Abstract
Site-specific recombinases are potent tools to regulate gene expression. In particular, the Cre (cyclization recombination) and FLP (flipase) enzymes are widely used to either activate or inactivate genes in a precise spatiotemporal manner. Both recombinases work efficiently in the popular model organism Caenorhabditis elegans, but their use in this nematode is still only sporadic. To increase the utility of the FLP system in C. elegans, we have generated a series of single-copy transgenic strains that stably express an optimized version of FLP in specific tissues or by heat induction. We show that recombination efficiencies reach 100% in several cell types, such as muscles, intestine, and serotonin-producing neurons. Moreover, we demonstrate that most promoters drive recombination exclusively in the expected tissues. As examples of the potentials of the FLP lines, we describe novel tools for induced cell ablation by expression of the PEEL-1 toxin and a versatile FLP-out cassette for generation of GFP-tagged conditional knockout alleles. Together with other recombinase-based reagents created by the C. elegans community, this toolkit increases the possibilities for detailed analyses of specific biological processes at developmental stages inside intact animals.
Keywords: baf-1, DamID, FLP-out gene inactivation, genome engineering, MosSCI, PEEL-1 toxin, tissue-specific gene expression
PRECISE manipulation of gene expression is central to understand most biological processes. Strategies such as gene knockouts (KO), expression of transgenes, and RNA-mediated interference are commonly used tools, but many experiments also require spatiotemporal regulation; that is, either activation or inactivation of (trans-) genes in specific cell types and/or at particular moments during the experiment. Efficient optogenetics methods have emerged to achieve this, but they require certain technical expertise and are most suitable for analysis of a relatively low number of cells and animals (Qi et al. 2012; Churgin et al. 2014; Glock et al. 2015; Xu and Chisholm 2016). Genetic approaches based on site-specific recombinases, such as FLP (flipase) and Cre (cyclization recombination) derived from the yeast Saccharomyces cerevisiae and the bacteriophage P1, respectively, have been adapted to induce DNA excisions, inversions, or translocations in a variety of organisms, including several plants (Liu et al. 2013), mice (Anastassiadis et al. 2010), fish (Trinh le et al. 2011; Wong et al. 2011), fruit flies (Xu and Rubin 2012), and nematodes (Hubbard 2014). FLP and Cre recognize specific 34-bp recombination target (RT) sequences known as Frt (FLP recognition target) and loxP (locus of crossing over in P1), respectively. The RT sites consist of a central, nonpalindromic 8-bp sequence flanked by 13-bp inverted perfect (wild-type loxP) or imperfect (Frt) repeats. Two molecules of the recombinase bind cooperatively to an RT site and if two compatible RTs are within the vicinity of each other, recombination will occur between the central 8-bp sequences. If the two RT sites are positioned on the same DNA molecule and in the same orientation, the recombination reaction will excise the intervening sequence and leave a single RT site (Figure 1). If the RTs are oriented toward each other, the intervening sequence will be inverted, whereas reactions involving two pairs of RT sites can be used for recombinase-mediated cassette exchange. Normally, the recombination events catalyzed by FLP and Cre are reversible, but variation in Frt and loxP sequences has been explored to make reactions irreversible and to increase the control of multiple recombination events between homotypic and heterotypic recognition sites (Anastassiadis et al. 2010; Hubbard 2014).
Figure 1.
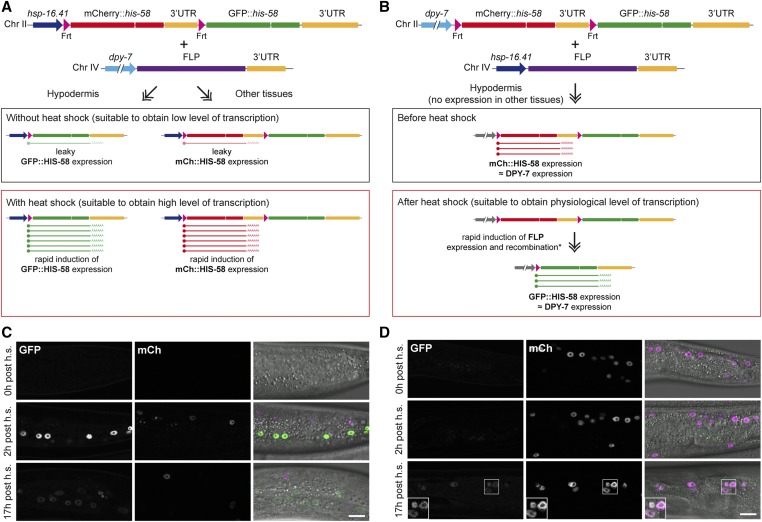
Comparison of strategies for spatiotemporal control of gene expression. (A and B) Schematic representation of dual color reporters integrated by MosSCI into chromosome II, and constructs expressing FLP recombinase inserted into chromosome IV. The dual color reporters contain a Frt-flanked mCherry::his-58 cassette with a let-858 3′UTR and a downstream GFP::his-58 cassette with a unc-54 3′UTR, whereas the glh-2 3′UTR is used in the FLP constructs. (A) Placing the FLP and Frt transgenes under control of a tissue-specific promoter (dpy-7 is used as example) and the heat-inducible hsp-16.41 promoter, respectively, enables very rapid, strong, and transient induction of a downstream transgene (GFP::his-58 in this example) in the target tissue. In other tissues, induction of the hsp-16.41 promoter leads to expression of mCherry::HIS-58. Because of the leaky expression of the hsp-16.41 promoter in the absence of induction, the system can be utilized to obtain low expression in the target tissue. (B) In the opposite configuration, in the absence of heat induction, the Frt-flanked transgene is expressed in the target tissue at levels equivalent to the activity of the tissue-specific promoter. A heat shock will lead to high expression of FLP in all tissues and efficient recombination. After recombination (i.e., after a short time delay indicated by an asterisk), the downstream transgene is under control of the tissue-specific promoter. In (A), the target tissue includes all cells derived from lineages in which the tissue-specific promoter has induced recombination; in (B), the downstream gene will only be expressed when and where the tissue-specific promoter is active. (C and D) Single confocal sections of animals carrying the transgenes described in (A and C) and (B and D). Shown are examples 0 hr (top), 2 hr (middle), and 17 hr (bottom) after heat induction. Note the rapid and transient induction of GFP::HIS-58 (green in merge; mCherry in magenta) in (C), which is in contrast to the slow induction in (D). Images were acquired with identical microscope settings; inserts in (D) represent contrast adjusted images to facilitate visualization of GFP::HIS-58. Bar, 10 µm. Chr, chromosome; h.s., heat shock; MosSCI, Mos1-mediated single-copy insertion.
There are multiple considerations for optimal design of a recombination system. FLP and Cre both work efficiently in mammalian cells, and the latter in particular has gained popularity in the design of transgenic mice, although high expression levels of Cre can be toxic to mammalian cells (see Anastassiadis et al. (2010) and Takata et al. (2011) for references). The original FLP enzyme has optimal activity at 30°, whereas recombination by Cre is highest at ∼37° (Buchholz et al. 1996). This might have favored the creation of Cre-related tools in mammalian systems, although FLP variants with increased temperature stability have been developed (Buchholz et al. 1998). A direct comparison of FLP and Cre activities suggests that the optimal recombinase for a particular reaction might differ between recombination strategies (Takata et al. 2011). The combined use of Frt and loxP sites in allele construction can provide additional layers of control of gene expression (Anastassiadis et al. 2010).
Whereas, in most organisms, the use of recombinases to control gene expression involves stable lines where FLP or Cre expression constructs are inserted into the genome and crossed to lines carrying Frt or loxP-containing transgenes, experiments in Caenorhabditis elegans have generally been performed with expression of recombinases from extrachromosomal arrays or injected mRNA (Hubbard 2014). Extrachromosomal arrays are easy to generate in C. elegans and their variable stability during mitotic divisions can be explored through analyses of mosaic animals. Nevertheless, for many experiments, it is desirable that recombination occurs in most or all cells of a particular tissue and in a reproducible manner between individual animals in a population. The Hubbard and Jorgensen laboratories were the first to report the utility of the FLP/Frt system in C. elegans (Davis et al. 2008; Voutev and Hubbard 2008). Both studies found that FLP expressed from extrachromosomal arrays was able to excise (“flip out”) transgenes that were also present in multiple copies. For concatenated multi-copy Frt reporter constructs different outcomes can be expected, ranging from no excision to partial excision (i.e., some but not all Frt-flanked cassettes are flipped out from the multi-copy transgene) and complete excision. Generally, partial excision was observed in the aforementioned studies although some cells underwent complete excision (Davis et al. 2008; Voutev and Hubbard 2008). Voutev and Hubbard also quantified FLP activity in a single cell by expressing FLP from integrated constructs carrying the hsp-16.2 heat shock promoter and observed recombination of an extrachromosomal reporter in the dorsal cell of gonadal sheath pair 1 in 11–20% of the animals. Possibly, the complexity of the multi-copy reporter was partly responsible for this relatively low efficiency. Importantly, expression of FLP was reported not to affect viability, fecundity, or behavior (Voutev and Hubbard 2008).
Direct comparison of FLP and Cre in the same cell types and on identical reporters is still limited in C. elegans. Intersectional restriction of gene expression involves two different cell type-specific promoters to drive expression of the recombinase and its target construct: the gene downstream of the RT cassette is expressed only in tissues where both promoters are active. A recent study tested the FLP and Cre systems to achieve intersectional restriction of expression and concluded that it depends on the promoter combinations which recombinase works best (Schmitt et al. 2012). Remarkably, highly restricted expression in one or two neurons with either FLP/Frt or Cre/loxP can be achieved in close to 100% of tested animals, although extensive optimization might be required (Macosko et al. 2009; Ezcurra et al. 2011; Schmitt et al. 2012). Insertion of Frt or loxP sites into endogenous loci has also proven to be feasible for either complete gene KO (Vazquez-Manrique et al. 2010) or conditional deletion of a genomic ∼1-kb fragment (Lo et al. 2013). In addition, Cre has successfully been implemented for removal of selection markers after clustered regularly interspaced short palindromic repeats (CRISPR)-mediated genome engineering (Dickinson and Goldstein 2016; Schwartz and Jorgensen 2016), as well as for control of loxP-flanked, integrated single-copy transgenes (Flavell et al. 2013; Ruijtenberg and van den Heuvel 2015).
To facilitate the use of the FLP/Frt recombination system to manipulate gene expression in a highly efficient, versatile, and reproducible manner, we decided to generate a collection of stable FLP lines. We provide a characterization of 13 integrated, single-copy FLP lines that cover all major tissue types in C. elegans and report their efficiencies at single-cell resolution. We demonstrate that the system offers tight spatiotemporal control of expression of a fluorescent reporter protein, and as further examples of their utility we developed tools for conditional cell ablation and KO of CRISPR-engineered GFP-tagged alleles. Importantly, the tools presented here are compatible with other reagents created by the C. elegans community, and together they increase the potentials of the gene expression toolbox.
Materials and Methods
Plasmid construction
We initially PCR-amplified a FLP::3′UTRglh-2 sequence from the plasmid pWD79 (Davis et al. 2008) with primers B441 and B551 (Supplemental Material, Table S1) and inserted it into KpnI/XmaI of plasmid pBN9 designed for Mos1-mediated single-copy insertion (MosSCI) into chromosome IV (Morales-Martinez et al. 2015). This FLP version, which we here name FLP pWD79, encodes a glycine residue at position 5 and contains a single artificial intron (see File S1 for complete sequences of FLP versions used in this study). Next, we inserted a 983-bp PstI/KpnI Pmyo-2 fragment from pBN41 (Morales-Martinez et al. 2015) upstream of the FLP pWD79 sequence to generate plasmid pBN155 Pmyo-2::FLP pWD79. Plasmids pBN158 Pmyo-3::FLP pWD79, and pBN159 Punc-119::FLP pWD79 were made by substituting the PstI/KpnI Pmyo-2 sequence of pBN155 by the corresponding promoter sequences amplified from Bristol N2 genomic DNA [or plasmid pUP1 (Rodenas et al. 2012) for unc-119] using a KAPA HiFi DNA polymerase (KAPA Biosystems, Wilmington, MA). Throughout this study, we used conventional restriction enzyme-mediated cloning and Gibson assembly technology. By introducing relatively few modifications, our plasmids can be made Gateway compatible, but others have observed that the extra sequences inherent to Gateway recombination sites might decrease recombination efficiencies (Schmitt et al. 2012). The primers used to clone promoters in this study as well as promoter lengths and sequence verification are described in Table S2 in the Supplemental Material.
To increase FLP expression levels we used the C. elegans codon adapter tool (Redemann et al. 2011) with the parameters “Optimize for weak mRNA structure at ribosome binding site” and “Avoid splice sites in coding region” resulting in a codon adaptation index of 0.9808. The sequence, which we termed FLP G5, was synthesized by Eurofins Genomics (Ebersberg, Germany). The initial design contained three artificial introns, but due to cloning problems the final sequence contains two introns as well as the glh-2 3′UTR. The FLP G5::3′UTRglh-2 sequence is flanked by KpnI and XmaI sites and was used to substitute the KpnI/XmaI fragment of pBN155 to create plasmid pBN168 Pmyo-2::FLP G5. Derivatives of pBN168 were made by substituting the myo-2 promoter by other promoters as either PstI/KpnI or NotI/KpnI fragments from PCR amplicons or plasmids to generate pBN172 Pmyo-3::FLP G5, pBN173 Punc-119::FLP G5, pBN176 Pnhr-82::FLP G5, and pBN219 Pdpy-7::FLP G5.
Substitution of glycine for aspartic acid at position 5 to generate FLP D5 was made by PCR amplification using FLP G5 as the template and primers B572 and B826. The forward primer introduced two nucleotide changes in the fifth FLP codon; the resulting fragment was digested with KpnI and SalI and used to replace the KpnI/SalI fragment of pBN176 to generate pBN237 Pnhr-82::FLP D5. Derivatives of pBN237 were made by substituting the nhr-82 promoter with other promoters by either conventional cloning of NotI/KpnI fragments (from PCR amplicons or subcloning) or Gibson assembly to generate pBN258 Punc-119::FLP D5, pBN260 Pmyo-3::FLP D5, pBN262 Phsp-16.41::FLP D5, pBN263 Pmyo-2::FLP D5, pBN266 Pdpy-7::FLP D5, pBN267 Prgef-1::FLP D5, pBN278 Pmec-7::FLP D5, pBN279 Phlh-8::FLP D5, pBN280 Ptph-1::FLP D5, pBN282 Pelt-2::FLP D5, pBN283 Pdat-1::FLP D5, pBN303 Punc-47::FLP D5, and pBN310 Plag-2::FLP D5. Sequence verification revealed that pBN282 carries a mutation that introduces an amino acid substitution at position 6 of FLP D5 (I6L), which does not prevent enzymatic activity.
Finally, FLAG-NLS-FLP D5 was made by Gibson assembly using a 222-nucleotide gBlock (Integrated DNA Technologies, Leuven, Belgium) encoding 3xFLAG and a nuclear localization sequence (NLS) from EGL-13, a PCR fragment encoding FLP D5 (primers B827 + B828), a PCR fragment containing the tbb-2 3′UTR (primers B829 + B830), and pBN176 digested with KpnI and XmaI to generate pBN238 Pnhr-82::FLAG-NLS-FLP D5. These modifications were inspired by results with the Cre recombinase (Ruijtenberg and van den Heuvel 2015). Derivatives of pBN238 were made by substituting the nhr-82 promoter by other promoters by either conventional cloning of NotI/KpnI fragments from plasmids described above or Gibson assembly to generate pBN252 Pdat-1::FLAG-NLS-FLP D5, pBN254 Punc-47::FLAG-NLS-FLP D5, pBN261 Phsp-16.41::FLAG-NLS-FLP D5, pBN268 Prgef-1::FLAG-NLS-FLP D5, pBN269 Punc-119::FLAG-NLS-FLP D5, and pBN270 Pmyo-3::FLAG-NLS-FLP D5.
Plasmid pBN338 Pdat-1::FLP D5::SL2::mNG was designed to provide visualization of FLP D5 expression. First, an intergenic gpd-2/-3 fragment and a mNeonGreen sequence were amplified with primers B1012 + B1013 and B1014 + B1015 using pMLS268 (Schwartz and Jorgensen 2016) and dg357 (Hostettler et al. 2017) as templates, respectively. Next, the two products were stitched by PCR with primers B1012 and B1015 and inserted by Gibson assembly into MluI of pBN283 followed by sequence verification.
Whereas the FLP expression plasmids described here all are designed for MosSCI integration into chromosome IV, most promoter::FLP::3′UTR cassettes (including cassettes with dat-1, myo-2, nhr-82, rgef-1, unc-47, or unc-119 promoters) can be excised by digestion with NotI and XmaI and inserted into NotI/NgoMIV of pBN8 (Rodenas et al. 2012). Plasmid pBN8 was originally designed for insertion into chromosome II by MosSCI, but can be used for insertion into any of the universal MosSCI sites generated on chromosomes I–V (see http://www.wormbuilder.org/ and (Frokjaer-Jensen et al. 2014).
The heat inducible, dual color reporter plasmid pBN154 Phsp-16.41 > mCherry::his-58::3′UTRlet-858 > gfp::his-58::3′UTRunc-54 (Frt recombination sites are indicated with “>” symbols) was generated through multiple steps. (1) A > mCherry::3′UTRlet-858 > gfp cassette was amplified from pWD178 (Davis et al. 2008) with primers B445 and B446 and inserted into the cloning vector pSPARK I (Canvax Biotech, Cordoba, Spain) to produce plasmid #1151. (2) Plasmid #1151 was digested with NotI and religated to produce plasmid #1231. (3) The > mCherry::3′UTRlet-858 > gfp cassette was purified from plasmid #1231 by digestion with SalI and XmaI and inserted into XhoI/NgoMIV of pBN16 (Rodenas et al. 2012) to generate plasmid pBN152. (4) A his-58-containing XhoI/BamHI fragment from plasmid pJH4.52 (Strome et al. 2001) was inserted into XhoI/BglII of pBN152 to produce pBN153. (5) Two overlapping PCR fragments containing Phsp-16.41 > mCherry 5′half (amplified with primers B211 and B440 using pBN153 as template) and mCherry 3′half::his-58 [amplified with primers B220 and B355 using pBN1 (Rodenas et al. 2012) as template] were PCR-stitched with primers B211 and B355 and inserted into pSPARK I. (6) Finally, a SphI/StuI fragment encoding Phsp-16.41 > mCherry::his-58 from this plasmid was inserted into SphI/NaeI of pBN153 to generate pBN154. All inserts were sequence verified. The hypodermal dual color reporter plasmid pBN368 Pdpy-7 > mCherry::his-58::3′UTRlet-858 > gfp::his-58::3′UTRunc-54 was generated in two steps. (1) The > mCherry::his-58::3′UTRlet-858 > gfp::his-58::3′UTRunc-54 sequence was amplified from pBN154 with primers B1064 and B1065, and inserted by Gibson assembly into pBN266 cut with KpnI and XmaI to generate pBN360. (2) A 2.5-kb SphI/XhoI fragment from pBN360 was inserted into SphI/XhoI of pBN154 to produce pBN368.
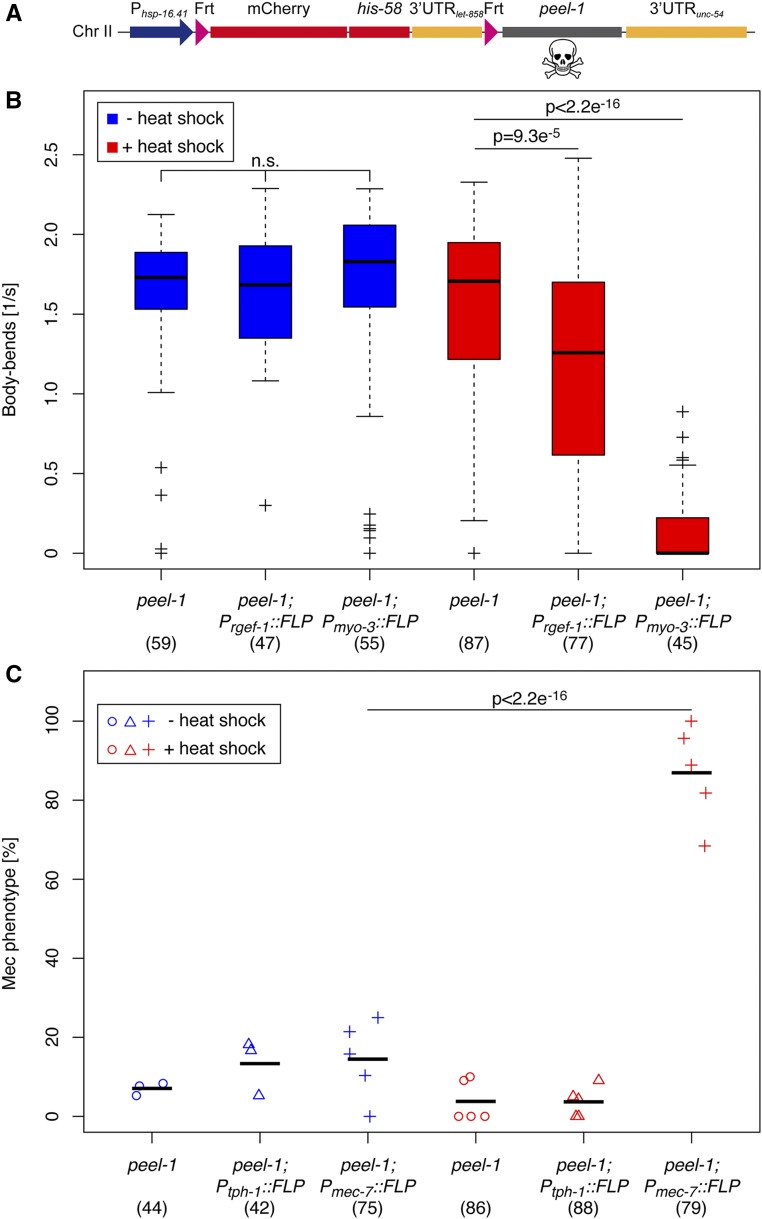
Plasmid pBN189 Phsp-16.41 > mCherry::his-58::3′UTRlet-858 > Dam::mel-28::3′UTRunc-54 for FLP-induced expression of Dam::MEL-28 was generated by Gibson assembly using two overlapping PCR fragments containing Phsp-16.41 > mCherry::his-58::3′UTRlet-858> (amplified with primers B662 and B676 using pBN154 as template) and Dam [amplified with primers B674 and B675 using pBN69 Phsp-16.41::Dam::mel-28::3′UTRunc-54 (Sharma et al. 2014) as template], together with pBN69 digested with NotI and NgoMIV. Next, pBN189 was digested with NgoMIV and NheI to substitute the mel-28 sequence by a NgoMIV/NheI fragment from pBN79 Phsp-16.41::Dam::emr-1::3′UTRunc-54 (Gonzalez-Aguilera et al. 2014) to produce pBN209 Phsp-16.41 > mCherry::his-58::3′UTRlet-858 > Dam::emr-1::3′UTRunc-54. Finally, pBN209 was digested with XhoI and NheI to substitute the Dam::emr-1 sequence by a XhoI/NheI peel-1 cDNA fragment amplified by PCR with primers B871 and B872, and pMA122 (Frokjaer-Jensen et al. 2012) as template, producing plasmid pBN255 Phsp-16.41 > mCherry::his-58::3′UTRlet-858 > peel-1::3′UTRunc-54. All inserts were sequence verified.
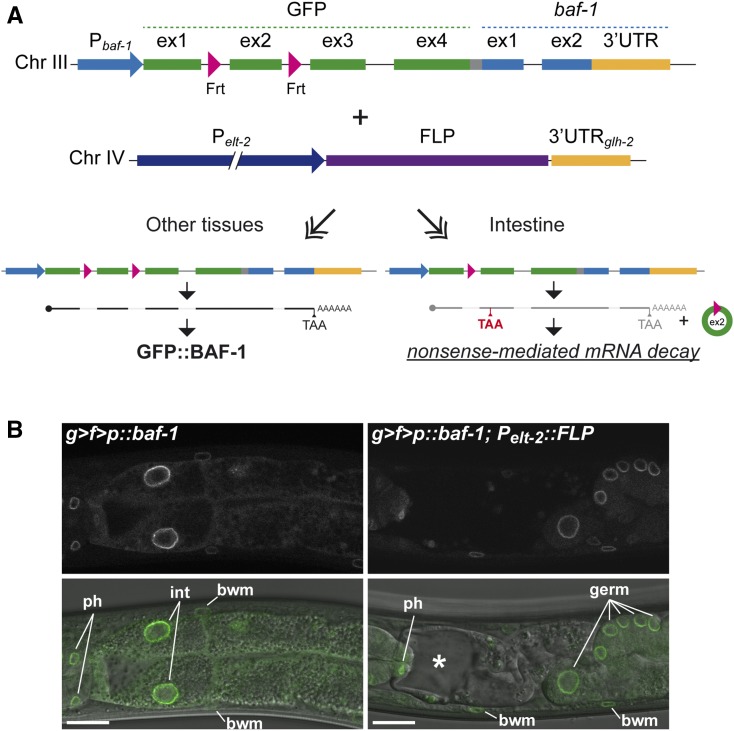
Plasmid pBN312 GFP KO for FLP-mediated gene KO was generated by insertion of two synthetic DNA fragments Integrated DNA Technologies (IDT) into NcoI+XbaI of pMLS252 (a gift from Erik Jorgensen; Addgene plasmid # 73720). pBN312 contains 34-bp Frt sequences in the first and second intron of GFP as well as an unc-119-rescuing fragment flanked by loxP sites in the third GFP intron. pBN312 can be used as “Tag + Marker Donor Plasmid” in the recent SapTrap protocol for CRISPR/Cas9 genome engineering (Schwartz and Jorgensen 2016). The targeting vector pBN313 for insertion of the GFP KO cassette into the baf-1 locus was made by SapTrap cloning with destination plasmid pMLS256, N-tagging connector plasmid pMLS288, and tag + donor plasmid pBN312, as well as the annealed primer pairs 5′HA (B963–B964), 3′HA (B965–B966), and sg (B961–B962). All inserts were sequence verified.
Plasmid pBN306 Pmyo-2::GFP::3′UTRunc-54 was generated by subcloning a PstI/SpeI fragment from pBN41 into PstI/SpeI of pBN9.
Strains and transgenesis
Strains were maintained on standard NGM plates at 16° (Stiernagle 2006). Except where noted, all transgenic strains were made by MosSCI with unc-119(+) recombination plasmids together with pCFJ601 (transposase) and three red (pCFJ90, pCFJ104, and pBN1) or green (pBN40, pBN41, and pBN42) co-injection markers (Frokjaer-Jensen et al. 2012; Dobrzynska et al. 2016). Plasmids were microinjected into the gonads of uncoordinated adults and successful integrants were identified after 7–14 days based on wild-type locomotion and absence of fluorescent markers. FLP transgenes were inserted into the cxTi10882 locus in chromosome IV of strain EG5003 unc-119(ed3) III or EG6700 unc-119(ed3) III. Frt constructs were inserted into the ttTi5605 locus in chromosome II of strain EG4322 unc-119(ed9) III. Strains BN225 (Morales-Martinez et al. 2015), BN243 (Gonzalez-Aguilera et al. 2014), and BN578, carrying fluorescent markers inserted into the cxTi10882 and ttTi5605 loci, were used to facilitate crosses. Strains OH7193 (Flames and Hobert 2009) and XE1375 (Firnhaber and Hammarlund 2013) were used to mark dopaminergic and GABA (γ-aminobutyric acid)-ergic neurons, respectively.
Strain BN552, which carries a GFP KO cassette and an unc-119-rescuing gene inserted into the baf-1 locus, was generated by injection of HT1593 unc-119(ed3) with plasmids pBN313 (65 ng/µl), #1286 Peft-3::cas9-SV40-NLS (25 ng/µl; Friedland et al. 2013), pBN1 (10 ng/µl), pCFJ90 (2.5 ng/µl), and pCFJ104 (5 ng/µl), and selecting for nonred, wild-type moving animals. Next, the unc-119-rescuing gene in the third intron of GFP in BN552 was removed to generate BN565. This was achieved by injection with Cre plasmid pMLS328 (50 ng/µl; Schwartz and Jorgensen 2016), pBN1 (10 ng/µl), pCFJ90 (2.5 ng/µl), pCFJ104 (5 ng/µl), and an irrelevant plasmid to reach a total DNA concentration of 100 ng/µl, followed by selection for nonred, Unc animals. Finally, BN565 was crossed with wild-type N2 to produce BN580, which is unc-119(+), and with BN508 Pelt-2::FLP D5 to produce BN582, in which baf-1 expression is disrupted specifically in intestinal cells.
Microscopy
Evaluation of FLP activity on the dual color reporter was assayed by heat induction at 34° for 15 min followed by recovery at 20° for ∼3 hr, unless indicated otherwise. In experiments with two heat shocks, these were performed 24-hr apart. For confocal microscopy, nematodes were mounted in 3 μl 10 mM levamisole HCl (Sigma [Sigma Chemical], St. Louis, MO) on 2% agarose pads (Askjaer et al. 2014) and observed on a Nikon Eclipse Ti microscope equipped with Plan Fluor 40×/1.3 and Plan Apo VC 60×/1.4 objectives and a A1R scanner (Nikon, Garden City, NY) using a pinhole of 1.2–1.4 airy unit. Quantification of GFP-positive nuclei was done by direct observation at the microscope or, in most cases, after acquisition of z-stack images with integrated Nikon NIS software. Image stacks were opened in Fiji/ImageJ 2.0.0-rc-43 (Schindelin et al. 2012), which was also used for postacquisition processing, such as cropping, merging, and maximum projection.
peel-1-induced toxicity
For strains with FLP D5 expressed in body wall muscles (Pmyo-3) or broadly in neurons (Prgef-1), plates with 20 synchronized L3–L4 larvae raised at 16° were heat shocked at 34° for 1 hr followed by recovery at 20°. After 24 hr, 20 animals were transferred to 0.5 ml M9 in 24-well plates and videos were immediately acquired at 20 frames per second on an Olympus SZX16 stereoscope equipped with a PLAPO 1× lens and an Olympus DP73 camera. Unbiased quantification of locomotion (expressed as body bends per second) was performed using the wrMTrck plugin for ImageJ/Fiji (Nussbaum-Krammer et al. 2015). For strains with FLP D5 expressed in mechanosensory neurons (Pmec-7) or serotonin-producing neurons (Ptph-1), synchronized L4 larvae were raised at 16° and heat shocked at 33° for 1 hr followed by recovery at 20°. Mechanosensation was evaluated after 24 hr by gently touching animals laterally on their head 1–3 times with an eyelash mounted on a toothpick. Animals that did not respond immediately were scored as Mec (Mechanosensation defective). Rate of egg laying was determined during a 3-hr time window 24 hr after the heat shock.
Data availability
See Table S1, Table S2, and Table S3 for complete lists of primers, promoters, and strains, respectively. At least one strain for each FLP D5 line will be made immediately available through the Caenorhabditis Genetics Center; others can be requested from the authors. Most plasmids produced in this study and their sequences are available at Addgene (www.addgene.org); the remaining plasmids can be requested from the authors. File S6 contains legends for File S2 – S5.
Results and Discussion
Evaluation of FLP variants
Previous reports on FLP activity in C. elegans were mainly based on expression from extrachromosomal arrays containing typically hundreds of copies of the recombinase. To achieve completely stable meiotic and mitotic inheritance of the FLP transgene, we decided to optimize FLP activity from single-copy integrations into known sites of the C. elegans genome (MosSCI; Frokjaer-Jensen et al. 2012). We first designed a MosSCI dual color reporter integrated on chromosome II to enable direct visual scoring. The reporter contains the hsp-16.41 promoter, which, in the absence of FLP activity, drives widespread and strong expression of mCherry (mCh) fused to the histone HIS-58 upon a short heat shock (Figure 1A; see also Figure 3). The mCh::his-58 fusion gene is also regulated by the let-858 3′UTR, which serves as transcriptional terminator. The mCh::his-58::let-858 3′UTR cassette is flanked by two Frt sequences, which trigger excision of the cassette in the presence of the FLP enzyme. Recombination between the two Frt sequences juxtaposes the hsp-16.41 promoter with a GFP::his-58::unc-54 3′UTR fusion gene, which upon heat shock produces bright green nuclei. Thanks to the transparency of C. elegans, the number of green nuclei can be counted in live, anesthetized animals on a compound microscope, providing a measure on the efficiency of FLP activity. We never observed GFP::HIS-58 expression in the absence of FLP, which indicates that the let-858 3′UTR sequence efficiently prevents transcriptional readthrough (Davis et al. 2008; Voutev and Hubbard 2008). However, one report based on a multi-copy extrachromosomal array has suggested that a gene placed downstream of the let-858 3′UTR might be expressed at low levels (Ruaud et al. 2011). Thus, depending on the particular experiment, it is advisable to investigate potential effects caused by the downstream gene in the absence of recombination.
Figure 3.
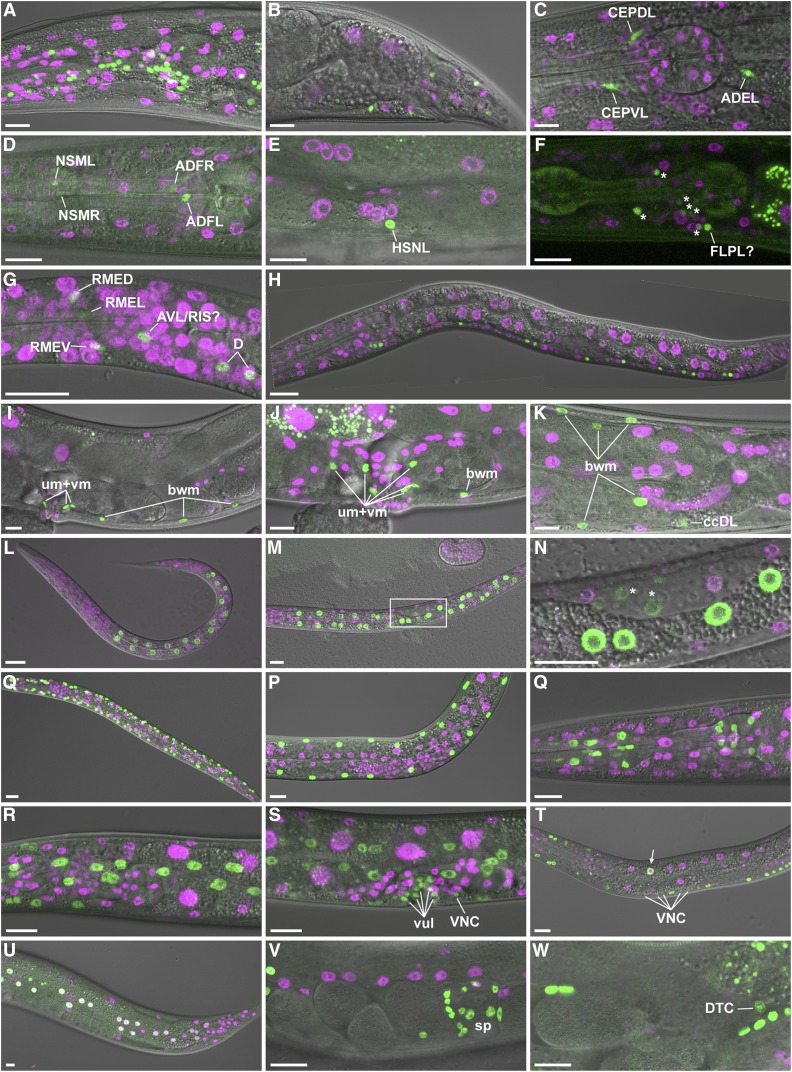
Tissue-specific FLPase activity. Transgenic animals expressing FLP D5 in specific cell types and the dual color reporter were observed by live confocal microscopy. (A and B) Promoter of rgef-1 (Prgef-1); projection of eight confocal sections [(A) adult head lateral view] or single confocal section [(B) adult tail lateral view]. (C) Pdat-1; projection of four confocal sections of head of L4 larva. Note that the green channel represents coexpressed GFP::HIS-58 and mNeonGreen in this strain (see Figure S3). (D and E) Ptph-1; projection of three confocal sections [(D); adult head lateral view] or a single confocal section [(E); adult vulva lateral view]. (F) Pmec-7; projection of seven confocal sections of head of L4 larva. To ease visualization of the weak GFP signal, the corresponding DIC image was not included in the merge. Asterisks indicate six unidentified neurons, whereas a seventh neuron might be FLPL. (G and H) Punc-47; late L1; projection of four confocal sections [(G); head lateral view] or stitch of single confocal sections (H). (I–K) Phlh-8; single confocal section [(I); adult vulva lateral view] or max projections [(J); vulva region, 8 confocal sections and (K); posterior gonad loop region; 15 confocal sections]. Examples of body wall muscle (bwm), coelomocyte (cc), uterine muscle (um), and vulval muscle (vm) cells are indicated. (L–N) Pelt-2; L1 [(L); projection of seven confocal sections] and L2 larvae [(M); projection of seven confocal sections, (N); single confocal section corresponding to boxed area in (M); note weak GFP expression in the gonad primordium indicated by *]. (O and P) Pmyo-3; projection of seven confocal sections of L2 (O) and L4 (P) larvae. (Q) Pmyo-2; projection of 12 confocal sections of adult head. (R–T) Pdpy-7; projection of 8 [(R); head lateral view] or 10 [(S); vulva lateral view] confocal sections of L4 larva or single confocal section of L3 larva (T). Examples of vulva cells (vul) and ventral nerve cord neurons (VNC) are indicated. Arrow points to intestinal cell that expresses both markers. (U) Pnhr-82; projection of four confocal sections of tail of young adult. Note that white nuclei indicate simultaneous expression of mCh::HIS-58 and GFP::HIS-58. (V and W). Plag-2; projection of six confocal sections (V) or single confocal section (W) of central region of young adult. Spermatheca (sp) and distal tip cell (DTC) are indicated. All micrographs are oriented with anterior to the left and ventral down. Bar, 10 µm.
Recombination efficiency can be expressed either as the percentage of animals expressing the downstream gene in at least a single cell or as the frequency of nuclei in which recombination has occurred. The former is the most feasible measurement for tissues with a high number of nuclei, whereas the latter is a more precise predictor of how many cells of a particular cell type will be affected in a given experiment (Table 1). Importantly, it should be noted that we might falsely score nuclei as GFP negative if the induction of the hsp-16.41 promoter is weak, particularly if these nuclei are positioned deeper into the animal relative to the microscope objective. Thus, FLP activity might be underestimated for some tissues.
Table 1. Efficiency of FLP-mediated recombination.
| Promoter | Cell Typea | Stageb | Observedc | Expected | Efficiency (%) |
|---|---|---|---|---|---|
| dat-1 | Dopaminergic neurons | L2-adult | 8.0 ± 0.0 (15) | 8 | 100 |
| dpy-7 | Hypodermal (vulval cells) | L2 | 6.0 ± 0.0 (3) | 6d | 100 |
| L3 | 12.0 ± 0.0 (5) | 12d | 100 | ||
| L4 | 22.0 ± 0.0 (2) | 22d | 100 | ||
| elt-2 | Intestine | L1 | 20.2 ± 1.0 (14) | 20 (19–22) | 101 |
| L2 | 31.3 ± 1.1 (9) | 32 (30–34) | 98 | ||
| hlh-8 | M lineage | Adult | 19.2 ± 4.9 (18) | 32 | 60 |
| hsp-16.41 | Ubiquitous | L4–adult | Overlap with mCh::HIS-positive nuclei (20) | 92e | |
| lag-2 | Multiple (distal tip cell) | L3–adult | 1.0 ± 0.0 (8) | 1f | 100 |
| mec-7 | Mechanosensory neurons | L4–adult | > 12 (8) | 12 | NAg |
| myo-2 | Pharyngeal muscle | Adult | 33.5 ± 1.6 (14) | 37 | 91 |
| myo-3 | Body wall muscle | L1–L2 | 80.4 ± 1.3 (9) | 81 | 99 |
| nhr-82 | Seam cell lineage | Adult | 86.2 ± 22.2 (25) | 130 | 66 |
| rgef-1 | Neurons (dopaminergic and GABAergic) | L2–L4 | Overlap with unc-47 and dat-1 markers (15) | 99h | |
| tph-1 | Serotonin-producing neurons | Adult | 6.0 ± 0.0 (14) | 6 | 100 |
| unc-47 | GABAergic motor neurons | L2-adult | 19.2 ± 4.3 (27) | 26 | 74 |
Quantification of FLP D5 activity in different tissues using the dual color reporter (see Figure 1). For all strains, we observed multiple recombination events in all animals. mCh, mCherry; NA, not applicable; GABA, γ-aminobutyric acid.
Cell types where the promoter is active; cell types used for quantification of recombination efficiency are indicated in brackets.
Developmental stage that was used for determination of recombination efficiency; recombination was also observed in other stages.
Average number of GFP-positive nuclei ± SD; number in brackets: n.
Efficiencies were calculated based on recombination in central Pn.p cells and their descendants: recombination was also observed throughout the hypodermis.
Experiments performed with NLS-FLAG-FLP D5; recombination efficiency was calculated as percentage of nuclei coexpressing mCh::HIS-58 and GFP::HIS-58 after two rounds of heat induction (see Figure 4).
Efficiency was calculated based on recombination in the distal tip cell most proximal to the microscope objective; recombination was also observed in other cell types.
Not applicable; we did not attempt to identify and evaluate the 12 expected nuclei because expression levels were low and because we observed GFP in unexpected neurons.
Efficiency was calculated based on recombination in the dopaminergic neurons in the head and GABAergic motor neurons in the ventral nerve cord; recombination was also observed in other types of neurons.
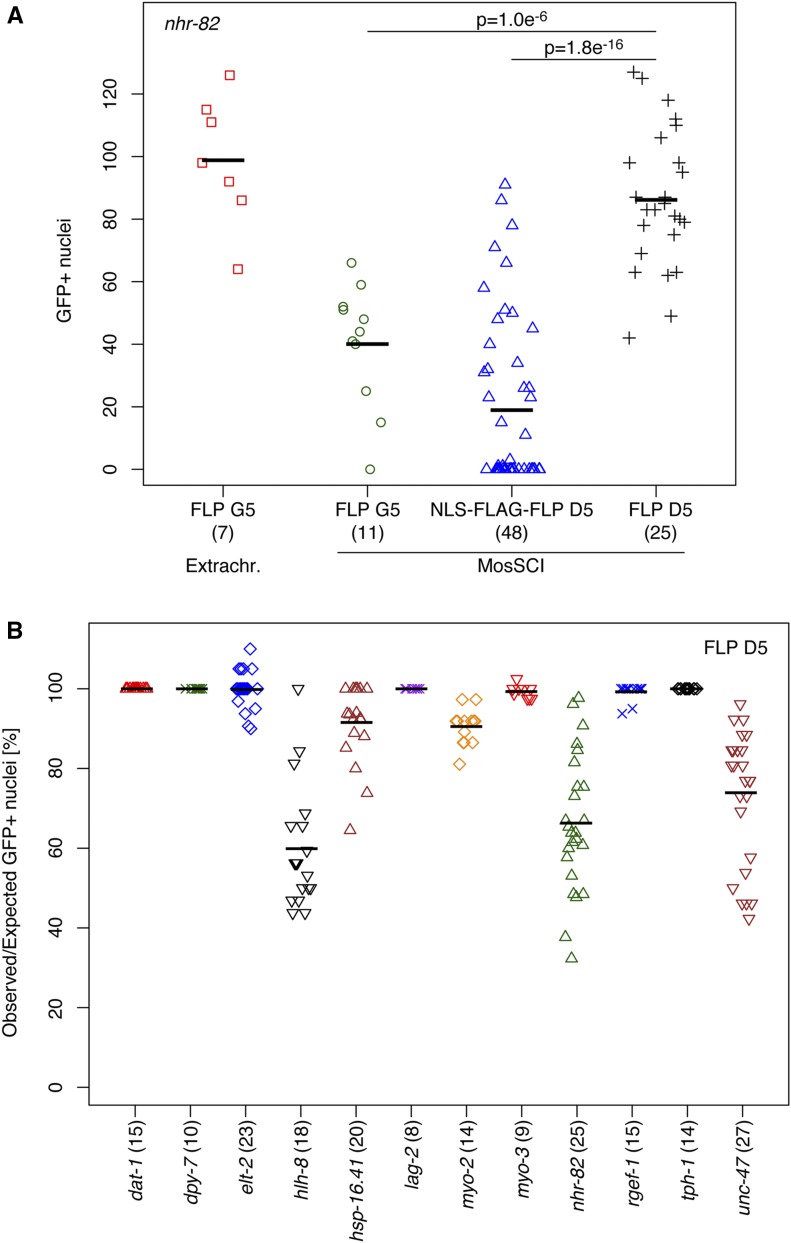
We assumed that MosSCI FLP transgenes would be expressed at lower levels than transgenes on multicopy arrays, so we engineered a novel FLP-encoding sequence with optimized codon usage for C. elegans and artificial introns (Redemann et al. 2011; see Materials and Methods). Using the nhr-82 promoter and the glh-2 3′UTR to control expression of FLP containing a glycine residue at position 5 (FLP G5; Davis et al. 2008), we first compared the activities of extrachromosomal and MosSCI lines on the dual color reporter. nhr-82 is expressed in the seam cell lineage, which gives rise to a total of 130 seam and hypodermal nuclei in adults (Miyabayashi et al. 1999). We found that animals expressing FLP G5 from an extrachromosomal array had 99 ± 21 GFP-positive nuclei (average ± SD; n = 7), whereas animals expressing the same FLP version from an integrated single copy had 40 ± 20 GFP-positive nuclei (n = 11; Figure 2A).
Figure 2.
Optimization and quantification of FLP activity. FLP activity was evaluated based on the number of nuclei expressing GFP::HIS-58 in animals carrying the integrated dual color reporter described in Figure 1. (A) Comparison of strains expressing FLP from an extrachromosomal array (FLP G5; red squares) or from integrated single-copy transgenes (MosSCI): FLP G5 (green circles), NLS-FLAG-FLP D5 (blue triangles), or FLP D5 (black crosses). The same nhr-82 promoter sequence was used in all strains. Shown are absolute numbers of GFP-positive nuclei; numbers in brackets indicate the number of adults evaluated for each strain; black horizontal lines represent the means. P values were obtained through unpaired, two-sided t-tests, indicating that FLP D5 has the highest activity among the MosSCI strains. (B) Comparison of MosSCI strains expressing FLP D5 under control of different cell type-specific or inducible promoters: dopaminergic neurons (dat-1; red triangles), hypodermis (dpy-7; green crosses), intestine (elt-2; blue diamonds), M lineage (hlh-8; black triangles), heat inducible (hsp-16.41; brown triangles), notch signaling (lag-2; purple crosses), pharynx (myo-2; orange diamonds), body wall muscles (myo-3; red triangles), seam cell lineage (nhr-82; green triangles), pan-neuronal (rgef-1; blue crosses), serotonin-producing neurons (tph-1; black diamonds), and GABAergic motor neurons (unc-47; brown triangles). Shown are the percentages of expected nuclei expressing GFP::HIS-58; numbers in brackets indicate the number of animals evaluated for each strain; black horizontal lines represent the means. For three promoters, on average, FLP activity was observed in 60–70% of the expected nuclei; for the remaining promoters, the mean activity was > 90% (see Table 1). GABA, γ-aminobutyric acid; MosSCI, Mos1-mediated single-copy insertion; NLS, nuclear localization signal.
The relative low efficiency of the MosSCI FLP G5 line led us to test a FLP variant with an aspartic acid residue at position 5, which was reported to be much more active in Drosophila melanogaster (FLP D5; Nern et al. 2011). Moreover, inspired by modifications of the Cre recombinase reported by Ruijtenberg and van den Heuvel (2015), we also generated a FLP D5 variant with a heterologous nuclear localization signal and a 3xFLAG epitope, both at the N-terminus (FLAG-NLS-FLP D5). Surprisingly, animals expressing FLAG-NLS-FLP D5 had a highly variable number of GFP-positive nuclei with many animals showing no recombination (19 ± 27; n = 48), whereas untagged FLP D5 was much more active (86 ± 22 GFP-positive nuclei; n = 25), equaling the multicopy FLP G5 array (P = 0.19 by unpaired, two-sided t-test; Figure 2A). It should be noted that, in all experiments, animals were maintained at 16° to minimize transcription from the hsp-16.41 promoter of our reporters prior to heat induction and recovery. Because FLP has an optimum temperature of 30° (Buchholz et al. 1996), we speculated that higher recombination efficiencies could be obtained if animals were raised at 25°. However, we obtained similar efficiencies at 16 and 25° (Figure S1). Importantly, we only observed GFP-positive nuclei in seam cells and the hypodermis, suggesting that the nhr-82 promoter activates the FLP recombinase exclusively in the expected cell types. Therefore, we conclude that the combination of a hsp-16.41-controlled Frt construct and expression of FLP D5 in the seam cell lineage from integrated single-copy transgenes provides efficient spatiotemporal control of gene expression.
Generation of tissue-specific FLP lines
We next generated multiple MosSCI lines expressing FLP D5 in specific cell types. In particular, to demonstrate the versatility of the FLP/Frt system, we were interested in developing tools to control expression either broadly in the entire nervous system or restricted to specific neuronal subtypes. As pan-neuronal promoters we tested unc-119 and rgef-1. Although expression of FLP D5 from the unc-119 promoter induced recombination in neurons, we also detected several green nonneuronal nuclei (Figure S2A). The activity of unc-119 outside the nervous system has been reported in WormBase (http://www.wormbase.org) and we did not characterize this construct further. Instead, we focused on rgef-1, which is expressed in postmitotic neurons (Altun-Gultekin et al. 2001). In all Prgef-1::FLP D5 animals (n > 40), we observed numerous green nuclei throughout the nervous system (Figure 3, A–C; note that several of the images represent single confocal sections, hence only subsets of nuclei are visible; Table 1). We did not attempt to count the absolute number of GFP-positive nuclei, but when crossed to markers of dopaminergic and GABAergic motor neurons, we observed recombination in 99% of the cells expressing the marker (n = 15; Figure S2, B and C and Table 1). Thus, we conclude that the rgef-1 promoter is highly active and specific for induction of recombination in C. elegans neurons. As examples of specific neuronal subtypes, we tested four promoters: dat-1 (dopaminergic neurons, Nass et al. 2002), mec-7 (mechanosensory and other neurons, Mitani et al. 1993), tph-1 (serotonin-producing neurons, (Sze et al. 2000), and unc-47 (GABAergic motor neurons, McIntire et al. 1997). In all 14 Ptph-1::FLP D5 animals analyzed, we observed GFP expression exclusively in six neurons that were located in the expected positions (NSML/R, ADFL/R, and HSNL/R; Figure 2B, Figure 3, D and E, and Table 1). For the unc-47 promoter, we observed 19 ± 4 GFP-positive nuclei in the head and along the ventral nerve cord, which corresponds to 74% of the expected nuclei (Figure 2B, Figure 3, G and H, and Table 1). Although the nhr-82 and unc-47 promoters produced similar recombination efficiencies in terms of percentage of GFP-positive nuclei (Figure 2B and Table 1), they showed a remarkable difference: Most green nuclei in Pnhr-82 animals coexpressed mCh::HIS-58, which suggests that only one of the two alleles in the genome had undergone FLP-recombination (Figure 3U). In contrast, none of the green nuclei in Punc-47 animals had expression of mCh::HIS-58, indicating that either both alleles had recombined or, less likely, only a single allele was expressed. Similar to the results with the tph-1 and unc-47 promoters, the Pmec-7::FLP strain also produced recombination specifically in the nervous system, but in all eight animals analyzed we observed more GFP-positive nuclei than expected. For instance, we expected recombination only in two neurons in the head (FLPL/R) but we detected up to seven GFP-positive nuclei (Figure 3F). Because the FLP/Frt system also tracks cell lineages (Voutev and Hubbard 2008), we speculate that these “extra” cells might reflect transient activation of the mec-7 promoter earlier in development. An alternative, and not mutually exclusive, possibility is that current expression annotations are incomplete (Schmitt et al. 2012). The first line carrying a Pdat-1::FLP D5 transgene did not produce recombination of the dual color reporter (data not shown), possibly due to insufficient expression of the recombinase. Therefore, we generated a second line, this time including a downstream transsplicing cassette encoding mNeonGreen to indirectly visualize FLP D5 expression. This line had robust mNeonGreen expression in six head neurons and two posterior neurons of all animals, as expected for the dat-1 promoter (n = 37; Figure S3). When crossed with the dual color reporter, coexpression of GFP::HIS-58 and mNeonGreen was observed in all dopaminergic neurons (Figure 3C, Figure S3, and Table 1). Based on these results, we conclude that the C. elegans nervous system is amenable to spatiotemporal control of gene expression using integrated single-copy FLP/Frt transgenes, in some cases with ∼65–100% efficiency.
The M lineage is derived from a single embryonic mesodermal blast cell, which generates 14 striated body wall muscle cells, 16 nonstriated uterine and vulval muscle cells, and 2 coelomocytes during larval development (Sulston and Horvitz 1977). Thus, the M lineage offers an attractive system to study postembryonic control of cell proliferation and differentiation. The hlh-8 gene is a marker of the M lineage (Harfe et al. 1998) and we tested if the hlh-8 promoter can be used to induce specific expression of FLP D5 and recombination in this cell lineage. In adults, we observed GFP expression in 19 ± 5 nuclei, all belonging to the expected cell types (Figure 2B, Figure 3, I–K, and Table 1). This demonstrates that the M lineage is susceptible to lineage-specific, FLP-mediated recombination via the hlh-8 promoter. A similar conclusion was recently made based on the Cre/lox system, although a low frequency of recombination outside the M lineage was also observed (Ruijtenberg and van den Heuvel 2015).
The C. elegans intestine consists of 20, or occasionally 19, 21, or 22, cells (Sulston and Horvitz 1977; McGhee 2007). At the L1/L2 larval molt, 10–14 of the ∼20 nuclei divide to produce a total of 30–34 nuclei; for calculation of recombination frequencies, we assumed 20 and 32 nuclei in L1 and L2 larvae, respectively, as expected values. To drive expression of the recombinase in the intestine, we cloned the elt-2 promoter (Fukushige et al. 1998) and crossed Pelt-2::FLP D5 animals with the dual color reporter strain. In L1 and L2 larvae we observed 20 ± 1 and 31 ± 1 GFP-positive nuclei, respectively, which corresponds to 98–100% efficiency (Figure 2B, Figure 3, L–N, and Table 1). We also found expression of GFP::HIS-58 in a few cells of the gonad primordium (asterisks in Figure 3N). Analyzing adult animals consistently revealed GFP-positive nuclei in the somatic gonad, including the spermathecae and gonadal sheet cells (Figure S2B). Activity of the elt-2 promoter in the somatic gonad has been reported (Ruijtenberg and van den Heuvel 2015), and this should be taken into account when designing FLP or Cre-based recombination experiments to analyze biological processes in the intestine. Nevertheless, in L1 and young L2 larvae before proliferation of the gonad primordium, the contribution from nonintestinal cells is likely to be negligible.
To achieve recombination in nonstriated pharyngeal muscles and striated body wall muscles, we tested the well-established myo-2 and myo-3 promoters (Okkema et al. 1993). Both promoters induced specific recombination in the expected cell types at 91–99% efficiency with 34 ± 2 and 80 ± 1 GFP-positive nuclei in the pharynx and the body wall muscles, respectively (Figure 2B, Figure 3, O–Q, and Table 1). As described above, using the nhr-82 promoter to control FLP D5 expression leads to recombination in nuclei that will later form part of the hypodermis. However, many hypodermal nuclei are not derived from the seam cell lineage, so to target the hypodermis more broadly we tested the dpy-7 promoter (Gilleard et al. 1997; Page and Johnstone 2007). All animals analyzed (n > 50) had very high density of GFP-positive nuclei in the hypodermis, as well as in the ventral nerve cord and in vulval cells (Figure 3, R–T and Table 1). The expression of GFP in these latter cell types suggests that Pdpy-7::FLP D5-mediated recombination happens relatively early in development before the P cells migrate and produce neurons and vulva precursor cells (Sulston and Horvitz 1977). Moreover, we also observed partial recombination in a few polyploid intestinal cells in some animals (1.4 ± 2.2 nuclei, n = 50; arrow in Figure 3T). Finally, we included the lag-2 promoter in our battery of FLP D5 lines to explore the potential of the FLP/Frt system in the context of a classical cell signaling pathway. LAG-2 is a ligand for GLP-1 and LIN-12 Notch-like receptors and is expressed in many different cell types to influence the proliferation and differentiation of neighboring cells (Henderson et al. 1994; Wilkinson et al. 1994; Greenwald 2005). In all Plag-2::FLP D5 animals (n > 50), we observed a large number of GFP-positive nuclei, in particular in the somatic gonads including the spermathecae and the distal tip cells (Figure 3, V and W and Table 1).
Based on the results above, we conclude that single-copy FLP D5 transgenes are highly efficient and specific tools to excise Frt-flanked DNA fragments from the C. elegans genome. Because the FLP D5 transgenes are integrated into the genome, they can easily be crossed to relevant Frt strains and show complete stability during meiotic and mitotic divisions, ensuring that all cells in individual animals and all animals in a population carry the FLP D5 transgene.
Temporal control of leaky transcription
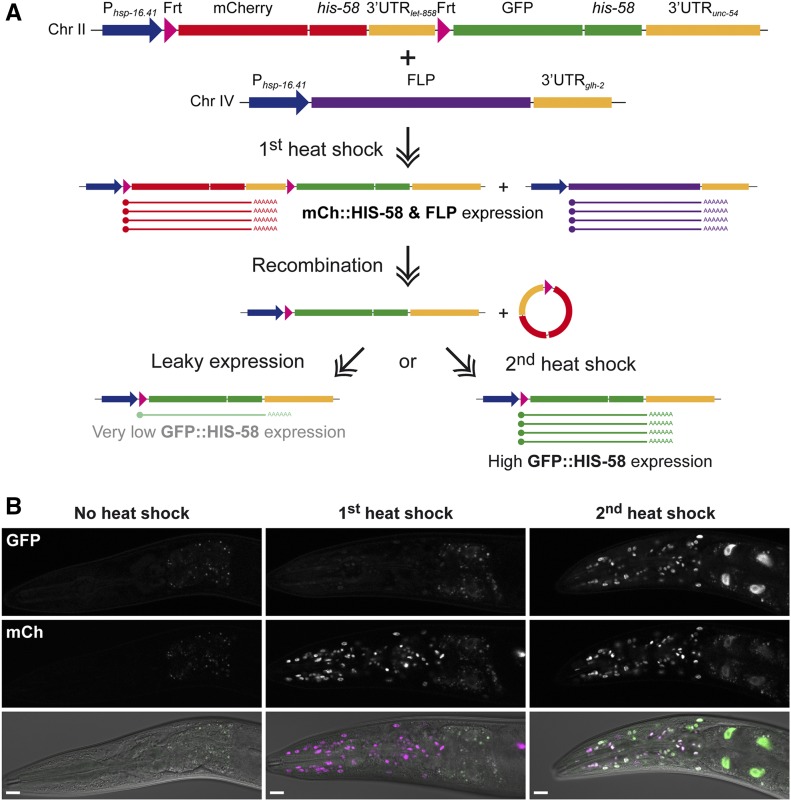
Some protocols utilize the basal or “leaky” transcription of the hsp-16.41 promoter in noninduced conditions to achieve low expression levels. This is typically the case in DNA adenine methyltransferase (Dam)ID (identification) experiments, where low amounts of a chromatin-interacting protein fused to Escherichia coli Dam are used to identify protein–DNA interactions (Gomez-Saldivar et al. 2016). This low expression level is typically not detectable by fluorescent reporters or western blotting but is sufficient to generate specific DamID signals [Figure 1C, 0 hr post heat shock; see Figure S1 in Gonzalez-Aguilera et al. (2014)]. However, because methylation marks made by Dam are stable in postmitotic cells, DamID profiles obtained from old animals will represent the sum of methylation accumulated since their cells were born. Therefore, to determine protein–DNA interactions taking place in old animals, expression of the Dam fusion protein should be completely avoided at earlier time points. To this end, we tested if the hsp-16.41 promoter can be used to temporally control FLP-mediated recombination (Figure 4A). Using again the dual color reporter to evaluate recombination efficiency, we observed that an initial heat shock led to ubiquitous and strong expression of mCh::HIS-58, whereas similar GFP::HIS-58 expression was only observed after a second heat shock (Figure 4B). These results demonstrate that recombination requires induction of the hsp-16.41 promoter to produce sufficient FLP enzyme and that, once this occurs, recombination is efficient throughout the animal. We have implemented this system to analyze nuclear organization in old animals and preliminary data suggest both local and global changes when compared to young animals (C. Muñoz-Jiménez and P. Askjaer, unpublished data). Moreover, by substituting the hsp-16.41 promoter in the Frt construct with a tissue-specific promoter, the system can easily be adapted to stimulation within a target tissue using heat shock (Figure 1B). For instance, if a BirA fusion gene is inserted after the Frt-flanked cassette, the system can be used for spatiotemporal control of proximity-dependent biotin identification (BioID) of protein interaction partners (Kim and Roux 2016). Whether the best configuration to achieve spatiotemporal control is by expression of FLP from a tissue-specific promoter and the Frt construct from a heat-inducible promoter or vice versa depends on the particular experiment. In the former configuration, potentially the entire cell lineage(s) in which FLP has been transiently expressed will rapidly produce a boost of temporal expression of the downstream gene upon heat shock (Figure 1, A and C). In contrast, if the recombinase is placed under control of the hsp-16.41 promoter, a heat shock will induce expression of the downstream gene only in cells in which the tissue-specific promoter is currently active and with a time delay reflecting the kinetics of recombination (Figure 1, B and D). Typically, the expression levels in the latter case will be lower than the ones obtained with the induced hsp-16.41 promoter, although this will naturally depend on the particular tissue-specific promoter.
Figure 4.
Temporal control of FLP recombination. Frt-flanked cassettes can be efficiently excised by heat shock-induced FLP expression. (A) Schematic representation of experimental setup and (B) single confocal sections of adult heads. The first time the animals are heat shocked they express mCh::HIS-58 and FLP. The FLP recombinase excises the mCh::his-58 cassette but efficient expression of the downstream GFP::his-58 fusion gene requires a second heat shock, suggesting that recombination is completed after the transient activation of the hsp-16.41 promoter ends. Bar, 10 µm. Chr, chromosome; mCh, mCherry.
Spatiotemporal control of cell ablation
Killing of specific cells or cell types represents an important tool to understand their physiological role. Different systems have been proposed to achieve temporal and/or spatial control, including expression of reconstituted caspases (Chelur and Chalfie 2007), tetanus toxin (Davis et al. 2008), mini Singlet Oxygen Generator (miniSOG; Qi et al. 2012; Xu and Chisholm 2016), and histamine-gated chloride channels (Pokala et al. 2014). Each of these methods has its advantages and drawbacks, and we reasoned that our collection of stable FLP lines could potentially serve as an efficient and fast way to target a variety of specific tissues without the need of multiple cloning steps or experience with optogenetic equipment. To test this, we used a cDNA encoding the PEEL-1 toxin (Seidel et al. 2011) to replace the GFP::his-58 sequence of our dual color reporter and inserted this construct as single-copy transgene on chromosome II (Figure 5A). We first crossed the peel-1 line to the Prgef-1::FLP D5 and Pmyo-3::FLP D5 lines to broadly target neurons and muscles, respectively. For unbiased assessment of locomotion, we transferred the animals to liquid media and quantified the number of body bends per second using the wrMTrck software (Nussbaum-Krammer et al. 2015). In the absence of heat shock all lines behaved like wild-type animals, indicating that leaky expression of PEEL-1 in neurons or muscles has no obvious consequence (blue boxes in Figure 5B; video in File S2 and File S3). In contrast, when PEEL-1 expression was induced in muscles, all animals became uncoordinated (Unc phenotype) and were paralyzed within 2 hr (video in File S4). Although the penetrance was less severe, peel-1 Prgef-1::FLP D5 animals (video in File S5) had a significant Unc phenotype, whereas the heat shock did not affect peel-1 animals in the absence of FLP recombinase (red boxes in Figure 5B).
Figure 5.
Spatiotemporal control of cell ablation. (A) Schematic representation of FLP-regulated peel-1 cell killing construct. Heat shock activation of the construct produces mCh::HIS-58 in the absence of FLP and PEEL-1 toxin in cells positive for FLP activity. (B) Locomotion of animals carrying the peel-1 construct alone or together with either Prgef-1::FLP or Pmyo-3::FLP was quantified before (blue boxes) or after heat shock (red boxes). Top and bottom of boxes indicate the first and third quartiles, respectively, whereas lines inside boxes correspond to the medians; whiskers cover the 1.5× interquartile range. (C) Strains carrying the peel-1 construct alone (circles), or together with either Ptph-1::FLP (triangles) or Pmec-7::FLP (crosses), were observed for mechanosensation defects (Mec) in the absence of (blue symbols) or after heat shock (red symbols). Shown are the percentages of Mec animals in three to nine independent experiments; numbers in brackets indicate the total number of animals evaluated for each condition; and black horizontal lines represent the means. P values were obtained through Mann–Whitney–Wilcoxon (B) and Fisher’s exact (C) tests, indicating that heat-induced ablation of neurons (Prgef-1::FLP), body wall muscles (Pmyo-3::FLP), or mechanosensory neurons (Pmec-7::FLP) induce Unc or Mec phenotypes. Chr, chromosome; mCh, mCherry.
Next, we tested the cell ablation system in specific neuronal subtypes using the tph-1 and mec-7 promoters. As described above, mec-7 is expressed in mechanosensory neurons and mec-7 mutants have Mec phenotypes (Mitani et al. 1993). In concordance with this, we observed 87 ± 12% Mec animals in heat-shocked peel-1 Pmec-7::FLP D5 animals (n = 79), whereas FLP expression in serotonin-producing neurons (from the tph-1 promoter) did not affect mechanosensation (red symbols Figure 5C). As expected, in the absence of heat shock, the expression of the recombinase in any of the two neuronal subtypes did not produce Mec phenotypes (P = 0.25–0.48 by Fisher’s exact test) (blue symbols in Figure 5C). Mutation of tph-1 interferes with egg laying (Sze et al. 2000) and peel-1 Ptph-1::FLP D5 animals produced fewer eggs per hour than peel-1 animals after heat shock (P = 0.02 by unpaired, two-sided t-test; Figure S4). However, the transgene Ptph-1::FLP D5 reduced egg laying also without heat shock (P = 0.003), which could reflect that the serotonin-producing neurons are potentially already affected by the leaky expression of PEEL-1 from the hsp-16.41 promoter (Figure S4). In conclusion, the tools developed here to induce killing of cells in a spatiotemporal manner produce, in most cases, the expected behavioral phenotypes. More detailed microscopy studies are required to determine the precise damage at the tissue level, and future experiments under comparable conditions are needed to determine which effector (e.g., PEEL-1, tetanus toxin, caspases, and histamine-gated chloride channels, etc.) might be most suitable for recombination-driven cell ablation.
Tissue-specific gene disruption by combined FLP and CRISPR tools
The development of CRISPR/Cas9 methods to precisely engineer the genome has revolutionized genetics experiments in a wide range of organisms, including C. elegans (Dickinson and Goldstein 2016). We decided to explore the combination of CRISPR and FLP-based recombination to interfere with gene expression in specific cells. As a proof-of-principle, we modified the endogenous baf-1 locus. BAF-1 is a small chromatin-binding protein that is essential for nuclear envelope assembly (Gorjanacz et al. 2007). Homozygous baf-1 mutants complete embryogenesis thanks to maternally contributed protein but arrest as larvae with defects in multiple tissues, whereas BAF-1-depleted embryos die during early embryogenesis (Gorjanacz et al. 2007; Margalit et al. 2007). We first generated a SapTrap-compatible (Schwartz and Jorgensen 2016) GFP KO cassette containing Frt sites in introns 1 and 2 (indicated as g > f > p in genotype descriptions). The two Frt sites are separated by 205 bp, which should be compatible with optimal recombination efficiency (Ringrose et al. 1999). When expressed in C. elegans cells without FLP activity, the introns are removed from the primary transcript by splicing and the processed mRNA will encode GFP::BAF-1, which localizes to the nuclear envelope (Figure 6A; “Other tissues”). In contrast, in the presence of FLP, the second GFP exon is excised by recombination between the two Frt sites, which causes a reading frame shift in the third GFP exon and the appearance of a premature termination codon (PTC; Figure 6A; “Intestine”). This will most likely lead to degradation of the aberrant GFP::baf-1 transcript by nonsense-mediated mRNA decay, which is highly efficient in C. elegans (Zahler 2012). Even if a few mRNA molecules escape decay, expression of baf-1 downstream of the PTC would require that ribosomes are not released at the PTC but instead reinitiate translation (He and Jacobson 2015).
Figure 6.
Tissue-specific inhibition of gene expression. (A) Schematic representation of experimental design and (B) single confocal sections of anterior part of intestine in L4 larvae. The endogenous baf-1 locus was modified by insertion of a GFP KO cassette immediately upstream of the coding region. The GFP KO cassette contains Frt sites in the first and second intron [indicated as g > f > p in (B)]: these sites do not interfere with pre-mRNA splicing and, in the absence of FLP, the locus expresses GFP::BAF-1. In cells expressing FLP, GFP exon 2 is excised and the spliced mRNA contains a premature termination codon (red TAA), which triggers nonsense-mediated mRNA decay. (B) The left animal expresses GFP::BAF-1 in all cells, whereas the right animal lack expression in intestinal cells due to the specific expression of FLP in this tissue. Nuclei of different tissues are indicated: ph: pharynx; int: intestine; bwm: body wall muscle nuclei; germ: germ line. An asterisk indicates an enlarged intestinal lumen surrounded by abnormally thin cellular processes. Bar, 10 µm. Chr, chromosome; KO, knockout.
Animals homozygous for the GFP KO cassette in the baf-1 locus did not show any obvious phenotypes and a bright fluorescent signal was detected at the periphery of nuclei throughout the body at all developmental stages (Figure 6B left and data not shown). However, when crossed with the Pelt-2::FLP D5 line, the intestine showed clear signs of atrophy and lack of GFP expression, whereas other tissues as expected expressed GFP::BAF-1 (Figure 6B right). Concordantly, animals were sick with reduced body size and slow movements. A more detailed characterization of the role of BAF-1 in the development and/or maintenance of the intestine goes beyond the scope of this study, but our data clearly demonstrate the potential of this new collection of FLP lines to address gene function in specific tissues. We have focused on an example with insertion of the GFP KO cassette into an endogenous locus, but the same strategy can easily be adapted for generation of Frt-flanked transgenes for MosSCI into the genome, as has recently been demonstrated for the Cre/lox system (Flavell et al. 2013; Ruijtenberg and van den Heuvel 2015). In the example presented here, we inserted the GFP KO cassette at the translational start codon and, as explained above, this is likely to efficiently prevent synthesis of the tagged proteins in FLP-expressing cells. We are currently testing the efficiency of conditionally controlling expression of loci in which the GFP KO cassette, or a novel red variant (wrmScarlet; El Mouridi et al. 2017) KO cassette, is inserted immediately in front of the endogenous translational termination codon. We expect that the mRNA produced in FLP-expressing cells will be unstable due to the presence of a premature termination codon specifically in these cells, but this needs to be tested empirically.
Conclusions
We have provided quantitative evidence that stable FLP lines can drive specific and efficient recombination between Frt sites in multiple cell types in C. elegans. Because the FLP transgenes are integrated into a known locus in the genome, genetic crosses to different Frt-flanked constructs are straightforward. We plan to expand the collection of FLP lines in the future and we hope that other members of the community will participate to increase the number of specific cell types amenable to FLP-mediated recombination. Site-specific recombination can be utilized in many experimental strategies [see Hubbard (2014) for a comprehensive review] and the FLP lines reported here can easily be combined with existing tools for tissue-specific GFP tagging (Schwartz and Jorgensen 2016) or expression of RNAi hairpins (Voutev and Hubbard 2008). We have demonstrated that the FLP system can be used tissue-specifically to knock out gene function or ablate cells. Our GFP KO cassette is compatible with the efficient SapTrap protocol for preparation of repair templates for genome engineering (Schwartz and Jorgensen 2016) and can therefore be readily applied to other loci. Finally, we envision that other advances in CRISPR technology will further facilitate the insertion of Frt sites into endogenous loci to make them controllable by FLP.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.201012/-/DC1.
Acknowledgments
We are grateful to Alejandro Ferreiro Morales, Carmen Serrano Tasset, Cristina González-Aguilera, Eric Jorgensen, and Esther Dalfo for plasmids, as well as to Nuria Flames for suggestions on neuron-specific promoters and Anita Fernandez for critical reading of the manuscript. We also wish to acknowledge Katherina García from the CABD Imaging Facility; Addgene for sequence verifications; the Caenorhabditis Genetics Center (CGC), which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD-010440); and WormBase, which is supported by a grant from the National Human Genome Research Institute at the NIH (grant number U41 HG-002223) and the British Medical Research Council. This project was funded by the Spanish Ministry of Economy and Competitiveness (BFU2013-42709P and BFU2016-79313-P) and the European Regional Development Fund.
Footnotes
Communicating editor: O. Hobert
Literature Cited
- Altun-Gultekin Z., Andachi Y., Tsalik E. L., Pilgrim D., Kohara Y., et al. , 2001. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development 128: 1951–1969. [DOI] [PubMed] [Google Scholar]
- Anastassiadis K., Glaser S., Kranz A., Berhardt K., Stewart A. F., 2010. A practical summary of site-specific recombination, conditional mutagenesis, and tamoxifen induction of CreERT2. Methods Enzymol. 477: 109–123. [DOI] [PubMed] [Google Scholar]
- Askjaer P., Ercan S., Meister P., 2014. Modern techniques for the analysis of chromatin and nuclear organization in C. elegans (April 2, 2014), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.169.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz F., Ringrose L., Angrand P. O., Rossi F., Stewart A. F., 1996. Different thermostabilities of FLP and Cre recombinases: implications for applied site-specific recombination. Nucleic Acids Res. 24: 4256–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz F., Angrand P. O., Stewart A. F., 1998. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 16: 657–662. [DOI] [PubMed] [Google Scholar]
- Chelur D. S., Chalfie M., 2007. Targeted cell killing by reconstituted caspases. Proc. Natl. Acad. Sci. USA 104: 2283–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churgin M. A., He L., Murray J. I., Fang-Yen C., 2014. Construction of a system for single-cell transgene induction in Caenorhabditis elegans using a pulsed infrared laser. Methods 68: 431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. W., Morton J. J., Carroll D., Jorgensen E. M., 2008. Gene activation using FLP recombinase in C. elegans. PLoS Genet. 4: e1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Goldstein B., 2016. CRISPR-based methods for Caenorhabditis elegans genome engineering. Genetics 202: 885–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrzynska A., Askjaer P., Gruenbaum Y., 2016. Lamin-binding proteins in Caenorhabditis elegans. Methods Enzymol. 569: 455–483. [DOI] [PubMed] [Google Scholar]
- El Mouridi S., Lecroisey C., Tardy P., Mercier M., Leclercq-Blondel A., et al. , 2017. Reliable CRISPR/Cas9 genome engineering in Caenorhabditis elegans using a single efficient sgRNA and an easily recognizable phenotype. G3 (Bethesda) 7: 1429–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezcurra M., Tanizawa Y., Swoboda P., Schafer W. R., 2011. Food sensitizes C. elegans avoidance behaviours through acute dopamine signalling. EMBO J. 30: 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firnhaber C., Hammarlund M., 2013. Neuron-specific feeding RNAi in C. elegans and its use in a screen for essential genes required for GABA neuron function. PLoS Genet. 9: e1003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N., Hobert O., 2009. Gene regulatory logic of dopamine neuron differentiation. Nature 458: 885–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell S. W., Pokala N., Macosko E. Z., Albrecht D. R., Larsch J., et al. , 2013. Serotonin and the neuropeptide PDF initiate and extend opposing behavioral states in C. elegans. Cell 154: 1023–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiacovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C., Davis M. W., Ailion M., Jorgensen E. M., 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9: 117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C., Davis M. W., Sarov M., Taylor J., Flibotte S., et al. , 2014. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat. Methods 11: 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T., Hawkins M. G., McGhee J. D., 1998. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol. 198: 286–302. [PubMed] [Google Scholar]
- Gilleard J. S., Barry J. D., Johnstone I. L., 1997. cis regulatory requirements for hypodermal cell-specific expression of the Caenorhabditis elegans cuticle collagen gene dpy-7. Mol. Cell. Biol. 17: 2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glock C., Nagpal J., Gottschalk A., 2015. Microbial rhodopsin optogenetic tools: application for analyses of synaptic transmission and of neuronal network activity in behavior. Methods Mol. Biol. 1327: 87–103. [DOI] [PubMed] [Google Scholar]
- Gomez-Saldivar G., Meister P., Askjaer P., 2016. DamID analysis of nuclear organization in Caenorhabditis elegans. Methods Mol. Biol. 1411: 341–358. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Aguilera C., Ikegami K., Ayuso C., de Luis A., Iniguez M., et al. , 2014. Genome-wide analysis links emerin to neuromuscular junction activity in Caenorhabditis elegans. Genome Biol. 15: R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjanacz M., Klerkx E. P., Galy V., Santarella R., Lopez-Iglesias C., et al. , 2007. Caenorhabditis elegans BAF-1 and its kinase VRK-1 participate directly in post-mitotic nuclear envelope assembly. EMBO J. 26: 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I., 2005. LIN-12/Notch signaling in C. elegans (August 4, 2005), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.10.1, http://www.wormbook.org. [Google Scholar]
- Harfe B. D., Vaz Gomes A., Kenyon C., Liu J., Krause M., et al. , 1998. Analysis of a Caenorhabditis elegans twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. 12: 2623–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Jacobson A., 2015. Nonsense-mediated mRNA decay: degradation of defective transcripts is only part of the story. Annu. Rev. Genet. 49: 339–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S. T., Gao D., Lambie E. J., Kimble J., 1994. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120: 2913–2924. [DOI] [PubMed] [Google Scholar]
- Hostettler L., Grundy L., Kaser-Pebernard S., Wicky C., Schafer W. R., et al. , 2017. The bright fluorescent protein mNeonGreen facilitates protein expression analysis in vivo. G3 (Bethesda) 7: 607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard E. J., 2014. FLP/FRT and Cre/lox recombination technology in C. elegans. Methods 68: 417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. I., Roux K. J., 2016. Filling the void: proximity-based labeling of proteins in living cells. Trends Cell Biol. 26: 804–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Yuan J. S., Stewart C. N., Jr., 2013. Advanced genetic tools for plant biotechnology. Nat. Rev. Genet. 14: 781–793. [DOI] [PubMed] [Google Scholar]
- Lo T. W., Pickle C. S., Lin S., Ralston E. J., Gurling M., et al. , 2013. Precise and heritable genome editing in evolutionarily diverse nematodes using TALENs and CRISPR/Cas9 to engineer insertions and deletions. Genetics 195: 331–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko E. Z., Pokala N., Feinberg E. H., Chalasani S. H., Butcher R. A., et al. , 2009. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature 458: 1171–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalit A., Neufeld E., Feinstein N., Wilson K. L., Podbilewicz B., et al. , 2007. Barrier to autointegration factor blocks premature cell fusion and maintains adult muscle integrity in C. elegans. J. Cell Biol. 178: 661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., 2007. The C. elegans intestine (March 27, 2007), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.133.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire S. L., Reimer R. J., Schuske K., Edwards R. H., Jorgensen E. M., 1997. Identification and characterization of the vesicular GABA transporter. Nature 389: 870–876. [DOI] [PubMed] [Google Scholar]
- Mitani S., Du H., Hall D. H., Driscoll M., Chalfie M., 1993. Combinatorial control of touch receptor neuron expression in Caenorhabditis elegans. Development 119: 773–783. [DOI] [PubMed] [Google Scholar]
- Miyabayashi T., Palfreyman M. T., Sluder A. E., Slack F., Sengupta P., 1999. Expression and function of members of a divergent nuclear receptor family in Caenorhabditis elegans. Dev. Biol. 215: 314–331. [DOI] [PubMed] [Google Scholar]
- Morales-Martinez A., Dobrzynska A., Askjaer P., 2015. Inner nuclear membrane protein LEM-2 is required for correct nuclear separation and morphology in C. elegans. J. Cell Sci. 128: 1090–1096. [DOI] [PubMed] [Google Scholar]
- Nass R., Hall D. H., Miller D. M., III, Blakely R. D., 2002. Neurotoxin-induced degeneration of dopamine neurons in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 99: 3264–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A., Pfeiffer B. D., Svoboda K., Rubin G. M., 2011. Multiple new site-specific recombinases for use in manipulating animal genomes. Proc. Natl. Acad. Sci. USA 108: 14198–14203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum-Krammer C. I., Neto M. F., Brielmann R. M., Pedersen J. S., Morimoto R. I., 2015. Investigating the spreading and toxicity of prion-like proteins using the metazoan model organism C. elegans. J. Vis. Exp. 8: 52321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema P. G., Harrison S. W., Plunger V., Aryana A., Fire A., 1993. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135: 385–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A. P., Johnstone I. L., 2007. The cuticle (March 19, 2007), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.138.1, http://www.wormbook.org. [Google Scholar]
- Pokala N., Liu Q., Gordus A., Bargmann C. I., 2014. Inducible and titratable silencing of Caenorhabditis elegans neurons in vivo with histamine-gated chloride channels. Proc. Natl. Acad. Sci. USA 111: 2770–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y. B., Garren E. J., Shu X., Tsien R. Y., Jin Y., 2012. Photo-inducible cell ablation in Caenorhabditis elegans using the genetically encoded singlet oxygen generating protein miniSOG. Proc. Natl. Acad. Sci. USA 109: 7499–7504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redemann S., Schloissnig S., Ernst S., Pozniakowsky A., Ayloo S., et al. , 2011. Codon adaptation-based control of protein expression in C. elegans. Nat. Methods 8: 250–252. [DOI] [PubMed] [Google Scholar]
- Ringrose L., Chabanis S., Angrand P. O., Woodroofe C., Stewart A. F., 1999. Quantitative comparison of DNA looping in vitro and in vivo: chromatin increases effective DNA flexibility at short distances. EMBO J. 18: 6630–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenas E., Gonzalez-Aguilera C., Ayuso C., Askjaer P., 2012. Dissection of the NUP107 nuclear pore subcomplex reveals a novel interaction with spindle assembly checkpoint protein MAD1 in Caenorhabditis elegans. Mol. Biol. Cell 23: 930–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruaud A. F., Katic I., Bessereau J. L., 2011. Insulin/Insulin-like growth factor signaling controls non-Dauer developmental speed in the nematode Caenorhabditis elegans. Genetics 187: 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijtenberg S., van den Heuvel S., 2015. G1/S Inhibitors and the SWI/SNF complex control cell-cycle exit during muscle differentiation. Cell 162: 300–313. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., et al. , 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt C., Schultheis C., Pokala N., Husson S. J., Liewald J. F., et al. , 2012. Specific expression of channelrhodopsin-2 in single neurons of Caenorhabditis elegans. PLoS One 7: e43164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. L., Jorgensen E. M., 2016. SapTrap, a toolkit for high-throughput CRISPR/Cas9 gene modification in Caenorhabditis elegans. Genetics 202: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., Ailion M., Li J., van Oudenaarden A., Rockman M. V., et al. , 2011. A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. PLoS Biol. 9: e1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R., Jost D., Kind J., Gomez-Saldivar G., van Steensel B., et al. , 2014. Differential spatial and structural organization of the X chromosome underlies dosage compensation in C. elegans. Genes Dev. 28: 2591–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T., 2006. Maintenance of C. elegans (February 11, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.101.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S., Powers J., Dunn M., Reese K., Malone C. J., et al. , 2001. Spindle dynamics and the role of gamma-tubulin in early Caenorhabditis elegans embryos. Mol. Biol. Cell 12: 1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R., 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. [DOI] [PubMed] [Google Scholar]
- Sze J. Y., Victor M., Loer C., Shi Y., Ruvkun G., 2000. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 403: 560–564. [DOI] [PubMed] [Google Scholar]
- Takata Y., Kondo S., Goda N., Kanegae Y., Saito I., 2011. Comparison of efficiency between FLPe and Cre for recombinase-mediated cassette exchange in vitro and in adenovirus vector production. Genes Cells 16: 765–777. [DOI] [PubMed] [Google Scholar]
- Trinh le A., Hochgreb T., Graham M., Wu D., Ruf-Zamojski F., et al. , 2011. A versatile gene trap to visualize and interrogate the function of the vertebrate proteome. Genes Dev. 25: 2306–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Manrique R. P., Legg J. C., Olofsson B., Ly S., Baylis H. A., 2010. Improved gene targeting in C. elegans using counter-selection and Flp-mediated marker excision. Genomics 95: 37–46. [DOI] [PubMed] [Google Scholar]
- Voutev R., Hubbard E. J., 2008. A “FLP-Out” system for controlled gene expression in Caenorhabditis elegans. Genetics 180: 103–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson H. A., Fitzgerald K., Greenwald I., 1994. Reciprocal changes in expression of the receptor lin-12 and its ligand lag-2 prior to commitment in a C. elegans cell fate decision. Cell 79: 1187–1198. [DOI] [PubMed] [Google Scholar]
- Wong A. C., Draper B. W., Van Eenennaam A. L., 2011. FLPe functions in zebrafish embryos. Transgenic Res. 20: 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Chisholm A. D., 2016. Highly efficient optogenetic cell ablation in C. elegans using membrane-targeted miniSOG. Sci. Rep. 6: 21271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Rubin G. M., 2012. The effort to make mosaic analysis a household tool. Development 139: 4501–4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler A. M., 2012. Pre-mRNA splicing and its regulation in Caenorhabditis elegans (March 21, 2012), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.31.2, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
See Table S1, Table S2, and Table S3 for complete lists of primers, promoters, and strains, respectively. At least one strain for each FLP D5 line will be made immediately available through the Caenorhabditis Genetics Center; others can be requested from the authors. Most plasmids produced in this study and their sequences are available at Addgene (www.addgene.org); the remaining plasmids can be requested from the authors. File S6 contains legends for File S2 – S5.