Understanding evolution requires the comparison of more than a few model species, and exploration of the genotype/phenotype relationship is limited...
Keywords: Oscheius tipulae, genome assembly, mapping-by-sequencing, vulva development, mig-13
Abstract
Mapping-by-sequencing has become a standard method to map and identify phenotype-causing mutations in model species. Here, we show that a fragmented draft assembly is sufficient to perform mapping-by-sequencing in nonmodel species. We generated a draft assembly and annotation of the genome of the free-living nematode Oscheius tipulae, a distant relative of the model Caenorhabditis elegans. We used this draft to identify the likely causative mutations at the O. tipulae cov-3 locus, which affect vulval development. The cov-3 locus encodes the O. tipulae ortholog of C. elegans mig-13, and we further show that Cel-mig-13 mutants also have an unsuspected vulval-development phenotype. In a virtuous circle, we were able to use the linkage information collected during mutant mapping to improve the genome assembly. These results showcase the promise of genome-enabled forward genetics in nonmodel species.
A few model organisms have greatly contributed to biological research in the last decades, among them the nematode Caenorhabditis elegans. However, to tell conserved from specific features and understand the evolutionary process that gave rise to extant diversity, comparisons between different species at key phylogenetic positions are necessary. Genome sequences have been instrumental in model organism research, and the ongoing revolution in new genome sequencing and assembly technologies eases the once-daunting task of building such resources for any species. Draft assemblies can now be achieved within a few months at a reasonable cost, even by individual teams. Once a reference genome and gene annotation are available, other high-throughput sequencing techniques, such as RNA sequencing, can be used to explore genotype–phenotype interactions (Liu et al. 2015; Roux et al. 2015). Classical forward genetic approaches, i.e., phenotype-based mutagenesis screens, are easy to perform and universal, provided the species can be cultured and crossed. Forward genetics has the huge advantage of identifying genes without prior knowledge or bias, which is particularly important in evolutionary comparisons. Such forward screens have been applied to many nonmodel species, but a remaining challenge is the identification of causative mutations and thus the function of target genes at the molecular level.
Massively parallel sequencing permits rapid identification of phenotype-causing mutations through “mapping-by-sequencing.” Mapping-by-sequencing has become a standard forward genetic approach in most model organisms, including Arabidopsis thaliana (James et al. 2013), Saccharomyces cerevisiae, Danio rerio, Drosophila melanogaster, and C. elegans (Schneeberger 2014). Mapping-by-sequencing strategies are generally based on the sequencing of bulk-segregant populations (Michelmore et al. 1991). Starting with a cross between the mutant strain of interest to a phenotypically wild-type but genetically different outcrossing strain, F2 grand-progeny individuals showing the recessive mutant phenotype are selected, and their DNA is pooled and sequenced. The recessive causative allele is necessarily homozygous for all individuals in this bulk mutant sample. For any polymorphic position between the two backgrounds, allele frequencies can be measured from mapped sequencing reads. If unlinked to the selected mutation, polymorphisms that distinguish the two parental strains will be found in the mutant pool at equal frequencies. However, if these markers are genetically linked to the causative mutation, the proportion of the allele from the wild-type background will decrease close to the mutated locus and approach zero in its immediate vicinity. Plotting allele frequencies along a reference genome will thus define a region of low wild-type allele frequency, surrounding the location of the causative mutation. The physical size of this interval will decrease with the number of meiotic recombination events in the F2 population (and, if limiting, with the number of available polymorphic positions).
In theory, if a mutant strain only differs from a nonmutant strain by a single mutation, comparing whole-genome sequencing data should reveal this polymorphism with no need for mapping (Nordström et al. 2013). In practice, genetic mapping information is required because a mutagenized strain and its nonmutagenized reference will have many spurious fixed differences: nonphenotype-causing mutations due to mutagenesis, or de novo spontaneous mutations fixed by drift in each strain. Technical noise, such as sequencing or mapping errors, can also contribute to observed variation. After one or several crosses, most of these variations can be excluded because they fall outside the mapping interval. Hence, sequencing bulk-segregant F2 populations and mapping allele frequencies on a reference genome is key to identifying the phenotype-causing mutation. Recently, mapping-by-sequencing has been applied to crop plants like rice (Abe et al. 2012; Fekih et al. 2013; Takagi et al. 2013), maize (Liu et al. 2012; Li et al. 2013), or barley (Mascher et al. 2014; Pankin et al. 2014), in which the genome is incomplete but high-resolution physical and genetic maps exist for each chromosome. Whether mapping-by-sequencing can be efficient in organisms with a fragmented or incomplete reference genome and no genetic map has not been tested.
The phylum Nematoda, which includes the model organism C. elegans, is an ideal target for the development of new species as model organisms amenable to forward genetics, because many species combine easy genomics and easy genetics. First, the relatively small size of nematode genomes, ranging from 20 to 400 Mb, ensures reasonable costs and good quality for most genome projects (Kumar et al. 2012). Second, the powerful genetics of C. elegans comes from its short life cycle and its androdioecious mode of reproduction, with selfing XX hermaphrodites that mate with X0 males in a facultative and controllable manner. Other nematode species sharing these features have therefore been chosen to perform forward genetics: C. briggsae (Gupta et al. 2007), Oscheius tipulae (Félix 2006), and Pristionchus pacificus (Sommer 2006). With C. elegans, these three free-living bacteriovorous nematodes belong to the Rhabditinae (De Ley and Blaxter 2002) [also referred as clade V (Blaxter et al. 1998) or 9 (Holterman et al. 2006; van Megen et al. 2009); see Figure 1]. Besides its mode of reproduction and easy culture, O. tipulae has been chosen for several reasons: the two-step, anchor-cell induction of vulval-precursor-cell fates and its simple vulval cell lineage (Félix and Sternberg 1997), its easy isolation from various regions of the world (Baïlle et al. 2008), and its phylogenetic position compared to C. elegans as an outgroup to Caenorhabditis species but an ingroup to P. pacificus (Blaxter et al. 1998). High-quality genome assemblies have been generated for C. briggsae (Stein et al. 2003) and P. pacificus (Dieterich et al. 2008). Classical techniques have been employed to map and identify genes involved in different traits with particular emphasis on the convergent evolution of self-fertile hermaphroditism, reviewed in Ellis and Lin (2014), and vulva development (Seetharaman et al. 2010; Sharanya et al. 2012, 2015; Sommer 2012; Kienle and Sommer 2013). In P. pacificus, genetic analyses of the evolution of sex muscles (Photos et al. 2006), gonad development (Rudel et al. 2008), dauer formation (Ogawa et al. 2009, 2011), and buccal tooth dimorphism (Bento et al. 2010) have been published. Inspired by the versatile and robust pipelines of mapping-by-sequencing routinely used for C. elegans (Minevich et al. 2012), we generated a genome assembly for O. tipulae and here test mapping-by-sequencing in this species.
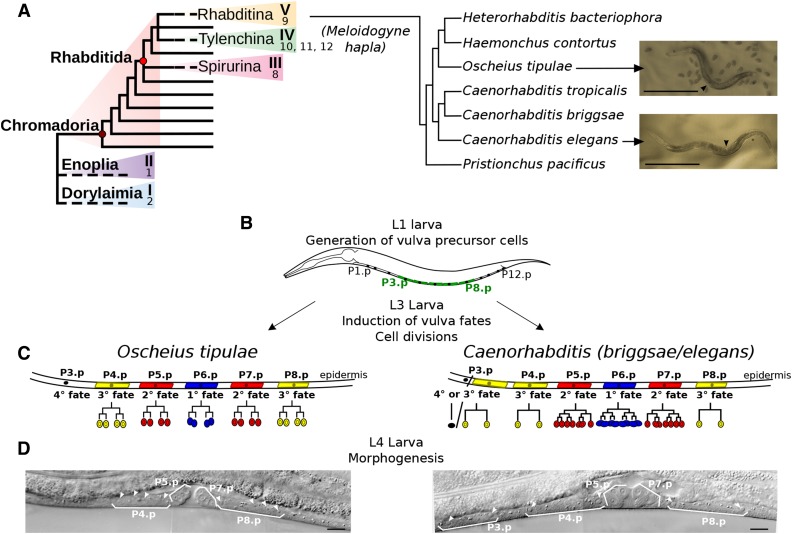
Figure 1.
Phylogenetic relationships of O. tipulae and comparison of vulva development with C. elegans. (A) Cartoons showing (left) the systematic structure phylum Nematoda [clades I–V or 1–12 defined according to Blaxter et al. (1998), p. 199, and van Megen et al. (2009), respectively] and (right) the relationship of O. tipulae to key Rhabditina species discussed in the text. Pictures of young adult hermaphrodites of O. tipulae and C. elegans are shown on the right (arrowhead pointing to the vulva). Bar, 0.5 mm. (B–D) Vulva development in O. tipuale and C. elegans. (B) Ventral epidermal cells called P3.p to P8.p are specified as vulva precursor cells in young L1 larvae. (C) Conserved anteroposterior pattern of cell fates, despite variation in cell lineages: P6.p occupies the central position and adopts a 1° fate, P5.p and P7.p are induced to follow a 2° fate, and their daughters will form the border of the vulval invagination. Other cells (P3,4,8.p) differentiate into nonvulval epidermal fates, either with or without one round of division (3° or 4° fates, respectively). (D) Nomarski images of wild-type, L4-stage hermaphrodites (left, O. tipulae; right, C. elegans), highlighting the daughter cells of the Pn.p cells (arrowheads). Bar, 10 µm.
The available O. tipulae mutants have been obtained in forward genetic screens for mutations affecting vulva development and egg laying (Dichtel et al. 2001; Louvet-Vallée et al. 2003; Dichtel-Danjoy and Félix 2004). The nematode vulva connects the uterus to the outside and is required for egg laying and copulation. Vulva precursor cell specification is one of the best known developmental systems in C. elegans (Sternberg 2005), and the development of the vulva has become an important system for comparative studies in nematodes (Sommer and Bumbarger 2012). The C. elegans vulva develops from a group of six precursor cells aligned along the ventral midline of the animal, called P3.p to P8.p (Figure 1). While the vulva is under strong selective pressure for egg laying and mating (especially in obligate outcrossing species), a complex mosaic of change and stasis of different vulval developmental traits is observed among Rhabditinae (Kiontke et al. 2007). To date, most studies have focused on the conservation of the anteroposterior pattern of fates expressed by the three central precursor cells P5.p, P6.p, and P7.p across wide phylogenetic distances (Figure 1). Progeny of these three cells have specific division patterns and play specific roles in forming the mature vulva, with (usually) P6.p taking a central, primary (1°) fate, and P5.p and P7.p taking a secondary (2°) fate. Nematode vulval development is one of the best examples of how pervasive but cryptic evolution modifies the mechanisms of development despite an invariable output; a phenomenon known as “developmental system drift” (True and Haag 2001; Burgess 2011; Robinson 2011). Interestingly, the phenotypic spectrum of O. tipulae vulva mutants suggests substantial underlying evolutionary differences in specification and interaction compared to other species. In O. tipulae, all but 1 of the 34 vulval mutations so far isolated in forward genetic screens await molecular identification. Uncovering the molecular nature of the 33 mutants that are still uncharacterized in O. tipulae will help to unveil the mode and tempo of the vulva development system drift and possible innovations among Rhabditinae.
Here, we integrate a range of genomic tools to rapidly and cost effectively build a draft genome assembly and annotate genes in the nematode O. tipulae. Despite its fragmentation, we show that this draft assembly is a suitable platform for mapping-by-sequencing. As a proof of concept, we identify the vulva mutant gene cov-3 (Louvet-Vallée et al. 2003) as the O. tipulae homolog of C. elegans mig-13. Finally, we show that linkage information collected during mapping of mutant alleles can be further used to detect mis-scaffolding and to group scaffolds into chromosome-scale linkage groups, improving the initial assembly, and providing useful information for further genetic mapping and mutant gene identification.
Materials and Methods
Nematode strains and culture
The O. tipulae reference strain CEW1, originally from Brazil, was used for forward mutant screens (Félix et al. 2001). The second wild strain that we used as a source of molecular polymorphisms is JU170, a strain sampled from soil in Sevilla, Spain, in 2000. This latter strain was chosen as it has a high genetic distance compared to CEW1, based on prior amplified fragment length polymorphisms (AFLP) analysis of a set of 63 wild O. tipulae isolates (Baïlle et al. 2008). O. tipulae mutant strains were generated as previously described (Félix et al. 2000; Dichtel et al. 2001). See Supplemental Material, Table S3, for the list of mutant alleles used in this study. These alleles were described in Dichtel et al. (2001), Louvet-Vallée et al. (2003), and Dichtel-Danjoy and Félix (2004), except for mf33, whose phenotype is only weakly penetrant and will be described elsewhere. O. tipulae strains were thawed from frozen stocks and cultured at 23° on NGM-plates seeded with Escherichia coli OP50, as previously described (Félix et al. 2000). The C. elegans strains used were Bristol strain N2 and the CF726 strain carrying the mig-13(mu225) X mutation, and they were cultured according to standard protocols (Brenner 1974) at 20°.
Library preparation and next-generation sequencing
For genome assembly, genomic DNA was extracted from a mixed-stage growing population using reagents from the Puregene Core Kit A (QIAGEN, Valencia, CA). Data were generated from a 400-bp library and a 3-kb mate-pair or jumping library, following manufacturer’s instructions. The mate-pair library was constructed by the Centre for Genome Research, Liverpool. The 400-bp library was sequenced on the Illumina MiSeq platform (6.5 million 100-base read pairs and 20.7 million 300-bp read pairs). The paired-end library was sequenced on an Illumina HiSeq2500 (145.5 million 100-bp paired reads) (Table S1). Raw data have been submitted to the International Nucleotide Sequence Database Consortium (INSDC) under project accession no. PRJEB15512.
RNA was extracted from a mixed-stage growing population of O. tipulae CEW1, cultured in standard laboratory conditions. Poly(A)-enriched complementary DNA was prepared from the RNA by GATC (Konstanz, Germany) and normalized using reassociation kinetics. RNA sequencing was performed by GATC on the Roche GS FLX Titanium platform. A total of 592,650 reads (average length 369 bp) remained after filtering for quality. Raw transcriptome data are available in INSDC under project no. PRJEB15512).
For mapping-by-sequencing, genomic libraries and sequencing data from O. tipulae JU170 (INSDC project accession no. PRJEB19969) and mutant F2 pools (see project accessions in Table S3) used were generated by BGI. Short insert libraries (<800 bp) were paired-end sequenced on Illumina Hiseq2000, Hiseq2500, or Hiseq4000 with 100-bp reads to obtain 2.2 GB (∼40× coverage) of clean data per sample after manufacturer’s data filtering (removing adaptor sequences, contamination, and low-quality reads).
Genome assembly
All software tools used (including versioning and command line main options) are summarized in Table S4. Raw reads were trimmed for adaptors using Cutadapt (Martin 2011) and low-quality bases, then corrected for sequencing errors based on k-mer content using Quake (Kelley et al. 2010) and JELLYFISH (Marçais and Kingsford 2011). Raw data were checked with FastQC (Andrews 2010) and a preliminary assembly generated with CLC Assembly Cell (CLC bio 2017) (Table S5). The CLC assembly was screened for contaminants using taxon-annotated, GC-coverage (TAGC) plots (Kumar et al. 2013). Only data deriving from E. coli, the food source, was identified as contaminant and the corresponding reads were removed. The optimal k-mer size for assembly of the cleaned read set was estimated using KmerGenie (Chikhi and Medvedev 2014). Nine different assemblers (Table 1) were used to generate preliminary assemblies and these were assessed using basic metrics, correctness of read alignment using ALE (Clark et al. 2013) and REAPR (Hunt et al. 2013), and biological completeness using Core Eukaryotic Genes Mapping Appoach (CEGMA) (Parra et al. 2007) and direct identification of ribosomal RNA genes and mitochondrial genome sequences. SPAdes (Bankevich et al. 2012) outperformed the other assembly tools in almost all aspects and was chosen as draft assembly nOt.1.0. An improved assembly limited to the nuclear genome (nOt.2.0) was generated by removing mitochondrial contigs and contigs of abnormally low coverage and by breaking all scaffolds where mis-assembly had been indicated from analysis of mapping plots and REAPR fragment coverage distribution (FCD) scores.
Table 1. Statistics of the assemblies generated by different assemblers.
| MaSuRCA (Zimin et al. 2013) | SGA (Simpson and Durbin 2010) | Velvet (Zerbino and Birney 2008) | AbySS (Simpson et al. 2009) | CLC (CLC bio 2017) | Ray (Boisvert et al. 2010) | SOAPdenovo (Luo et al. 2012) | ALLPATHS-LG (Gnerre et al. 2011) | SPAdes (Bankevich et al. 2012) | |
|---|---|---|---|---|---|---|---|---|---|
| Span (bp) | 89,577,071 | 61,516,197 | 58,979,055 | 59,511,073 | 59,525,064 | 59,264,893 | 60,512,111 | 59,578,117 | 59,698,979 |
| No. of scaffolds | 6,646 | 5,619 | 786 | 782 | 520 | 147 | 313 | 151 | 203 |
| Longest scaffold | 134,185 | 836,405 | 1,580,233 | 1,581,252 | 2,304,523 | 3,256,501 | 2,993,798 | 3,772,824 | 4,597,891 |
| N50 | 18,584 | 78,455 | 213,759 | 258,755 | 659,741 | 1,269,874 | 1,466,413 | 1,567,404 | 1,573,002 |
| No. N’s | 3,067,902 | 276,207 | 1,238 | 160,214 | 480,962 | 153,992 | 1,918,481 | 492,105 | 17,036 |
| GC content (%) | 44.5 | 44.5 | 44.5 | 44.5 | 44.5 | 44.5 | 44.5 | 44.5 | 44.5 |
| Absolute REAPR scorea | 3.7 | 8.1 | 9.1 | 9.4 | 6.4 | 2.8 | 1.5 | 5.2 | 8.4 |
| ALE scoreb | −2,909 × 106 | −934 × 106 | −482 × 106 | −467 × 106 | −976 × 106 | −1,110 × 106 | −982 × 106 | −978 × 106 | −291 × 106 |
REAPR absolute score measures the frequency of error-free bases and contigs, ranging from 0 to 1.
ALE score is computed from the logarithm of the probability that the assembly is correct. ALE scores are negative: the closer to zero, the larger is the probability of the assembly of being correct.
Gene prediction and orthogroup inference
Genes were predicted using a two-pass pipeline (Koutsovoulos et al. 2014) (see Figure S3) based on MAKER2 (Holt and Yandell 2011) and Augustus (Stanke et al. 2006), and using the transcriptome data as evidence. Repeats were identified in the assembly using RepeatModeler (Smit et al. 2013–2015). MAKER2 was run in an SGE cluster using the SNAP (Korf 2004) ab initio gene finder trained by CEGMA (Parra et al. 2007) output models, the GeneMark-ES ab initio finder, SwissProt proteins, and O. tipulae transcripts. The transcriptome data were filtered so that only reads >300 bases that had significant basic-local-alignment-search-tool (BLAST) similarity to C. elegans protein databases were kept. The MAKER2 predictions were used to train Augustus to generate a custom gene-finding profile for O. tipulae. Finally, Augustus was used with the gene-finder profile and O. tipulae transcripts to predict the final gene set. Not enough transcript evidence was available to train a model of untranslated regions (UTRs), and therefore no UTRs were annotated. Protein sets for C. elegans (C. elegans Sequencing Consortium 1998), Dictyocaulus viviparus (Koutsovoulos et al. 2014), Meloidogyne hapla (Opperman et al. 2008), and P. pacificus (Dieterich et al. 2008), downloaded from WormBase (http://www.wormbase.org/), were clustered with Orthofinder (Emms and Kelly 2015) using an inflation value of three.
Synteny
O. tipulae scaffolds containing >100 predicted protein-coding genes were selected to perform synteny analyses between O. tipulae and C. elegans. Predicted proteins from these 36 scaffolds were compared to the C. elegans protein data set with BLAST to identify orthologous pairs. For each pair, the chromosome location of the C. elegans ortholog was identified. Hierarchical clustering was performed to group the scaffolds into groups based on the proportions of C. elegans chromosomal attributions (Figure S9).
Mapping crosses
JU170 males were crossed with young mutant hermaphrodites of the desired recessive mutant genotype (in the CEW1 background) and several F1 cross-progeny were singled. In the F2 progeny, mutant animals were isolated based on the observation of the mutant phenotype (checked under Nomarski microscopy if necessary) and singled onto individual plates. These lines were amplified by selfing and allowed to grow until the E. coli food was just exhausted. For cov-3 F2 mutant pool sequencing, independent mutant F2-derived populations (21 for mf35 and 51 for sy463) were washed several times in M9 buffer (Stiernagle 2006) and stored at −80° as pellets. A similar mass of nematodes from each F2 line was then pooled for DNA extraction. For the F2 sequencing of other mutations, each F2-derived population was checked for the presence of the mutation in the homozygous state and allowed to grow until the E. coli food was just exhausted. Nematodes were directly pooled from different plates and washed in M9 buffer. DNA was then extracted using the Puregene Core Kit A (QIAGEN).
Variant analysis, gene mapping, and identification
JU170 whole-genome sequencing data were analyzed to identify SNPs compared to the CEW1 reference genome. These variants were then used for genetic mapping of the mutants (listed in Table S3). Reads were mapped with bwa (Li and Durbin 2009) to the CEW1 assembly, mappings processed with the GATK tool suite (McKenna et al. 2010) version 3.3-0 and variants called with HaplotypeCaller using default parameters. We followed HaplotypeCaller’s authors’ recommendations of best practice (DePristo et al. 2011; Van der Auwera et al. 2013), realigning reads around indels and performing BQSR by bootstrapping a first call made with HaplotypeCaller. We analyzed the 300-bp CEW1 MiSeq data used for genome assembly with the same pipeline, after E. coli decontamination, as a control for variant calling. We then hard filtered a list of high-confidence SNPs of JU170 with conservative criteria, retaining homozygous positions covered by at least three reads in each strain, with a sequencing and mapping quality higher than 100 and 55, respectively, and a position noted as reference in CEW1 and variant in JU170. Sequencing data from pooled F2 mutants were analyzed with the same pipeline, except that variant calling was restricted to a list of JU170 SNPs previously established for faster computing (using the HaplotypeCaller option genotyping_mode GENOTYPE_GIVEN_ALLELES). Output VCF files were used to compute allele frequencies for each SNP on the JU170 list as the ratio of the number of reads with the JU170 allele over the total number of reads. These frequencies were plotted along each scaffold using custom R scripts. Scaffolds displaying a mean JU170 allele frequency <10% were selected as possibly linked to the candidate locus and retained for a second, unrestricted variant call. JU170 variants were filtered out from the output at this stage. We also systematically added for analysis the 47 scaffolds that do not carry SNPs between JU170 and CEW1 (0.1% of the genome). The functional impact of identified variants was assessed using snpEff (Cingolani et al. 2012) and used to prioritize candidate genes. Where two alleles of the same gene were analyzed, candidate gene lists were compared to exclude identical variations (likely initial background variations) and were inspected for independent hits to the same gene with a different noncomplementing mutation. When necessary, visual inspections of variations in aligned reads was performed with Tablet (Milne et al. 2013). Scripts used to automate this pipeline are available at: https://github.com/fabfabBesnard/Andalusian_Mapping.
Sanger sequencing and gene validation
Four primers were designed to cover the Oti-mig-13 coding region. Oti-mig-13 fragments were amplified from strains homozygous for the four alleles of cov-3 [mf35, mf79, mf80, and sy463 (Louvet-Vallée et al. 2003)]. PCR products were verified on agarose gels, cleaned on columns, and sent for Sanger sequencing to Eurofins.
X chromosome linkage of scaffolds and pyrosequencing
Linkage of scaffolds to the X chromosome was determined using directed pyrosequencing of F1 males from crosses (in both directions) between the CEW1 and JU170 strains. For each scaffold, we selected one polymorphic nucleotide position in the middle of the scaffold in an otherwise conserved context (no other variations in the 300 bp surrounding the variant) to ensure unbiased PCR. For each position, two PCR primers and one sequencing primer were designed (Table S6) using the pyrosequencer’s companion design software (PyroMark Q96 ID instrument from Biotage, Uppsala, Sweden). The primers were tested on parental strains. PCR using universal biotinylated primers and single-stranded PCR amplicon purification was performed as previously described (Duveau and Félix 2010). For each genotyping assay, in a successful mating plate, three individual adult F1 males and three individual adult F1 hermaphrodites were transferred individually into 10 µl worm lysis buffer containing proteinase K (200 mg/ml) and frozen at −20°. Nematodes were then thawed and lysed at 60° followed by 15 min at 95° to inactivate the proteinase. A total of 4 µl of worm lysate was used as PCR template. Pyrosequencing reactions were performed in the sequencing mode. High-quality DNA extracts from the parental strains were used as positive controls (separately and mixed to mimic a heterozygote) for each assay. Linkage to autosomes or the X chromosome was made if at least two male genotypes were concordant.
Scaffold linkage analysis
For each mutant strain (see Table S3) and each scaffold, the mean frequency of alleles in the F2 mapping population whole-genome sequencing was extracted from the previous pipeline. All data sets included frequencies for the 144 scaffolds (over a total of 191 in nOt.2.0) that contained polymorphic positions between the strains CEW1 and JU170. Scaffolds and F2 mapping populations were clustered using the “heatmap.2” function of the “gplots” package in R.
Microscopy
The vulval-cell phenotypes were determined during the early to mid-L4 larval stage using Nomarski microscopy.
Data availability
All raw sequencing data supporting the conclusions of this article have been submitted to the INSDC. Sequencing data of genomic DNA and RNA from reference strain CEW1 (used for assembly) are available under project accession no. PRJEB15512. Whole-genome resequencing data of the mapping strain JU170 is under project accession no. PRJEB19969. Project accession numbers corresponding to the sequencing data of F2 mapping populations of the different strains used in this study are listed in Table S3. Annotations have also been submitted to the database European Molecular Biology Laboratory–European Bioinformatics Institute under the accession no. PRJEB15512.
All scripts used for mapping-by-sequencing and gene identification are available in https://github.com/fabfabBesnard/Andalusian_Mapping.
The genome assembly and annotation is available for browsing, exploration, and download at http://ensembl.caenorhabditis.org/index.html; and will be uploaded soon to WormBase (http://www.wormbase.org/).
Results
Assembly and annotation of the O. tipulae genome
We sequenced the genome of O. tipulae strain CEW1, which was used as the reference strain in all previous molecular and genetic studies (Blaxter et al. 1998; Félix et al. 2001; Ahn and Winter 2006; Félix 2006). Following the strategy suggested by the 959 Nematode Genomes project (Kumar et al. 2012), we generated data from two libraries (see Materials and Methods and Table S1). Paired-end 300-base Illumina MiSeq reads (∼50-fold genome coverage) were generated from a short insert library, and mate-pair 100-base Illumina HiSeq2500 reads (∼150-fold coverage) from a 3-kb virtual insert library. Before assembly, we cleaned the raw data by removing adaptor and low-quality bases, and performed error correction (see Materials and Methods and Figure S1). E. coli contaminating data were identified using TAGC plots and removed (Kumar et al. 2013) (Figure S2). We compared the performance of nine different assemblers (Table 1). All but one of the assemblers agreed on a genome size of ∼60 Mb, confirming evaluations based on k-mer counting. The assembly generated by SPAdes (Bankevich et al. 2012) had the highest quality metrics, with only 203 scaffolds >500 bp, N50 of >1.5 Mb, and only ∼17,000 undetermined bases. The SPAdes assembly also had the highest accuracy as assessed by ALE (Clark et al. 2013) and the third best by REAPR (Hunt et al. 2013), which are two reference-independent programs designed to track assembly errors (Table 1). From this first version, named nOt.1.0, we derived the nuclear genome assembly nOt.2.0 by removing three mitochondrial scaffolds and discarding 32 contigs which had very low coverage. We also broke 16 scaffolds that showed evidence for overscaffolding (see below). The final draft assembly spans 59,468,623 bases and includes 191 contigs >500 bp. Gene finding was performed as previously described (Koutsovoulos et al. 2014), using both ab initio predictions and evidence from RNA-sequencing data (Figure S3 and see Materials and Methods), and resulted in 14,938 gene predictions (Table 2). We assessed the completeness of the nOt.2.0 assembly using the CEGMA pipeline, and identified 97.6% complete and 99.2% partial genes out of the set of 248 conserved and likely essential genes (Parra et al. 2007). The O. tipulae genome is surprisingly small, with a span only 59% of that of the model species C. elegans. This reduction likely results from different factors (Table S2). Genes are less numerous (74% that of C. elegans) and have a denser packing (251 genes per Mb compared to 202 genes per Mb in C. elegans). They are also shorter overall (mean gene length is 81% of C. elegans) with shorter introns (160 bp vs. 339 bp), despite more introns per gene (8.5 vs. 6). The reduction in genome size is mirrored in the reduction in the span of intergenic DNA, and the repeat content of this intergenic DNA is much reduced. Overall repeat content in O. tipulae is 8.4% of the genome compared to 18.7% in C. elegans, mostly due to a much lower span of DNA elements (1.0% in O. tipulae compared to 9.9% in C. elegans). Comparing gene orthologies revealed no particular patterns of gene losses but indicated a reduced amount of species-specific genes, suggesting lower frequency of gene expansion and diversification in O. tipulae (Figure S4).
Table 2. Genomic characteristics of O. tipulae.
| O. tipulae (assembly nOt.2.0) | |
|---|---|
| Genome size (Mb) | 59 |
| Total intergenic span (Mb) (% of total genome) | 21 (35.5%) |
| Total genic span (Mb) (% of total genome) | 38 (64.5%) |
| Number of genes | 14,938 |
| Number of genes/Mbp | 251 |
| Mean/median transcript length (bp) | 1,368/1,032 |
| Mean/median exon length (bp) | 160/132 |
| Mean/median number of exons per gene | 8.5/7 |
| Mean intron length (bp) | 160 |
Mapping-by-sequencing identifies the cov-3 locus
Our motivation to assemble the O. tipulae genome was to identify the molecular lesions affecting vulva development in a set of previously characterized mutant strains (Dichtel et al. 2001; Louvet-Vallée et al. 2003; Dichtel-Danjoy and Félix 2004). To establish this proof of concept, we chose the cov-3 (cov standing for competence and centering of vulva) mutant. cov3-mutant O. tipulae display a partial loss of vulval competence with a highly penetrant anterior shift of the vulval fate pattern (Louvet-Vallée et al. 2003). This phenotype has only recently been described in C. elegans, occurring at low penetrance in some Wnt pathway mutants (Milloz et al. 2008; Grimbert et al. 2016); suggesting that the coupling of competence and centering differs between the two species. Four alleles of cov-3 were available, permitting independent confirmation of candidate genes.
For genetic mapping, we chose the strain JU170. This wild O. tipulae isolate is genetically distant from CEW1 (the genetic background of all mutants) based on AFLP (Baïlle et al. 2008). We resequenced JU170 to ∼40-fold coverage and identified SNPs and indels that distinguish CEWI and JU170. We found one SNP every 95 bp on average, and a total of 632,027 SNPs (see Figure S5). For comparison, the genetically distant strain used routinely in C. elegans mapping-by-sequencing (CB4856) has an average SNP density of 1 SNP every 1000 bp (Hillier et al. 2008; Minevich et al. 2012), which is 10-fold less.
We crossed cov-3 hermaphrodites (alleles mf35 and sy465) with JU170 males and selected F2 grand-progeny displaying the recessive cov-3 phenotype. After amplification by selfing, F2-derived populations were pooled and genomic DNA extracted and sequenced (see Materials and Methods and Figure 2). To identify the cov-3 mutations, a first variant call was performed only on known JU170 SNPs, and their frequencies were plotted genome wide. Scaffolds containing the lowest JU170 allele frequency were retained. A second-pass call scanned these scaffolds for all other variants (excluding JU170 and other background variations). This analysis retrieved only eight candidate mutations which were prioritized according to their functional impact. In the sy463 pool, a putative 38-bp insertion was predicted to cause a frameshift in the nOt.2.0.1.t01002 gene, homologous to Cel-mig-13. However, inspection of read pairing revealed a much larger deletion (1888 bp) in this gene. No SNP variants were called at this locus for the mf35 pool, but inspection revealed a 283-bp deletion affecting the same gene (Figure S6). Such large deletions are expected to be less frequent than SNPs following EMS mutagenesis (Flibotte et al. 2010) and the variant toolkit we used is not optimal for identification of large indels.
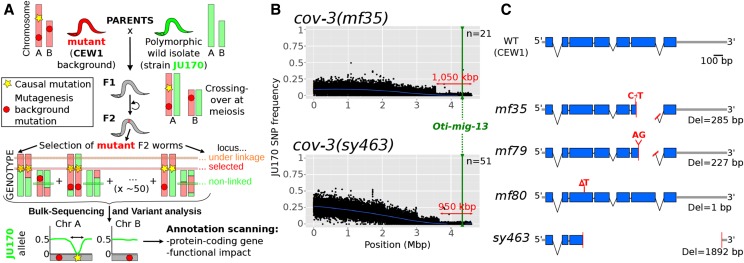
Figure 2.
Mapping-by-sequencing of O. tipulae vulva mutants and identification of cov-3 mutations in the Oti-mig-13 gene. (A) Principle of the mapping-by-sequencing approach, involving the wild isolate JU170 as a mapping strain and whole-genome resequencing of a bulk of mutant F2 grand-progeny (see text for details). The phenotype-causing mutation is mapped genetically by the cross as the region of low frequency of JU170 alleles. Final identification requires scanning of this interval for variations specific to the mutant background. (B) JU170 allele frequency plots in scaffold 1 (genome version nOt.2.0.) in bulk-sequencing data generated with the independent cov-3 alleles mf35 and sy463 (upper and lower plots respectively). On each plot, the blue line is a local regression of the JU170 allele frequency, the red arrow indicates the mapping interval size, the green line the position of the mig-13 gene, and n the number of F2 lines pooled in each mapping population. (C) Cartoon depicting the structure of the wild-type Oti-mig-13 gene and the alterations found in all independent alleles isolated so far. Mutations are indicated in red, exons are blue boxes, introns thin black lines, and intergenic regions thick gray lines. Del, deletion; WT, wild type.
Both mutations are predicted to result in truncation of the expressed protein, and thus are likely to be loss-of-function alleles. In the F2 bulk-segregant data, JU170 allele frequency displayed a clear drop in the ∼1 Mb at the end of scaffold nOt.2.0.scaf00001, an interval that contains ∼200 gene predictions including Oti-mig-13 (Figure 2). We conclude that Oti-mig-13 (nOt.2.0.1.t01002) is the best candidate gene for the cov-3 locus. To confirm this, the Oti-mig-13 locus was amplified and directly sequenced from all four available, independently isolated cov-3 alleles: sy463, mf35, mf79, and mf80. The deletions predicted from whole-genome resequencing were confirmed in sy463 and mf35. In the trimethylpsoralen-ultraviolet (TMP-UV)-induced alleles mf79 and mf80, two new frameshift-causing deletions were found, and these are thus also likely to be loss-of-function alleles (Figure 2). We have thus been able to use a draft genome assembly to identify a locus by mapping-by-sequencing. The O. tipulae cov-3 locus is orthologous to C. elegans mig-13. Following standard nematode genetics nomenclature procedures, we redesignate Oti-cov-3 as Oti-mig-13.
The role of mig-13 in vulva development has changed between C. elegans and O. tipulae
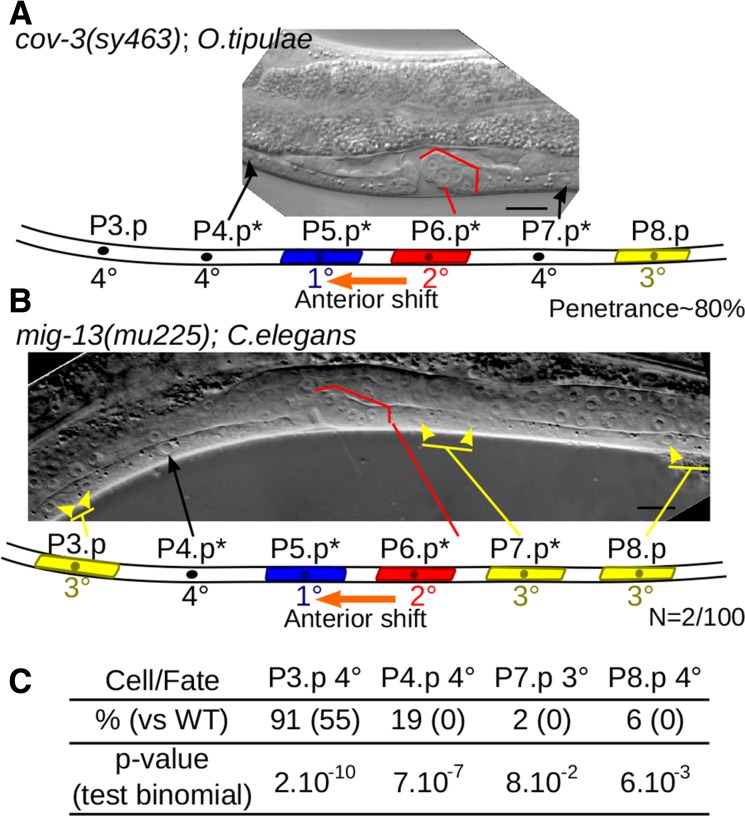
The MIG-13 protein is predicted to be a single-pass, trans-membrane protein that contains two protein–protein interaction domains (a CUB domain and an LDL-receptor repeat), both extracellular. C. elegans and O. tipulae MIG-13 proteins are quite similar (48% aa identity and 63.5% similarity), with higher identity in CUB and LDL functional domains (Figure S7). Similar organization is observed in MIG-13 homologs in other nematodes. In C. elegans, mig-13 is necessary for the anterior migration of the neuroblasts of the QR lineage (Sym et al. 1999), but Cel-mig-13 was not known to play any role in vulva development. While O. tipulae cov-3 mutants display a partially penetrant egg laying-defective phenotype, the C. elegans mig-13 mutants are not defective in egg laying. We further investigated the Cel-mig-13(mu225) null mutant using Nomarski microscopy on a large number of animals. We observed an anterior shift of the 1° fate on P5.p, associated with decreased P4.p competence, a phenotype identical to the O. tipulae mig-13 phenotype, with very low (2%) penetrance. Penetrance of this phenotype is 80% in O. tipulae (Louvet-Vallée et al. 2003) (Figure 3, A and B). We also observed a more penetrant reduction in competence of vulval equivalence group cells in Cel-mig13(mu255), where P(3,4,8).p adopt the noncompetent fused 4° fate more frequently than the competent uninduced 3° fate (Figure 3C). Thus, mig-13 does play a role in vulva development in C. elegans, but the impact of its loss of function is reduced compared to O. tipulae, especially as far as the centering defect is concerned. The difference in the mutational phenotypic spectrum between the two species is thus likely to be due to a quantitative rather than a qualitative evolution of the contributions of mig-13 to the vulva genetic network. This example demonstrates the power of streamlined forward genetics in O. tipulae, as it both uncovers the evolution of developmental mechanisms hidden by a highly conserved cell-fate pattern, and also reveals new aspects of C. elegans development, even in a well-studied system such as vulva formation.
Figure 3.
Conservation and evolution of MIG-13 between C. elegans and O. tipulae. (A) Typical phenotype of a Oti-cov-3 mutant, shown in a Nomarski picture and an interpretation cartoon below: an anterior shift (1° fate shifted from P6.p to P5.p) is coupled to a reduced competence of vulva precursor cells. * indicates Pn.p cells with a modified fate compared to the wild type. (B) A cov-3-like phenotype can be observed at very low frequency in a mig-13 null mutant, Cel-mig13(mu225), of C. elegans. (C) Reduced competence of P3,4,8.p cells in Cel-mig-13(mu225) is indicated by increased frequency of 4° fate vs. 3° fate. WT, wild type.
Improvements of the O. tipulae genome assembly using genetic linkage data
The JU170 allele frequency plots provide genome-wide information about genetic linkage that can be used to improve the genome assembly, both in identifying errors in assembly and in superscaffolding into linkage groups. Since many F2 lines are pooled in each data set, recombination events are averaged out and JU170 allele frequency (with mean of ∼0.5) should vary continuously, especially along scaffolds unlinked to the selected mutations. Following precedents (Leshchiner et al. 2012), we used abrupt breaks in JU170 allele frequency in the mf35 and sy463 data sets as indications of mis-assembly. We confirmed overscaffolding by direct inspection of the aligned reads in 11 cases (Figure 4A). From the REAPR FCD score for each broken scaffold, we estimated heuristically an FCD cutoff value, and broke five additional scaffolds that were not highlighted in the allele frequency plots (Figure 4B). Detection of mis-assemblies using allele-frequency plots is highly dependent on the position of the mapped mutant locus and the number of pooled F2 animals. However, allele frequency plots provide evidence-based criteria to inform cutoff parameters to assess the correctness of the whole assembly. The modifications outlined above were integrated in assembly version nOt.2.0.
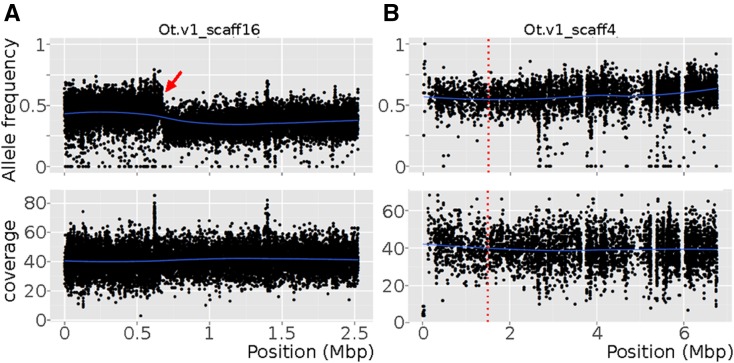
Figure 4.
Mapping-by-sequencing detects mis-assembly. Variant analysis was performed using the preliminary assembly (nOt.1.0). Plots obtained from mapping-by-sequencing data set from the cov-3(sy463) mapping population. JU170 allele frequency is plotted in upper graphs for each SNP along (A) contig 16 or (B) contig 4 and the coverage is indicated below. Blue lines are local regressions of the allele frequency or of the coverage. (A) Contig 16: ← indicates a sudden shift in JU170 allele frequencies in the scaffold. (B) Contig 4: a REAPR FCD threshold score was deduced from plots with obvious breaks and applied to all scaffolds. Overscaffolding was detected in scaffold 4 (red vertical dotted line) in the absence of any conspicuous break in SNP frequency.
A second kind of information present in JU170 allele frequencies is genetic linkage between scaffolds. In an F2 mapping population, a scaffold with a JU170 allele frequency significantly <0.5 indicates linkage to the mutation of interest. Scaffolds displaying a low mean JU170 allele frequency in the sy463 data set were also consistently low in the mf35 data set (see Figure 2B, Figure 5A, and Figure S8). We reasoned that we could infer linkage over the whole genome by analyzing similar data for several independent loci distributed over the genome. Following the same strategy as for cov-3, we generated and sequenced F2 mapping populations from a cross between JU170 and 16 additional strains from our O. tipulae mutant collection, corresponding to 14 different genetic loci (see Table S3). Using the same pipeline as for cov-3, the genes mutated in these strains were almost all identified and they will be described in a future article focusing on the evolutionary changes in vulva development between O. tipuale and C. elegans. Here, we only extracted the mean JU170 allele frequency of all scaffolds from each independent mapping data set, which does not require the causal mutations to be found. Using this approach, we sorted the genome into large chromosome-scale clusters, but which remain unordered (Figure S8).
Figure 5.
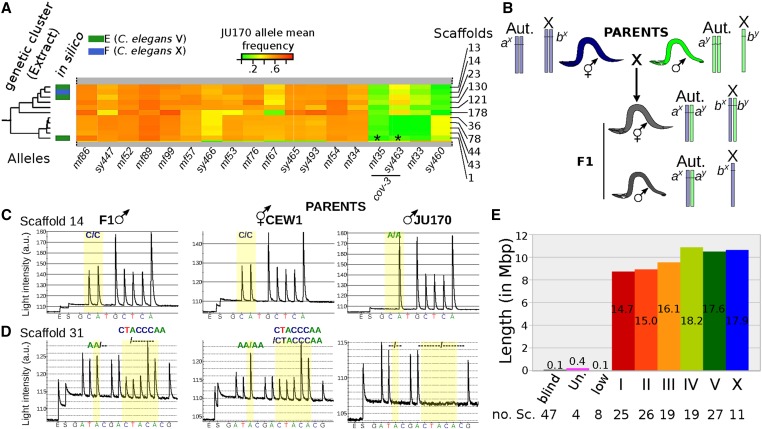
Building a chromosome-scale assembly of the O. tipulae draft assembly. (A) The O. tipulae cluster of scaffolds that correspond to the X chromosome. This cluster was selected from a global clustering based on JU170 allele mean frequency across 18 F2 mapping populations. The entire heatmap is shown in Figure S8. Each column of the heatmap corresponds to an independent F2 mapping population (Table S3) and each rectangle to the mean frequency of JU170 alleles in a scaffold. Scaffold identifiers are listed on the right side of the heatmap and the color scale is given above. If available, the prediction made from orthology-driven clusters (Figure S9) is indicated on the left side of the heat map. * indicates scaffolds where the causative cov-3 mutation has been found. (B) Method to distinguish autosome-linked vs. X-linked loci. a and b loci are on an autosome (Aut.) and the X chromosome, respectively, and are polymorphic between two strains (alleles x and y). While F1 hermaphrodites are heterozygotes for both loci, F1 males are homozygotes for the X-linked locus and bear the maternal allele. (C and D) Pyrograms of an F1 male progeny (left) from a CEW1 hermaphrodite (middle) and a JU170 male (right): genotyping was performed with polymorphic markers in scaffold 14 and 31 (assembly nOt.2.0.), which are placed in genetic clusters (C and D), respectively. Scaffold 14 is linked to X while scaffold 31 is linked to an autosome. (E) Cumulative length of each cluster of scaffolds. Numbers within bars are percent of the whole assembly, numbers below the chart count the number of scaffolds in each cluster. ♀, female; ♂, male; blind, scaffolds bearing no genomic variants between JU170 and CEW1; low, scaffolds with consistent low mean JU170 allele frequency; roman numbers, chromosomes; Un., unplaced scaffolds.
While nematode genomes evolve rapidly, retention of chromosomal linkage in the absence of close synteny has been observed in several comparisons (Stein et al. 2003; Ghedin et al. 2007; Dieterich et al. 2008). We produced a C. elegans chromosome homology profile for each of the 36 largest scaffolds (>100 kb, 79% of the assembly span) (Figure S9). For each scaffold, we observed that a majority of the genes had orthologs located on a single C. elegans chromosome. This pattern was strongest for genes mapping to C. elegans chromosomes I, II, III, IV; and weaker for chromosomes V and X. Clustering of these orthology-based profiles generated six groups (labeled A–F in Figure S8 and Figure S9) that are likely to represent the six O. tipulae chromosomes observed by microscopy (Ahn and Winter 2006). We reasoned that if these orthology-driven clusters represent real chromosomes, they should also cluster by linkage. Indeed, orthology-driven clusters A, B, C, and D were fully preserved in the linkage clusters. We thus labeled by homology the corresponding clusters as part of O. tipulae chromosomes I, II, III, and IV, respectively, because the majority of genes in these linkage groups have their C. elegans ortholog in the corresponding chromosome (Figure S9).
In contrast, two scaffolds assigned in silico to group E and one assigned to group F were not genetically linked to other scaffolds of the respective group. Consideration of their content of orthologs suggested they should instead be swapped between these two chromosomes (Figure 5A and Figure S8). It was not possible to assign a clear C. elegans chromosome homology for these two remaining genetic clusters since they each have many orthologs in both C. elegans chromosomes V and X (Figure S9). To determine which O. tipulae chromosome is the sex chromosome, we directly genotyped F1 males from a cross between the reference strain CEW1 and the polymorphic strain JU170. Markers were designed from the larger scaffolds of groups E and F and animals were genotyped by pyrosequencing. X0-males will be hemizygous for any X-linked markers, but diploid and thus heterozygous for autosomal markers that distinguish CEWI and JU170 (Figure 5, B–D) (Srinivasan et al. 2002). This strategy identified group E as representing the X chromosome, confirming previous observations of X-linkage for cov-3 (Louvet-Vallée et al. 2003) and mf33 (M.-A. Félix, unpublished data). Cel-mig-13 is on the C. elegans X chromosome. This assignment suggests that there have been substantial rearrangements involving what are now the C. elegans and O. tipulae V and X chromosomes. The genotyping also identified an additional mis-assembled scaffold (scaffold 8, Figure S10). We further annotated our preliminary chromosome assembly with the scaffold position of telomeres (File S1).
The scaffolds assigned to a chromosome corresponded to 99.5% of the genome. The remaining scaffolds were all small (Figure 5E). A group of 47 scaffolds that bear no genomic variants between JU170 and CEW1 cannot be mapped in this cross. Four scaffolds had a consistently high JU170 allele frequency across all genetic mapping data sets and are likely to be regions that happened to be genetically unlinked to all loci that were mapped in this study. Eight scaffolds had a consistently rather low mean JU170 allele frequency (20–25%) and clustered together. Their low frequency of JU170 alleles could be an artifact due to problems of read mapping in divergent, repeated, or mis-assembled regions, or may be due to transmission distortion in the cross between two wild isolates as seen in C. elegans (Seidel et al. 2008) or C. briggsae (Ross et al. 2011). For our mapping-by-sequencing approach, we flagged putative causative mutations associated with these scaffolds, but will return to them to attempt linkage attribution in the future.
Discussion
A draft assembly with a high quality provides a useful and versatile resource for a new nematode species
We assembled a rapid draft assembly for the reference CEWI strain of O. tipulae from relatively inexpensive Illumina short-read data. The assembly has good contiguity and gene content metrics, and appears to represent the O. tipulae genome well. It ranks among the better nematode genome assemblies alongside the complete C. elegans and Onchocerca volvulus assemblies and the almost complete Strongyloides ratti, C. briggsae, and Candida tropicalis assemblies, on which considerable finishing effort has been expended (C. elegans Sequencing Consortium 1998; Stein et al. 2003; Hillier et al. 2007; Koboldt et al. 2010; Ross et al. 2011; Cotton et al. 2016; Hunt et al. 2016). The high quality of the O. tipulae assembly is likely to be a result of the high homozygosity in the inbred strain sequenced (Barrière et al. 2009) and the small genome size (and concomitant reduced contribution of repeats, Table S2). The total genome length in our assembly is 40% shorter than a previous estimate from reassociation kinetics (Ahn and Winter 2006). We found no evidence for missing genetic content, or a large span of overcollapsed repeats. It is perhaps more likely that the reassociation kinetics-based estimate is in error, as was observed for A. thaliana [actual genome span of ∼135 Mb (The Arabidopsis Genome Initiative 2000), flow cytometry estimate of ∼150 Mb (Bennett et al. 2003), reassociation kinetics estimate of 70–80 Mb (Leutwiler et al. 1984)].
Regardless of the specific advantages of O. tipulae, achieving high-quality draft assemblies at reasonable cost is now feasible for a wide range of species. Advances in sequencing technologies and assembly algorithms can be combined to enable high contiguity assemblies, even from highly heterozygous organisms. For example, Fierst et al. (2015) recently produced a 131-Mb assembly of the highly polymorphic outcrossing C. remanei in only 1600 scaffolds, using a mix of short insert and mate-pair libraries, as we did here. We also found marked differences between assemblers, even those based on the same underlying algorithm. Comparisons of assembly toolkits have been made in several “Assemblathon” competitions, and from these it is clear that, while some assemblers do perform better consistently, customization of approach is key to optimal assembly (Earl et al. 2011; Bradnam et al. 2013). As the assumptions made in the coding of different assemblers may interact differently with the particular patterns of genome structure and diversity present in a target species, it may still be useful to assess several different assemblers in parallel for each new genome.
The O. tipulae assembly could still be improved. Long-read sequence data from Pacific Biosciences, Oxford Nanopore, or 10× Genomics platforms could be used to further contiguate the genome. Long-range physical mapping using the BioNano or OpGen optical-mapping platforms could be used to superscaffold the existing assembly. Both of these approaches could yield chromosomal-sized scaffolds. Traditional genetic map production from a large mapping cross could also be used to bin and order scaffolds in a linkage map, and validate the sequence- or physical-based assembly. Genotyping-by-sequencing approaches such as restriction site-associated DNA sequencing (Baird et al. 2008; Baxter et al. 2011; Fierst et al. 2015) or other reduced-representation sequencing methods would provide the density of markers required at minimal cost. Low-coverage, whole-genome skimming; for example, of recombinant inbred lines as applied in C. elegans (Li et al. 2006; Doroszuk et al. 2009; Rockman and Kruglyak 2009), C. briggsae (Hillier et al. 2007; Ross et al. 2011), and P. pacificus (Srinivasan et al. 2002); would serve the same goal. We were able to leverage the data generated from bulk-segregant identification of selected markers in a “virtuous circle” to also improve the assembly, binning scaffolds representing 99.5% of the assembly into putative chromosomal groups and will continue to do so with further mutations.
A complete assembly of the O. tipulae genome will require significant additional effort, but until that goal is achieved, we have shown that the existing assembly is a sufficient substrate for mapping of mutants and identification of genes. It is also highly informative concerning genome evolutionary dynamics in the Rhabditinae. The genome is available for browsing and download at http://ensembl.caenorhabditis.org/Oscheius_tipulae_not2/Info/Index.
Toward universal forward genetics?
For the first time, we successfully identified without a candidate-gene approach a phenotype-causing gene in O. tipulae, namely the Oti-mig-13 gene corresponding to the previously described cov-3 vulva mutants (Louvet-Vallée et al. 2003). Our pipeline is similar to those developed for model species (Schneeberger et al. 2009; Minevich et al. 2012; James et al. 2013). The fragmentation of the genome into ∼200 contigs did not impair our ability to identify causative lesions. Three of the four mutations we identified in Oti-mig-13 were large deletions, including two derived from EMS mutagenesis screens. While TMP-UV mutagenesis is chosen because of its propensity to induce deletions, mutations induced by EMS are thought to mainly comprise G/C to A/T transitions (Anderson 1995); but they can also include a significant proportion of deletions, especially after screens for strong loss-of-function mutations (Flibotte et al. 2010; C. elegans Deletion Mutant Consortium 2012).
Our results suggest that most good-quality draft genome assemblies will be sufficient to allow the identification of loci identified by forward genetics. Methods have been developed that directly identify fixed differences in raw whole-genome sequencing data without mapping to an assembly (Nordström et al. 2013). This linkage- and reference-free strategy is particularly useful in organisms, such as plants, with large and repetitive genomes, because assembling such genomes is still challenging (Schneeberger 2014). However, this approach tends to output more candidate variants, and cannot exclude candidates by linkage. This is critical, as downstream validation of candidates is time consuming and occasionally not technically possible in nonmodel species. For Nematoda, where genomes are generally small, a pipeline with a de novo reference assembly and mapping by bulk-segregant analysis is very efficient.
Many nematode species are attractive subjects for laboratory research, thanks to their small size, fast life cycle, large broods, and simplicity of culture. The fact that many species can be cryopreserved simplifies genetic approaches. Many genetically diverse isolates are available from wild sampling collections for each species for generation of mapping populations. Facultative selfing has evolved multiple independent times in the phylum, and this, as in C. elegans, significantly simplifies genetic analyses. The homozygosing effect of selfing also ensures easier genome assembly, but high-quality genomic resources can still be built for obligate outcrossers, such as C. remanei (Fierst et al. 2015). Thus, many new nematode species, selfing or outcrossing, could now be turned into genetic model systems. The real bottleneck will be the development of functional genetic tools to validate the identified candidate genes. However, the revolution brought by the CRISPR-Cas9 system for genome editing may solve this issue, as shown by the recent successful implementations of this versatile technique in various nematode species other than C. elegans, including P. pacificus (Witte et al. 2015, p. 201) and C. briggsae (Culp et al. 2015).
Evolution of development of the vulva in nematodes
We exemplify the relevance of the comparative genetic approach with our analyses of the evolution of the role of mig-13 in nematode vulva development. First, this gene would not have been investigated in a targeted reverse genetic study: phenotype-based screens ensure unexpected findings about genetic innovation. Second, although C. elegans vulva development has been studied in exquisite detail, the findings in O. tipulae allowed us to uncover a role for mig-13 in C. elegans vulval development. The precise involvement of mig-13 in vulva development remains an open question. MIG-13 in C. elegans was known to be required for the anterior migration of the QR neuroblast (Sym et al. 1999, p. 1). Lrp12, a mouse trans-membrane protein containing CUB and LDL repeats, is expressed in populations of migrating and polarized neurons during corticogenesis (Schneider et al. 2011) and can partially rescue a C. elegans mig-13 mutant (Wang et al. 2013). Although the mechanism of action of mig-13 in neuroblast migration remains elusive, it likely acts cell autonomously to polarize the actin skeleton at the leading edge of the migrating cell (Wang et al. 2013). As reported previously (Sym et al. 1999), we were unable to detect mig-13 expression in the Pn.p vulva precursor cells in C. elegans, but this could be due to a weak expression [as in QR neurons (Wang et al. 2013)]. A cell-autonomous role of MIG-13 in migration of Pn.p cells could explain the centering phenotype observed in O. tipulae, although the mechanism of its effect on competence is less clear. In C. elegans, although vulval precursor cells move (Grimbert et al. 2016), this movement could be of reduced importance for proper centering of the cell fate pattern on P6.p. This reduced role would explain why cov phenotypes are not penetrant. It is tempting to speculate that MIG-13 acts as a receptor, but no ligand or protein partner has been identified. Finally, since a mixture of competence and centering phenotypes have been reported in Wnt mutants of C. elegans (Eisenmann et al. 1998; Milloz et al. 2008; Grimbert et al. 2016), it will be important to test whether the Wnt and the mig-13 pathways interact during vulva development, and whether the Wnt pathway is disrupted in other O. tipulae cov mutants. Interestingly, mig-13 appears to be absent from the P. pacificus genome. Comparing vulva development in the three species will provide a useful framework to polarize evolutionary changes and understand the genetic basis of phenotypic change and stasis despite pervasive developmental system drift.
In conclusion, we have shown how a draft de novo genome assembly can be used to identify phenotype-causing mutations in a nonmodel species. Our method does not require physical or genetic maps of the genome. In addition to further understanding of key developmental mechanisms in nematodes, the new assembly will be a useful resource for phylum-wide nematode genome analyses. We validated our approach by successfully identifying Oti-mig-13 as the gene responsible for the vulva mutant phenotypes described in cov-3 mutants (Louvet-Vallée et al. 2003). Our results pave the way for the identification of further O. tipulae mutations. Mapping-by-sequencing provides further linkage information, creating a virtuous circle between genome assembly and mutant mapping. More broadly, this work shows that the combination of better assembly techniques and mapping-by-sequencing now makes forward genetics realistic in nonmodel species.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.203521/-/DC1.
Acknowledgments
We thank Sujai Kumar for a preliminary analysis of the Oscheius tipulae genome. We thank the staff of Edinburgh Genomics and the Centre for Genome Research for support. We thank Amhed Vargas Velazquez for computer help and for pointing out a duplication in the assembly. We thank Lewis Stevens for adding O. tipulae nOt.2.0 to http://ensembl.caenorhabditis.org. We thank two anonymous reviewers for their comments and suggestions. We acknowledge WormBase. We gratefully acknowledge support from the Pôle Scientifique de Modélisation Numérique computing center of École Normale Supérieure de Lyon. G.K. was supported by a Biotechnological and Biological Sciences Research Council Ph.D. studentship. This work was funded by grants from the Agence Nationale de la Recherche (ANR12-BSV2-0004-01 and ANR10-LABX-54 MEMOLIFE). We also acknowledge the support of the Bettencourt Schueller Foundation (Coup d’Elan 2011) and the support of European Molecular Biology Organization for a short-term fellowship awarded to F.B. (ASTF 491–2014). Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD-010440). The authors declare that they have no competing interests.
Author contributions: M.-A.F. designed the project. F.B., M.-A.F., and S.D. performed the experiments. M.B. designed the sequencing scheme and supervised the downstream analysis. G.K. carried out the assembly and genomic analysis. F.B. implemented the mapping-by-sequencing workflow, conducted assembly improvements, and the genetic linkage analysis. F.B., M.-A.F., and S.D. mapped, identified, and confirmed the mutants. F.B., M.A.F., M.B., and G.K. wrote the article. All authors read and approved the final manuscript.
Footnotes
Communicating editor: O. Hobert
Literature Cited
- Abe A., Kosugi S., Yoshida K., Natsume S., Takagi H., et al. , 2012. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30: 174–178. [DOI] [PubMed] [Google Scholar]
- Ahn I. Y., Winter C. E., 2006. The genome of Oscheius tipulae: determination of size, complexity, and structure by DNA reassociation using fluorescent dye. Genome 49: 1007–1015. [DOI] [PubMed] [Google Scholar]
- Anderson P., 1995. Mutagenesis. Methods Cell Biol. 48: 31–58. [PubMed] [Google Scholar]
- Andrews, S., 2010 FastQC A Quality Control Tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Baïlle D., Barrière A., Félix M.-A., 2008. Oscheius tipulae, a widespread hermaphroditic soil nematode, displays a higher genetic diversity and geographical structure than Caenorhabditis elegans. Mol. Ecol. 17: 1523–1534. [DOI] [PubMed] [Google Scholar]
- Baird N. A., Etter P. D., Atwood T. S., Currey M. C., Shiver A. L., et al. , 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3: e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., et al. , 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrière A., Yang S.-P., Pekarek E., Thomas C. G., Haag E. S., et al. , 2009. Detecting heterozygosity in shotgun genome assemblies: lessons from obligately outcrossing nematodes. Genome Res. 19: 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter S. W., Davey J. W., Johnston J. S., Shelton A. M., Heckel D. G., et al. , 2011. Linkage mapping and comparative genomics using next-generation RAD sequencing of a non-model organism. PLoS One 6: e19315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M. D., Leitch I. J., Price H. J., Johnston J. S., 2003. Comparisons with Caenorhabditis (∼100 Mb) and Drosophila (∼175 Mb) using flow cytometry show genome size in Arabidopsis to be ∼157 Mb and thus ∼25% larger than the Arabidopsis genome initiative estimate of ∼125 Mb. Ann. Bot. 91: 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento G., Ogawa A., Sommer R. J., 2010. Co-option of the hormone-signalling module dafachronic acid-DAF-12 in nematode evolution. Nature 466: 494–497. [DOI] [PubMed] [Google Scholar]
- Blaxter M. L., De Ley P., Garey J. R., Liu L. X., Scheldeman P., et al. , 1998. A molecular evolutionary framework for the phylum Nematoda. Nature 392: 71–75. [DOI] [PubMed] [Google Scholar]
- Boisvert S., Laviolette F., Corbeil J., 2010. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J. Comput. Biol. 17: 1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam K. R., Fass J. N., Alexandrov A., Baranay P., Bechner M., et al. , 2013. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience 2: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, D. J., 2011 Evo-devo: hidden rewiring comes to light. Nat. Rev. Genet. 12: 586. [DOI] [PubMed] [Google Scholar]
- C. elegans Deletion Mutant Consortium, 2012 Large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda) 2: 1415–1425. [DOI] [PMC free article] [PubMed]
- C elegans Sequencing Consortium, 1998 Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018 [corrigenda: Science 283: 35 (1999); Science 283: 2103 (1999); Science 285: 1493 (1999)]. [DOI] [PubMed] [Google Scholar]
- Chikhi R., Medvedev P., 2014. Informed and automated k-mer size selection for genome assembly. Bioinformatics 30: 31–37. [DOI] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. C., Egan R., Frazier P. I., Wang Z., 2013. ALE: a generic assembly likelihood evaluation framework for assessing the accuracy of genome and metagenome assemblies. Bioinformatics 29: 435–443. [DOI] [PubMed] [Google Scholar]
- CLC bio, 2014 CLC Assembly Cell. https://www.qiagenbioinformatics.com/products/clc-assembly-cell/.
- Cotton J. A., Bennuru S., Grote A., Harsha B., Tracey A., et al. , 2016. The genome of Onchocerca volvulus, agent of river blindness. Nat. Microbiol. 2: 16216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culp E., Richman C., Sharanya D., Gupta B., 2015. Genome editing in Caenorhabditis briggsae using the CRISPR/Cas9 System. bioRxiv 021121. DOI https//.org/10.1101/021121. [Google Scholar]
- De Ley P., Blaxter M. L., 2002. Systematic position and phylogeny, pp. 1–30 in The Biology of Nematodes, edited by Lee D. Taylor and Francis, London. [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtel M. L., Louvet-Vallée S., Viney M. E., Félix M. A., Sternberg P. W., 2001. Control of vulval cell division number in the nematode Oscheius/Dolichorhabditis sp. CEW1. Genetics 157: 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtel-Danjoy M.-L., Félix M.-A., 2004. The two steps of vulval induction in Oscheius tipulae CEW1 recruit common regulators including a MEK kinase. Dev. Biol. 265: 113–126. [DOI] [PubMed] [Google Scholar]
- Dieterich C., Clifton S. W., Schuster L. N., Chinwalla A., Delehaunty K., et al. , 2008. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat. Genet. 40: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroszuk A., Snoek L. B., Fradin E., Riksen J., Kammenga J., 2009. A genome-wide library of CB4856/N2 introgression lines of Caenorhabditis elegans. Nucleic Acids Res. 37: e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duveau F., Félix M.-A., 2010. Evolution of sex determination in C. elegans. hidden variation mapped. Heredity 105: 423–425. [DOI] [PubMed] [Google Scholar]
- Earl D., Bradnam K., St John J., Darling A., Lin D., et al. , 2011. Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome Res. 21: 2224–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann D. M., Maloof J. N., Simske J. S., Kenyon C., Kim S. K., 1998. The beta-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin-39 during Caenorhabditis elegans vulval development. Development 125: 3667–3680. [DOI] [PubMed] [Google Scholar]
- Ellis R. E., Lin S.-Y., 2014. The evolutionary origins and consequences of self-fertility in nematodes. F1000Prime Rep. 6: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D. M., Kelly S., 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekih R., Takagi H., Tamiru M., Abe A., Natsume S., et al. , 2013. MutMap+: genetic mapping and mutant identification without crossing in rice. PLoS One 8: e68529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix, M.-A., 2006 Oscheius tipulae (August 16, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.119.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Félix M. A., Sternberg P. W., 1997. Two nested gonadal inductions of the vulva in nematodes. Development 124: 253–259. [DOI] [PubMed] [Google Scholar]
- Félix M.-A., Delattre M., Dichtel M.-L., 2000. Comparative developmental studies using Oscheius/Dolichorhabditis sp. CEW1 (Rhabditidae). Nematology 2: 89–98. [Google Scholar]
- Félix M. A., Vierstraete A., Vanfleteren J., 2001. Three biological species closely related to Rhabditis (Oscheius) pseudodolichura Körner in Osche, 1952. J. Nematol. 33: 104–109. [PMC free article] [PubMed] [Google Scholar]
- Fierst J. L., Willis J. H., Thomas C. G., Wang W., Reynolds R. M., et al. , 2015. Reproductive mode and the evolution of genome size and structure in Caenorhabditis nematodes. PLoS Genet. 11: e1005323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flibotte S., Edgley M. L., Chaudhry I., Taylor J., Neil S. E., et al. , 2010. Whole-genome profiling of mutagenesis in Caenorhabditis elegans. Genetics 185: 431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E., Wang S., Spiro D., Caler E., Zhao Q., et al. , 2007. Draft genome of the Filarial nematode parasite brugia malayi. Science 317: 1756–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S., MacCallum I., Przybylski D., Ribeiro F. J., Burton J. N., et al. , 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. USA 108: 1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimbert S., Tietze K., Barkoulas M., Sternberg P. W., Félix M.-A., et al. , 2016. Anchor cell signaling and vulval precursor cell positioning establish a reproducible spatial context during C. elegans vulval induction. Dev. Biol. 416: 123–135. [DOI] [PubMed] [Google Scholar]
- Gupta, B. P., R. Johnsen, and N. Chen, 2007 Genomics and biology of the nematode Caenorhabditis briggsae (May 3, 2007), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.136.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Hillier L. W., Miller R. D., Baird S. E., Chinwalla A., Fulton L. A., et al. , 2007. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 5: e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier L. W., Marth G. T., Quinlan A. R., Dooling D., Fewell G., et al. , 2008. Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods 5: 183–188. [DOI] [PubMed] [Google Scholar]
- Holt C., Yandell M., 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holterman M., van der Wurff A., van den Elsen S., van Megen H., Bongers T., et al. , 2006. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Mol. Biol. Evol. 23: 1792–1800. [DOI] [PubMed] [Google Scholar]
- Hunt M., Kikuchi T., Sanders M., Newbold C., Berriman M., et al. , 2013. REAPR: a universal tool for genome assembly evaluation. Genome Biol. 14: R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt V. L., Tsai I. J., Coghlan A., Reid A. J., Holroyd N., et al. , 2016. The genomic basis of parasitism in the Strongyloides clade of nematodes. Nat. Genet. 48: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James G. V., Patel V., Nordström K. J., Klasen J. R., Salomé P. A., et al. , 2013. User guide for mapping-by-sequencing in Arabidopsis. Genome Biol. 14: R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D. R., Schatz M. C., Salzberg S. L., 2010. Quake: quality-aware detection and correction of sequencing errors. Genome Biol. 11: R116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienle S., Sommer R. J., 2013. Cryptic variation in vulva development by cis-regulatory evolution of a HAIRY-binding site. Nat. Commun. 4: 1714. [DOI] [PubMed] [Google Scholar]
- Kiontke K., Barrière A., Kolotuev I., Podbilewicz B., Sommer R., et al. , 2007. Trends, stasis, and drift in the evolution of nematode vulva development. Curr. Biol. CB 17: 1925–1937. [DOI] [PubMed] [Google Scholar]
- Koboldt D. C., Staisch J., Thillainathan B., Haines K., Baird S. E., et al. , 2010. A toolkit for rapid gene mapping in the nematode Caenorhabditis briggsae. BMC Genomics 11: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I., 2004. Gene finding in novel genomes. BMC Bioinformatics 5: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsovoulos G., Makepeace B., Tanya V. N., Blaxter M., 2014. Palaeosymbiosis revealed by genomic fossils of Wolbachia in a strongyloidean nematode. PLoS Genet. 10: e1004397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Koutsovoulos G., Kaur G., Blaxter M., 2012. Toward 959 nematode genomes. Worm 1: 42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Jones M., Koutsovoulos G., Clarke M., Blaxter M., 2013. Blobology: exploring raw genome data for contaminants, symbionts and parasites using taxon-annotated GC-coverage plots. Front. Genet. 4: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshchiner I., Alexa K., Kelsey P., Adzhubei I., Austin-Tse C. A., et al. , 2012. Mutation mapping and identification by whole-genome sequencing. Genome Res. 22: 1541–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutwiler L., Hough-Evans B., Meyerowitz E. M., 1984. The DNA of Arabidopsis thaliana. Mol. Gen. Genet. 194: 15–23. [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li D., Liu S., Ma X., Dietrich C. R., et al. , 2013. The maize glossy13 gene, cloned via BSR-Seq and Seq-walking encodes a putative ABC transporter required for the normal accumulation of epicuticular waxes. PLoS One 8: e82333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Alvarez O. A., Gutteling E. W., Tijsterman M., Fu J., et al. , 2006. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet. 2: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Yeh C.-T., Tang H. M., Nettleton D., Schnable P. S., 2012. Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS One 7: e36406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Yu L., Liu L., Li H., Li Y., 2015. Comparative transcriptomes and EVO-DEVO studies depending on next generation sequencing. Comput. Math. Methods Med. 2015: 896176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvet-Vallée S., Kolotuev I., Podbilewicz B., Félix M.-A., 2003. Control of vulval competence and centering in the nematode Oscheius sp. 1 CEW1. Genetics 163: 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R., Liu B., Xie Y., Li Z., Huang W., et al. , 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G., Kingsford C., 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27: 764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. [Google Scholar]
- Mascher M., Jost M., Kuon J.-E., Himmelbach A., Aßfalg A., et al. , 2014. Mapping-by-sequencing accelerates forward genetics in barley. Genome Biol. 15: R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore R. W., Paran I., Kesseli R. V., 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milloz J., Duveau F., Nuez I., Félix M.-A., 2008. Intraspecific evolution of the intercellular signaling network underlying a robust developmental system. Genes Dev. 22: 3064–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I., Stephen G., Bayer M., Cock P. J. A., Pritchard L., et al. , 2013. Using tablet for visual exploration of second-generation sequencing data. Brief. Bioinform. 14: 193–202. [DOI] [PubMed] [Google Scholar]
- Minevich G., Park D. S., Blankenberg D., Poole R. J., Hobert O., 2012. CloudMap: a cloud-based pipeline for analysis of mutant genome sequences. Genetics 192: 1249–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K. J. V., Albani M. C., James G. V., Gutjahr C., Hartwig B., et al. , 2013. Mutation identification by direct comparison of whole-genome sequencing data from mutant and wild-type individuals using k-mers. Nat. Biotechnol. 31: 325–330. [DOI] [PubMed] [Google Scholar]
- Ogawa A., Streit A., Antebi A., Sommer R. J., 2009. A conserved endocrine mechanism controls the formation of dauer and infective larvae in nematodes. Curr. Biol. CB 19: 67–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa A., Bento G., Bartelmes G., Dieterich C., Sommer R. J., 2011. Pristionchus pacificus daf-16 is essential for dauer formation but dispensable for mouth form dimorphism. Development 138: 1281–1284. [DOI] [PubMed] [Google Scholar]
- Opperman C. H., Bird D. M., Williamson V. M., Rokhsar D. S., Burke M., et al. , 2008. Sequence and genetic map of Meloidogyne hapla: a compact nematode genome for plant parasitism. Proc. Natl. Acad. Sci. USA 105: 14802–14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankin A., Campoli C., Dong X., Kilian B., Sharma R., et al. , 2014. Mapping-by-sequencing identifies HvPHYTOCHROME C as a candidate gene for the early maturity 5 locus modulating the circadian clock and photoperiodic flowering in barley. Genetics 198: 383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G., Bradnam K., Korf I., 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23: 1061–1067. [DOI] [PubMed] [Google Scholar]
- Photos A., Gutierrez A., Sommer R. J., 2006. sem-4/spalt and egl-17/FGF have a conserved role in sex myoblast specification and migration in P. pacificus and C. elegans. Dev. Biol. 293: 142–153. [DOI] [PubMed] [Google Scholar]
- Robinson R., 2011. Different paths, same structure: “developmental systems drift” at work. PLoS Biol. 9: e1001113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., Kruglyak L., 2009. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 5: e1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. A., Koboldt D. C., Staisch J. E., Chamberlin H. M., Gupta B. P., et al. , 2011. Caenorhabditis briggsae recombinant inbred line genotypes reveal inter-strain incompatibility and the evolution of recombination. PLoS Genet. 7: e1002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux J., Rosikiewicz M., Robinson-Rechavi M., 2015. What to compare and how: comparative transcriptomics for Evo-Devo. J. Exp. Zoolog. B Mol. Dev. Evol. 324: 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel D., Tian H., Sommer R. J., 2008. Wnt signaling in Pristionchus pacificus gonadal arm extension and the evolution of organ shape. Proc. Natl. Acad. Sci. USA 105: 10826–10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K., 2014. Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat. Rev. Genet. 15: 662–676. [DOI] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A. H., et al. , 2009. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6: 550–551. [DOI] [PubMed] [Google Scholar]
- Schneider S., Gulacsi A., Hatten M. E., 2011. Lrp12/Mig13a reveals changing patterns of preplate neuronal polarity during Corticogenesis that are absent in reeler mutant mice. Cereb. Cortex 21: 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharaman A., Cumbo P., Bojanala N., Gupta B. P., 2010. Conserved mechanism of Wnt signaling function in the specification of vulval precursor fates in C. elegans and C. briggsae. Dev. Biol. 346: 128–139. [DOI] [PubMed] [Google Scholar]
- Seidel H. S., Rockman M. V., Kruglyak L., 2008. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science 319: 589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharanya D., Thillainathan B., Marri S., Bojanala N., Taylor J., et al. , 2012. Genetic control of vulval development in Caenorhabditis briggsae. G3 (Bethesda) 2: 1625–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharanya D., Fillis C. J., Kim J., Zitnik E. M., Ward K. A., et al. , 2015. Mutations in Caenorhabditis briggsae identify new genes important for limiting the response to EGF signaling during vulval development. Evol. Dev. 17: 34–48. [DOI] [PubMed] [Google Scholar]
- Simpson J. T., Durbin R., 2010. Efficient construction of an assembly string graph using the FM-index. Bioinformatics 26: i367–i373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J. T., Wong K., Jackman S. D., Schein J. E., Jones S. J. M., et al. , 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19: 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A. F. A., R. Hubley, and P. Green, 2013–2015 RepeatMasker Open-4.0. http://www.repeatmasker.org.
- Sommer R. J., 2006. Pristionchus pacificus (August 14, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.102.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer R. J., 2012. Evolution of regulatory networks: nematode vulva induction as an example of developmental systems drift. Adv. Exp. Med. Biol. 751: 79–91. [DOI] [PubMed] [Google Scholar]
- Sommer R. J., Bumbarger D. J., 2012. Nematode model systems in evolution and development. Wiley Interdiscip. Rev. Dev. Biol. 1: 389–400. [DOI] [PubMed] [Google Scholar]
- Srinivasan J., Sinz W., Lanz C., Brand A., Nandakumar R., et al. , 2002. A bacterial artificial chromosome-based genetic linkage map of the nematode Pristionchus pacificus. Genetics 162: 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Tzvetkova A., Morgenstern B., 2006. AUGUSTUS at EGASP: using EST, protein and genomic alignments for improved gene prediction in the human genome. Genome Biol. 7: S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L. D., Bao Z., Blasiar D., Blumenthal T., Brent M. R., et al. , 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 1: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg P. W., 2005. Vulval development (June 25, 2005), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.6.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T., 2006. Maintenance of C. elegans (February 11, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.101.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M., Robinson N., Kenyon C., 1999. MIG-13 positions migrating cells along the anteroposterior body axis of C. elegans. Cell 98: 25–36. [DOI] [PubMed] [Google Scholar]
- Takagi H., Uemura A., Yaegashi H., Tamiru M., Abe A., et al. , 2013. MutMap-Gap: whole-genome resequencing of mutant F2 progeny bulk combined with de novo assembly of gap regions identifies the rice blast resistance gene Pii. New Phytol. 200: 276–283. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative , 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- True J. R., Haag E. S., 2001. Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev. 3: 109–119. [DOI] [PubMed] [Google Scholar]
- Van der Auwera G. A., Carneiro M. O., Hartl C., Poplin R., del Angel G., et al. , 2013. From fastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics 11: 11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Megen H., van den Elsen S., Holterman M., Karssen G., Mooyman P., et al. , 2009. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology 11:927–950 [Google Scholar]
- Wang X., Zhou F., Lv S., Yi P., Zhu Z., et al. , 2013. Transmembrane protein MIG-13 links the Wnt signaling and Hox genes to the cell polarity in neuronal migration. Proc. Natl. Acad. Sci. USA 110: 11175–11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte H., Moreno E., Rödelsperger C., Kim J., Kim J.-S., et al. , 2015. Gene inactivation using the CRISPR/Cas9 system in the nematode Pristionchus pacificus. Dev. Genes Evol. 225: 55–62. [DOI] [PubMed] [Google Scholar]
- Zerbino D. R., Birney E., 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin A. V., Marçais G., Puiu D., Roberts M., Salzberg S. L., et al. , 2013. The MaSuRCA genome assembler. Bioinformatics 29: 2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequencing data supporting the conclusions of this article have been submitted to the INSDC. Sequencing data of genomic DNA and RNA from reference strain CEW1 (used for assembly) are available under project accession no. PRJEB15512. Whole-genome resequencing data of the mapping strain JU170 is under project accession no. PRJEB19969. Project accession numbers corresponding to the sequencing data of F2 mapping populations of the different strains used in this study are listed in Table S3. Annotations have also been submitted to the database European Molecular Biology Laboratory–European Bioinformatics Institute under the accession no. PRJEB15512.
All scripts used for mapping-by-sequencing and gene identification are available in https://github.com/fabfabBesnard/Andalusian_Mapping.
The genome assembly and annotation is available for browsing, exploration, and download at http://ensembl.caenorhabditis.org/index.html; and will be uploaded soon to WormBase (http://www.wormbase.org/).