Sumoylation is required to repair protein-linked DNA damage, but its presence can limit the use of alternative repair pathways. Through a suppressor...
Keywords: SUMO, Smt3, SUMO-targeted Ub ligase, SLX5-SLX8, synthetic lethality
Abstract
Protein modification by the small ubiquitin-like modifier (SUMO) plays important roles in genome maintenance. In Saccharomyces cerevisiae, proper regulation of sumoylation is known to be essential for viability in certain DNA repair mutants. Here, we find the opposite result; proper regulation of sumoylation is lethal in certain DNA repair mutants. Yeast cells lacking the repair factors TDP1 and WSS1 are synthetically lethal due to their redundant roles in removing Top1-DNA covalent complexes (Top1ccs). A screen for suppressors of tdp1∆ wss1∆ synthetic lethality isolated mutations in genes known to control global sumoylation levels including ULP1, ULP2, SIZ2, and SLX5. The results suggest that alternative pathways of repair become available when sumoylation levels are altered. Curiously, both suppressor mutations that were isolated in the Slx5 subunit of the SUMO-targeted Ub ligase created new lysine residues. These “slx5-K” mutations localize to a 398 amino acid domain that is completely free of lysine, and they result in the auto-ubiquitination and partial proteolysis of Slx5. The decrease in Slx5-K protein leads to the accumulation of high molecular weight SUMO conjugates, and the residual Ub ligase activity is needed to suppress inviability presumably by targeting polysumoylated Top1ccs. This “lysine desert” is found in the subset of large fungal Slx5 proteins, but not its smaller orthologs such as RNF4. The lysine desert solves a problem that Ub ligases encounter when evolving novel functional domains.
THE cell’s genome is under constant assault from extrinsic and intrinsic sources of DNA damage. To orchestrate a quick and effective response to DNA damage, a variety of protein modifications are employed, including sumoylation (Hoege et al. 2002; Pfander et al. 2005; Sacher et al. 2006; Cremona et al. 2012a,b; Jalal et al. 2017; Zilio et al. 2017). SUMO, also known as Smt3 in Saccharomyces cerevisiae, is processed from an inactive precursor by the SUMO protease Ulp1. Ulp1 is tethered to the nuclear envelope by components of the nuclear pore including Nup60 (Li and Hochstrasser 2003; Zhao et al. 2004; Lewis et al. 2007). Like ubiquitin (Ub), mature Smt3 is conjugated to lysine residues via an ATP-dependent enzyme cascade of E1 (Aos1/Uba2), E2 (Ubc9), and E3 (Siz1, Siz2, and Mms21) enzymes (Johnson et al. 1997; Johnson and Gupta 2001; Zhao and Blobel 2005). Sumoylation is dynamic, and SUMO can be decoupled from its target proteins by the Ulp1 and Ulp2 deconjugases (Li and Hochstrasser 2000).
Because SUMO bears a sumoylation consensus motif, it can be coupled to itself. This results in poly-SUMO chains analogous to those of Ub (Tatham et al. 2001; Bylebyl et al. 2003). Polysumoylation has been implicated in multiple functions (Zhang et al. 2008; Srikumar et al. 2013). It can be induced by stresses such as heat shock and proteasomal inhibition, and is associated with ubiquitination and protein turnover (Uzunova et al. 2007; Schimmel et al. 2008; Golebiowski et al. 2009; Tatham et al. 2011; Zhou et al. 2012).
One of poly-SUMO’s best characterized functions is to mark target proteins for destruction via the SUMO-targeted Ub ligase (STUbL). STUbLs are RING-finger type Ub E3 ligases including RNF4 in higher cells, Slx5–Slx8 in budding yeast, and Rfp1-2/Slx8 in fission yeast (Prudden et al. 2007; Sun et al. 2007; Uzunova et al. 2007; Xie et al. 2007; Mullen and Brill 2008; Tatham et al. 2008). These orthologs differ in size and subunit structure. RNF4 functions as a homodimer of 190 amino acids (aa), whereas the fungal STUbLs exist as heterodimers whose homologs Slx5 (619 aa) and Rfp1-2 (254 aa) also differ in size. It is unknown whether these size differences are functionally important, although it is known that RNF4 complements some yeast phenotypes (Prudden et al. 2007; Mullen et al. 2011; Sriramachandran and Dohmen 2014). The Slx5, Rfp1-2, and RNF4 proteins achieve substrate specificity through four SUMO Interacting Motifs (SIMs) that bind noncovalently to SUMO moieties within a chain (Keusekotten et al. 2014). Binding stimulates the ubiquitination of target proteins bearing the poly-SUMO chain, as well as the chain itself (Uzunova et al. 2007; Mullen and Brill 2008; Tatham et al. 2008; Wang and Prelich 2009; Plechanovova et al. 2011; Rojas-Fernandez et al. 2014). Yeast cells lacking STUbLs are slow growing, and display several phenotypes including hyper-recombination, sensitivity to hydroxyurea, and accumulation of high molecular weight SUMO conjugates (Zhang et al. 2006; Burgess et al. 2007; Kosoy et al. 2007; Prudden et al. 2007).
Sumoylation is important for DNA repair (Hoege et al. 2002; Potts and Yu 2005; Zhao and Blobel 2005; Branzei et al. 2006; Motegi et al. 2006; Burgess et al. 2007; Prudden et al. 2007; Galanty et al. 2012; Yin et al. 2012; Jalal et al. 2017; Zilio et al. 2017). In budding yeast, regulators of sumoylation show synthetic-lethal interactions with different pathways of DNA repair. For example, yeast cells lacking the Srs2 DNA helicase require Ulp1, those lacking the Sgs1 DNA helicase require Slx5–Slx8, and those lacking the Rad27 flap endonuclease require Siz1 or Siz2 (Mullen et al. 2001; Soustelle et al. 2004; Chen et al. 2007). Such synthetic-lethal interactions suggest that proper regulation of SUMO is essential for recruiting repair factors in alternative repair pathways (Cremona et al. 2012a; Psakhye and Jentsch 2012).
The covalent linkage of proteins to DNA is a source of intrinsic DNA damage whose repair is not completely understood. The transient cleavage cycle of type I DNA topoisomerases is a major source of such damage. Stabilization of the cleavage intermediate, by prior damage or drugs, for example, leads to a nicked-DNA protein complex. Studies in different systems have identified a variety of pathways to repair such Top1-DNA covalent complexes (Top1ccs) (Pommier et al. 2006). These pathways involve a disparate group of proteins including the tyrosyl-DNA phosphodiesterase Tdp1 (Pouliot et al. 1999; Hoa et al. 2016), the protease Wss1/SPRTN (Stingele et al. 2014, 2016; Balakirev et al. 2015; Vaz et al. 2016), and endonucleases such as MRX, Rad1-10, Slx1–4, and Mus81-Mms4 (Liu et al. 2002; Vance and Wilson 2002; Deng et al. 2005; Hartsuiker et al. 2009; Regairaz et al. 2011). Originally identified as a phosphodiesterase specific for topoisomerase I-induced DNA damage, Tdp1 can hydrolyze the phosphodiester bond between DNA and substrates such as 3′-and 5′-phosphotyrosine (Pouliot et al. 1999; Interthal et al. 2005; Nitiss et al. 2006; Huang et al. 2013; Pommier et al. 2014). The neurodegenerative disorder spinocerebellar ataxia with axonal neuropathy (SCAN1) is due to a mutation in TDP1, and mice with this mutation are sensitive to the topoisomerase I poison camptothecin (Takashima et al. 2002; Hirano et al. 2007). Wss1/SPRTN is a recently characterized metalloprotease that acts on DNA-protein crosslinks. In yeast, Wss1 forms a complex with Cdc48/p97 to target sumoylated proteins bound to DNA, including Top1 (Stingele et al. 2014; Balakirev et al. 2015). Mutations in SPRTN are associated with Ruijs-Aalfs syndrome, a progeroid and cancer-predisposition disease in humans. Sumoylation has been implicated in the repair of topoisomerase-linked DNA damage (Mao et al. 2000; Chen et al. 2007; Heideker et al. 2011). Some models suggest that sumoylation of Top1 may lead to the ubiquitin-dependent degradation of Top1ccs prior to removal of the 3′-phosphotyrosine-linked peptide by Tdp1 (Desai et al. 2001; Mao et al. 2001; Horie et al. 2002; Lin et al. 2008; Interthal and Champoux 2011).
In budding yeast, TDP1 and WSS1 have been shown to define redundant pathways for the repair of Top1ccs. The tdp1∆ wss1∆ double mutant is synthetically lethal, and suppressible by loss of TOP1 (Dixon et al. 2008; Stingele et al. 2014; Balakirev et al. 2015). To identify alternative pathways for the repair of Top1-dependent DNA damage, we searched for additional suppressors of tdp1∆ wss1∆ synthetic lethality. We found that tdp1∆ wss1∆ cells can survive if the cell’s sumoylation is altered. The results suggest that sumoylation inhibits alternative pathways for DNA repair, and they revealed a novel functional domain in Slx5 whose sequence is constrained by its tendency to self-destruct.
Materials and Methods
Yeast strain construction and manipulation
The yeast strains used in this study are RAD5 derivatives of strain W303-1a (Thomas and Rothstein 1989), and are listed in Supplemental Material, Table S1. Strains were maintained on 1% yeast extract, 2% peptone, and 2% dextrose (YPD) or synthetic complete (SC) medium, unless otherwise stated, and standard techniques were used for their manipulation (Adams et al. 1997). Gene disruptions were carried out using PCR-generated cassettes that replaced entire open reading frames (ORFs) with antibiotic resistance markers as described (Guldener et al. 1996; Goldstein and McCusker 1999). When necessary, chromosomally marked strains were obtained from the commercially available knock-out collection (Open Biosystems). To construct tdp1∆ wss1∆ double mutants, single mutants containing the balancer plasmid pNJ7478 (WSS1/ADE3/URA3/CEN) were mated, and diploid cells were sporulated and microdissected. Only spore clones that germinated with the balancer plasmid were used throughout the study to eliminate spontaneous suppressor mutations.
DNA manipulation
Oligonucleotides were from IDT (Integrated DNA Technologies, Coralville, IA). Phusion DNA polymerase was used to amplify genes by the polymerase chain reaction (PCR). Point mutations were created by site-directed mutagenesis using specific primer sets and two-rounds of PCR.
Screen for tdp1∆ wss1∆ suppressors
Screens for spontaneous and induced suppressors were performed. To identify spontaneous suppressors, 107 cells from an overnight culture of NJY3292 [tdp1∆ wss1∆ pNJ7478 (WSS1/ADE3/URA3/CEN)] were spread onto each of 10 SC plates containing (5-Fluoroorotic Acid) (FOA). Plates were incubated at 30° for 7 days, and the fastest growing colonies were selected as potential suppressor mutants. Loss of pNJY7478 was confirmed by patching these cells onto YPD plates. Since the parent strain is white (ade2ade3), cells forming red colonies indicate the presence of the ADE3 balancer plasmid. Two mutants grew white on YPD plates and were chosen for further analysis. To isolate induced mutations, an isogenic strain of opposite mating type, PSY3288, was treated with ethyl methansulfonate (EMS) as described (Mullen et al. 2001). A 30 min EMS treatment resulted in ∼70% lethality, and these cells were used to screen for fast-growing colonies on FOA as described above.
Cloning the suppressor genes
All suppressor strains were backcrossed to the parent strain of the opposite mating type to identify recessive mutations and for tetrad analysis. The two spontaneous suppressors (sup92-z and sup92-h) and four induced suppressors (sup88-2, sup88-3, sup88-9, and sup88-y) were recessive and segregated in tetrads as single mutations (Table 1). To identify these six suppressor mutations, we took advantage of the fact that they all generated nibbled spore clones (including sup88-3 [TOP1], Figure S3A in File S1), and five of the six were judged to be centromere-linked based on their segregation pattern with respect to TRP1 (sup88-y [NUP60]; sup88-3 [TOP1]; sup88-2 [SLX5]; sup92-h [SLX5]; and sup88-9 [ULP1]).
Table 1. Suppressors of tdp1Δ wss1Δ synthetic lethality.
| Suppressor name | Centromere-linkage | Gene | Allele |
|---|---|---|---|
| sup92-z | No | SIZ2 | L301-STOP frameshift at E289 due to ΔntG868 |
| sup92-h | Yes/ChrIV | SLX5 | Q362K |
| sup88-y | Yes/ChrI | NUP60 | V48-STOP frameshift at Q42 due to ΔntC124 |
| sup88-2 | Yes/ChrIV | SLX5 | E350K |
| sup88-3 | Yes/ChrXV | TOP1 | Q668-STOP |
| sup88-9 | Yes/ChrXVI | ULP1 | W163-STOP |
The centromere-linked mutations sup88-y and sup88-3 were mapped to specific chromosomes by crossing these mutants to a set of knockout strains bearing mutations in nonessential centromere-proximal genes, and identifying the ones that yielded parental di-type tetrads (Table 1). Very tight linkage of sup92-h and sup88-2 to TRP1 placed these genes at, or near, SLX5 on Chr IV. These four genes were cloned by complementation using a tiling library of LEU2-marked plasmids (Jones et al. 2008). Specifically, a suppressed strain (e.g., tdp1∆ wss1∆ supX) was transformed with the balancer pNJ7478, and then with a set of LEU2 plasmids from the tiling library that was known to cover the chromosomal region of interest. When the suppressor mutation was complemented by one of the plasmids, transformants retained pNJ7478 (WSS1/ADE3/URA3/CEN), and were inviable on medium lacking leucine and containing FOA. Mutant alleles were then PCR amplified and sequenced.
There are several genes that are known to cause a nibbled colony phenotype and are not centromere linked. Sequencing some of these from the sup92-z strain (SLX8, SIZ1, SIZ2, ULP1, and ULP2) revealed a frame-shift mutation in SIZ2 that caused a premature stop codon at aa L301.
DNA from backcrossed sup88-9 progeny was subjected to whole genome sequencing (Genwiz). Sequence analysis by a custom pipeline consisting of SNP detection by Crossbow version 1.2.0 (Langmead et al. 2009), and variant annotation with the ANNOVAR package (Wang et al. 2010) revealed a mutation in ULP1 (nt G489A) that was confirmed by complementing the suppressor phenotype with a wt ULP1 plasmid. ULP1 is an essential gene, and this mutation creates a premature stop codon at aa 163, so we presume the cell survives because of translation initiation at an internal methionine codon (e.g., M295) resulting in an N-terminal truncation. This idea is supported by the fact that a viable N-terminal truncation of Ulp1 (Ulp1347–654) (Li and Hochstrasser 2003) suppressed tdp1∆ wss1∆ synthetic lethality (strain NJY4285). Both Nup60 and the N-terminus of Ulp1 are needed to localize Ulp1 to the nuclear envelope (Li and Hochstrasser 2003; Zhao et al. 2004; Lewis et al. 2007) so mutations in NUP60 and ULP1 were probably isolated due to the same mechanism of mis-localizing Ulp1.
Yeast extracts for immunoblotting
Total yeast cell extracts were prepared under denaturing conditions as follows. Cells equivalent to 10 OD600 units were harvested from exponentially growing cultures by centrifugation. The pellet was washed with cold water once, resuspended in 1 ml water, transferred to a microfuge tube, and centrifuged again. The cell pellet was resuspended in a lysis solution [1.85 N NaOH, 7.5% beta-mercaptoethanol (BME), 5 µg/ml leupeptin, 10 µg/ml pepstatin A, and 20 mM iodoacetamide], whose volume was twice that of the cell pellet. Following incubation on ice for 10 min, an equal amount of 50% trichloroacetic acid (TCA) solution was added, and the sample was incubated on ice for another 10 min. Precipitated protein was collected by high-speed centrifugation at 4°, followed by incubation with 90% acetone at −20° for 20 min. The precipitate was dried, resuspended in 400 µl of solution E [0.5 M Tris base, 3% SDS, and 0.1 M dithiothreitol (DTT)] with gentle sonication, heated at 65° for 20 min and centrifuged to remove insoluble material. The soluble portion (350 µl) was moved to a new tube and made 1× in Laemmli buffer lacking SDS prior to subjecting the sample to SDS-PAGE and immunoblotting.
Ni-NTA affinity pull down
Denaturing extracts were prepared from yeast cultures containing His6-tagged proteins as described above. However, the pellet obtained after TCA precipitation was resolubilized in 2 ml Buffer G [6 M guanidine, 0.1 M sodium phosphate (pH 8.5), and 75 mM Tris (pH 8.5)], with light sonication followed by rocking at room temperature for 1 hr. Solubilized proteins were separated away from the insoluble fraction by high-speed centrifugation, and a portion used for immunoblotting. The remainder of the sample was made 5 mM in imidazole, and incubated with 40 µl Ni-NTA agarose beads (Qiagen) equilibrated in Buffer G. Following incubation for 3 hr at 4°, the unbound fraction was removed by a 15 sec low-speed centrifugation spin. Beads were washed twice with buffer U [8.0 M urea, 0.1 M sodium phosphate (pH 6.5), 10 mM Tris (6.5), 10 mM imidazole, and 0.05% Tween 20], and resuspended in 40 µl gel loading buffer [8.0 M urea, 0.2 M Tris (pH 6.5), 1 mM EDTA, 5% SDS, 5% glycerol, 0.1 M DTT, and 0.1% bromophenol blue]. Samples were heated at 60° for 20 min, and a portion was analyzed by immunoblotting.
Protein ubiquitination assay
Proteins were expressed, purified, and assayed essentially as described (Ii et al. 2007a; Mullen and Brill 2008). In vitro ubiquitination assays were performed in a buffer consisting of 20 mM HEPES (pH 7.5), 5 mM MgCl2, 0.1 mM DTT, and 5 µM ZnSO4. The reactions contained 20 nM Ube1 (Boston Biochem), 200 nM Ubc4, 1 μM Ub, 1 mM creatine phosphate, 0.1 mg/ml creatine phosphate kinase, 2 mM ATP, and 500 ng of the indicated Slx5/8 dimer. Reactions were incubated at 30° for 30 min, terminated with 3× Laemmli’s buffer, after which the samples were heated at 95° for 5 min and analyzed by immunoblotting. Under these conditions Ubc4/Ubc5 has been shown to generate K48-linked Ub chains (Ii et al. 2007a).
Mouse embryo fibroblasts (MEFs)
Standard methods were used to isolate homozygous wild type (Rnf4+/+) and mutant (Rnf4−/−) MEFs using an Rnf4+/− mouse bearing a gene-trap insertion mutation in the first intron of Rnf4 (Hu et al. 2010; Jozefczuk et al. 2012). MEFs were immortalized by transduction with pBABE-SV40 TAg/Neo (Hahn et al. 2002), maintained in DMEM containing 15% FBS plus penicillin/streptomycin, and transduced with retroviruses containing wild-type (wt) and mutant rat RNF4/SNURF genes. RNF4-2CS and RNF4-4CS mutations encode C to S changes at residues 177/180, or 136/139/177/180, respectively (Hakli et al. 2004). RNF4 and its mutant derivatives were cloned into the retroviral transfer vector pMX-pie (Kitamura 1998), and packaged into retroviruses using 293T cells essentially as described (Robbiani et al. 2008). Transduced MEFs were selected in 2 µg/ml puromycin, and maintained in 1 µg/ml puromycin. Cell extracts were prepared from trypsinized cells by lysing them in 50 mM Tris (pH 7.5), 0.2 M NaCl, 1% Tween 20, 0.2% Igepal, 50 mM glycerol-2-phosphate plus protease inhibitors.
Anti-RNF4 antibody
Recombinant His6-tagged rat RNF4/SNURF protein was expressed from plasmid pNJ6891 in E. coli BL21(DE3) RIL cells. Cells were grown at 37° in 300 ml LB containing ampicillin to OD = 0.15, and then shifted to 17°. At OD = 0.5, IPTG was added to 0.4 mM, and growth was continued overnight at 17°. Cells were harvested and resuspended in 10 ml N500 buffer [25 mM Tris (8.3), 500 mM NaCl, 0.01% NP-40, 10% glycerol, 0.1 mM PMSF, and 1 mM DTT] containing the following protease inhibitors: pepstatin, 10 mg/ml; leupeptin, 5 mg/ml; benzamidine, 10 mM; bacitracin 100 mg/ml; and aprotinin, 20 mg/ml. Cells were placed on ice, treated with 0.1 mg/ml lysozyme for 15 min, sonicated, and centrifuged at 13,000 × g for 15 min. The supernatant was filtered, made 10 mM in imidazole, and loaded onto a 1 ml HIS-TRAP column (GE Healthcare). The column was washed with 10 ml of N500 buffer containing 10 mM imidazole, and then with phosphate buffered saline (PBS) containing 10 mM imidazole. The protein was eluted with a 10–500 mM gradient of imidazole in PBS. Peak fractions were pooled (9.3 mg), and dialyzed against PBS containing 0.1 mM PMSF and 1 mM DTT. A portion of this protein (total 875 µg) was used to produce a commercial rabbit antiserum using a 118 days protocol (Covance, Denver, PA). Serum proteins were precipitated from 30 ml of serum by dilution with 30 ml 0.12 M NaOAc (pH 4.8), and addition of 2.25 ml caprylic acid while stirring at room temperature. The sample was centrifuged at 10,000 × g for 15 min, and the supernatant was brought to pH 7 with 1 M Tris base. IgG was preciptated by addition of 60 ml saturated (NH4)2SO4 with stirring on ice. This sample was centrifuged as above, the precipitate was resuspended in 10 ml PBS, and then dialyzed against PBS and filtered. Anti-RNF4 antibody was isolated by binding the IgG fraction to an RNF4 affinity column that was prepared by incubating 0.4 g dry NHS-activated agarose beads (Pierce) with 5.5 ml PBS containing 10 mg RNF4 as per manufacturer’s instructions. Bound antibody was eluted with 0.1 M glycine (pH 2), and rapidly made 33 mM in Tris (pH 8.5) and 50 µg/ml in BSA. This antibody preparation was dialyzed against PBS and stored at −80°.
Data availability
All yeast strains and plasmids used in this study are presented in Table S1. Strains, plasmids and antibodies are available on request.
Results
Isolation of suppressors of tdp1∆ wss1∆ synthetic lethality
To identify new pathways of DNA repair, we searched for suppressors of the synthetic-lethal interaction between the two repair factors TDP1 and WSS1 (Dixon et al. 2008; Stingele et al. 2014). We confirmed that yeast cells lacking TDP1 and WSS1 fail to form spore clones after tetrad dissection (Figure S1 in File S1). Haploid tdp1∆ wss1∆ cells were rescued by a complementing “balancer” plasmid (pNJ7478 [WSS1/ADE3/URA3/CEN]), and these cells were unable to proliferate when diluted and spotted onto medium containing FOA, which selects against Ura3+ cells (Figure 1A). As described in the Materials and Methods, we used this strain to isolate extragenic suppressor mutants. As summarized in Table 1, the six suppressors were shown to be due to mutations in five genes. With the exception of TOP1, all of the genes are involved in regulating sumoylation. Mutations in TOP1 were expected since loss of TOP1 was previously shown to suppress lethality by eliminating topoisomerase-induced DNA damage (Stingele et al. 2014).
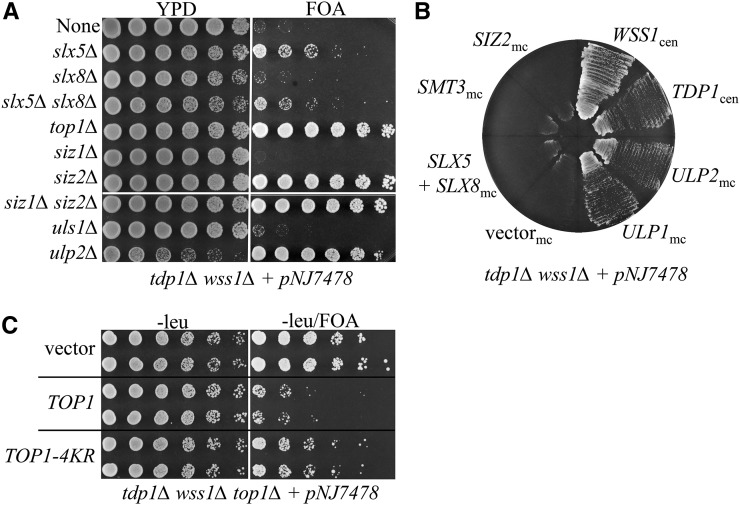
Figure 1.
Suppression of tdp1∆ wss1∆ synthetic lethality by altered SUMO regulation. (A) Haploid tdp1∆ wss1∆ strains containing the balancer plasmid pNJ7478 (WSS1/ADE3/URA3/CEN) and the additional gene deletion(s), as indicated, were serially diluted in 10-fold increments starting at OD600 = 3.0. Five microliters were pinned onto nonselective medium (YPD), or onto SC medium containing FOA. (B) Strain NJY3288 (tdp1∆ wss1∆ plus pNJ7478 [WSS1/ADE3/URA3/CEN]) was transformed with plasmids in which the indicated gene was under the control of the GPD1 promoter in a multi-copy 2µ vector (mc) or its natural promoter in a CEN/ARS (cen) vector. Transformants were streaked onto SC medium containing FOA. (C) Strain NJY4142 (tdp1∆ wss1∆ top1∆ plus pNJ7478 [WSS1/ADE3/URA3/CEN]) was transformed with the indicated TOP1 allele in vector pRS415 (CEN/ARS/LEU2) or vector alone. Transformants were treated as in (A) and pinned onto selective medium in the presence or absence of FOA. TOP1-4KR = TOP1-K65,91,92,600R (Chen et al. 2007).
To confirm the results of the suppressor screen, we tested complete deletions of these and other genes implicated in SUMO metabolism. Complete deletions of SIZ2 and SLX5 suppressed lethality (Figure 1A). However, slx5∆ was not as good a suppressor as siz2∆, nor as good as the original slx5 suppressors (see below). Loss of ULP2 strongly suppressed lethality, whereas deletion of SIZ1 or ULS1 or the mms21-11 mutation (data not shown) did not (Figure 1A). Interestingly, loss of SLX8 did not suppress lethality, even though Slx8 is thought to be required for Slx5–Slx8 Ub ligase activity in vivo. To our knowledge, this is the first case where the phenotypes of slx5∆ and slx8∆ differ. The slx5∆ slx8∆ tdp1∆ wss1∆ quadruple mutant displayed weak suppression like slx5∆ (Figure 1A), suggesting that Slx5 is functioning independently of Slx8. However, based on results below, it is possible that Slx5 antagonizes slx8∆ cells due to its ability to bind poly-SUMO.
Surprisingly, suppression was obtained by mutations that would be expected to either decrease sumoylation (siz2), increase or stabilize sumoylation (ulp2 and slx5), or alter sumoylation patterns (ulp1 and nup60). To further test this idea, we used high-copy expression of various SUMO regulators to test for suppression. Multi-copy plasmids containing the ULP1 or ULP2 SUMO isopeptidase genes suppressed tdp1∆ wss1∆ lethality, whereas those containing SLX5–SLX8, SMT3 or SIZ2 did not (Figure 1B). The smt3-3KR allele, which reduces inter-SUMO linkages, was also found to suppress inviability (strain PSY3621; data not shown). One interpretation of these results is that tdp1∆ wss1∆ cells accumulate harmful sumoylated proteins, and that viability is restored by eliminating the Siz2 SUMO ligase or by overexpressing SUMO isopeptidases.
Because Top1 is a target for sumoylation, and a causal factor in tdp1∆ wss1∆ synthetic lethality, we suspected that a nonsumoylated version of Top1 might be an effective suppressor. The TOP1-K65,91,92,600R allele (here called TOP1-4KR) mutates the major sumoylation sites of Top1 by changing the critical lysine residues to arginine. This allele was previously shown to eliminate most, but not all, Top1 sumoylation (Chen et al. 2007). This plasmid-borne allele was transformed into a tdp1∆ wss1∆ top1∆ tester strain containing the pNJ7478 balancer plasmid, and subjected to the spot dilution assay. As shown in Figure 1C, cells containing TOP1-4KR showed improved growth compared to wt TOP1, but reduced growth compared to top1∆. The partial suppression obtained by TOP1-4KR indicates that Top1 sumoylation contributes to the lethality of tdp1∆ wss1∆ cells. The failure to obtain full suppression may be due to the sumoylation of other proteins, or to residual Top1 sumoylation, which is difficult to completely eliminate by point mutation (Chen et al. 2007).
Suppression by novel alleles of SLX5
The isolation of SLX5 alleles in this screen was unexpected given that TDP1 and SLX5–SLX8 have been reported to show negative genetic interactions by Synthetic Genetic Array (SGA) analysis in S. cerevisiae and synthetic lethality in Schizosaccharomyces pombe (Collins et al. 2007; Dixon et al. 2008; Heideker et al. 2011). However when spores were microdissected and germinated on YPD medium, we found that tdp1∆ slx5∆ and tdp1∆ slx8∆ double-mutant spore clones were viable with no synergistic slow-growth defect (Figure S1A in File S1). Interestingly, spore clones of the double mutants typically lacked the nibbled morphology of slx5∆ and slx8∆ single mutants. Some of this variance in results may be due to the different strain backgrounds or assay conditions used in the two methods. We also observed no obvious growth defects in tdp1∆ mms21-11 or tdp1∆ esc2∆ double mutants (Figure S1B in File S1), which contrasts with the case in S. pombe where tdp1∆ nse2∆ and tdp1∆ rad60∆ double mutants are synthetically lethal (Heideker et al. 2011). These results indicate that, although the pathways for the repair of Top1ccs are highly conserved between the two yeasts, the pathways appear to differ in their relative importance.
The two SLX5 alleles were sequenced, and each was found to encode a single amino acid change. These alleles, slx5-E350K and slx5-Q362K (referred to below as the “slx5-K” alleles) were individually subcloned into a single-copy LEU2-based vector and transformed into a tdp1∆ wss1∆ slx5∆ tester strain that carried the WSS1/URA3 balancer plasmid. The transformants were then subjected to the spot dilution assay in the presence and absence of FOA. As expected, a plasmid carrying wt SLX5 strongly inhibited growth on FOA, while the empty vector allowed slow growth (Figure 2A), which is more obvious at later times (Figure 5B). In contrast to growth inhibition by SLX5, both slx5-K alleles provided good growth. Suppression by these alleles is unlikely to be due simply to elimination of Slx5–Slx8 Ub ligase activity, since an allele that inactivates the Slx5 RING domain failed to suppress inviability (ring−, Figure 2A). Interestingly, good growth was obtained with N-terminal truncations of Slx5, including a 200 aa deletion that eliminates the four SIMs of Slx5 (∆N200, Figure 2A) (see Figure 5 for a schematic of the Slx5 protein). Larger N-terminal truncations failed to suppress inviability presumably because they destabilized the ∆N300 and ∆N400 proteins (Figure S2 in File S1). Since the N-terminal truncations of Slx5 suppressed tdp1∆ wss1∆ lethality, but slx5-ring− and slx8∆ did not, we conclude that optimal suppression requires altered, or reduced, STUbL activity.
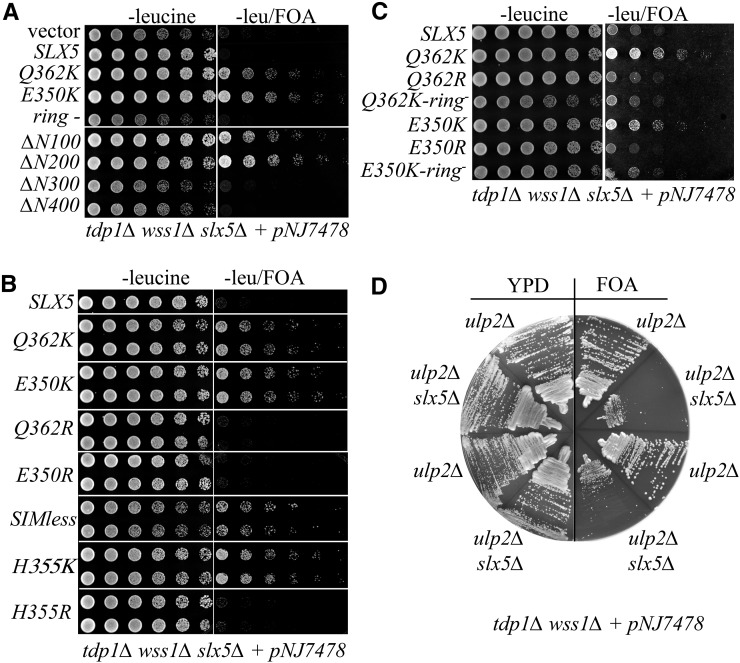
Figure 2.
Role of SLX5 in the suppression of tdp1∆ wss1∆ synthetic lethality. (A) Strain NJY4113 (tdp1∆ wss1∆ slx5∆ plus pNJ7478) was transformed with a LEU2/CEN/ARS plasmid containing either no insert (vector) or the indicated SLX5 allele. Transformants were serially diluted in half-log increments starting at OD600 = 3.0 and 5 µl were spotted onto medium lacking leucine in the presence or absence of FOA. The ring− allele refers to slx5–8, which encodes the RING domain mutations C556S, H558A, and C561S (Ii et al. 2007b), and is otherwise wt. (B) NJY4113 was transformed with single-copy plasmids containing indicated slx5 alleles, and treated as in (A). SIMless refers to the sim-1234 allele with point mutations in the four SIMs as described (Xie et al. 2010). (C) NJY4113 was transformed with single-copy plasmids containing the indicated slx5 alleles, and treated as in (A). (D) Strain PSY3494 (wss1∆ tdp1∆ slx5∆ + pNJ7478) was crossed to NJY4167 (wss1Δ tdp1Δ ulp2-1::HIS3), and the indicated haploid progeny, all containing the pNJ7478 balancer plasmid, were streaked onto YPD or SC medium containing FOA.
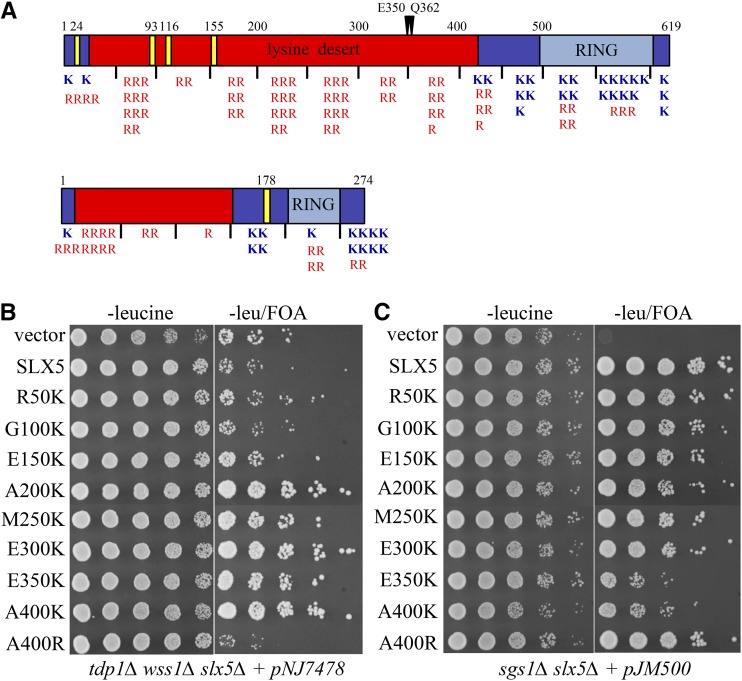
Figure 5.
Identification of a functionally important lysine desert in Slx5. (A) Presented are schematic illustrations of the Slx5 (upper) and Slx8 (lower) proteins with amino acid numbering shown above each schematic. Domains entirely lacking in lysine residues are shown in red. All other domains are shown in blue, including the RING domains shown in light blue. Each lysine (blue) and arginine (red) residue in a protein is depicted by a letter below the schematic within 50-aa bins. Vertical lines below the illustrations mark every 50th residue. The four SIMs of Slx5, and one in Slx8, are indicated as yellow bars. The positions of the E350 and Q362 mutations are indicated above the schematic. (B) The ability of slx5-K variants to suppress tdp1∆ wss1∆ lethality was assayed by transforming strain NJY4113 (tdp1∆ wss1∆ slx5∆ plus pNJ7478) with LEU2/CEN/ARS plasmids containing either no insert, SLX5, or the indicated slx5 variants containing lysine substitutions at 50 aa intervals. Transformants were diluted in fivefold increments and treated as in Figure 2A. (C) Strain JMY2462 (sgs1∆ slx5∆ plus pJM500 [SGS1/ADE3/URA3/CEN]) was treated as in (B) except that transformants were diluted in 10-fold increments.
The fact that the slx5-K alleles create new lysine residues within 12 residues of each other suggested that modification of Slx5 by Ub or SUMO may be responsible for their suppressor function. To test this idea, we transformed the tdp1∆ wss1∆ slx5∆ tester strain described above with plasmids containing other mutations in this region of SLX5, and compared their growth by spot dilution assay. As shown in Figure 2B, wt SLX5 was lethal while alleles bearing E350K, Q362K, or point mutations that eliminate the four SIMs (SIMless) promoted good growth. In contrast, genes with the nonmodifiable slx5-E350R and slx5-Q362R mutations failed to confer growth. Consistent with these results, changing a random, closely placed basic residue to lysine (H355K) suppressed inviability, while H355R did not. These results strongly suggest a role for protein modification by Ub or SUMO in the regulation of the slx5-K alleles. We next tested whether suppression by the slx5-K alleles was dependent on Ub ligase activity by creating compound slx5-K-ring− alleles. These compound alleles lost the ability to suppress (Figure 2C). This confirms that optimal suppression requires both the slx5-K mutations, and at least some Ub ligase activity.
As mentioned above, SLX5 and ULP2 may have been isolated in this screen because mutations in these genes are known to increase global sumoylation levels. It is also known that ulp2∆ slx5∆ double mutants show cosuppression including the loss of distinct ulp2∆ phenotypes, such as temperature and DNA damage sensitivities (Mullen et al. 2011; Gillies et al. 2016). A potential explanation for the negative effect of wt Slx5–Slx8 in ulp2∆ cells is that the ulp2∆ strain accumulates polysumoylated proteins that are inappropriately ubiquitinated and degraded by the STUbL. We therefore hypothesized that the good growth of tdp1∆ wss1∆ ulp2∆ cells should be further improved, or at least unaffected, by the loss of SLX5. Surprisingly, tdp1∆ wss1∆ ulp2∆ slx5∆ cells reverted to poor growth (Figure 2D). Thus, suppression of tdp1∆ wss1∆ by ulp2∆ is dependent on SLX5. The simplest explanation for this result is that the Slx5–Slx8 STUbL targets polysumoylated Top1ccs for proteolytic destruction.
The slx5-K alleles display a hypomorphic phenotype
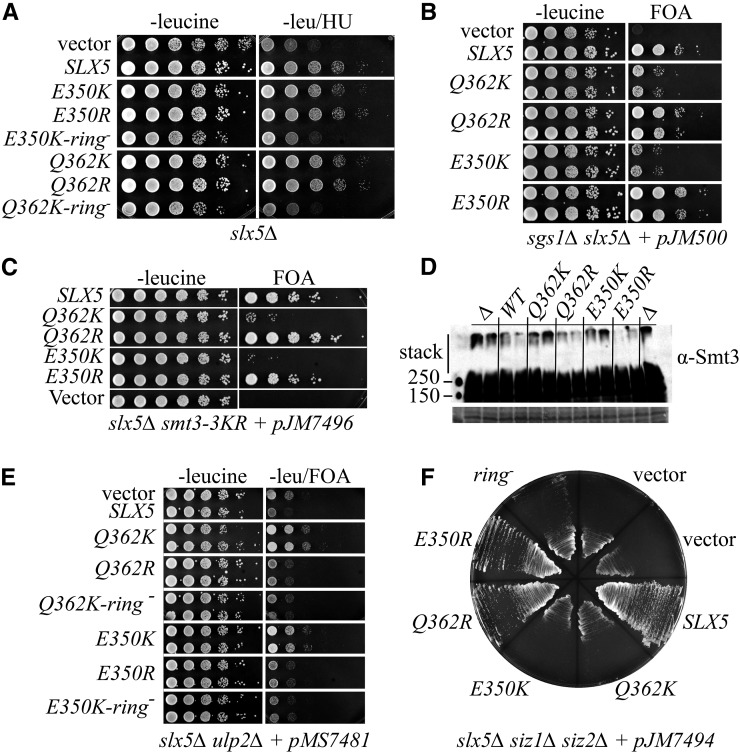
To determine the level of activity retained in the slx5-K alleles, we assayed them for known slx5∆ phenotypes. Compared to slx5∆, the slx5-K strains exhibited only a slight sensitivity to hydroxyurea (HU), indicating that they retain near wt levels of activity based on this assay (Figure 3A). However, the slx5-K alleles only partially complemented the synthetic lethality of slx5∆ sgs1∆ (Figure 3B) and slx5∆ smt3-3KR (Figure 3C) (Mullen et al. 2001, 2011). In both cases, the levels of growth were intermediate to that of wt and null. The slx5-R alleles functioned like wt in both of these synthetic-lethal assays. Another phenotype of slx5∆ cells is the accumulation of high molecular weight poly-SUMO chains that can be detected by immunoblot. Strains bearing slx5-K alleles contained elevated levels of poly-SUMO chains that migrated in the stacking gel following SDS-PAGE, while those bearing slx5-R alleles did not (Figure 3D). While this assay is difficult to quantify, it is consistent with a defect in the slx5-K alleles but not the slx5-R alleles.
Figure 3.
slx5-K suppressor alleles are hypomorphic. (A) Strain PSY3884 (slx5∆) was transformed with the indicated SLX5 allele in a LEU2/CEN/ARS plasmid. Transformants were subjected to 10-fold serial dilutions, and pinned onto selective medium in the presence or absence of 0.1 M HU. (B) Strain JMY2462 (sgs1∆ slx5∆ plus pJM500 [SGS1/ADE3/URA3/CEN]) was treated as in (A) except that transformants were pinned onto selective medium or SC medium containing FOA. (C) Strain JMY3694 (slx5∆ smt3-3KR plus pJM7496 [SLX5/SMT3/ADE3/URA3/CEN]) was treated as in (B). (D) Whole-cell extracts were prepared under denaturing conditions from strains with the indicated SLX5 genotype. Extracts were subjected to SDS-PAGE and immunoblotted with anti-Smt3 antibody. Shown is the high molecular-weight region of the blot including the stacking gel. The lower panel shows a portion of the membrane stained with ponceau S as loading control. (E) Strain PSY3868 (ulp2∆ slx5∆ + pMS7481 [ULP2/ADE3/URA3/CEN]) was treated as in (B) except cells were pinned onto medium lacking leucine in the presence or absence of FOA. (F) Strain PSY3866 (siz1∆ siz2∆ slx5∆ + pJM7494 [SLX5/ADE3/URA3/CEN]) was treated as in (B) except that transformants were streaked onto SC medium containing FOA.
One of the hallmarks of an slx5∆ mutant is its nibbled-colony morphology (Mullen et al. 2001). Following sporulation and tetrad dissection, both slx5-E350K and slx5-Q362K strains revealed this phenotype, while the slx5-R alleles gave rise to spore clones that were round (Figure S3B in File S1). The same result was obtained when slx5∆ cells were transformed with the respective plasmids and spread on agar plates (Figure S3C in File S1). The nibbled phenotype is characteristic of slx5 strains lacking Ub ligase activity (ring−), as well as alleles lacking SUMO binding sites (SIMless) (Figure S3C in File S1) (Xie et al. 2007). Based on colony morphology, the slx5-K alleles are at least partially compromised for function.
Given that slx5∆ suppresses the phenotypes of ulp2∆ cells, we asked whether ulp2∆ slow growth can be suppressed by the slx5-K alleles. When the tester strain ulp2∆ slx5∆ carrying pULP2/URA3 is spotted on FOA, it displays the double mutant phenotype (Figure 3E, top row) in which it grows slightly faster than the same strain forced to carry a wt SLX5/LEU2 plasmid (ulp2∆ phenotype). Interestingly, both slx5-E350K and slx5-Q362K improved the growth of this strain on FOA better than vector alone. The corresponding slx5-R alleles slowed growth like wt SLX5. The improved growth provided by the slx5-K alleles was also dependent on their Ub ligase activity since compound alleles, such as slx5-Q362K/ring−, resembled vector alone. We conclude that the slx5-K alleles display reduced activity, as in other assays, and that the residual STUbL activity of the slx5-K alleles is beneficial to ulp2∆ cells.
An slx5∆ null strain also lacking the SIZ1 and SIZ2 SUMO ligases is very slow growing (Ii et al. 2007b). The siz1∆ siz2∆ slx5∆ triple mutant (complemented with an SLX5/URA3 balancer plasmid) was transformed with various SLX5 alleles on a LEU2/CEN vector, and streaked onto medium containing FOA. This strain grew poorly with vector alone (vector, Figure 3F), but grew well with SLX5. The slx5-K alleles were severely compromised in their ability to complement the siz1∆ siz2∆ slx5∆ slow-growth phenotype, whereas the slx5-R versions complemented like wt. Slx5–Slx8 Ub ligase activity is required for good growth in the siz1∆ siz2∆ background because the slx5-ring− allele failed to promote growth on FOA. Thus, in siz1∆ siz2∆ cells, the slx5-K alleles appear to be essentially null. Taken together, the above assays lead us to conclude that the slx5-K alleles have reduced, but not null, activity. However, it is clear that different assays and genetic backgrounds can vary markedly in their response to limiting amounts of Slx5–Slx8 activity.
The slx5-K alleles display enhanced auto-ubiquitination activity in vitro
The Slx5–Slx8 heterodimer is a Ub ligase that displays auto-ubiquitination activity in vitro (Ii et al. 2007a; Xie et al. 2007). To test whether the slx5-K alleles retain this activity, we purified Slx5–Slx8 complexes containing the slx5-K or slx5-R mutations and assayed them directly. Following the in vitro reactions, the products were analyzed by SDS-PAGE and immunoblotting with antibody against Slx5 (Figure 4). Both slx5-K complexes generated multiple high molecular-weight forms of Slx5 that were not observed with the wt heterodimer or the slx5-R complexes. We conclude that the mutant Slx5-K complexes display enhanced auto-ubiquitination activity of the Slx5 subunit on the respective 350 and 362 K residues.
Figure 4.
Auto-ubiquitination of Slx5/Slx8 is enhanced due to mutant lysine residues. Wild-type and mutant Slx5–Slx8 complexes were incubated under ubiquitination conditions as described in Materials and Methods. Following the reaction, the products were resolved by 10% SDS-PAGE and immunoblotted with antibody against Slx5. The Slx5 subunit was either wt (+) or contained the indicated mutation at position 350 or 362.
Slx5 contains a functional lysine desert
The yeast Slx5 protein contains a C-terminal RING domain at aa 494–619. Its N-terminal domain (1–493 aa) contains four SIMs in the first 160 aa (Figure 5A). Analysis of the basic residues in Slx5 revealed a region of 398 aa within the N-terminal domain in which 15% of the amino acids are arginine residues (32–429 aa). Interestingly, this region is devoid of lysine residues, and is hereafter referred to as the “lysine desert.” The fact that the slx5-K alleles introduce lysines into this domain suggests that the lysine desert is biologically important.
To define the endpoints of the lysine desert functionally, we changed every 50th aa in the Slx5 N-terminus to lysine, and tested whether the resulting mutant suppressed tdp1∆ wss1∆ synthetic lethality. As shown in Figure 5B, lysine residues at positions 50, 100, and 150 repressed growth of tdp1∆ wss1∆ cells, similar to wt SLX5. However, lysines at positions 200, 250, 300, 350, and 400 resulted in better growth, suggesting lower levels of SLX5 activity. As with the original slx5-K alleles, all of these mutants promoted better growth than slx5∆ (vector). As a control, the same plasmids were tested for complementation of sgs1∆ slx5∆ synthetic lethality where SLX5 function is required for viability. Here, Slx5 mutants with lysine residues at positions 50–300 promoted growth that was almost as robust as SLX5 (Figure 5C). But, when lysines were present at aa 350 and 400, SLX5 function was noticeably compromised in the sgs1∆ slx5∆ cells (Figure 5C). Based on this data, we conclude that the C-terminal portion of the lysine desert (200–400 aa) is functionally important, and that lysine residues between aa 350 and 400 have the strongest negative effect on SLX5 function.
Amino acid sequence analysis suggests that the lysine desert is conserved in fungal species where the Slx5 homolog is >∼350 aa, including Saccharomyces, Candida, and Ustilago (Figure S4A in File S1). Less clear are the smaller fungal Slx5 homologs, such as the Rfp proteins of S. pombe (205–254 aa). Many of these smaller Slx5 homologs contain arginine-rich (10–15%) domains that lack lysine and comprise a large portion (50–80%) of the full-length proteins. Similarly, almost all Slx8 homologs are smaller than their Slx5 counterparts (∼250 aa), and many also contain a domain lacking in lysine that comprises a significant proportion (34–66%) of the full length protein (Figure S4B in File S1). Given their small size and the small number of arginines, it is unknown whether any of these smaller proteins contain functional lysine deserts. The homodimeric RNF4 proteins of higher cells are similar to Rfp and Slx8 homologs in that they are small (∼190 aa), and contain relatively large domains with arginine but without lysine. As shown in Figure S4C in File S1, these potential lysine deserts comprise between 43 and 78% of the full-length RNF4 proteins.
To test whether lysine deserts are common in other budding yeast Ub ligases, a similar sequence analysis was performed on yeast SIM-containing proteins, RING-domain Ub ligases, HECT-domain Ub ligases, and SCF subunits (data not shown). Most of the lysine-free domains within this group comprised only ∼10% of the full-length proteins. The only exception is San1 (54% of the full-length protein), which was previously shown to employ a functional lysine desert to prevent auto-ubiquitination (Fredrickson et al. 2013). Thus, large lysine-free domains are rare in budding yeast Ub E3 ligases, with San1 and Slx5 being the only ones known to have functionally important lysine deserts.
Lysine mutations affect the steady-state levels of Slx5 protein
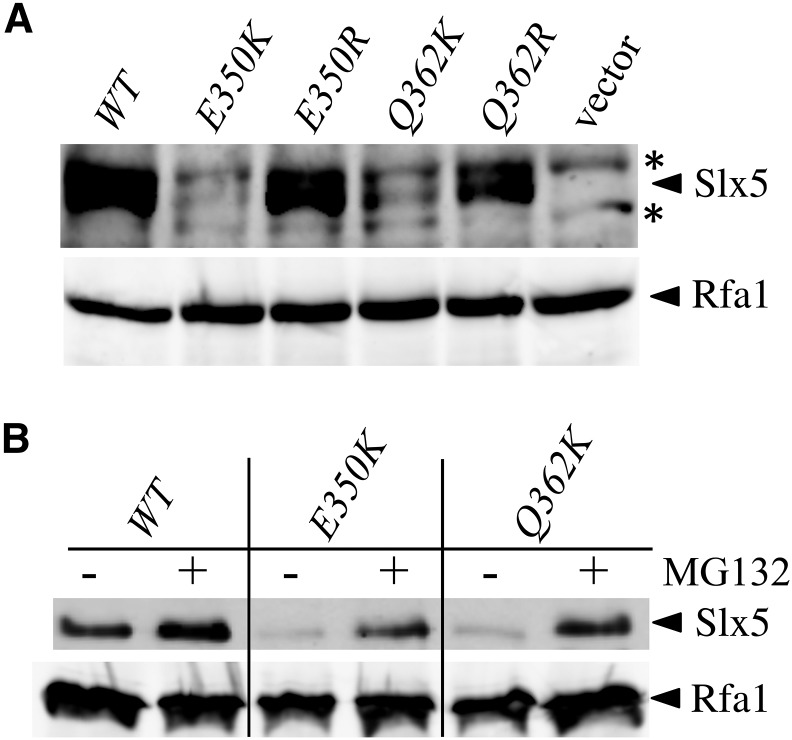
To test the idea that the Slx5-K proteins were regulated by ubiquitin-mediated proteolysis, we used immunoblotting to determine the effect of the slx5-K mutations on protein levels. Extracts were prepared from yeast cells carrying plasmid-borne His6-tagged versions of each allele as their only copy of SLX5. The proteins were then concentrated by binding to Ni-NTA beads and immunoblotted for Slx5. As shown in Figure 6A, Slx5-E350K and Slx5-Q362K proteins were detectable, but at levels that were dramatically reduced compared to wt Slx5. The corresponding Slx5-R proteins were expressed at wt levels consistent with their wt function in genetic assays. To determine whether the levels of the Slx5-K proteins were regulated by the ubiquitin-proteasome system, we repeated this experiment in a strain that is permeable to the proteasome inhibitor MG132. Treatment of these cells with MG132 stabilized the Slx5-K proteins (Figure 6B). The simplest interpretation of these results is that mutant Slx5-K proteins are susceptible to ubiquitination and degradation by the Ub-proteasome system.
Figure 6.
The Slx5-K mutant proteins are ubiquitinated and degraded by the proteasome. (A) Yeast strain PSY3884 (slx5∆) was transformed with LEU2/CEN/ARS plasmids containing either no insert (vector), wt SLX5, or the indicated mutant allele of SLX5 and grown in liquid medium lacking leucine. All inserts express N-terminal His6-tagged Slx5 protein. Extracts were prepared and incubated with Ni-NTA agarose beads under denaturing conditions. Bound proteins were then immunoblotted with anti-Slx5 antiserum (upper). Equal volumes of extract were separately immunoblotted and probed with anti-Rfa1 antibody as a control for the pulldown (lower). * indicates nonspecific cross-reacting bands. (B) Yeast strain NJY3912 (slx5∆ pdr5∆ erg6∆) was transformed with the indicated plasmids and treated as in (A), but, prior to harvesting the cells, they were incubated for 1 hr in the presence of 75 µM MG132 in DMSO (+), or in DMSO alone (−).
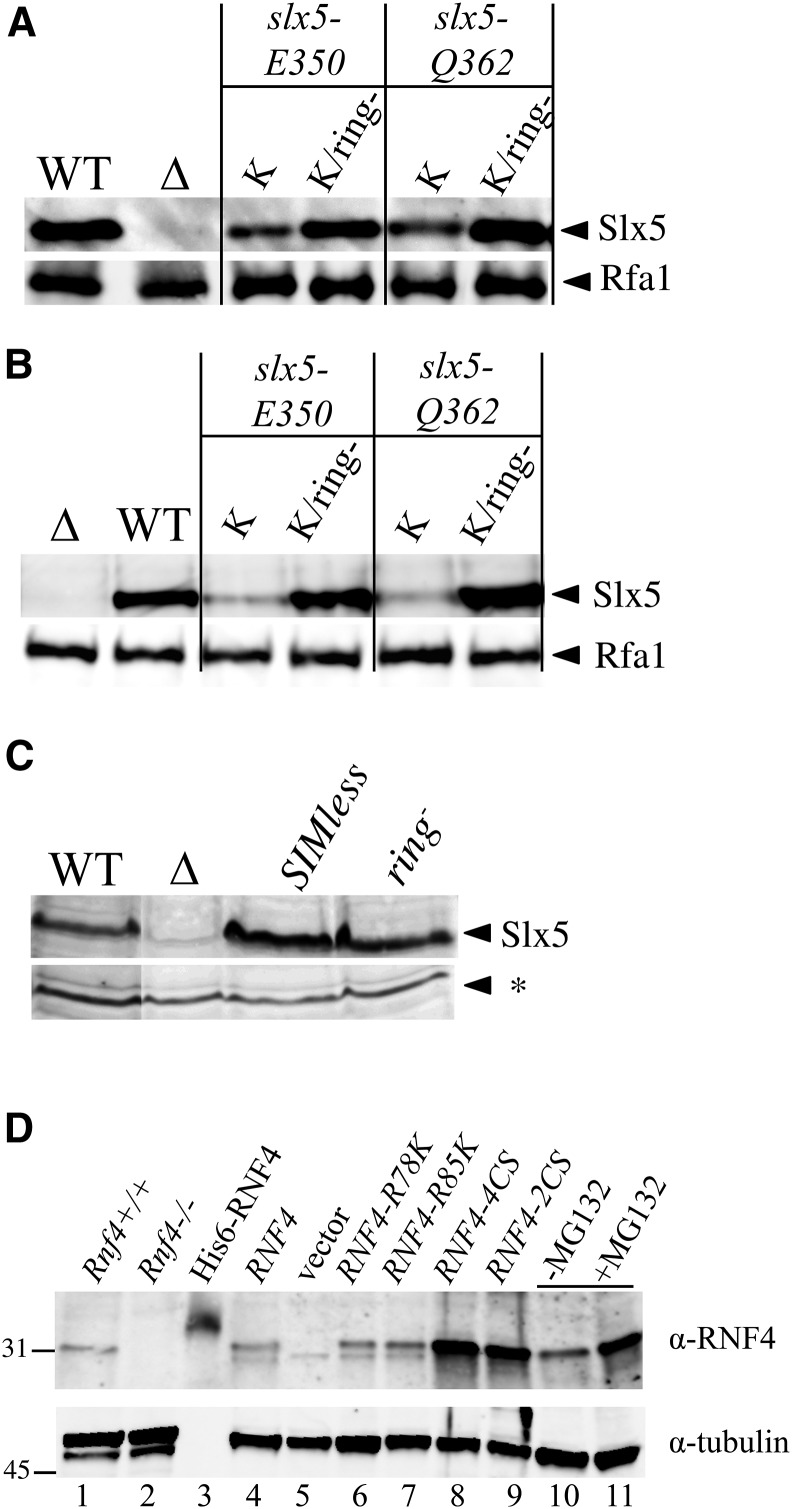
If the low levels of the Slx5-K proteins are due to auto-ubiquitination, then a compound allele encoding Slx5-K but lacking Ub ligase activity would be expected to restore its abundance. As shown in Figure 7A, the intensities of the two mutant Slx5-K proteins were increased to wt levels when they also contained the ring− mutation. This suggests that the Slx5-K protein is undergoing auto-ubiquitination in cis, but it does not rule out the possibility that ubiquitination is happening in trans. We therefore performed the same experiment in a strain expressing both untagged wt Slx5 and His6-tagged Slx5-K-ring− to test whether the Slx5-K protein is susceptible to ubiquitination in trans. Following Ni pull-down, the His6-Slx5-K-ring− proteins were again found to be elevated to wt levels (Figure 7B). This indicates that wt STUbL is unable to ubiquitinate Slx5-K-ring− in trans.
Figure 7.
Slx5-K proteins undergo auto-ubiquitination in cis, and are unique to the yeast STUbL. (A) Strain PSY3884 (slx5∆) was transformed with LEU2/CEN/ARS plasmids that contained either SLX5 (WT), no insert (∆), the indicated slx5-K mutant, or a compound allele also containing the ring− mutation. All inserts express N-terminal His6-tagged Slx5 protein. Cell extracts were incubated with Ni-NTA agarose beads, and bound proteins were immunoblotted as in Figure 6A. (B) Wild-type strain JMY3107 (SLX5) was treated as in (A). (C) Strain PSY3884 (slx5∆) was transformed with plasmids containing the indicated SLX5 alleles and treated as in (A), except that extracts were directly immunoblotted for Slx5. A nonspecific band (*) on the blot is used as an internal loading control. (D) Mammalian cell extracts were prepared from wt MEFs (lane 1), Rnf4−/− MEFs (lane 2), or from Rnf4−/− MEFs that were stably transduced with empty vector or the indicated RNF4 alleles (lanes 4–9). Cell extracts (25 µg) were immunoblotted with anti-RNF4 antibodies (upper), and the blot was reprobed with antibody to α-tubulin as control (lower). Extracts were also prepared from wt RNF4 transductants that had been treated for 4 hr with 20 µM MG132 in DMSO (+) or with DMSO alone (−) (lanes 10 and 11). Purified His6-RNF4 (80 pg) was included as positive control (lane 3).
Several results suggest that the lysine desert does not completely protect wt Slx5 from auto-ubiquitination. First, wt Slx5 levels were seen to increase upon treatment with MG132 (Figure 6C). Second, the abundance of Slx5-ring− protein is elevated relative to wt Slx5, as judged by a simple immunoblotting (Figure 7C). Finally, the abundance of Slx5-SIMless protein is elevated relative to wt Slx5 (Figure 7C), suggesting that auto-ubiquitination occurs when it is bound to poly-SUMO chains. In the case of mammalian RNF4, SUMO chains have been shown to stimulate its homo-dimerization and auto-ubiquitination (Rojas-Fernandez et al. 2014), so this characteristic appears to be conserved between yeast and mammalian homologs despite their different subunit composition. Taken together, these results suggest that the lysine desert of Slx5 plays an important role in protecting it from excessive degradation.
While it is known that RNF4 is similarly subject to auto-ubiquitination in vivo (Hakli et al. 2004; Rojas-Fernandez et al. 2014), it was of interest to test whether the lysine-poor N-terminus of mammalian RNF4 was functionally important to protect it from excessive degradation. We therefore introduced lysines into this region of the mammalian RNF4 protein to test its stability. We first established an Rnf4−/− MEF cell line from the previously described gene-trap mouse (Hu et al. 2010). Immunoblotting with a rabbit antiserum against RNF4 demonstrated that this cell line lacked detectable RNF4 compared to wild-type MEFs (Figure 7D, lanes 1 and 2). Mutant and wt versions of RNF4 were then stably introduced into the cell line by retroviral transduction. Transductants expressing wt RNF4 or versions containing the R78 or R85 K mutations produced similar levels of protein (Figure 7D, lanes 4–7). The levels of RNF4-H67K protein were also unchanged from wt (data not shown). In contrast, the abundance of RNF4 proteins deficient in Ub ligase activity (RNF4-4CS or RNF4-2CS) was elevated compared to wt RNF4, and transduced wt RNF4 was stabilized by the proteasome inhibitor MG132 (Figure 7D, lanes 8–11). The fact that RNF4 protein levels were unaffected by the introduction of lysine residues indicates that it lacks the functional lysine desert found in the yeast ortholog.
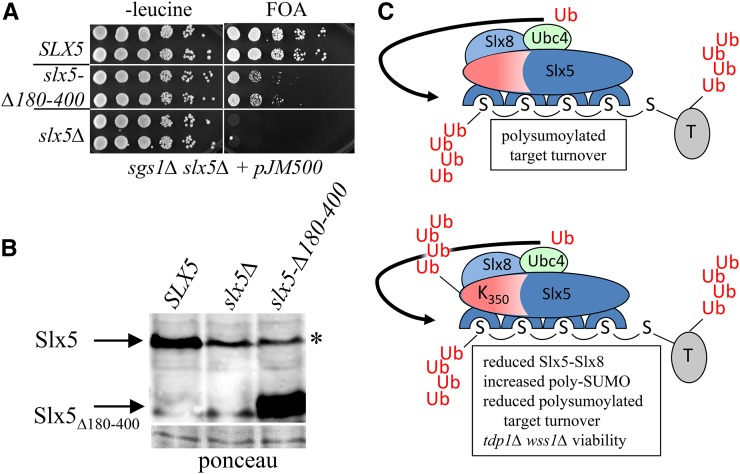
Because the lysine desert is not found in mammalian RNF4, and does not appear to be present in other small orthologs, we asked whether it was important for Slx5 function in budding yeast. A deletion of residues 180–400 removes just over half of the lysine desert while leaving intact the four N-terminal SIMs and C-terminal RING finger. To ensure optimal expression, the slx5-∆180-400 allele was cloned downstream of the strong constitutive GPD1 promoter and shown to rescue sgs1∆ slx5∆ synthetic lethality (Figure 8A). This suggests that the Slx5∆180–400 protein interacts productively with Slx8 to provide STUbL activity. However, compared to wt SLX5, the rescued strain is slow growing, which suggests that the deleted region is important for Slx5–Slx8 function. The slow-growth phenotype cannot be due to an unstable protein because the Slx5∆180–400 protein was well expressed (Figure 8B). We conclude that the lysine desert exists to protect a functionally important domain of Slx5 that is not conserved in smaller versions of this STUbL.
Figure 8.
The lysine desert is important for Slx5 function. (A) Strain JMY2462 (sgs1∆ slx5∆ plus pJM500 [SGS1/ADE3/URA3/CEN]) was transformed with a LEU2/CEN/ARS plasmid carrying either SLX5, no insert (slx5∆) or slx5-∆180-400 under the control of the GPD1 promoter. Transformants were serially diluted in 10-fold increments, and 5 µl was pinned onto selective medium or SC medium containing FOA. (B) PSY3884 (slx5∆) was transformed with the same plasmids as in (A), and cell extracts were immunoblotted with antibody against Slx5. A portion of the ponceau S-stained filter is shown as a loading control. (C) The upper panel illustrates a model of wt Slx5-Slx8 binding to a poly-SUMO chain as a heterodimer via the four SIMs of Slx5. The Ub ligase stimulates Ubc4 to ubiquitinate the poly-SUMO chain at its N-terminus as well as the sumoylated target protein (T). Auto-ubiquitination of Slx5 is minimized by the lysine desert in red. The lower panel illustrates how the Slx5-E350K (or Slx5-Q352) mutation provides a target for auto-ubiquitination within the lysine desert leading to polyubiquitination.
Discussion
The synthetic lethality of tdp1∆ wss1∆ cells is most easily explained by the redundancy of these factors in removing Top1ccs (Stingele et al. 2014; Balakirev et al. 2015). Our isolation of suppressor mutations in genes known to affect sumoylation strongly suggests that these factors are regulated by sumoylation. This is consistent with previous studies implicating sumoylation in Top1-dependent DNA damage repair. The synthetic growth defects observed in rad52siz1siz2 or rad52ulp1 mutants in budding yeast, or pli1 rhp51 mutants in S. pombe, are all relieved by deletion of TOP1, as is the hyper-recombination phenotype of pli1 mutants (chen et al. 2007; Prudden et al. 2011; Steinacher et al. 2013). The implication of these studies is that Top1 is the direct target of sumoylation; however, sumoylation of other proteins, such as Tdp1 (Hudson et al. 2012), may also play a role. In mammalian cells, it is thought that Top1 sumoylation leads to the proteasomal degradation of Top1ccs prior to Tdp1 cleavage (Mao et al. 2000; Lin et al. 2008; Interthal and Champoux 2011). In budding yeast, Top1 sumoylation has been implicated in Wss1-dependent proteolysis (Balakirev et al. 2015), while in fission yeast it has been implicated in the Rad16-Swi10/Rad1-10 pathway that relies on Rad60, Nse2, and the Slx8 STUbL (Heideker et al. 2011).
Based on the above studies, it is tempting to speculate that Top1 is the critical SUMO target that mediates tdp1∆ wss1∆ cell inviability. In this model, wt levels of Top1 sumoylation constrain the repair of Top1ccs to the Tdp1 or Wss1 pathways such that lethality results in their absence. By altering Top1 sumoylation, other DNA repair factors such as the nucleases MRX, Mus81-Mms4, or Slx1–Slx4 could be recruited to sites of damage. Consistent with this model, we showed that mutation of Top1’s major sumoylation sites partially suppressed tdp1∆ wss1∆ synthetic lethality. In this case, the TOP1-4KR suppressed strain did not grow as well as top1∆. This could be due to residual sumoylation occurring at one of the other 100 lysine residues in Top1, or to the sumoylation of Top1-associated proteins such as Tri1 (Chen et al. 2007).
The suppression of tdp1∆ wss1∆ inviability by decreased sumoylation was extremely effective. In fact, suppression by siz2∆ rivaled that of top1∆. This suppression likely occurs via the same mechanism as mutating Top1 sumoylation sites, although siz2∆ is clearly more efficient. As suggested above, sumoylation of Top1ccs in otherwise wt tdp1∆ wss1∆ cells would be expected to play a negative role by blocking alternative pathways. This could be due to SUMO recruiting SIM-containing inhibitory proteins. By eliminating the Siz2 SUMO ligase, Top1cc sumoylation is reduced, inhibitory proteins do not bind, and Top1ccs are accessible to other repair pathways.
Like siz2∆, the nup60 and ulp1 mutations isolated here probably suppress tdp1∆ wss1∆ inviability by reducing Top1cc sumoylation. Ulp1 is tethered to the nuclear envelope by its N-terminus and intact nuclear pores (Li and Hochstrasser 2003; Zhao et al. 2004; Lewis et al. 2007), so we expect that the nup60 and ulp1 mutations delocalize Ulp1. It is difficult to quantify the effect of Ulp1 delocalization on global sumoylation levels. But because Ulp1 is needed to generate mature Smt3, ULP1 mutations could negatively affect sumoylation. Indeed, it has been observed that ulp1 phenotypes closely resemble those of the siz1siz2 mutant (Chen et al. 2007).
Suppression by increased or stabilized sumoylation was very effective in the case of ulp2∆, and it is plausible that Top1ccs are the target of this hypersumoylation. If the poly-SUMO chains observed in ulp2∆ cells accumulate on Top1ccs, they might recruit novel repair factors. One of the best known functions of poly-SUMO chains is to recruit STUbLs, and, in certain cases, the poly-SUMO chains that accumulate in ulp2∆ cells have been shown to be substrates of the Slx5–Slx8 STUbL (Gillies et al. 2016). This suggests that the Slx5–Slx8 STUbL is recruited to these chains to ubiquitinate and proteolyze Top1ccs. Such an idea is consistent with the fact that suppression by ulp2∆ was dependent on SLX5 (Figure 2C). Moreover, it has previously been shown that the loss of SLX5 is associated with an increase in the level of sumoylated Top1 (Albuquerque et al. 2013).
The above model may seem inconsistent with the fact that mutant alleles of Slx5 were isolated as suppressors in this screen. However, the slx5∆ null was a poor suppressor. Although the slx5-K alleles led to the accumulation of poly-SUMO chains, optimal suppression by these alleles was dependent on Ub ligase activity. We suggest that the slx5-K’s are Goldilocks alleles whose Slx5–Slx8 levels are low enough to stimulate polysumoylation, and whose Ub ligase levels are high enough to target polysumoylated Top1ccs for proteolysis. More surprising was the ability of slx5∆ to suppress tdp1∆ wss1∆ at all. This indicates the presence of yet another pathway, perhaps one involving a minor STUbL such as Uls1. Clearly, additional experiments will be needed to test these models. Biochemical experiments have the potential to confirm whether Top1ccs are the target of Slx5–Slx8 Ub ligase in ulp2∆ cells, and epistasis experiments could identify the repair pathways that eliminate Top1ccs in siz2∆ and slx5∆ cells.
Slx5-Slx8 differs from RNF4 in several ways that are likely to be meaningful. First, the Slx5 subunit is large (∼600 aa) compared to RNF4 (∼200 aa). The disparity in size between Slx5 and RNF4 is largely accounted for by the 400 aa lysine desert domain identified here. This lysine desert is highly conserved among heterodimeric STUbLs, with large Slx5 subunits and is especially common in fungi. Fungi with smaller Slx5 homologs (∼200 aa), including the versions found in S. pombe (Rfp1 and Rfp2), lack the domain. This suggests that the domain may confer unique activities on the budding yeast STUbL.
A second distinction is that poly-SUMO chains activate RNF4 by dimerizing RNF4 monomers that individually bind the chain (Rojas-Fernandez et al. 2014). In budding yeast, poly-SUMO chains are not needed for formation of the Slx5–Slx8 heterodimer. These subunits are found stably associated in yeast extracts and in recombinant systems. As shown in Figure 8C, the SIMs of Slx5 interact with the poly-SUMO chain, which stimulates the ubiquitination of the chain (Mullen and Brill 2008) and presumably the target protein. Why employ a lysine desert when eliminating the domain entirely would achieve the same goal? One reason is that the SIMs of Slx5 are dispersed over 155 aa instead of 50 aa in its smaller orthologs. This larger, functionally important, region may require protection from auto-ubiquitination. Perhaps the remaining portion of the domain (Slx5180–400), is retained to assist Slx5–Slx8 dimerization by interacting with Slx8. If deletion of this domain diminished the Slx5–Slx8 interaction it would be expected to compromise STUbL activity, which is what we observed.
A final distinction is that budding yeast SLX5–SLX8 displays genetic interactions that are not observed in other model systems. For example, although STUbL mutants in fission and budding yeast display similar phenotypes, including accumulation of poly-SUMO conjugates and genome instability, these phenotypes are suppressed in fission yeast by eliminating poly-SUMO chains with a nonconjugable SUMO, or by deletion of the SUMO E3 ligase (Prudden et al. 2007, 2011). But, in budding yeast STUbL mutants, the corresponding mutations, smt3-3KR and siz1∆ siz2∆, result in synthetic lethality and synthetic sickness, respectively (Ii et al. 2007b; Mullen et al. 2011). These results have led to the paradoxical conclusion that polysumoylation is essential in the absence of budding yeast STUbL activity. This suggests that Slx5 shares a function with poly-SUMO. It will be of interest to test the role of the lysine desert in this and other unique aspects of the budding yeast STUbL.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.202697/-/DC1.
Acknowledgments
We thank Anupama Sureshchandra and Letzibeth Mendez-Rivera for technical assistance, Erica Johnson and Xiaolan Zhao for yeast strains, Jorma Palvimo for plasmids, and Xiaolu Yang for providing Rnf4+/− mice. This work was supported by grants from the National Institutes of Health to S.J.B. (GM101613), S.F.B. (CA190858), and M.Z. (GM105831).
Footnotes
Communicating editor: N. Hunter
Literature Cited
- Adams A., Gottschling D. E., Kaiser C. A., Stearns T., 1997. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Albuquerque C. P., Wang G., Lee N. S., Kolodner R. D., Putnam C. D., et al. , 2013. Distinct SUMO ligases cooperate with Esc2 and Slx5 to suppress duplication-mediated genome rearrangements. PLoS Genet. 9: e1003670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakirev M. Y., Mullally J. E., Favier A., Assard N., Sulpice E., et al. , 2015. Wss1 metalloprotease partners with Cdc48/Doa1 in processing genotoxic SUMO conjugates. Elife 4: 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Sollier J., Liberi G., Zhao X., Maeda D., et al. , 2006. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509–522. [DOI] [PubMed] [Google Scholar]
- Burgess R. C., Rahman S., Lisby M., Rothstein R., Zhao X., 2007. The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol. Cell. Biol. 27: 6153–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylebyl G. R., Belichenko I., Johnson E. S., 2003. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 278: 44113–44120. [DOI] [PubMed] [Google Scholar]
- Chen X. L., Silver H. R., Xiong L., Belichenko I., Adegite C., et al. , 2007. Topoisomerase I-dependent viability loss in Saccharomyces cerevisiae mutants defective in both SUMO conjugation and DNA repair. Genetics 177: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S. R., Miller K. M., Maas N. L., Roguev A., Fillingham J., et al. , 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810. [DOI] [PubMed] [Google Scholar]
- Cremona C. A., Sarangi P., Yang Y., Hang L. E., Rahman S., et al. , 2012a Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol. Cell 45: 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona C. A., Sarangi P., Zhao X., 2012b Sumoylation and the DNA damage response. Biomolecules 2: 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C., Brown J. A., You D., Brown J. M., 2005. Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics 170: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai S. D., Li T. K., Rodriguez-Bauman A., Rubin E. H., Liu L. F., 2001. Ubiquitin/26S proteasome-mediated degradation of topoisomerase I as a resistance mechanism to camptothecin in tumor cells. Cancer Res. 61: 5926–5932. [PubMed] [Google Scholar]
- Dixon S. J., Fedyshyn Y., Koh J. L., Prasad T. S., Chahwan C., et al. , 2008. Significant conservation of synthetic lethal genetic interaction networks between distantly related eukaryotes. Proc. Natl. Acad. Sci. USA 105: 16653–16658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrickson E. K., Clowes Candadai S. V., Tam C. H., Gardner R. G., 2013. Means of self-preservation: how an intrinsically disordered ubiquitin-protein ligase averts self-destruction. Mol. Biol. Cell 24: 1041–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y., Belotserkovskaya R., Coates J., Jackson S. P., 2012. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 26: 1179–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies J., Hickey C. M., Su D., Wu Z., Peng J., et al. , 2016. SUMO pathway modulation of regulatory protein binding at the ribosomal DNA locus in Saccharomyces cerevisiae. Genetics 202: 1377–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H., 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Golebiowski F., Matic I., Tatham M. H., Cole C., Yin Y., et al. , 2009. System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2: ra24. [DOI] [PubMed] [Google Scholar]
- Guldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. H., 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24: 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn W. C., Dessain S. K., Brooks M. W., King J. E., Elenbaas B., et al. , 2002. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol. Cell. Biol. 22: 2111–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakli M., Lorick K. L., Weissman A. M., Janne O. A., Palvimo J. J., 2004. Transcriptional coregulator SNURF (RNF4) possesses ubiquitin E3 ligase activity. FEBS Lett. 560: 56–62. [DOI] [PubMed] [Google Scholar]
- Hartsuiker E., Neale M. J., Carr A. M., 2009. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell 33: 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heideker J., Prudden J., Perry J. J., Tainer J. A., Boddy M. N., 2011. SUMO-targeted ubiquitin ligase, Rad60, and Nse2 SUMO ligase suppress spontaneous Top1-mediated DNA damage and genome instability. PLoS Genet. 7: e1001320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano R., Interthal H., Huang C., Nakamura T., Deguchi K., et al. , 2007. Spinocerebellar ataxia with axonal neuropathy: consequence of a Tdp1 recessive neomorphic mutation? EMBO J. 26: 4732–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoa N. N., Shimizu T., Zhou Z. W., Wang Z. Q., Deshpande R. A., et al. , 2016. Mre11 is essential for the removal of lethal topoisomerase 2 covalent cleavage complexes. Mol. Cell 64: 580–592. [DOI] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S., 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141. [DOI] [PubMed] [Google Scholar]
- Horie K., Tomida A., Sugimoto Y., Yasugi T., Yoshikawa H., et al. , 2002. SUMO-1 conjugation to intact DNA topoisomerase I amplifies cleavable complex formation induced by camptothecin. Oncogene 21: 7913–7922. [DOI] [PubMed] [Google Scholar]
- Hu X. V., Rodrigues T. M., Tao H., Baker R. K., Miraglia L., et al. , 2010. Identification of RING finger protein 4 (RNF4) as a modulator of DNA demethylation through a functional genomics screen. Proc. Natl. Acad. Sci. USA 107: 15087–15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. Y., Murai J., Dalla Rosa I., Dexheimer T. S., Naumova A., et al. , 2013. TDP1 repairs nuclear and mitochondrial DNA damage induced by chain-terminating anticancer and antiviral nucleoside analogs. Nucleic Acids Res. 41: 7793–7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson J. J., Chiang S. C., Wells O. S., Rookyard C., El-Khamisy S. F., 2012. SUMO modification of the neuroprotective protein TDP1 facilitates chromosomal single-strand break repair. Nat. Commun. 3: 733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ii T., Fung J., Mullen J. R., Brill S. J., 2007a The yeast Slx5-Slx8 DNA integrity complex displays ubiquitin ligase activity. Cell Cycle 6: 2800–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ii T., Mullen J. R., Slagle C. E., Brill S. J., 2007b Stimulation of in vitro sumoylation by Slx5-Slx8: evidence for a functional interaction with the SUMO pathway. DNA Repair (Amst.) 6: 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Interthal H., Champoux J. J., 2011. Effects of DNA and protein size on substrate cleavage by human tyrosyl-DNA phosphodiesterase 1. Biochem. J. 436: 559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Interthal H., Chen H. J., Kehl-Fie T. E., Zotzmann J., Leppard J. B., et al. , 2005. SCAN1 mutant Tdp1 accumulates the enzyme–DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 24: 2224–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalal D., Chalissery J., Hassan A. H., 2017. Genome maintenance in Saccharomyces cerevisiae: the role of SUMO and SUMO-targeted ubiquitin ligases. Nucleic Acids Res. 45: 2242–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. S., Gupta A. A., 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106: 735–744. [DOI] [PubMed] [Google Scholar]
- Johnson E. S., Schwienhorst I., Dohmen R. J., Blobel G., 1997. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16: 5509–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G. M., Stalker J., Humphray S., West A., Cox T., et al. , 2008. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat. Methods 5: 239–241. [DOI] [PubMed] [Google Scholar]
- Jozefczuk J., Drews K., Adjaye J., 2012. Preparation of mouse embryonic fibroblast cells suitable for culturing human embryonic and induced pluripotent stem cells. J. Vis. Exp. 64: pii: 3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keusekotten K., Bade V. N., Meyer-Teschendorf K., Sriramachandran A. M., Fischer-Schrader K., et al. , 2014. Multivalent interactions of the SUMO-interaction motifs in RING finger protein 4 determine the specificity for chains of the SUMO. Biochem. J. 457: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T., 1998. New experimental approaches in retrovirus-mediated expression screening. Int. J. Hematol. 67: 351–359. [DOI] [PubMed] [Google Scholar]
- Kosoy A., Calonge T. M., Outwin E. A., O’Connell M. J., 2007. Fission yeast Rnf4 homologs are required for DNA repair. J. Biol. Chem. 282: 20388–20394. [DOI] [PubMed] [Google Scholar]
- Langmead B., Schatz M. C., Lin J., Pop M., Salzberg S. L., 2009. Searching for SNPs with cloud computing. Genome Biol. 10: R134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A., Felberbaum R., Hochstrasser M., 2007. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J. Cell Biol. 178: 813–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. J., Hochstrasser M., 2000. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20: 2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. J., Hochstrasser M., 2003. The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J. Cell Biol. 160: 1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. P., Ban Y., Lyu Y. L., Desai S. D., Liu L. F., 2008. A ubiquitin-proteasome pathway for the repair of topoisomerase I-DNA covalent complexes. J. Biol. Chem. 283: 21074–21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Pouliot J. J., Nash H. A., 2002. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc. Natl. Acad. Sci. USA 99: 14970–14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Sun M., Desai S. D., Liu L. F., 2000. SUMO-1 conjugation to topoisomerase I: a possible repair response to topoisomerase-mediated DNA damage. Proc. Natl. Acad. Sci. USA 97: 4046–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Desai S. D., Ting C. Y., Hwang J., Liu L. F., 2001. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J. Biol. Chem. 276: 40652–40658. [DOI] [PubMed] [Google Scholar]
- Motegi A., Kuntz K., Majeed A., Smith S., Myung K., 2006. Regulation of gross chromosomal rearrangements by ubiquitin and SUMO ligases in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 1424–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J. R., Brill S. J., 2008. Activation of the Slx5-Slx8 ubiquitin ligase by poly-SUMO conjugates. J. Biol. Chem. 283: 19912–19921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J. R., Kaliraman V., Ibrahim S. S., Brill S. J., 2001. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157: 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J. R., Das M., Brill S. J., 2011. Genetic evidence that polysumoylation bypasses the need for a SUMO-targeted Ub ligase. Genetics 187: 73–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss K. C., Malik M., He X., White S. W., Nitiss J. L., 2006. Tyrosyl-DNA phosphodiesterase (Tdp1) participates in the repair of Top2-mediated DNA damage. Proc. Natl. Acad. Sci. USA 103: 8953–8958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B., Moldovan G. L., Sacher M., Hoege C., Jentsch S., 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433. [DOI] [PubMed] [Google Scholar]
- Plechanovova A., Jaffray E. G., McMahon S. A., Johnson K. A., Navratilova I., et al. , 2011. Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat. Struct. Mol. Biol. 18: 1052–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y., Barcelo J. M., Rao V. A., Sordet O., Jobson A. G., et al. , 2006. Repair of topoisomerase I-mediated DNA damage. Prog. Nucleic Acid Res. Mol. Biol. 81: 179–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y., Huang S. Y., Gao R., Das B. B., Murai J., et al. , 2014. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair (Amst.) 19: 114–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts P. R., Yu H., 2005. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell. Biol. 25: 7021–7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouliot J. J., Yao K. C., Robertson C. A., Nash H. A., 1999. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 286: 552–555. [DOI] [PubMed] [Google Scholar]
- Prudden J., Pebernard S., Raffa G., Slavin D. A., Perry J. J., et al. , 2007. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 18: 4089–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J., Perry J. J., Nie M., Vashisht A. A., Arvai A. S., et al. , 2011. DNA repair and global sumoylation are regulated by distinct Ubc9 noncovalent complexes. Mol. Cell. Biol. 31: 2299–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye I., Jentsch S., 2012. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151: 807–820. [DOI] [PubMed] [Google Scholar]
- Regairaz M., Zhang Y. W., Fu H., Agama K. K., Tata N., et al. , 2011. Mus81-mediated DNA cleavage resolves replication forks stalled by topoisomerase I-DNA complexes. J. Cell Biol. 195: 739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D. F., Bothmer A., Callen E., Reina-San-Martin B., Dorsett Y., et al. , 2008. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell 135: 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Fernandez A., Plechanovova A., Hattersley N., Jaffray E., Tatham M. H., et al. , 2014. SUMO chain-induced dimerization activates RNF4. Mol. Cell 53: 880–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M., Pfander B., Hoege C., Jentsch S., 2006. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat. Cell Biol. 8: 1284–1290. [DOI] [PubMed] [Google Scholar]
- Schimmel J., Larsen K. M., Matic I., van Hagen M., Cox J., et al. , 2008. The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol. Cell. Proteomics 7: 2107–2122. [DOI] [PubMed] [Google Scholar]
- Soustelle C., Vernis L., Freon K., Reynaud-Angelin A., Chanet R., et al. , 2004. A new Saccharomyces cerevisiae strain with a mutant Smt3-deconjugating Ulp1 protein is affected in DNA replication and requires Srs2 and homologous recombination for its viability. Mol. Cell. Biol. 24: 5130–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikumar T., Lewicki M. C., Costanzo M., Tkach J. M., van Bakel H., et al. , 2013. Global analysis of SUMO chain function reveals multiple roles in chromatin regulation. J. Cell Biol. 201: 145–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriramachandran A. M., Dohmen R. J., 2014. SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta 1843: 75–85. [DOI] [PubMed] [Google Scholar]
- Steinacher R., Osman F., Lorenz A., Bryer C., Whitby M. C., 2013. Slx8 removes Pli1-dependent protein-SUMO conjugates including SUMOylated topoisomerase I to promote genome stability. PLoS One 8: e71960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele J., Schwarz M. S., Bloemeke N., Wolf P. G., Jentsch S., 2014. A DNA-dependent protease involved in DNA-protein crosslink repair. Cell 158: 327–338. [DOI] [PubMed] [Google Scholar]
- Stingele J., Bellelli R., Alte F., Hewitt G., Sarek G., et al. , 2016. Mechanism and regulation of DNA-protein crosslink repair by the DNA-dependent metalloprotease SPRTN. Mol. Cell 64: 688–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Leverson J. D., Hunter T., 2007. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 18: 4102–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima H., Boerkoel C. F., John J., Saifi G. M., Salih M. A., et al. , 2002. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 32: 267–272. [DOI] [PubMed] [Google Scholar]
- Tatham M. H., Jaffray E., Vaughan O. A., Desterro J. M., Botting C. H., et al. , 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276: 35368–35374. [DOI] [PubMed] [Google Scholar]
- Tatham M. H., Geoffroy M. C., Shen L., Plechanovova A., Hattersley N., et al. , 2008. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10: 538–546. [DOI] [PubMed] [Google Scholar]
- Tatham M. H., Matic I., Mann M., Hay R. T., 2011. Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci. Signal. 4: rs4. [DOI] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R., 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56: 619–630. [DOI] [PubMed] [Google Scholar]
- Uzunova K., Gottsche K., Miteva M., Weisshaar S. R., Glanemann C., et al. , 2007. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 282: 34167–34175. [DOI] [PubMed] [Google Scholar]
- Vance J. R., Wilson T. E., 2002. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl. Acad. Sci. USA 99: 13669–13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz B., Popovic M., Newman J. A., Fielden J., Aitkenhead H., et al. , 2016. Metalloprotease SPRTN/DVC1 orchestrates replication-coupled DNA-protein crosslink repair. Mol. Cell 64: 704–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M., Hakonarson H., 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Prelich G., 2009. Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol. Cell. Biol. 29: 1694–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Kerscher O., Kroetz M. B., McConchie H. F., Sung P., et al. , 2007. The yeast HEX3–SLX8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 282: 34176–34184. [DOI] [PubMed] [Google Scholar]
- Xie Y., Rubenstein E. M., Matt T., Hochstrasser M., 2010. SUMO-independent in vivo activity of a SUMO-targeted ubiquitin ligase toward a short-lived transcription factor. Genes Dev. 24: 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Seifert A., Chua J. S., Maure J. F., Golebiowski F., et al. , 2012. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 26: 1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Roberts T. M., Yang J., Desai R., Brown G. W., 2006. Suppression of genomic instability by SLX5 and SLX8 in Saccharomyces cerevisiae. DNA Repair (Amst.) 5: 336–346. [DOI] [PubMed] [Google Scholar]
- Zhang X.-D., Goeres J., Zhang H., Yen T. J., Porter A. C. G., et al. , 2008. SUMO-2/3 Modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol. Cell 29: 729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Wu C. Y., Blobel G., 2004. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 167: 605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Blobel G., 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 102: 4777–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Hannoun Z., Jaffray E., Medine C. N., Black J. R., et al. , 2012. SUMOylation of HNF4alpha regulates protein stability and hepatocyte function. J. Cell Sci. 125: 3630–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilio N., Eifler-Olivi K., Ulrich H. D., 2017. Functions of SUMO in the maintenance of genome stability. Adv. Exp. Med. Biol. 963: 51–87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All yeast strains and plasmids used in this study are presented in Table S1. Strains, plasmids and antibodies are available on request.