How do cells ensure everything is set for cell division before they start the process? Piccirillo et al. demonstrate that a checkpoint in budding....
Keywords: actin-organizing center, Paracoccidioides, satellite-cell group, Swe1, cytokinesis
Abstract
The Rlm1 transcription factor is a target of the cell wall integrity pathway. We report that an rlm1Δ mutant grown on a nonfermentable carbon source at low osmolarity forms cell groups in which a mother cell is surrounded by smaller “satellite-daughter” cells. Mother cells in these groups progressed through repeated rounds of cell division with normal rates of bud growth and genetic stability; however, these cells underwent precocious START relative to wild-type mothers. Thus, once activated, Rlm1 delays the transition from G1 to S, a mechanism we term the cell wall/START (CW/START) checkpoint. The rlm1Δ satellite-cell phenotype is suppressed by deletion of either SLT2, which encodes the kinase that activates Rlm1, or SWI4, which is also activated by Slt2; suggesting that Slt2 can have opposing roles in regulating the START transition. Consistent with an Rlm1-dependent CW/START checkpoint, rlm1Δ satellite daughters were unable to grow or divide further even after transfer to rich medium, but UV irradiation in G1 could partially rescue rlm1Δ satellite daughters in the next division. Indeed, after cytokinesis, these satellite daughters shrank rapidly, displayed amorphous actin staining, and became more permeable. As a working hypothesis, we propose that duplication of an “actin-organizing center” in late G1 may be required both to progress through START and to reestablish the actin cytoskeleton in daughter cells.
CHECKPOINTS are fundamental biological mechanisms that coordinate sequential events in the cell division cycle. Checkpoints function to ensure that later events in the cell cycle depend on completion of earlier events and/or repair of earlier damage (reviewed in Hartwell and Weinert 1989; Paulovich et al. 1997). For example, DNA-damage checkpoints cause the cell cycle to pause in response to DNA damage long enough for this damage to be repaired (reviewed in Harrison and Haber 2006; Putnam et al. 2009). Similarly, the spindle assembly checkpoint arrests cells prior to anaphase until every chromosome is connected to both spindle poles (London and Biggins 2014; Musacchio 2015; Etemad and Kops 2016). Checkpoint components are revealed by mutants that fail in these cell-cycle delays and hence display genetic instability and/or reduced viability.
In the budding yeast Saccharomyces cerevisiae, checkpoints connecting the cell division cycle with the actin cytoskeleton are poorly understood. The yeast actin cytoskeleton, which underlies the plasma membrane, is comprised of actin patches, which drive endocytosis; actin cables, which drive exocytosis; and an actin ring, which drives cytokinesis. This cytoskeleton is organized differently at different stages of the cell cycle (reviewed in Bi and Park 2012; Howell and Lew 2012). From bud emergence to mitosis, actin patches and cable are “polarized” to direct cell-wall components to the bud neck and bud cortex. In contrast, in late stages of mitosis and throughout most of G1, the actin cytoskeleton is not polarized and hence cell-wall growth is isotropic. Polarized actin distribution requires the polarisome, a protein complex that forms at the site of bud emergence and remains associated with the growing bud tip (reviewed in Mishra et al. 2014).
One mechanism for coordinating the organization of the actin cytoskeleton with cell-cycle progression is the “morphogenesis checkpoint.” This checkpoint senses disruption of the actin cytoskeleton (Kang and Lew 2017) to inhibit Clb2/Cdk activity and arrest in G2/M (Sia et al. 1996; McMillan et al. 1999; Martinez-Anaya et al. 2003; King et al. 2013). The morphogenesis checkpoint may also be directly or indirectly activated by cell-wall damage, through the cell wall integrity (CWI) pathway (Harrison et al. 2001). However, known targets of the CWI pathway, such as the Rlm1 transcription factor, are not required for this checkpoint (reviewed in Negishi and Ohya 2010; Levin 2011).
Although Rlm1 is not required for the morphogenesis checkpoint, it is a key player in a second major role of the CWI pathway: responding to cell-wall damage by activating cell-wall repair. For example, treatments causing cell-wall damage culminate in the phosphorylation and activation of Rlm1 by the CWI MAPK, Mpk1/Slt2 (Dodou and Treisman 1997; Watanabe et al. 1997). Rlm1, in turn, activates transcription of cell-wall synthesis genes (Jung and Levin 1999; Garcia et al. 2004; Kim et al. 2008). In addition to activating cell-wall synthesis, the CWI pathway regulates the actin cytoskeleton through multiple mechanisms. For example, an upstream component of the CWI pathway, the Rho1 GTP-binding protein, activates the formin Bni1, which in turn stimulates actin cable formation mediating bud growth (reviewed in Levin 2005; Bi and Park 2012).
The triple role of the CWI pathway in the morphogenesis checkpoint, cell-wall repair, and the actin cytoskeleton informs the current study. This investigation initiated with the discovery that under some growth conditions, the rlm1Δ mutant formed cell groups consisting of a mother cell surrounded by many smaller daughter cells. This “satellite-group” phenotype is caused by a defective G1 checkpoint. The G1/S transition is a key regulatory point during the cell division cycle (reviewed in Johnson and Skotheim 2013; Fisher 2016) and in yeast this regulatory point (termed “START”) is inhibited by mating pheromone, by insufficient nutrients, and by DNA damage (Charvin et al. 2010; Truman et al. 2012; Pope and Pryciak 2013). START is also the point at which yeast cells choose between the alternative fates of quiescence, mating (in haploids), meiosis (in diploids), and filamentous growth (reviewed in Honigberg 2016). In the current study, we characterize the Rlm1-dependent G1 checkpoint, which we term the “cell wall/START” (CW/START) checkpoint, with respect to cell size, viability, cell permeability, and actin cytoskeleton organization. We also investigate the influence on the CW/START checkpoint of environmental conditions, other components of the CWI pathway, the Cdk regulators Swe1 and Mih1, and a UV-induced G1 delay. We propose that Rlm1 may connect this checkpoint to the reestablishment of the actin cytoskeleton after cytokinesis.
Materials and Methods
Strains
All strains used in this study (Supplemental Material, Table S1) are in the W303 strain background (SH3881) and are prototrophs. Deletion alleles were constructed in yeast by transformation with PCR fragments to delete >90% of the ORF and verified by diagnostic PCR using primers flanking the targeted region (Baudin et al. 1993). Primers used to amplify the PCR disruption fragment were chosen as described (Gray and Honigberg 2001). The swi4Δ rlm1Δ double mutants were constructed by tetrad dissection (Kaiser et al. 1994) because deleting RLM1 in swi4Δ mutants was inefficient.
Media and growth
Except as noted, yeast growth was as follows. Spot colonies were grown by inoculating 1 × 105 cells in 0.5 μl H2O on low acetate (LA) agar medium (50 mM potassium acetate, 0.5% yeast extract, 2% agar, pH 7.0) or on the same medium containing 40 μg/ml X-gal. A total of 20 equally spaced spot colonies were arranged in a circle ∼1 cm from the edge of the plate, and colonies were grown for 24 hr at 30° before analysis (Piccirillo et al. 2010). For measurements of the timing of bud emergence, cytokinesis, and cell size, suspended cultures were grown for 72 hr in YPD with adenine (YPDA) medium at 30°, harvested, washed two times in water, sonicated, and resuspended at 2.5 × 105 cells/ml in synthetic low acetate (SLA) medium (0.25% potassium acetate, 0.17% yeast nitrogen base, 0.5% ammonium sulfate, pH 7.0). A 50 μl volume of this resuspension was then inoculated in the wells of a 96-well microtiter plate, the plate was sealed (InterMountain Sci., T-3021-8), and cells were allowed to settle. To ensure that cells remained static for time-lapse microscopy, microtiter wells were incubated with 50 μl 1 mg/ml concanavalin A at 30° overnight, then washed two times with distilled H2O, and dried before adding the sample. For staining of cultures with trypan blue or phalloidin, SLA-suspended cultures were inoculated at 1 × 106 cells/ml and then grown for 7 or 24 hr at 30°. Other media used in this study have been described (Rose et al. 1990; Piccirillo and Honigberg 2010).
Cytological assays
To determine the population distribution (number of cells per group), we resuspended spot colonies in water and examined by microscope to count the total number of cells per group for 250–300 cell groups. Rlm1 activity was assayed in strains carrying the UASRlm1-LacZ allele and expression of the fusion allele on medium containing X-gal was calculated from the intensity of blue color in images of the colony surface (Piccirillo et al. 2015).
To stain nuclei with DAPI, resuspended colonies were first fixed by adding 2 vol of 95% ethanol and then spotted on slides, allowed to dry, and covered with 50% glycerol containing 0.5 μg/ml DAPI. To stain nuclei with propidium iodide (PI), cells were fixed by the addition of 2 vol of 95% ethanol, dried on a slide, then treated with a 10 μg/ml PI solution in 2 M sorbitol, and examined directly (Pringle et al. 1991). To test cell permeability, unfixed cells were stained with 0.1% trypan blue (Thermo-Fisher) and examined immediately according to the supplier’s directions, except that dye was diluted in 100 mM sorbitol. For the images shown, cells were stained with 0.4% trypan blue for increased contrast. To examine the polarity of actin patches, cells were resuspended, sonicated for 10 pulses as described above, fixed in 4% paraformaldehyde, and stained with 0.5 μg/ml tetramethylrhodamine B isothiocyanate (TRITC)-phalloidin (Sigma-Aldrich). To examine diffuse staining of daughter cells, cells were fixed as above and stained with 5 μg/ml TRITC-phalloidin. Visualization of actin cables employed 0.7 μm Rh-phalloidin (Molecular Probes, Eugene, OR) as described (Haarer et al. 2011). To visualize both the mitotic spindle and the actin cytoskeleton, cells were inoculated at 1 × 106 cells/ml in 2% galactose medium and incubated with shaking for 3 hr to induce GAL-GFP-TUB3, then harvested, washed, sonicated to disrupt cells, resuspended in LA medium, incubated for a further 3 hr, and stained with 0.5 μg/ml TRITC-phalloidin as above.
The timing of both bud emergence and cytokinesis was determined from time-lapse videos using images captured every 60 sec (White et al. 2011). Both the first and second bud emergence were easily observable for most cell groups, and the average length of one cell division cycle was inferred from the average length of time from the first to the second bud emergence. The time of cytokinesis was indicated by the sudden shift in the spatial positioning of the bud relative to the mother. Bud and daughter cell size was estimated by manually tracing still images of the bud/daughter from these videos at various times, and the area of these traces determined using ImageJ (Schneider et al. 2012).
To confirm cell-cycle length, bud emergence was monitored over time by visual inspection of 250–300 cell groups in resting cultures. For this analysis, each cell group was classified as containing one, two, or three or more cells.
For time-lapse visualization of UV-irradiated cells, cells were grown as above and after sonication 10 ml of cells were irradiated with constant agitation for 15 sec in a 100-mm uncovered plastic petri dish with the UV source (4 W, 254 nm), 15 cm from the cells. Cells were then placed in microtiter wells and bud emergence monitored by time-lapse microscopy as above.
Cytokinesis in resuspended colonies was verified by sonication using a probe sonicator. Sonication was delivered from a probe sonicator in 10 or 20 1-sec pulses to 200 μl of 5 × 106 cells/ml in a 600 μl microfuge tube. The sample was cooled after every five pulses.
Genetic stability/viability assays
Genetic stability was measured as loss of heterozygosity (LOH) at the URA3+/ ura3-1 locus [chromosome (Chr.) V] to generate Ura− strains. Ura− strains were detectable because they are resistant to the drug 5-fluoroorotic acid (5-FOA). Specifically, spot colonies were grown under standard conditions, resuspended in 1 M sorbitol, transferred to YPDA liquid cultures for 24 hr, then harvested, washed, sonicated, and plated on synthetic medium containing 1 g/liter 5-FOA. To ensure that rare haploid spores were not detected by the assay, the FOA medium lacked histidine and lysine so that these lys2Δ::HIS3+/LYS2+ strains only form colonies when they remain diploid.
LOH can occur by recombination or chromosome loss, but these mechanisms are distinguishable because of the presence of the duplication allele can1-1r:ADE2:CAN1s on both copies of Chr. V. In the case of chromosome loss, the resulting Ura− monosomes can undergo a single recombination event to yield Canr isolates at relatively high frequencies. In contrast, diploid recombinants require two sequential recombination events to yield Canr isolates, and this will occur only at relatively low frequencies. Thus, monosomes were distinguished from recombinants by patching FOA colonies to a YPDA master, and then replica plating to synthetic media containing canavanine (60 μg/ml) and lacking arginine. Monosomic, haploid, and diploid strains were included on each plate as controls.
Overall viability in 1-day colonies grown on LA medium was determined by plating on His− Lys− medium (Piccirillo et al. 2015) both in the absence of sonication, which leaves cell aggregates intact, and after sonication, which disrupts mother/daughter contacts.
Viability specifically in mother or daughter cells was determined by growing spot colonies under standard conditions, resuspending into YPDA medium at 2.5 × 105 cells/ml, placing 50 μl of this culture in a 96-well microtiter plate, capturing images over time, and assembling a time-lapse video as above to follow bud formation in mother and daughter cells.
Statistics and reproducibility
Still images were adjusted for brightness and contrast; images being compared were adjusted identically. Images used for the time-lapse videos were adjusted for brightness, contrast, and sharpness, and assembled into video using Adobe Premiere Pro CS6. All quantitative data in the study is expressed as the mean ± SEM of at least three biological replicas with error bars representing the SEM. Except where noted, P-values are from unpaired Student’s t-test (raw). All experiments were replicated on at least two separate dates, and all experiments comparing mutant to wild type were performed with at least two independently derived mutant isolates. Data based on scoring cells in the microscope (e.g., number of satellite-daughter cells) involved scoring at least 250 cells. Cells were scored in double-blind experiments unless precluded by the characteristic morphology of the rlm1Δ mutant.
Data availability
Strains together with their genotypes are shown in Table S1 and are available on request. All data necessary for the conclusions from the current study are represented either in the manuscript or supplemental files; data from individual trials and repeat trials are available on request. File S1, File S2, and File S3 are videos of dividing yeast cells.
Results
The rlm1Δ satellite morphology
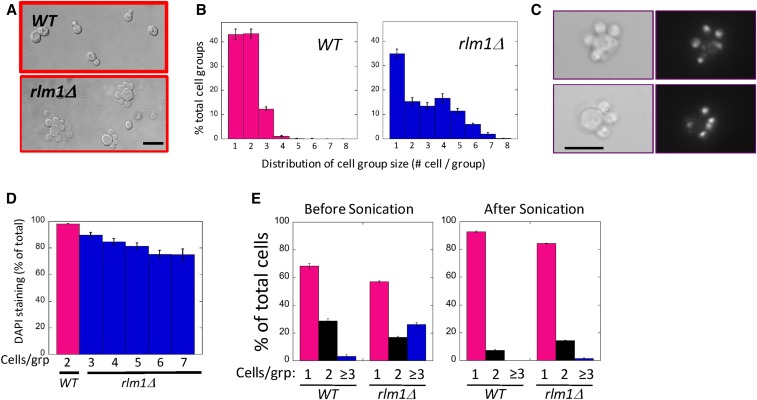
In the course of characterizing the role of the Rlm1 transcription factor during colony development (Piccirillo et al. 2015, 2016), we noticed that rlm1Δ colonies growing on 50 mM potassium acetate medium contained many cells displaying an unusual morphology: a large central cell surrounded by two or more smaller cells (or buds) (Figure 1A). To characterize these satellite groups, we resuspended colonies and determined the distribution of cell groups (i.e., aggregates) containing one, two, three, etc. cells (Figure 1B). As expected, wild-type colonies mostly contained groups of just one or two cells, e.g., a mother cell with a bud. In contrast, in the mutant approximately half of the groups contained three or more cells. Furthermore, wild-type groups of more than two cells often contained cells of different sizes; whereas in mutant groups, the satellite cells surrounding the mother were usually of equal size. Importantly, although satellite morphology was relatively common in cells from an rlm1Δ colony, many other cells from the same colony displayed normal morphologies.
Figure 1.
The rlm1Δ mutant forms multi-budded cells. Wild-type (WT) (SH3881) and rlm1Δ (SH4708) spot colonies grown on LA medium for 24 hr, then resuspended and examined by microscope. (A) Representative wild-type (top) and rlm1Δ (bottom) cell groups visualized using Nomarski optics. Bar, 10 μm. (B) Distribution of the percentage of the total cell groups that contain indicated number of cells per group for both wild type (left) and rlm1Δ (right). n = 3. (C) Two examples of rlm1Δ satellite-cell groups stained with DAPI and visualized by bright-field (left) and fluorescent (right) microscopy. Bar, 10 μm. (D) Percentage of daughter cells that display nuclear DAPI staining for the wild type (magenta, n = 4) or rlm1Δ mutant (blue, n = 3) with the indicated number of cells / group. Wild-type groups containing more than two cells and rlm1Δ groups containing less than three cells are not shown. (E) Percentage of groups with one, two, or three or more cells for wild type or rlm1Δ before sonication (left panel) or after sonication (right panel), n = 3.
rlm1Δ satellite daughters undergo both nuclear division and cytokinesis
To detect nuclear division in satellite groups, we stained cells with DAPI and visualized nuclei by fluorescent microscopy (Figure 1C). Both the mother and bud stained in most rlm1Δ satellite groups, indicating that nuclear division occurred in these cell groups. Indeed, the frequency of daughter cells with stained nuclei was nearly as high in the mutant as the wild type (Figure 1D). However, the frequency of rlm1Δ satellite daughters with stained nuclei did decrease gradually as the number of satellite daughters in the group increased. Thus satellite groups in which the mother had undergone more satellite-type divisions were somewhat more likely to contain anucleate satellite daughters.
To determine whether rlm1Δ mutants undergo cytokinesis as well as mitosis, we compared the number of cells in aggregates before and after sonication. Sonication disrupts affinity between separate cells, but will not separate a mother from a bud (i.e., before cytokinesis). As expected, prior to sonication the rlm1Δ mutant displayed significantly more aggregates containing more than two cells than did the wild type (Figure 1E, left). In contrast, sonication almost completely eliminated aggregates of more than two cells in the rlm1Δ mutant as well as in the wild type (Figure 1E, right), indicating that most cell associations in either strain were between mother and daughter rather than between mother and bud. Thus, rlm1Δ satellite cells are daughters rather than buds, and the mutant is able to complete cytokinesis.
Genome stability is not affected by the rlm1Δ mutant
We next examined whether satellite groups had high levels of genome stability. As a first test of genetic stability, we grew the wild type and the rlm1Δ mutant as spot colonies, and measured LOH at the URA3+/ura3Δ locus (see Materials and Methods). We found that LOH at this allele was not significantly greater in the rlm1Δ mutant (1.1 ± 0.1 × 10−4) than in the wild type (2.0 ± 0.2 × 10−4).
LOH at the URA3+/ura3Δ locus could result from either nondisjunction leading to monosomy or mitotic recombination. To distinguish between these possibilities, we used a CAN1S-ADE2-can1r duplication allele present on the same chromosome as the URA3+/ura3Δ locus (see Materials and Methods). We found that <5% of the cells displaying LOH were Chr. V monosomes in either the wild type or the rlm1Δ mutant, indicating that most LOH in either strain derived from recombination rather than nondisjunction. In summary, under conditions in which the rlm1Δ mutant formed satellite groups, this strain did not display dramatically increased genome instability (either recombination or nondisjunction) relative to the wild type.
Satellite-daughter phenotype is not specific to colonies or to diploids
To determine if the rlm1Δ satellite-daughter morphology was specific to colonies, we grew suspended cultures in the same media contained in the plates. We found that the characteristic satellite morphology was easily visualized in these cultures (Figure S1A in File S4), and many cell groups contained more than two cells, though the difference between wild-type and rlm1Δ cell-group distributions in cultures may be less than in colonies (compare Figure 1B and Figure S1B in File S4).
To determine if the rlm1Δ satellite-daughter morphology was specific to diploid cells, we also examined rlm1Δ haploids grown in spot colonies. Satellite daughters were present in haploids (either a or α type) under the same growth conditions as in the diploids, but at much lower frequency than in diploids. However, haploid satellite daughters were relatively common in the rlm1Δ mutant when acetate concentration was decreased from 50 to 25 mM (Figure S1, C and D, in File S4). Indeed, cell groups containing at least three cells were significantly more frequent in the mutant than the wild type (Kolmogorov–Smirnov test, P = 8.2 × 10−6). Thus, the rlm1Δ satellite-daughter morphology is not specific to diploid cells.
Because rlm1Δ satellite groups are more frequent in diploids than in haploids, and these groups are somewhat more frequent in colonies than in suspended cultures, the remaining experiments in this study, except as noted, used diploid spot colonies.
Satellite-daughter morphology requires nonfermentable carbon sources and low osmolarity
Satellite groups were initially observed when an rlm1Δ mutant is grown on LA medium, which contains acetate as the carbon source. However, this morphology is absent when this same mutant is grown on standard glucose growth media (YPDA or SC). To examine the dependency of carbon source on this phenotype, we compared the morphology of the mutant on a range of fermentable and nonfermentable carbon (NFC) sources, keeping the carbon-source concentration and other media components constant (Table 1, column 2). We found that in either fermentable carbon source tested (50 mM glucose or galactose), very few cell groups contained more than two cells. In contrast, in three of the four NFC sources tested (50 mM acetate, pyruvate, or ethanol), many groups containing more than two cells were observed, and many of these groups displayed the characteristic satellite-group morphology. As discussed further below, glycerol, although it is an NFC source, did not promote the formation of rlm1Δ satellite groups at the tested concentration.
Table 1. Effect of carbon source on satellite formation and Rlm1 activity.
| Carbon sourcea | More than two cells (% of total cells)b | UASRlm1-LacZ expressionc |
|---|---|---|
| Acetate (NFC) | 41 ± 3 | 113 ± 3 |
| Pyruvate (NFC) | 18 ± 1 | 111 ± 3 |
| Ethanol (NFC) | 21 ± 1 | 135 ± 3 |
| Glycerol (NFC) | 2 ± 0.4 | 99 ± 3 |
| 10 mM glycerol (FC) | 13 ± 1 | 99 ± 2 |
| Galactose (FC) | 0.8 ± 0.2 | 2 ± 3 |
| Glucose (FC) | 0.7 ± 0.3 | 2 ± 2 |
The table lists the fraction of cell groups that contain more than two cells and the expression of the UASRlm1-LacZ promoter fusion in media containing a range of carbon sources.
Except as indicated, all media were identical and contained 50 mM of a carbon source. Each carbon source is indicated as fermentable (FC) or nonfermentable (NFC).
Percentage of the total cell groups that contain more than two cells per group, n = 4.
LacZ expression in 48-hr spot colonies as a measure of Rlm1 activity. LacZ expression (arbitrary units) is calculated as the difference in signal between SH5065 (UASRlm1-LacZ) colonies and SH5067 (uasD-LacZ) colonies, n = 10.
We compared the same six carbon sources with respect to Rlm1 activity in wild-type cells. For this purpose, we used the UASRlm1-Cyc1-LacZ promoter fusion, which requires active Rlm1 for LacZ expression (Jung et al. 2002). We found that media containing any of the four NFCs tested (including glycerol) activated the reporter allele, whereas media containing either fermentable carbon source failed to express this allele (Table 1, column 3). This result is consistent with our earlier study demonstrating that phosphorylation of the CWI pathway MAPK (Mpk1/Slt2), a marker of CWI pathway activation, occurs in colonies grown on acetate but not in colonies grown on glucose (Piccirillo et al. 2015). Thus, any NFC source tested induced the CWI pathway, but only some NFC sources promoted rlm1Δ satellite-group formation.
To investigate why the rlm1Δ mutant formed satellite daughters in other respiratory carbon sources but not glycerol, we compared growth rates in rlm1Δ-suspended cultures containing the same glycerol and acetate media used above (Figure S2 in File S4). Growth rates of the mutants in these two media were indistinguishable, demonstrating that the difference in cell morphology in glycerol and acetate is not a result of dramatically different metabolic rates in the two carbon sources.
One clue as to why glycerol does not promote formation of rlm1Δ satellite groups is that the media used in this study all have relatively low osmolarity, which can also induce the CWI pathway (Zu et al. 2001). Although the concentration of glycerol tested was the same as the other NFCs, different carbon sources may be more or less effective at maintaining osmotic balance depending on their rate of transport into or out of the cell. Consistent with this idea, when we tested the rlm1Δ mutant in a medium containing a lower concentration of glycerol (10 mM), satellite groups formed efficiently (Table 1, row 5). In contrast, the same concentration of glucose or galactose formed only relatively few satellite groups even in the mutant (1.8 ± 0.4% in glucose, and 4.1 ± 05% in galactose), and it is possible that even these rare satellite groups only formed after metabolism of glucose or galactose to NFC.
To test the idea that low osmolarity, in addition to NFC, is necessary for the rlm1Δ satellite morphology, we measured the effect of adding a low concentration of solute (50 mM) to LA medium (Table 2). Indeed, we found that adding 50 mM of either glycerol or NaCl suppressed satellite-group formation in the mutant (column 3), though it did not inhibit Rlm1 activity in the wild type (column 4). Similarly, adding sorbitol or erythritol at this same concentration, neither of which are metabolized by S. cerevisiae, also suppressed satellite-group formation. Interestingly, only erythritol, of all solutes tested, also inhibited Rlm1 activity. Indeed, even increasing the acetate concentration from 50 to 100 mM blocked satellite-daughter formation. Thus, the formation of rlm1Δ satellite groups required both an NFC source and low osmolarity, whereas Rlm1 activation in wild-type cells required only the NFC.
Table 2. Effect of solutes on morphology of rlm1Δ mutant.
| More than two cells (% of total cells)b | |||
|---|---|---|---|
| Media (50 mM acetate)a | Wild type | rlm1Δ | UASRlm1-LacZ expressionc |
| No addition | 10 ± 1 | 41 ± 1 | 139 ± 1 |
| +50 mM glycerol | 8 ± 1 | 10 ± 2 | 157 ± 1 |
| +50 mM NaCl | 10 ± 2 | 14 ± 2 | 145 ± 2 |
| +50 mM sorbitol | 9 ± 1 | 10 ± 1 | 138 ± 3 |
| +50 mM erythritol | 13 ± 2 | 12 ± 2 | 33 ± 2 |
| +50 mM acetate | 3 ± 1 | 7 ± 1 | 150 ± 3 |
The table lists the percentage of cell groups that contain more than two cells and the expression of the UASRlm1-LacZ promoter fusion in acetate media to which the indicated solutes were added.
All experiments conducted in LA medium with indicated solutes added.
Percentage of total cell groups containing more than two cells, calculated as in Table 1 (n = 3).
Rlm1 activity based on UASRlm1-LacZ expression as in Table 1 (n = 10).
Timing of cell division for satellite daughters
The smaller size of satellite daughters compared to wild-type daughters suggests that the timing of division or the rate of bud growth may be different in the mutant than in the wild type. As a first step to monitoring these two parameters, we captured time-lapse video of cell division in rlm1Δ and wild-type cells. For this purpose, we grew cells in glucose medium for 72 hr, sonicated to get primarily single cells, and then inoculated these cells into standing cultures at 25° in microtiter wells. Unexpectedly, satellite-daughter groups were much more frequent in SLA standing cultures than on LA plates. SLA contains 25 mM potassium acetate, rather than the 50 mM in LA, and contains a synthetic rather than rich source of nitrogen.
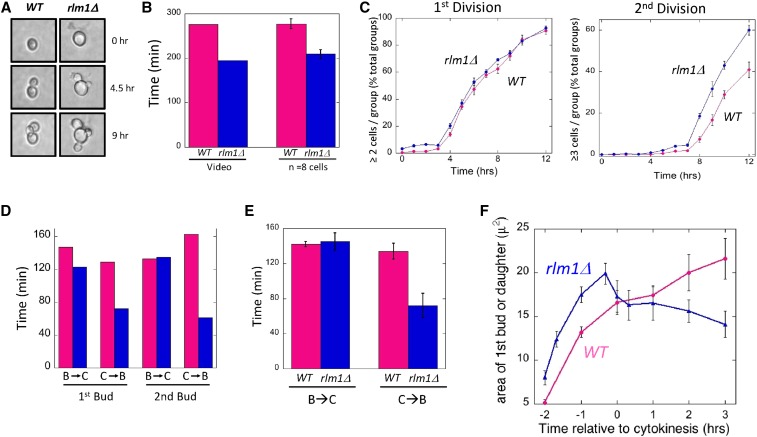
In a representative video of a wild-type cell and an rlm1Δ cell (File S1), the rlm1Δ mother undergoes cell division more rapidly than the wild type (Figure 2A). In particular, the time from the first bud emergence to the second bud emergence is shorter for the rlm1Δ mother than the wild-type mother (Figure 2B, left). To determine whether this difference is significant, we examined video from eight wild-type and eight rlm1Δ cell groups and averaged the time between the first and second bud emergence. As expected, the average time from the first to second bud emergence in SLA was significantly faster (P = 0.004) in the rlm1Δ mutant than in the wild type (Figure 2B, right), confirming that the video in File S1 is typical with respect to timing.
Figure 2.
Timing of cell division in wild-type and rlm1Δ cells. Wild-type (WT) (SH3881) and rlm1Δ (SH4708) strains were grown as standing cultures (SLA medium) in microtiter wells at 25°. (A) Images of cell division in wild-type (left) and rlm1Δ (right) cells shown in the video in File S1 at the indicated times. (B) The time from the first bud emergence to the second bud emergence for the cell groups shown in the video in File S1 (left two bars), and the average time between these two events based on time-lapse video of eight cell groups (right two bars) for both wild-type (magenta bars) and rlm1Δ (blue bars) mother cells. (C) The timing of the first bud to emerge (left panel) and the second bud to emerge (right panel) for both wild type (magenta circles) or rlm1Δ (blue circles); 240–300 cells counted at each time point, n = 4. (D) For the cells shown in video in File S1, the time from bud emergence to cytokinesis (B → C) and from cytokinesis to bud emergence (C → B) for both the wild type (magenta bars) and rlm1Δ (blue bars). (E) The average time from bud emergence to cytokinesis (B → C) and from cytokinesis to bud emergence (C → B) for both the wild type (magenta bars, n = 8) and rlm1Δ (blue bars, n = 8). (F) Average change in bud size over time during the first division for wild type (magenta, n = 6) and rlm1Δ (blue, n = 4). Indicated times are shown relative to cytokinesis, which is set as T = 0.
As an independent method for estimating the timing of divisions, we inoculated microtiter plates with 5 × 104 cells/ml in SLA as above and measured the average number of cell groups over time containing one, two, or more than two cells. This method allowed the average timing of first and second bud emergence to be estimated from 250 to 300 cell groups (Figure 2C). No significant difference between mutant and wild type was observed in the timing of emergence of the first bud, but the second bud emerged an average of 1.75 hr earlier in the mutant than in the wild type based on x-intercepts from linear regression analysis of natural log-transformed data (P = 0.02). The timing of the first division may be approximately the same in the mutant and wild type as a result of a delay in the response to the low-osmolarity media. In summary, rlm1Δ satellite-daughter groups have an increased rate of cell division relative to the wild type when measured by either of the two approaches.
Shortened G1 phase in the rlm1Δ mutant implicates G1/S checkpoint
To determine what stage of the cell cycle is accelerated in the mutant, we measured the timing of bud emergence and cytokinesis from the same video (File S1). We noted that in an earlier time-lapse video created by J. Bean and F. Cross (Rockefeller University) (http://www.rockefeller.edu/labheads/crossf/tubulin.php), two indicators of cytokinesis, namely dissolution of the mitotic spindle and diminished Cdc10 (septin) localization at the bud site, strongly correlated with a rapid rotational or lateral movement of the daughter cell relative to the mother. We also noted this relative motion in our videos, and hence estimated the time of cytokinesis based on the timing of this movement. Comparing video of wild-type and rlm1Δ cells (File S1) through two cell divisions, the time elapsed from bud emergence to cytokinesis (S-G2-M) is approximately the same in the two cells. In contrast, G1 phase (cytokinesis to bud emergence) was shorter for each of the first two divisions in the rlm1Δ cell than in the wild type (Figure 2D).
To confirm that G1 phase was shortened in the rlm1Δ mutant, video of eight wild-type cells and eight mutant cells were used to measure the average time between emergence of the first bud and cytokinesis, and between this cytokinesis and emergence of the second bud. Consistent with the video in File S1, the average time period between the first bud emergence and cytokinesis (i.e., S-G2-M) was essentially the same in the mutant as in the wild type (Figure 2E, left), but the average time period between this cytokinesis and the second bud emergence (i.e., G1) was significantly less in the mutant than in the wild type (Figure 2E, right; P = 0.01).
As a control, we examined the timing of G1 under conditions where the rlm1Δ mutant does not form satellite daughters (SLA plus 200 mM sorbitol). Analysis of eight wild-type and eight rlm1Δ cell groups in this medium revealed no significant difference between the two strains either for the timing from first bud emergence to first cytokinesis or for the timing from this cytokinesis to the next bud emergence (Figure S3 in File S4). Indeed, G1 in the wild type was significantly shorter in the high-osmolarity media relative to the low-osmolarity medium. In contrast, osmolarity had little effect on the length of G1 in rlm1Δ satellite groups (compare Figure 2E and Figure S3 in File S4). This environment-specific G1 delay in the wild type but not the rlm1Δ mutant indicates an Rlm1-dependent G1/S checkpoint. We term this checkpoint the CW/START checkpoint.
Effect of rlm1Δ allele on bud growth and daughter cell size
A defining characteristic of rlm1Δ satellite daughters is that, unlike wild-type daughters, rlm1Δ satellite daughters did not continue to grow after cell division, but instead remained small (Figure 2A and video in File S1). Although the time between bud emergence and cytokinesis was approximately the same in the mutant as in the wild type, it is possible that the smaller size reflects abnormal bud growth in the mutant. However, for the cell groups shown in the video, bud growth was not detectably slower in the mutant than the wild type for either the first or second divisions (video in File S1). Instead, rlm1Δ daughters appear to shrink rapidly after cytokinesis.
To confirm these observations in a larger number of cells, we used still images derived from video to calculate the area of the first bud/daughter in images of six wild-type and four rlm1Δ cells over time. To compile data from asynchronous cell groups, we set the time of cytokinesis of each cell group as T = 0. This experiment confirmed the observations from the video (File S1) that the initial rlm1Δ bud grew slightly faster and to a larger size than did the wild type (Figure 2F). As expected, wild-type daughters resume growth shortly after cytokinesis is complete. In contrast, rlm1Δ satellite daughters not only failed to grow after cytokinesis, they rapidly shrank after cell division and then maintained this diminished size.
In the course of examining videos of 20 wild-type cell groups undergoing multiple cell divisions in SLA medium, we identified two wild-type cell groups that appeared to yield satellite daughters as well as normal daughters. One example is shown in the video in File S2. We measured the size of three normal and four satellite daughters in these two wild-type cell groups over time and confirmed that these wild-type satellite daughters shrank after cytokinesis, whereas normal daughters resumed growth (Figure S4 in File S4). Thus, while satellite daughters were much more common in the rlm1Δ mutant than in the wild type, low frequencies of satellite daughters formed even in the wild type under the respiration plus low-osmolarity condition.
Involvement of upstream kinases in satellite-group phenotype
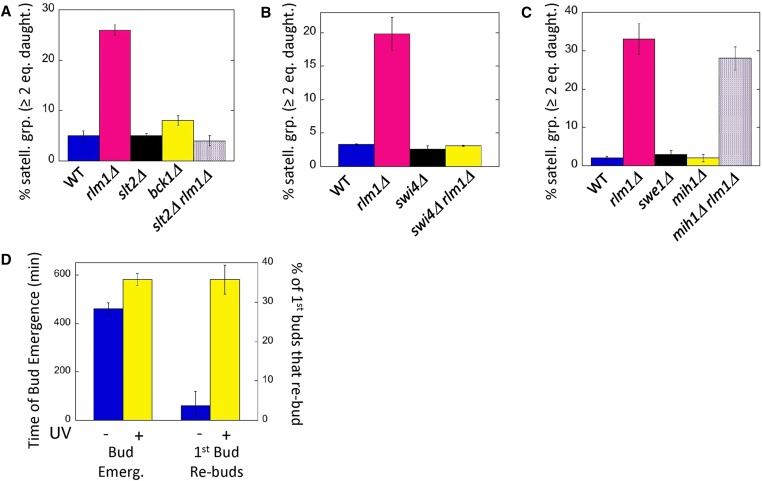
We next asked whether mutants in genes acting upstream of Rlm1 in the CWI MAPK pathway also displayed a satellite-group phenotype. Surprisingly, given that this pathway is the principal known regulator of Rlm1, neither a deletion allele of the MAPK that directly phosphorylates/ activates Rlm1, Slt2 (Mpk1), nor a deletion allele of the MEK kinase acting upstream of Slt2, Bck1, resulted in satellite daughters (Figure 3). Kdx1/Mlp1 kinase may activate Rlm1 independently of Slt2 (Watanabe et al. 1997; Kim and Levin 2010; Kahana-Edwin et al. 2013), but satellite groups (measured as two or more buds per group) were not seen in either a kdx1Δ mutant (3 ± 0.3%, n = 3) or an slt2Δ kdx1Δ double mutant (6 ± 0.3%, n = 3). However, images from an earlier study reveal that a particular point-mutation allele of SLT2 (slt2-1), does form satellite groups at high temperature (Mazzoni et al. 1993).
Figure 3.
Role of CWI and Swe1/Mih1 pathways on satellite-daughter formation: Satellite-daughter cell formation is assayed in resuspended colonies as the percentage of cell groups that contain two or more satellite-daughter cells of equal size. (A) Effect of deletion mutations in the indicated CWI-pathway genes on satellite-daughter formation in diploid colonies grown as in Figure 1 except on LA (0.25% potassium acetate, pH 7.5) medium. Wild type (WT) (SH3881), rlm1Δ (SH4767), slt2Δ (SH5520), bck1Δ (SH5260), and rlm1Δ slt2Δ (SH5523). (B) Effect of deleting SWI4 and/or RLM1 on satellite-daughter formation in haploid colonies grown as in (A). Wild type (SH5480), rlm1Δ (SH5474), swi4Δ (SH5600), and rlm1Δ swi4Δ (SH5602). (C) Effect of deleting the indicated Cdc28 (Y19) checkpoint regulators, MIH1 and SWE1, on satellite-daughter formation. Diploid colonies were grown as in Figure 1; wild type (SH3881), rlm1Δ (SH4708), swe1Δ (SH5419), mih1Δ (SH5344), and mih1Δ rlm1Δ (SH5377). (D) Effect of UV irradiation on microcolony growth in SLA medium. rlm1Δ mutants (SH4708 and SH4767) were either exposed to UV irradiation for 15 sec (+, yellow) or not exposed (−, blue). Microcolonies were monitored by time-lapse microscopy for the time after transfer that the first bud emerges (left) and also for the fraction of daughters that themselves had formed a bud within 6 hr after emerging (right), n = 4.
One clue as to why rlm1Δ and slt2-1 mutants, but not slt2Δ or bck1Δ mutants, can form satellite groups is that Slt2 has multiple targets. In particular, under conditions in which satellite groups are formed, different Slt2 targets may have opposite effects on START. For example, our study indicates that Rlm1 can delay START, but another target activated by Slt2 (the Swi4/Swi6 transcription activator) is known to activate START (Nasmyth and Dirick 1991; Horak et al. 2002). To test the idea that Slt2 has both positive and negative effects on START whereas Rlm1 has primarily negative effects, we examined the phenotype of the slt2Δ rlm1Δ double mutant. We found that this mutant, like the slt2Δ single mutant and the wild type, failed to form satellite daughters, i.e., slt2Δ suppressed formation of rlm1Δ satellite groups (Figure 3A). This result suggests that Slt2 both activates and inhibits the G1/S transition, whereas Rlm1 only inhibits this transition.
To test whether suppression of the rlm1Δ satellite-daughter phenotype by slt2Δ involved the Slt2 target, Swi4, we examined cell morphology in the swi4Δ and swi4Δ rlm1Δ double mutant. We were unable to obtain diploid swi4Δ rlm1Δ double mutants after crossing MATa and MATα double mutants, suggesting a defect either in mating or in zygote viability; indeed, swi4Δ/swi4Δ diploids are inviable in some strain backgrounds (Ogas et al. 1991). For this reason, we examined cell morphology in wild-type, rlm1Δ, swi4Δ, and swi4Δ rlm1Δ haploid strains. As expected, the rlm1Δ single mutant formed satellite daughters whereas the wild type and the swi4Δ single mutant did not (Figure 3B). Interestingly, the swi4Δ rlm1Δ haploid also did not form satellite daughters. Thus, swi4Δ, like slt2Δ, suppresses the rlm1Δ satellite-daughter phenotype. This result is consistent with Slt2 both advancing START in LA medium by activating Swi4 and delaying START by activating Rlm1.
Satellite-daughter formation does not require bypass of Swe1/Mih1 controls
We next considered the relationship of the CW/START checkpoint to the G2/M morphogenesis checkpoint described in the Introduction. Both checkpoints involve the CWI pathway, but the CW/START checkpoint requires Rlm1, whereas the morphogenesis checkpoint does not (Harrison et al. 2001). As described in the Introduction, the morphogenesis checkpoint primarily arrests the cell cycle by activating Swe1 kinase, so we asked if the CW/START checkpoint also requires this kinase. As an initial test, we examined cell morphology of a swe1Δ mutant in LA media. No satellite daughters were observed in this mutant, indicating that Swe1 is not required to maintain daughter cell size (Figure 3C).
Although Swe1 is the major regulator of Cdc28-Tyr19 at the morphogenesis checkpoint, the CWI pathway may regulate this checkpoint by decreasing Mih1 phosphatase rather than by increasing Swe1 kinase (see Introduction). Thus to determine the role (if any) of Mih1 in the CW/START checkpoint, we examined cell groups in mih1Δ and rlm1Δ mih1Δ mutants grown under the same conditions that led to rlm1Δ satellite groups. As expected, mih1Δ did not display satellite daughters; although, as reported earlier, this mutant can sometimes form chains of cells (Ahn et al. 1999). Furthermore, the rlm1Δ mih1Δ double mutant displayed the same satellite-group morphology as the rlm1Δ single mutant (Figure 3C). Taken together, our results with swe1Δ and rlm1Δ mih1Δ mutants indicate that satellite-daughter cell formation does not result from a defect in Cdk Tyr19-PO4 regulation.
UV arrest partially rescues satellite-daughter defect
An established test of whether a particular mutant is defective in a checkpoint function is to determine whether introducing a cell-cycle delay into this mutant rescues the lethality (Hartwell and Weinert 1989). To apply this test to the rlm1Δ mutant, we grew rlm1Δ cells in YPDA to stationary phase, then sonicated to generate single unbudded cells. These cells were then UV irradiated to generate a DNA-damage delay in G1, and the irradiated cells used to inoculate standing SLA cultures. We employed time-lapse video to monitor the time that the first bud emerged and also the ability of the resulting daughter cell to form a bud itself (Figure 3D and video in File S3). As expected, bud emergence was significantly delayed in irradiated cells relative to the unirradiated control (P = 0.003, n = 34). Interestingly, approximately one-third of first daughters formed in the irradiated sample were able to themselves form buds in the same media. In contrast, significantly fewer first daughters were able to bud in unirradiated samples (P = 0.004, n =3). Because UV irradiation of the rlm1Δ mutant both causes a G1 delay and rescues the ability of daughters formed after the delay to progress through the cell cycle, the checkpoint bypass in the mutant is very likely responsible for satellite-daughter formation.
Satellite daughters are inviable
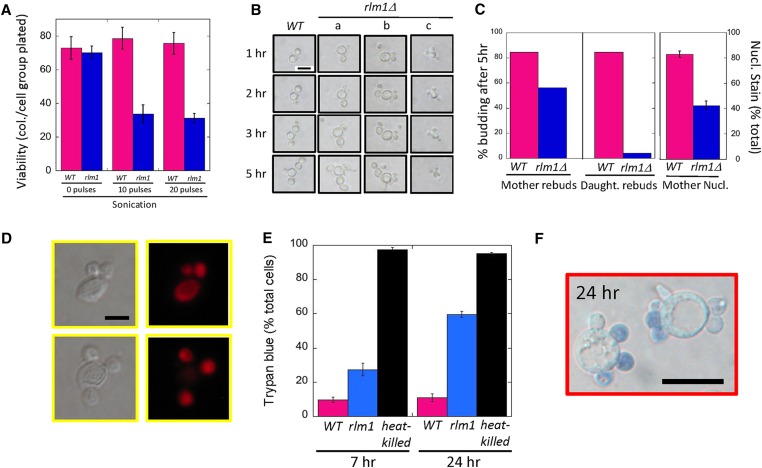
As shown above, satellite daughters fail to rebud in low-osmolarity medium (Figure 3D and video in File S1), so we next asked whether these daughters could grow if transferred to more optimal growth conditions. As a first test, we resuspended rlm1Δ and wild-type colonies and plated on His− Lys− glucose medium. From this experiment we determined that rlm1Δ cell groups had a similar group viability (colonies/cell groups plated) as wild-type cell groups (Figure 4A, left side). However, because rlm1Δ satellite groups contain many more cells than wild-type groups, these results do not give an accurate comparison of rlm1Δ vs. wild-type cell viability. For a direct cell-to-cell comparison of viability, we compared wild-type to rlm1Δ colonies after either 10 pulses of sonication (Figure 4A, center) or 20 pulses of sonication (Figure 4A, right). As shown previously (Figure 1E), even the milder sonication treatment completely disrupts satellite-daughter groups. After sonication, viability remained high in wild-type cells, but decreased by ∼50% in mutant cells. These results indicate that satellite-daughter groups contain both viable and inviable cells.
Figure 4.
Viability of wild-type (WT) (SH3881) and rlm1Δ (SH4708 and SH4767) cell groups: (A) Viability of wild-type (magenta) and rlm1Δ (blue) cell groups grown under standard conditions on LA medium before sonication (left panel), after 10 pulses of sonication (center panel), and after 20 pulses of sonication (right panel). (B) Cell division of mother and bud cells after transfer from LA to YPDA medium. A typical wild-type and three typical rlm1Δ cell groups are shown at the indicated times after transfer. For the wild-type cell group shown in the left column and the rlm1Δ cell group shown in column (a), both the mother and daughter bud. For the rlm1Δ cell group in column (b), the mother cell buds but not the daughter cells. For the rlm1Δ cell group in column (c), neither the mother nor the daughters form buds. Bar, 10 μm. (C) Left panel: the percentage of mother cells that have budded by 5 hr for wild type (11/13) and rlm1Δ (9/16). Center panel: the percentage of buds that have budded by 5 hr for wild type (11/13) and rlm1Δ (2/49). Right panel: percentage of mother cells in which the nucleus stains with PI for wild-type and rlm1Δ strains, n = 4. (D) rlm1Δ satellite-daughter groups fixed and stained with PI. Bar, 5 μm. (E) Permeability: wild-type (magenta) and rlm1Δ (blue) strains were inoculated at 1 × 106 cells/ml and grown for 7 hr at 30° in shaking cultures containing SLA medium (left), or these same strains were grown for 24 hr as colonies grown on LA medium (right). Cells from either growth condition were stained with trypan blue and examined by light microscope. As a positive control, wild type were incubated at 100° for 10 min (heat-killed, black). (F) Representative rlm1Δ satellite groups after 24 hr growth on LA medium stained with trypan blue. Bar, 10 μm.
To determine the viability of mother cells and daughter cells separately, we resuspended wild-type or rlm1Δ colonies (without sonication) into YPDA (rich glucose) media, allowed cells to settle in microtiter wells, and monitored bud emergence by time-lapse video as above for 5 hr. In particular, we followed budding in 13 wild-type cell groups, each containing one daughter, and 16 rlm1Δ satellite groups, each containing at least two daughters of equal size (Figure 4B).
We first assessed the ability of mother cells to continue to bud after transfer. As expected, the majority of mother cells from both the wild-type and rlm1Δ strain continued to bud (Figure 4B, first three columns; and Figure 4C, left). Both strains formed only normal daughters from the mother in the YPDA medium (not satellite daughters), and given enough time these daughters themselves formed buds. Although cells in which the mother failed to bud by 5 hr were somewhat more common in the mutant than in the wild type (Figure 4B, column c; and Figure 4C left), this difference is not statistically significant (Fisher’s exact test, P = 0.13). To independently assess the viability of rlm1Δ mothers relative to wild-type mothers, we stained fixed cells with PI (Figure 4C, right). Indeed, we detected significantly more anucleate mothers in the rlm1Δ mutant than in the wild type, a result that is consistent with the somewhat diminished frequency of nucleated rlm1Δ daughters relative to wild-type daughters (Figure 1D).
In contrast to the ability of most rlm1Δ mothers to form buds after transfer to rich medium, almost all rlm1Δ satellite daughters failed to form buds in rich medium (Figure 4B, column a–c; and Figure 4C, center). For example, we were able to monitor 15 rlm1Δ satellite-cell groups through 5 hr, with these cells initially containing an average of 4.3 cells per group. At the completion of these 5 hr only one daughter cell had formed its own bud. In contrast, in the wild type, most buds (10 of 13) grew and themselves formed buds during the same time period (Figure 4C, center; Fisher’s exact test, P < 10−4). Considering these results, the viability drop we observed in the rlm1Δ mutant after sonication (Figure 4A) likely results from the very low viability of satellite daughters. Indeed, in rlm1Δ standing cultures, likely only mothers and rare normal daughters remain viable.
Satellite daughters become permeable
We next asked if rlm1Δ satellite daughters become permeable to the dye, trypan blue, which is commonly used to stain inviable cells. For example, it is possible that satellite daughters form as the result of cell lysis after cytokinesis, in which case, satellite daughters would always become permeable to the dye upon forming. To address this possibility, we first examined daughters and buds from an early stage of growth (7 hr) in SLA cultures as in Figure 2. In these cultures, most cell groups in both the mutant and wild-type cultures contained at least one bud (67 ± 5% in the wild type and 69 ± 3% in the mutant). Two-cell groups can be mother plus bud or mother plus daughter, but only mother–bud groups will remain after sonication. As a result, we determined the relative frequencies of these two types of two-cell groups in these 7-hr cultures by sonicating and then tabulating the remaining two-cell groups. After sonication, 31 ± 2% of the wild-type culture and 32 ± 3% of the mutant cultures were mother–bud groups. This comparison reveals that approximately half of two-cell groups in the mutant cultures were mother–daughter pairs.
As a positive control for trypan-blue staining of permeable cells, we verified that almost all heat-killed cells were permeable to the dye (Figure 4E, black bars). The frequency of trypan-blue staining in the daughters (or buds) of unsonicated samples was significantly higher in rlm1Δ than in the wild type (Figure 4E, left), indicating that rlm1Δ mutant cells have increased permeability in these cultures. Nevertheless, most rlm1Δ satellites (∼75%) were not permeable to the dye (Figure 4E, left), indicating that cell permeability is unlikely to be the cause of satellite-daughter formation. Instead, satellite daughters often become permeable as they form or after they form.
To further investigate the timing of daughter-cell permeability, we examined trypan-blue permeability in later stages of growth (24 hr colonies). In these colonies, many rlm1Δ satellite groups contained three or more cells. As with the 7-hr cultures, rlm1Δ satellite daughters from groups of three or more cells were significantly more permeable to the dye than wild-type daughters from two-cell groups (Figure 4E, right). Nevertheless, almost half of satellite daughters were still impermeable to the dye (Figure 4, E and F). The experiments in Figure 4, E and F, together indicate that many rlm1Δ satellite daughters remain impermeable after cytokinesis is complete, but most satellite daughters eventually become permeable.
Satellite daughters display diffuse actin staining
Although the relationship between cell size and actin cytoskeleton in yeast is still unclear (Jorgensen et al. 2002; Goranov and Amon 2010; Turner et al. 2012; Sattlegger et al. 2014; Kopecka et al. 2015), the connection between the CWI pathway and the actin cytoskeleton (see Introduction) and the rapid shrinking of daughters at the time of cytokinesis suggests that satellite daughters result from an actin cytoskeleton defect. Thus, we asked whether satellite daughters displayed defects in the actin cytoskeleton. Actin cables were only weakly visible by Rh-phalloidin staining in 7-hr SLA cultures relative to rapidly growing YPD cultures (Figure S5, A and B, in File S4), as previously reported for other conditions where respiration is limited (Vasicova et al. 2016). However, similar frequencies of cables were observed in the rlm1Δ mutant as in the wild type.
The polarity of actin patches in mother–bud pairs was also similar in the rlm1Δ mutant as in the wild type (sonicated 7-hr SLA cultures, Figure S5B in File S4). Actin patch polarity in mitotic cells was also the same in the rlm1Δ mutant as the wild type (mitotic spindle visualized by GFP-tubulin; Figure S5, C and D, in File S4). Thus, we did not detect any effect of the rlm1Δ mutant on the actin cytoskeleton prior to cytokinesis, and this finding is consistent with the relatively normal rates of bud growth observed in the mutant (Figure 2F).
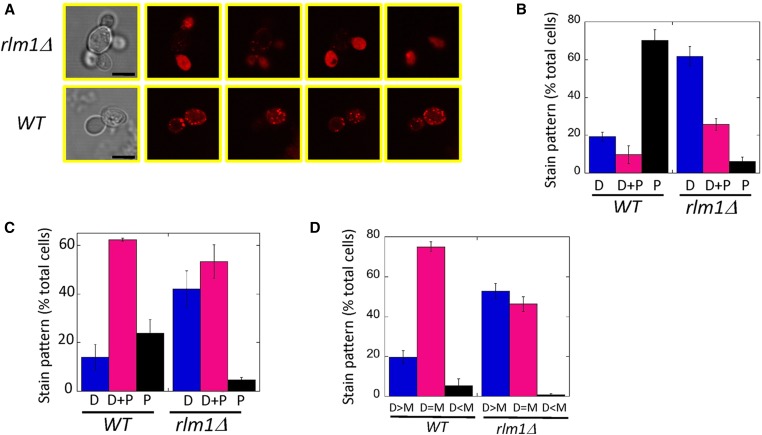
In contrast to the normal actin cytoskeleton seen in dividing rlm1Δ cells, rlm1Δ satellite daughters displayed an abnormal actin cytoskeleton. As expected, most wild-type daughter cells from 24-hr colonies displayed punctate staining of actin patches. In contrast, most rlm1Δ satellite daughters grown under the same condition displayed diffuse (amorphous) actin staining (Figure 5, A and B). Similar results were seen when comparing wild-type and rlm1Δ two-cell groups from 7-hr SLA cultures (Figure 5C). As an independent method of quantifying diffuse actin staining in these 7-hr cultures, we compared two-cell groups in wild type and rlm1Δ mutant with respect to whether diffuse staining was brighter in the mother, brighter in the daughter, or similar in mother and daughter. As expected for the wild type, diffuse actin staining, when it occurred, was usually equal in the mother and daughter cells. In contrast, in the rlm1Δ mutant, more than half of satellite daughters displayed brighter diffuse staining than the mother cell (Figure 5D). Thus, both methods of quantifying actin staining in daughter cells indicate increased diffuse actin staining of daughters in the rlm1Δ mutant relative to the wild type.
Figure 5.
Actin deposition in wild-type (WT) and rlm1Δ daughters. (A) Representative wild-type or rlm1Δ cell groups from 24-hr colonies stained with TRITC-phalloidin and visualized by bright-field (left column) and confocal scanning laser microscopy (right four columns are Z-series). Bar, 10 μm. (B) Percentage of total wild-type (SH3881) or rlm1Δ (SH4708 and SH4767) daughter cells from 24-hr colonies scored as displaying diffuse (amorphous) staining only (D), both diffuse and punctate staining (D+P), and punctate staining only (P), n = 3. In these experiments 99 ± 1% of wild-type and 94 ± 2% of rlm1Δ cells stained with phalloidin. Wild-type and rlm1Δ distributions are significantly different (chi-square distribution comparison, P = 2 × 10−28). (C) percentage of total wild-type (SH3881) or rlm1Δ (SH4708 and SH4767) daughter cells from 7-hr SLA cultures scored as displaying diffuse (amorphous) staining only (D), both diffuse and punctate staining (D+P), or punctate staining only (P), n = 3. In these experiments 48 ± 5% of wild-type and 65 ± 6% of rlm1Δ cells stained with phalloidin. Wild-type and rlm1Δ distributions are significantly different (chi-square distribution comparison, P = 2 × 10−146). (D) Percentage of total phalloidin-stained rlm1Δ two-cell groups from 7-hr SLA cultures as in (C) scored as displaying brighter diffuse staining in the daughter cell than in the mother (D > M), equal intensity staining (D = M), and less intense staining in the daughter (D < M), n =3. In these experiments 63 ± 2% of wild-type and 63 ± 3% of rlm1Δ cells stained with phalloidin. Wild-type and rlm1Δ distributions are significantly different (chi-square distribution comparison, P = 1 × 10−27).
Discussion
The principal results reported in this study are as follows (diagrammed in Figure 6): First, under conditions of low osmolarity and an NFC source, the rlm1Δ mutant underwent premature START but an apparently normal mitosis and cell division. Second, after cytokinesis, rlm1Δ daughter cells shrank rapidly and lost viability. As a result, this mutant forms satellite-cell groups, which consist of a single mother cell surrounded by multiple small, inviable daughters.
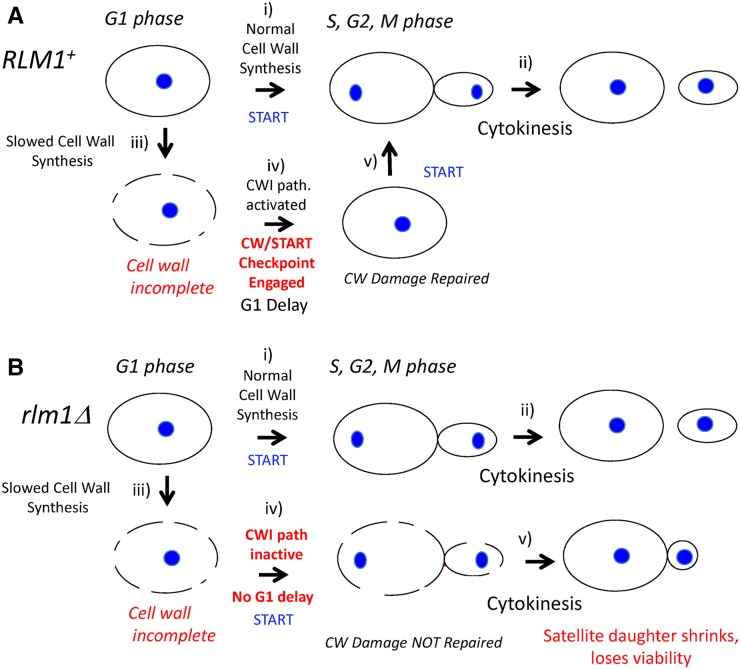
Figure 6.
Working model of cell-wall checkpoint at G1/S (A) RLM1+ strains have a functional CW/START checkpoint: the cell cycle from G1 to cytokinesis (i and ii) proceeds without delay when cell-wall synthesis is not limiting (e.g., glucose growth medium) as shown in the top row of figures. Nuclei are shown as blue ovals. In the second row, the cell wall is shown as a dotted line to represent incomplete cell-wall synthesis (iii); this damage induces the Rlm1-dependent cell-wall checkpoint leading to G1 arrest (iv), and completion of cell-wall synthesis during this delay allows the G1/S transition (v). (B) The rlm1Δ mutant is defective in the CW/START checkpoint. Cell division is normal when cell-wall synthesis is not limiting (top row, i and ii). As in the wild type, incomplete cell wall (dotted line) occurs in LA medium (iii); but in contrast to the wild type, the defective checkpoint allows the G1/S transition in the absence of complete cell wall (iv), resulting in daughter cells that shrink after cytokinesis and are inviable (v).
The satellite-cell morphology described in this study is reminiscent of the pathogenic form of the fungus Paracoccidioides brasiliensis (reviewed in Borges-Walmsley et al. 2002; Bocca et al. 2013). This fungus is typically found in soil in filamentous form, but in the warmer temperatures of the human lung, the species instead forms a yeast-like mother cell surrounded by many smaller buds/daughters. The similarity in morphology between S. cerevisiae rlm1Δ satellite daughters and the pathogenic form of P. brasiliensis (and other Paracoccidioides species), suggests that the pathogenicity in this species may be connected to the CWI pathway. Indeed, cell-wall synthesis genes are strongly induced in the pathogenic form relative to the filamentous form (Tavares et al. 2015). Furthermore, Paracoccidioides infections occur in women much less frequently than in men, and one mechanism proposed for this gender-specific difference is that estrogen inhibits the CWI pathway/cell-wall synthesis (Shankar et al. 2011; Tavares et al. 2015). For this reason, antifungal drugs that inhibit formation or maintenance of rlm1Δ satellite-group formation may be candidates for treatment of Paracoccidioides infections.
Several lines of evidence link the satellite-group phenotype to a defective cell-wall stress response. In the first place, the Rlm1 transcription factor is a target of the CWI pathway that responds to cell-wall stress. In the second place, low osmolarity and NFC sources, the conditions that trigger satellite-daughter formation in the rlm1Δ mutant, activate the Rlm1 branch of the CWI pathway (Merchan et al. 2004; Krasley et al. 2006; Jin et al. 2014; Piccirillo et al. 2015). In the third place, rlm1Δ satellite daughters are more permeable to the dye trypan blue than are wild-type daughters.
The role of other components of the CWI pathway in the satellite-cell morphology is surprisingly complex given that the CWI pathway is the principal activator of Rlm1. In particular, deleting genes for either of two essential components of the CWI/MAPK pathway (SLT2 or BCK1) did not result in the satellite-group phenotype. Indeed, deleting either SLT2 or the Slt2-target SWI4 in the rlm1Δ mutant suppressed the satellite phenotype. One explanation for these results is that under LA conditions, Slt2 has opposite roles in regulating START; it can either delay START by activating Rlm1 or advance this transition by activating SWI4. Interestingly, several other MAPK pathways (Hog1, Fus3, and Kss1) inhibit the START transition in response to other signals (Cherkasova et al. 1999; Escote et al. 2004).
It was initially puzzling that of the four NFCs we tested (acetate, ethanol, glycerol, and pyruvate), only glycerol failed to promote satellite daughters. Two subsequent experiments indicated that this result reflects the requirement for osmotic imbalance in triggering satellite-daughter formation. First, decreasing the concentration of glycerol further allowed efficient rlm1Δ satellite formation. Second, augmenting the standard low-osmolarity medium used in this study with an additional 50 mM solute inhibits satellite-group formation in the mutant. A straightforward interpretation of these results is that, at the low concentrations of NFC source used, glycerol is more effective than these other NFCs at osmo-stabilizing cells. For example, at low concentrations glycerol may be less efficiently transported into the cell than other NFCs, making it more effective at maintaining osmotic balance.
The CW/START checkpoint meets the following criteria for checkpoints (Figure 6): (1) the CW/START checkpoint responds to cellular damage to delay the cell cycle at a specific stage (G1); (2) this checkpoint is abolished by a specific mutation (rlm1Δ) resulting in precocious START, cell damage, and death; and (3) the rlm1Δ mutant can be rescued by a treatment that delays G1 (UV irradiation). Thus, we suggest respiration at low osmolarity results in cell-wall stress that triggers the CW/START checkpoint, an Rlm1-dependent delay in G1, until the cell wall is repaired. In the rlm1Δ mutant, repeated rounds of bypassing this checkpoint results in satellite groups because the mother cell, but not the daughters, continue to divide.
Both the CW/START checkpoint and the morphogenesis checkpoint connect cell-wall stress to cell-cycle progression through the CWI pathway. However, the morphogenesis checkpoint acts at G2 and is Rlm1 independent; whereas the CW/START checkpoint acts at G1 and is Rlm1 dependent. Also the morphogenesis checkpoint depends on Swe1/Mih1 controls on Cdc28 phosphorylation, whereas the CW/START checkpoint does not. Thus, the CWI pathway may be linked to cell-cycle progression at both G1/S and G2/M, but if this is so then this pathway acts at these two stages through separate mechanisms.
Several lines of evidence connect the CW/START checkpoint to the actin cytoskeleton. First, the rlm1Δ bypass of the CW/START checkpoint at G1/S did not result in chromosomal instability or in any detectable defect in bud growth, mitosis, or cytokinesis. Second, a connection between the CWI pathway and the actin cytoskeleton is well established; for example, multiple components of the CWI pathway regulate the actin cytoskeleton (Mazzoni et al. 1993, 2003; Delley and Hall 1999; Harrison et al. 2001; Gao and Bretscher 2009; Guo et al. 2009). Third, the specific phenotypes of rlm1Δ satellite daughters implicate a defect in the actin cytoskeleton; satellite daughters shrink rapidly, fail to grow or rebud, increase in permeability, and accumulate high levels of diffuse actin cytoskeleton.
The connections between the Rlm1-dependent CW/START checkpoint, the formation of satellite daughters, and the actin cytoskeleton are unknown. However, our finding that rlm1Δ daughter (but not mother) cells lose viability soon after cytokinesis suggests the working hypothesis that an “actin-organizing center” (AOC) must duplicate before cells can transit G1, and one copy of this AOC must segregate into the daughter cell for this daughter to thrive. Thus the AOC would be a G1 analog of the polarisome, which organizes the polarized actin cytoskeleton during bud growth (reviewed in Sagot et al. 2002; Bidlingmaier and Snyder 2004; Kadota et al. 2004). For example, the AOC might function to distribute actin patches evenly over the plasma membrane in G1 and hence promote isomorphic growth.
In this working hypothesis, AOC duplication depends on adequate cell-wall synthesis. Hence, in low-osmolarity NFC media, cell-wall stress results in an unduplicated AOC. In wild-type cells, the lack of a duplicated AOC would activate the Rlm1-dependent CW/START checkpoint until cell-wall synthesis and hence AOC duplication recovers. In the rlm1Δ mutant, bypass of this CW/START checkpoint results in bud emergence without AOC duplication in the low-osmolarity condition. In this mutant, the lack of a duplicated AOC does not prevent bud growth, mitosis, or cytokinesis, but it does prevent actin reorganization after cytokinesis. The mammalian centrosome was recently proposed as an AOC in mammalian cells (Farina et al. 2016), but in yeast the spindle pole body (SPB) functions normally in mitosis even when the CW/START checkpoint is bypassed. Association between a duplicated AOC and the duplicated SPB provides a mechanism by which the duplicated AOC could segregate efficiently to mother and daughter cells in M phase.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.204206/-/DC1.
Acknowledgments
We thank Rong Li (Johns Hopkins University) and Sue Jaspersen (Stowers Institute of Medical Research) for helpful discussions, David Amberg (State University of New York Upstate Medical University) for the pGAL-GFP-TUB3 plasmid and for advice on staining actin cables, and David Levin (Boston University) for the UASRlm1-LacZ plasmid. We are grateful to James Bean and Frederick R. Cross (The Rockefeller University) for allowing us to cite their unpublished work. Research reported in this publication was supported by the National Institutes of General Medical Sciences of the National Institutes of Health under award number R15 GM-094770. We acknowledge use of the confocal microscope in the University Missouri, Kansas City School of Dentistry, Confocal Microscopy Core. This facility is supported by the University of Missouri–Kansas City (UMKC) Office of Research Services, UMKC Center of Excellence in Dental and Musculoskeletal Tissues, and National Institutes of Health grant S10 RR-027668. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Communicating editor: D. J. Lew
Literature Cited
- Ahn S. H., Acurio A., Kron S. J., 1999. Regulation of G2/M progression by the STE mitogen-activated protein kinase pathway in budding yeast filamentous growth. Mol. Biol. Cell 10: 3301–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C., 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21: 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Park H. O., 2012. Cell polarization and cytokinesis in budding yeast. Genetics 191: 347–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidlingmaier S., Snyder M., 2004. Regulation of polarized growth initiation and termination cycles by the polarisome and Cdc42 regulators. J. Cell Biol. 164: 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocca A. L., Amaral A. C., Teixeira M. M., Sato P. K., Shikanai-Yasuda M. A., et al. , 2013. Paracoccidioidomycosis: eco-epidemiology, taxonomy and clinical and therapeutic issues. Future Microbiol. 8: 1177–1191. [DOI] [PubMed] [Google Scholar]
- Borges-Walmsley M. I., Chen D., Shu X., Walmsley A. R., 2002. The pathobiology of Paracoccidioides brasiliensis. Trends Microbiol. 10: 80–87. [DOI] [PubMed] [Google Scholar]
- Charvin G., Oikonomou C., Siggia E. D., Cross F. R., 2010. Origin of irreversibility of cell cycle start in budding yeast. PLoS Biol. 8: e1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova V., Lyons D. M., Elion E. A., 1999. Fus3p and Kss1p control G1 arrest in Saccharomyces cerevisiae through a balance of distinct arrest and proliferative functions that operate in parallel with Far1p. Genetics 151: 989–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delley P. A., Hall M. N., 1999. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 147: 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E., Treisman R., 1997. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17: 1848–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escote X., Zapater M., Clotet J., Posas F., 2004. Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat. Cell Biol. 6: 997–1002. [DOI] [PubMed] [Google Scholar]
- Etemad B., Kops G. J., 2016. Attachment issues: kinetochore transformations and spindle checkpoint silencing. Curr. Opin. Cell Biol. 39: 101–108. [DOI] [PubMed] [Google Scholar]
- Farina F., Gaillard J., Guerin C., Coute Y., Sillibourne J., et al. , 2016. The centrosome is an actin-organizing centre. Nat. Cell Biol. 18: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. P., 2016. Getting to S: CDK functions and targets on the path to cell-cycle commitment. F1000 Res. 5: 2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Bretscher A., 2009. Polarized growth in budding yeast in the absence of a localized formin. Mol. Biol. Cell 20: 2540–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia R., Bermejo C., Grau C., Perez R., Rodriguez-Pena J. M., et al. , 2004. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 279: 15183–15195. [DOI] [PubMed] [Google Scholar]
- Goranov A. I., Amon A., 2010. Growth and division–not a one-way road. Curr. Opin. Cell Biol. 22: 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M., Honigberg S. M., 2001. Effect of chromosomal locus, GC content and length of homology on PCR-mediated targeted gene replacement in Saccharomyces. Nucleic Acids Res. 29: 5156–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Shen X., Yan G., Ma D., Bai X., et al. , 2009. A MAP kinase dependent feedback mechanism controls Rho1 GTPase and actin distribution in yeast. PLoS One 4: e6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer B., Aggeli D., Viggiano S., Burke D. J., Amberg D. C., 2011. Novel interactions between actin and the proteasome revealed by complex haploinsufficiency. PLoS Genet. 7: e1002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J. C., Haber J. E., 2006. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40: 209–235. [DOI] [PubMed] [Google Scholar]
- Harrison J. C., Bardes E. S., Ohya Y., Lew D. J., 2001. A role for the Pkc1p/Mpk1p kinase cascade in the morphogenesis checkpoint. Nat. Cell Biol. 3: 417–420. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A., 1989. Checkpoints: controls that ensure the order of cell cycle events. Science 246: 629–634. [DOI] [PubMed] [Google Scholar]
- Honigberg S. M., 2016. Similar environments but diverse fates: responses of budding yeast to nutrient deprivation. Microb. Cell 3: 302–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak C. E., Luscombe N. M., Qian J., Bertone P., Piccirrillo S., et al. , 2002. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 16: 3017–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell A. S., Lew D. J., 2012. Morphogenesis and the cell cycle. Genetics 190: 51–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Strich R., Cooper K. F., 2014. Slt2p phosphorylation induces cyclin C nuclear-to-cytoplasmic translocation in response to oxidative stress. Mol. Biol. Cell 25: 1396–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A., Skotheim J. M., 2013. Start and the restriction point. Curr. Opin. Cell Biol. 25: 717–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Nishikawa J. L., Breitkreutz B. J., Tyers M., 2002. Systematic identification of pathways that couple cell growth and division in yeast. Science 297: 395–400. [DOI] [PubMed] [Google Scholar]
- Jung U. S., Levin D. E., 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 34: 1049–1057. [DOI] [PubMed] [Google Scholar]
- Jung U. S., Sobering A. K., Romeo M. J., Levin D. E., 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46: 781–789. [DOI] [PubMed] [Google Scholar]
- Kadota J., Yamamoto T., Yoshiuchi S., Bi E., Tanaka K., 2004. Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell 15: 5329–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana-Edwin S., Stark M., Kassir Y., 2013. Multiple MAPK cascades regulate the transcription of IME1, the master transcriptional activator of meiosis in Saccharomyces cerevisiae. PLoS One 8: e78920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., Mitchell A., 1994. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Kang H., Lew D. J., 2017. How do cells know what shape they are? Curr. Genet. 63: 75–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. Y., Levin D. E., 2010. Transcriptional reporters for genes activated by cell wall stress through a non-catalytic mechanism involving Mpk1 and SBF. Yeast 27: 541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. Y., Truman A. W., Levin D. E., 2008. Yeast Mpk1 mitogen-activated protein kinase activates transcription through Swi4/Swi6 by a noncatalytic mechanism that requires upstream signal. Mol. Cell. Biol. 28: 2579–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K., Kang H., Jin M., Lew D. J., 2013. Feedback control of Swe1p degradation in the yeast morphogenesis checkpoint. Mol. Biol. Cell 24: 914–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecka M., Yamaguchi M., Kawamoto S., 2015. Effects of the F-actin inhibitor latrunculin A on the budding yeast Saccharomyces cerevisiae. Microbiology 161: 1348–1355. [DOI] [PubMed] [Google Scholar]
- Krasley E., Cooper K. F., Mallory M. J., Dunbrack R., Strich R., 2006. Regulation of the oxidative stress response through Slt2p-dependent destruction of cyclin C in Saccharomyces cerevisiae. Genetics 172: 1477–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E., 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69: 262–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E., 2011. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189: 1145–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N., Biggins S., 2014. Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol. 15: 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Anaya C., Dickinson J. R., Sudbery P. E., 2003. In yeast, the pseudohyphal phenotype induced by isoamyl alcohol results from the operation of the morphogenesis checkpoint. J. Cell Sci. 116: 3423–3431. [DOI] [PubMed] [Google Scholar]
- Mazzoni C., Zarov P., Rambourg A., Mann C., 1993. The SLT2 (MPK1) MAP kinase homolog is involved in polarized cell growth in Saccharomyces cerevisiae. J. Cell Biol. 123: 1821–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni C., Mancini P., Verdone L., Madeo F., Serafini A., et al. , 2003. A truncated form of KlLsm4p and the absence of factors involved in mRNA decapping trigger apoptosis in yeast. Mol. Biol. Cell 14: 721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan J. N., Sia R. A., Bardes E. S., Lew D. J., 1999. Phosphorylation-independent inhibition of Cdc28p by the tyrosine kinase Swe1p in the morphogenesis checkpoint. Mol. Cell. Biol. 19: 5981–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchan S., Bernal D., Serrano R., Yenush L., 2004. Response of the Saccharomyces cerevisiae Mpk1 mitogen-activated protein kinase pathway to increases in internal turgor pressure caused by loss of Ppz protein phosphatases. Eukaryot. Cell 3: 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M., Huang J., Balasubramanian M. K., 2014. The yeast actin cytoskeleton. FEMS Microbiol. Rev. 38: 213–227. [DOI] [PubMed] [Google Scholar]
- Musacchio A., 2015. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Biol. 25: R1002–R1018. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Dirick L., 1991. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell 66: 995–1013. [DOI] [PubMed] [Google Scholar]
- Negishi T., Ohya Y., 2010. The cell wall integrity checkpoint: coordination between cell wall synthesis and the cell cycle. Yeast 27: 513–519. [DOI] [PubMed] [Google Scholar]
- Ogas J., Andrews B. J., Herskowitz I., 1991. Transcriptional activation of CLN1, CLN2, and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell 66: 1015–1026. [DOI] [PubMed] [Google Scholar]
- Paulovich A. G., Toczyski D. P., Hartwell L. H., 1997. When checkpoints fail. Cell 88: 315–321. [DOI] [PubMed] [Google Scholar]
- Piccirillo S., Honigberg S. M., 2010. Sporulation patterning and invasive growth in wild and domesticated yeast colonies. Res. Microbiol. 161: 390–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo S., White M. G., Murphy J. C., Law D. J., Honigberg S. M., 2010. The Rim101p/PacC pathway and alkaline pH regulate pattern formation in yeast colonies. Genetics 184: 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo S., Morales R., White M. G., Smith K., Kapros T., et al. , 2015. Cell differentiation and spatial organization in yeast colonies: role of cell-wall integrity pathway. Genetics 201: 1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo S., Kapros T., Honigberg S. M., 2016. Phenotypic plasticity within yeast colonies: differential partitioning of cell fates. Curr. Genet. 62: 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope P. A., Pryciak P. M., 2013. Functional overlap among distinct G1/S inhibitory pathways allows robust G1 arrest by yeast mating pheromones. Mol. Biol. Cell 24: 3675–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J. R., Adams A. E. M., Drubin D. G., Haarer B. K., 1991. Guide to yeast genetics and molecular biology, pp. 565–602 in Methods in Enzymology, edited by Guthrie C., Fink G. Academic Press, San Diego. [Google Scholar]
- Putnam C. D., Jaehnig E. J., Kolodner R. D., 2009. Perspectives on the DNA damage and replication checkpoint responses in Saccharomyces cerevisiae. DNA Repair 8: 974–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P., 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sagot I., Klee S. K., Pellman D., 2002. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4: 42–50. [DOI] [PubMed] [Google Scholar]
- Sattlegger E., Chernova T. A., Gogoi N. M., Pillai I. V., Chernoff Y. O., et al. , 2014. Yeast studies reveal moonlighting functions of the ancient actin cytoskeleton. IUBMB Life 66: 538–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar J., Wu T. D., Clemons K. V., Monteiro J. P., Mirels L. F., et al. , 2011. Influence of 17beta-estradiol on gene expression of Paracoccidioides during mycelia-to-yeast transition. PLoS One 6: e28402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia R. A., Herald H. A., Lew D. J., 1996. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol. Biol. Cell 7: 1657–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares A. H., Fernandes L., Bocca A. L., Silva-Pereira I., Felipe M. S., 2015. Transcriptomic reprogramming of genus Paracoccidioides in dimorphism and host niches. Fungal Genet. Biol. 81: 98–109. [DOI] [PubMed] [Google Scholar]
- Truman A. W., Kristjansdottir K., Wolfgeher D., Hasin N., Polier S., et al. , 2012. CDK-dependent Hsp70 Phosphorylation controls G1 cyclin abundance and cell-cycle progression. Cell 151: 1308–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J. J., Ewald J. C., Skotheim J. M., 2012. Cell size control in yeast. Curr. Biol. 22: R350–R359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasicova P., Rinnerthaler M., Haskova D., Novakova L., Malcova I., et al. , 2016. Formaldehyde fixation is detrimental to actin cables in glucose-depleted S. cerevisiae cells. Microb. Cell 3: 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Takaesu G., Hagiwara M., Irie K., Matsumoto K., 1997. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17: 2615–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. G., Piccirillo S., Dusevich V., Law D. J., Kapros T., et al. , 2011. Flo11p adhesin required for meiotic differentiation in Saccharomyces cerevisiae minicolonies grown on plastic surfaces. FEMS Yeast Res. 11: 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T., Verna J., Ballester R., 2001. Mutations in WSC genes for putative stress receptors result in sensitivity to multiple stress conditions and impairment of Rlm1-dependent gene expression in Saccharomyces cerevisiae. Mol. Genet. Genomics 266: 142–155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains together with their genotypes are shown in Table S1 and are available on request. All data necessary for the conclusions from the current study are represented either in the manuscript or supplemental files; data from individual trials and repeat trials are available on request. File S1, File S2, and File S3 are videos of dividing yeast cells.