Abstract
Previously expressed inducible genes can remain poised for faster reactivation for multiple cell divisions, a conserved phenomenon called epigenetic transcriptional memory. The GAL genes in Saccharomyces cerevisiae show faster reactivation for up to seven generations after being repressed. During memory, previously produced Gal1 protein enhances the rate of reactivation of GAL1, GAL10, GAL2, and GAL7. These genes also interact with the nuclear pore complex (NPC) and localize to the nuclear periphery both when active and during memory. Peripheral localization of GAL1 during memory requires the Gal1 protein, a memory-specific cis-acting element in the promoter, and the NPC protein Nup100. However, unlike other examples of transcriptional memory, the interaction with NPC is not required for faster GAL gene reactivation. Rather, downstream of Gal1, the Tup1 transcription factor and growth in glucose promote GAL transcriptional memory. Cells only show signs of memory and only benefit from memory when growing in glucose. Tup1 promotes memory-specific chromatin changes at the GAL1 promoter: incorporation of histone variant H2A.Z and dimethylation of histone H3, lysine 4. Tup1 and H2A.Z function downstream of Gal1 to promote binding of a preinitiation form of RNA Polymerase II at the GAL1 promoter, poising the gene for faster reactivation. This mechanism allows cells to integrate a previous experience (growth in galactose, reflected by Gal1 levels) with current conditions (growth in glucose, potentially through Tup1 function) to overcome repression and to poise critical GAL genes for future reactivation.
Keywords: transcriptional memory, GAL genes, gene positioning, epigenetic, RNA polymerase II, chromatin, nuclear pore complex
TRANSIENT stimuli can lead to changes in gene expression that are inherited for several cell divisions and play an important role in development and adaptation to the environment (Koornneef et al. 1994; Lee et al. 1994; Livingstone et al. 1995; Sung and Amasino 2004; Sung et al. 2006; Hansen et al. 2008; Seong et al. 2011; Nestorov et al. 2013). When a previous experience/stimulus leads to heritable changes in the transcriptional response/output, such phenomena are called “epigenetic memory” (Riggs 1996; Suganuma and Workman 2011; D’Urso and Brickner 2017). Epigenetic transcriptional memory enables certain inducible genes to respond much faster upon reexposure to the same stimulus in yeast, plants, and humans (Brickner et al. 2007; D’Urso and Brickner 2017). Although such genes are repressed between the two inductions, they are poised for faster reactivation. Transcriptional memory is inherited through multiple cell divisions, and serves as a model to study how cells can benefit from their previous experiences and how that information can be stored and inherited.
A well-characterized example of memory in budding yeast following inositol starvation leads to poising of the INO1 gene for faster reactivation (Brickner et al. 2007; Light et al. 2010, 2013; D’Urso et al. 2016; D’Urso and Brickner 2017). INO1 memory requires both a physical interaction of the promoter with the nuclear pore complex (NPC) as well as changes in chromatin structure to permit binding of a preinitiation form of RNA polymerase II (RNAPII; Light et al. 2010, 2013; D’Urso et al. 2016). Although INO1 interacts with the NPC both when active and during memory, these interactions are independently regulated by distinct mechanisms (Brickner and Walter 2004; Brickner et al. 2007; Light et al. 2010). During memory, interaction with the NPC is mediated by the Sfl1 transcription factor binding to a cis-acting promoter element called the Memory Recruitment Sequence (MRS). Sfl1 is required for the interaction of INO1 with the NPC protein Nup100, and this interaction is necessary and sufficient to induce increased incorporation of H2A.Z and dimethylation of histone 3, lysine 4 (H3K4) (Light et al. 2010; D’Urso et al. 2016). Both H3 dimethyl K4 (H3K4me2) and H2A.Z persist during memory at the INO1 promoter and are themselves required for peripheral localization and binding of poised RNAPII. Thus, loss of Sfl1, Nup100, and H2A.Z, or mutations in the MRS, disrupts peripheral localization and binding of poised RNAPII, leading to slower reactivation of INO1 (Light et al. 2010; Brickner et al. 2015; D’Urso et al. 2016).
Critical aspects of INO1 transcriptional memory are conserved and widespread (Light et al. 2013). Hundreds of genes in HeLa cells exhibit interferon γ (IFN-γ)-induced transcriptional memory (Gialitakis et al. 2010; Light et al. 2013). IFN-γ memory requires the interaction of genes with the nuclear pore protein Nup98 (homologous to yeast Nup100), leading to H3K4 dimethylation and binding of poised RNAPII to the promoter (Gialitakis et al. 2010; Light et al. 2013). Likewise, yeast genes induced by oxidative stress show faster induction in cells previously exposed to high salt (Guan et al. 2012). This type of memory also leads to H3K4me2 and binding of poised RNAPII but requires Nup42 instead of Nup100 (Guan et al. 2012; D’Urso et al. 2016). This suggests that both gene-specific and general mechanisms promote epigenetic transcriptional memory.
Galactose-induced transcriptional memory leads to faster reactivation of yeast GAL genes (GAL1, GAL10, GAL7, and GAL2) for up to seven generations (∼12 hr) after shifting from activating to repressing conditions (Brickner et al. 2007; Zacharioudakis et al. 2007; Kundu and Peterson 2010). However, GAL memory is more complex than INO1 memory as it exhibits two distinct phases with different molecular requirements. During the first ∼4 hr of repression, the NPC-associated protein Mlp1 facilitates looping between the 5′- and 3′- ends of the GAL1 gene and this looping, combined with the SWItching deficient/Sucrose NonFermentable (SWI/SNF) chromatin remodeler, is required for faster reactivation (Kundu et al. 2007; Laine et al. 2009; Tan-Wong et al. 2009). Short-term GAL transcriptional memory is distinct from long-term GAL memory, which occurs between 4 and 12 hr of repression and is epigenetically inherited. Long-term memory requires the Gal1 protein, correlates with localization to the nuclear periphery and is independent of the SWI/SNF complex (Zacharioudakis et al. 2007; Kundu and Peterson 2010). Gal1 produced during activation acts as a coactivator by interfering with Gal80 repression during memory and is both necessary and sufficient to enhance the rate of reactivation (Bhat and Hopper 1992; Zacharioudakis et al. 2007).
Here, we have focused on understanding the molecular and cellular consequences of Gal1 expression during long-term, epigenetic GAL gene memory. Like INO1, GAL1 and GAL2 localize at the nuclear periphery during memory (Brickner et al. 2007). We defined cis- and trans-acting factors that control GAL gene targeting to the nuclear periphery during epigenetic GAL memory. We find that Gal1 protein is necessary and sufficient to promote targeting of GAL genes to the nuclear periphery. A cis-acting DNA element (MRSGAL1) in the GAL1 promoter is necessary for targeting of the GAL1 locus to the periphery during memory. Further, targeting by MRSGAL1 is both dependent on Nup100 and responsive to ectopic expression of Gal1. Although loss of Nup100 or mutations in the MRSGAL1 block peripheral localization, they do not affect GAL1 transcription rates. Thus, although GAL gene transcriptional memory leads to interaction with the NPC, this interaction is not required to enhance transcriptional reactivation rates under these conditions.
GAL1 memory also leads to increased incorporation of histone variant H2A.Z, dimethylation of H3K4, and binding of poised RNAPII at the promoter. Furthermore, long-term GAL memory is most beneficial to cells that are currently growing in glucose and, in other carbon sources, memory is not observed. Both growth in glucose and the Tup1 transcription factor function upstream of peripheral localization and downstream of Gal1 during memory. Tup1 is necessary for incorporation of both H2A.Z and H3K4me2 at the GAL1 promoter, leading to binding of poised RNAPII. Thus, future rates of GAL gene reactivation depend both on a previous experience (growth in galactose) and current conditions (growth in glucose).
Materials and Methods
Reagents
Unless noted otherwise, all chemicals were from Sigma ([Sigma Chemical], St. Louis, MO). Yeast media components were from Sunrise Science Products (San Diego, CA). Restriction enzymes were from New England Biolabs (Beverly, MA). Dynabeads, rabbit anti-GFP, goat anti-mouse-Alexafluor 594, and goat anti-rabbit Alexafluor 488 were from Invitrogen (Carlsbad, CA), mouse anti-Myc (9E10) was from Santa Cruz Biotechnology, mouse anti-RNAPII (8WG16) was from Covance, mouse anti-Nsp1 was from EnCor Biotechnology (Gainesville, FL), and rabbit anti-H2A.Z (4626) and rabbit anti-H3K4me2 (32356) were from Abcam. Rapamycin was from Millipore (Bedford, MA).
Plasmids, yeast strains, and molecular biology
Plasmids pAFS144 (Straight et al. 1996), p6LacO128-GAL1, and p6LacO128-GAL1-10prom have been described previously (Brickner and Walter 2004; Brickner et al. 2007, 2016). p6LacO128-GAL2 was created by amplifying the 3′ region of GAL2 using PCR with the GAL2 3′ F and GAL2 3′ R primers. The PCR product was digested using NotI and BamHI and cloned into p6LacO128 (Brickner and Walter 2004). pRS304-ADH1-GAL1 was created by ligating PADH1-GAL1, excised from pGREG700, into SacI- and KpnI-digested pRS304 (Sikorski and Hieter 1989). pGREG700 in turn was generated from pGREG600 (Jansen et al. 2005) by swapping the GAL1 promoter with the ADH1 promoter using the SacI and SpeI sites. Promoter fragments and MRS variants were integrated at URA3:p6LacO128 using the pZIPKan plasmid (Egecioglu et al. 2014) or by cloning in p6LacO128 (Ahmed et al. 2010; Light et al. 2010). The plasmids were linearized by digestion and integrated at the desired locus.
Yeast strains used in this study appear in Supplemental Material, Table S1 in File S1. Except for cells containing Nup2–TAP, Nup100-TAP, and Gal1-GFP (Ghaemmaghami et al. 2003; Huh et al. 2003), all strains were constructed from CRY1 or CRY2 (Brickner and Fuller 1997), derived from the W303 background (ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1). Cells were grown in Synthetic Dextrose Complete (SDC), Synthetic Galactose Complete (SGC), or Synthetic Raffinose Complete (SRC) medium at 30° (Burke et al. 2000) for localization, RT- quantitative (q) PCR, and chromatin immunoprecipitation (ChIP) experiments. For flow cytometry of the Gal1-mCherry cells, cells were grown in either Yeast Peptone Dextrose (YPD) or Yeast Peptone Galactose (YPG).
A PCR-based system was used for deletion (Longtine et al. 1998) and C-terminally tagging genes with fluorophore or FRB tags. The mutant form of mrsGAL1 at the endogenous GAL1 locus was generated by first replacing the promoter with the Kanr marker and then transforming with the mutant promoter and selecting on galactose plates. Strains used for the chromatin localization assay using immunofluorescence (IF) were transformed with either pAFS144 (Straight et al. 1996) or pRS305-GFP-LacI for GFP-LacI expression, pRS304-Sec63myc for immunolabeling the nuclear envelope, and derivatives of the p6LacO128 plasmid to tag the locus of interest (Brickner et al. 2007). For live cell localization assays, the endoplasmic reticulum (ER)/nuclear envelope was visualized by tagging PHO88 with mCherry-His5+ cassette. For flow-cytometric study of GAL1 expression, GAL1 was C-terminally tagged with mCherry-KanMx cassette and PTDH-CFP-NATmx cassette was inserted at the HO locus. For all Anchor-Away experiments the parent strain, HHY168, was adapted for live cell chromatin localization assay (Haruki et al. 2008). Cells were treated with 1 µg/μl rapamycin for depletion of FRB-tagged proteins for 1 hr before imaging.
Chromatin localization assay
Chromatin localization experiments using IF with fixed cells (Brickner et al. 2010) and with live cells (Egecioglu et al. 2014) were performed as described. Cells were imaged using SP5 Line Scanning Confocal Microscope (Leica Biosystems) at the Northwestern University Biological Imaging Facility. Gene localization was scored in stacks of images using LAS AF Lite software: in the z-slice with brightest and most focused LacO dot, if the center of the dot overlapped with the nuclear membrane the gene position was scored as peripheral. Localization was not scored in cells where the dot was either on the top or bottom of the nucleus. Error bars represent the SEM for three biological replicates of 30–50 cells each.
ChIP
Cells were fixed in 1% formaldehyde for 15 min at room temperature, 150 mM glycine was added to quench the formaldehyde reaction, and ChIP was performed as described previously (Brickner and Walter 2004; Ahmed et al. 2010; Light et al. 2010; Egecioglu et al. 2014). For Nup2 and Nup100 ChIP, cells were fixed at room temperature for 1 hr. RNAPII, H2A.Z, and H3K4me2 were recovered with respective antibodies coupled with either anti-pan-mouse (RNAPII) or sheep anti-rabbit IgG (H2A.Z and H3K4me2) Dynabeads, while Nup2 and Nup100 were recovered directly using anti-pan-mouse IgG Dynabeads. Recovery of the DNAs from GAL1, BUD3, and PRM1 promoter by ChIP was quantified by qPCR as described previously (Brickner and Walter 2004) using primers listed in Table S2 in File S1. Error bars represent the SEM from three biological replicates.
RT-qPCR
For activation experiments, cells were grown in SDC to an OD600 0.7–1. For reactivation experiments, cells were grown in SGC overnight, diluted to OD600 ∼0.01 in SDC, and grown for 12 hr. After shifting from glucose to galactose medium, cells were harvested at various times, pelleted, and frozen in liquid nitrogen. RNA was isolated and RT-qPCR was performed as described previously (Brickner et al. 2007). GAL1, GAL2, and GAL7 mRNA levels were quantified relative to ACT1 levels using the GAL1 coding sequence (CDS), GAL2 CDS, and GAL7 CDS primers, respectively (Table S2 in File S1). For experiments using the gal1∆ strain, cells were grown in SRC, shifted to SGC for 4 hr, and then shifted to SDC for 12 hr. Error bars represent the SEM of three biological replicates.
Flow cytometry
Cells with GAL1-mCherry were induced in YPG and maintained at OD600 ≤ 0.3 throughout the induction. Next, 1 ml of culture was harvested at different times of induction, the cells were frozen in 10% glycerol, and stored at −80°. For flow cytometry, cells were thawed on ice and analyzed a BD LSRII flow-cytometer. mCherry and cyan fluorescent protein (CFP) were excited with 561 and 405 nm lasers, respectively. For detecting mCherry emission, a 600-nm long pass dichroic mirror and 610/20-nm band pass filter set was used, while for CFP emission a 505-nm long pass dichroic mirror and 525/50-band pass filter set was used. Roughly 5000 cells were analyzed to obtain the average intensity of Gal1-mCherry and CFP. The constitutively expressed CFP (PTDH-CFP) served as a normalization control for Gal1-mCherry fluorescence; Gal1 expression levels were expressed as ratio of Gal1-mCherry to CFP fluorescence intensity.
Statistical analysis
To evaluate the significance of differences between peripheral localization scores or ChIP signals between strains or treatments with respect to the reference, an unpaired, two-tailed Student’s t-test was performed.
Data availability
All the data necessary to support the conclusions of this study are presented within this article. Raw data are available upon request.
Results
Gal1 promotes targeting of GAL genes to the nuclear periphery during transcriptional memory
The Gal1 protein is necessary for faster reactivation of GAL genes during memory and ectopically expressed Gal1 is sufficient to promote faster GAL gene expression (Zacharioudakis et al. 2007; Kundu and Peterson 2010). Following 12 hr of repression in glucose, the rate of reactivation of GAL2 was much faster than the initial activation and this effect is lost in cells lacking Gal1 (Figure 1A). Furthermore, ectopic expression of Gal1 (ADH1 promoter driving Gal1, PADH-GAL1, integrated at the TRP1 locus) leads to faster activation of GAL7 mRNA (Figure 1B) or Gal1-mCherry protein (Figure 1C). Cells ectopically expressing mutant Gal1 lacking galactokinase activity (deletion of amino acids 171 and 172; gal1-∆SA; Platt et al. 2000) also showed faster activation of Gal1-mCherry (Figure 1C). Thus, GAL1 is necessary and sufficient to enhance the rate of GAL gene induction, suggesting that the production of Gal1 during activating conditions produces a trans-acting, cytoplasmically inherited factor that enhances reactivation rates (Zacharioudakis et al. 2007; Kundu and Peterson 2010).
Figure 1.
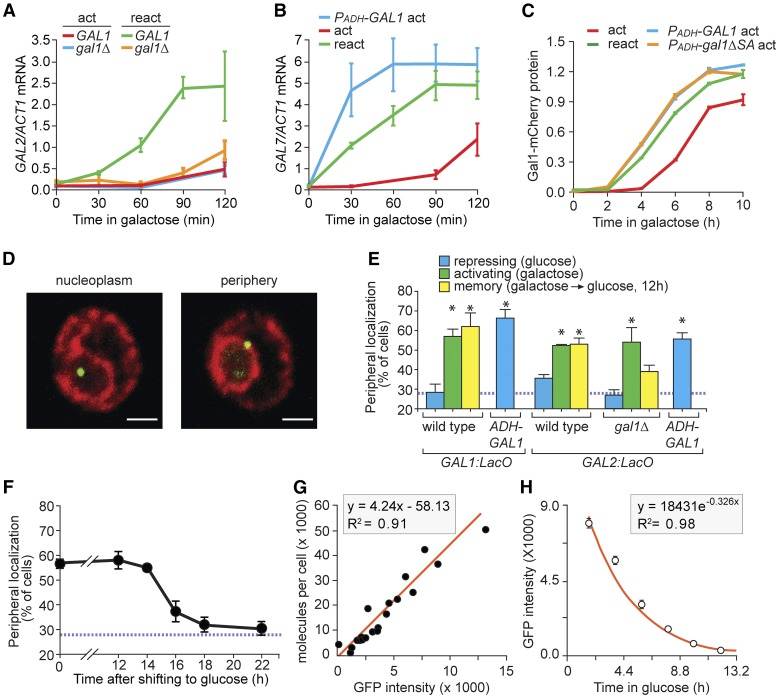
Gal1 promotes GAL gene localization at the nuclear periphery during memory. (A–C) Cells were shifted from glucose to galactose (act; activation) or grown overnight in galactose, shifted to glucose for 12 hr and then shifted to galactose (react; reactivation). Cells were harvested at the indicated times, RNA was prepared and mRNA levels were quantified relative to ACT1 by reverse-transcriptase quantitative PCR (RT qPCR) (A and B) or fluorescence was quantified using flow cytometry (C). (A) GAL2 activation and reactivation in wild-type and gal1∆ cells. (B) GAL7 activation and reactivation or activation with PADH-GAL1. (C) Gal1-mCherry levels, normalized to constitutively expressed cyan fluorescent protein (CFP) (PTDH-CFP) during activation, reactivation, and activation in cells with ectopically expressed wild-type GAL1 (PADH-GAL1) or catalytically inactive mutant (PADH-gal1-∆SA). (D) Immunofluorescence images of cells having the LacO array integrated downstream of the GAL1 gene, stained for GFP-LacI (green) and Sec-63myc (red) and scored as either nucleoplasmic or peripheral. Bar, 1 µm. (E) Peripheral localization of GAL1 and GAL2 under repressing (glucose), activating (galactose), and memory (galactose → glucose, 12 hr) conditions in wild-type or gal1∆ cells and in the presence of PADH-GAL1. (F) Cells with the LacO array downstream of GAL1 were shifted from galactose to glucose media for the indicated length of times and the percentage of cells in which GAL1 colocalized with the nuclear envelope was plotted. The hatched blue line in (E and F) represents the baseline colocalization predicted by chance (Brickner and Walter 2004). (G) Plot of the fluorescence intensities of 20 GFP-tagged proteins (Ghaemmaghami et al. 2003; Huh et al. 2003), measured by flow cytometry, against protein copy number per cell (Newman et al. 2006). (H) Gal1-GFP fluorescence decay after shifting from galactose to glucose. Note: to avoid potential effects of continued translation and maturation of GFP, the initial point for curve fitting was 2 hr after repression. Error bars represent SEM for ≥ three biological replicates. Each replicate for localization (E and F) consisted of 30–50 cells and for fluorescence estimation using flow cytometer (C, G, and H) consisted of ≥ 5000 cells, respectively. * P ≤ 0.05 (Student’s t-test) relative to the repressing condition.
To assess the effect of Gal1 on GAL gene positioning at the nuclear periphery during memory, GAL1 and GAL2 were tagged using an array of 128 Lac-repressor binding-sites (LacO array) in strains expressing the GFP-Lac repressor (Robinett et al. 1996; Brickner and Walter 2004). The fraction of the population in which the gene of interest colocalizes with the nuclear envelope can be determined either by immunofluorescence (IF) with fixed cells or directly in live cells using confocal microscopy (Brickner and Walter 2004; Brickner et al. 2010; Egecioglu et al. 2014). Genes that localize in the nucleoplasm colocalize with the nuclear envelope in ∼30% of cells, corresponding to the baseline for this assay (shown as a blue hatched line throughout), whereas genes that interact with the NPC colocalize with the nuclear envelope in 50–65% of the population (Figure 1D; Brickner and Walter 2004; Casolari et al. 2004; Brickner et al. 2007). By IF, GAL1 and GAL2 localized at the nuclear periphery both when active and for up to 12 hr after repression, but not in glucose (Figure 1E; Brickner et al. 2007; Light et al. 2010). Consistent with previous studies, the fraction of the population that scored as colocalized with the nuclear periphery was lower for GAL2 (∼50%; Dieppois et al. 2006; Gard et al. 2009; Brickner et al. 2012) than for GAL1 (∼60%; Brickner et al. 2007). However, the increase in peripheral localization from repressing to either activating or memory conditions was clear and statistically significant (P = 0.002; two tailed t-test).
In the gal1∆ strain, the GAL2 locus was targeted to the nuclear periphery under activating conditions, but not during memory (Figure 1E). Furthermore, PADH-GAL1 caused both GAL1 and GAL2 to reposition to the nuclear periphery under repressing conditions (Figure 1E). Thus, Gal1 protein plays a critical role in controlling peripheral localization of GAL genes during memory.
GAL1 remained localized at the nuclear periphery for up to ∼14 hr, or ∼7.6 cell divisions, before returning to the nucleoplasm (Figure 1F). To approximate the concentration of Gal1 protein that is sufficient to promote peripheral localization, we quantified the steady-state amount of Gal1-GFP under activating conditions, as well as its rate of decay after repression. Using a standard curve of fluorescence intensity for 20 GFP-tagged proteins of known abundance (Newman et al. 2006), we estimated the abundance of Gal1 protein to be ∼28,000 molecules per cell in cells grown overnight in galactose (Figure 1G). GFP fluorescence was measured over time after shifting the Gal1-GFP strain from galactose to glucose to measure the rate of Gal1 decay after repression (Figure 1H). The t1/2 of Gal1-GFP fluorescence was ∼130 min, somewhat longer than the cell division time in this experiment (∼90 min). Because budding yeast cells divide asymmetrically, producing smaller daughters than mothers, this suggests that the rate of Gal1 decay reflects dilution by cell growth without any appreciable degradation. This may explain how GAL gene memory persists for so many generations. From these estimates, we calculate that ∼300 Gal1 molecules per cell are sufficient to promote peripheral localization (Figure 1F) and faster reactivation of GAL genes (Figure S1 in File S1) after 14 hr of repression. This concentration is comparable to that of Gal80 under these conditions (∼800 molecules per cell; Ghaemmaghami et al. 2003; Huh et al. 2003).
Peripheral localization of GAL genes during memory was observed using IF in which the ER/nuclear envelope was marked with the membrane protein Sec63-myc. However, in both live cells and fixed cells, GAL gene localization at the nuclear periphery was disrupted by overexpression of certain red fluorescent ER/nuclear membrane proteins (Figure S2C in File S1; Green et al. 2012). We do not yet understand the reason for this effect. Fortunately, we found that tagging the endogenous ER/nuclear envelope-resident protein Pho88 with mCherry did not disrupt peripheral localization during GAL memory (Figure S2D in File S1) or INO1 memory (D’Urso et al. 2016). This system permitted both IF and live cell experiments to study the localization of GAL genes during memory.
Peripheral localization of GAL1 during transcriptional memory requires a cis-acting DNA element and Nup100
Localization of INO1 to the nuclear periphery during memory requires a specific cis-acting element (the MRS) and the nuclear pore protein Nup100, neither of which are required for localization of active INO1 to the nuclear periphery. This element functions as a DNA zip code that is sufficient to reposition an ectopic locus to the nuclear periphery (Light et al. 2010). We asked if targeting of GAL1 to the nuclear periphery during memory also requires a specific cis-acting DNA zip code or Nup100. To identify DNA zip codes, we exploited the URA3 locus, which normally localizes in the nucleoplasm (Figure 2A). Insertion of the full-length GAL1 promoter at URA3 (URA3:PGAL1) causes URA3 to localize at the nuclear periphery under both activating (Brickner et al. 2016) and memory (Figure 2A) conditions, supporting the hypothesis that this promoter possesses DNA zip code activity. Using this assay, we mapped a 63-bp MRS (MRSGAL1; Figure S2A in File S1). The MRSGAL1 did not overlap with two other zip codes in the GAL1 promoter (GRS4 and GRS5; Brickner et al. 2016) that mediate peripheral localization of active GAL1 (Figure S2A in File S1). Inserting the MRSGAL1 alone at URA3 led to peripheral localization specifically during memory (Figure 2A). Furthermore, mutations in this element (Figure S2B in File S1) disrupted targeting to the periphery of URA3:MRSGAL1, URA3:PGAL1, and the endogenous GAL1 locus during memory (Figure 2A). Thus, the MRSGAL1 is necessary and sufficient to control targeting to the nuclear periphery during GAL memory.
Figure 2.
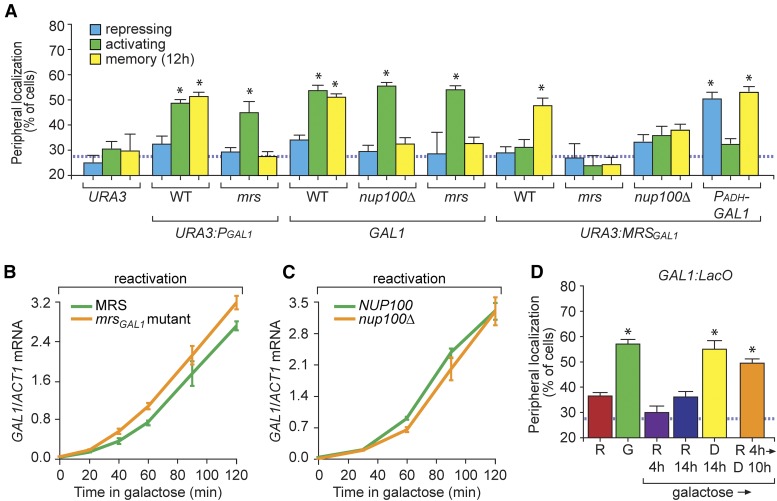
Memory Recruitment Sequence (MRS)GAL1-dependent peripheral localization of GAL1 during memory requires growth in glucose and Tup1. (A) Peripheral localization of URA3, GAL1, URA3:PGAL1, or URA3:MRSGAL1 was quantified under repressing (glucose), activating (galactose), and memory (galactose → glucose, 12 hr) conditions in wild-type (WT) or nup100∆ cells using immunofluorescence or live cell microscopy. The full-length GAL1 promoter (PGAL1, 667 bp) or the 63-bp MRSGAL1 were inserted at URA3 along with a LacO array as described (Egecioglu et al. 2014). The mrs mutation is shown in Figure S2B in File S1. (B and C) Cells were grown in galactose overnight, shifted to glucose for 12 hr, and then shifted to galactose (reactivation) to assay GAL1 expression using RT-quantitative PCR in WT, mrsGAL1 (B), and nup100∆ (C) mutant cells. (D) Peripheral localization of GAL1 in cells grown in raffinose (R), galactose (G), and upon shift from galactose to: raffinose for 4 hr (R 4 hr), raffinose for 14 hr (R 14 hr), glucose for 14 hr (D 14 hr), or raffinose 4 hr followed by glucose 10 hr (R 4 hr → D 10 hr). The hatched line represents the level of colocalization with the nuclear envelope predicted by chance (A and D). Error bars represent SEM for at least three biological replicates. * P ≤ 0.05 (Student’s t-test) relative to the repressing condition.
Loss of Nup100 also specifically disrupted GAL1 peripheral localization during memory, but had no effect on GAL1 peripheral localization during activating conditions (Figure 2A). Likewise, targeting of URA3:MRSGAL1 to the nuclear periphery during memory required Nup100 (Figure 2A). ChIP against nuclear pore proteins Nup2 and Nup100 showed that, while Nup2 interacted with the GAL1 promoter under both activating and memory conditions, Nup100 interacted with the GAL1 promoter only during memory (Figure S3A in File S1). Finally, while inactivation of a conditional allele of Nup2 using the Anchor-Away technique (Haruki et al. 2008) led to rapid loss of peripheral localization under both activating and memory conditions, inactivation of Nup100 disrupted peripheral localization only during memory (Figure S3, B and C in File S1). Thus, while Nup2 plays a general role in GAL1 peripheral localization, the molecular mechanism of GAL1 targeting to the NPC during memory specifically requires the cis-acting MRSGAL1 and the nuclear pore protein Nup100.
Although mutations in the MRSGAL1 or loss of Nup100 blocked targeting of GAL1 to the nuclear periphery during memory, these mutations did not alter the rate of reactivation of GAL1 following 12 hr of repression (Figure 2, B and C). This suggests that targeting to the nuclear periphery is a product of GAL memory, but that the interaction with the NPC is not essential to promote faster GAL gene reactivation.
Targeting GAL1 to the nuclear periphery during memory requires both Gal1 protein and growth in glucose
Ectopic expression of Gal1 was sufficient to cause URA3:MRSGAL1 localization to the nuclear periphery under repressing conditions (Figure 2A). Thus, like the native GAL1, MRSGAL1-mediated targeting to the nuclear periphery is stimulated by expression of Gal1. Therefore, peripheral localization serves as a useful single-cell assay for long-term GAL transcriptional memory. Unexpectedly, ectopic expression of Gal1 did not lead to peripheral targeting of URA3:MRSGAL1 in galactose medium (activating, Figure 2A). This suggested that MRSGAL1-mediated peripheral localization during GAL transcriptional memory either required growth in glucose or is inhibited in galactose.
If glucose is necessary for the peripheral localization of GAL1 and potentially other aspects of memory, we expected that recently-repressed GAL1 would localize in the nucleoplasm in raffinose medium, a nonrepressing and nonactivating condition. Whereas induced GAL1 in cells grown in galactose localized at the nuclear periphery, uninduced GAL1 in cells grown in raffinose localized to the nucleoplasm (Figure 2D). This result conflicts with previous work showing that GAL1 localizes at the nuclear periphery in cells growing in raffinose (Green et al. 2012). However, we find that expression of the ER/nuclear envelope marker used in that study, red fluorescent protein-HDEL (RFP-HDEL) is responsible for the discrepancy (data not shown).
Unlike GAL1 in cells shifted from galactose to glucose, which remained at the periphery (D 14 hr, Figure 2D), GAL1 in cells shifted from galactose to raffinose for either 4 or 14 hr localized in the nucleoplasm (R, Figure 2D). This was not due to lower Gal1 protein levels in cells shifted to raffinose; 4 hr after shifting from galactose to raffinose, Gal1-mCherry levels were slightly higher than in cells shifted from galactose to glucose for 4 hr (Figure S4 in File S1). Furthermore, cells shifted from galactose to raffinose retain the ability to target repressed GAL1 to the nuclear periphery; in cells shifted from galactose to raffinose for 4 hr and then shifted to glucose for 10 hr, GAL1 relocalized to nuclear periphery (R 4 hr → D 10 hr; Figure 2D). Therefore, Gal1 and glucose together promote targeting of GAL genes to the nuclear periphery during memory.
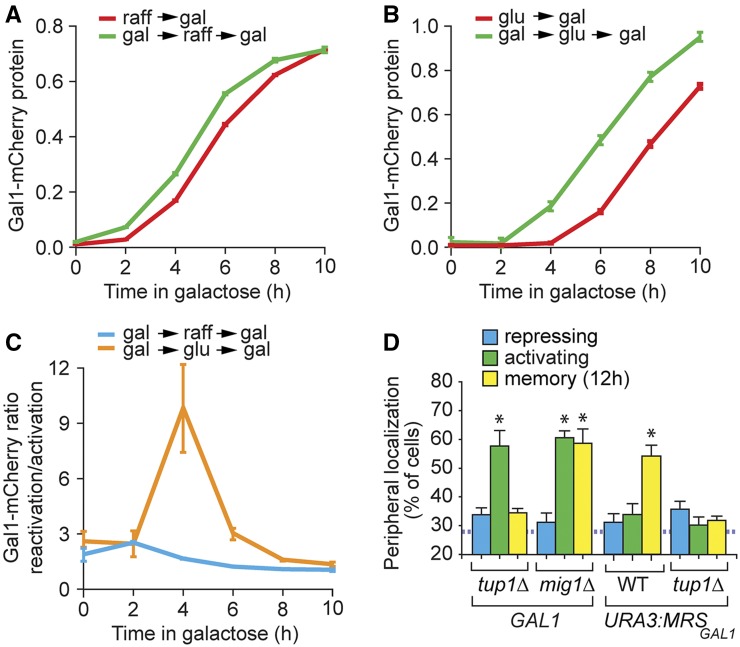
The rate of activation of GAL genes is much slower in cells shifted from glucose than in cells shifted from a nonrepressing carbon source like raffinose (Biggar and Crabtree 2001; Kundu and Peterson 2010). Cells shifted from galactose to glucose, upon returning to galactose, induce GAL1 more rapidly than cells that have not previously grown in galactose. We hypothesized that memory is only evident in glucose because it only provides an adaptive advantage in cells growing in glucose. If so, then cells shifted from galactose to raffinose would, upon returning to galactose, induce GAL1 with similar kinetics as naïve cells. We tested this idea by quantifying the effect of previous growth in galactose on the rate of induction of Gal1-mCherry when cells were shifted either from raffinose to galactose or from glucose to galactose (Figure 3). In cells shifted from raffinose to galactose, the rates of activation (raff → gal) and reactivation (gal → raff, seven divisions → gal) were similar (Figure 3A). In contrast, in cells shifted from glucose to galactose, the rate of activation (glu → gal) was significantly slower than the rate of reactivation (gal → glu, seven divisions → gal; Figure 3B). The difference between these two repressive sugars was also evident from the reactivation:activation ratio of Gal1-mCherry during induction (Figure 3C). This ratio was maximal (∼11) in cells shifted from glucose back to galactose for 4 hr, illustrating the much greater impact of memory in cells grown in glucose.
Figure 3.
The adaptive value of memory in cells grown in nonrepressing and repressing carbon sources. (A and B) Gal1-mCherry expression during activation and reactivation, measured by flow cytometry. Activation: cells were shifted to galactose (gal) from either a nonrepressing carbon source, raffinose (raff) (A), or a repressing carbon source, glucose (glu) (B). Reactivation: cells were shifted from gal to either raff (A) or glu (B) for around seven cell divisions and then reactivated in galactose. (C) Gal1-mCherry reactivation:activation ratio at the indicated time points after shifting cells from raffinose to galactose or glucose to galactose. (D) Peripheral localization of GAL1 or URA3:MRSGAL1 in tup1∆ and mig1∆ mutant strains. The hatched line represents the level of colocalization with the nuclear envelope predicted by chance. WT, wild type. * P ≤ 0.05 (Student’s t-test) relative to the repressing condition. Error bars represent SEM for at least three biological replicates.
In glucose, the Mig1 repressor and the corepressors Tup1 and Cyc8 bind to the GAL gene promoters to repress transcription (Santangelo 2006; Broach 2012). Therefore, we asked if these factors played a role in GAL1 localization during transcriptional memory by scoring GAL1 localization in mig1Δ and tup1Δ cells. The cyc8Δ mutant showed a severe growth defect, so it was not included in this analysis. While loss of Mig1 had no effect on GAL1 localization, loss of Tup1 led to a specific defect in the targeting of GAL1 to the nuclear periphery during memory and disrupted peripheral localization of URA3:MRSGAL1 (Figure 3D). Thus, Tup1 is required for MRSGAL1-mediated peripheral localization of GAL1 during memory.
Tup1 regulates binding of poised RNAPII to the GAL1 promoter and faster reactivation of GAL genes
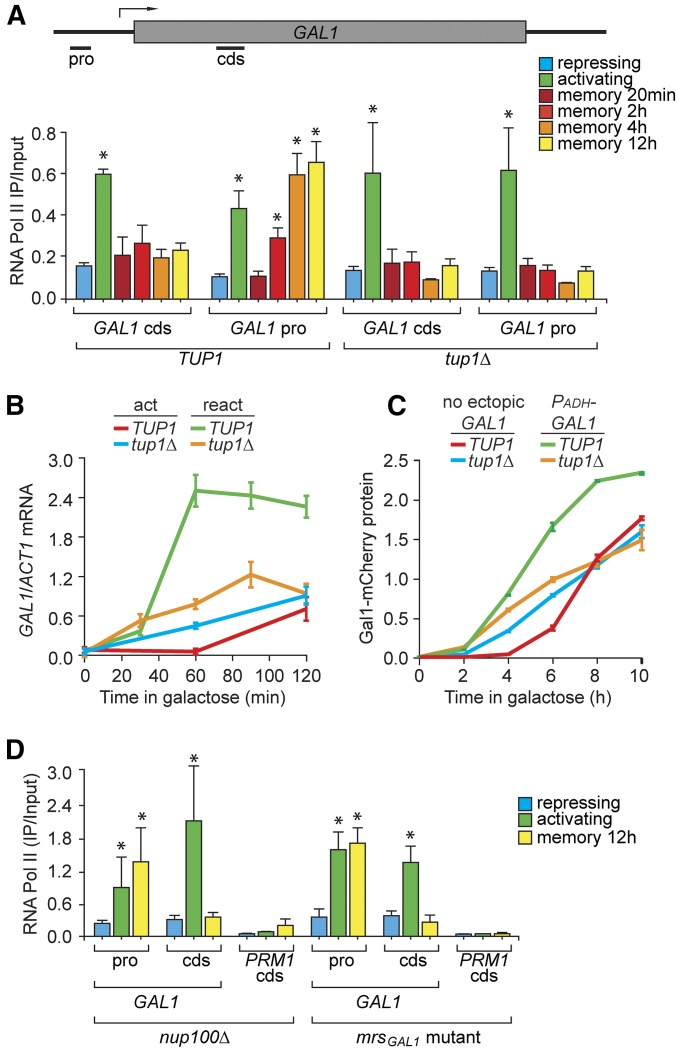
Faster reactivation during memory in yeast and humans is associated with binding of preinitiation RNAPII to the promoter (Light et al. 2010, 2013; D’Urso et al. 2016). To test if GAL1 transcriptional memory involves a similar mechanism, we used ChIP to monitor binding of RNAPII at the GAL1 locus under repressing and activating conditions and at different times after repression. Recovery of both the GAL1 promoter and the 5′-end of the GAL1 CDS was quantified by qPCR (Figure 4A). RNAPII occupancy was low over both the GAL1 promoter and CDS under repressing conditions and was high over both under activating conditions (Figure 4A). Shortly after shifting the cells from activating to repressing conditions (memory 20 min), RNAPII occupancy returned to background levels at both the promoter and the CDS (Figure 4A). However, between 2 and 4 hr of repression, RNAPII association with the promoter increased (Figure 4A). Binding of RNAPII during memory was unaffected by loss of Nup100 or mutations in the MRSGAL1 (Figure 4D). However, loss of Tup1 specifically blocked RNAPII binding to the GAL1 promoter during memory (Figure 4A). This suggests that long-term GAL1 memory leads to binding of poised RNAPII to the promoter.
Figure 4.
Tup1 functions downstream of Gal1 to promote binding of RNAPII to the promoter and faster reactivation of GAL1 during memory. (A) RNAPII ChIP from wild-type and tup1∆ cells under repressing (glucose), activating (galactose), and at different times during memory (galactose → glucose, 20 min to 12 hr) conditions. Recovery of the GAL1 promoter and cds was quantified relative to input by qPCR. (B) Time course of RT-qPCR for GAL1 expression relative to ACT1 during activation (act; glucose → galactose) and reactivation (react; galactose → glucose 12 hr → galactose) in WT and tup1Δ cells. (C) Gal1-mCherry expression during activation in wild-type and tup1Δ cells with or without PADH-GAL1 integrated at the TRP1 locus. (D) RNAPII ChIP under repressing (glucose), activating (galactose), and memory (galactose → glucose, 12 hr) conditions for mrsGAL1 and nup100∆ mutant. Error bars represent SEM for at least three biological replicates. * P ≤ 0.05 (Student’s t-test) relative to the repressing condition. cds, coding sequence; ChIP, chromatin immunoprecipitation; pro, promoter; qPCR, quantitative PCR; RNAPII, RNA polymerase II; WT, wild type.
We next assessed the effects of Tup1 on GAL1 activation and reactivation using RT-qPCR to measure mRNA levels (Figure 4B). In the wild-type strain, the rate of reactivation of GAL1 was much faster than the rate of initial activation (Figure 4B, green vs. red). Consistent with a role in glucose repression, the rate of GAL1 activation was slightly faster in absence of Tup1 (Figure 4B, cyan). However, following 12 hr of repression, the rate GAL1 reactivation was significantly slower in the tup1∆ strain (Figure 4B, orange) and the rates of GAL1 activation and reactivation were quite similar. This was not true under conditions of short-term GAL1 memory; after 1 hr of repression in glucose, tup1∆ cells showed very rapid reactivation that was faster than the wild-type cells (Figure S5 in File S1). During osmotic stress, the Hog1 kinase converts the Tup1-Cyc8-Sko1 repressor complex into an activator (Rep et al. 2001; Proft and Struhl 2002). However, loss of Sko1 had no effect on GAL memory (Figure S6 in File S1). Thus, Tup1 plays a role in both glucose repression and in long-term GAL gene memory.
To establish the order of function of Tup1 and Gal1 in GAL1 memory, we asked if loss of Tup1 is epistatic to ectopic expression of Gal1. Gal1-mCherry protein levels were measured using flow-cytometry in wild-type and tup1∆ cells in the presence and absence of PADH-GAL1 (Figure 4C). In wild-type cells, PADH-GAL1 led to a dramatic increase in the rate of activation of Gal1-mCherry (Figure 4C, green vs. red). As observed with mRNA quantification, activation of Gal1-mCherry was slightly faster in the tup1Δ strain (Figure 4C, cyan vs. red). However, loss of Tup1 blocked the effect of ectopic expression of Gal1 (Figure 4C, orange vs. cyan). This suggests that Tup1 functions downstream of Gal1 to promote faster GAL gene reactivation.
H2A.Z functions downstream of Gal1 to promote GAL memory
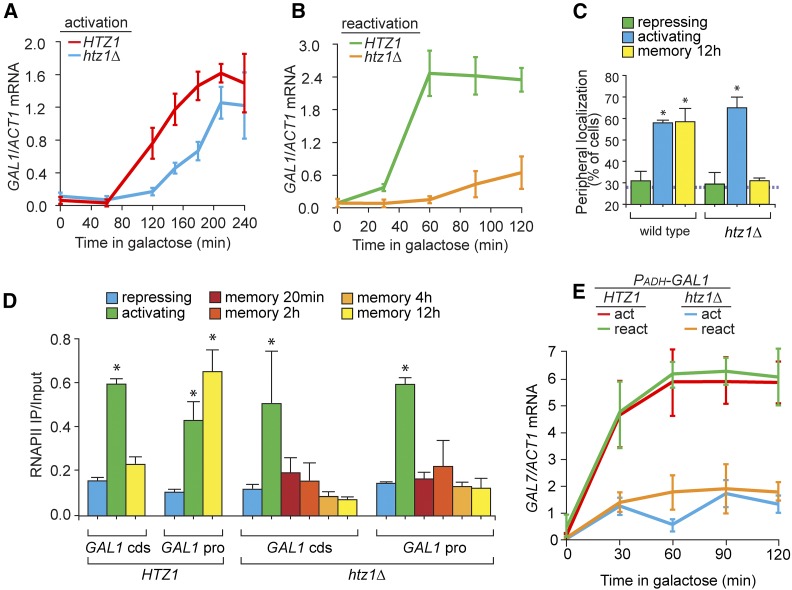
In addition to its role in glucose repression, Tup1 also promotes incorporation of H2A.Z into the GAL1 promoter after repression (Gligoris et al. 2007). H2A.Z incorporation into the INO1 promoter is essential for INO1 transcriptional memory and loss of H2A.Z also leads to a strong, specific defect in the rate of INO1 reactivation during memory (Brickner et al. 2007; Light et al. 2010). However, understanding the role of H2A.Z in GAL gene memory has been challenging because loss of H2A.Z leads to a defect in both activation and reactivation (Figure 5, A and B; Halley et al. 2010). To explore the role of H2A.Z in GAL1 memory, we determined the effect of loss of H2A.Z using assays that are specific to memory: GAL1 localization to the nuclear periphery and RNAPII binding after repression. Loss of H2A.Z disrupted both GAL1 localization to the nuclear periphery (Figure 5C) and binding of poised RNAPII to the promoter during memory (Figure 5D), but did not affect GAL1 localization to the nuclear periphery or RNAPII recruitment under activating conditions. Furthermore, loss of H2A.Z blocked the effect of ectopic expression of GAL1 on the rate of induction of GAL7 (Figure 5E). Thus, in addition to its role(s) in promoting GAL gene activation, H2A.Z plays an important role downstream of Gal1 in promoting GAL gene transcriptional memory.
Figure 5.
H2A.Z functions downstream of Gal1 to promote GAL transcriptional memory. (A and B) GAL1 expression, relative to ACT1, measured by RT-qPCR over time in wild-type and htz1∆ cells during activation (A) and reactivation after 12 hr of repression (B). (C) Peripheral localization of GAL1 under repressing (glucose), activating (galactose), and memory (galactose → glucose, 12 hr) conditions in wild-type and htz1∆ cells. The hatched line represents the level of colocalization with the nuclear envelope predicted by chance. (D) RNAPII ChIP from wild-type and htz1∆ cells under repressing, activating, and at different times during memory (galactose → glucose, 20 min to 12 hr) conditions. (E) GAL7 expression, relative to ACT1, measured by RT-qPCR during activation or reactivation in wild-type and htz1∆ cells transformed with PADH-GAL1. Error bars represent SEM from at least three independent replicates. * P ≤ 0.05 (Student’s t-test) relative to the repressing condition. cds, coding sequence; ChIP, chromatin immunoprecipitation; pro, promoter; qPCR, quantitative PCR; RNAPII, RNA polymerase II.
Tup1 promotes incorporation of H2A.Z and H3K4me2 chromatin modification at the GAL1 promoter during memory
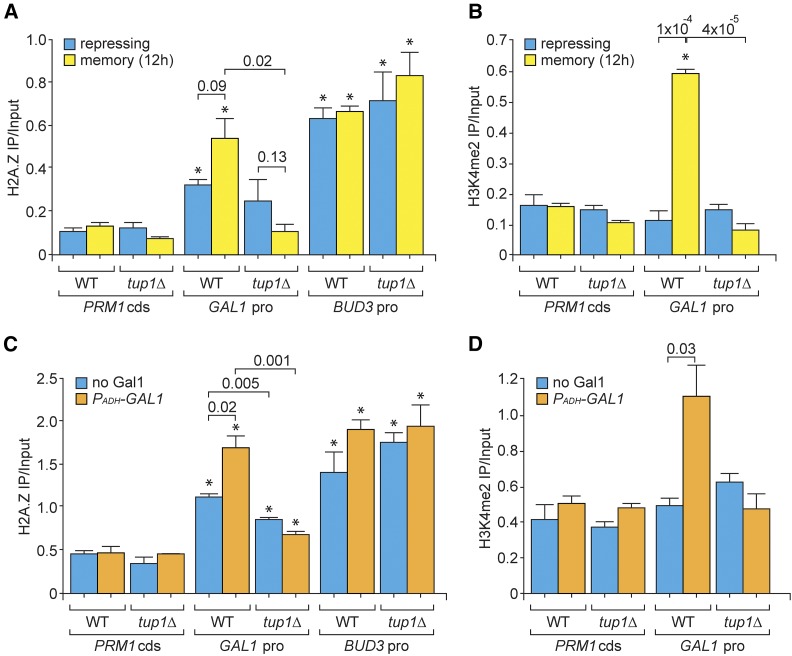
INO1 memory requires both persistent H2A.Z incorporation and H3K4me2 chromatin modification at the promoter (Light et al. 2010, 2013; D’Urso et al. 2016). Therefore, we tested if GAL gene transcriptional memory is associated with these chromatin alterations. The recovery of the CDS of the repressed PRM1 gene served as a negative control for these ChIP experiments, and the recovery of the BUD3 promoter served as a positive control for H2A.Z incorporation (Light et al. 2010; D’Urso et al. 2016). During memory, both H2A.Z occupancy and dimethylation of H3K4 increased significantly at the GAL1 promoter, relative to the repressed condition (Figure 6, A and B). Likewise, expression of PADH-GAL1 under repressing conditions also led to an increase in both H2A.Z occupancy and H3K4me2 (Figure 6, C and D). Thus, Gal1-mediated transcriptional memory leads to increased incorporation of H2A.Z and dimethylation of H3K4.
Figure 6.
Tup1 promotes H2A.Z incorporation and H3K4me2 modification during GAL memory. (A and C) H2A.Z ChIP in WT and tup1∆ cells under repressing (glucose) and memory (galactose → glucose, 12 hr) conditions (A) or under repressing conditions with PADH-GAL1 (C). The recovered DNA fragments in IP were analyzed for sequences arising from the GAL1 promoter, PRM1 coding sequence (negative control), and BUD3 promoter (positive control) and plotted relative to input fraction. (B and D) H3K4me2 ChIP in WT and tup1∆ cells, performed as described in (A and C). Error bars represent SEM from at least three independent replicates. * P ≤ 0.05 (Student’s t-test) relative to the repressing condition. cds, coding sequence; ChIP, chromatin immunoprecipitation; H3K4me2, histone 3 dimethyl lysine 4; IP, immunoprecipitation; pro, promoter; qPCR, quantitative PCR; WT, wild type.
The increased H2A.Z incorporation and the dimethylation of H3K4me2 over the GAL1-10 promoter associated with memory or ectopic expression of Gal1 was lost in strains lacking Tup1 (Figure 6). This effect was specific; loss of Tup1 had no effect on the H2A.Z incorporation into the BUD3 promoter. Thus, Tup1 functions downstream of Gal1 to promote the changes in chromatin structure or modification associated with memory.
Discussion
The yeast GAL genes localize to the nuclear periphery and physically interact with the NPC during both activation and memory (Brickner et al. 2007). During activation, peripheral localization of GAL1 requires the GRS4 and GRS5 DNA zip codes and is necessary for full expression (Brickner et al. 2016). We find that a different DNA zip code, the MRSGAL1, controls the persistent localization to the nuclear periphery during GAL1 memory. Targeting to the nuclear periphery is downstream of Gal1 protein; loss of Gal1 disrupts peripheral retention during memory and ectopic expression of Gal1 leads to MRSGAL1 zip code- dependent targeting of GAL1 to the nuclear periphery even under repressing conditions. However, the association of GAL genes with the NPC is not necessary for faster reactivation, suggesting that it is a product, rather than a driver, of memory. Because localization to the nuclear periphery during memory required growth in glucose, this led us to uncover a critical role for the Tup1 transcription factor in GAL memory. Tup1 contributes to repression of GAL genes in the presence of glucose. However, during transcriptional memory, Tup1 functions downstream of Gal1 to promote changes in chromatin structure and binding of RNAPII to the GAL1 promoter.
Among yeast genes that exhibit transcriptional memory, the GAL genes show the strongest increase in reactivation kinetics and the longest duration (∼8 generations). The GAL genes remain associated with the nuclear periphery during this period. Although faster reactivation of GAL1 does not require peripheral localization, peripheral localization requires all of the factors that are required for faster reactivation (Gal1, Tup1, and H2A.Z). Thus, the NPC association reflects the memory state and serves as a useful assay for this phenomenon. It is possible that, under conditions distinct from those that we have tested, interaction with the NPC contributes to the rate of GAL gene reactivation. Alternatively, interaction with the NPC might impact events that we have not assessed. Finally, interaction with the NPC may be functionally redundant with another pathway that promotes GAL gene reactivation, both of which are downstream of Gal1.
Exploring the conditions under which the MRSGAL1 leads to peripheral localization highlighted the role of glucose in GAL transcriptional memory. Peripheral localization mediated by MRSGAL1 requires growth in the presence of glucose, even in cells expressing ectopic Gal1. Furthermore, the benefit of previous growth in galactose is most apparent when cells are shifted from glucose to galactose, where memory provides a large adaptive benefit. Glucose regulates expression of GAL genes via the Mig1-Tup1-Cyc8 repressor complex (Treitel and Carlson 1995). Although Mig1 recruits the Tup1-Cyc8 corepressor to the GAL1 promoter in glucose (Nehlin et al. 1991), Tup1 is also recruited to the active GAL1 promoter in a Mig1-independent manner (Papamichos-Chronakis et al. 2004). This suggests that Tup1 has function(s) in addition to glucose repression. Consistent with this notion, loss of Mig1 had different effects than loss of Tup1. While loss of Mig1 did not affect GAL1 localization and accelerated both activation and reactivation (Figure S7 in File S1), loss of Tup1 specifically disrupted GAL1 peripheral localization during memory, and led to slightly faster activation and significantly slower reactivation. This suggests that Tup1 plays distinct roles during activation and reactivation. Tup1-Cyc8 is mostly characterized as a corepressor (Smith and Johnson 2000) that masks activation domains (Wong and Struhl 2011), binds hypoacetylated histones (Davie et al. 2002), recruits histone deacetylases (Wu et al. 2001), interacts with mediator subunits (Lee et al. 2000; Papamichos-Chronakis et al. 2000), and repositions nucleosomes (Cooper et al. 1994). However, Tup1 can also function as a coactivator, facilitating recruitment of Spt-Ada-Gcn5 acetyltransferase SAGA or SWI/SNF to promote transcription (Zhang and Guarente 1994; Ozcan et al. 1997; Conlan et al. 1999; Papamichos-Chronakis et al. 2002; Hickman and Winston 2007). Thus, the different effects of Tup1 on active GAL1 and recently-repressed GAL1 may reflect different activities of Tup1 at the GAL1 promoter during repression and memory.
Our current model for Tup1 function in memory is that this protein alters the chromatin of the promoter by promoting H2A.Z incorporation and H3K4me2 modification, allowing both peripheral localization and RNAPII binding. Tup1-Cyc8 promotes H2A.Z incorporation into the active GAL1 promoter and SAGA recruitment (Papamichos-Chronakis et al. 2002; Gligoris et al. 2007). Loss of H2A.Z leads to a defect in the rate of activation and reactivation of GAL1, but leads to specific defects in RNAPII binding at the GAL1 promoter and GAL1 peripheral localization during memory (Brickner et al. 2007; Halley et al. 2010). Furthermore, H2A.Z is required for Gal1-mediated faster reactivation of GAL7. Thus, we propose that Tup1 promotes transcriptional memory through increasing H2A.Z incorporation and, potentially, enhancing dimethylation of H3K4.
Because only a few hundred Gal1 molecules are sufficient to induce GAL transcriptional memory, memory persists through at least seven cell divisions, providing a very long adaptive benefit to previous growth in galactose. However, memory is most adaptive when cells are switched from glucose and glucose is required for features of memory. Although we do not yet understand how growth in glucose impinges upon GAL memory, it is plausible that Tup1 function requires the presence of glucose. Because Gal1 requires Tup1 to mediate memory, these two factors may function to integrate prior growth in galactose with current growth in glucose to regulate memory. Such a mechanism would allow cells to induce memory only when it would be most beneficial.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.201632/-/DC1.
Acknowledgments
The authors thank members of the Brickner laboratory for helpful comments on the manuscript. This work was supported by National Institutes of Health grants R01 GM-080484 and R01 GM-118712, an American Heart Association predoctoral fellowship (to V.S.), a Cellular and Molecular Basis of Disease training grant T32 GM-008061 (to A.D.), and a Rappaport Fellowship for Research Excellence (to W.H.L.).
Footnotes
Communicating editor: A. Hinnebusch
Literature Cited
- Ahmed S., Brickner D. G., Light W. H., Cajigas I., McDonough M., et al. , 2010. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat. Cell Biol. 12: 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat P. J., Hopper J. E., 1992. Overproduction of the GAL1 or GAL3 protein causes galactose-independent activation of the GAL4 protein: evidence for a new model of induction for the yeast GAL/MEL regulon. Mol. Cell. Biol. 12: 2701–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggar S. R., Crabtree G. R., 2001. Cell signaling can direct either binary or graded transcriptional responses. EMBO J. 20: 3167–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D. G., Cajigas I., Fondufe-Mittendorf Y., Ahmed S., Lee P. C., et al. , 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5: e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D. G., Light W., Brickner J. H., 2010. Quantitative localization of chromosomal loci by immunofluorescence. Methods Enzymol. 470: 569–580. [DOI] [PubMed] [Google Scholar]
- Brickner D. G., Ahmed S., Meldi L., Thompson A., Light W., et al. , 2012. Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Dev. Cell 22: 1234–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D. G., Coukos R., Brickner J. H., 2015. INO1 transcriptional memory leads to DNA zip code-dependent interchromosomal clustering. Microb. Cell 2: 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D. G., Sood V., Tutucci E., Coukos R., Viets K., et al. , 2016. Subnuclear positioning and interchromosomal clustering of the GAL1–10 locus are controlled by separable, interdependent mechanisms. Mol. Biol. Cell 27: 2980–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner J. H., Fuller R. S., 1997. SOI1 encodes and novel, conserved protein that promotes TGN-endosome cycling of Kex2 and other membrane proteins by modulating the function of two TGN localization signals. J. Cell Biol. 139: 23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner J. H., Walter P., 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2: e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., 2012. Nutritional control of growth and development in yeast. Genetics 192: 73–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D., Dawson D., Stearns T., 2000. Methods in Yeast Genetics, 2000 Edition: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Casolari J. M., Brown C. R., Komili S., West J., Hieronymus H., et al. , 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439. [DOI] [PubMed] [Google Scholar]
- Conlan R. S., Gounalaki N., Hatzis P., Tzamarias D., 1999. The Tup1-Cyc8 protein complex can shift from a transcriptional co-repressor to a transcriptional co-activator. J. Biol. Chem. 274: 205–210. [DOI] [PubMed] [Google Scholar]
- Cooper J. P., Roth S. Y., Simpson R. T., 1994. The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev. 8: 1400–1410. [DOI] [PubMed] [Google Scholar]
- Davie J. K., Trumbly R. J., Dent S. Y., 2002. Histone-dependent association of Tup1-Ssn6 with repressed genes in vivo. Mol. Cell. Biol. 22: 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieppois G., Iglesias N., Stutz F., 2006. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol. Cell. Biol. 26: 7858–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Urso A., Brickner J. H., 2017. Epigenetic transcriptional memory. Curr. Genet. 63: 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Urso A., Takahashi Y. H., Xiong B., Marone J., Coukos R., et al. , 2016. Set1/COMPASS and mediator are repurposed to promote epigenetic transcriptional memory. Elife 5: e16691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu D. E., D’Urso A., Brickner D. G., Light W. H., Brickner J. H., 2014. Approaches to studying subnuclear organization and gene-nuclear pore interactions. Methods Cell Biol. 122: 463–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard S., Light W., Xiong B., Bose T., McNairn A. J., et al. , 2009. Cohesinopathy mutations disrupt the subnuclear organization of chromatin. J. Cell Biol. 187: 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., et al. , 2003. Global analysis of protein expression in yeast. Nature 425: 737–741. [DOI] [PubMed] [Google Scholar]
- Gialitakis M., Arampatzi P., Makatounakis T., Papamatheakis J., 2010. Gamma interferon-dependent transcriptional memory via relocalization of a gene locus to PML nuclear bodies. Mol. Cell. Biol. 30: 2046–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gligoris T., Thireos G., Tzamarias D., 2007. The Tup1 corepressor directs Htz1 deposition at a specific promoter nucleosome marking the GAL1 gene for rapid activation. Mol. Cell. Biol. 27: 4198–4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green E. M., Jiang Y., Joyner R., Weis K., 2012. A negative feedback loop at the nuclear periphery regulates GAL gene expression. Mol. Biol. Cell 23: 1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q., Haroon S., Bravo D. G., Will J. L., Gasch A. P., 2012. Cellular memory of acquired stress resistance in Saccharomyces cerevisiae. Genetics 192: 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halley J. E., Kaplan T., Wang A. Y., Kobor M. S., Rine J., 2010. Roles for H2A.Z and its acetylation in GAL1 transcription and gene induction, but not GAL1-transcriptional memory. PLoS Biol. 8: e1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K. H., Bracken A. P., Pasini D., Dietrich N., Gehani S. S., et al. , 2008. A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 10: 1291–1300. [DOI] [PubMed] [Google Scholar]
- Haruki H., Nishikawa J., Laemmli U. K., 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell 31: 925–932. [DOI] [PubMed] [Google Scholar]
- Hickman M. J., Winston F., 2007. Heme levels switch the function of Hap1 of Saccharomyces cerevisiae between transcriptional activator and transcriptional repressor. Mol. Cell. Biol. 27: 7414–7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., et al. , 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Jansen G., Wu C., Schade B., Thomas D. Y., Whiteway M., 2005. Drag&Drop cloning in yeast. Gene 344: 43–51. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Blankestijn‐de Vries H., Hanhart C., Soppe W., Peeters T., 1994. The phenotype of some late‐flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild‐type. Plant J. 6: 911–919. [Google Scholar]
- Kundu S., Peterson C. L., 2010. Dominant role for signal transduction in the transcriptional memory of yeast GAL genes. Mol. Cell. Biol. 30: 2330–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu S., Horn P. J., Peterson C. L., 2007. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 21: 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine J. P., Singh B. N., Krishnamurthy S., Hampsey M., 2009. A physiological role for gene loops in yeast. Genes Dev. 23: 2604–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Aukerman M. J., Gore S. L., Lohman K. N., Michaels S. D., et al. , 1994. Isolation of LUMINIDEPENDENS: a gene involved in the control of flowering time in Arabidopsis. Plant Cell 6: 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Chatterjee S., Struhl K., 2000. Genetic analysis of the role of Pol II holoenzyme components in repression by the Cyc8-Tup1 corepressor in yeast. Genetics 155: 1535–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light W. H., Brickner D. G., Brand V. R., Brickner J. H., 2010. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol. Cell 40: 112–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light W. H., Freaney J., Sood V., Thompson A., D’Urso A., et al. , 2013. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol. 11: e1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone C., Patel G., Jones N., 1995. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 14: 1785–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Nehlin J. O., Carlberg M., Ronne H., 1991. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 10: 3373–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestorov P., Tardat M., Peters A. H., 2013. H3K9/HP1 and Polycomb: two key epigenetic silencing pathways for gene regulation and embryo development. Curr. Top. Dev. Biol. 104: 243–291. [DOI] [PubMed] [Google Scholar]
- Newman J. R., Ghaemmaghami S., Ihmels J., Breslow D. K., Noble M., et al. , 2006. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846. [DOI] [PubMed] [Google Scholar]
- Ozcan S., Vallier L. G., Flick J. S., Carlson M., Johnston M., 1997. Expression of the SUC2 gene of Saccharomyces cerevisiae is induced by low levels of glucose. Yeast 13: 127–137. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M., Conlan R. S., Gounalaki N., Copf T., Tzamarias D., 2000. Hrs1/Med3 is a Cyc8-Tup1 corepressor target in the RNA polymerase II holoenzyme. J. Biol. Chem. 275: 8397–8403. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M., Petrakis T., Ktistaki E., Topalidou I., Tzamarias D., 2002. Cti6, a PHD domain protein, bridges the Cyc8-Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol. Cell 9: 1297–1305. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M., Gligoris T., Tzamarias D., 2004. The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tup1 co-repressor. EMBO Rep. 5: 368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A., Ross H. C., Hankin S., Reece R. J., 2000. The insertion of two amino acids into a transcriptional inducer converts it into a galactokinase. Proc. Natl. Acad. Sci. USA 97: 3154–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M., Struhl K., 2002. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 9: 1307–1317. [DOI] [PubMed] [Google Scholar]
- Rep M., Proft M., Remize F., Tamas M., Serrano R., et al. , 2001. The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol. Microbiol. 40: 1067–1083. [DOI] [PubMed] [Google Scholar]
- Riggs A. D, Porter T. N., 1996. Overview of epigenetic mechanisms. Cold Spring Harb. Monogr. Arch. 32: 29–45. [Google Scholar]
- Robinett C. C., Straight A., Li G., Willhelm C., Sudlow G., et al. , 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135: 1685–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo G. M., 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70: 253–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong K. H., Li D., Shimizu H., Nakamura R., Ishii S., 2011. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell 145: 1049–1061. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. L., Johnson A. D., 2000. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25: 325–330. [DOI] [PubMed] [Google Scholar]
- Straight A. F., Belmont A. S., Robinett C. C., Murray A. W., 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6: 1599–1608. [DOI] [PubMed] [Google Scholar]
- Suganuma T., Workman J. L., 2011. Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. 80: 473–499. [DOI] [PubMed] [Google Scholar]
- Sung S., Amasino R. M., 2004. Vernalization and epigenetics: how plants remember winter. Curr. Opin. Plant Biol. 7: 4–10. [DOI] [PubMed] [Google Scholar]
- Sung S., He Y., Eshoo T. W., Tamada Y., Johnson L., et al. , 2006. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat. Genet. 38: 706–710. [DOI] [PubMed] [Google Scholar]
- Tan-Wong S. M., Wijayatilake H. D., Proudfoot N. J., 2009. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 23: 2610–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treitel M. A., Carlson M., 1995. Repression by SSN6–TUP1 is directed by MIG1, a repressor/activator protein. Proc. Natl. Acad. Sci. USA 92: 3132–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K. H., Struhl K., 2011. The Cyc8-Tup1 complex inhibits transcription primarily by masking the activation domain of the recruiting protein. Genes Dev. 25: 2525–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Suka N., Carlson M., Grunstein M., 2001. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell 7: 117–126. [DOI] [PubMed] [Google Scholar]
- Zacharioudakis I., Gligoris T., Tzamarias D., 2007. A yeast catabolic enzyme controls transcriptional memory. Curr. Biol. 17: 2041–2046. [DOI] [PubMed] [Google Scholar]
- Zhang L., Guarente L., 1994. Evidence that TUP1/SSN6 has a positive effect on the activity of the yeast activator HAP1. Genetics 136: 813–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data necessary to support the conclusions of this study are presented within this article. Raw data are available upon request.