waals contact distance. As mentioned above, one can detect conformational changes by alterations in fluorescence caused by movement of the domain to which the fluorophore is attached (the voltage sensor; Fig. 1 A) or caused by some other part of the protein moving toward an immobile fluorophore and thereby altering its local environment (Fig. 1 B). Therefore, to obtain further evidence that β3 is close to one of the S4s, the authors also engineered a cysteine at the extracellular region of β3 and labeled it with the fluorophore. When this fluorophore-labeled β3 subunit was coexpressed with the WT NaV1.5 α subunit, they similarly found changes in fluorescence upon channel activation. Although this experiment is consistent with β3 being close to one S4, one cannot conclude which one of the four S4s caused the fluorescence quenching (but one can presume that it was caused by the outward movement of DIII-S4).
Model of β1 and β3 modulation of NaV1.5
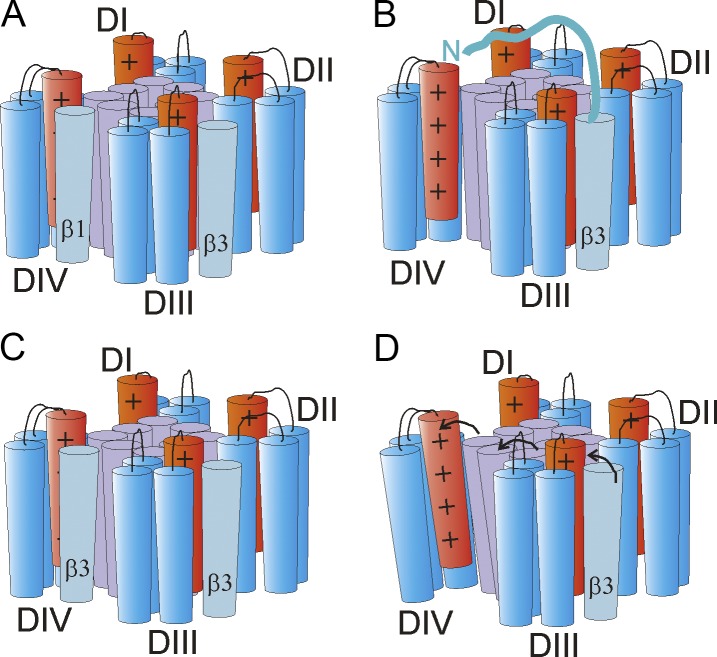
Zhu et al. (2017) propose that β1 binds to the cleft formed between DIII-VSD and DIV-VSD (Fig. 3 A). From that position, β1 may interact directly with DIV-VSD to make it easier for this particular S4 to move back to its resting position. That, in turn, would provide a molecular explanation for the β1-induced increase in the rate of recovery from inactivation and the shift in steady-state inactivation toward more positive voltages. Although this proposed model provides a mechanistic basis for the effect of β1 on the macroscopic NaV1.5 current, there are still some questions that remain unsolved. In particular, the inability of a tryptophan introduced in β1 to quench the fluorescence signal from DIV-S4 raises the question of whether β1 is in close enough proximity to DIV-VSD to interact directly with this voltage sensor domain. In the future, one could try to place a tryptophan at other places in β1 to further detect putative interactions between β1 and DIV-VSD.
Figure 3.
Proposed localizations of β1 and β3 subunits and their putative interactions with NaV1.5. (A) Location of β1 and β3 according to Zhu et al. (2017), with β1 located between DIII-VSD and DIV-VSD, close to DIV-S4, and β3 located between DII-VSD and DIII-VSD, close to DIII-S4. From these locations, it is easy to imagine how β1 would affect DIV-S4 and β3 would affect DIII-S4. However, it is not clear how β3 affects DIV-S4 from this location. (B–D) Three possible β3 DIV-S4 interactions. (B) The N terminus of β3 reaches over to DIV-S4 and directly affects DIV-S4. (C) Two β3s bind to NaV1.5, one at DIII-S4 and one at DIV-S4. (D) β3 affects DIV-S4 indirectly through an effect via the pore that does not use the S4–S5 linker to S6 coupling. Here shown as a β3-induced rotation of the external end of S5–S6 that is transmitted to DIV-S4 (arrows).
In contrast, Zhu et al. (2017) propose that β3 localizes in the cleft between DII-VSD and DIII-VSD (Fig. 3 A) and, from this position, regulates ionic current kinetics of activation and inactivation of the NaV1.5 channel. In particular, as shown by β1 and β3 chimeras, the extracellular and transmembrane domains of β3 are crucial for the right shift of the fluorescence versus voltage (F(V)) curve of DIII-S4. However, as depicted in Fig. 3 A, it is hard to imagine how β3 would also affect a distant DIV-VSD to exert a depolarizing shift in the F(V) curve of DIV-S4 from this spatial localization. One possibility is that, for example, the extreme end of the N terminus of β3 reaches over to DIV-VSD and modulates DIV-S4 directly (Fig. 3 B), even if the β3 transmembrane segment is located between DII-VSD and DIII-VSD. A second possibility is that two β3s bind to NaV1.5 simultaneously, one to DIII-VSD and one to DIV-VSD (Fig. 3 C). The absence of quenching of DIV-S4 fluorescence by a tryptophan introduced in β3 seems to speak against β3 binding to DIV-VSD. However, there was also no quenching of DIV-S4 fluorescence by a tryptophan introduced in β1. Maybe a tryptophan introduced in β1 or β3 is not close enough to induce quenching of DIV-S4 fluorescence, even if both β1 and β3 can bind to DIV-VSD. A third possibility is that β3 allosterically affects DIV-VSD. However, Zhu et al. (2017) showed that any putative allosteric effect was not caused by voltage sensor domain–pore coupling by the S4–S5 linker to S6 of NaV1.5 channel, as mentioned above. However, this result does not rule out that β3 also indirectly interacts with the DIV-VSD through alternative voltage sensor domain–pore coupling mechanisms. For instance, as proposed in Fig. 3 D, β3 would also be close to DIV-S5 or DIV-S6, which could underlie an allosteric modulation of β3 on DIV-S4 (the authors propose that the DIII–DIV linker might be a putative coupling domain). Clearly, more experimental data would be needed to fully understand the underlying mechanism by which β3 modulates DIV-S4 and steady-state inactivation.
It has been proposed that β1 and β3 expression vary during heart development (Domínguez et al., 2005; Okata et al., 2016). Moreover, even in different regions of the heart, such as the atria and ventricles, β1 and β3 subunits have different expression patterns (Fahmi et al., 2001; Calhoun and Isom, 2014). These spatial-temporal differences in expression suggest that these two subunits may play distinct physiological roles in regulating NaV channel function in different cell types and at different times (Calhoun and Isom, 2014). The stoichiometry of α, β1, and β3 in native cardiomyocytes is unknown; anything from 1:1 to 1:4 α/β stoichiometries have been proposed in earlier work (Namadurai et al., 2014, 2015). It seems plausible that, because of the nonadditive effects of β1 and β3 and the proposed different localizations of β1 and β3 by Zhu et al. (2017), β1 and β3, at least in heterologous systems, can both bind to and modulate NaV1.5 at the same time. Thus, a NaV1.5–β channel complex might have a 1:1 α/β stoichiometry (i.e., either one β1 or one β3) or a 1:1:1 α/β1/β3 stoichiometry. However, as Zhu et al. (2017) show when overexpressing β3, several β3 subunits can be associated to the NaV1.5 channel. Because both β1 and β3 bind to the channel noncovalently, the α–β subunit interaction could be dynamic and regulated according to the need of the cell under different conditions and/or in different cell types. The α–β subunit interaction could also be modulated by external factors, such as drugs, or in different disease states. The results of Zhu et al. (2017) on the differential interactions of β1 and β3 with various structural regions of NaV1.5 channels will shed light on the molecular basis of how mutations in those β subunits can cause disease and, importantly, will help to facilitate the development of novel drugs to treat these diseases.
Acknowledgments
This work was partly funded by an NINDS KO1 award (1K01NS096778-01A1) to R. Barro-Soria and grants from the Swedish Society for Medical Research to S.I. Liin and from NIGMS (R01 GM109762) and NHLBI (R01 HL131461) to H.P. Larsson.
The authors declare no competing financial interests.
Richard W. Aldrich served as editor.
References
- Abriel H. 2010. Cardiac sodium channel Nav1.5 and interacting proteins: Physiology and pathophysiology. J. Mol. Cell. Cardiol. 48:2–11. 10.1016/j.yjmcc.2009.08.025 [DOI] [PubMed] [Google Scholar]
- An R.H., Wang X.L., Kerem B., Benhorin J., Medina A., Goldmit M., and Kass R.S.. 1998. Novel LQT-3 mutation affects Na+ channel activity through interactions between α- and β1-subunits. Circ. Res. 83:141–146. 10.1161/01.RES.83.2.141 [DOI] [PubMed] [Google Scholar]
- Calhoun J.D., and Isom L.L.. 2014. The role of non-pore-forming β subunits in physiology and pathophysiology of voltage-gated sodium channels. Handb. Exp. Pharmacol. 221:51–89. 10.1007/978-3-642-41588-3_4 [DOI] [PubMed] [Google Scholar]
- Capes D.L., Goldschen-Ohm M.P., Arcisio-Miranda M., Bezanilla F., and Chanda B.. 2013. Domain IV voltage-sensor movement is both sufficient and rate limiting for fast inactivation in sodium channels. J. Gen. Physiol. 142:101–112. 10.1085/jgp.201310998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall W.A., Wisedchaisri G., and Zheng N.. 2017. The chemical basis for electrical signaling. Nat. Chem. Biol. 13:455–463. 10.1038/nchembio.2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda B., and Bezanilla F.. 2002. Tracking voltage-dependent conformational changes in skeletal muscle sodium channel during activation. J. Gen. Physiol. 120:629–645. 10.1085/jgp.20028679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez J.N., Navarro F., Franco D., Thompson R.P., and Aránega A.E.. 2005. Temporal and spatial expression pattern of beta1 sodium channel subunit during heart development. Cardiovasc. Res. 65:842–850. 10.1016/j.cardiores.2004.11.028 [DOI] [PubMed] [Google Scholar]
- Fahmi A.I., Patel M., Stevens E.B., Fowden A.L., John J.E. III, Lee K., Pinnock R., Morgan K., Jackson A.P., and Vandenberg J.I.. 2001. The sodium channel beta-subunit SCN3b modulates the kinetics of SCN5a and is expressed heterogeneously in sheep heart. J. Physiol. 537:693–700. 10.1113/jphysiol.2001.012691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko S.H., Lenkowski P.W., Lee H.C., Mounsey J.P., and Patel M.K.. 2005. Modulation of Nav1.5 by β1- and β3-subunit co-expression in mammalian cells. Pflugers Arch. 449:403–412. 10.1007/s00424-004-1348-4 [DOI] [PubMed] [Google Scholar]
- Mannuzzu L.M., Moronne M.M., and Isacoff E.Y.. 1996. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 271:213–216. 10.1126/science.271.5246.213 [DOI] [PubMed] [Google Scholar]
- Namadurai S., Balasuriya D., Rajappa R., Wiemhöfer M., Stott K., Klingauf J., Edwardson J.M., Chirgadze D.Y., and Jackson A.P.. 2014. Crystal structure and molecular imaging of the Nav channel β3 subunit indicates a trimeric assembly. J. Biol. Chem. 289:10797–10811. 10.1074/jbc.M113.527994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namadurai S., Yereddi N.R., Cusdin F.S., Huang C.L., Chirgadze D.Y., and Jackson A.P.. 2015. A new look at sodium channel β subunits. Open Biol. 5:140192 10.1098/rsob.140192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okata S., Yuasa S., Suzuki T., Ito S., Makita N., Yoshida T., Li M., Kurokawa J., Seki T., Egashira T., et al. 2016. Embryonic type Na+ channel β-subunit, SCN3B masks the disease phenotype of Brugada syndrome. Sci. Rep. 6:34198 10.1038/srep34198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazis A., and Olcese R.. 2012. Relative transmembrane segment rearrangements during BK channel activation resolved by structurally assigned fluorophore-quencher pairing. J. Gen. Physiol. 140:207–218. 10.1085/jgp.201210807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z., Zhu W., Schubert A.R., Pardieck J.L., Krumholz A., Hsu E.J., Zaydman M.A., Cui J., and Silva J.R.. 2015. Direct Measurement of Cardiac Na+ Channel Conformations Reveals Molecular Pathologies of Inherited Mutations. Circ Arrhythm Electrophysiol. 8:1228–1239. 10.1161/CIRCEP.115.003155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerman C.C., Wilde A.A., and Lodder E.M.. 2015. The cardiac sodium channel gene SCN5A and its gene product NaV1.5: Role in physiology and pathophysiology. Gene. 573:177–187. 10.1016/j.gene.2015.08.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Voelker T.L., Varga Z., Schubert A.R., Nerbonne J.M., and Silva J.R.. 2017. Mechanisms of noncovalent β subunit regulation of NaV channel gating. J. Gen. Physiol. 149 10.1085/jgp.201711802 [DOI] [PMC free article] [PubMed] [Google Scholar]