Abstract
Heparan sulfates (HS) are linear polysaccharides with complex modification patterns, which are covalently bound via conserved attachment sites to core proteins to form heparan sulfate proteoglycans (HSPGs). HSPGs regulate many aspects of the development and function of the nervous system, including cell migration, morphology, and network connectivity. HSPGs function as cofactors for multiple signaling pathways, including the Wnt-signaling molecules and their Frizzled receptors. To investigate the functional interactions among the HSPG and Wnt networks, we conducted genetic analyses of each, and also between these networks using five cellular migrations in the nematode Caenorhabditis elegans. We find that HSPG core proteins act genetically in a combinatorial fashion dependent on the cellular contexts. Double mutant analyses reveal distinct redundancies among HSPGs for different migration events, and different cellular migrations require distinct heparan sulfate modification patterns. Our studies reveal that the transmembrane HSPG SDN-1/Syndecan functions within the migrating cell to promote cellular migrations, while the GPI-linked LON-2/Glypican functions cell nonautonomously to establish the final cellular position. Genetic analyses with the Wnt-signaling system show that (1) a given HSPG can act with different Wnts and Frizzled receptors, and that (2) a given Wnt/Frizzled pair acts with different HSPGs in a context-dependent manner. Lastly, we find that distinct HSPG and Wnt/Frizzled combinations serve separate functions to promote cellular migration and establish position of specific neurons. Our studies suggest that HSPGs use structurally diverse glycans in coordination with Wnt-signaling pathways to control multiple cellular behaviors, including cellular and axonal migrations and, cellular positioning.
Keywords: heparan sulfate, cell migration, development, axon guidance, wnt signaling
CELL migrations occur in metazoans and are essential for embryonic development and throughout life. Consequently, pathological conditions can arise from aberrant migrations. Some of these pathologies include neurodevelopmental diseases, tumor formation, and metastasis (Lauffenburger and Horwitz 1996; Gleeson and Walsh 2000; Sahai 2007). Cell migrations are especially important for nervous system development, as a substantial number of neurons in the human brain are born distant from the location they will adopt later in life (Jessell 2000; Marin et al. 2010). Migrating cells utilize molecular and cellular information in the extracellular milieu to determine position and direction (Lauffenburger and Horwitz 1996). The information received by the moving cell is integrated and intracellular changes result in directed migration to the appropriate anatomical position (Reig et al. 2014).
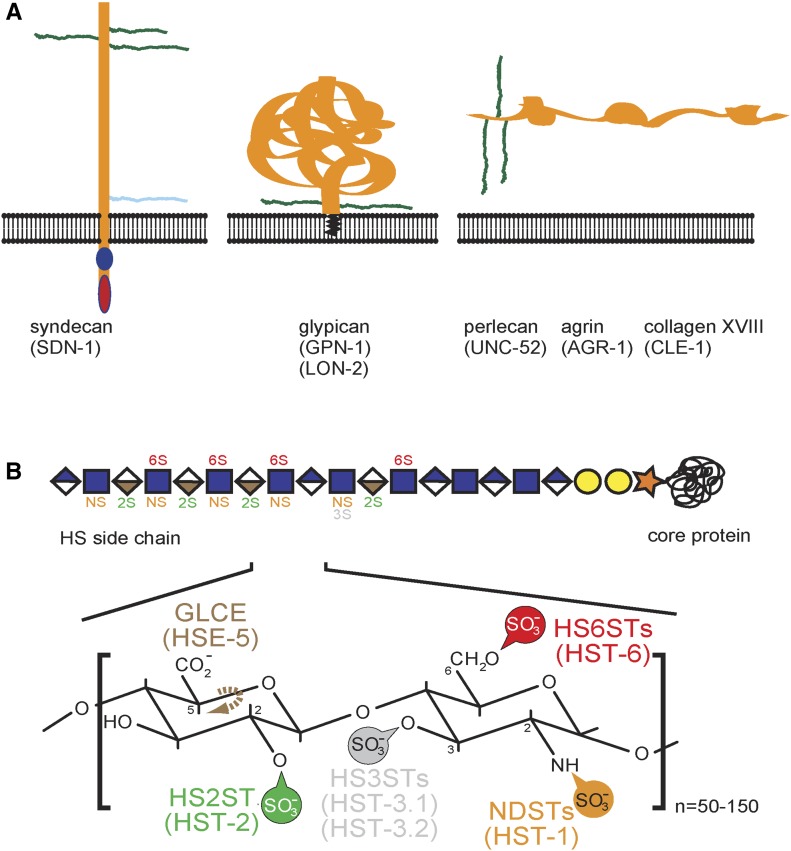
Heparan sulfates (HS) are diverse polysaccharides, which are conjugated to membrane-associated and secreted extracellular proteins to form heparan sulfate proteoglycans (HSPGs). HSPGs contribute to multiple aspects of the development of the nervous system, including cell migration, protein–protein interactions, and cellular responses to protein signaling (Holt and Dickson 2005; Bülow and Hobert 2006; Van Vactor et al. 2006; Poulain and Yost 2015). Proteoglycans can be categorized into structural classes with different functional roles (Figure 1A). These classes include the membrane spanning syndecans with a conserved intracellular domain that interacts with the actin cytoskeleton, and the glycophosphatidylinositol (GPI)-linked glypicans that function in lipid rafts to promote macromolecular trafficking between and into cells. A third class includes secreted forms such as perlecan, agrin, or collagen XVIII (Figure 1A) (Bernfield et al. 1999; Häcker et al. 2005).
Figure 1.
Schematic of heparan sulfate proteoglycans and heparan sulfate structure. (A) Heparan sulfate proteoglycans (HSPGs) are classified as either membrane-bound forms, such as syndecans (sdn-1) and glypicans (lon-2 and gpn-1), or secreted forms, which include perlecan (unc-52), agrin (agr-1), and collagen XVIII (cle-1). C. elegans proteins in all panel shown in parentheses. The HS chains are indicated in green and chondroitin sulfate chains in blue. After Díaz-Balzac et al. (2014). (B) Schematic of the typical disaccharide repeat. HS modification enzymes and the specific positions they modify are shown: NDST, N-deacetylase-N-sulfotransferase; GLCE, C-5-glucuronyl-epimerase; HS2ST, HS-2-O-sulfotransferase; HS3STs, HS-3-O-sulfotransferases; HS6ST, HS-6-O-sulfotransferase. After Díaz-Balzac et al. (2014).
Heparan sulfate glycan chains are unbranched macromolecules comprised of 50–150 disaccharide units of hexuronic acid and glucosamine. A network of enzymes known as heparan sulfate modification enzymes (Figure 1B) can alter and diversify the glycosaminoglycan chains through sulfation, acetylation, and epimerization (Lindahl et al. 1998; Esko and Lindahl 2001; Lindahl and Li 2009). An increasing body of literature has demonstrated the functional importance of HS modification patterns in the development of the nervous system in vertebrates (Yamaguchi 2001; Pratt et al. 2006; Conway et al. 2011; Poulain and Chien 2013; Tillo et al. 2016), as well as in invertebrates such as Caenorhabditis elegans (Bülow and Hobert 2004; Kinnunen et al. 2005; Rhiner et al. 2005; Díaz-Balzac et al. 2014; Wang et al. 2015) and Drosophila melanogaster (Lin and Perrimon 1999; Baeg et al. 2001; Johnson et al. 2004, 2006; Ren et al. 2009; Dani et al. 2012). An important function of HS is its involvement as a cofactor for protein signaling pathways, such as fibroblast growth factor (FGF) signaling, the Slit/Robo axon guidance pathway, and Wnt signaling, among others (Inatani et al. 2003; Bülow and Hobert 2006; Sarrazin et al. 2011). Most, if not all, of these pathways play significant roles in neuronal patterning. Particularly, genes of the Wnt-signaling family have been determined necessary for these cellular processes in different organisms (Lyuksyutova et al. 2003; Ciani and Salinas 2005; Zinovyeva et al. 2008). Wnt proteins are morphogens believed to organize cells and orchestrate formation of body patterns (Zecca et al. 1996). The Frizzled seven-pass transmembrane receptors are responsible for activating the intracellular Wnt-signaling cascade by physically interacting with Wnt ligands on the cellular membrane (Schulte and Bryja 2007; Angers and Moon 2009). Studies in C. elegans have established genetic interactions between the transmembrane HSPG syndecan and Wnt-signaling genes to promote mitotic spindle orientation and distal-tip cell migration, respectively (Schwabiuk et al. 2009; Dejima et al. 2014). Whether and how these interactions influence the development of the nervous system, particularly neuronal migrations and axon guidance in C. elegans, remains largely unknown.
To understand the role of HSPGs and their glycan chains in neuronal patterning in conjunction with Wnt signaling, we first focused on HSPGs. Building on earlier work (Rhiner et al. 2005; Kinnunen 2014; Sundararajan et al. 2015), we performed comprehensive genetic analyses of five cellular and axonal migration events, including ones that differ in their anterior–posterior (A-P) directionality and developmental timing. We then focused on a particular neuronal migration (HSN neurons) and axonal guidance process (PDB neuron) to determine the genetic relationship of HSPGs with Wnt ligands and Frizzled receptors. Our studies show that distinct combinations of proteoglycan core proteins are required for each of the cellular migrations studied. Importantly, the transmembrane-type HSPGs function in the migrating cells, whereas GPI-linked HSPGs are required in surrounding tissues to determine cell position. Each migration event tested requires a unique combination of HS modification(s) for proper cell positioning; that is, each migration is governed by a particular combinatorial code (an HS code) that allows cells to move and find their correct location. Moreover, we find that egl-20/Wnt requires the presence of sdn-1/Syndecan to promote migration of HSN neurons. Our epistasis analysis including the Wnt-signaling system indicates that different Wnt/Frizzled receptors genetically act with different HSPGs to mediate cell migration and axonal guidance in a redundant and context-dependent fashion.
Materials and Methods
C. elegans strains
All strains were maintained using standard methods (Brenner 1974). Experiments were performed at 20° unless otherwise mentioned. All rescue experiments conducted in a pha-1 mutant background, were performed at 22°. Mutant alleles used included: LGI: mig-1(e1787); lin-17(n671, n3091); lin-44(n1792); LGII: hst-3.1(tm734); unc-52(e998); LGIII: hse-5(tm472); LGIV: egl-20(n585); LGV: vab-8(e1017); LGX: hst-3.2(tm3006); sul-1(gk151); lon-2(e678); hst-6(ok273); sdn-1(zh20); hst-2(ok595); gpn-1(ok377). Fluorescent reporter strains included: zdIs5 I [Pmec-4::GFP, lin-15 (+)] (Clark and Chiu 2003), dzIs75 II [Pkal-1p9::GFP, Pttx-3::mCherry] (this study), mgIs18 IV [Pttx-3::GFP] (Altun-Gultekin et al. 2001), otIs76 IV [Pttx-3::kal-1] (Bülow et al. 2002), kyIs262 IV [Punc-86::myr GFP, odr-1::RFP (Adler et al. 2006), zdIs13 IV [Ptph-1::GFP] (Clark and Chiu 2003), otIs33 IV [Pkal-1::GFP] (Bülow et al. 2002), muIs49 [Punc-22(+), Pegl-20::egl-20::GFP] (gift from Gian Garriga; Pan et al. 2006), opIs170 [Psdn-1::sdn-1::GFP] (Rhiner et al. 2005).
Plasmids and transgenesis
To assemble tissue and cell-specific expression plasmids, the sdn-1 complementary DNA (cDNA) was cloned under control of the following promoters: hypodermal dpy-7 (Gilleard et al. 1997), body wall muscle myo-3 (Okkema et al. 1993), pan-neuronal unc-119 (Maduro and Pilgrim 1995), mechanosensory neuron-specific promoter mec-7 (Driscoll and Chalfie 1991), HSN-promoter unc-86 (Shen and Bargmann 2003), and coelomocytes-specific promoter unc-122 (Loria et al. 2003). All plasmids contained the unc-54 3′UTR, and plasmid sequences are available upon request. Point mutant versions of sdn-1/Syndecan were created by site-directed mutagenesis (Quickchange). For rescue experiments, tissue and cell-specific expression plasmids were injected into EB614 [otIs76mgIs18 IV; sdn-1(zh20) X], EB2507 [dzIs75 II; sdn-1(zh20) X] and EB657 [zdIs5 I; pha-1 (e2123)III; sdn-1(zh20) X], at 5 ng/μl together with myo-3::mCherry or ttx-3::mCherry as an injection marker at 25 or 50 ng/μl. pBX (pha-1(+)) was injected at 40 ng/µl in EB657 to rescue the pha-1(e2123ts) defects in all cases. pBS was added to all injection mixes to a final DNA concentration of 100 ng/µl unless otherwise noted. For a complete list of all transgenic strains used in this study see Supplemental Material Table S1 in File S1.
Quantification of cell migration events
Anterior lateral microtubule (ALM) and anterior ventral microtubule (AVM) neurons were visualized using the zdIs5 I [Pmec-4::gfp] strain. To quantify ALM and AVM position, we divided the distance from the cell bodies to the anus by the length of the animal from the nose to the anus. Null mutations in sdn-1/Syndecan and lon-2/Glypican lead to shorter or longer animals, respectively. To determine whether these mutants affect body proportions, we determined vulva location in relation to the whole animal (Figure S1, I and J in File S1). Vulva location was not affected in these mutants and the length from vulva to anus was not statistically different when compared against controls in L4-stage animals and young adults. These experiments suggest that positioning defects observed in sdn-1 and lon-2 animals are not due to secondary effects as a result of changes in size or proportion of the animals. To illustrate cell migration results, five bins labeled U2 (undermigration), U1, N (normal), O1, O2 (overmigration) were created using the median of the control as reference and the following range of ratios for ALM, respectively: >0.59 (U2), 0.59–0.55 (U1), 0.55–0.49 (N), 0.49–0.43 (O1), <0.43 (O2). For AVM, the following range of ratios was used, respectively: >0.64 (O2), 0.64–0.60 (O1), 0.60–0.56 (N), 0.56–0.52 (U1), <0.52 (U2). Bars represent the percentage of cells located in each bin along the A-P axis of the worm (see Figure 2, A and B).
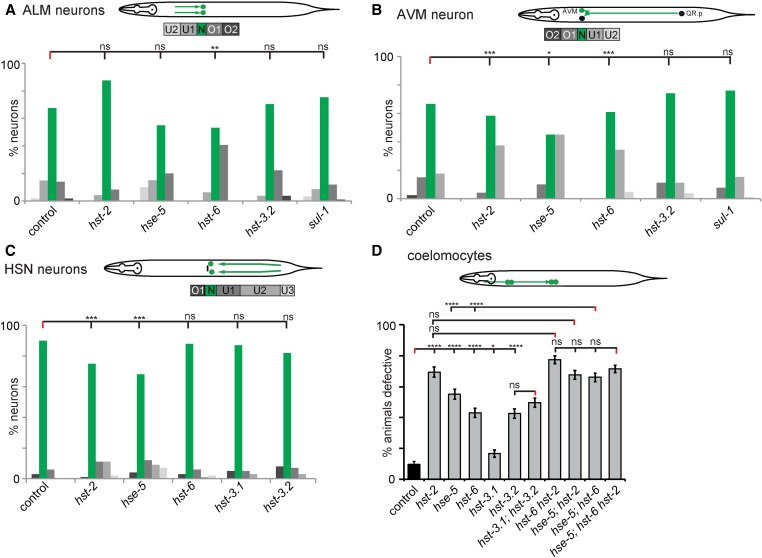
Figure 2.
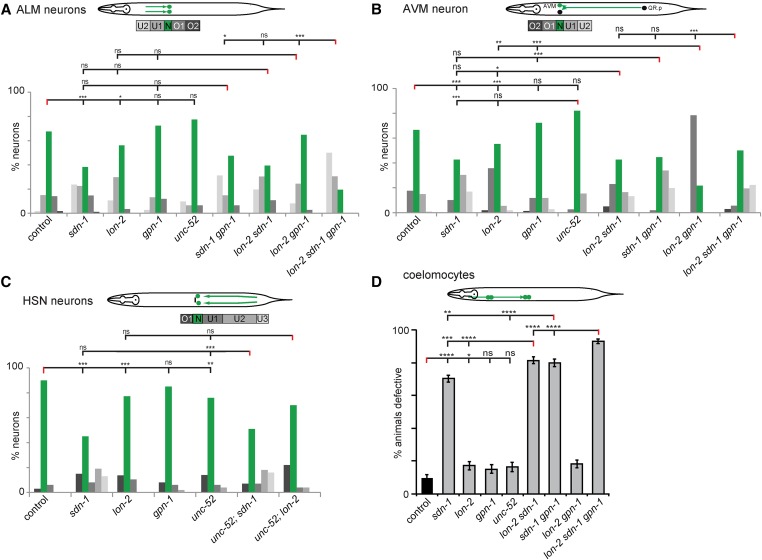
Genetic analysis of heparan sulfate proteoglycans in cell migration. Genetic analysis of ALM (A) and AVM (B) migration in heparan sulfate proteoglycans mutants at the L4/young adult stage as indicated. A schematic of the stereotypical migration of ALM and QR.p (AVM) neurons in C. elegans is shown (top). To illustrate the results, five bins labeled U2 (undermigration), U1, N (normal), and O1 and O2 (overmigration) were created. Bars represent the percentage of cells that are located in each bin along the anterior–posterior (A-P) axis of the worm, with green bars highlighting the normal position and an increase in gray color intensity correlating with the distance from the area the cell(s) were born. Statistical comparisons were performed using the Kolmogorov–Smirnov test. Asterisks in figures represent statistical significance: * P < 0.05, ** P < 0.005, *** P < 0.0005; ns, not significant. N ≥ 36 for all strains measured. (C) Genetic analysis of HSN migration in heparan sulfate proteoglycans mutants of young adults as indicated. A schematic of the stereotypical migration of the HSN neurons in C. elegans is shown (top). HSNs were visualized using the zdIs13 IV [Ptph-1::GFP] strain. Five bins were labeled as O1 (overmigration), N (normal), U1, U2, U3 (undermigration) along the body of the worm. Bars represent the percentage of cells that are located in each bin along the A-P axis of the worm, with the green bars highlighting the normal position and increased intensity of gray correlating with the distance from the area the cell(s) were born. For statistical comparisons, Fisher’s exact test was utilized: * P < 0.05, ** P < 0.005, *** P < 0.0005; ns, not significant. N ≥ 74 for all strains analyzed. (D) Genetic analysis of coelomocytes’ migration in heparan sulfate proteoglycan mutants at the L1 stage as indicated. A schematic of the stereotypical migration of coelomocytes (CC) in C. elegans is shown (top). Coelomocytes’ migration was scored as the percentage of animals that were defective and are demonstrated with error bars representing the SE of proportion. Statistical comparisons were performed using the z-test. Asterisks denote statistical significance: * P < 0.05, ** P < 0.005, *** P < 0.0005, **** P < 0.0001; ns, not significant. N ≥ 172 in all strains analyzed.
For HSN migration, three reporter strains were utilized: zdIs13 [Ptph-1::GFP] IV, dzIs75 [Pkal-1p9::gfp; Pttx-3::mCherry] II, and kyIs262 [Punc-86:: myr GFP; odr-1::RFP] IV, all of which gave comparable results where tested. Five bins labeled as O1 (overmigration), N, U1, U2, U3 (undermigration) were created representing five sections along the body of the worm. Bars represent the percentage of cells located in each section along the A-P axis of the worm (see Figure 2C). Position O1 represents any cell at or anterior to the vulva that is positioned at least two cell body diameters away from the normal position. Position N is the wild-type position of HSN and comprises a section of 0–3 cell body diameters posterior to the vulva. Position U1 is three cell body diameters posterior to the wild-type position (two cell bodies from the center of the gonad primordium in larval animals). Position U2 represents cells that are positioned between the region posterior to the gonad arms and before the preanal ganglion. Position U3 represents the region at or posterior to the preanal ganglion.
Four coelomocyte cells are already present at hatching, while two generate during the first larval stage in hermaphrodites. One pair migrates close to the posterior end of the pharynx and a second pair migrates posteriorly to the midbody next to the gonad primordium. The third pair, which generates in the posterior of the animal during the first larval stage, does not migrate. A defect was scored when any one pair of the migrating coelomocytes fails to move posteriorly. Coelomocytes’ migration was scored using the otIs76mgIs18 [Pttx-3::kal-1, Pttx- 3::GFP] IV strain (Bülow et al. 2002) and is shown as the percentage of animals that were defective with error bars representing the SE of proportion.
Microscopy and imaging
For live microscopy and imaging of C. elegans, animals were mounted on 5% agarose pads in M9 buffer containing 10 mM sodium azide as an anesthetic to immobilize worms. Fluorescent images were acquired using a Zeiss AxioCam MRm camera mounted on an AxioImager Z1 compound microscope equipped with ×16, ×40, and ×63 objectives. Confocal-like images were acquired for some worms using the Zeiss Apotome, which utilizes structured illumination to acquire optical sections. All images were obtained using Axiovision Image acquisition and analysis software from Zeiss. To scale and assemble captured images into figures, Adobe Photoshop and Adobe Illustrator were utilized.
Fluorescent microscopy on embryos and quantification of early HSN defects
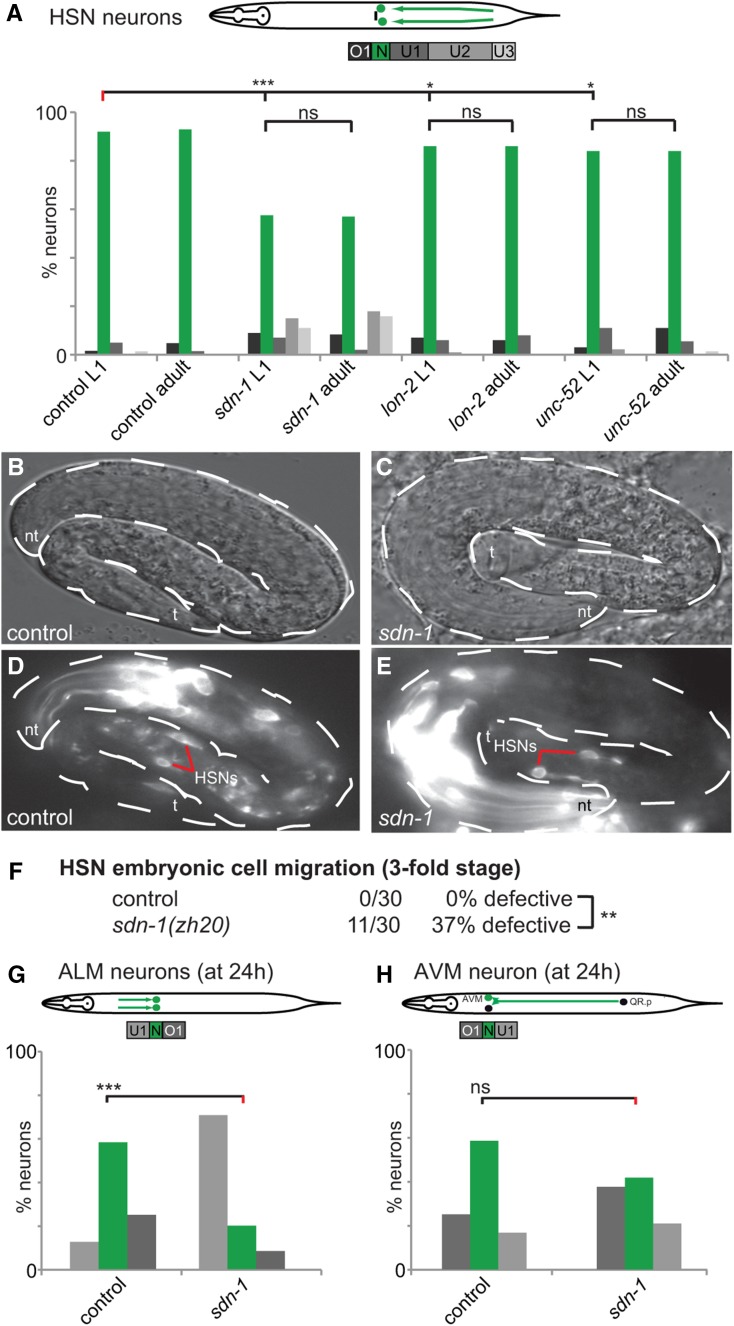
To determine the role of sdn-1/Syndecan in embryos, egg prep synchronization techniques were used as previously described (Stiernagle 1999). Briefly, all adult and larval worms were washed from plates using M9 buffer, and remaining eggs were grown until the desired time point and picked directly onto agar slides. HSN neurons were visualized using the (Punc-86::myr GFP) transgene in wild-type and sdn-1(zh20) mutant embryos starting at the 1.5-fold stage. Embryos showed staining in the anterior portion (that eventually becomes the pharynx/head of the animal) and the tail, where at least two and up to four cells, including the HSNs, could be distinguished. By the 3-fold embryonic stage, we observed that the pair of HSN neurons had migrated to the midbody region, in close proximity to the gonad primordium.
Statistical analysis
For statistical calculations of ALM and AVM cell position, which comprise absolute measurements on a continuous scale, we used the nonparametric Kolmogorov–Smirnov test to analyze differences in the variance and distribution between two samples. Computations were performed using the Prism 7 software package from GraphPad. For the analysis of HSN cell position, five bins – created using anatomical landmarks of the worm – were compared between genotypes on a 5 × 2 contingency table and by applying Fisher’s exact test between two samples using an online calculator provided at http://www.quantitativeskills.com/sisa/index.htm (Uitenbroek 1997). For statistical analysis related to coelomocyte migration and PDB axon guidance phenotypes, which were recorded as proportions of affected animals, we applied the z-test. To correct for multiple comparisons, Bonferroni post hoc analysis was performed when necessary.
Data availability
All strains and reagents are available upon request. Figure S1 in File S1 shows additional schematics and genetic data. Figure S2 in File S1 shows correlations between HSN neuron and CAN position in different mutant backgrounds. Figure S3 in File S1 provides additional rescue data with tissue-specific promoters. Figure S4 in File S1 provides additional genetic data pertaining to the genetic interactions between lon-2/Glypican and egl-20/Wnt. Figure S5 in File S1 shows additional data for PDB patterning. Table S1 in File S1 provides a complete list of transgenic strains created for this study. Complete raw data and details on statistical analyzes are provided in supplemental files. File S2 comprises all data used to create figures. File S3 comprises all statistical analyses, and File S4 contains raw measurements for all AVM- and ALM-related analyses.
Results
Heparan sulfate proteoglycans function in a combinatorial fashion for cell migration and positioning of neuronal and nonneuronal cells
To determine the functional interactions within the HSPG network during cell migration, we first performed a comparative genetic analysis of HSPG mutants in four different migration events in C. elegans. These included the migration of the ALM and the precursor of the AVM neurons, the pair of hermaphrodite-specific (HSN) motor neurons, and two pairs of the nonneuronal coelomocytes (Figure S1, A–D in File S1) (reviewed in Hedgecock et al. 1987).
ALM neurons migrate posteriorly from their birth place to a lateral position close to the midbody region during embryogenesis (Sulston and Horvitz 1977). During growth into adulthood, this position along the A-P axis is maintained relative to the surrounding tissue. We found that in L4/young adult animals, mutations in genes encoding the HSPG core proteins sdn-1/Syndecan and lon-2/Glypican changed the distribution of ALM position with one or both ALM neurons being more anteriorly displaced, i.e., undermigrating compared to controls (Figure 2A), indicating that both genes are individually required for correct positioning of ALM neurons. In contrast, a null mutant in gpn-1/Glypican or a mutant allele of unc-52/Perlecan that harbors a nonsense mutation affecting several splice variants, displayed no significant defects indicating that gpn-1/Glypican and the unc-52/Perlecan splice variants are dispensable for proper ALM position (Figure 2A). All double mutant combinations of sdn-1/Syndecan, lon-2/Glypican, and gpn-1/Glypican failed to show enhancement when compared to the single mutants, whereas a lon-2sdn-1gpn-1 triple mutant showed a significantly enhanced undermigration defect when compared to the more severe of the double mutants. These findings indicate that all three genes act in a partially redundant fashion to establish ALM position (Figure 2A).
The AVM neuron is the descendant of QR.p, a cell on the right side of the animal, which is born postembryonically and migrates anteriorly during the first larval stage (Sulston and Horvitz 1977). Toward the end of the migration, the QR.p cell undergoes a series of stereotyped cell divisions to give rise to the AVM neuron. We found the AVM cell body was displaced posteriorly in sdn-1/Syndecan mutants at the L4/young adult stage. Like the ALM neurons, lon-2/Glypican mutants showed an anterior displacement defect in AVM neurons, which was enhanced in a double mutant with the second glypican gpn-1/Glypican (Figure 2B). These results suggest that sdn-1/syndecan is required to establish AVM position while lon-2/Glypican serves a partially redundant function with gpn-1/Glypican. None of the other double mutants (sdn-1gpn-1, lon-2sdn-1) or the lon-2sdn-1gpn-1 triple mutant showed a more severe phenotype than the sdn-1/Syndecan single mutant. These findings indicate that sdn-1/Syndecan function is epistatic to the role of the two glypicans lon-2 and gpn-1. Collectively, the results of ALM and AVM genetic analyses suggest that sdn-1/Syndecan has a role in promoting the migration or maintaining the position of ALM and AVM, while the GPI-linked HSPGs may function in providing absolute positional information. Alternatively, and not mutually exclusive, GPI-linked HSPGs may serve an inhibitory role during certain migrations.
Next, we analyzed the HSN neurons, which are born in the tail and migrate anteriorly to the midbody region during embryogenesis (Desai et al. 1988). Similar to previous reports (Rhiner et al. 2005; Kinnunen 2014), we found that the HSN neurons display severe undermigration defects in sdn-1/Syndecan mutants whereas lon-2/Glypican mutants show a broader distribution of HSN position (Figure 2C). We found that unc-52/Perlecan mutant animals also displayed positional defects (not statistically different from lon-2 but different from sdn-1 mutant animals), which were not enhanced in double mutants with lon-2/Glypican or sdn-1/Syndecan (Figure 2C). It should be kept in mind that CAN cells (canal-associated neurons) can provide a “stop signal” for migrating HSNs (Forrester and Garriga 1997), and hence the defects in HSPG mutants could be secondary effects due to CAN cell positioning defects. We do not believe this to be the case, because overmigrating HSNs show poor correlation with CAN cell position, with even occasional inversions where the HSN neurons are located anterior to CAN cells (Figure S2 in File S1). Thus, our data collectively show that sdn-1/Syndecan, lon-2/Glypican, and unc-52/Perlecan may act together to mediate HSN position (Figure 2C).
To investigate a nonneuronal cell migration, we focused on two pairs of coelomocytes, which are born in the head of the animals and migrate posteriorly to the midbody region during embryogenesis (Sulston and Horvitz 1977) in a sdn-1/Syndecan-dependent manner (Rhiner et al. 2005) (Figure 2D). With the possible exception of lon-2/Glypican we found no defects in any of the other proteoglycan mutants tested at the L1 stage (Figure 2D). However, we found significantly enhanced defects when sdn-1 mutants were combined with mutations in lon-2 or gpn-1 (Figure 2D). Moreover, the lon-2sdn-1gpn-1 triple mutant further enhanced the defects, consistent with a role for both glypicans in parallel genetic pathways with sdn-1/Syndecan during coelomocyte migration (Figure 2D).
HSPGs serve developmental and maintenance functions in a context-dependent manner
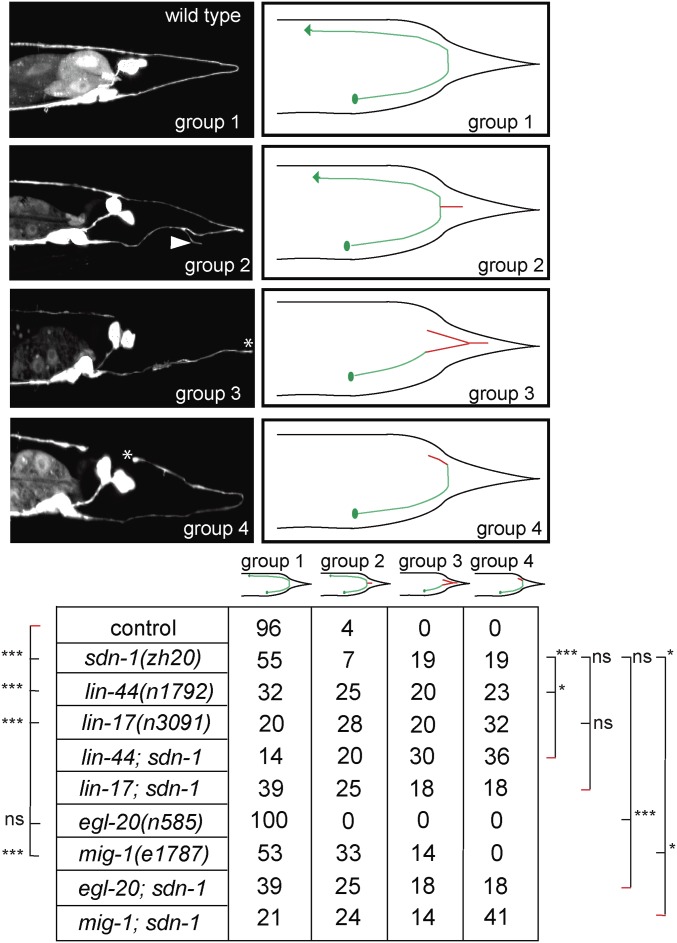
The defects in neuron position in various HSPG mutants could be the result of a bona fide embryonic migration defect or a failure to maintain the correct cellular position during postembryonic growth. To distinguish between these possibilities, we analyzed HSN migration and determined whether HSPG mutants displayed a similar range of migration defects at early developmental stages. We found that defects in sdn-1/Syndecan, lon-2/Glypican, and unc-52/Perlecan mutants at the early L1 larval stage were statistically indistinguishable from defects in adult animals (Figure 3A). Moreover, we saw a similar range of defects in sdn-1 mutants already during embryogenesis, ∼2 hr after completion of HSN migration (Figure 3, B–F). We conclude that the defects in HSN position in sdn-1/Syndecan as well as, likely, lon-2/Glypican and unc-52/Perlecan mutants are bona fide migration/positioning rather than defects in maintenance of cell position.
Figure 3.
Genetic analysis of neuronal cell migration defects in HSPG mutants at early developmental stages. (A) Analysis of heparan sulfate proteoglycan mutants at the L1 stage as indicated. HSN neurons were visualized using the dzIs75 II [Pkal-1p9::GFP, Pttx-3::mCherry] strain. Statistical analyses were performed to compare L1 stage mutant defects against young adult defects of the same strain using Fisher’s exact test. Asterisks denote statistical significance: * P < 0.05, *** P < 0.0005, ns, not significant. N ≥ 49 for all strains analyzed. (B–E) Representative images of HSN neuron positions at the threefold embryonic stage. Control (B and D) and sdn-1(zh20) mutant animals (C and E) as indicated. HSNs were visualized using the kyIs262 IV [Punc-86::myr GFP; odr-1::RFP] strain. nt and t denote the nose tip and tail, respectively. Red lines highlight the position of the HSN neurons in D and E. At this early stage (roughly 2 hr after starting its migration), the HSN neurons display a posterior displacement from each other in sdn-1 mutants. (F) Quantification of HSN migration defects in control and sdn-1(zh20) mutants in 3-fold stage embryos. Statistical significance is indicated: ** P < 0.005. (G–H) Quantification of ALM (G) and AVM (H) migration defects at 24 hr after egg laying of control and sdn-1(zh20) mutant animals. A schematic of the stereotypical migration of ALM and QR.p (AVM) neurons in C. elegans is shown (top), including three bins labeled U1 (undermigration), N (normal), O1 (overmigration) for ALM migration and three bins labeled O1 (overmigration), N, U1 (undermigration) for QR.p (AVM) migration. Bars represent the percentage of cells that are located in each bin along the A-P axis of the worm, with the green bars highlighting the normal position and increased intensity of gray correlating with the distance from the area the cell(s) were born. *** P < 0.0005; ns, not significant. N ≥ 23 for strains analyzed.
We also analyzed neuronal positioning of ALM and AVM neurons at earlier larval time points in sdn-1/Syndecan mutants. Interestingly, we found that while ALM migration was already defective at 24 hr after egg laying, we observed no statistically significant differences for AVM migration in sdn-1/Syndecan mutant animals compared to control animals at this time point (Figure 3, G and H). These results imply that sdn-1/Syndecan may be required for maintenance of AVM position in addition to possible functions in migration. The latter result resembles the phenotype observed in AQR migration (a descendant of QR.a) where sdn-1 is also necessary to maintain cell position (Sundararajan et al. 2015). Collectively, our analyses argue that HSPGs may serve functions both during development and maintenance of cellular migration/positioning events in a gene- and context-dependent manner.
SDN-1/Syndecan functions cell-autonomously to promote cell migration
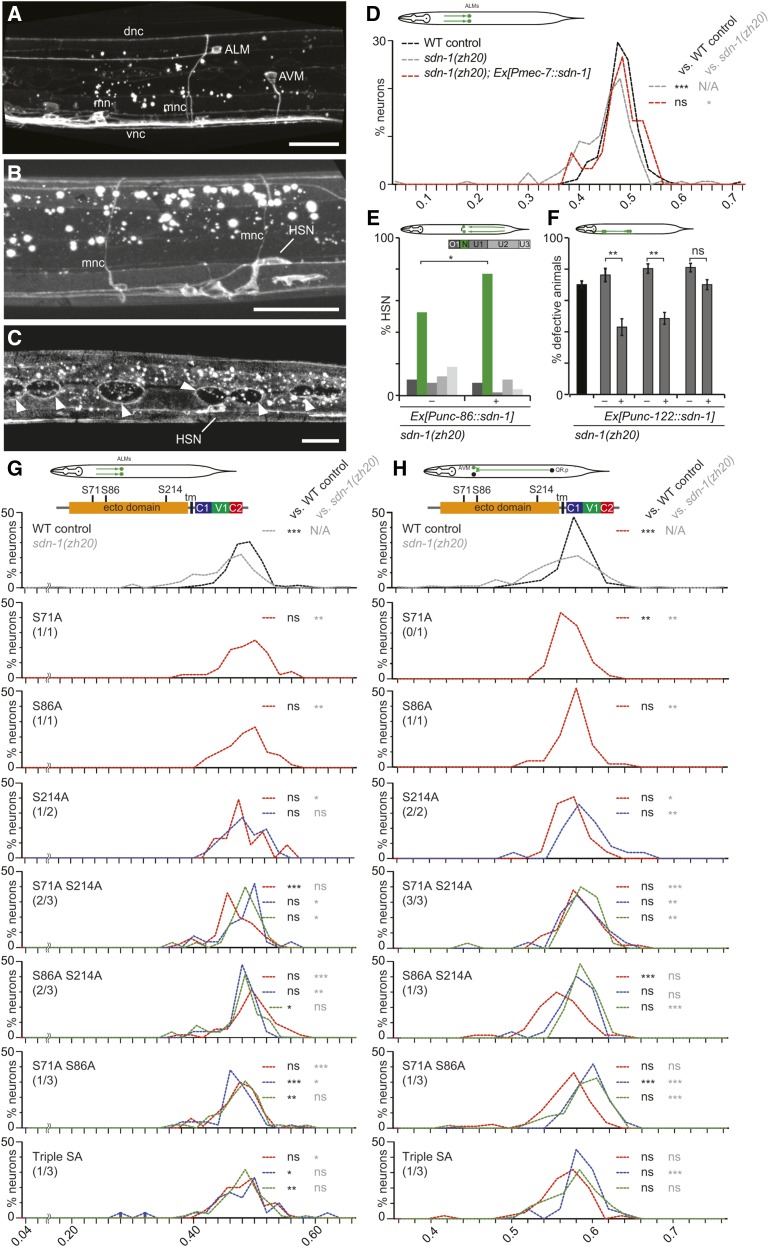
Several lines of evidence suggest that sdn-1/Syndecan functions primarily in a cell-autonomous fashion during cell migration. First, a syndecan translational reporter is expressed in the AVM, ALM, and HSN neurons (Rhiner et al. 2005) (Figure 4, A–C). Second, transgenic rescue experiments by us and others with pan-neuronal promoters driving expression of SDN-1/Syndecan rescue migration defects of ALM, AVM, or HSN neurons (Rhiner et al. 2005) (Figure S3 in File S1). Third, expression of SDN-1/Syndecan under control of a touch neuron-specific (Pmec-7) or an HSN neuron-specific promoter (Punc-86) rescued the ALM and HSN migration defects, respectively (Figure 4, D and E and Figure S3 in File S1). Similarly, coelomocyte migration defects were rescued by expression of sdn-1/Syndecan specifically in coelomocytes (Figure 4F). In addition, AQR migration defects (another QR descendant) have been rescued by transgenic expression with a promoter that is expressed in the Q cells (Pegl-17) (Sundararajan et al. 2015). However, we observed some partial rescue upon expression of sdn-1/Syndecan under control of nonautonomously acting promoters (Figure S3A in File S1). On the other hand, rescue under control of autonomously acting promoters is sometimes not complete (Rhiner et al. 2005). Since a sdn-1/Syndecan reporter is also expressed in other tissues (Figure 4C) (Rhiner et al. 2005), these findings raise the possibility that in addition to its cell-autonomous function, SDN-1/Syndecan may also serve nonautonomous functions.
Figure 4.
Rescue of cell migration defects in sdn-1 mutants. (A–C) Fluorescent images of animals expressing the translational reporter Psdn-1::sdn-1::GFP. (A) Expression of SDN-1::GFP is seen in the ALM and AVM mechanosensory neurons. Expression was also observed in the motor neurons (mn), motor neuron commissures (mnc), dorsal nerve cord (dnc), and ventral nerve cord (vnc). (B and C) SDN-1::GFP expression is seen in the HSN neurons. Expression was also detected in the motor neuron commissures (mnc) and in the hypodermis around the seam cells (arrowheads). Bars indicate 20 µm. (D) Distribution of ALM cell positions in wild-type (WT) control (black dashed line), sdn-1(zh20) mutant (gray dashed line), and sdn-1(zh20) mutant animals with a representative rescuing transgene (red dashed line). The X axis shows the ratio of the distance from the cell body to the anus divided by the length of the animal from the nose to the anus to indicate cell position along the anterior–posterior axis of the animal. All animals contained the zdIs5 I [Pmec-4::GFP] transgene to visualize ALM neurons, a pha-1(e2123) mutant allele, and transgene containing pha-1 rescuing DNA plus additional transgenes as indicated. Statistical comparisons were calculated using the Kolmogorov–Smirnov test and are indicated in black (for comparison against WT) and gray (for comparison against sdn-1(zh20) mutant animals). Asterisks denote statistical significance: *** P < 0.0005; ns, not significant. N ≥ 22 for all lines analyzed. Data for two additional transgenic lines is shown in File S4. (E) Distribution of HSN cell positions (shown as the percentage of HSNs in bins as indicated) in sdn-1(zh20) mutant animals containing a representative rescuing transgene (+) or their nontransgenic siblings (−). For a comparable WT and mutant distribution, see Figure 2C. Statistical comparisons were calculated using Fisher’s exact test and are indicated as: * P < 0.05. N ≥ 50. Data for seven additional transgenic lines is shown in File S2. (F) The percentage of animals with defects in coelomocyte migration in three independent transgenic lines carrying a rescuing transgene (+) or their nontransgenic siblings (−) (gray bars) are shown with error bars representing the SE of proportion. The defect in sdn-1(zh20) mutant animals (black bar) is shown for comparison only and identical to data shown in Figure 2D. Asterisks denote statistical significance: ** P < 0.005, ns, not significant. N ≥ 86. (G and H) Distributions of ALM (G) and AVM (H) cell positions in sdn-1(zh20); pha-1 mutant animals. Schematics show the cell migration within the animal as well as the SDN-1 primary protein structure with domains indicated (tm: transmembrane). All animals carry transgenes containing the pha-1 rescuing DNA (pBX) alone (labeled “sdn-1(zh20)” in gray), or a construct with the unc-119 promoter driving a wild-type sdn-1 cDNA (labeled “WT control” in black) or mutant versions of the sdn-1 cDNA as indicated (labeled e.g., “S71A,” in red, blue, or green for independent transgenic lines). The number of rescuing lines (defined as statistically different from “sdn-1(zh20)” mutant but not “WT control”) out of the total number of lines is indicated below each point mutant. The X axis shows the ratio of the distance from the cell body to the anus divided by the length of the animal from the nose to the anus to indicate cell position along the anterior–posterior axis of the animal. All animals contained the zdIs5 I [Pmec-4::GFP] transgene to visualize ALM and AVM neurons. Statistical comparisons between individual lines and “WT control” or “sdn-1(zh20)” mutant data (shown in the respective top panels) were calculated using the Kolmogorov–Smirnov test. Asterisks denote statistical significance: * P < 0.05, ** P < 0.005, *** P < 0.0005; ns, not significant, and are color coded in black (for comparisons with “WT control”) and gray (for comparisons with “sdn-1(zh20)”). N ≥ 22 for all lines analyzed. Data for all transgenic lines is shown in File S4.
SDN-1/Syndecan glycosaminoglycan attachment sites have distinct functions to promote neuronal migration
The SDN-1/Syndecan protein is predicted to contain three potential glycosaminoglycan attachment sites (i.e., Ser-Gly motifs) at amino acids position S71, S86, and S214 (Minniti et al. 2004). To determine the function of these attachment sites for correct positioning of ALM and AVM cell bodies, we performed rescue experiments where we mutated the putative glycosaminoglycan attachment sites of SDN-1/Syndecan individually or in combination. We found significant transgenic rescue of AVM and ALM cell positioning when each of the three putative attachment sites were mutated individually (Figure 4, G and H). Similarly, removing the S71 or S86 attachment sites in combination with the S214 site showed significant rescuing activity (Figure 4, G and H), suggesting that the glycosaminoglycan chains attached to S71 or S86 retain substantial activity individually. Consistent with this interpretation, removing both S71 and S86 attachment sites resulted in less rescuing activity, suggesting that the chain on S214 provides less activity (Figure 4, G and H). Surprisingly, the triple mutant versions displayed some rescuing activity for both ALM and AVM positioning (Figure 4, G and H). We cannot exclude indirect effects of these point mutations on folding or trafficking of SDN-1/Syndecan to the cell surface; nor can we formally exclude differences in expression levels in individual transgenic lines. With these caveats in mind, our findings suggest that glycosaminoglycan chains on different attachment sites of SDN-1/Syndecan may be of different importance for SDN-1/Syndecan function during ALM and AVM cell migrations.
Unique combinations of HS epitopes and HSPGs are required for distinct cellular migrations
To gain insight into the HS epitopes that mediate each of the cell migration events reported here, we analyzed the effect of mutations in HS-modifying enzymes (Figure 1B). We found that ALM neurons are posteriorly displaced (i.e., overmigrate) in a mutant of the hst-6/HS 6-O-sulfotransferase, whereas mutations in the hse-5/GLCE glucuronyl epimerase, the hst-2/HS 2-O-sulfotransferase, or the hst-3.2/HS 3-O-sulfotransferase display no statistically significant defects (Figure 5A). These findings indicate a function of HS 6-O-sulfation in restricting migration of ALM neurons whereas the other HS-modifying enzymes are not needed individually. In sdn-1hst-6, or lon-2hst-6 double mutants, the ALM neuron displayed the anterior displacement (i.e., undermigration phenotype) seen in the HSPG core protein single mutants, rather than the hst-6/HS 6-O-sulfotransferase mutant phenotype (Figure S1E in File S1). In other words, the HSPG mutants proved epistatic to the hst-6 mutant. These results suggest that in the hst-6 mutant, migration-promoting HS devoid of 6-O-sulfation may be primarily attached to SDN-1/Syndecan and LON-2/Glypican.
Figure 5.
Genetic analysis of heparan sulfate modification enzymes in cell migration. Quantification of ALM (A) and AVM (B) migration defects in heparan sulfate modifying enzyme mutants at the L4 stage/young adult stage as indicated. A schematic of the stereotypical migration of ALM and AVM neurons in C. elegans is shown (top). Control data are identical to Figure 2, A and B and shown here for comparison only. Bins, bar graphs, and statistical comparisons in all panels are as described in corresponding panels of Figure 2. N ≥ 20 for all strains. (C) Quantification of HSN migration defects in heparan sulfate modification enzyme mutants at the young adult stage as indicated. A schematic of the stereotypical migration of HSN neurons in C. elegans is shown (top). HSN neurons were visualized using the zdIs13 IV [Ptph-1::GFP] strain. Control data are identical to Figure 2C and shown here for comparison only. N ≥ 100 for all strains analyzed. (D) Quantification of coelomocyte migration defects in heparan sulfate modifying enzyme mutants at the L1 stage as indicated. A schematic of the stereotypical migration of coelomocytes in C. elegans is shown (top). Control data are identical to Figure 2D and shown here for comparison only. N ≥ 195 for all strains analyzed.
The AVM neuron displayed posterior displacement phenotypes in mutants of the hst-2/HS 2-O-sulfotransferase, hse-5/GLCE glucuronyl epimerase, or hst-6/HS 6-O-sulfotransferase (Figure 5B). As in the case of ALM positioning, a double mutant between sdn-1/Syndecan and hst-6/HS 6-O-sulfotransferase was not enhanced for the AVM phenotype, suggesting that sdn-1 and hst-6 act in the same pathway and that, by inference, SDN-1/Syndecan may be decorated with 6-O-sulfated HS (Figure S1F in File S1). In contrast, a double mutant between lon-2/Glypican and the hst-6/HS 6-O-sulfotransferase (which have opposing phenotypes), appeared similar yet stronger than the lon-2/Glypican mutant phenotype (Figure S1F in File S1). This observation is consistent with a scenario where the normal function of lon-2/Glypican is to negatively regulate migration mediated by an HSPG modified by the hst-6/HS 6-O-sulfotransferase (Figure S1F in File S1). A possible candidate could be sdn-1/syndecan, which appears epistatic to lon-2/Glypican in the presence of the hst-6/HS 6-O-sulfotransferase (Figure 2B).
In accordance with previous reports (Kinnunen 2014), we found that anterior migration of HSN neurons requires the hse-5/GLCE glucuronyl epimerase and the hst-2/HS 2-O-sulfotransferase (Figure 5C). Previous work also showed that lon-2/Glypican required the HS chains to control HSN position (Pedersen et al. 2013). To elucidate the relationship between HS modifications and lon-2 in this context, we created double mutants between lon-2 and the HS-modifying enzymes. The hse-5; lon-2 double mutant was statistically indistinguishable from either single mutant (Figure S1G in File S1). However, the undermigration defects (of hst-2 and hst-6) appeared epistatic to the positioning defects (of lon-2) in lon-2hst-2 and lon-2hst-6 double mutants. These results argue that hse-5 and lon-2 act in the same genetic pathway and, by inference, that LON-2/Glypican glycans may carry HS chains modified by hse-5. In contrast, the HST-2/HS 2-O-sulfotransferase and the HST-6/HS 6-O-sulfotransferase may be acting on LON-2/Glypican or a different HSPG to promote HSN migration.
Posterior migration of the nonneuronal coelomocytes was dependent on all HS-modifying genes tested (Figure 5D). Whereas mutations in hst-2 failed to enhance the mutant phenotype of either the hse-5 or the hst-6 mutant (Figure 5D), the hse-5; hst-6 double mutant showed a more severe phenotype compared to the hse-5 or hst-6 single mutants, arguing that they act in parallel genetic pathways. Lastly, the hse-5; hst-2hst-6 triple mutant failed to enhance the phenotypes of any doubles (Figure 5D). Thus, at least two HS epitopes may act in parallel for coelomocyte migration: one containing 2-O-sulfation and 6-O-sulfation, the other containing 2-O-sulfation and C-5 epimerization. Double mutants between syndecan and the HS modification enzymes hse-5, hst-2, and hst-6 showed that the latter two acted in a pathway with syndecan whereas hse-5 acted in parallel (Figure S1H in File S1), as previously reported for HSN migration (Rhiner et al. 2005). In summary, our genetic experiments reveal that (1) both neuronal and nonneuronal cell migrations require unique heparan sulfate epitopes, which (2) may be attached to individual or combinations of core proteins. Specifically, sdn-1/Syndecan is necessary to promote cell migration (independently of the direction), while lon-2/Glypican and gpn-1/Glypican may convey positional information along the A-P axis to ensure proper cell positioning.
Distinct HSPG/Wnt gene combinations mediate HSN migration and positioning
Previous research in C. elegans has demonstrated a role of the Wnt family of signaling molecules in controlling A-P-directed neuronal migration and axon guidance (Pan et al. 2006; Silhankova and Korswagen 2007). The C. elegans genome encodes five Wnts including EGL-20, MOM-2, LIN-44, CWN-1, and CWN-2, and four Frizzled-like receptors (Fzs), MIG-1, LIN-17, MOM-5, and CFZ-2 (Sawa and Korswagen 2013). While much is known about the role of Wnt signaling in HSN migration (Forrester et al. 2004; Pan et al. 2006), little is known about the interaction between Wnt signaling and HSPG function in this migration.
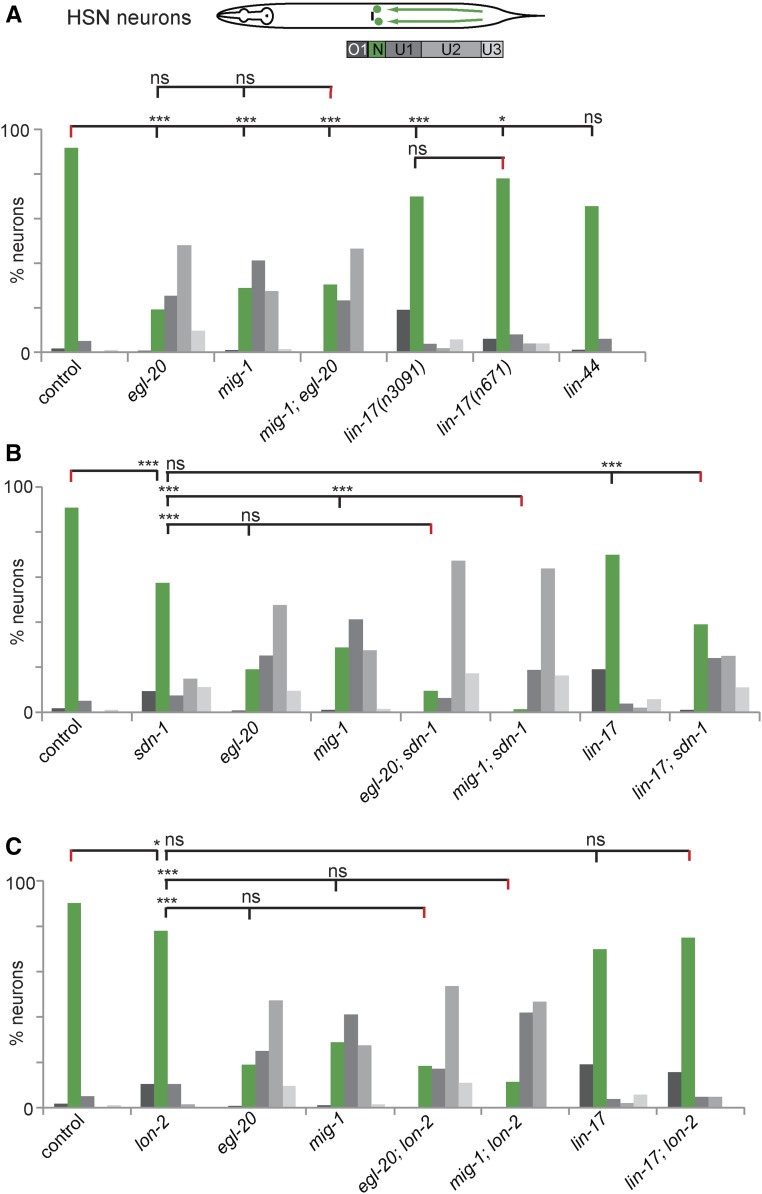
To understand the functional interaction between these two genetic networks, we performed single and double mutant analyses. As previously reported (Pan et al. 2006), egl-20/Wnt and mig-1/Fz mutants exhibited strong HSN migration defects (Figure 6A). Interestingly, the mig-1; egl-20 double mutants did not show a more severe phenotype compared to the single mutants suggesting that these genes act in a genetic pathway (Figure 6A). Thus, MIG-1/Fz is likely the bona fide receptor for the EGL-20/Wnt ligand, while other Wnts may act through alternate, redundantly acting Frizzleds in HSN migration. Because sdn-1/Syndecan and egl-20/Wnt mutants display similar defects in HSN migration, we created a double mutant and found that the distribution of HSN positions in the egl-20; sdn-1 double mutant was not statistically different from an egl-20 single mutant (Figure 6B). In contrast, the mig-1; sdn-1 double mutant showed an enhanced phenotype compared to the single mutants (Figure 6B), suggesting that these genes serve parallel functions, without precluding that they also act in the same genetic pathway. One interpretation of these findings could be that egl-20/Wnt and sdn-1/Syndecan act together during the early stages with possible parallel functions for sdn-1/Syndecan, e.g., in conjunction with other Wnts, at later stages.
Figure 6.
Wnt signaling and HSPGs genetically interact to promote HSN migration and positioning. (A–C) Quantification of HSN migration defects (visualized using the dzIs75 II [Pkal-1p9::GFP, Pttx-3::mCherry] transgene) at the L1 first larval stage. Control and sdn-1/Syndecan and lon-2/Glypican single mutants data are identical to Figure 3C (L1 stage) and shown for comparison only. The lin-17 (n3091) allele was used for double mutant analyses. Bins, bar graphs, and statistical comparisons in all panels are as described in corresponding panels of Figure 2. (A) N ≥ 53, (B) N ≥ 54, and (C) N ≥ 64 for all strains analyzed.
Interestingly, we found that mutations in a second Frizzled receptor, LIN-17/Fz, displayed an overmigration phenotype in two independently isolated null alleles (Figure 6A), in addition to the previously reported undermigration phenotype (Pan et al. 2006; Zinovyeva et al. 2008). This overmigration phenotype was not shared in mutants of lin-44/Wnt, which have been shown in other instances to act in concert with lin-17/Fz (Hilliard and Bargmann 2006; Kirszenblat et al. 2011; Zheng et al. 2015). Since previous work showed that a lin-44; egl-20 double mutant enhanced the HSN undermigration phenotype of egl-20/Wnt (Pan et al. 2006), we cannot exclude that lin-44/Wnt also serves redundant functions in other aspects of HSN patterning. The lin-17/Fz phenotype was reminiscent of the lon-2/Glypican phenotype, raising the possibility of a functional interaction between both genes. We found the lin-17; lon-2 double mutant not statistically different from the lin-17 or lon-2 single mutants, suggesting that both genes act in the same genetic pathway to effect cell positioning (Figure 6C). Even though a lin-17; sdn-1 double mutant was not statistically different from the sdn-1 single mutant (Figure 6B), loss of sdn-1 suppressed the overmigration caused by the lin-17/Fz mutant, suggesting that sdn-1 is epistatic to lin-17. On the other hand, both egl-20/Wnt and mig-1/Fz were epistatic to lon-2/Glypican, because the double mutants were different from the lon-2 single mutant, but statistically indistinguishable from the egl-20/Wnt or mig-1/Fz single mutant phenotypes (Figure 6C). Together with the observation that lin-17/Fz may be a negative regulator of mig-1/Fz function (Pan et al. 2006; Zinovyeva et al. 2008), our results suggest that SDN-1/Syndecan functions with EGL-20/Wnt–MIG-1/Fz whereas LON-2/Glypican functions with LIN-17/Fz to control HSN positioning.
SDN-1/Syndecan is required for EGL-20/Wnt-induced overmigration of HSN neurons
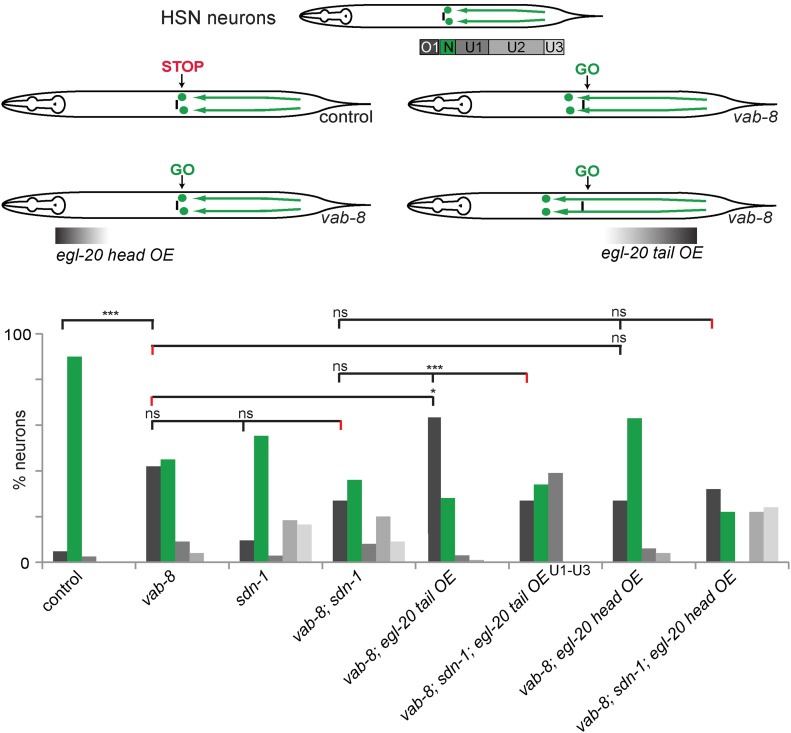
To gain further insight into the functional interaction between EGL-20/Wnt and SDN-1/Syndecan during HSN migration, we used a previously described paradigm, where HSN migration can be manipulated by ectopically expressing EGL-20/Wnt in the head or tail of the animal (Pan et al. 2006). As described above, the CAN cells provide a “stop signal,” which prevents the HSNs from overmigrating (Forrester and Garriga 1997). This impediment to HSN overmigration can be circumvented by removing the activity of vab-8, which results in mis-localization of the CAN cells (Forrester and Garriga 1997). HSN overmigration in this background can be suppressed or enhanced by ectopic expression of EGL-20/Wnt in the head or tail, respectively (Figure 7) (Pan et al. 2006). We predicted that loss of sdn-1/Syndecan activity should suppress HSN migration defects caused by removing vab-8 activity or by ectopic EGL-20/Wnt expression in the vab-8 mutant background. Indeed, we found that the vab-8; sdn-1 double mutant suppressed the phenotypes of vab-8 single mutants (Figure 7), demonstrating that sdn-1 is required for the HSN overmigration caused by mis-localization of the CAN cells. Interestingly, genetically removing sdn-1 activity in a vab-8 mutant background or in a background with ectopic EGL-20/Wnt expression results in a variable distribution of HSN, with a significant number of cells overmigrating and undermigrating, respectively (Figure 7). We interpret these results to mean that SDN-1/Syndecan is required for the standard response of HSN neurons to EGL-20/Wnt, while in the absence of SDN-1/Syndecan, parallel pathway(s) respond to exogenously provided EGL-20/Wnt. Consistent with this interpretation, posterior or anterior overexpression of EGL-20/Wnt in sdn-1/Syndecan mutant animals has no effect on HSN migration (Figure S4B in File S1). Finally, we performed similar overexpression experiments with EGL-20/Wnt in lon-2/Glypican mutants and found no effects in either a wild-type or vab-8 mutant background (Figure S4, C and D in File S1), suggesting LON-2/Glypican serves no direct function in EGL-20/Wnt-dependent HSN migration.
Figure 7.
EGL-20/Wnt requires the presence of SDN-1/Syndecan for HSN neurons to migrate to their final position. (Top) Schematics showing the effects of removing vab-8 and transgenic expression of EGL-20/Wnt in the HSN neurons. (Bottom) Quantification of HSN migration defects in the genotypes indicated as sdn-1/Syndecan, vab-8, and EGL-20 overexpression (OE) in the head (Plim-4::egl-20::GFP, [gmEx365]) or tail of the animals (Pegl-20::egl-20::GFP, [muIs49]) at the young adult stage. We were unable to classify the undermigration defects in the vab-8; sdn-1; muIs49 (EGL-20 tail OE) strain into different bins and therefore grouped them in one bar as U1–U3. HSNs were visualized using the dzIs75 II [Pkal-1p9::GFP, Pttx-3::mCherry] transgene. Control and sdn-1 single mutant data are identical to Figure 3C (adult) and shown here for comparison only. Statistical comparisons were performed against cells that migrated and finished at position O1 (overmigration) unless otherwise noted. Bins, bar graphs, and statistical analyses were done as described in Materials and Methods. N ≥ 46 for all strains analyzed.
sdn-1/Syndecan acts together with Wnt-Frizzled signaling to control axon guidance of the PDB neuron
In addition to the role of Wnt-signaling genes in cell migrations, these molecules have been implicated in the process of axon patterning in C. elegans and vertebrates (Lyuksyutova et al. 2003; Hilliard and Bargmann 2006; Kirszenblat et al. 2011; White et al. 2015). We, therefore, asked whether genetic interactions observed in cellular migration between sdn-1/Syndecan and Wnt-signaling genes were also present in the context of axon guidance. We performed our analysis in a motor neuron residing near the tail of the animal named PDB. This neuron is born postembryonically and sends a process toward the tail, where it loops around to join the dorsal cords in hermaphrodites (Hall and Altun 2008). We found that sdn-1/Syndecan mutant animals showed defects in turning as well as in extending the axon (Figure 8). None of the other proteoglycans tested seem to serve nonredundant functions in this process (Figure S4 in File S1). We next tested different Wnt-signaling genes in this assay and found that lin-44/Wnt, but not egl-20/Wnt, showed a highly penetrant defect in PDB axon guidance, similar to defects observed in sdn-1/Syndecan mutants. Similarly, mutations in the Frizzled receptors lin-17/Fz and mig-1/Fz also showed defects in PDB patterning. To establish any genetic interaction between sdn-1 and Wnt-signaling genes, we examined the following double mutants: egl-20; sdn-1, mig-1; sdn-1, lin-44; sdn-1 and lin-17; sdn-1. Both the egl-20; sdn-1 and the mig-1; sdn-1 double mutants showed enhancement of the phenotypes compared to each of the single mutants, suggesting that sdn-1/Syndecan acts in a parallel genetic pathway to egl-20/Wnt and mig-1/Fz (Figure 8). In contrast, the lin-17; sdn-1 double was not statistically different from the lin-17/Fz single mutant (the more severe of the single mutants), suggesting that both genes act in the same pathway. In support of this conclusion, while the lin-44; sdn-1 double is different from either of the single mutants, this double was also not different from the phenotype observed in the lin-17/Fz single mutant (Figure 8). Thus, sdn-1/Syndecan may function in different cellular contexts in concert with distinct Wnt/Fz signaling systems to control neuronal migration or axon patterning in the nervous system of C. elegans.
Figure 8.
Genetic analysis of PDB axon guidance defects in sdn-1/Syndecan and Wnt-signaling mutants. (Top) Representative images of PDB axon morphology in control (group 1) and mutant (groups 2–4) strains are shown in the top panels, with schematics to the right. The PDB neuron was visualized using the dzIs75 II [Pkal-1p9::GFP, Pttx-3::mCherry] transgene. (Bottom) Quantification of PDB patterning defects at the young adult stage in the genotypes indicated. Axon guidance defects in mutant animals were classified as: group 2, axons that show short ectopic branching, with the majority of cases occurring near the area where the axon turns dorsally; group 3, axons that extended to the tip of the tail instead of its stereotypic trajectory toward the dorsal side of the animal; group 4, axons that were misrouted or stop prematurely and do not join the dorsal cord. Some animals of the Wnt-signaling mutants did not contain a PDB neuron, possibly due to cell-lineage defects in the P12 cell (Jiang and Sternberg 1998). Statistical comparisons were performed against the total amount of defects unless otherwise noted. For statistical significance, the z-test was utilized: * P < 0.05, *** P < 0.0005; ns, not significant. N ≥ 26 in all strains analyzed.
Discussion
We report here a systematic genetic analysis of the functional interactions of HSPGs and Wnt-signaling pathways in cellular migrations, using five different cellular contexts in C. elegans. We found all cellular migration events to require distinct combinations of HSPGs and glycans for correct migration, proper positioning, and maintenance of anatomical location throughout life. Importantly, different HSPGs served different functions. Whereas sdn-1/Syndecan functions primarily in cellular migration, lon-2/Glypican is required for correct positioning of the cells. Moreover, we found that these different functions are mirrored in distinct, context-dependent genetic interactions of sdn-1 and lon-2 with Wnt ligands and Frizzled receptors, suggesting a network of complex interactions between HSPGs and the Wnt-signaling pathway during cellular migrations. The molecular heterogeneity encapsulated in HS chains may be utilized to direct interactions between HSPGs and proteins of the Wnt-signaling cascade during cellular migration.
Multilayered HS codes specify individual cellular migration events
Given the great diversity in structure and function of glycosaminoglycans, and HS in particular, it has been suggested that these glycans encode information and, possibly, constitute an “HS code” (Holt and Dickson. 2005; Bülow and Hobert 2006; Van Vactor et al. 2006; Poulain and Yost 2015). Still, how does the HS code regulate a complex process, such as cellular migration? Our findings reveal several important concepts regarding the HS code(s) that guide cells during normal development.
First, we show that different HSPGs serve distinct functions during cellular migration. We find that sdn-1/Syndecan functions in promoting migration in most cells tested, including neuronal and nonneuronal cellular contexts, irrespective of the directionality of the migration (Table 1). These data are consistent with the hypothesis that SDN-1/Syndecan is part of the machinery that detects cues and executes directed migrations. In contrast, LON-2/Glypican appears to serve a role in providing positional information during migration. That is, in all of the migrations reported here LON-2/Glypican mutants are anteriorly placed regardless of the direction of migration. For example, in posterior–anterior migrations LON-2/Glypican serves an inhibitory role whereas in anterior–posterior migrations LON-2 functions to promote migration (Table 1). One simple interpretation of these results is that glypicans can positively and negatively regulate the process of cell migration in a context-dependent manner. Alternatively, LON-2 could function as a “posteriorizing signal” for a migrating cell rather than being involved in the execution of the migration itself. The mechanism of LON-2/Glypican function could be to bind and sequester signaling molecules to shape gradients throughout the animal, resulting in tight regulation of positioning migrating cells (see below the discussion of Glypican function in Drosophila). Specifically, LON-2/Glypican could limit the diffusion of an anteriorizing morphogen, e.g., EGL-20/Wnt, or promote diffusion of a posteriorizing morphogen, e.g., the secreted Frizzled-like protein SFRP-1, which acts as a Wnt antagonist (Harterink et al. 2011). Recent publications have shown that lon-2/Glypican mediates neuronal development in different regions of the animal by acting from the hypodermis (skin) (Pedersen et al. 2013; Blanchette et al. 2015). On the other hand, LON-2/Glypican may be expressed in a locally restricted manner to create signals that are read by cells or simply provide an adhesive substrate.
Table 1. Summary of HS functions in cell migration and positioning.
| Anterior–posterior migrations | Posterior–anterior migrations | |||
|---|---|---|---|---|
| Gene | ALM neuron | Coelomocytes | AVM neuron | HSN neurons |
| sdn-1/Syndecan | +++ | ++++ | ++ | +++ |
| lon-2/Glypican | +++ | + | − − − | +/− |
| gpn-1/Glypican | ns | ns | ns | ns |
| unc-52/Perlecan | ns | ns | ns | +/− |
| hse-5/GLCE | ns | ++++ | + | +++ |
| hst-2/HS2ST | ns | ++++ | +++ | +++ |
| hst-6/HS6ST | − − − | ++++ | +++ | ns |
| hst-3.2/HS3ST | ns | ++++ | ns | ns |
Statistical comparisons are against the control strain in all cases. Number of pluses or minuses corresponds to the P-value when compared to control; (+) P < 0.05, (++) P < 0.005, (+++) P < 0.0005, (++++) P < 0.0001; ns, not significant. Note: +/− indicates that this gene is required for correct cell positioning (mutants show both overmigration and undermigration defects).
Second, we find HSPGs to act in a combinatorial fashion for different migrations. While sdn-1/Syndecan and lon-2/Glypican represent the major proteoglycans required for migration and positioning in all cellular contexts tested (summarized in Table 1), gpn-1/Glypican and unc-52/Perlecan act mostly in a redundant fashion, possibly in compensatory functions in the absence of sdn-1/Syndecan or lon-2/Glypican. Nonetheless, we observe different degrees of redundancy between them, suggesting differences in the combinatorial use of these factors in a given migration event.
Third, we find that different HS attachment sites on the SDN-1/Syndecan core protein may serve different functions during the migration/positioning of cells. A possible explanation is that interacting proteins require the HS glycans to be attached in particular positions for optimal stereochemical interactions. Alternatively, but not mutually exclusive, different attachment sites could be decorated with different HS glycan chains. Regardless, our findings related to attachment sites in SDN-1/Syndecan are also consistent with multivalency experiments performed in cell culture and zebrafish, which showed that HSPG function can correlate with the amount of linked HS chains (Gopal et al. 2010; Nguyen et al. 2013). Similar to findings by Dejima et al. (2014), we were able to partially rescue migration defects when removing all three putative attachment sites in SDN-1. A plausible explanation is that overexpression of the core protein alone is sufficient to rescue aspects of the mutant phenotype since HSPGs core proteins can confer functions independently of their glycan chains (Kirkpatrick et al. 2006; Yan et al. 2009, 2010). We also cannot rule out the possibility that there are cryptic attachment sites in SDN-1/Syndecan or that a different HSPG is functioning redundantly.
Fourth, all migration events utilize different combinations of HS modifications, similar to other developmental scenarios investigated over the years in vertebrates and invertebrates (reviewed in Poulain and Yost 2015). Together, these findings demonstrate a previously unappreciated, multilayered HS code to ensure correct localization of migrating cells in C. elegans. Fully understanding this HS code will require a better understanding of the glycan structures that mediate function in vivo. We propose that the genetic analyses performed in the present study, coupled with future structural studies of glycosaminoglycans in vivo, will provide important insights toward fully deciphering the HS code in live animals.
Functional interactions between HSPGs and Wnt signaling mediate cell migration and axon guidance
Our findings of distinct functions for syndecans and glypicans in cellular migrations beg the question of how such multilayered HS codes are translated during development. The Wnt-signaling system provided an excellent candidate, for several reasons. First, glypicans and syndecans act in noncanonical Wnt signaling during convergent extensions in vertebrates, where both types of proteoglycans can biochemically and functionally interact with Frizzled receptors and Dishevelled (Topczewski et al. 2001; Muñoz et al. 2006). Second, glypicans are known for their role as positive modulators of Wnts; this regulation has been described in experiments performed with Drosophila Dally and Dally-like (Lin and Perrimon 1999; Baeg et al. 2001). Third, studies of distal-tip cell migration in C. elegans uncovered a possible role of sdn-1/Syndecan as a negative regulator of EGL-20/Wnt in nematodes (Schwabiuk et al. 2009). Lastly, mutations in genes encoding Wnt-signaling components display strikingly similar phenotypes in HSN migration (Forrester et al. 2004; Pan et al. 2006; Zinovyeva et al. 2008). Our genetic loss-of-function and gain-of-function experiments established that egl-20/Wnt requires the presence of sdn-1/Syndecan to promote cell migration through mig-1/Fz. How could this process be mediated? A plausible explanation is that SDN-1/Syndecan retains EGL-20/Wnt (together with MIG-1/Fz) at the cellular membrane, resulting in endocytosis of the signaling complex. There is precedence for this cellular behavior in vertebrates, where binding of Rspo3/R-spondin to Syndecan-4 leads to endocytosis in a Wnt-dependent manner in Xenopus embryos (Ohkawara et al. 2011). Our findings further argue that egl-20/Wnt specifically interacts with sdn-1/Syndecan in HSN migration, since the EGL-20/Wnt overexpression phenotypes in HSN were independent of lon-2/Glypican. Taken together, these findings are consistent with the hypothesis that the syndecan/Wnt axis is important for cellular migration. However, our genetic analysis also suggested that lon-2/Glypican and lin-17/Fz act in a common genetic pathway to control cell positioning of HSN neurons, indicating a genetic interaction between lon-2/Glypican and a distinct set of components of the Wnt-signaling pathway to affect a different aspect of HSN migration. Interestingly, in PDB axon patterning, the interactions are different still. Here, sdn-1/Syndecan acts in a pathway with lin-44/Wnt, in parallel with egl-20/Wnt. Thus, the same HSPGs may engage different proteins of a signaling pathway in different cellular contexts, possibly as a result of molecular differences in their attached glycans. Taken together, we propose HSPGs interact with Wnt ligands in a context-dependent manner by acting as coreceptor(s) with different Frizzled receptors. Future work should focus on determining functional HS domains required for binding of Wnt ligands in vivo and their role in development and possibly disease.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.198739/-/DC1.
Acknowledgments
We thank A. Jenny, R. Pocock, and members of the Bülow laboratory for comments on the manuscript and helpful discussions; the Caenorhabditis Genetics Center (which is funded by the National Institutes of Health (NIH; P40 OD010440) and G. Garriga for strains; and M. Lázaro-Peña for injections. We are grateful to S. Cook for discussions and advice regarding statistical approaches. This work was supported in part by grants from the NIH (T32 GM007491 to K.S.-S. and E.T.; T32 NS07098 to R.A.T.; T32 GM007288 and F31 HD066967 to C.A.D.-B.; RC1 GM090825 and R01 GM01313 to H.E.B.; P30 HD071593 and P30 CA013330 to Albert Einstein College of Medicine). D.S.d.C. is the recipient of a Ph.D. exchange fellowship from Coordenação de Aperfeicoamento de Pessoal de Nível Superior, Brazil. H.E.B. is an Alfred P. Sloan and an Irma T. Hirschl/Monique Weill-Caulier Research Fellow.
Author contributions: K.S.-S., R.A.T., and H.E.B. designed the experiments. K.S.-S., R.A.T., J.D.A., and D.S.d.C. conducted the experiments and analyzed the data with H.E.B. C.A.D.-B. and E.T. provided crucial reagents. K.S.-S. and H.E.B. wrote the paper with editorial input from the other authors.
Footnotes
Communicating editor: D. I. Greenstein
Literature Cited
- Adler C. E., Fetter R. D., Bargmann C. I., 2006. UNC-6/Netrin induces neuronal asymmetry and defines the site of axon formation. Nat. Neurosci. 9: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun-Gultekin Z., Andachi Y., Tsalik E. L., Pilgrim D., Kohara Y., et al. , 2001. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development 128: 1951–1969. [DOI] [PubMed] [Google Scholar]
- Angers S., Moon R. T., 2009. Proximal events in Wnt signal transduction. Nat. Rev. Mol. Cell Biol. 10: 468–477. [DOI] [PubMed] [Google Scholar]
- Baeg G. H., Lin X., Khare N., Baumgartner S., Perrimon N., 2001. Heparan sulfate proteoglycans are critical for the organization of the extracellular distribution of wingless. Development 128: 87–94. [DOI] [PubMed] [Google Scholar]
- Bernfield M., Götte M., Park P. W., Reizes O., Fitzgerald M. L., et al. , 1999. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 68: 729–777. [DOI] [PubMed] [Google Scholar]
- Blanchette C. R., Perrat P. N., Thackeray A., Benard C. Y., 2015. Glypican is a modulator of netrin-mediated axon guidance. PLoS Biol. 13: e1002183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bülow H. E., Hobert O., 2004. Differential sulfations and epimerization define heparan sulfate specificity in nervous system development. Neuron 41: 723–736. [DOI] [PubMed] [Google Scholar]
- Bülow H. E., Hobert O., 2006. The molecular diversity of glycosaminoglycans shapes animal development. Annu. Rev. Cell Dev. Biol. 22: 375–407. [DOI] [PubMed] [Google Scholar]
- Bülow H. E., Berry K. L., Topper L. H., Peles E., Hobert O., 2002. Heparan sulfate proteoglycan-dependent induction of axon branching and axon misrouting by the Kallmann syndrome gene kal-1. Proc. Natl. Acad. Sci. USA 99: 6346–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciani L., Salinas P. C., 2005. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat. Rev. Neurosci. 6: 351–362. [DOI] [PubMed] [Google Scholar]
- Clark S. G., Chiu C., 2003. C. elegans ZAG-1, a Zn-finger-homeodomain protein, regulates axonal development and neuronal differentiation. Development 130: 3781–3794. [DOI] [PubMed] [Google Scholar]
- Conway C. D., Price D. J., Pratt T., Mason J. O., 2011. Analysis of axon guidance defects at the optic chiasm in heparan sulphate sulphotransferase compound mutant mice. J. Anat. 219: 734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani N., Nahm M., Lee S., Broadie K., 2012. A targeted glycan-related gene screen reveals heparan sulfate proteoglycan sulfation regulates WNT and BMP trans-synaptic signaling. PLoS Genet. 8: e1003031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejima K., Kang S., Mitani S., Cosman P. C., Chisholm A. D., 2014. Syndecan defines precise spindle orientation by modulating Wnt signaling in C. elegans. Development 141: 4354–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai C., Garriga G., McIntire S. L., Horvitz H. R., 1988. A genetic pathway for the development of the Caenorhabditis elegans HSN motor neurons. Nature 336: 638–646. [DOI] [PubMed] [Google Scholar]
- Díaz-Balzac C. A., Lázaro-Peña M. I., Tecle E., Gomez N., Bülow H. E., 2014. Complex cooperative functions of heparan sulfate proteoglycans shape nervous system development in Caenorhabditis elegans. G3 4: 1859–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll M., Chalfie M., 1991. The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration. Nature 349: 588–593. [DOI] [PubMed] [Google Scholar]
- Esko J. D., Lindahl U., 2001. Molecular diversity of heparan sulfate. J. Clin. Invest. 108: 169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W. C., Garriga G., 1997. Genes necessary for C. elegans cell and growth cone migrations. Development 124: 1831–1843. [DOI] [PubMed] [Google Scholar]
- Forrester W. C., Kim C., Garriga G., 2004. The Caenorhabditis elegans Ror RTK CAM-1 inhibits EGL-20/Wnt signaling in cell migration. Genetics 168: 1951–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard J. S., Barry J. D., Johnstone I. L., 1997. cis regulatory requirements for hypodermal cell-specific expression of the Caenorhabditis elegans cuticle collagen gene dpy-7. Mol. Cell. Biol. 17: 2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson J. G., Walsh C. A., 2000. Neuronal migration disorders: from genetic diseases to developmental mechanisms. Trends Neurosci. 23: 352–359. [DOI] [PubMed] [Google Scholar]
- Gopal S., Bober A., Whiteford J. R., Multhaupt H. A., Yoneda A., et al. , 2010. Heparan sulfate chain valency controls syndecan-4 function in cell adhesion. J. Biol. Chem. 285: 14247–14258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häcker U., Nybakken K., Perrimon N., 2005. Heparan sulphate proteoglycans: the sweet side of development. Nat. Rev. Mol. Cell Biol. 6: 530–541. [DOI] [PubMed] [Google Scholar]
- Hall D. H., Altun Z. F., 2008. Nervous system, pp. 57–130 in C. elegans Atlas. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Harterink M., Kim D. H., Middelkoop T. C., Doan T. D., van Oudenaarden A., et al. , 2011. Neuroblast migration along the anteroposterior axis of C. elegans is controlled by opposing gradients of Wnts and a secreted Frizzled-related protein. Development 138: 2915–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock E. M., Culotti J. G., Hall D. H., Stern B. D., 1987. Genetics of cell and axon migrations in Caenorhabditis elegans. Development 100: 365–382. [DOI] [PubMed] [Google Scholar]
- Hilliard M. A., Bargmann C. I., 2006. Wnt signals and frizzled activity orient anterior-posterior axon outgrowth in C. elegans. Dev. Cell 10: 379–390. [DOI] [PubMed] [Google Scholar]
- Holt C. E., Dickson B. J., 2005. Sugar codes for axons? Neuron 46: 169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inatani M., Irie F., Plump A. S., Tessier-Lavigne M., Yamaguchi Y., 2003. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science 302: 1044–1046. [DOI] [PubMed] [Google Scholar]
- Jessell T. M., 2000. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1: 20–29. [DOI] [PubMed] [Google Scholar]
- Jiang L. I., Sternberg P. W., 1998. Interactions of EGF, Wnt and HOM-C genes specify the P12 neuroectoblast fate in C. elegans. Development 125: 2337–2347. [DOI] [PubMed] [Google Scholar]
- Johnson K. G., Ghose A., Epstein E., Lincecum J., O’Connor M. B., et al. , 2004. Axonal heparan sulfate proteoglycans regulate the distribution and efficiency of the repellent slit during midline axon guidance. Curr. Biol. 14: 499–504. [DOI] [PubMed] [Google Scholar]
- Johnson K. G., Tenney A. P., Ghose A., Duckworth A. M., Higashi M. E., et al. , 2006. The HSPGs Syndecan and Dallylike bind the receptor phosphatase LAR and exert distinct effects on synaptic development. Neuron 49: 517–531. [DOI] [PubMed] [Google Scholar]
- Kinnunen T. K., 2014. Combinatorial roles of heparan sulfate proteoglycans and heparan sulfates in Caenorhabditis elegans neural development. PLoS One 9: e102919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnunen T., Huang Z., Townsend J., Gatdula M. M., Brown J. R., et al. , 2005. Heparan 2-O-sulfotransferase, hst-2, is essential for normal cell migration in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 102: 1507–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick C. A., Knox S. M., Staatz W. D., Fox B., Lercher D. M., et al. , 2006. The function of a Drosophila glypican does not depend entirely on heparan sulfate modification. Dev. Biol. 300: 570–582. [DOI] [PubMed] [Google Scholar]
- Kirszenblat L., Pattabiraman D., Hilliard M. A., 2011. LIN-44/Wnt directs dendrite outgrowth through LIN-17/Frizzled in C. elegans neurons. PLoS Biol. 9: e1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger D. A., Horwitz A. F., 1996. Cell migration: a physically integrated molecular process. Cell 84: 359–369. [DOI] [PubMed] [Google Scholar]
- Lin X., Perrimon N., 1999. Dally cooperates with Drosophila Frizzled 2 to transduce Wingless signalling. Nature 400: 281–284. [DOI] [PubMed] [Google Scholar]
- Lindahl U., Li J. P., 2009. Interactions between heparan sulfate and proteins-design and functional implications. Int. Rev. Cell Mol. Biol. 276: 105–159. [DOI] [PubMed] [Google Scholar]
- Lindahl U., Kusche-Gullberg M., Kjellen L., 1998. Regulated diversity of heparan sulfate. J. Biol. Chem. 273: 24979–24982. [DOI] [PubMed] [Google Scholar]
- Loria P. M., Duke A., Rand J. B., Hobert O., 2003. Two neuronal, nuclear-localized RNA binding proteins involved in synaptic transmission. Curr. Biol. 13: 1317–1323. [DOI] [PubMed] [Google Scholar]
- Lyuksyutova A. I., Lu C. C., Milanesio N., King L. A., Guo N., et al. , 2003. Anterior-posterior guidance of commissural axons by Wnt-frizzled signaling. Science 302: 1984–1988. [DOI] [PubMed] [Google Scholar]
- Maduro M., Pilgrim D., 1995. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141: 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O., Valiente M., Ge X., Tsai L. H., 2010. Guiding neuronal cell migrations. Cold Spring Harb. Perspect. Biol. 2: a001834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minniti A. N., Labarca M., Hurtado C., Brandan E., 2004. Caenorhabditis elegans syndecan (SDN-1) is required for normal egg laying and associates with the nervous system and the vulva. J. Cell Sci. 117: 5179–5190. [DOI] [PubMed] [Google Scholar]
- Muñoz R., Moreno M., Oliva C., Orbenes C., Larrain J., 2006. Syndecan-4 regulates non-canonical Wnt signalling and is essential for convergent and extension movements in Xenopus embryos. Nat. Cell Biol. 8: 492–500. [DOI] [PubMed] [Google Scholar]
- Nguyen T. K., Tran V. M., Sorna V., Eriksson I., Kojima A., et al. , 2013. Dimerized glycosaminoglycan chains increase FGF signaling during zebrafish development. ACS Chem. Biol. 8: 939–948. [DOI] [PubMed] [Google Scholar]
- Ohkawara B., Glinka A., Niehrs C., 2011. Rspo3 binds syndecan 4 and induces Wnt/PCP signaling via clathrin-mediated endocytosis to promote morphogenesis. Dev. Cell 20: 303–314. [DOI] [PubMed] [Google Scholar]
- Okkema P. G., Harrison S. W., Plunger V., Aryana A., Fire A., 1993. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135: 385–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C. L., Howell J. E., Clark S. G., Hilliard M., Cordes S., et al. , 2006. Multiple Wnts and frizzled receptors regulate anteriorly directed cell and growth cone migrations in Caenorhabditis elegans. Dev. Cell 10: 367–377. [DOI] [PubMed] [Google Scholar]
- Pedersen M. E., Snieckute G., Kagias K., Nehammer C., Multhaupt H. A., et al. , 2013. An epidermal microRNA regulates neuronal migration through control of the cellular glycosylation state. Science 341: 1404–1408. [DOI] [PubMed] [Google Scholar]
- Poulain F. E., Chien C. B., 2013. Proteoglycan-mediated axon degeneration corrects pretarget topographic sorting errors. Neuron 78: 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulain F. E., Yost H. J., 2015. Heparan sulfate proteoglycans: a sugar code for vertebrate development? Development 142: 3456–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt T., Conway C. D., Tian N. M., Price D. J., Mason J. O., 2006. Heparan sulphation patterns generated by specific heparan sulfotransferase enzymes direct distinct aspects of retinal axon guidance at the optic chiasm. J. Neurosci. 26: 6911–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reig G., Pulgar E., Concha M. L., 2014. Cell migration: from tissue culture to embryos. Development 141: 1999–2013. [DOI] [PubMed] [Google Scholar]
- Ren Y., Kirkpatrick C. A., Rawson J. M., Sun M., Selleck S. B., 2009. Cell type-specific requirements for heparan sulfate biosynthesis at the Drosophila neuromuscular junction: effects on synapse function, membrane trafficking, and mitochondrial localization. J. Neurosci. 29: 8539–8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhiner C., Gysi S., Fröhli E., Hengartner M. O., Hajnal A., 2005. Syndecan regulates cell migration and axon guidance in C. elegans. Development 132: 4621–4633. [DOI] [PubMed] [Google Scholar]
- Sahai E., 2007. Illuminating the metastatic process. Nat. Rev. Cancer 7: 737–749. [DOI] [PubMed] [Google Scholar]
- Sarrazin S., Lamanna W. C., Esko J. D., 2011. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 3: a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, H., and H. C. Korswagen, 2013 Wnt signaling in C. elegans, (December 9, 2013), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.7.2, http://www.wormbook.org.. 10.1895/wormbook.1.7.2 [DOI] [PMC free article] [PubMed]
- Schulte G., Bryja V., 2007. The Frizzled family of unconventional G-protein-coupled receptors. Trends Pharmacol. Sci. 28: 518–525. [DOI] [PubMed] [Google Scholar]
- Schwabiuk M., Coudiere L., Merz D. C., 2009. SDN-1/syndecan regulates growth factor signaling in distal tip cell migrations in C. elegans. Dev. Biol. 334: 235–242. [DOI] [PubMed] [Google Scholar]
- Shen K., Bargmann C. I., 2003. The immunoglobulin superfamily protein SYG-1 determines the location of specific synapses in C. elegans. Cell 112: 619–630. [DOI] [PubMed] [Google Scholar]
- Silhankova M., Korswagen H. C., 2007. Migration of neuronal cells along the anterior–posterior body axis of C. elegans: Wnts are in control. Curr. Opin. Genet. Dev. 17: 320–325. [DOI] [PubMed] [Google Scholar]
- Stiernagle T., 1999. Maintenance of C. elegans, pp. 51–67 in C. elegans: a Practical Approach, edited by Hope I. Oxford University Press, Oxford. [Google Scholar]
- Sulston J. E., Horvitz H. R., 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. [DOI] [PubMed] [Google Scholar]
- Sundararajan L., Norris M. L., Lundquist E. A., 2015. SDN-1/Syndecan acts in parallel to the transmembrane molecule MIG-13 to promote anterior neuroblast migration. G3 5: 1567–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo M., Charoy C., Schwarz Q., Maden C. H., Davidson K., et al. , 2016. 2- and 6-O-sulfated proteoglycans have distinct and complementary roles in cranial axon guidance and motor neuron migration. Development 143: 1907–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topczewski J., Sepich D. S., Myers D. C., Walker C., Amores A., et al. , 2001. The zebrafish glypican knypek controls cell polarity during gastrulation movements of convergent extension. Dev. Cell 1: 251–264. [DOI] [PubMed] [Google Scholar]
- Uitenbroek D. G., 1997. SISA Binomial. D.G. Uitenbroek, Southampton. [Google Scholar]
- Van Vactor D., Wall D. P., Johnson K. G., 2006. Heparan sulfate proteoglycans and the emergence of neuronal connectivity. Curr. Opin. Neurobiol. 16: 40–51. [DOI] [PubMed] [Google Scholar]
- Wang X., Liu J., Zhu Z., Ou G., 2015. The heparan sulfate-modifying enzyme glucuronyl C5-epimerase HSE-5 controls Caenorhabditis elegans Q neuroblast polarization during migration. Dev. Biol. 399: 306–314. [DOI] [PubMed] [Google Scholar]
- White R. R., Milholland B., MacRae S. L., Lin M., Zheng D., et al. , 2015. Comprehensive transcriptional landscape of aging mouse liver. BMC Genomics 16: 899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., 2001. Heparan sulfate proteoglycans in the nervous system: their diverse roles in neurogenesis, axon guidance, and synaptogenesis. Semin. Cell Dev. Biol. 12: 99–106. [DOI] [PubMed] [Google Scholar]
- Yan D., Wu Y., Feng Y., Lin S. C., Lin X., 2009. The core protein of glypican Dally-like determines its biphasic activity in wingless morphogen signaling. Dev. Cell 17: 470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Wu Y., Yang Y., Belenkaya T. Y., Tang X., et al. , 2010. The cell-surface proteins Dally-like and Ihog differentially regulate Hedgehog signaling strength and range during development. Development 137: 2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca M., Basler K., Struhl G., 1996. Direct and long-range action of a wingless morphogen gradient. Cell 87: 833–844. [DOI] [PubMed] [Google Scholar]
- Zheng C., Diaz-Cuadros M., Chalfie M., 2015. Dishevelled attenuates the repelling activity of Wnt signaling during neurite outgrowth in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 112: 13243–13248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinovyeva A. Y., Yamamoto Y., Sawa H., Forrester W. C., 2008. Complex network of Wnt signaling regulates neuronal migrations during Caenorhabditis elegans development. Genetics 179: 1357–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All strains and reagents are available upon request. Figure S1 in File S1 shows additional schematics and genetic data. Figure S2 in File S1 shows correlations between HSN neuron and CAN position in different mutant backgrounds. Figure S3 in File S1 provides additional rescue data with tissue-specific promoters. Figure S4 in File S1 provides additional genetic data pertaining to the genetic interactions between lon-2/Glypican and egl-20/Wnt. Figure S5 in File S1 shows additional data for PDB patterning. Table S1 in File S1 provides a complete list of transgenic strains created for this study. Complete raw data and details on statistical analyzes are provided in supplemental files. File S2 comprises all data used to create figures. File S3 comprises all statistical analyses, and File S4 contains raw measurements for all AVM- and ALM-related analyses.