Abstract
An extended meiotic prophase is a hallmark of oogenesis. Hormonal signaling activates the CDK1/cyclin B kinase to promote oocyte meiotic maturation, which involves nuclear and cytoplasmic events. Nuclear maturation encompasses nuclear envelope breakdown, meiotic spindle assembly, and chromosome segregation. Cytoplasmic maturation involves major changes in oocyte protein translation and cytoplasmic organelles and is poorly understood. In the nematode Caenorhabditis elegans, sperm release the major sperm protein (MSP) hormone to promote oocyte growth and meiotic maturation. Large translational regulatory ribonucleoprotein (RNP) complexes containing the RNA-binding proteins OMA-1, OMA-2, and LIN-41 regulate meiotic maturation downstream of MSP signaling. To understand the control of translation during meiotic maturation, we purified LIN-41-containing RNPs and characterized their protein and RNA components. Protein constituents of LIN-41 RNPs include essential RNA-binding proteins, the GLD-2 cytoplasmic poly(A) polymerase, the CCR4-NOT deadenylase complex, and translation initiation factors. RNA sequencing defined messenger RNAs (mRNAs) associated with both LIN-41 and OMA-1, as well as sets of mRNAs associated with either LIN-41 or OMA-1. Genetic and genomic evidence suggests that GLD-2, which is a component of LIN-41 RNPs, stimulates the efficient translation of many LIN-41-associated transcripts. We analyzed the translational regulation of two transcripts specifically associated with LIN-41 which encode the RNA regulators SPN-4 and MEG-1. We found that LIN-41 represses translation of spn-4 and meg-1, whereas OMA-1 and OMA-2 promote their expression. Upon their synthesis, SPN-4 and MEG-1 assemble into LIN-41 RNPs prior to their functions in the embryo. This study defines a translational repression-to-activation switch as a key element of cytoplasmic maturation.
Keywords: oocyte meiotic maturation, translational regulation, RNA-binding proteins, ribonucleoprotein particle purification, cytoplasmic polyadenylation
AN ancient reproductive strategy shared by most sexually reproducing animals is that oocytes arrest in the diplotene or diakinesis stage of meiotic prophase I and resume meiosis in response to hormonal signaling and soma-germline interactions (reviewed by Masui and Clarke 1979; Kim et al. 2013; Li and Albertini 2013). This conserved regulatory mechanism may serve to ensure that oocytes, which are often among the largest cells in an organism, have sufficient time to stockpile the cytoplasmic organelles, cellular constituents, and maternal determinants needed to produce healthy and fertile progeny after fertilization. The final stage of oocyte development, oocyte meiotic maturation, involves the transition to metaphase I (M phase), which is triggered by maturation-promoting factor (MPF), consisting of the CDK1 catalytic subunit and the cyclin B regulatory subunit (Masui and Markert 1971; Dunphy et al. 1988; Gautier et al. 1988, 1990; Lohka et al. 1988; reviewed by Nurse 1990; Masui 2001).
It was recognized nearly 100 years ago that the final stages of oogenesis involve nuclear and cytoplasmic events (Wilson 1925), referred to as nuclear and cytoplasmic maturation, respectively. Nuclear maturation is crucial for ensuring the faithful segregation of meiotic chromosomes. Hallmarks of nuclear maturation include the visually striking events of meiotic chromosome condensation and remodeling, nuclear envelope breakdown (NEBD), and meiotic spindle assembly. Active MPF phosphorylates protein substrates to promote chromosome condensation, NEBD, and meiotic spindle assembly. Cytoplasmic maturation, though less saliently observable and biochemically defined, is no less important. Cytoplasmic maturation includes accumulation and reorganization of cytoplasmic organelles and ribonucleoprotein (RNP) particles, cytoskeletal rearrangements, and changes in protein translation (Li and Albertini 2013; Ivshina et al. 2014). Studies in Drosophila and mice elucidated large-scale changes in protein translation occurring during oocyte meiotic maturation and the oocyte-to-embryo transition (Potireddy et al. 2006; Chen et al. 2011; Kronja et al. 2014a,b). Yet how post-transcriptional remodeling of the oocyte proteome is regulated and contributes to developmental transitions is incompletely understood.
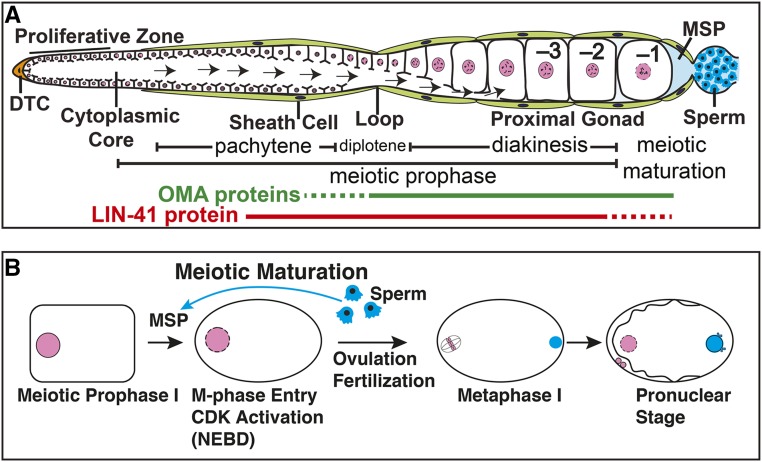
Species-specific signals and soma-germline interactions play a key role in exerting coordinate control of oocyte growth and meiotic maturation. In Caenorhabditis elegans, sperm use the major sperm protein (MSP) as an unconventional hormone to trigger meiotic maturation (Figure 1) (Miller et al. 2001; Kosinski et al. 2005). MSP also stimulates the actomyosin-dependent cytoplasmic flows that drive oocyte growth (Govindan et al. 2009). Oocytes grow and develop in close association with the follicle-like gonadal sheath cells, which appear to serve as the main MSP sensors. Cyclic AMP signaling through protein kinase A in the sheath cells is required for all germline responses to MSP (Govindan et al. 2006, 2009; Kim et al. 2012). Gap-junctional communication between sheath cells and oocytes negatively regulates the MSP-dependent oocyte growth and meiotic maturation responses (Hall et al. 1999; Govindan et al. 2006, 2009; Whitten and Miller 2007; Nadarajan et al. 2009; Starich et al. 2014).
Figure 1.
Representation of (A) adult hermaphrodite gonad arm and (B) oocyte meiotic maturation. The most proximal (−1) oocyte undergoes meiotic maturation in response to MSP secreted from sperm in a process that requires the function of LIN-41 and the OMA proteins. DTC, distal tip cell; arrows in panel (A), cytoplasmic flow for oocyte growth.
Large RNP complexes containing the TIS11 zinc-finger RNA-binding proteins OMA-1 and OMA-2 (collectively referred to as the OMA proteins) and the TRIM-NHL RNA-binding protein LIN-41 appear to represent chief downstream targets of MSP signaling (Spike et al. 2014a,b). The OMA proteins are redundantly required for oocyte meiotic maturation and function upstream of the conserved cell-cycle regulator CDK-1 (Detwiler et al. 2001). The OMA proteins function antagonistically to LIN-41. Genetic analysis established that LIN-41 functions as an essential regulator of the oogenesis program by promoting oocyte growth, inhibiting M-phase entry, and maintaining oocyte quality independently of its role in the heterochronic gene-regulatory pathway that controls developmental timing of somatic cell fates (Spike et al. 2014a). Null mutations in lin-41 cause pachytene-stage oocytes to prematurely cellularize, activate CDK-1, disassemble the synaptonemal complex, enter M phase, assemble spindles, and attempt to segregate chromosomes, resulting in sterility (Spike et al. 2014a). Abnormal oocytes in LIN-41 null mutants aberrantly express genes normally expressed by a variety of differentiated cell types, likely as an indirect consequence of premature CDK-1 activation (Spike et al. 2014a; Tocchini et al. 2014). LIN-41 was shown to inhibit CDK-1 activation in part through the 3′-untranslated region (UTR)-dependent translational repression of the CDK-1 activator, CDC-25.3 phosphatase (Spike et al. 2014a,b). The OMA proteins are also required for translational repression of cdc-25.3 in oocytes (Spike et al. 2014b). This finding raised the question of how the OMA proteins and LIN-41 could copurify in RNPs and mediate the translational repression of common messenger RNA (mRNA) targets despite having diametrically opposed null mutant phenotypes.
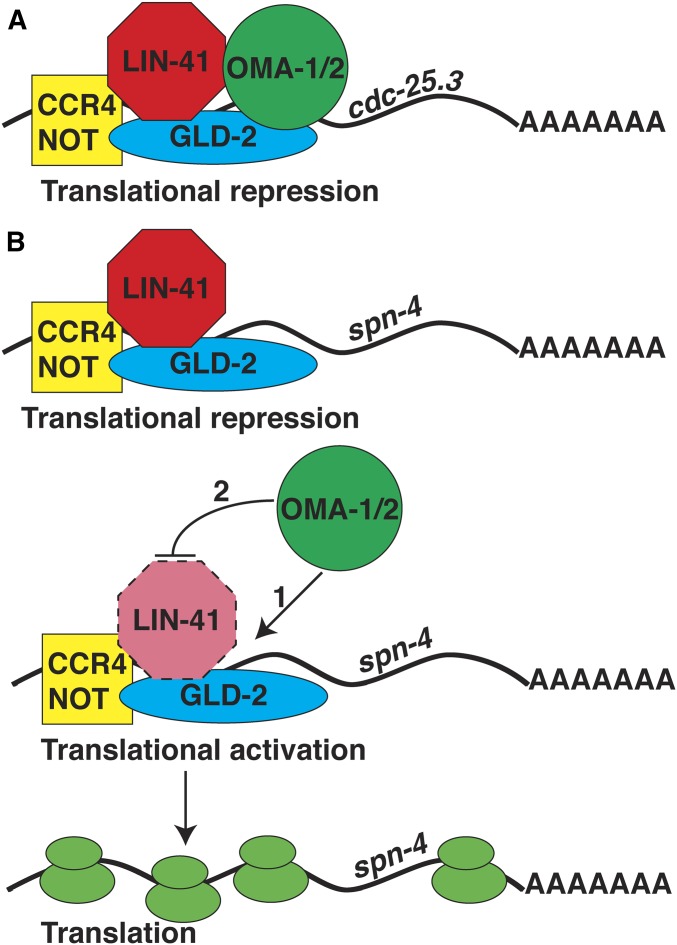
In this study, we investigate the biochemical and genetic mechanisms by which LIN-41 and the OMA proteins regulate oocyte development and the oocyte-to-embryo transition. We show that OMA-1 and LIN-41 are found together in large molecular weight complexes and we employed tandem affinity purification and mass spectrometry to characterize the proteins that copurify with them using an epitope-tagged version of LIN-41. This analysis identified 15 proteins previously shown to copurify with OMA-1 in the presence of RNase A and exhibit RNA-related functions, including several developmental regulatory RNA-binding proteins, the GLD-2 cytoplasmic poly(A) polymerase, the CCR4-NOT deadenylase complex, and translation initiation factors. This analysis is consistent with a major role for LIN-41 and the OMA proteins in regulating protein translation in oocytes. We used RNA sequencing (RNA-seq) to compare the mRNAs associated with LIN-41 and OMA-1. This analysis defined mRNAs that associate with both LIN-41 and OMA-1, as well as sets of mRNAs that selectively associate with LIN-41 or OMA-1. Prior work suggests that GLD-2 cytoplasmic poly(A) polymerases function as translational activators in invertebrates and vertebrates (Wang et al. 2002; Hansen et al. 2004; Kwak et al. 2004; Suh et al. 2006; Benoit et al. 2008; Cui et al. 2008, 2013; reviewed by Ivshina et al. 2014). Consistent with this view, we found that many transcripts that selectively associate with LIN-41 exhibit shortened poly(A) tails in a gld-2 null mutant, suggesting that gld-2 might stimulate the efficient translation of a subset of LIN-41-associated mRNAs. Through an analysis of the translation of two transcripts that associate selectively with LIN-41, spn-4 and meg-1, we provide evidence that LIN-41 and the OMA proteins mediate a translational repression-to-activation switch. LIN-41 represses translation of spn-4 and meg-1, whereas the OMA proteins promote their expression. Both spn-4 and meg-1 transcripts are substrates of the GLD-2 cytoplasmic poly(A) polymerase, and we present a model in which LIN-41, the OMA proteins, and GLD-2 might “toggle” specific mRNA targets between repression and activation. spn-4 and meg-1 encode protein and mRNA components of LIN-41-containing RNPs, and both proteins play important roles as RNA regulators during early embryonic development (Gomes et al. 2001; Huang et al. 2002; Ogura et al. 2003; Jadhav et al. 2008; Leacock and Reinke 2008; Wang et al. 2014). Further, we provide genetic evidence that LIN-41 and SPN-4 function together in the oocyte to ensure that the germline in the next generation develops and functions properly. This study defines a translational repression-to-activation switch mediated by LIN-41 and the OMA proteins as a key element of the cytoplasmic maturation of C. elegans oocytes. This regulatory mechanism is critical for promoting the assembly and function of oocyte RNPs and for controlling protein expression levels during meiotic maturation and the oocyte-to-embryo transition.
Materials and Methods
Strains
The genotypes of strains used in this study are reported in Supplemental Material, Table S1 in File S4. Genes and mutations are described in WormBase (http://www.wormbase.org; Harris et al. 2013) or in the indicated references. All analyses were conducted at 20° unless specified otherwise. The mutations used were as follows. Linkage group I (LGI): rnp-8(tm2435), fog-1(q253ts), gld-2(q497), gld-2(tn1688), unc-13(e1091), lin-41(n2914), lin-41(tn1487ts), lin-41(tn1541[gfp::tev::s::lin-41]), lin-41(tn1749[mkate2::3xflag::lin-41]), and lin-11(n566). LGII: gld-3(q730), lfor-1(tn1652), and lfor-2(tn1653). LGIII: cdc-25.3(tn1712[gfp::3xflag::cdc-25.3]), orc-1(tn1732[mng::3xflag::orc-1]), and unc-119(ed3). LGIV: oma-1(zu405te33). LGV: acy-4(ok1806), spn-4(tm291), spn-4(or191ts), spn-4(tn1699[spn-4::gfp::3xflag]), spn-4(tn1718[spn-4::gfp::3xflag]), spn-4(tn1722[spn-4::gfp::3xflag]), oma-2(te51), oma-2(cp145[mng::3xflag::oma-2]) (Dickinson et al. 2015), cyb-3(tn1755[gfp::3xflag::cyb-3]), and fog-2(oz40). LGX: meg-1(tn1724[gfp::3xflag::meg-1]). The following rearrangements were used: hT2[bli-4(e937) let-?(q782) qIs48] (I; III), mIn1[dpy-10(e128) mIs14] II, and nT1[qIs51] (IV; V). The following transgenes and transgene insertions were used: axIs1688[pCM5.50(pie-1p::gfp::histoneH2B::spn-4 3′-utr); pDPMM0016B(unc-119(+))] (Merritt et al. 2008), tnIs17[pCS410 (oma-1p::oma-1::s::tev::gfp)]; pDPMM0016B(unc-119(+))] V (Spike et al. 2014b), and tnEx37[acy-4(+); sur-5::gfp] (Govindan et al. 2009).
RNA interference
Gene-specific RNA interference (RNAi) was performed by feeding C. elegans with double-stranded RNA (dsRNA)-expressing Escherichia coli (Timmons and Fire 1998) using the RNAi culture media described by Govindan et al. (2006). RNAi clones were obtained from a C. elegans RNAi feeding library (Source BioScience). The identity of RNAi clones was verified by DNA sequencing. The empty vector L4440 was used as a control. Exposure to dsRNA-expressing E. coli was initiated at the beginning of the first larval stage in oma-1(RNAi); oma-2(RNAi), and lin-41(RNAi) experiments, and during the fourth larval stage in cdk-1(RNAi) experiments. RNAi experiments were conducted at 22°, except for RNAi experiments using JH2311 which were performed at 25° to minimize the potential for silencing of the spn-4 3′-UTR reporter. RNAi phenotypes were assessed on the second day of adulthood.
Immunofluorescence, fluorescent labeling, and microscopy
Dissected gonads were fixed in 3% paraformaldehyde as described (Rose et al. 1997), except that 1% paraformaldehyde (1 hr) without a methanol postfixation step was used for anti-GFP antibody staining experiments. Fixed gonads were stained with monoclonal anti-FLAG M2 antibody (Sigma Chemical, St. Louis, MO) at 1:500 or a mixture of monoclonal anti-GFP 12A6 antibody at 1.5 μg/ml and monoclonal anti-GFP 4C9 antibody at 4.2 μg/ml. Secondary antibodies were Cy3-conjugated goat anti-mouse (Jackson ImmunoResearch) used at 1:500 dilution. VECTASHIELD Mounting Medium for fluorescence (Vector Laboratories, Burlingame, CA) containing 4′,6-diamidino-2-phenylindole (DAPI) was used to detect DNA. DIC and fluorescent images were acquired on a Carl Zeiss (Thornwood, NY) motorized Axioplan 2 microscope with either a 40× Plan-Neofluar (numerical aperture of 1.3) or a 63× Plan-Apochromat (numerical aperture of 1.4) objective lens using an AxioCam MRm camera and AxioVision software (Carl Zeiss). For the fluorescence photomicrographs shown in the figures, identical exposures and camera settings were used for the images being directly compared unless noted otherwise. Typically, many animals (>100) of each genotype were examined by fluorescence microscopy. In some cases, exact n values for specific trials are reported. Several confocal photomicrographs in the supporting information were obtained on a Marianas 200 Microscopy Workstation (Intelligent Imaging Innovations) built on an AxioObserver Z.1 stand (Zeiss) and driven by SlideBook 6.0 software (Intelligent Imaging Innovations). The imaging was performed using a 40× oil Carl Zeiss Plan-Apochromat objective lens (numerical aperture of 1.4) and an Evolve electron-multiplying charge-coupled device camera (Photometrics). The Z-stack images were three-dimensionally projected with the mean value method using ImageJ version 2.0.0 software. lin-41(n2914) germline phenotypes were analyzed in adults that had recovered from the dauer stage of development because of their increased somatic health (Spike et al. 2014a). Wild-type hermaphrodites recovered from the dauer stage were analyzed in parallel as controls. None of the phenotypes discussed here depend on the passage through the dauer stage. Entry into M phase in lin-41(n2914); spn-4(tm291) animals was assessed using anti-phosphohistone H3 antibody and anti-lamin antibody as described (Spike et al. 2014a).
Fluorescent protein tagging in genomic contexts and genome editing
CRISPR-Cas9 genome editing used pRB1017 to express single guide RNA (sgRNA) under control of the C. elegans U6 promoter (Arribere et al. 2014). An sgRNA to target lin-41 (lin-41 sgRNA1) was previously described (Spike et al. 2014a). The sequences of all oligonucleotides used are listed in File S1. To generate sgRNA clones, annealed oligonucleotides were ligated to BsaI-digested pRB1017 plasmid vector, and the resulting plasmids were verified by Sanger sequencing. pDD162 served as the source of Cas9 expressed under control of the eef-1A.1/eft-3 promoter (Dickinson et al. 2013). The genes producing the lfor-1 and lfor-2 noncoding RNAs were deleted from the genome using a mixture of pCS539 lfor-2 sgRNA1, pCS541 lfor-1/2 sgRNA2, pCS543 lfor-1 sgRNA3, and the repair oligonucleotides C38C6.8 RP1 and C38C6.7b RP1, using a modification of the method of Paix et al. (2014). The injection mix contained pCS539 sgRNA1, pCS541 sgRNA2, and pCS543 sgRNA3 plasmids each at 25 μg/ml; pDD162 at 50 μg/ml; C38C6.8 RP1 at 500 nM; C38C6.7b RP1 at 500 nM; and Pmyo-2::tdTomato at 4 μg/ml. A total of 318 transgenic F1 progeny of injected wild-type parents were screened by PCR. The primer pairs of sre-13.F and sre-13.R, and of atgp-2.F and atgp-2.R, were used to detect deletions in lfor-1 and lfor-2, respectively. Worms homozygous for the edited deletion of both genes were recovered, verified by Sanger sequencing, and outcrossed to wild-type males three times.
N-terminal or C-terminal translational fluorescent-protein gene fusions to endogenous loci were constructed using the method of Dickinson et al. (2015). The oligonucleotides listed in File S1 were used to generate the sgRNA plasmids and the repair templates. The edited loci were validated by PCR and DNA sequencing. The sequences of the PCR primers and the DNA sequences of the edited loci are available upon request. The following gene fusions were generated and are described and analyzed in some detail herein, with the order of the markers indicating whether an N- or C-terminal fusion was used: gfp::3xflag::cdc-25.3, mkate2::3xflag::lin-41, gfp::3xflag::meg-1, mng::3xflag::orc-1, and spn-4::gfp::3xflag. Strains containing these edited loci were homozygous viable and fertile. To generate double mutants of spn-4::gfp::3xflag with the linked genes acy-4 and oma-2, we independently targeted spn-4 in acy-4(ok1806); tnEx37 and oma-2(te51) strains. We also generated C-terminal TagRFP-T and mKate2 fusions to SPN-4. Although the resulting strains were homozygous viable and fertile, we did not observe TagRFP-T fluorescence in oocytes or embryos, or mKate2 fluorescence in oocytes (some weak mKate2 fluorescence was detectable in early embryos). Anti-FLAG antibody staining of dissected gonads from the DG4121 strain detected SPN-4::TagRFP-T::3xFLAG in the two most proximal oocytes (T. Tsukamoto and D. Greenstein, unpublished results). The cellular dynamics or oxidative environment during oocyte meiotic maturation may be suboptimal for maturation of the TagRFP-T or mKate2 fluorophores in some contexts. For this reason, we used GFP fluorescence whenever possible. For technical reasons related to repair-template construction, expression analysis of ORC-1 used an mNeonGreen (mNG) (Allele Biotechnology) fusion (Shaner et al. 2013).
In addition, we also generated a series of other gene fusions. Although these are not featured in the Results section, they are briefly described here because of their potential interest for the field. These fusions are: mng::3xflag::cpg-1, gfp::3xflag::cyb-3, gfp::3xflag::gla-3a, gfp::3xflag::mex-3, picc-1::gfp::3xflag, gfp::3xflag::pos-1, gfp::3xflag::pqn-59, gfp::3xflag::puf-5, trcs-1::gfp::3xflag, and gfp::3xflag::zyg-11. Some brief comments on these strains are now provided. The GFP::3xFLAG::POS-1 expression pattern was expanded to the loop region following lin-41(RNAi) but not after oma-1(RNAi); oma-2(RNAi), which was similar to that observed in the wild-type genetic background (Figure S1 in File S4). The GFP::3xFLAG::MEX-3 expression pattern was expanded to the loop region in both the lin-41(n2914) and oma-1(zu405te33); oma-2(te51) genetic backgrounds (Figure S1 in File S4). The gfp::3xflag::cyb-3 strain, although maintainable as a homozygote (brood size of 24.6 ± 18.4, n = 10), produced many dead embryos (82.2% embryonic lethality, n = 1373). GFP::3xFLAG::CYB-3 is expressed in all germline nuclei in adults and its expression is not substantially affected by RNAi depletion of lin-41 or oma-1/2 (Figure S2 in File S4). GFP::3xFLAG::GLA-3A is expressed in the germline cytoplasm from the distal tip to near the bend region of the gonad. GFP::3xFLAG::GLA-3A expression was not observed to change in a oma-1(zu405te33); oma-2(te51) genetic background. mNG::3xFLAG::CPG-1 is expressed in the cytoplasm of oocytes and in the eggshell of embryos. GFP::3xFLAG::PUF-5 is expressed in the cytoplasm of oocytes, in the distal gonad, and in the cytoplasm and nuclei of embryos. GFP::3xFLAG::PQN-59 expression was ubiquitous and cytoplasmic. We did not observe fluorescence expression of PICC-1::GFP::3xFLAG and TRCS-1::GFP::3xFLAG in a wild-type background, or after lin-41(RNAi) or oma-1(RNAi); oma-2(RNAi). GFP::3xFLAG::ZYG-11 was nonfunctional.
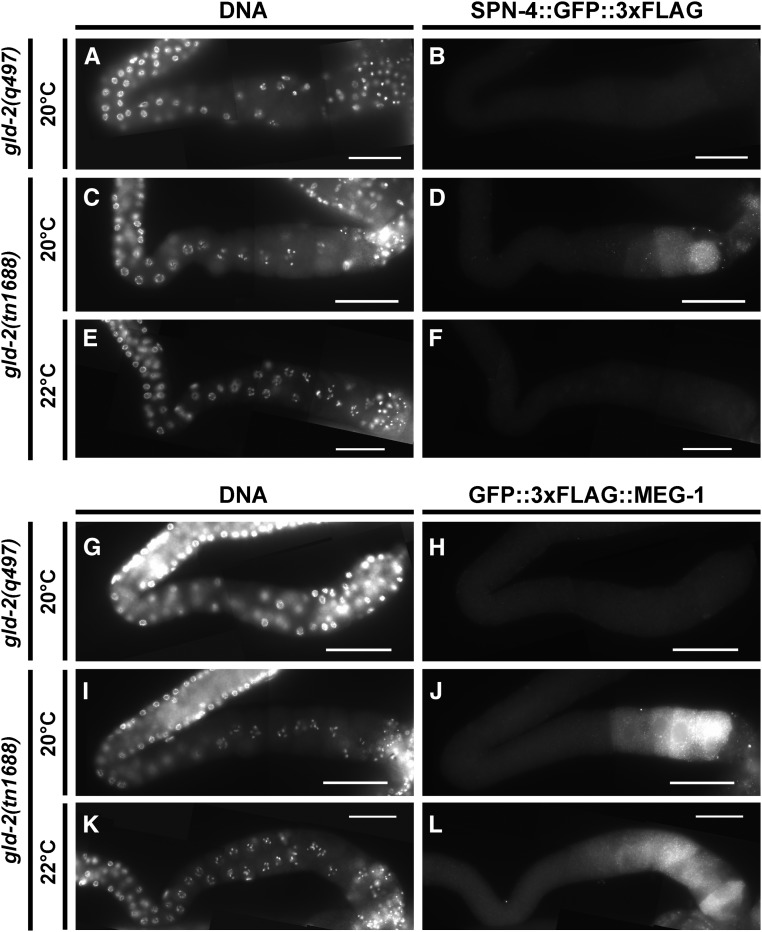
In an attempt to generate a temperature-sensitive gld-2 allele, we sought to use CRISPR-Cas9 genome editing to introduce targeted mutations in the hydrophobic core of the protein, as suggested by theoretical and empirical investigations (Chakshusmathi et al. 2004; Lockwood et al. 2011). We used University of California, San Francisco Chimera (Pettersen et al. 2004) to assess residue solvent accessibility in the GLD-2 crystal structure (Nakel et al. 2015). We identified two residues near the catalytic core (L590 and V592) with a predicted solvent accessibility of <1%, corresponding to a region of the gene with a nearby sgRNA (sg5Gld2sgRNA) predicted (Xu et al. 2015) to be efficient. We used a repair oligonucleotide (sg5_2deg592,590) containing degeneracies at these positions. The repair oligonucleotide also introduced a BamHI site with synonymous coding changes to facilitate screening. The injection mix contained 25 μg/ml of sg5Gld2sgRNA, 50 μg/ml of pDD162, 20 μg/ml of sg5_2deg592,590, 7.5 μg/ml of pJA58.2 (Arribere et al. 2014), and 16 μg/ml of AF-ZF-827 (Arribere et al. 2014). While screening was not exhaustive, we succeeded in isolating a mutation, gld-2(tn1688[L590A, V592H]), which was maternal-effect lethal at 15° and sterile at 25°; resembling gld-2(q497) at 25°. gld-2(tn1688) was outcrossed and balanced with hT2[qIs48]. We also isolated gld-2(tn1689[L590A, V592T]), which is homozygous viable and fertile.
Monoclonal antibody production and purification
Hybridoma cell lines producing anti-GFP monoclonal antibodies 12A6 and 4C9 (Sanchez et al. 2014) were obtained from the Developmental Studies Hybridoma Bank and cultured in T75 flasks (BD Falcon) containing RPMI-1640 medium (Gibco, Grand Island, NY) with the addition of 10% fetal bovine serum (Gibco), 1% HB101 supplement (Irvine Scientific), and 1% penicillin streptomycin (pen-strep) (Gibco) at 37° and 5% CO2. For high-yield antibody production, hybridoma cells were grown in a CELLine 350 Bioreactor (Wheaton) in RPMI-1640 medium with the addition of 15% fetal bovine serum, 1.5% HB101 supplement, and 1% pen-strep. The reservoir for medium exchange contained RPMI-1640 medium and 1% pen-strep. The CELLine 350 Bioreactors were generally reused up to 30 times, harvesting cultures every 7 days. Antibodies were purified using Nab Protein L Spin Columns (Thermo Scientific) according to the manufacturer’s instructions.
Western blots
Proteins were separated using NuPAGE 4–12% Bis-Tris Gels (Invitrogen, Carlsbad, CA) and visualized after Western blotting. Blots were blocked with 5% nonfat dried milk. Primary antibodies used to detect proteins were affinity-purified rabbit anti-LIN-41(203–420) R214 antibody (100 ng/ml) (Spike et al. 2014a) and rabbit anti-GFP NB600-308 antibody (250 ng/ml) (Novus Biologicals). The secondary antibody used for Western blots was peroxidase-conjugated donkey anti-rabbit antibody (1:30,000) (Jackson ImmunoResearch). Detection was performed using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific).
LIN-41 and OMA-1 immunopurifications
GFP::TEV::S::LIN-41 and OMA-1::S::TEV::GFP were purified from strains DG3923 and DG2566, respectively (see Table S1 in File S4). The series of immunopurifications performed in this work are diagrammed in Figure 2. Worms were grown at 15° on peptone-enriched plates seeded with bacterial strain NA22. Embryos were isolated by alkaline hypochlorite treatment (20% bleach and 0.5 N NaOH), washed in M9 buffer, and allowed to hatch overnight in the absence of food at 25° (hatch rates were typically >90%). A total of 30,000 L1-stage animals were cultured on each 150- by 15-mm petri dish on peptone-enriched medium with NA22 as a food source (lysate preparation used 65–70 petri dishes per experiment). Because lysates were prepared by pooling many individually grown cultures, reproducibility was typically assessed using a variety of technical replicates. Animals were fed again in the L4 stage with concentrated NA22 and collected for lysate preparation as young adults, ∼48 hr after being placed on food at 25°. Lysate preparation, GFP::TEV::S::LIN-41 and OMA-1::S::TEV::GFP immunopurification, and tobacco etch virus (TEV) protease digestion was performed as described for OMA-1::S::TEV::GFP immunopurifications (Spike et al. 2014b) and quantitative RT-PCR of OMA-1 target mRNAs (Oldenbroek et al. 2013), with some modifications. Basic IP buffer was 50 mM HEPES (pH 7.5), 1 mM MgCl2, 100 mM KCl. IP wash buffer was basic IP buffer with 300 mM KCl (final concentration), 10% glycerol, 0.05% NP-40, 5 mM 2-mercaptoethanol, 5 mM sodium citrate, 10 μM ZnCl2, and cOmplete Mini EDTA-free protease inhibitors (Roche). For experiment VI and experiment VII, the IP wash buffer contained 1 mM EGTA and no ZnCl2, and similar results were obtained. A high-speed supernatant fraction (∼16 mg/ml protein) was prepared by ultracentrifugation at 100,000 × g for 1 hr at 4°. Dynabeads Protein G (Life Technologies) were coupled and cross-linked to a 1:1 mixture of anti-GFP 12A6 and anti-GFP 4C9 with a ratio of 36 μg of antibodies per 150 μl of Dynabeads according to the manufacturer’s instructions. For tandem affinity GFP::TEV::S::LIN-41 immunopurifications, the second immunopurification step started with the first TEV eluate and used affinity-purified rabbit anti-LIN-41(203–420) R214 antibody coupled to Dynabeads Protein G (36 μg of antibodies per 150 μl of Dynabeads). Immunopurified proteins were eluted from the anti-LIN-41 Dynabeads with 100 mM glycine (pH 2.5) and neutralized immediately with an equal volume of 2 M Tris-Cl (pH 8.5). For analysis of small-scale immunopurifications (Figure 3C), protein gels were stained with SYPRO Ruby Protein Gel Stain (Invitrogen) according to the manufacturer’s instructions. Digestion with 5 μg/ml RNase A (Sigma Chemical) was performed for 15 min at room temperature in IP wash buffer containing 10 μM ZnCl2. Immunopurified proteins were precipitated with 16.7% trichloroacetic acid (TCA), washed with acetone at −20°, and briefly separated on a 12% NuPAGE Bis-Tris Gel stained with Colloidal Blue Staining Kit (Invitrogen). Lanes were subdivided into eight gel slices and mass spectrometry was performed at the Taplin Biological Mass Spectrometry Facility (Harvard Medical School) using an LTQ Orbitrap Velos Pro Ion Trap Mass Spectrometer (Thermo Fisher Scientific). Protein identification used the Sequest software program (Thermo Fisher Scientific) to match the fragmentation pattern of tryptic peptides to the C. elegans proteome. The data were filtered to a 1–2% peptide false discovery rate. File S2 reports the mass spectrometry results and the additional filtering criteria for identifying nonspecific interactions as described in the Results section below.
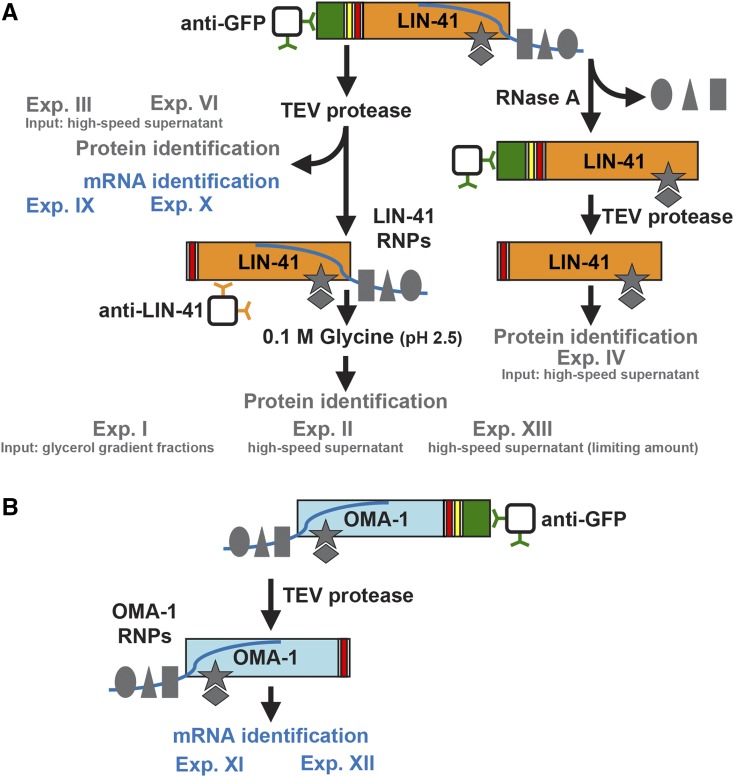
Figure 2.
Overview of purification methods used to characterize (A) LIN-41-interacting proteins and RNAs and (B) OMA-1-interacting RNAs. Endogenous LIN-41 and an integrated OMA-1 transgene were tagged with GFP (green), a TEV protease cleavage site (yellow), and an S-tag (red). Controls (exp. V, exp. VII, and exp. VIII) and filtering criteria are described in the main text and in File S2, which contains the mass spectrometry data. Exp, experiment; gray shapes, proteins; blue lines, mRNA.
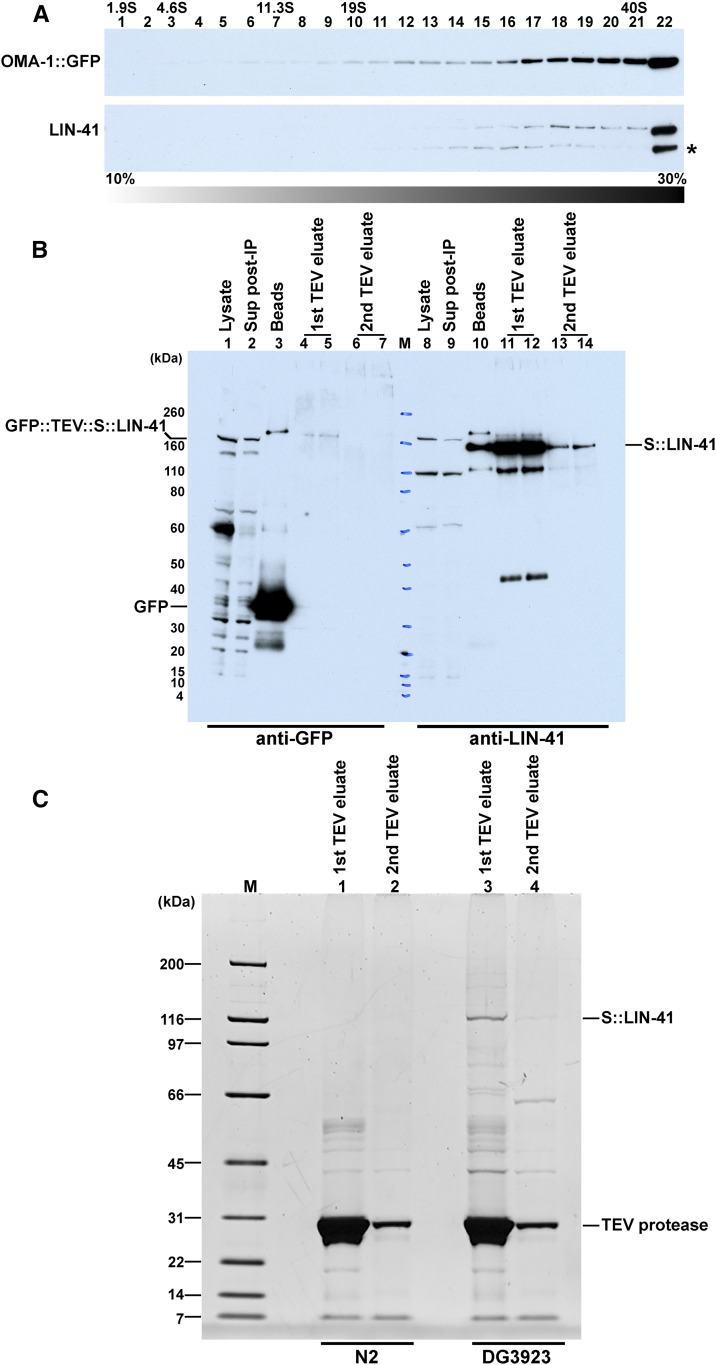
Figure 3.
Immunopurification of LIN-41 RNP complexes. (A) Glycerol-gradient sedimentation profile of LIN-41 and OMA-1::GFP indicate that they are found in large molecular weight complexes. A high-speed supernatant extract from the DG2566 fog-1(q253ts); oma-1(zu405te33) IV; tnIs17[oma-1p::oma-1::s::tev::gfp, unc-119(+)] strain was applied to the gradient. Proteins were detected with anti-GFP (NB600-308) and anti-LIN-41 antibodies. (B) Detection of tagged LIN-41 proteins and GFP in the first step of a tandem affinity purification. TEV cleavage releases S::LIN-41 complexes for a second step of purification. (C) Detection of S::LIN-41 and associated proteins using SYPRO Ruby staining following the first step of a tandem affinity purification. DG3923 lin-41(tn1541[gfp::tev::s::lin-41)] fog-1(q253ts) strain used for affinity purifications, and N2 is the control wild-type strain, which lacks any of the affinity tags.
Glycerol gradients (10–30%) were prepared in basic IP buffer using a Gradient Master 107 (BioComp Instruments). For analytical gradients, 250–350 μl of high-speed supernatant protein extract was applied to the top of 10-ml gradients and spun at 4° in a SW 41 Ti Rotor (Beckman Coulter, Fullerton, CA) at 29,000 rpm (100,000 × g) for 18 hr. Molecular weight standards for glycerol gradients were: cytochrome C (1.9 S), bovine serum albumin (4.6 S), catalase (11.3 S), and thyroglobulin (19 S). Protein fractions of 0.5 ml were collected, TCA precipitated, and analyzed by Western blotting as described above. For preparative gradients, 1 ml of high-speed supernatant protein extract (100,000 × g, 1 hr) was applied to the top of a 35 ml 10–30% glycerol gradient and spun at 4° in a SW 32 Ti Rotor (Beckman Coulter) at 29,100 rpm (100,000 × g) for 18 hr. Protein fractions of 1.64 ml were collected. For tandem affinity purification from glycerol-gradient fractions, 18 ml of protein extract was purified on 18 preparative gradients, corresponding to three centrifuge runs. Protein fractions in the 19–40 S range were pooled and subjected to tandem affinity purification.
RNA-seq
The RNasin RNase Inhibitor (Promega, Madison, WI) was added to GFP::TEV::S::LIN-41 and OMA-1::S::TEV::GFP immunopurification lysates and buffers at a final concentration of 20 units/ml to inhibit RNA degradation. RNPs were isolated using anti-GFP monoclonal antibodies 12A6 and 4C9 linked to Dynabeads as described above. Total RNA was isolated from immunopurified RNPs from ∼5 ml input high-speed supernatant lysate (80 mg of total C. elegans protein) using the RNAeasy Micro Kit (QIAGEN, Valencia, CA). RNA was quantified using Qubit fluorometric quantitation (Thermo Fisher). For the GFP::TEV::S::LIN-41 immunopurification (experiment IX and X; Figure 2A), 81.6 and 53.6 ng of total RNA were obtained in each of two technical replicates. For the OMA-1::S::TEV::GFP immunopurification (experiment XI and XII; Figure 2B), 163.4 and 207.0 ng of total RNA were obtained in each of two technical replicates. Total RNA was also extracted from 200 μl of untreated extract as a control. A Ribo-Zero Kit (Illumina) was used to remove ribosomal RNA (rRNA) from the total RNA sample. For sequencing we submitted the IP RNA, total RNA, and Ribo-Zero-depleted total RNA to the University of Minnesota Genomics Center (UMGC). The UMGC prepared standard TruSeq (Illumina) RNA libraries starting at the eluate, fragment, prime step, using random primers. Single-end reads of 50 bp were obtained on an Illumina HiSeq 2500 instrument in high-output mode. Eight samples were multiplexed in one lane to yield 287 million reads with an average depth of 36 million reads per sample.
Bioinformatics
Sequenced reads were trimmed (Trimmomatic v0.32) to remove Illumina adapter sequences and quality was assessed using FastQC (version 0.10.1). Trimmed reads were mapped to the WBcel235/ce11 genome using STAR (version 2.4.2a_modified) and sorted and indexed with Samtools (version 1.2). Annotation data were downloaded from Ensembl (release 87) and used to estimate gene-level abundance in R/Bioconductor using the GenomicAlignments package (version 1.8.4). On average across the samples, 41 and 25 million reads were found to map to annotated genes without or with the mapping quality filter (MAPQ ≥ 4), respectively. Analysis of the gene type from these two estimates shows that low map quality reads are mostly found in rRNA and rRNA pseudogenes (Figure S3A in File S4). The lysate samples that were not depleted of rRNA sequences contained the largest fraction of rRNA reads. Reads that did not pass the mapping quality filter or mapped uniquely to rRNA genes were excluded from further analyses. Sample normalization and enrichment calculations for the immunopurification experiments were performed using DESeq2 (version 1.12.3) by specifying “greater” as an alternative hypothesis to look for gene-level enrichment. Enrichment values for the previous work (Spike et al. 2014b) were estimated as the log2 difference between the reads in the published OMA-1 IP and the average of the log2 expression in the ribosomal sequence-depleted lysate samples. Germline enrichment values were obtained by processing the raw data from GSE57109 (Ortiz et al. 2014) using the method described above. Gene ontology (GO) data were obtained from WormBase release WS257 and analyzed taking length bias into account using the Goseq (version 1.24.0) package in Bioconductor (Young et al. 2010). The GO analysis is reported in Figure S4 in File S4. Motif analyses were performed using Biostrings (version 2.40.2), and the poly(A)-length competitive-gene-set enrichment test was performed using Camera in the Limma package (version 3.28.14). A reproducible workflow with complete details is available at https://github.com/micahgearhart/lin41.
Data availability
Strains (see Table S1 in File S4) and reagents are available upon request. RNA-seq data have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus and are accessible through accession number GSE98130. Proteomic data are available through Proteome Xchange through accession number PXD006726 (http://www.proteomexchange.org).
Results
Identification of LIN-41-associated proteins
We sought to identify LIN-41-associated proteins to understand how LIN-41 inhibits oocyte M-phase entry, promotes oocyte growth, and maintains oocyte quality. Although LIN-41 also functions in the heterochronic pathway that controls developmental timing of somatic cell fates (Reinhart et al. 2000; Slack et al. 2000), in the adult LIN-41 is expressed in the oogenic germline with somatic expression below the limit of detection (Spike et al. 2014a). Prior work established that LIN-41 copurifies in RNPs with the TIS11 zinc-finger RNA-binding proteins OMA-1 and OMA-2 (Spike et al. 2014b). To evaluate the biochemical association between LIN-41 and OMA-1, we conducted glycerol-gradient sedimentation analysis. High-speed (100,000 × g, 1 hr) supernatant lysate fractions prepared from DG2566 fog-1(q253ts); oma-1(zu405te33); tnIs17[oma-1p::oma-1::s::tev::gfp, unc-119(+)] females were analyzed on 10–30% glycerol gradients. We prepared extracts from unmated sterile females because LIN-41 is rapidly degraded upon the onset of meiotic maturation (Spike et al. 2014a), and we reasoned that the inclusion of early embryos in the extract might enhance the degradative activity toward LIN-41. In prior work, OMA-1 was observed to associate with the same set of proteins, including LIN-41, in the presence or absence of sperm (Spike et al. 2014b). We observed that OMA-1::GFP and LIN-41 are found in large molecular weight complexes (Figure 3A), consistent with the finding that LIN-41 is a component of OMA-1-containing RNPs.
To identify LIN-41-associated proteins and address their relationship to OMA-1 RNP components, we conducted tandem affinity purification using an epitope-tagged version of LIN-41 in the strain DG3923 lin-41(tn1541[gfp::tev::s::lin-41)] fog-1(q253ts) (Figure 2A). For tandem affinity purification, the first immunopurification step used a mixture of monoclonal antibodies to native GFP (Figure 2A and Figure 3, B and C). TEV protease digestion, which cleaves between GFP and LIN-41 (Figure 2A), released LIN-41 and associated proteins (Figure 3B, lanes 11–14; Figure 3C, lanes 3 and 4); whereas GFP remained bound to the purification matrix (Figure 3B, lane 3). Some LIN-41 remained associated with the affinity matrix and was only recoverable using denaturing conditions (70°, 10 min; Figure 3B, lane 10). LIN-41 was recovered from extracts made from the DG3923 strain bearing the affinity tag but not from N2 control extracts (Figure 3C and File S2). In the second purification step, we used affinity-purified anti-LIN-41 antibody (Figure 2A).
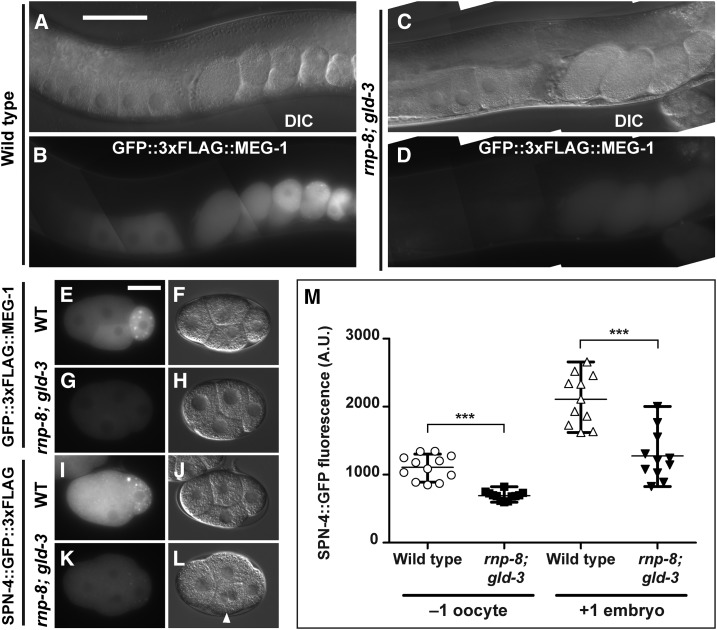
We began by conducting a large-scale tandem affinity purification from glycerol-gradient fractions in the ∼19–40 S range starting with 300 mg of total C. elegans protein (Figure 2A, experiment I). We used mass spectrometry to identify proteins that copurify with LIN-41 (File S2 and Table 1). LIN-41 was well represented in this purification (48% protein coverage; Table 1) and identified by a large number of peptides (63 peptides; File S2). Both OMA-1 and OMA-2 were identified in the large-scale purification, exhibiting ∼17% protein coverage in experiment I (File S2 and Table 1). In addition to OMA-1 and OMA-2, we identified 15 other proteins shown previously to copurify with OMA-1 in the presence of RNase A and exhibit RNA-related functions (indicated by bold font in Table 1). These include RNA-binding proteins (MEX-1, MEX-3, GLD-1, SPN-4, and POS-1), translation initiation factors (IFE-3/eIF4E and IFG-1/eIF4G), regulators of cytoplasmic polyadenylation [GLD-2 poly(A) polymerase and its accessory factors, GLD-3 and RNP-8], and subunits of the CCR4-NOT deadenylase complex (LET-711, NTL-9, and CCF-1). In total, we identified all nine subunits of the CCR4-NOT deadenylase complex in the LIN-41 immunopurified sample, including the eight previously described subunits (Nousch et al. 2013) and TAG-153, which is an NTL-2 paralog.
Table 1. Many proteins that copurify with LIN-41 associate with OMA-1 or regulate mRNA translation, cytoplasmic polyadenylation, or deadenylation.
| Proteina | Protein coverage (%)b | |||
|---|---|---|---|---|
| Experiment I | Experiment II | Experiment III | Experiment IV | |
| Tandem IP from glycerol-gradient fractions | Tandem IP from high-speed supernatant | GFP IP from high-speed supernatant | GFP IP from high-speed supernatant with RNase Ac | |
| LIN-41 | 48.2 | 40.4 | 51.8 | 48.0 (0.9) |
| mRNA translation | ||||
| MEX-1d | 43.5 | 48.0 | 59.9 | 62.6 (1.0) |
| MEX-3d | 29.1 | 40.9 | 55.8 | 66.8 (1.2) |
| GLD-1d | 41.3 | 39.7 | 45.8 | 43.4 (0.9) |
| MEX-5d | 17.5 | 30.6 | 25.0 | 22.9 (0.9) |
| TIAR-1d | 11.5 | 28.9 | 28.9 | 24.5 (0.8) |
| OMA-1d | 17.2 | 26.5 | 42.0 | 36.4 (0.9) |
| IFE-3 | 17.1 | 25.1 | 43.0 | 43.0 (1.0) |
| PUF-11d | 12.5 | 22.8 | 27.5 | 25.9 (0.9) |
| IFG-1d | 1.9 | 22.2 | 32.6 | 24.5 (0.8) |
| SPN-4d | 2.9 | 21.1 | 36.8 | 34.8 (0.9) |
| OMA-2d | 17.6 | 20.6 | 36.1 | 33.3 (0.9) |
| MEG-1 | 8.5 | 17.6 | 32.1 | 6.8 (0.2) |
| POS-1d | 7.2 | 11.0 | 39.8 | 13.3 (0.3) |
| VBH-1 | 2.6 | 10.8 | 20.6 | 11.1 (0.5) |
| PUF-6 | 2.6 | 10.5 | 12.2 | 5.0 (0.4) |
| NHL-2d | 4.8 | 10.5 | 17.6 | 11.5 (0.7) |
| FBF-1/FBF-2e | 2.3 | 9.6 | 17.9 | 6.2 (0.3) |
| Cytoplasmic polyadenylation | ||||
| GLD-2d | 21.1 | 34.2 | 35.9 | 32.2 (0.9) |
| GLD-3d | 23.2 | 33.2 | 50.2 | 47.2 (0.9) |
| RNP-8 | 6.5 | 26.2 | 43.1 | 32.4 (0.8) |
| Deadenylationf | ||||
| LET-711d | 19.9 | 31.2 | 32.3 | 25.6 (0.8) |
| TAG-153d | 5.5 | 22.1 | 37.0 | 27.8 (0.8) |
| NTL-4d | 22.4 | 20.3 | 38.9 | 30.8 (0.8) |
| NTL-9d | 7.8 | 17.5 | 32.7 | 23.7 (0.7) |
| CCF-1 | 10.0 | 16.1 | 39.0 | 35.2 (0.9) |
| NTL-3 | 4.7 | 13.7 | 18.5 | 17.1 (0.9) |
| NTL-11 | 7.3 | 11.7 | 16.9 | 18.9 (1.1) |
| CCR-4d | 9.9 | 8.3 | 23.4 | 16.8 (0.7) |
| NTL-2 | 4.7 | 4.7 | 28.8 | 21.6 (0.8) |
| Other | ||||
| NCL-1d | 8.1 | 20.8 | 31.1 | 25.6 (0.8) |
| D1081.7 | 3.7 | 20.5 | 36.1 | 31.2 (0.9) |
| CLIK-1 | 61.6 | 19.7 | 33.6 | 3.5 (0.1) |
| LST-3/CCAR-1 | 13.2 | 16.5 | 27.9 | 23.5 (0.8) |
| C05C10.2d | 3.7 | 16.5 | 16.7 | 10.8 (0.6) |
| RLE-1 | 6.8 | 15.6 | 23.2 | 20.2 (0.9) |
| C30G12.2 | 12.1 | 15.5 | 11.3 | 5.7 (0.5) |
| C35A5.8 | 5.4 | 15.5 | 17.4 | 14.3 (0.8) |
| ADR-1d | 9.9 | 15.2 | 17.7 | 17.1 (1.0) |
| LIN-66 | 3.8 | 14.7 | 26.6 | 14.4 (0.5) |
| GLS-1d | 8.3 | 13.6 | 13.6 | 12.1 (0.9) |
| PQN-87d | 2.2 | 13.4 | 18.8 | 14.3 (0.8) |
| NMY-1 | 5.4 | 13.1 | 32.1 | 12.0 (0.4) |
| ERGO-1g | 3.0 | 12.7 | 18.5 | 7.4 (0.4) |
| KIN-2 | 9.3 | 12.0 | 35.3 | 20.2 (0.6) |
| NASP-2d | 5.0 | 11.9 | 35.2 | 42.3 (1.2) |
| H05L14.2 | 1.0 | 10.7 | 16.7 | 10.1 (0.6) |
| EGG-4/EGG-5e | 2.0 | 9.8 | 16.3 | 11.4 (0.7) |
| AIN-2 | 2.7 | 9.6 | 27.2 | 23.9 (0.9) |
IP, immunopurification.
Proteins previously reported to copurify with OMA-1 in the presence of RNase A (see Table 1 and File S2 from Spike et al. 2014b) are indicated in bold font. Proteins reported to directly interact with OMA-1 (Spike et al. 2014b) are underlined. mRNAs encoding the proteins indicated in italic font are enriched fourfold in LIN-41 IPs compared to the input lysate control (see File S3 and below). GLD-1 is reported in Table 1 despite its representation in the control SACY-1 tandem affinity purification because of its interactions with OMA-1 in yeast two-hybrid assays (Spike et al. 2014b). The copurification of GLD-1 and SACY-1 might reflect their involvement in germline sex determination (Francis et al. 1995; Kim et al. 2012). Similarly, IFG-1 and LET-711 are reported despite their representation in the control SACY-1 tandem affinity purification because they copurify with OMA-1 in the presence of RNase A. NTL-9 is reported despite its presence in the SACY-1 control purification because the results indicate that most other components of the CCR4-NOT deadenylase complex copurify with LIN-41.
Peptide coverage in a single gel slice assessed by mass spectrometry in the various LIN-41 purifications from DG3923 lin-41(tn1541[gfp::tev::s::lin-41]) fog-1(q253) extracts. Only proteins showing at least 10% coverage in the tandem IP from the high-speed supernatant are shown, with the exception that all CCR4-NOT coverage data are reported because the data indicate the complex copurifies with LIN-41. The complete data on which Table 1 is based are presented in File S2.
The ratio of the peptide coverage with and without RNase A digestion (experiment IV/experiment III) is indicated in parentheses.
These proteins were detected in a tandem IP from DG3923 high-speed supernatant using a limiting amount of extract (File S2, experiment XIII).
The mass spectrometry data do not differentiate between the indicated paralogs.
Components of the CCR4-NOT deadenylase complex (Nousch et al. 2013) that copurify with LIN-41. tag-153 is a paralog of ntl-2 (http://www.wormbase.org).
In addition to the Argonaute protein ERGO-1 (Gent et al. 2010; Vasale et al. 2010), WAGO-1 and CSR-1 were represented in the LIN-41 tandem affinity purification from the high-speed supernatant. The representation of WAGO-1 (9.1% coverage) was just below the 10% cutoff threshold and CSR-1 (14.8% coverage in the LIN-41 tandem IP) was also represented (7.4% coverage) in the SACY-1 control IP. Previously, Duchaine et al. (2006) reported the recovery of LIN-41 in DCR-1 IPs (7% coverage). In our tandem affinity LIN-41 IPs DCR-1 exhibited 1% coverage.
Since the GLD-2 cytoplasmic poly(A) polymerase exhibits mutually exclusive interactions with its accessory factors GLD-3 and RNP-8 (Kim et al. 2009; Nakel et al. 2016), the proteomic data likely report the aggregate composition of distinct LIN-41 RNP complexes. We detected four LIN-41-associated RNA-binding proteins, MEX-3, GLD-1, SPN-4, and OMA-2 (underlined in Table 1), shown previously to interact directly with OMA-1 in yeast two-hybrid experiments (Spike et al. 2014b). Among these, MEX-3 was previously shown to interact with SPN-4 and POS-1 (Huang et al. 2002), which were also shown to bind each other (Ogura et al. 2003). Thus, LIN-41 and OMA-1 associate with an interacting network of RNA-binding proteins. To assess the replicability of the LIN-41 associations, we conducted a tandem affinity purification from DG3923 high-speed supernatants starting with 160 mg of total C. elegans protein (Figure 2A, experiment II; File S2 and Table 1). For the majority of proteins shown in Table 1, the percent peptide coverage was markedly higher in the direct tandem affinity purification (experiment II), as compared to the tandem affinity purification following glycerol-gradient fractionation (experiment I). This difference possibly reflects the fact that a modest portion of LIN-41 exhibits proteolysis during glycerol-gradient centrifugation (indicated by an asterisk in Figure 3A).
Because mass spectrometry is very sensitive, we used multiple negative controls to recognize likely contaminants among LIN-41-associated proteins. To this end, we conducted single (experiment VII; see Figure 3C, lanes 1 and 2) and tandem (experiment V) affinity purifications from lysates of N2 wild-type hermaphrodites to filter out proteins identified by these approaches (File S2). We also filtered out abundant background contaminants from several control purifications done previously in the course of identifying OMA-1-associated proteins (Spike et al. 2014b). The largest source of background in our tandem affinity LIN-41 immunopurifications apparently derives from proteins that nonspecifically adsorb to the RNPs and are present at low but detectable levels in the immunopurified samples. To identify such proteins, we conducted a tandem affinity purification (experiment VIII) of a spliceosomal RNP protein (GFP::SACY-1) from the fog-1(q253ts) genetic background using the same primary anti-GFP monoclonal antibodies (File S2; T. Tsukamoto and D. Greenstein, unpublished results). This proved to be a useful step for eliminating contaminants, especially those exhibiting a low but comparable number of peptide hits in the LIN-41 and SACY-1 purifications. Nevertheless, six abundant mitochondrial and metabolic proteins persisted in passing these filtering criteria (Table S2 in File S4). As a final step, we conducted a tandem affinity purification from DG3923 high-speed supernatants starting with a limiting amount (80 mg) of total C. elegans protein (experiment XIII; File S2). This approach succeeded in identifying 25 of 50 LIN-41-associated proteins listed in Table 1, but none of the likely mitochondrial or metabolic protein contaminants listed in Table S2 in File S4. As additional validation for the identification of LIN-41-associated proteins, we conducted single affinity purifications using anti-GFP antibodies and TEV protease elution (Figure 2A) using a buffer condition compatible with RNase A digestion (experiment III; our standard buffer conditions) and a slightly modified buffer (experiment VI). These experiments confirmed the reproducibility and reliability of the protein identifications. Finally, we conducted an affinity purification in which immunopurified proteins were subjected to RNase A digestion prior to the TEV protease elution (Figure 2A, experiment IV; File S2 and Table 1). The majority of proteins (42/50) were observed to maintain their association (≥50%) with LIN-41 following RNase A treatment, suggesting that these interactions were less dependent on the presence of RNA. By contrast, RNase A treatment reduced or eliminated the association of several (three out of six) likely contaminating mitochondrial and metabolic proteins with LIN-41-containing RNPs (Table S2 in File S4). One notable exception of a LIN-41-associated protein whose association with the RNP appears largely dependent on the presence of RNA is MEG-1. This observation is consistent with the recent finding that MEG-1, along with its paralogs, appears to play a scaffolding function in the higher order organization of RNPs within larger granules (Wang et al. 2014). Taken together, these biochemical studies define LIN-41 as an integral component of OMA-1-containing RNPs and identified translational regulators that might function with LIN-41 to control oogenesis or the oocyte-to-embryo transition.
LIN-41 and OMA-1 associate with shared and distinct mRNAs
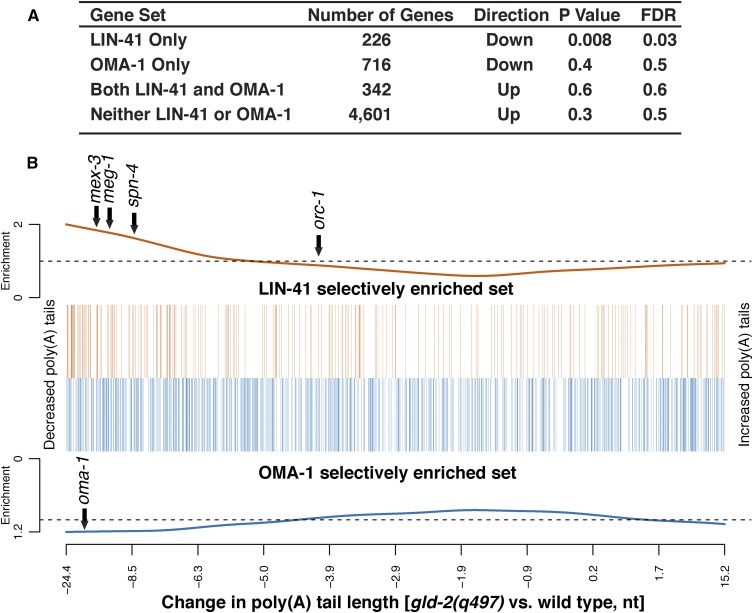
The OMA proteins and LIN-41 are found together in oocyte RNPs (Spike et al. 2014b; this work), and they function in part via the 3′-UTR-mediated translational repression of shared mRNA targets, including cdc-25.3, zif-1, and rnp-1 (Spike et al. 2014b). Paradoxically, the lin-41 and oma-1; oma-2 null mutant phenotypes are polar opposites. Pachytene-stage oocytes in lin-41 null mutants cellularize and enter M phase prematurely. By contrast, oocytes in oma-1; oma-2 null mutants grow abnormally large in the presence of sperm and fail to undergo meiotic maturation and its hallmark and subsequent events [e.g., M-phase entry, NEBD, cortical rearrangement, ovulation, fertilization, and meiotic spindle assembly (Detwiler et al. 2001)]. Potentially mitigating this conundrum, OMA-1, OMA-2, and LIN-41 copurify with GLD-2, GLD-3, and RNP-8 (Spike et al. 2014b; this work), which are respectively the catalytic subunit of a cytoplasmic poly(A) polymerase and its accessory factors (Eckmann et al. 2002; Wang et al. 2002; Kim et al. 2009). Since GLD-2 cytoplasmic poly(A) polymerases are thought to function primarily as translational activators (Hansen et al. 2004; Suh et al. 2006; Benoit et al. 2008; reviewed by Ivshina et al. 2014), we previously proposed a model in which OMA-1/2–LIN-41-containing RNPs might toggle specific mRNAs between repression and activation (Spike et al. 2014a). To begin to test this model, we sought to identify mRNAs that selectively associate with LIN-41 or OMA-1 using high-throughput sequencing, the idea being that selective association might reflect differential regulation.
LIN-41-associated mRNAs were purified and sequenced in duplicate from adult females from the strain DG3923 lin-41(tn1541[gfp::tev::s::lin-41)] fog-1(q253ts) (Figure 2A, experiment IX and experiment X; File S3). We immunopurified mRNAs using extracts prepared from adult females because LIN-41 is active in inhibiting M-phase entry in all oocytes (distal and proximal) in the absence of sperm and because LIN-41 is destabilized in the presence of sperm (Spike et al. 2014a). We also sequenced the total RNA in the lysate both before and after depleting rRNA (File S3). In parallel, we isolated OMA-1-associated mRNAs by repeating our previous mRNA purification and sequencing experiments (Spike et al. 2014b) in duplicate with minor modifications. OMA-1-associated mRNAs were isolated from extracts prepared from adult females from the strain DG2566 fog-1(q253ts); oma-1(zu405te33); tnIs17[oma-1p::oma-1::s::tev::gfp, unc-119(+)] (Figure 2B, experiment XI and experiment XII; File S3). Our prior work established that OMA-1 stably associates with the same set of mRNAs in the presence or absence of sperm-dependent MSP signaling (Spike et al. 2014b).
We mapped the sequencing reads from the LIN-41-associated RNAs, the OMA-1-associated RNAs, and the RNAs in the total RNA or rRNA-depleted starting lysates to the WBcel235/ce11 build of the C. elegans genome and enumerated reads that map unambiguously to the annotated Ensembl 87 gene set, excluding ∼30% of the reads that map to rRNA genes from further analysis (Figure S3A in File S4). We used principal component analysis (PCA) to reduce this multidimensional data set to two dimensions such that the distance between two samples reflects their relatedness. The PCA analysis revealed that the respective technical replicates clustered together (Figure 4A), which is indicative of their similarity. By these criteria, the method of lysate preparation and immunopurification is reproducible, including the published OMA-1 immunopurification (Spike et al. 2014b) and OMA-1 immunopurifications in this study. By contrast, the LIN-41-associated mRNAs and the OMA-1-associated mRNAs are distinctly separated along PC1, which accounts for 52% of the variance in the sample; indicating that they comprise distinct sets of mRNAs (Figure 4A).
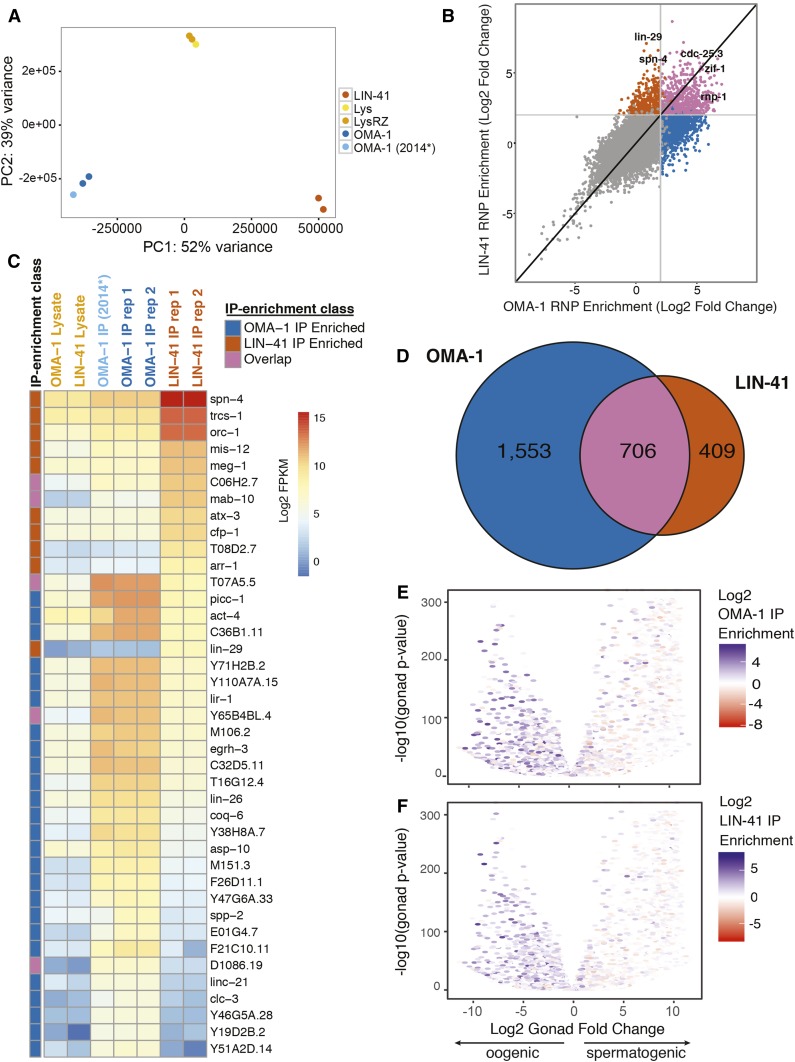
Figure 4.
Characterization of LIN-41- and OMA-1-associated RNAs. (A) PCA comparison of LIN-41- and OMA-1-associated RNA purifications. The Ribo-Zero-depleted lysates (LysRZ) and untreated lysates (Lys) are indicated. OMA-1 (2014*) is the RNA-seq data set of OMA-1-associated mRNAs reported previously (Spike et al. 2014b). (B) Distribution of mRNAs according to their enrichment in the LIN-41 and OMA-1 immunopurifications. (C) Heat map showing the abundance of the 40 most differentially enriched transcripts in the immunopurifications above a threshold of 50 FPKM in one of the immunopurifications. (D) Venn diagram showing the overlap between LIN-41- and OMA-1-associated RNAs with a fourfold enrichment cutoff. (E and F) The distribution of (E) OMA-1-associated and (F) LIN-41-associated transcripts according to their enrichment when the gonad is oogenic vs. spermatogenic. Negative and positive values indicate enrichment in the oogenic and spermatogenic gonad, respectively. The volcano plots show the Log2FC in the ratio of expression in fem-3(q96gf) spermatogenic gonads vs. fog-2(q71) oogenic gonads on the x-axis and statistical significance on the y-axis using the data from Ortiz et al. (2014). PC, principal component.
To identify transcripts primarily associated with OMA-1 and LIN-41 immunopurifications, we used DESeq2 to calculate moderated log2 fold change (Log2FC) estimates (Love et al. 2014) of the enrichment in the OMA-1 and LIN-41 immunopurified samples relative to their abundance in the rRNA-depleted inputs. To gauge the replicability of the purification of OMA-1-associated mRNAs, we compared the Log2FCs obtained in experiments XI and XII to the previously reported results (Spike et al. 2014b) and found that the results to be well correlated (Spearman correlation coefficient r = 0.841; Figure S3B in File S4). Consistent with the PCA analysis, the Spearman correlation coefficient comparing the Log2FC values from the OMA-1 and LIN-41 immunopurifications in this study was 0.642 (Figure 4B and Figure S3C in File S4).
Plotting C. elegans mRNAs according to their enrichment in the LIN-41 and OMA-1 immunopurifications indicates that we reproducibly identified mRNAs associated with either LIN-41 or OMA-1, as well as mRNAs that associate with both RNA-binding proteins (Figure 4B). We identified 1115 mRNAs exhibiting at least a statistically significant fourfold enrichment in the LIN-41 immunopurification compared to the input (P < 0.05, Benjamini–Hochberg adjusted P-value; Figure 4, B and D, and File S3). In contrast, we identified 2259 mRNAs exhibiting at least a statistically significant fourfold enrichment in the OMA-1 immunopurification as compared to the input (P < 0.05, Benjamini–Hochberg adjusted P-value; Figure 4, B and D, and File S3). This analysis identified 706 mRNAs that displayed at least a statistically significant fourfold enrichment in both the LIN-41 and OMA-1 immunopurifications (P < 0.05, Benjamini–Hochberg adjusted P-value; Figure 4, B and D, and File S3). Notably, the class of mRNAs that associates with both LIN-41 and OMA-1 includes zif-1, cdc-25.3, and rnp-1 (Figure 4, B and D, and File S3), which are shared mRNA targets of OMA-1 and LIN-41 for 3′-UTR-mediated translational repression (Spike et al. 2014b). These data suggest that the set of LIN-41- and OMA-1-associated mRNAs are likely relevant to their biological activities.
The most enriched mRNA associated with LIN-41 but not OMA-1 was lin-29 (Figure 4, B and D, and File S3), which is a target of lin-41 regulation in the heterochronic pathway (Reinhart et al. 2000; Slack et al. 2000; Aeschimann et al. 2017; reviewed by Rougvie and Moss 2013). While lin-29 mRNA is highly enriched in the LIN-41 immunopurified samples (>100-fold; File S3), it is not abundantly represented (Figure 4C). We measured lin-29 mRNA representation in the LIN-41 immunopurified samples to be ∼176 complementary DNA fragments per kilobase of transcript per million mapped reads (FPKM) (Figure 4C and File S3). For comparison, spn-4 mRNA and orc-1 mRNA are both enriched (∼74-fold and ∼97-fold, respectively) and abundant (43,194 FPKM and 9817 FPKM, respectively) in the LIN-41 immunopurified samples (Figure 4C and File S3). Analysis of the distribution of sequencing reads of spn-4 and lin-29 transcripts associated with LIN-41 indicates the purified RNAs are largely intact and that the results are highly reproducible (Figure 5C and Figure S3D in File S4). Aeschimann et al. (2017) also provided evidence that mab-10, mab-3, and dmd-3 are LIN-41 targets. Among these, the mRNAs for mab-10 and mab-3 are enriched in the LIN-41 immunopurification (∼400-fold and ∼140-fold enrichment, respectively; File S3), likely reflecting their function in the heterochronic pathway in somatic cells.
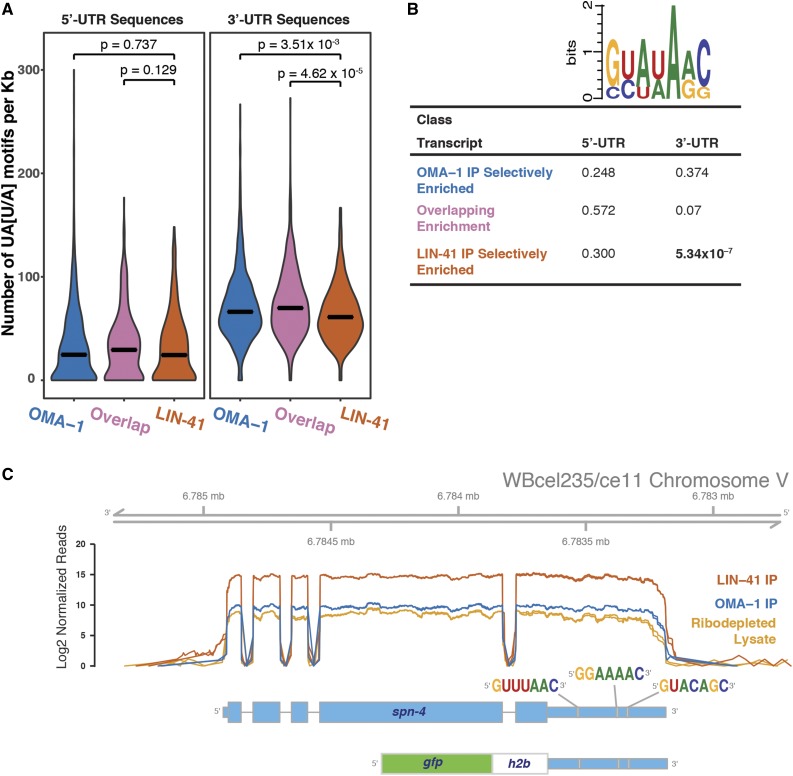
Figure 5.
Binding motifs in transcripts associated with LIN-41 and OMA-1. (A) Violin plots showing the mean (bar) and the probability density of the prevalence of the OMA-1-binding motif (UA[U/A]) per kilobase in the 5′-UTRs and 3′-UTRs of transcripts that associate with LIN-41 and/or OMA-1. Statistical significance between distributions (Kolmogorov–Smirnov test) is indicated for each comparison. (B) A WebLogo and P-values from a statistical analysis (Fisher’s exact test) of the prevalence of LIN-41-binding motifs in the 5′-UTRs and 3′-UTRs of transcripts that associate with LIN-41 and/or OMA-1. (C) The spn-4 3′-UTR contains three LIN-41-binding motifs. The spn-4 3′-UTR GFP::histone H2B reporter used in Figure 10 is diagrammed and the position of the LIN-41-binding motifs is shown. The plots show the distribution of sequencing reads of spn-4 transcripts in the immunopurifications and the starting lysate depleted of rRNA sequences. The data for the technical replicates are plotted separately, but because of the high replicability, the graphs are nearly coincident. The uniform coverage across the transcript suggests that the spn-4 mRNA is largely intact in the purified RNPs and includes the annotated 3′-UTR.
By contrast, our analysis in the germline found that many (513 of 1115) of the LIN-41-associated mRNAs are oocyte enriched by the criterion that they exhibit at least a twofold increase in gonadal expression when the germline is specified in the female mode (i.e., oocytes) as opposed to the male mode (i.e., sperm) (File S3) (Ortiz et al. 2014). mRNAs that are enriched and abundant in LIN-41 and OMA-1 immunopurified samples tend to be oocyte enriched (Figure 4, E and F). For example, of the 50 most abundant (FPKM > 1000) and enriched (more than fourfold) LIN-41-associated transcripts, 39 (78%) are oocyte enriched; exhibiting at least a twofold enrichment in oogenic vs. spermatogenic gonads (File S3 and Table S3 in File S4). Of these 39 oocyte-enriched transcripts, which are enriched and abundant in the LIN-41 immunopurified samples, 24 (62%) have documented requirements in oocyte or early embryonic development; whereas the functions of most of the others have not been determined (Table S3 in File S4). Thus, the transcripts most highly associated with LIN-41 are likely to be expressed in oocytes, which abundantly express LIN-41 in the adult stage. Because the LIN-41-containing RNPs are sufficiently stable such that they can be isolated on glycerol gradients (Figure 3A), it was formally possible that abundant mRNAs might enter the complex during lysate preparation or during purification. Among the 100 most abundant mRNAs in the lysate, only two (cpg-2 and clec-87) are at least fourfold enriched in LIN-41 immunopurified samples (File S3). Therefore, incorporation of transcripts into the RNPs in the lysate is unlikely to substantially contribute to the results. However, we cannot exclude the possibility that some mRNAs might enter the RNP complexes during their isolation.
Transcripts encoding several LIN-41 RNP components associate with LIN-41 and/or OMA-1
We identified 15 proteins in LIN-41 RNPs whose corresponding transcripts associate with LIN-41 and/or OMA-1 (Table 2). LIN-41 RNP components whose transcripts display at least a fourfold enrichment in the LIN-41 immunopurification but not the OMA-1 immunopurification include LIN-41, MEX-3, SPN-4, MEG-1, LIN-66, and EGG-4/5. By contrast, transcripts for the LIN-41 RNP components OMA-2, POS-1, and PUF-11 were associated with both LIN-41 and OMA-1 using the fourfold cutoff criterion; transcripts for GLD-1, OMA-1, PUF-6, LST-3/CCAR-1, and ERGO-1 were specifically enriched at this level only in the OMA-1 immunopurified samples. The LIN-41 RNP components whose transcripts associate with LIN-41 and/or OMA-1 include mainly RNA-binding proteins (Table 2). LIN-41 RNP components include proteins involved in the regulation of cytoplasmic polyadenlyation (GLD-2, GLD-3, and RNP-8) and deadenylation (subunits of the CCR4-NOT complex; Table 1). Interestingly, transcripts for none of these factors are found to be enriched in either LIN-41 or OMA-1 immunopurifications (Spike et al. 2014b; this work). These results are consistent with the possibility that for many transcripts, mRNA association with LIN-41 and/or OMA-1 may be functionally relevant for oocyte or early embryonic development. The analysis of biological processes correlating to the LIN-41- and OMA-1-associated mRNAs supports this conclusion (Figure S4 in File S4).
Table 2. LIN-41 RNP components whose transcripts associate with LIN-41 and/or OMA-1.
| Gene | mRNA abundance in LIN-41 IP (FPKM) | Log2 mRNA enrichment in LIN-41 IP compared to lysate | mRNA abundance in OMA-1 IP (FPKM) | Log2 mRNA enrichment in OMA-1 IP compared to lysate |
|---|---|---|---|---|
| lin-41 | 2,514 | 4.64 | 348 | 1.80 |
| mex-3 | 1,757 | 2.97 | 596 | 1.41 |
| gld-1 | 1,092 | 1.85 | 6,303 | 4.37 |
| oma-1 | 425 | 1.66 | 1,113 | 3.04 |
| puf-11 | 2,633 | 3.94 | 1,081 | 2.65 |
| spn-4 | 43,194 | 6.21 | 1,306 | 1.17 |
| oma-2 | 2,043 | 3.23 | 2,339 | 3.42 |
| meg-1 | 1,747 | 4.30 | 116 | 0.40 |
| pos-1 | 12,112 | 4.31 | 4,604 | 2.92 |
| puf-6 | 113 | 1.53 | 508 | 3.70 |
| lst-3/ccar-1 | 98 | −0.28 | 527 | 2.15 |
| lin-66 | 82 | 2.32 | 39 | 1.25 |
| ergo-1 | 117 | 0.35 | 583 | 2.67 |
| egg-4 | 152 | 2.17 | 60 | 0.84 |
| egg-5 | 254 | 2.77 | 102 | 1.46 |
Binding motifs in the 3′-UTRs of LIN-41 and OMA-1-associated mRNAs
RNAs that contain multiple copies of a short UA[A/U] consensus sequence bind OMA-1 with high affinity in vitro (Kaymak and Ryder 2013). We previously found that OMA-1-binding motifs are prevalent in the 3′-UTRs of OMA-1-associated mRNAs (Spike et al. 2014b). We extended this analysis by quantitating the number of UA[A/U] motifs that occur per kilobase in the 5′-UTRs and 3′-UTRs of the transcripts that are selectively associated with OMA-1, transcripts that are selectively associated with LIN-41, and transcripts in the overlapping set (Figure 5A). We find that UA[A/U] motifs are more common in the 3′-UTRs than in the 5′-UTRs after correcting for length in each set (Kolmogorov–Smirnov test, P < 2.2 × 10−16; Figure 5A), reflecting the genome-wide base-composition distribution (68% A and U in 3′-UTRs vs. 60% in 5′-UTRs). Interestingly, UA[A/U] motifs are also more prevalent in the 3′-UTRs of transcripts that associate with OMA-1 selectively or in transcripts in the overlapping set than in those that associate selectively with LIN-41 (Kolmogorov–Smirnov test, P = 3.51 × 10−3 and P = 4.62 × 10−5, respectively; Figure 5A). Thus, an increased prevalence of UA[A/U] motifs in the 3′-UTRs of these transcript classes correlates with their stable association with OMA-1.
Recently, RNAcompete analysis (Ray et al. 2009) was used to identify consensus LIN-41 binding sites (Loedige et al. 2015). Thus, we analyzed the prevalence of LIN-41-binding motifs in mRNAs associated with LIN-41 and/or OMA-1 (Figure 5B). We found that LIN-41-binding motifs were significantly enriched in the 3′-UTRs of transcripts that associate selectively with LIN-41 (Fisher’s exact test, P = 5.34 × 10−7). We also found that LIN-41-binding motifs were enriched in the 3′-UTR sequences of transcripts that associate with both LIN-41 and OMA-1, albeit at a lower level of significance (Fisher’s exact test, P = 0.07). LIN-41-binding motifs were not enriched in the 3′-UTRs of transcripts that associate selectively with OMA-1 (Fisher’s exact test, P = 0.374). We observed no enrichment of LIN-41-binding motifs in the 5′-UTRs of any of these three transcript classes (Figure 5B). This analysis suggests that the presence of LIN-41-binding motifs correlates with LIN-41 association. In fact, we observed three LIN-41-binding motifs in the 3′-UTR of spn-4 (Figure 5C), which is repressed by LIN-41 (see below).
Two LIN-41-associated noncoding RNAs
In addition to protein coding transcripts, we observed several likely noncoding RNAs in our immunopurifications (File S3). Two nearly identical noncoding RNAs, C38C6.8 (299 nt) and C38C6.7b (302 nt) were moderately enriched (∼3.7-fold) in the LIN-41 immunopurified samples but substantially depleted (∼30-fold) from the OMA-1 immunopurified samples (Figure S5 in File S4 and File S3). C38C6.8 and C38C6.7b were described as rapidly evolving nematode-specific RNA polymerase III transcripts (Gruber 2014). Because of the selective association of C38C6.8 and C38C6.7b with LIN-41, we named them lfor-1 and lfor-2 (lin forty one-associated noncoding RNA), respectively. We used CRISPR-Cas9 genome editing to delete both noncoding RNAs from the genome (Figure S5 in File S4). We found the double knockout lfor-1(tn1652) lfor-2(tn1653) strain was viable, fertile, and localized GFP::LIN-41 normally (Figure S5 in File S4). Further, oocytes in unmated lfor-1(tn1652) lfor-2(tn1653); fog-2(oz40) females arrest in diakinesis as they normally do in the absence of sperm. These data indicate that lfor-1 and lfor-2 noncoding RNAs are not required for oogenesis. Thus, the function of these noncoding RNAs and the significance of their association with LIN-41 remains enigmatic.
LIN-41 and OMA-1/2 regulate mRNA translation in a coordinate and antagonistic fashion
Having defined mRNAs that associate selectively with LIN-41 or OMA-1, as well as mRNAs that associate with both proteins, we sought to test whether LIN-41 and OMA-1/2 might differentially affect the expression of certain mRNAs as predicted by the model. We took the approach of using CRISPR-Cas9 genome editing (Dickinson et al. 2015) to fluorescently tag the endogenous loci of mRNAs that associate selectively with LIN-41, OMA-1, or both proteins. Here we focus on results that illustrate the capacity of LIN-41 and the OMA proteins to affect target gene expression in both a coordinate and antagonistic fashion.
LIN-41 and OMA-1/2 coordinately repress CDC-25.3 expression
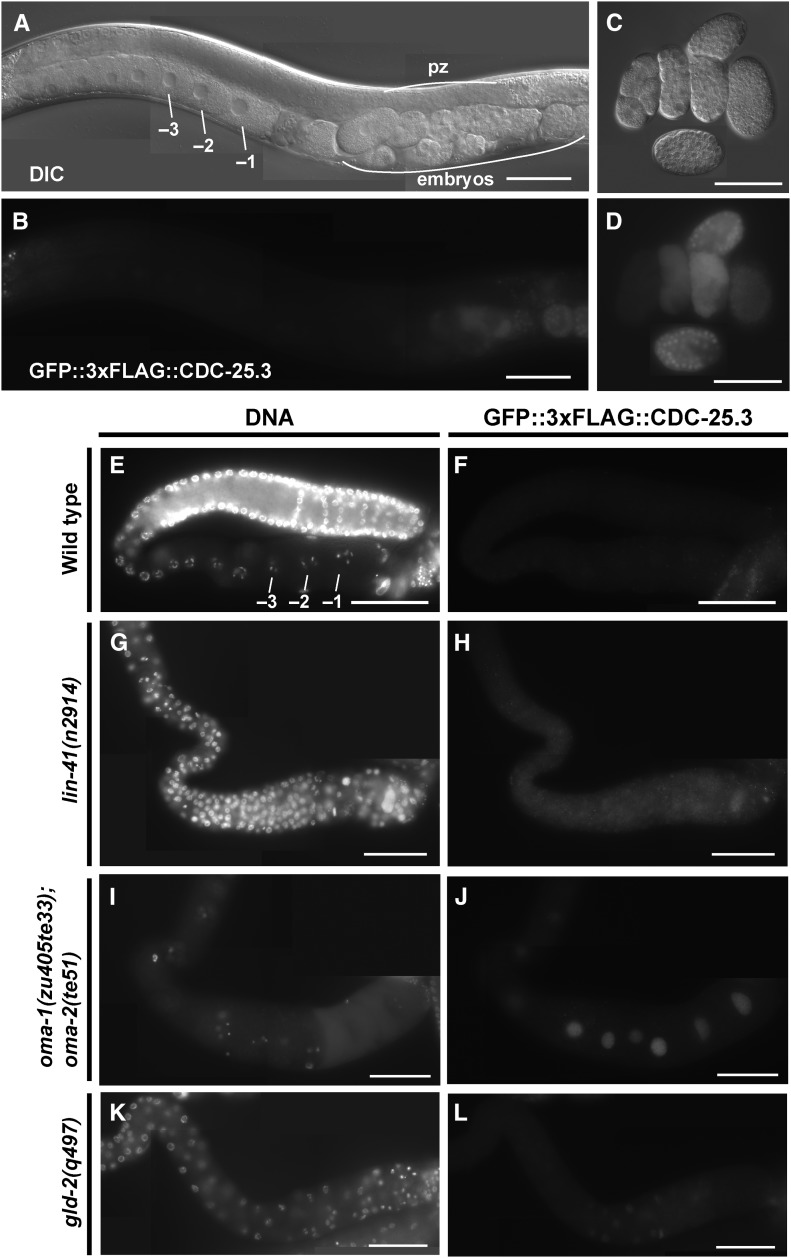
LIN-41 and OMA-1 associate with cdc-25.3 mRNA (Spike et al. 2014b; this work), and LIN-41 and the OMA proteins repress the translation of a cdc-25.3 3′-UTR reporter (Spike et al. 2014b). Genetic analysis suggests that derepression of cdc-25.3 contributes in part to the premature M-phase entry phenotype in lin-41(n2914) null mutant oocytes (Spike et al. 2014a). Consistent with these results, we observed that GFP::3xFLAG::CDC-25.3 is expressed in oocytes in both lin-41(n2914) (Figure 6, G and H; n = 42) and oma-1(zu405te33); oma-2(te51) (Figure 6, I and J; n = 46) mutants but not the wild-type background (Figure 6, E and F; n = 29). Using the lin-41(tn1487ts) temperature-sensitive allele, we also observed nuclear GFP::3xFLAG::CDC-25.3 expression in oocytes at 25° (strain DG4273; n = 10). By contrast, in the wild-type background, GFP::3xFLAG::CDC-25.3 expression is first observed in early embryos (Figure 6, A–D). Thus, LIN-41 and the OMA proteins coordinately repress expression of CDC-25.3. Inactivation of either LIN-41 or both OMA-1 and OMA-2 is sufficient to derepress expression.
Figure 6.
lin-41 and oma-1/2 repress expression of CDC-25.3. A GFP::3xFLAG::CDC-25.3 fusion generated by genome editing was examined in the indicated genetic backgrounds {all strains contained the cdc-25.3(tn1712[gfp::3xflag::cdc-25.3]) edit, which is referred to as wild type}. (A–F) The wild-type background (strain DG4190). (G and H) The lin-41(n2914) null background (from strain DG4206). (I and J) An oma-1; oma-2 null background (from strain DG4253). (K and L) A gld-2(q497) null background (from strain DG4242). GFP::3xFLAG::CDC-25.3 expression was detected by (B and D) GFP fluorescence or (F, H, J, and L) indirect immunofluoresence in dissected gonads stained with anti-FLAG antibody. (E, G, I, and K) DNA was detected with DAPI. In the wild type, CDC-25.3 was expressed in early embryos but not oocytes. In wild-type embryos, CDC-25.3 expression is found in the cytoplasm and nuclei, depending on the embryonic and cell-cycle stage. GFP::3xFLAG::CDC-25.3 is not detected in the distal proliferative zone (pz) or in meiotic germ cells in the adult stage. In lin-41(n2914) null mutants, pachytene-stage oocytes prematurely enter M phase and cycle in and out of M phase multiple times, which provides an explanation for why gonadal GFP::3xFLAG::CDC-25.3 expression is largely cytoplasmic in this genetic background. Bar, 50 μm.
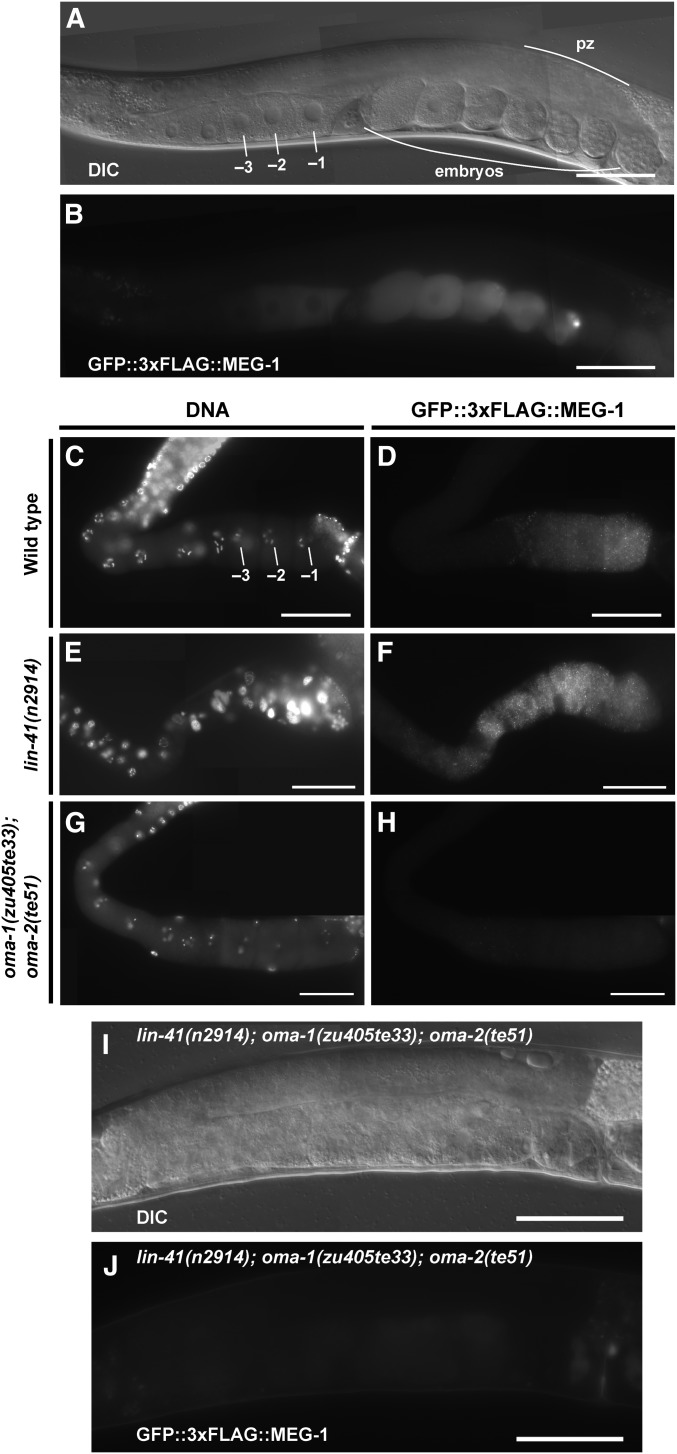
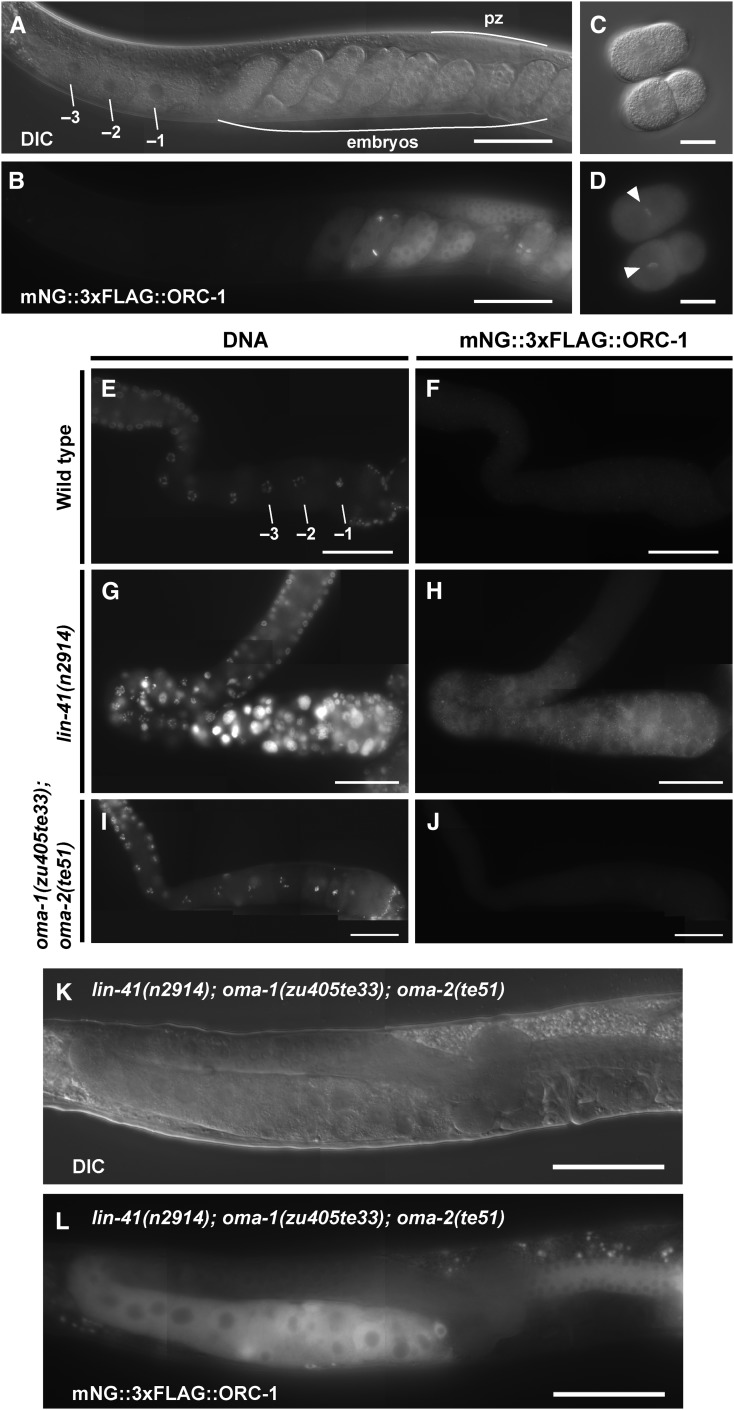
LIN-41 and OMA-1/2 display differential effects on the regulation of SPN-4, MEG-1, and ORC-1
Regulation of spn-4 translation:
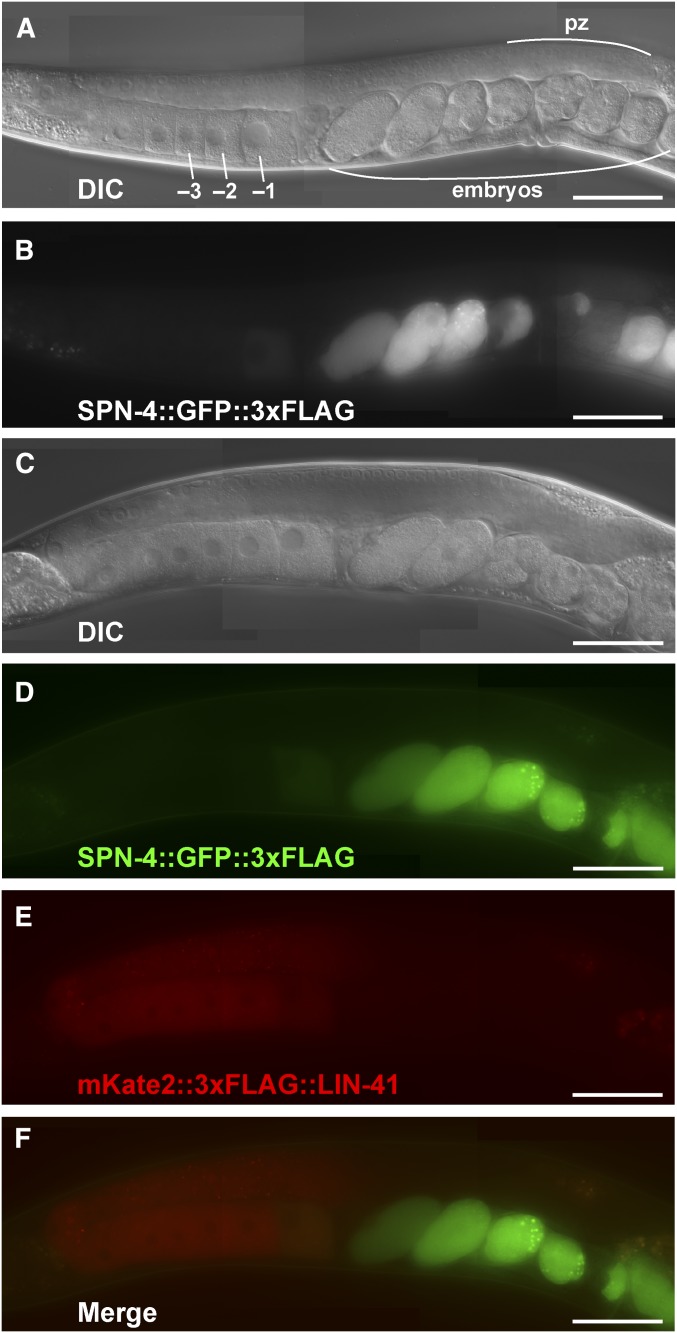
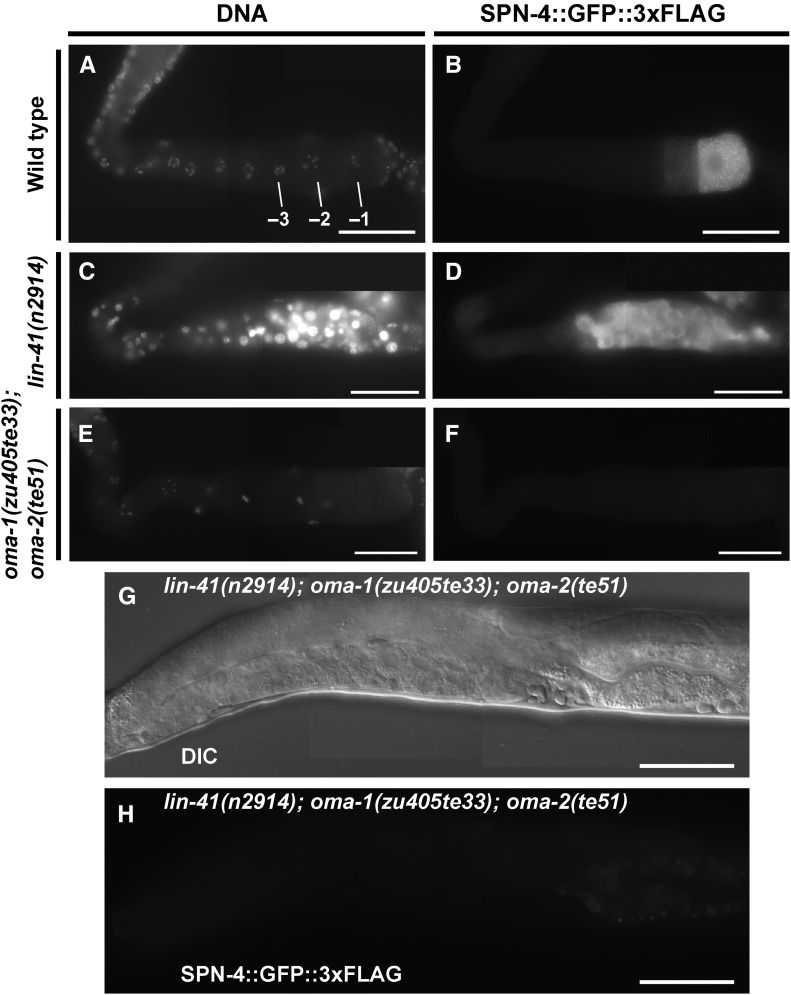
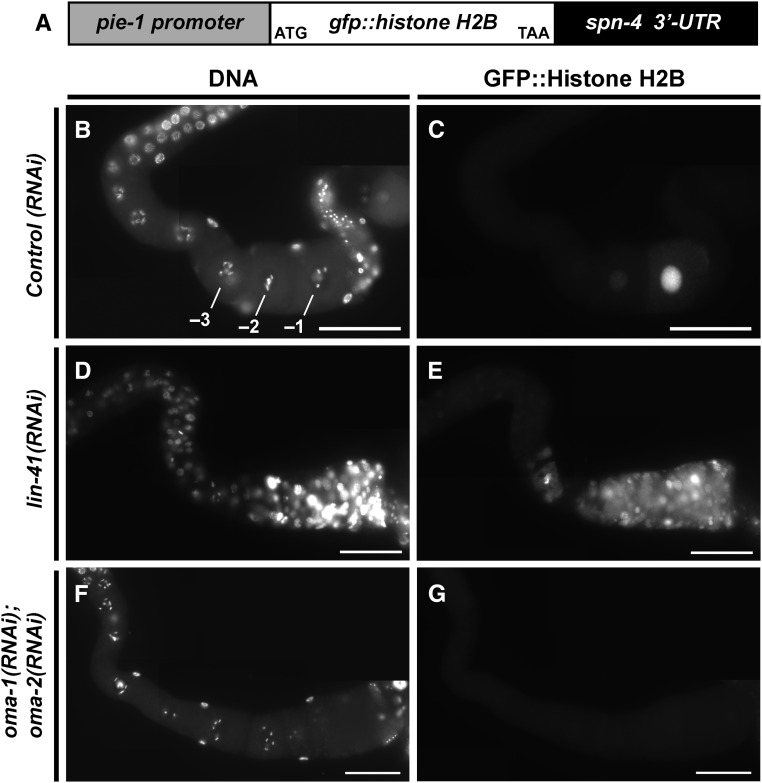
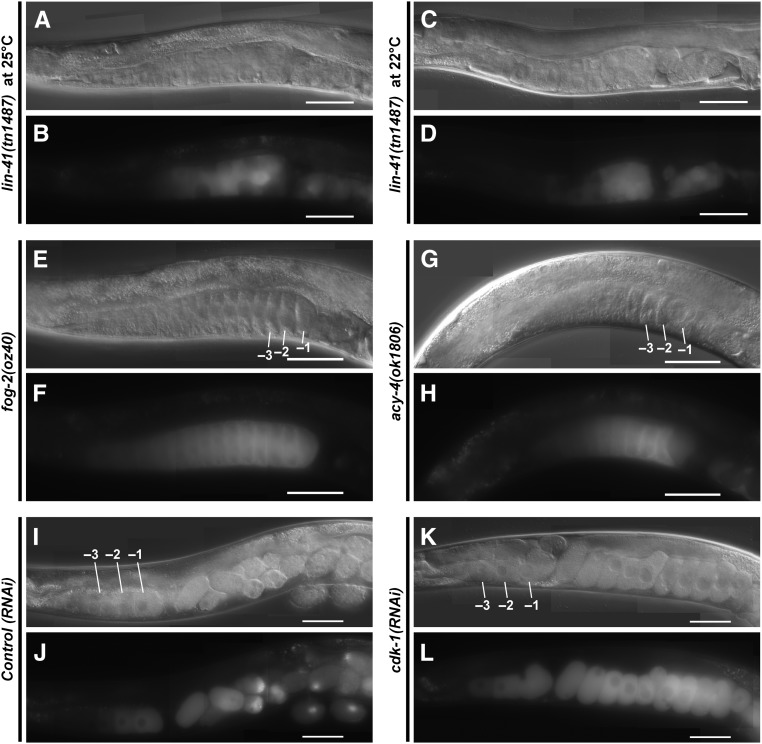
Of the mRNAs enriched in the LIN-41 immunopurified samples, spn-4 mRNA is by far the most abundant (Figure 4C). As mentioned above, spn-4 mRNA selectively associates with LIN-41 but not OMA-1, and SPN-4 protein associates with both LIN-41 and OMA-1 (Table 1). spn-4 is maternally required for specification of mesendoderm and for proper embryonic germline development (Gomes et al. 2001). We used CRISPR-Cas9 genome editing to generate a SPN-4::GFP::3xFLAG fusion, and the resulting spn-4(tn1699[spn-4::gfp::3xflag]) strain was viable and fertile. In addition to the distal proliferative zone, we observed SPN-4::GFP::3xFLAG fluorescence in the most proximal (−1) oocyte (Figure 7, A and B). SPN-4::GFP fluorescence increases substantially after fertilization, paralleling the elimination of LIN-41 (Figure 7, C–F), which commences upon the onset of meiotic maturation (Spike et al. 2014a). Because the rate at which the GFP fluorescent chromophore forms (Heim et al. 1995; Iizuka et al. 2011) is of the same order of the oocyte meiotic maturation rate (once every ∼23 min; McCarter et al. 1999), we examined SPN-4::GFP::3xFLAG expression in dissected gonads by indirect immunofluorescence with anti-FLAG antibody. Using this method, we detect SPN-4::GFP::3xFLAG in the two most proximal oocytes (−1 and −2; Figure 8, A and B; n = 38;), consistent with previous results obtained with anti-SPN-4 antibody (Ogura et al. 2003; Mootz et al. 2004). In a lin-41(n2914) null background, strong expression of SPN-4::GFP::3xFLAG is expanded in the proximal region of the gonad and weak staining extends to the loop region (Figure 8, C and D; n = 61), suggesting that LIN-41 has a repressive effect on SPN-4::GFP::3xFLAG expression. Similarly, we observed strong and expanded expression of SPN-4::GFP::3xFLAG after lin-41(RNAi) (n = 32; Figure S2 in File S4). We also observed expanded SPN-4::GFP::3xFLAG fluorescence in proximal oocytes in a lin-41(tn1487ts) mutant background at both the restrictive (25°; n = 21) and semipermissive (22°; n = 27) temperatures (Figure 9, A–D). In lin-41(n2914) null mutants, pachytene-stage oocytes prematurely enter M phase (Spike et al. 2014a). This phenotype is not prominently observed in lin-41(tn1487ts) mutants at the semipermissive temperature of 22° (Spike et al. 2014a). Thus the expanded expression of spn-4 in proximal oocytes in lin-41 mutant backgrounds is unlikely a secondary consequence of the premature M-phase entry phenotype. Previously, the germline pie-1 promoter was used to express GFP::histone H2B under control of the spn-4 3′-UTR (Merritt et al. 2008; see Figure 10A). In agreement with the SPN-4::GFP::3xFLAG expression results, this reporter is expressed in the −1 and −2 oocytes, in addition to early embryos and the distal proliferative zone (Figure 10, B and C; n = 68; Merritt et al. 2008). When examined following lin-41(RNAi), the spn-4 3′-UTR expression reporter was ectopically expressed in the proximal gonad (Figure 10, D and E; n = 41). Thus, LIN-41 mediates 3′-UTR-dependent translational repression of spn-4.
Figure 7.
SPN-4::GFP::3xFLAG expression increases after meiotic maturation. A functional SPN-4::GFP::3xFLAG fusion protein was generated by CRISPR-Cas9 genome editing of the spn-4 locus to examine protein localization in (A and B) otherwise wild-type hermaphrodites (strain DG4158) or (C–F) in hermaphrodites with the edited lin-41(tn1749[mkate2::3xflag::lin-41]) locus (strain DG4290). Both strains shown contain the spn-4(tn1699[spn-4::gfp::3xflag]) edit, which is referred to as wild type. (A and C) DIC and (B and D–F) fluorescent images of living animals are shown. Note, the mKate2::3xFLAG::LIN-41 fusion protein has some tendency to aggregate in hermaphrodites, which is not observed with GFP fusions or the native protein. pz, proliferative zone. Bar, 50 μm.
Figure 8.
LIN-41 and the OMA proteins regulate spn-4 expression in an antagonistic fashion. (A–F) The expression of spn-4 was analyzed in dissected gonads from wild type (strain DG4158), lin-41 null (from strain DG4176), and oma-1; oma-2 null (from strain DG4239) backgrounds. SPN-4::GFP::3xFLAG expression was detected by indirect immunofluorescence in dissected and fixed gonads stained with anti-FLAG antibody. DNA was detected with DAPI. (G) DIC and (H) GFP fluorescence images of a living adult hermaphrodite showing an absence of SPN-4::GFP::3xFLAG expression in proximal oocytes in a lin-41(n2914); oma-1(zu405te33); oma-2(te51) triple null mutant (from strain DG4313). SPN-4::GFP::3xFLAG expression is unaffected in the distal proliferative zone in this genetic background, but this is not shown in this image (T. Tsukamoto and D. Greenstein, unpublished results). All strains shown contain a spn-4(spn-4::gfp::3xflag) edit, which is (A–D) spn-4(tn1699) or (E–H) spn-4(tn1718). The tight linkage of spn-4 and oma-2 necessitated independent targeting of spn-4. spn-4(tn1718) and spn-4(tn1699) are the same at the DNA sequence level and exhibit identical expression patterns. oma-1(RNAi); oma-2(RNAi) prevents expression of spn-4(tn1699) (Figure S2 in File S4; n = 31). Bar, 50 μm.
Figure 9.
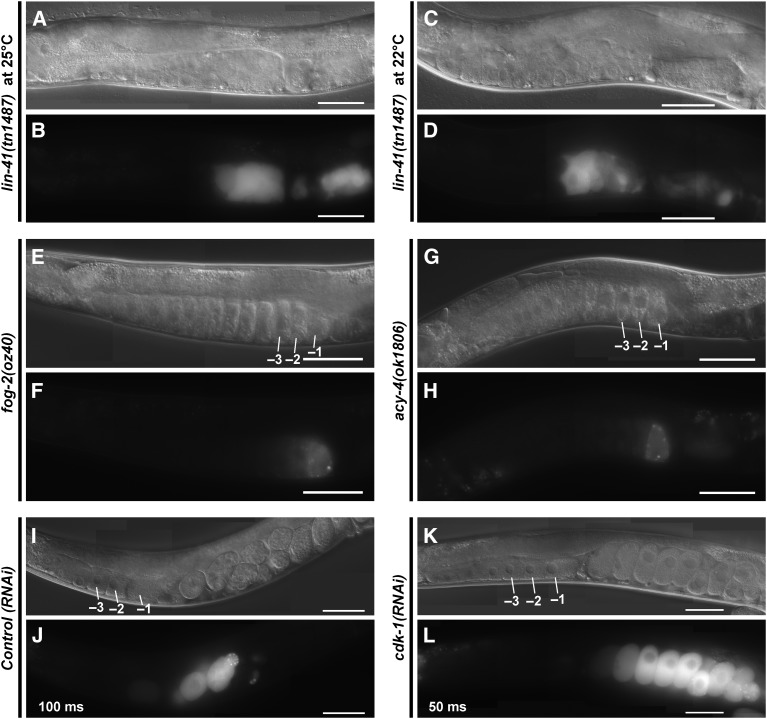
Oocyte M-phase entry is not required for expression of SPN-4::GFP::3xFLAG. The expression of SPN-4::GFP::3xFLAG was examined by fluorescence and DIC microscopy in (A–H) mutant backgrounds or (I–L) after RNAi treatments as indicated. Strong SPN-4::GFP::3xFLAG expression is observed in small abnormal oocytes in the lin-41(tn1487ts) mutant background (strain DG4192) at both (A and B) 25° and (C and D) 22°. A premature M-phase-entry phenotype characteristic of lin-41(n2914) null mutants is not observed in lin-41(tn1487ts) proximal oocytes at 22° (Spike et al. 2014a). SPN-4::GFP::3xFLAG expression is observed in proximal oocytes in (E and F) unmated fog-2(oz40) females (strain DG4185) and (G and H) acy-4(ok1806) hermaphrodites (from strain DG4212). M-phase entry occurs infrequently in these genetic backgrounds. (K and L) Strong SPN-4::GFP::3xFLAG expression is observed after cdk-1(RNAi). Note, different exposure times were used to image the effects of control(RNAi) [100 msec in (J)] and cdk-1(RNAi) [50 msec in (L)]. Expression levels are somewhat higher in the cdk-1(RNAi) one-cell arrested embryos, possibly because they continue to express SPN-4::GFP::3xFLAG. Bar, 50 μm.
Figure 10.
LIN-41 and the OMA proteins regulate the 3′-UTR-dependent translation of spn-4 in an antagonistic fashion. The expression of a spn-4 3′-UTR reporter [depicted in (A); strain JH2311] detected with (C, E, and G) anti-GFP antibodies following the indicated RNAi treatment. (B, D, and F) DNA was detected with DAPI. Bar, 50 μm.
Next we examined the effects of oma-1 and oma-2 on spn-4 expression. We did not detect SPN-4::GFP::3xFLAG expression in the oma-1(zu405te33); oma-2(te51) null mutant background using either GFP fluorescence (n = 24) or anti-FLAG antibody staining (n = 50) methods (Figure 8, E and F). In oma-1(zu405te33); oma-2(te51) mutants, oocytes grow abnormally large and arrest prior to undergoing meiotic maturation. The failure of oma-1(zu405te33); oma-2(te51) mutant oocytes to express SPN-4::GFP::3xFLAG is unlikely a secondary consequence of their terminally arrested state because we also did not observe SPN-4::GFP::3xFLAG expression in oocytes of the young adult mutant hermaphrodites shortly before or after the L4-to-adult molt. We also observed that oocytes failed to express the spn-4 3′-UTR reporter upon oma-1(RNAi); oma-2(RNAi) (Figure 10, F and G; n = 65). Importantly, we did not observe SPN-4::GFP::3xFLAG expression in the lin-41(n2914); oma-1(zu405te33); oma-2(te51) triple null mutant background (Figure 8, G and H; n = 43). This result indicates that the failure of oma-1; oma-2 null mutants to express spn-4 in oocytes is not due to the persistence of LIN-41-mediated repression. Taken together, we conclude that OMA-1 and OMA-2 promote translation of spn-4 in a 3′-UTR-dependent manner.
Several observations suggest that the lack of SPN-4::GFP::3xFLAG expression in oma-1(zu405te33); oma-2(te51) double mutants is not due to the inability of oocytes to undergo meiotic maturation in this genetic background. In the wild-type background, SPN-4::GFP::3xFLAG expression is detected in the −2 oocyte, prior to the onset of meiotic maturation, which occurs after oocytes reach the −1 position. Also, SPN-4::GFP::3xFLAG expression is observed in mutant backgrounds in which meiotic maturation occurs at very low rates, including an unmated fog-2(oz40) female background (Figure 9, E and F; n = 24) and an acy-4(ok1806) mutant background (Figure 9, G and H; n = 27) (McCarter et al. 1999; Govindan et al. 2009). The observation that SPN-4::GFP::3xFLAG expression is not observed in the lin-41(n2914); oma-1(zu405te33); oma-2(te51) triple null mutant background further suggests that the ectopic expression of SPN-4::GFP::3xFLAG in lin-41(n2914) mutants is not a secondary consequence of the premature entry of oocytes into M phase, as this phenotype is also observed in the triple null mutant background (Spike et al. 2014a). Consistent with this result, SPN-4::GFP::3xFLAG expression is observed in oocytes and embryos after cdk-1(RNAi) treatment (Figure 9, K and L; n = 29). Taken together these results suggest that LIN-41 and OMA-1/2 regulate spn-4 translation in an antagonistic fashion. LIN-41 represses spn-4 translation in oocytes until they reach the −2 position, apparently by counteracting the expression-promoting activity of OMA-1/2. In turn, the OMA proteins appear to attenuate some LIN-41 translational repressive functions in the most proximal oocytes.
LIN-41 and SPN-4 function together to ensure fertility of the next generation:
The affinity purification results (Table 1) suggest that upon its expression in the −2 and −1 oocytes, SPN-4 is incorporated into RNPs with LIN-41 and the OMA proteins. Thus, we assessed the functional relationship between lin-41 and spn-4. Since SPN-4::GFP::3xFLAG expression is derepressed in the lin-41(n2914) null mutant background, we tested whether ectopic SPN-4 expression affects lin-41(n2914) null mutant phenotypes. Consequently, we analyzed lin-41(n2914); spn-4(tm291) double null mutants. lin-41(n2914); spn-4(tm291) hermaphrodites are sterile and resemble lin-41(n2914) mutants in that they produce small abnormal oocytes. We attempted to analyze the double mutant in more detail by collecting dauer-recovered animals (see Materials and Methods). Unfortunately, double null mutants were dramatically underrepresented among dauer-recovered animals and thus difficult to analyze. However, we were able to verify that pachytene-stage oocytes prematurely enter M phase in 6 of 13 germlines analyzed. Interestingly, whereas diakinesis-stage oocytes are rarely observed in lin-41 strong loss-of-function mutants, they were frequently observed in the lin-41(n2914); spn-4(tm291) double-mutant germlines we analyzed (7 of 13). This result suggests that ectopic expression of SPN-4 might make a minor contribution to the lin-41(n2914) null mutant phenotype. To extend these results, we analyzed lin-41(tn1487ts); spn-4(or191ts) double mutants at a semipermissive temperature (20°). We observed enhanced infertility and embryonic lethality in the double-mutant background (Table 3). A major proportion of the infertility results from a maternal-effect sterile phenotype in which the germline fails to proliferate (Table 3). Because LIN-41 levels decline rapidly upon the onset of meiotic maturation and this decline is still observed in spn-4(tm291) null mutants (Figure S6 in File S4), LIN-41 likely functions in proximal oocytes to promote the activity of SPN-4 during oocyte or embryonic development. Importantly, LIN-41 function in the oocyte appears to have a major impact on a SPN-4 function needed for proper development of the germline in the next generation. Together, these genetic results suggest that LIN-41 and SPN-4 exhibit concerted and antagonistic functions. We suggest they may function in part as components of a shared RNP.
Table 3. Genetic interaction of lin-41 and spn-4 conditional alleles.
| Genotype | Fertilitya (%) | Brood sizeb |
|---|---|---|
| spn-4(or191ts) | 99 (n = 120) | 175 ± 43 (n = 52) |
| lin-41(tn1487ts) | 99 (n = 120) | 31 ± 22 (n = 60) |
| spn-4(or191ts); lin-41(tn1487ts) Mts Ztsc | 51 (n = 394) | 4 ± 7 (n = 160) |
| spn-4(or191ts); lin-41(tn1487ts) M+ Ztsd | 87 (n = 134) | 13 ± 12 (n = 134) |
The percentage of adult hermaphrodites producing viable progeny at 20°, a semipermissive temperature for both spn-4(or191ts) and lin-41(tn1487ts). Progeny were scored as viable if they hatched.
The average number of embryos hatching at 20°.
Infertile worms were sterile and maternal-effect lethal: 56% were sterile and had underproliferated germlines containing few germ cells and 44% exhibited defects in oogenesis (n = 98). To address the basis for the sterility, we examined spn-4(or191ts); lin-41(tn1487ts) Mts Zts adults in which the germline was marked using PGL-1::mTagRFP-T::3xFLAG. Adults with underproliferated germlines were agametic and their gonads often contained 0–20 germ cells that were frequently necrotic.
Progeny of spn-4(or191ts); lin-41(tn1487ts)/hT2[qIs48] parents grown at 20°. Infertile animals largely had defects in oogenesis but not underproliferated germlines.
Regulation of meg-1 expression:
Like spn-4, meg-1 encodes both an mRNA and protein component of LIN-41-containing RNPs (Table 1 and Table 2). GFP::3xFLAG::MEG-1 is detected in the −1 to −3 oocytes in the wild-type background (Figure 11, A–D; n = 81 for C and D). The expression of GFP::3xFLAG::MEG-1 increases after meiotic maturation and fertilization (Figure 11, A and B). As observed for spn-4, LIN-41 represses (Figure 11, E and F; n = 110) and OMA-1/2 promote (Figure 11, G and H; n = 94) the expression of GFP::3xFLAG::MEG-1. We observed very weak expression of GFP::3xFLAG::MEG-1 in the oma-1(zu405te33); oma-2(te51) null mutant background (Figure 11, G and H). Likewise, GFP::3xFLAG::MEG-1 expression is only very weakly observed in the lin-41(n2914); oma-1(zu405te33); oma-2(te51) triple null mutant background (Figure 11, I and J; n = 37). This result suggests that the OMA proteins promote meg-1 mRNA translation as found for spn-4 above. Likewise, we found that GFP::3xFLAG::MEG-1 is expressed in oocytes and embryos following cdk-1(RNAi) (Figure 12, K and L; n = 32). Thus, M-phase entry is neither necessary nor sufficient for GFP::3xFLAG::MEG-1 expression. We did, however, observe an expansion of GFP::3xFLAG::MEG-1 expression in unmated fog-2(oz40) females (n = 27) and in acy-4(ok1806) (n = 10) hermaphrodites, which both exhibit very low rates of meiotic maturation (Figure 12, E–H). This result suggests that OMA-1/2 are active in proximal oocytes to facilitate GFP::3xFLAG::MEG-1 expression even under situations in which their meiotic maturation-promoting function is not efficiently engaged, but rather is poised awaiting the reception of the MSP signal from sperm via the gonadal sheath cells.
Figure 11.
LIN-41 and the OMA proteins regulate expression of MEG-1. (A and I) DIC and (B and J) GFP fluorescence images of adult hermaphrodites showing expression of GFP::3xFLAG::MEG-1 in proximal oocytes and early embryos in (A and B) an otherwise wild-type adult hermaphrodite or (I and J) very weak expression in proximal oocytes in a lin-41(n2914); oma-1(zu405te33); oma-2(te51) triple null mutant. (C–H) GFP::3xFLAG::MEG-1 expression was detected by indirect immunofluoresence in dissected gonads stained with anti-FLAG antibody. DNA was detected with DAPI. All strains shown contain the meg-1(tn1724[gfp::3xflag::meg-1]) edit [strain DG4213 in (A–D), gonads from strain DG4261 in (E and F), gonads from strain DG4259 in (G and H), and strain DG4316 in (I and J)]. pz, proliferative zone. Bar, 50 μm.
Figure 12.
Oocyte M-phase entry is not required for expression of GFP::3xFLAG::MEG-1. The expression of GFP::3xFLAG::MEG-1 was examined by fluorescence and DIC microscopy in (A–H) mutant backgrounds or (I–L) after RNAi treatments as indicated. GFP::3xFLAG::MEG-1 expression is observed in small abnormal oocytes in the lin-41(tn1487ts) mutant background (strain DG4274) at both (A and B) 25° and (C and D) 22°. A spatial expansion of GFP::3xFLAG::MEG-1 expression is observed in proximal oocytes in unmated fog-2(oz40) females [(E and F); strain DG4251] and acy-4(ok1806) hermaphrodites [(G and H); from strain DG4260]. (K and L) GFP::3xFLAG::MEG-1 expression is observed after cdk-1(RNAi). Bar, 50 μm.
Regulation of orc-1 expression:
mRNA encoding ORC-1, an essential component of the origin recognition complex required for DNA replication (Sonneville et al. 2012; reviewed by Parker et al. 2017), is one of the most abundant and enriched mRNAs specifically associated with LIN-41 (Figure 4C and File S3 and Table S3 in File S4). An N-terminal mNeonGreen fusion to ORC-1, mNG::3xFLAG::ORC-1, is visibly expressed in the distal proliferative zone, but not elsewhere in the adult germline (Figure 13, A and B). mNG::3xFLAG::ORC-1 is first clearly detectable in the embryo on chromatin in mitotic metaphase at the one-cell stage (Figure 13, C and D). This localization is consistent with the fact that mitotic and premeiotic S phase are restricted to the proliferative zone of the adult hermaphrodite gonad, and the next S phase only ensues in the embryo upon completion of the second meiotic division. We observed that lin-41 has a strong repressive effect on mNG::3xFLAG::ORC-1 expression through the examination of the lin-41(n2914) null allele (Figure 13, G and H; n = 55) and also using lin-41(RNAi) (n = 16). The examination of mNG::3xFLAG::ORC-1 expression in an oma-1(zu405te33); oma-2(te51) null mutant background was uninformative; mNG::3xFLAG::ORC-1 expression was not observed in proximal oocytes (Figure 13, I and J; n = 56), similar to that observed in the wild type (Figure 13, E and F; n = 78;). Our data do not address whether OMA-1/2 might have a role in promoting mNG::3xFLAG::ORC-1 expression after the completion of meiosis because elimination of oma-1/2 activity blocks meiotic maturation. We did, however, observe robust expression of mNG::3xFLAG::ORC-1 in the lin-41(n2914); oma-1(zu405te33); oma-2(te51) triple null mutant background (Figure 13, K and L; n = 28). This result suggests that, unlike that observed for spn-4 and meg-1, orc-1 expression in the absence of LIN-41 is independent of the OMA proteins. These examples highlight the differential gene-specific regulation of LIN-41- and OMA-1-associated mRNAs.
Figure 13.
LIN-41 represses expression of ORC-1. A functional mNG::3xFLAG::ORC-1 fusion protein was generated by CRISPR-Cas9 genome editing to examine protein localization in otherwise wild-type hermaphrodites; all strains shown contain the orc-1(tn1732[mng::3xflag::orc-1]) edit. (A) DIC and (B) fluorescence images of an adult hermaphrodite showing the expression of mNG::3xFLAG::ORC-1 in early embryos and the distal proliferative zone (pz). (C) DIC and (D) fluorescence images showing that mNG::3xFLAG::ORC-1 is first detectable in the embryo at the one-cell stage; arrowheads indicate mNG::3xFLAG::ORC-1 on mitotic chromatin [strain DG4228 is shown in (A–D)]. (E–J) mNG::3xFLAG::ORC-1 expression was detected by indirect immunofluoresence in dissected gonads stained with anti-FLAG antibody. DNA was detected with DAPI. Gonads from strain (E and F) DG4228, (G and H) DG4284, and (I and J) DG4293 are shown. (K) DIC and (L) fluorescence images of an orc-1(tn1732); lin-41(n2914); oma-1(zu405te33); oma-2(te51) (from strain DG4318) adult hermaphrodite showing the expression of mNG::3xFLAG::ORC-1 in most or all developing oocytes in addition to the distal proliferative zone. (A, B, and E–L) Bar, 50 μm. (C and D) Bar, 20 μm.
Many transcripts that selectively associate with LIN-41 are substrates of the GLD-2 cytoplasmic poly(A) polymerase
LIN-41 and GLD-2 share common mRNA targets:
Because GLD-2 and its accessory factors GLD-3 and RNP-8 associate in RNPs with OMA-1 and LIN-41 (Spike et al. 2014b; this work), we tested whether LIN-41- and OMA-1-associated transcripts might be substrates of the GLD-2 cytoplasmic poly(A) polymerase. We undertook this line of investigation to address the mechanism by which the OMA proteins might promote the translation of several transcripts that selectively associate with LIN-41. Recently, Harrison et al. (2015) developed a poly(A)-test RNA-seq (PAT-seq) method to determine native poly(A) tail lengths in transcripts from total RNA samples. This method was used to compare poly(A) tail lengths of transcriptomes of adult hermaphrodites from gld-2(q497) null mutants and the wild type (P. R. Boag, A. Barugahare, and T. H. Beilharz, unpublished results). We used a competitive-gene-set test (Wu and Smyth 2012) to ask if the transcripts that selectively associate with LIN-41 or OMA-1, as well as transcripts that associate with both proteins (see Figure 4D), are enriched among those that have poly(A) tail lengths that are affected by the gld-2(q497) mutation. We found that as a gene-set class, transcripts that associate selectively with LIN-41 are more likely to be among those that exhibit a reduction in poly(A) tail lengths in gld-2(q497) mutants (Figure 14A). As notable examples, spn-4 and meg-1 exhibit an average poly(A) tail length reduction of 9 and 14 adenosine residues in gld-2(q497) mutants, respectively (Figure 14B and Table S4 in File S4). The effect on orc-1 was milder; a reduction of 4.5 adenosine residues was observed (Figure 14B). By contrast, the sets of transcripts that selectively associate with OMA-1 or with both proteins were not enriched among those transcripts that had altered poly(A) tail length in gld-2(q497) mutants (Figure 14A). We also observed that the set of transcripts not associated with either LIN-41 or OMA-1 also showed no enrichment for transcripts with altered poly(A) tail lengths in gld-2(q497) mutants (Figure 14A). Yet, specific members of all three RNP-associated gene classes were observed to have shorter poly(A) tail lengths in gld-2(q497) mutants (Figure 14B and Table S4 in File S4). At a global level, the mean change in poly(A) length in gld-2(q497) mutants among all genes measured was −3.1 adenosine residues and the SD was 4.1 adenosine residues (P. R. Boag, A. Barugahare, and T. H. Beilharz, unpublished results). Notably, transcripts coding for several LIN-41 RNP components, including mex-3, mex-5, oma-1, puf-11, spn-4, oma-2, meg-1, pos-1, gld-3, gls-1, ergo-1, nasp-2, and egg-4 were observed to exhibit substantially shorter poly(A) tail lengths in gld-2(q497) mutants (>1 SD from the mean; Table S4 in File S4). Thus, GLD-2 may exert its oogenic functions in part by contributing to the efficient translation of LIN-41-associated transcripts and a subset of LIN-41 RNP components. Such a role might explain the strong genetic interaction observed between gld-2 and lin-41: female germ cells in gld-2(q497) lin-41(n2914) double null mutants exhibit pachytene arrest, a phenotype not observed in either single mutant (Spike et al. 2014a). In this model, the lin-41 null mutant phenotype would result in part through the translational derepression of associated transcripts, and in turn the efficient translation of many of these transcripts would depend on gld-2 function.
Figure 14.
Transcripts that associate selectively with LIN-41 are enriched for GLD-2 targets. The lengths of poly(A) tails were measured using a PAT-seq assay in the wild type and gld-2(q497) mutants (P. R. Boag, A. Barugahare, and T. H. Beilharz, unpublished results). (A) A competitive gene-set enrichment test (Wu and Smyth 2012) was used to derive P-values and false discovery rates (FDRs) for each gene set shown in the Venn diagram in Figure 4D. The number of genes shown in (A) is smaller than in Figure 4D because every gene was not well represented in the PAT-seq data set. The genes selectively enriched in the LIN-41 immunopurification are more likely to be among genes that have shortened poly(A) tail lengths (direction, down) in gld-2(q497) mutants. (B) A bar-code plot was used to visualize this enrichment. Genes in the LIN-41 and OMA-1 selectively enriched sets were ordered based on the nucleotide change in poly(A) length in gld-2(q497) mutants compared to the wild type, with those that have decreased tail lengths on the left and those with increased tail lengths on the right. Orange and blue vertical bars were placed at the position of each gene selectively associated with either LIN-41 (top set of bars) or OMA-1 (bottom set of bars), respectively. Note that the x-axis is not linear but rather provides an evenly spaced representation of each transcript for which PAT-seq data are available. The traces above and below represent a rescaled running average tracking the distribution of genes in each set relative to a random distribution (dotted lines). Note the distribution of genes in the LIN-41 selectively enriched set is skewed to the left, whereas the genes in the OMA-1 selectively enriched set are more evenly distributed. The genes mex-3, meg-1, spn-4, and oma-1 exhibit a reduction in poly(A) tail length >1 SD from the mean change.
GLD-2 promotes the expression of spn-4 and meg-1:
To begin to test this model, we sought to investigate the involvement of gld-2 in the translation of several LIN-41-associated transcripts, focusing on spn-4 and meg-1 which exhibit shortened poly(A) tails in gld-2 mutants. This assessment is complicated by the fact that oocytes in gld-2(q497) null mutants do not as a rule progress to diakinesis (Kadyk and Kimble 1998), the stage at which they express SPN-4::GFP::3xFLAG and GFP::3xFLAG::MEG-1. Accordingly, we did not observe expression of SPN-4::GFP::3xFLAG or GFP::3xFLAG::MEG-1 in gld-2(q497) mutants using anti-FLAG antibody staining (Figure 15, B and H; n = 56 and n = 55, respectively). Similarly, we did not observe expression of either SPN-4::GFP::3xFLAG or GFP::3xFLAG::MEG-1 in gld-2(q497) mutants using GFP fluorescence detection (n = 39 and n = 37, respectively). Consequently, we examined the expression of SPN-4 and MEG-1 in the gld-2(tn1688) reduction-of-function mutation (see Materials and Methods), which exhibits some temperature-sensitive character and progresses further in oogenesis at 20° than at 22° despite being infertile at both temperatures. For example, at 20° all day 1 adult gld-2(tn1688) dissected gonad arms assessed by DAPI staining (n = 31) contained diakinesis-stage oocytes, whereas no diakinesis-stage oocytes were observed in day 1 adult gld-2(q497) gonad arms (n = 23). We observed that GFP::3xFLAG::MEG-1 was expressed in gld-2(tn1688) mutants at 22° using both anti-FLAG antibody staining (Figure 15L; n = 61) and GFP fluorescence methods (n = 28). However, we did not observe SPN-4::GFP::3xFLAG expression in gld-2(tn1688) mutants at 22° using both anti-FLAG antibody staining (Figure 15F; n = 60) and GFP fluorescence methods (n = 40). Both SPN-4 and MEG-1 fusion proteins were expressed in gld-2(tn1688) mutants at 20° as detected by antibody staining (Figure 15, D and J; n = 43 and n = 57, respectively) and GFP fluorescence methods (n = 41 and n = 39, respectively). In a wild-type genetic background, GFP::3xFLAG::MEG-1 is expressed earlier during oogenesis than SPN-4::GFP::3xFLAG (i.e., the −3 oocyte vs. the −2 oocyte). Thus, we conclude that the defect in meiotic progression in gld-2 mutant oocytes likely contributes to the failure to express spn-4 and meg-1. However, we cannot exclude the possibility that the observed effect is directly caused by a reduction in translation efficiency (or some combination of the two).
Figure 15.
Effect of gld-2 mutations on expression of SPN-4 and MEG-1 detected using indirect immunofluorescence in dissected and fixed gonads stained with anti-FLAG antibody in the genetic backgrounds and at the temperatures indicated. DNA was detected with DAPI. SPN-4::GFP::3xFLAG and GFP::3xFLAG::MEG-1 expression used the spn-4(tn1699) and meg-1(tn1724) edits, respectively. gld-2(q497) is a likely null allele (Wang et al. 2002) and gld-2(tn1688) is a reduction-of-function allele with temperature-sensitive character (see Materials and Methods). Gonads from strains (A and B) DG4210, (C–F) DG4264, (G and H) DG4263, and (I–L) DG4262 are shown. Bar, 50 μm.
Since the expression of SPN-4::GFP::3xFLAG and GFP::3xFLAG::MEG-1 is affected by the function of OMA-1, OMA-2, and LIN-41, we also determined the effect of gld-2(q497) on their expression. We observed that OMA-1::GFP and GFP::LIN-41 are expressed in the gld-2(q497) null mutant background, though GFP::LIN-41 occasionally exhibited a punctate distribution in the proximal gonad (Figure S7 in File S4). By contrast, mNG::3xFLAG::OMA-2, which is expressed somewhat later in oogenesis than OMA-1::GFP and GFP::LIN-41, is not expressed in gld-2(q497) mutants (Figure S7 in File S4). Despite the expression of OMA-1::GFP and GFP::LIN-41 in gld-2(q497) mutants, translational repression may be somewhat compromised because GFP::3xFLAG::CDC-25.3 is weakly expressed in nuclei of gld-2(q497) mutant oocytes, as detected using GFP fluorescence (n = 39) and anti-FLAG antibody staining methods (Figure 6L; n = 43). Likewise, we detected weak GFP fluorescence of GFP::3xFLAG::CDC-25.3 in oocyte nuclei of gld-2(tn1688) mutants at 20° (n = 35). In the wild-type background, GFP::CDC-25.3 expression is observed in embryos but not oocytes (Figure 6, A–D).
To examine further the involvement of gld-2 in the expression of SPN-4 and MEG-1, we examined the less pleiotropic rnp-8(tm2435); gld-3(q730) double-mutant background. In rnp-8(tm2435); gld-3(q730) double mutants, oocytes progress to diakinesis, undergo meiotic maturation and fertilization, but arrest during embryogenesis (Kim et al. 2010). Although both SPN-4::GFP::3xFLAG and GFP::3xFLAG::MEG-1 were expressed in oocytes and early embryos in rnp-8(tm2435); gld-3(q730) double-mutant backgrounds, they exhibited moderately reduced expression levels (Figure 16). In embryos, GFP::3xFLAG::MEG-1 localized to P granules and accumulated to maximal levels in germline blastomeres in the wild-type background (Figure 16, E and F) but not in the rnp-8(tm2435); gld-3(q730) double-mutant background (Figure 16, G and H). Likewise, SPN-4::GFP::3xFLAG exhibited decreased but relatively uniform expression levels among embryonic blastomeres in the rnp-8(tm2435); gld-3(q730) double-mutant background (Figure 16, K and L) compared to that of the wild type (Figure 16, I and J). Similarly, the expression of GFP::3xFLAG::MEG-1 was reduced in oocytes in the rnp-8(tm2435); gld-3(q730) double-mutant background compared to the wild-type background (Figure 16, A–D). We quantified SPN-4::GFP::3xFLAG fluorescence levels in the −1 oocyte and the +1 embryo in the wild-type and rnp-8(tm2435); gld-3(q730) mutant backgrounds and observed a statistically significant difference of approximately twofold (Figure 16M; P < 0.0001, unpaired t-test). The reduction of SPN-4::GFP::3xFLAG and GFP::3xFLAG::MEG-1 expression in embryos likely originates from the lower oocyte expression levels. However, we are unable to rule out the possibility that early embryonic defects in rnp-8(tm2435); gld-3(q730) mutants, which include disruptions in cell division and cell fate specification (see Figure 16L), contribute to the reduction in embryonic expression levels. Taken together, the analysis of gld-2 single mutants and rnp-8; gld-3 double mutants suggests: (1) GLD-2 and its cofactors GLD-3 and RNP-8 promote efficient translation of mRNA targets; (2) GLD-2 activity may not be absolutely required for the expression of every one of its mRNA targets, the effects on expression might be largely quantitative; (3) GLD-2 might exert substantial function for promoting oogenesis in the absence of its known stimulatory cofactors GLD-3 and RNP-8; and (4) defects in meiotic progression in gld-2 mutants might contribute to the failure to express individual mRNA targets.
Figure 16.
MEG-1 and SPN-4 are expressed at lower levels in oocytes and embryos in rnp-8; gld-3 double mutants. Paired DIC and fluorescence images showing (A–D and E–H) GFP::3xFLAG::MEG-1 expression using the meg-1(tn1724) edit and (I–L) SPN-4::GFP::3xFLAG expression using the spn-4(tn1699) edit in wild-type genetic backgrounds or the rnp-8(tm2435); gld-3(q730) double null mutant background. (L) An arrowhead indicates an embryonic blastomere with multiple nuclei. (M) Quantification of SPN-4::GFP::3xFLAG fluorescence levels (arbitrary units) in the −1 oocyte and +1 embryo in a wild-type genetic background or the rnp-8(tm2435); gld-3(q730) double null mutant background as indicated. An unpaired t-test with Welch’s correction for unequal variance indicated *** P < 0.0001 for the comparisons. A similar level of significance was also observed without the Welch correction. The strains used in this figure are: DG4158, DG4363, DG4213, and DG4366. (A–D) Bar, 50 μm. (E–L) Bar, 20 μm.
Discussion
LIN-41 and the OMA proteins as master regulators of C. elegans oogenesis
Genetic and phenotypic analyses establish the OMA proteins and LIN-41 as key regulators of oocyte development and the oocyte-to-embryo transition (Detwiler et al. 2001; Nishi and Lin 2005; Shirayama et al. 2006; Stitzel et al. 2006; Guven-Ozkan et al. 2008; Spike et al. 2014a,b; Tocchini et al. 2014). Here we provide biochemical insights into potential mechanisms by which LIN-41 and the OMA proteins antagonistically control the prophase-to-metaphase transition and growth of oocytes, thereby enabling the production of viable and fertile progeny. The results reported here suggest that LIN-41 and the OMA proteins control and coordinate oocyte growth, meiotic cell-cycle progression, and the oocyte-to-embryo transition in large part through the concerted and antagonistic translational regulation of a battery of genes which include regulators of the cell cycle, protein translation, and membrane trafficking.
Null mutations in lin-41 result in catastrophic defects in oogenesis: pachytene-stage oocytes prematurely cellularize, activate CDK-1, disassemble the synaptonemal complex, enter M phase, assemble spindles, and attempt to segregate chromosomes (Spike et al. 2014a). The analysis of mutations that reduce but do not eliminate lin-41 function demonstrates that lin-41 also facilitates the production of high-quality oocytes and the faithful segregation of meiotic chromosomes (Spike et al. 2014a). The translational derepression of the CDK-1 activator, CDC-25.3, contributes in part to the premature M-phase entry phenotype of lin-41 null mutant oocytes (Spike et al. 2014a,b). However, misregulation of genes like cdc-25.3, which are targets of translational repression by both LIN-41 and the OMA proteins, provides an insufficient explanation for the lin-41 and oma-1/2 null phenotypes, which are in many respects polar opposites. For example, oocytes in oma-1; oma-2 null mutant hermaphrodites grow abnormally large and fail to activate CDK-1 and undergo meiotic maturation (Detwiler et al. 2001), in contrast to the situation in lin-41 null mutants in which oocytes prematurely cellularize and activate CDK-1 at the end of pachytene. The analysis of genetic interactions between lin-41 and oma-1/2 reveals a complex relationship. Pachytene-stage oocytes in lin-41(n2914); oma-1(zu405te33); oma-2(te51) triple null mutants prematurely enter M phase, exhibiting the lin-41 mutant phenotype, thereby establishing that lin-41 is epistatic to oma-1/2 for this phenotype (Spike et al. 2014a). However, analysis of the behavior of more developed diakinesis-stage oocytes in a lin-41(tn1487ts); oma-1(zu405te33); oma-2(te51) triple-mutant combination at a semipermissive temperature shows that these oocytes fail to enter M phase (Spike et al. 2014a). This result was interpreted to suggest that the OMA proteins function to inhibit LIN-41 to promote meiotic maturation of the most proximal (−1) oocyte (Spike et al. 2014a). Here we define a new class of mRNAs, exemplified by spn-4, meg-1, and orc-1, which preferentially associate with LIN-41 (Figure 17). Whereas LIN-41 has a repressive effect on spn-4, meg-1, and orc-1 expression; the OMA proteins promote their expression in the adult germline (spn-4 and meg-1) or have no apparent effect (orc-1). This contrasts with mRNA targets exemplified by cdc-25.3 which are translationally repressed by LIN-41 and the OMA proteins (Figure 17A). Our results suggest that the OMA proteins might function in part to counteract or attenuate LIN-41-mediated translational repression to facilitate expression of SPN-4 and MEG-1 in proximal oocytes. All these factors substantially increase in expression in the early embryo. Taken together, these results suggest the possibility that LIN-41 activity, which includes the prevention of M-phase entry, is inactivated sequentially during oocyte meiotic maturation (Figure 17B). In the first step (1), LIN-41 translational repression activity is attenuated in the most proximal oocytes; and in the second step (2), LIN-41 is inactivated and degraded. Genetic analysis is consistent with a model in which the OMA proteins antagonize LIN-41 activity in both steps. The OMA proteins may promote the second step in part through the activation of CDK-1, which is required for the elimination of LIN-41, commencing with the onset of meiotic maturation (Spike et al. 2014a). The nature of the first step is unclear, but may operate through another component of the complex because the OMA proteins are required for expression of spn-4 and meg-1, even in the absence of lin-41. Alternatively, the OMA proteins may promote expression of spn-4 and meg-1 independently of LIN-41 RNP complexes. The translation-promoting activity of the OMA proteins appears to occur in the absence of the MSP meiotic-maturation signal as SPN-4::GFP::3xFLAG is expressed in the proximal two oocytes in unmated fog-2(oz40) females. Thus the attenuation of LIN-41 repression and the synthesis of SPN-4 in the proximal two oocytes may define a sperm-independent step in cytoplasmic oocyte maturation. Interestingly, Lee et al. (2007) noted that when females are mated and analyzed soon afterward, meiotic maturation and ovulation are observed at ∼11-min intervals, compared to ∼23-min intervals in wild-type hermaphrodites. Thus, proximal oocytes in hermaphrodites require ∼12 additional minutes to complete a process that is already finished while oocytes arrest in unmated adult females. We suggest that this process may include protein translation of mRNA targets like spn-4 and meg-1 which are repressed distally and translated in the most proximal oocytes.
Figure 17.
A model for translational regulation by protein complexes containing LIN-41 and the OMA proteins. (A) Several transcripts that stably associate with LIN-41 and OMA-1 are translationally repressed in a 3′-UTR-dependent mechanism. For these mRNA targets, which include cdc-25.3, rnp-1, and zif-1, inactivation of either LIN-41 or OMA-1 and OMA-2 results in translational derepression. (B) Several transcripts that stably associate with LIN-41 but not OMA-1, including spn-4 and meg-1, are translationally repressed by LIN-41. In this model, association of the OMA proteins with transcripts of this class results in translational activation and their stable association with ribosomes. LIN-41 activity appears to be inactivated sequentially during the late stages of oogenesis: (1) the OMA proteins activate translation through another component of the complex, and (2) LIN-41 is inactivated and degraded. The OMA proteins may promote the degradation of LIN-41 upon the onset of meiotic maturation in part by promoting CDK-1 activation. The cartoon is schematic and is not meant to imply the actual position of proteins on the transcript or within the RNP complexes.
How might LIN-41 and OMA-1 “toggle” mRNA targets between repression and activation? The proteomic identification of OMA-1 and LIN-41 RNP components (Spike et al. 2014b; this work) suggests a possible model (Figure 17). OMA-1 and LIN-41 associate with a common set of translational regulators, including the GLD-2 cytoplasmic poly(A) polymerase (Wang et al. 2002; Barnard et al. 2004; Kwak et al. 2004; Cui et al. 2008, 2013) and the CCR4-NOT deadenylase complex (reviewed by Goldstrohm and Wickens 2008), which have been shown in many organisms to activate and repress protein translation, respectively. The finding that many transcripts that are selectively associated with LIN-41 have shortened poly(A) tails in gld-2 null mutants is consistent with the possibility that these transcripts are poised for efficient translation upon the disruption of LIN-41-mediated repression. For example, if spn-4 and meg-1 transcripts acquired long poly(A) tails when associated with LIN-41, the OMA proteins might then promote their translation at a step after cytoplasmic poly(A) elongation. Alternatively, the OMA proteins might promote the expression of spn-4 and meg-1 by stimulating GLD-2 activity and/or by inhibiting the CCR4-NOT deadenlyase. Detailed information on the timing of GLD-2 activity for specific mRNA targets will be important for distinguishing between these possibilities.
Upon their synthesis, both SPN-4 and MEG-1 are incorporated into RNPs containing LIN-41. Genetic data indicate that LIN-41 function in the oocyte facilitates SPN-4 function in the embryo. In this regard, it is notable that LIN-41 RNPs contain multiple RNA-binding proteins whose transcripts also associate with, and are regulated by, LIN-41 and/or OMA-1 (Table 2). Thus, in addition to their role in coordinating oocyte growth and meiotic cell-cycle progression, LIN-41 and OMA-1/2 facilitate the assembly of oocyte RNPs containing proteins and mRNAs with critical maternal roles in the early embryo, including mex-3, pos-1, meg-1, and spn-4 (Draper et al. 1996; Tabara et al. 1999; Gomes et al. 2001; Ogura et al. 2003; Leacock and Reinke 2008).
LIN-41 is conserved in evolution and it seems likely that the translational control mechanisms uncovered here in a study of the role of LIN-41 in C. elegans oogenesis will prove informative for other systems by analogy if not homology. The mammalian ortholog LIN-41/TRIM71 is required for embryonic viability and neural-tube closure in mice (Maller Schulman et al. 2008; Cuevas et al. 2015; Mitschka et al. 2015). LIN-41/TRIM71 was defined as an RNA-binding protein found in mouse embryonic stem (ES) cells (Kwon et al. 2013). The literature contains conflicting reports on the role of LIN-41/TRIM71 in ES cells. One study found that small-interfering-RNA knockdown of LIN-41/TRIM71 in mouse ES cells decreases their rate of proliferation (Chang et al. 2012); whereas another study using conditional gene targeting showed that LIN-41/TRIM71 −/− ES cells exhibited normal growth kinetics (Mitschka et al. 2015). The latter study also found that LIN-41/TRIM71 −/− ES cells retain pluripotency and stem cell characteristics; however, they displayed increased expression of neural genes upon treatment with retinoic acid (Mitschka et al. 2015). A study of reprogramming of human dermal fibroblasts into induced pluripotent stem cells found a role for LIN-41/TRIM71 in promoting reprogramming through the negative regulation-of-differentiation genes including the transcription factor EGR1 (Worringer et al. 2014). LIN-41/TRIM71 was shown to regulate EGR1 expression by directly binding to the EGR1 transcript (Worringer et al. 2014). Several studies show that LIN-41/TRIM71 associates with mRNAs and promotes translational repression and mRNA decay (Loedige et al. 2013). Detailed studies of the Drosophila LIN-41 homolog BRAT have provided a model for how the six-bladed β propeller NHL domain recognizes RNA through its positively charged top surface (Loedige et al. 2014, 2015). Our finding that LIN-41 associates in RNPs with conserved regulators of protein translation provides an impetus for assessing the generality of these results.
Contrasting strategies for translational regulation in the temporal control of gene expression in the germline and soma
Recent results on the role of LIN-41 in the heterochronic pathway which controls the developmental timing of somatic cell fates suggested that LIN-41 recognition of the 3′-UTR of three targets, mab-10, mab-3, and dmd-3 leads to mRNA destabilization (Aeschimann et al. 2017). By contrast, recognition of the 5′-UTR in lin-29 led to translational repression (Aeschimann et al. 2017). These results provide an interesting counterpoint to the results presented here, thereby highlighting the contrasting strategies for translational regulation deployed by the germline and soma. In the C. elegans germline, translational regulation by 3′-UTRs represents a major mode of gene regulation (Merritt et al. 2008). All of the examined mRNA targets of LIN-41-mediated translational repression (e.g., zif-1, cdc-25.3, rnp-1, spn-4, meg-1, and orc-1) are expressed in early embryos. In C. elegans, as in many species, full-grown oocytes are transcriptionally quiescent (Starck 1977; Gibert et al. 1984; Schisa et al. 2001; Walker et al. 2007). Thus, it would not make biological sense for LIN-41-mediated translational repression to result in mRNA destabilization in the germline. Consistent with this view, transcripts isolated from germline LIN-41 RNPs are largely intact. LIN-41 RNPs appear to preserve mRNAs that are maternally inherited and translationally activated late in oogenesis or during the early development of the embryo. Many LIN-41-associated transcripts appear to be poised for efficient translation given that they are substrates of the GLD-2 cytoplasmic poly(A) polymerase. By contrast, transcriptional regulation plays a dynamic role in the regulation of somatic cell fates. Thus, LIN-41-mediated mRNA destabilization may play a key role in developmental cell fate switches in the soma by reinforcing the dependence on new transcription. Indeed, the major targets of LIN-41-mediated translational regulation in the heterochronic pathway are all transcription factors with key roles in regulating cell fate specification (Rougvie and Ambros 1995; Mason et al. 2008; Harris and Horvitz 2011; Nelson et al. 2011). This contrasts with LIN-41’s germline role in which interacting proteins and mRNA targets include regulators of mRNA translation. Through their roles in regulating translation, LIN-41 and the OMA proteins control the dynamic maturation of RNPs needed for meiotic maturation and the oocyte-to-embryo transition.
Although the heterochronic pathway also relies extensively on translational regulation, this is achieved in large part through the action of microRNAs (Rougvie and Moss 2013). MicroRNAs use mRNA destabilization as a major mode by which they mediate post-transcriptional gene silencing (Jonas and Izaurralde 2015). In contrast, current data suggest that microRNAs play little or no role in translational regulation in the C. elegans germline. Genetic mosaic analysis of dcr-1, which encodes the Dicer endonuclease required for cytoplasmic processing of precursor microRNAs into mature microRNAs (Grishok et al. 2001; Hutvágner et al. 2001; Ketting et al. 2001), provided strong evidence that dcr-1 is not required cell autonomously in the germline for normal germline development (Drake et al. 2014). Although several microRNAs, notably those of the mir-35 family, are produced in the germline and are maternally provided to the embryo, they appear to exert their effects long after the oocyte-to-embryo transition (Lau et al. 2001; Alvarez-Saavedra and Horvitz 2010; McJunkin and Ambros 2014, 2017). The miR-309 cluster of microRNAs that regulates the transition from maternal to zygotic gene expression in Drosophila provides the exception that proves the rule. Although miR-309-cluster microRNAs degrade maternal mRNAs, these microRNAs are zygotically transcribed in a mechanism dependent on the Smaug RNA-binding protein (Bushati et al. 2008; Luo et al. 2016; reviewed by Laver et al. 2015). Instead of using microRNA regulation, translational control in the C. elegans germline is largely mediated by dynamic RNPs (Nousch and Eckmann 2013). The work herein documents a regulatory strategy employed in the germline in which LIN-41 and the OMA proteins act as foils to appropriately balance translational repression and activation during oogenesis. This translational repression-to-activation switch facilitates the proper spatial and temporal expression of many key genes and translational regulatory RNPs for the control of oogenesis and the oocyte-to-embryo transition.
Supplementary Material
Supplemental material is available online at http://www.genetics.org/content/early/2017/06/02/genetics.117.203174.supplemental.
Acknowledgments
We would like to thank Adele Barugahare and the Monash Bioinformatics Platform for contributing to the GLD-2 poly(A) length analysis. We are grateful to Joshua Arribere, Daniel Dickinson, Andrew Fire, Bob Goldstein, Kelly Liu, and Dustin Updike for providing strains or reagents. We thank Daniel Schmidt for guidance using Chimera and Sean Conner, Lorene Lanier, and Gant Luxton for the use of equipment. Swathi Arur, Ann Rougvie, Tim Schedl, Todd Starich, Dustin Updike, and Mariana Wolfner provided helpful suggestions for the manuscript. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by grant P40 OD010440 from the National Institutes of Health Office of Research Infrastructure Programs. We also thank WormBase for sequences and annotations. The monoclonal anti-GFP antibodies developed by the Developmental Studies Hybridoma Bank (DSHB) were obtained from the DSHB created by the National Institute of Child Health and Human Development of the National Institutes of Health and maintained at Department of Biology, The University of Iowa. We thank the University of Minnesota Genomics Center for high-throughput sequencing and the Taplin Mass Spectrometry Facility at Harvard University for generating the spectra used in this study. This work was supported by National Institutes of Health grant GM57173 (to D.G.) and National Health and Medical Research Council of Australia grants APP606575 (to P.R.B.), APP1042848 (to P.R.B. and T.H.B.), and APP1042851 (to T.H.B). T.H.B. is supported by a Monash Biomedicine Discovery Fellowship.
Footnotes
Communicating editor: T. Schupbach
Literature Cited
- Aeschimann F., Kumari P., Bartake H., Gaidatzis D., Xu L., et al. , 2017. LIN41 post-transcriptionally silences mRNAs by two distinct and position-dependent mechanisms. Mol. Cell 65: 476–489. [DOI] [PubMed] [Google Scholar]
- Alvarez-Saavedra E., Horvitz H. R., 2010. Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 20: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J. A., Bell R. T., Fu B. X., Artiles K. L., Hartman P. S., et al. , 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard D. C., Ryan K., Manley J. L., Richter J. D., 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119: 641–651. [DOI] [PubMed] [Google Scholar]
- Benoit P., Papin C., Kwak J. E., Wickens M., Simonelig M., 2008. PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development 135: 1969–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati S., Stark A., Brennecke J., Cohen S. M., 2008. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 18: 501–506. [DOI] [PubMed] [Google Scholar]
- Chakshusmathi G., Mondal K., Lakshmi G. S., Singh G., Roy A., et al. , 2004. Design of temperature-sensitive mutants solely from amino acid sequence. Proc. Natl. Acad. Sci. USA 101: 7925–7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.-M., Martinez N. J., Thornton J. E., Hagan J. P., Nguyen K. D., et al. , 2012. Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat. Commun. 3: 923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Melton C., Suh N., Oh J. S., Horner K., et al. , 2011. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 25: 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas E., Rybak-Wolf A., Rhode A. M., Nguyen D. T. T., Wulczyn F. G., 2015. Lin41/Trim71 is essential for mouse development and specifically expressed in postnatal ependymal cells of the brain. Front. Cell Dev. Biol. 3: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Sackton K. L., Horner V. L., Kumar K. E., Wolfner M., 2008. Wispy, the Drosophila homolog of GLD-2, is required during oogenesis and egg activation. Genetics 178: 2017–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Sartain C. V., Pleiss J. A., Wolfner M. F., 2013. Cytoplasmic polyadenylation is a major mRNA regulator during oogenesis and egg activation in Drosophila. Dev. Biol. 383: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detwiler M. R., Reuben M., Li X., Rogers E., Lin R., 2001. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev. Cell 1: 187–199. [DOI] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Pani A. M., Heppert J. K., Higgins C. D., Goldstein B., 2015. Streamlined genome engineering with a self-excising drug selection cassette. Genetics 200: 1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake M., Furuta T., Suen K. M., Gonzalez G., Liu B., et al. , 2014. A requirement for ERK-dependent dicer phosphorylation in coordinating oocyte-to-embryo transition in C. elegans. Dev. Cell 31: 614–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper B. W., Mello C. C., Bowerman B., Hardin J., Priess J. R., 1996. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell 87: 205–216. [DOI] [PubMed] [Google Scholar]
- Duchaine T. F., Wohlschlegel J. A., Kennedy S., Bei Y., Conte D., Jr, et al. , 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354. [DOI] [PubMed] [Google Scholar]
- Dunphy W. G., Brizuela L., Beach D., Newport J., 1988. The Xenopus cdc2 protein is a component of MPF, a cytoplasmic regulator of mitosis. Cell 54: 423–431. [DOI] [PubMed] [Google Scholar]
- Eckmann C. R., Kraemer B., Wickens M., Kimble J., 2002. GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev. Cell 3: 697–710. [DOI] [PubMed] [Google Scholar]
- Francis R., Barton M. K., Kimble J., Schedl T., 1995. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 139: 579–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier J., Norbury C., Lohka M., Nurse P., Maller J., 1988. Purified maturation-promoting factor contains the product of a Xenopus homolog of the fission yeast cell cycle control gene cdc2+. Cell 54: 433–439. [DOI] [PubMed] [Google Scholar]
- Gautier J., Minshull J., Lohka M., Glotzer M., Hunt T., et al. , 1990. Cyclin is a component of maturation-promoting factor from Xenopus. Cell 60: 487–494. [DOI] [PubMed] [Google Scholar]
- Gent J. I., Lamm A. T., Pavelec D. M., Maniar J. M., Parameswaran P., et al. , 2010. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol. Cell 37: 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibert M. A., Starck J., Beguet B., 1984. Role of the gonad cytoplasmic core during oogenesis of the nematode Caenorhabditis elegans. Biol. Cell 50: 77–85. [DOI] [PubMed] [Google Scholar]
- Goldstrohm A. C., Wickens M., 2008. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 9: 337–344. [DOI] [PubMed] [Google Scholar]
- Gomes J. E., Encalad S. E., Swan K. A., Shelton C. A., Carter J. C., et al. , 2001. The maternal gene spn-4 encodes a predicted RRM protein required for mitotic spindle orientation and cell fate patterning in early C. elegans embryos. Development 128: 4301–4314. [DOI] [PubMed] [Google Scholar]
- Govindan J. A., Cheng H., Harris J. E., Greenstein D., 2006. Gαo/i and Gαs signaling function in parallel with the MSP/Eph receptor to control meiotic diapause in C. elegans. Curr. Biol. 16: 1257–1268. [DOI] [PubMed] [Google Scholar]
- Govindan J. A., Nadarajan S., Kim S., Starich T. A., Greenstein D., 2009. Somatic cAMP signaling regulates MSP-dependent oocyte growth and meiotic maturation in C. elegans. Development 136: 2211–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli A. E., Conte D., Li N., Parrish S., et al. , 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34. [DOI] [PubMed] [Google Scholar]
- Gruber A. R., 2014. RNA polymerase III promoter screen uncovers a novel noncoding RNA family conserved in Caenorhabditis and other clade V nematodes. Gene 544: 236–240. [DOI] [PubMed] [Google Scholar]
- Guven-Ozkan T., Nishi Y., Robertson S. M., Lin R., 2008. Global transcriptional repression in C. elegans germline precursors by regulated sequestration of TAF-4. Cell 135: 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. H., Winfrey V. P., Blaeuer G., Hoffman L. H., Furuta T., et al. , 1999. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev. Biol. 212: 101–123. [DOI] [PubMed] [Google Scholar]
- Hansen D., Wilson-Berry L., Dang T., Schedl T., 2004. Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development 131: 93–104. [DOI] [PubMed] [Google Scholar]
- Harris D. T., Horvitz H. R., 2011. MAB-10/NAB acts with LIN-29/EGR to regulate terminal differentiation and the transition from larva to adult in C. elegans. Development 138: 4051–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. W., Baran J., Bieri T., Cabunoc A., Chan J., et al. , 2013. WormBase 2014: new views of curated biology. Nucleic Acids Res. 42: D789–D793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison P. F., Powell D. R., Clancy J. L., Preiss T., Boag P. R., et al. , 2015. PAT-seq: a method to study the integration of 3′-UTR dynamics with gene expression in the eukaryotic transcriptome. RNA 21: 1502–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R., Cubitt A. B., Tsien R. Y., 1995. Improved green fluorescence. Nature 737: 663–664. [DOI] [PubMed] [Google Scholar]
- Huang N. N., Mootz D. E., Walhout A. J., Vidal M., Hunter C. P., 2002. MEX-3 interacting proteins link cell polarity to asymmetric gene expression in Caenorhabditis elegans. Development 129: 747–759. [DOI] [PubMed] [Google Scholar]
- Hutvágner G., McLachlan J., Pasquinelli A. E., Balint É., Tuschl T., et al. , 2001. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838. [DOI] [PubMed] [Google Scholar]
- Iizuka I., Yamagishi-Shirasaki M., Funatsu T., 2011. Kinetic study of de novo chromophore maturation of fluorescent proteins. Anal. Biochem. 414: 173–178. [DOI] [PubMed] [Google Scholar]
- Ivshina M., Lasko P., Richter J. D., 2014. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu. Rev. Cell Dev. Biol. 30: 393–415. [DOI] [PubMed] [Google Scholar]
- Jadhav S., Rana M., Subramaniam K., 2008. Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development 135: 1803–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S., Izaurralde E., 2015. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 16: 421–433. [DOI] [PubMed] [Google Scholar]
- Kadyk L. C., Kimble J., 1998. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125: 1803–1813. [DOI] [PubMed] [Google Scholar]
- Kaymak E., Ryder S. P., 2013. RNA recognition by the Caenorhabditis elegans oocyte maturation determinant OMA-1. J. Biol. Chem. 288: 30463–30472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting R. F., Fischer S. E., Bernstein E., Sijen T., Hannon G. J., et al. , 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15: 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. W., Nykamp K., Suh N., Bachorik J. L., Wang L., et al. , 2009. Antagonism between GLD-2 binding partners controls gamete sex. Dev. Cell 16: 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. W., Wilson T. L., Kimble J., 2010. GLD-2/RNP-8 cytoplasmic poly(A) polymerase is a broad-spectrum regulator of the oogenesis program. Proc. Natl. Acad. Sci. USA 107: 17445–17450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Govindan J. A., Tu Z. J., Greenstein D., 2012. SACY-1 DEAD-Box helicase links the somatic control of oocyte meiotic maturation to the sperm-to-oocyte switch and gamete maintenance in Caenorhabditis elegans. Genetics 192: 905–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Spike C., Greenstein D., 2013. Control of oocyte growth and meiotic maturation in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 277–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski M., McDonald K., Schwartz J., Yamamoto I., Greenstein D., 2005. C. elegans sperm bud vesicles to deliver a meiotic maturation signal to distant oocytes. Development 132: 3357–3369. [DOI] [PubMed] [Google Scholar]
- Kronja I., Yuan B., Eichhorn S. W., Dzeyk K., Krijgsveld J., et al. , 2014a Widespread changes in the posttranscriptional landscape at the Drosophila oocyte-to-embryo transition. Cell Rep. 7: 1495–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronja I., Whitfield Z. J., Yuan B., Dzeyk K., Kirkpatrick J., et al. , 2014b Quantitative proteomics reveals the dynamics of protein changes during Drosophila oocyte maturation and the oocyte-to-embryo transition. Proc. Natl. Acad. Sci. USA 111: 16023–16028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J. E., Wang L., Ballantyne S., Kimble J., Wickens M., 2004. Mammalian GLD-2 homologs are poly(A) polymerases. Proc. Natl. Acad. Sci. USA 101: 4407–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S. C., Hyerim Y., Eichelbaum K., Föhr S., Fischer B., et al. , 2013. The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 20: 1122–1130. [DOI] [PubMed] [Google Scholar]
- Lau N. C., Lim L. P., Weinstein E. G., Bartel D. P., 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Laver D., Marsolais A. J., Smibert C. A., Lipshitz H. D., 2015. Regulation and function of maternal gene products during the maternal-to-zygotic transition in Drosophila. Curr. Top. Dev. Biol. 113: 43–84. [DOI] [PubMed] [Google Scholar]
- Leacock S. W., Reinke V., 2008. MEG-1 and MEG-2 are embryo-specific P-granule components required for germline development in Caenorhabditis elegans. Genetics 178: 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-H., Ohmachi M., Arur S., Nayak S., Francis R., et al. , 2007. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics 177: 2039–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Albertini D. F., 2013. The road to maturation: somatic cell interaction and self-organization of the mammalian oocyte. Nat. Rev. Mol. Cell Biol. 14: 141–152. [DOI] [PubMed] [Google Scholar]
- Lockwood S., Krishnamoorthy B., Ye P., 2011. Neighborhood properties are important determinants of temperature sensitive mutations. PLoS One 6: e28507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loedige I., Gaidatzis D., Sack R., Meister G., Filipowicz W., 2013. The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic Acids Res. 41: 518–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loedige I., Stotz M., Qamar S., Kramer K., Hennig J., et al. , 2014. The NHL domain of BRAT is an RNA-binding domain that directly contacts the hunchback mRNA for regulation. Genes Dev. 28: 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loedige I., Jakob L., Treiber T., Ray D., Stotz M., et al. , 2015. The crystal structure of the NHL domain in complex with RNA reveals the molecular basis of Drosophila Brain-Tumor-mediated gene regulation. Cell Rep. 13: 1206–1220. [DOI] [PubMed] [Google Scholar]
- Lohka M. J., Hayes M. K., Maller J. L., 1988. Purification of maturation-promoting factor, an intracellular regulator of early mitotic events. Proc. Natl. Acad. Sci. USA 85: 3009–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M., Huber W., Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Li X., Claycomb J. M., Lipshitz H. D., 2016. The Smaug RNA-binding protein is essential for microRNA synthesis during the Drosophila maternal-to-zygotic transition. G3 (Bethesda) 6: 3451–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller Schulman B. R., Liang X., Stahlhut C., DelConte C., Stefani G., et al. , 2008. The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle 7: 3935–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason D. A., Rabinowitz J. S., Portman D. S., 2008. dmd-3, a doublesex-related gene regulated by tra-1, governs sex-specific morphogenesis in C. elegans. Development 135: 2373–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui Y., 2001. From oocyte maturation to the in vitro cell cycle: the history of discoveries of maturation-promoting factor (MPF) and cytostatic factor (CSF). Differentiation 69: 1–17. [DOI] [PubMed] [Google Scholar]
- Masui Y., Clarke H. J., 1979. Oocyte maturation. Int. Rev. Cytol. 57: 185–282. [DOI] [PubMed] [Google Scholar]
- Masui Y., Markert C. L., 1971. Cytoplasmic control of nuclear behavior during meiotic maturation of frog oocytes. J. Exp. Zool. 177: 129–145. [DOI] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T., 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205: 111–128. [DOI] [PubMed] [Google Scholar]
- McJunkin K., Ambros V., 2014. The embryonic mir-35 family of microRNAs promotes multiple aspects of fecundity in Caenorhabditis elegans. G3 (Bethesda) 4: 1747–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McJunkin K., Ambros V., 2017. A microRNA family exerts maternal control on sex determination in C. elegans. Genes Dev. 31: 422–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C., Rasoloson D., Ko D., Seydoux G., 2008. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 18: 1476–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. A., Nguyen V. Q., Lee M. H., Kosinski M., Schedl T., et al. , 2001. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291: 2144–2147. [DOI] [PubMed] [Google Scholar]
- Mitschka S., Ulas T., Goller T., Schneider K., Egert A., et al. , 2015. Co-existence of intact stemness and priming of neural differentiation programs in mES cells lacking Trim71. Sci. Rep. 5: 11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz D., Ho D. M., Hunter C. P., 2004. The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in translational control of C. elegans PAL-1. Development 131: 3263–3272. [DOI] [PubMed] [Google Scholar]
- Nadarajan S., Govindan J. A., McGovern M., Hubbard E. J. A., Greenstein D., 2009. MSP and GLP-1/Notch signaling coordinately regulate actomyosin-dependent cytoplasmic streaming and oocyte growth in C. elegans. Development 136: 2223–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakel K., Bonneau F., Eckmann C. R., Conti E., 2015. Structural basis for the activation of C. elegans noncanonical cytoplasmic poly(A)-polymerase GLD-2 by GLD-3. Proc. Natl. Acad. Sci. USA 112: 8614–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakel K., Bonneau F., Basquin C., Habermann B., Eckmann C. R., et al. , 2016. Structural basis for the antagonistic roles of RNP-8 and GLD-3 in GLD-2 poly(A)-polymerase activity. RNA 22: 1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. D., Zhou E., Kiontke K., Fradin H., Maldonado G., et al. , 2011. A bow-tie genetic architecture for morphogenesis suggested by a genome-wide RNAi screen in Caenorhabditis elegans. PLoS Genet. 7: e1002010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi Y., Lin R., 2005. DYRK2 and GSK-3 phosphorylate and promote the timely degradation of OMA-1, a key regulator of the oocyte-to-embryo transition in C. elegans. Dev. Biol. 288: 139–149. [DOI] [PubMed] [Google Scholar]
- Nousch M., Eckmann C. R., 2013. Translational control in the Caenorhabditis elegans germ line. Adv. Exp. Med. Biol. 757: 205–247. [DOI] [PubMed] [Google Scholar]
- Nousch M., Techritz N., Hampel D., Millonig S., Eckmann C. R., 2013. The Ccr4–Not deadenylase complex constitutes the main poly(A) removal activity in C. elegans. J. Cell Sci. 126: 4274–4285. [DOI] [PubMed] [Google Scholar]
- Nurse P., 1990. Universal control mechanism regulating onset of M-phase. Nature 344: 503–508. [DOI] [PubMed] [Google Scholar]
- Ogura K., Kishimoto N., Mitani S., Gengyo-Ando K., Kohara Y., 2003. Translational control of maternal glp-1 mRNA by POS-1 and its interacting protein SPN-4 in Caenorhabditis elegans. Development 130: 2495–2503. [DOI] [PubMed] [Google Scholar]
- Oldenbroek M., Robertson S. M., Guven-Ozkan T., Spike C., Greenstein D., et al. , 2013. Regulation of maternal Wnt mRNA translation in C. elegans embryos. Development 140: 4614–4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz M. A., Noble D., Sorokin E. P., Kimble J., 2014. A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans. G3 (Bethesda) 4: 1765–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Wang Y., Smith H. E., Lee C. Y., Calidas D., et al. , 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics 198: 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M. W., Botchan M. R., Berger J. M., 2017. Mechanisms and regulation of DNA replication initiation in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 52: 107–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., et al. , 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Potireddy S., Vassena R., Patel B. G., Latham K. E., 2006. Analysis of polysomal mRNA populations of mouse oocytes and zygotes: dynamic changes in maternal mRNA utilization and function. Dev. Biol. 298: 155–166. [DOI] [PubMed] [Google Scholar]
- Ray D., Kazan H., Chan E. T., Peña Castillo L., Chaudhry S., et al. , 2009. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 27: 667–670. [DOI] [PubMed] [Google Scholar]
- Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., et al. , 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Rose K. L., Winfrey V. P., Hoffman L. H., Hall D. H., Furuta T., et al. , 1997. The POU gene ceh-18 promotes gonadal sheath cell differentiation and function required for meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 192: 59–77. [DOI] [PubMed] [Google Scholar]
- Rougvie A. E., Ambros V., 1995. The heterochronic gene lin-29 encodes a zinc finger protein that controls a terminal differentiation event in Caenorhabditis elegans. Development 121: 2491–2500. [DOI] [PubMed] [Google Scholar]
- Rougvie A. E., Moss E. G., 2013. Developmental transitions in C. elegans larval stages. Curr. Top. Dev. Biol. 105: 153–180. [DOI] [PubMed] [Google Scholar]
- Sanchez P., Daniels K. J., Park Y. N., Soll D. R., 2014. Generating a battery of monoclonal antibodies against native green fluorescent protein for immunostaining, FACS, IP, and ChIP using a unique adjuvant. Monoclon. Antib. Immunodiagn. Immunother. 33: 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schisa J. A., Pitt J. N., Priess J. R., 2001. Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development 128: 1287–1298. [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Lambert G. G., Chammas A., Ni Y., Cranfill P. J., et al. , 2013. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10: 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Soto M. C., Ishidate T., Kim S., Nakamura K., et al. , 2006. The conserved kinases CDK-1, GSK-3, KIN-19, and MBK-2 promote OMA-1 destruction to regulate the oocyte-to-embryo transition in C. elegans. Curr. Biol. 16: 47–55. [DOI] [PubMed] [Google Scholar]
- Slack F. J., Basson M., Liu Z., Ambros V., Horvitz H. R., et al. , 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5: 659–669. [DOI] [PubMed] [Google Scholar]
- Sonneville M., Querenet M., Craig A., Gartner A., Blow J. J., 2012. The dynamics of replication licensing in live Caenorhabditis elegans embryos. J. Cell Biol. 196: 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C. A., Coetzee D., Eichten C., Wang X., Hansen D., et al. , 2014a The NHL-TRIM protein LIN-41 and the OMA RNA-binding proteins antagonistically control the prophase-to-metaphase transition and growth of C. elegans oocytes. Genetics 198: 1535–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C. A., Coetzee D., Nishi Y., Guven-Ozkan T., Oldenbroek M., et al. , 2014b Translational control of the oogenic program by components of OMA ribonucleoprotein particles in Caenorhabditis elegans. Genetics 198: 1513–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck J., 1977. Radioautographic study of RNA synthesis in Caenorhabditis elegans (Bergerac variety) oogenesis. Biol. Cell. 30: 181–182. [Google Scholar]
- Starich T. A., Hall D. H., Greenstein D., 2014. Two classes of gap junction channels mediate soma-germline interactions essential for germline proliferation and gametogenesis in C. elegans. Genetics 198: 1127–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzel M. L., Pellettieri J., Seydoux G., 2006. The C. elegans DYRK kinase MBK-2 marks oocyte proteins for degradation in response to meiotic maturation. Curr. Biol. 16: 56–62. [DOI] [PubMed] [Google Scholar]
- Suh N., Jedamzik B., Eckmann C. R., Wickens M., Kimble J., 2006. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc. Natl. Acad. Sci. USA 103: 15108–15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H., Hill R. J., Mello C. C., Priess J. R., Kohara Y., 1999. pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development 126: 1–11. [DOI] [PubMed] [Google Scholar]
- Tocchini C., Keusch J. J., Miller S. B., Finger S., Gut H., et al. , 2014. The TRIM-NHL protein LIN-41 controls the onset of developmental plasticity in Caenorhabditis elegans. PLoS Genet. 10: e1004533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Fire A., 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Vasale J. J., Gu W., Thivierge C., Batista P. J., Claycomb J. M., et al. , 2010. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc. Natl. Acad. Sci. USA 107: 3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A. K., Boag P. R., Blackwell T. K., 2007. Transcription reactivation steps stimulated by oocyte maturation in C. elegans. Dev. Biol. 304: 382–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. T., Smith J., Chen B. C., Schmidt H., Rasoloson D., et al. , 2014. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. Elife 3: e04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Eckmann C. R., Kadyk L. C., Wickens M., Kimble J., 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419: 312–316. [DOI] [PubMed] [Google Scholar]
- Whitten S. J., Miller M. A., 2007. The role of gap junctions in Caenorhabditis elegans oocyte maturation and fertilization. Dev. Biol. 301: 432–446. [DOI] [PubMed] [Google Scholar]
- Worringer K. A., Rand T. A., Hayashi Y., Sami S., Takahashi K., et al. , 2014. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell 14: 40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E. B., 1925. The Cell in Development and Heredity. Macmillan, New York. [Google Scholar]
- Wu D., Smyth G. K., 2012. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 40: e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Xiao T., Chen C. H., Li W., Meyer C. A., et al. , 2015. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 25: 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. D., Wakefield M. J., Smyth G. K., Oshlack A., 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains (see Table S1 in File S4) and reagents are available upon request. RNA-seq data have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus and are accessible through accession number GSE98130. Proteomic data are available through Proteome Xchange through accession number PXD006726 (http://www.proteomexchange.org).