Abstract
The micronutrient boron is essential in maintaining the structure of plant cell walls and is critical for high yields in crop species. Boron can move into plants by diffusion or by active and facilitated transport mechanisms. We recently showed that mutations in the maize boron efflux transporter ROTTEN EAR (RTE) cause severe developmental defects and sterility. RTE is part of a small gene family containing five additional members (RTE2–RTE6) that show tissue-specific expression. The close paralogous gene RTE2 encodes a protein with 95% amino acid identity with RTE and is similarly expressed in shoot and root cells surrounding the vasculature. Despite sharing a similar function with RTE, mutations in the RTE2 gene do not cause growth defects in the shoot, even in boron-deficient conditions. However, rte2 mutants strongly enhance the rte phenotype in soils with low boron content, producing shorter plants that fail to form all reproductive structures. The joint action of RTE and RTE2 is also required in root development. These defects can be fully complemented by supplying boric acid, suggesting that diffusion or additional transport mechanisms overcome active boron transport deficiencies in the presence of an excess of boron. Overall, these results suggest that RTE2 and RTE function are essential for maize shoot and root growth in boron-deficient conditions.
Keywords: boron transport, RTE, BOR1, maize, gene duplication
BORON is an essential microelement for plant growth and development. The most well-known role of boron is the cross-linking of the pectic polysaccharide rhamnogalacturonan-II (RG-II), an essential structural component of the cell wall (Kobayashi et al. 1996; O’Neill et al. 2001). In general, monocot species have lower boron content than dicotyledonous species, a fact that correlates with an overall difference in pectin content in the cell wall (Hu et al. 1996). Nonetheless, actively growing tissues need a constant supply of exogenous boron given that the majority of endogenous boron in plants is trapped in the cell wall (Shelp et al. 1995; O’Neill et al. 1996). Boron has a narrow range of concentrations that span deficiency to toxicity levels, therefore its uptake needs to be carefully regulated.
It was originally believed that passive diffusion was the primary mechanism of boron transport in plants (Raven 1980; Shelp et al. 1995). However, more recent experiments have shown that boron uptake involves an active, carrier-mediated process (Dordas and Brown 2001; Stangoulis et al. 2001; Brown et al. 2002). Several members of the major intrinsic protein family have since been identified as boric acid channels (Takano et al. 2006; Tanaka et al. 2008; Durbak et al. 2014). These channel proteins facilitate the transport of boron from the soil into the root cells (Takano et al. 2008; Miwa and Fujiwara 2010). The first identified efflux-type active boron transporter, AtBOR1, was shown to play a major role in loading boron into the xylem in Arabidopsis thaliana (Noguchi et al. 1997; Takano et al. 2002). Under low boron conditions, bor1 mutants showed reduced rosette leaves, and loss of apical dominance and fertility. Overexpression of BOR1, on the other hand, improved seed production under boron-limiting conditions (Miwa et al. 2006). Six additional BOR1-like genes were identified in the Arabidopsis genome, and tissue- and cell-specific expression patterns indicated that these genes play distinct roles in boron transport (Miwa et al. 2006, 2007, 2013). BOR2, a close paralog of BOR1, is strongly expressed in the lateral root cap and elongation zones of the root epidermis. bor2 mutant roots showed reduced cell elongation and reduced levels of cross-linked RG-II under low boron conditions (Miwa et al. 2013). In contrast, BOR4 was found to mediate tolerance to high boron levels and its overexpression improved boron tolerance by removing excess boron from roots (Miwa et al. 2007; Miwa and Fujiwara 2010). In addition, in eudicots, functional BOR1-like genes have been studied in several species, such as grapes (Perez-Castro et al. 2012), Brassica napus (Sun et al. 2012), and Citrus macrophylla (Canon et al. 2013).
In cereals, boron deficiency and toxicity affect yield and constrain productivity (Gupta et al. 1985; Mickelbart et al. 2015). To address this problem, various boron channel proteins and transporters were characterized in cereals (Nakagawa et al. 2007; Reid 2007; Sutton et al. 2007; Schnurbusch et al. 2010; Leaungthitikanchana et al. 2013; Chatterjee et al. 2014; Durbak et al. 2014; Liu et al. 2015) and landraces that can grow in soil with wide ranges of boron concentration were identified (Reid 2007; Sutton et al. 2007; Pallotta et al. 2014; Hayes et al. 2015). For example, rice has four BOR1-like genes (Nakagawa et al. 2007). Among them, OsBOR1 is required for xylem loading and for efficient uptake of boron in roots under low boron conditions. OsBOR4 is instead a pollen-specific efflux transporter and is essential for normal pollen germination and pollen-tube elongation (Tanaka et al. 2013). In barley, Bot1/HvBOR2 is responsible for the high boron tolerance of the Sahara landrace. Compared to intolerant genotypes, Sahara has four tandem copies of the Bot1 gene and higher transcript levels, and a direct correlation exists between Bot1 expression levels and the degree of tolerance in various landraces (Hayes and Reid 2004; Reid 2007; Sutton et al. 2007; Mickelbart et al. 2015). Similarly, a study of bread and durum wheat showed that variation in Bot-B5/D5 alleles influenced the degree of boron tolerance in various cultivars and landraces (Pallotta et al. 2014). Determining the number and function of boron transporters in crop species and landraces therefore has practical implications for the development of varieties that can grow in soils with differing boron availability.
Among cereals, maize has a relatively small requirement for boron but it is nonetheless affected by boron deficiency around the world (Shorrocks 1997; Lordkaew et al. 2011). The most common boron-deficiency symptom in maize is the formation of small cobs with few kernels, resulting in lower yields. In general, reproductive tissues are more sensitive and plants with marginal boron deficiency show poor pollen germination (Agarwala et al. 1981; Lordkaew et al. 2011). Under severe boron-deficiency conditions, leaves develop white necrotic spots and streaking (Lordkaew et al. 2011; Chatterjee et al. 2014). Only recently have the first maize mutants affected in boron transport been reported. The maize tassel-less1 (tls1) mutant exhibited symptoms of boron deficiency in vegetative and inflorescence development. TLS1 encodes an aquaporin and is coorthologous to known Arabidopsis channel proteins (Durbak et al. 2014; Leonard et al. 2014). We recently characterized a maize mutant called rotten ear (rte) that displayed severe defects in inflorescence development, as well as necrotic lesions in leaves under boron-deficient conditions. RTE is a functional homolog of the boron efflux transporter BOR1 protein. Under low boron conditions, maize inflorescences exhibited widespread tissue death, likely due to loss of cell wall integrity (Chatterjee et al. 2014).
In this study, we identified five additional boron transporter-like genes in the maize genome (RTE2–RTE6). One of these genes, RTE2, is a close paralog of RTE. Functional characterization of RTE2 showed that the dual action of both RTE and RTE2 is required for maize vegetative and reproductive development in boron-deficient conditions.
Materials and Methods
Plant materials and phenotypic analysis
RTE2 transposon insertion lines were obtained from the Maize Genetics Cooperation Stock Center (UFMu-02112, UFMu-02812, and UFMu-01459) (Settles et al. 2007). The transposon insertions are located at position +303 (UFMu-02112; target site duplication ACGGTGCTC; rte2-1), +321 (UFMu-02812; target site duplication TTCATGTTC; rte2-2), and +1670 (UFMu-01459; target site duplication GTTGGTCTG; rte2-3) of the RTE2/GRMZM2G082203 coding sequence. These lines were backcrossed once in Mo17 and A619. For rte;rte2 double mutants, rte-1 and rte-2 alleles (Chatterjee et al. 2014) in Mo17 and A619 backgrounds were crossed with all three rte2 transposon insertion lines and self-fertilized. The resulting segregating F2 populations all showed the same rte;rte2 double mutant phenotype shown by representative plants in Figure 3.
Figure 3.
Genetic interaction between rte and rte2 mutants. (A) Phenotype of wild type, rte2-2, rte-1, and double rte-1;rte2-2 mutants grown in boron-deficient soil. (B and C) rte;rte2 double mutants are short in stature and showed rudimentary ear-like structures (arrowheads). (D) Longitudinal section of shoot tips from rte;rte2 double mutants stained with Toluidine blue, showing tissue anomalies (arrowhead) in the stem. (E) Longitudinal section of shoot stained with Safranin O-Alcian Blue. Lignified red patches (arrowhead) are visible. (F) Average plant height (n = 10, t-test, * P < 0.0001). (G) Average number of leaves (n = 10, t-test, * P < 0.0001). Error bars indicate SD. wt, wild type.
The vegetative phenotype was analyzed using 6-week-old plants grown in Rutgers University fields. Plant height was measured as the distance from the ground to the upper leaf node. For root length measurements, F2 seeds from a cross of rte-1 (BC3 A619) and UFMu-02812 (BC1 A619) were germinated in greenhouses using low-boron-content field soil (from Rutgers fields). Wild-type and mutant plants were genotyped and used for primary root length measurements in two separate experiments. Student’s t-test was used to determine statistical significance.
Boron measurements
Leaf samples (upper three leaves per each plant) were collected from ∼45-day-old, field-grown plants, and at least six plants were bulked per sample (Supplemental Material, Table S1 in File S1). For greenhouse samples (Figure 4), all leaves above the top elongated internode were collected from 50-day-old plants grown in pots containing Rutgers field soil for both treated and control samples. Each data point represents the average of two bulked samples (total number of individuals is listed). All samples were air dried and analyzed by inductively coupled plasma optical emission spectroscopy by the Missouri University Plant and Soil Analysis Facility. Data are expressed in micrograms of boron per gram of dry weight.
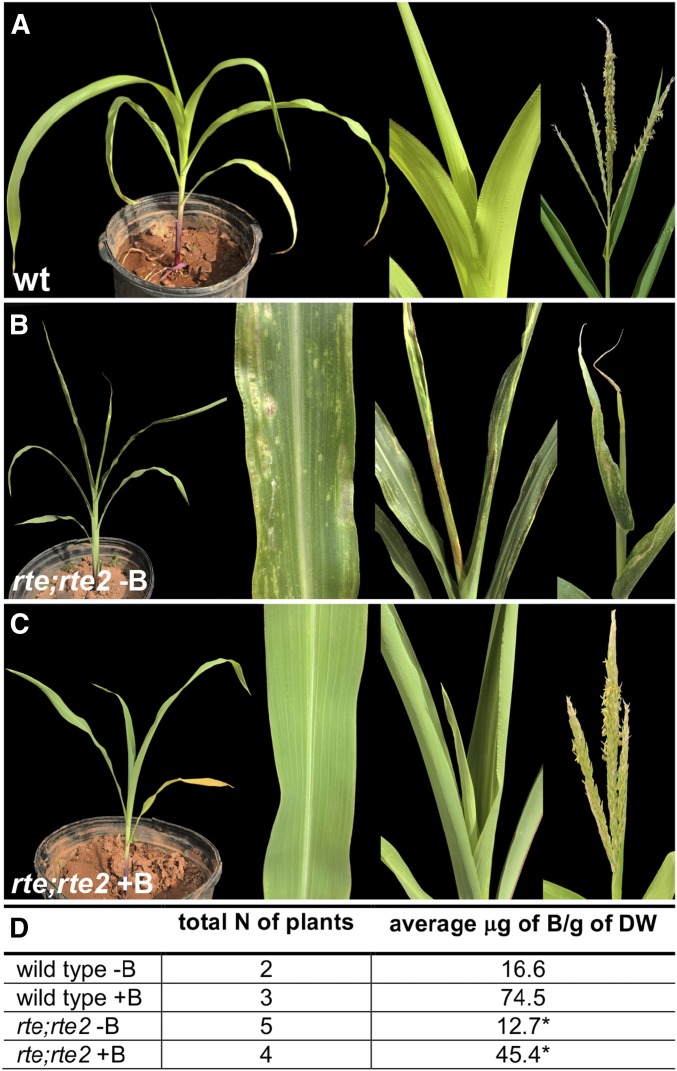
Figure 4 .
(A–C) Boric acid rescue of the double rte-1;rte2-2 mutant phenotype. Zoomed in portion of leaf lamina, top leaves, and mature tassels are visible. (D) Quantification of boron levels in treated and control plants (* P < 0.01, comparison within the same genotype). B/g, boron per gram; DW, dry weight.
For Arabidopsis measurements (Table S1 in File S1), rosette leaves from 4-week-old plants were bulked from at least three lines and subject to the same analysis highlighted above.
Boric acid rescue and soil analysis
Double rte-1;rte2-2 mutant and normal plants were germinated in pots containing soil from Rutgers fields and grown in standard greenhouse conditions. A stock solution of 100 mM boric acid (Sigma Chemical, St. Louis, MO) in Milli-Q water was diluted in tap water to a final concentration of 200 μM and used to regularly water plants. For the control experiment, Milli-Q water without the addition of boric acid was diluted in the same tap water. Plants were watered as required for growth.
Soil samples from treated and control pots were collected according to standard practices and analyzed at the New Jersey Agricultural Experiment Station, Rutgers University (Table S2 in File S1).
Phylogenetic tree construction
The amino acid sequences of RTE-like proteins were identified through searches at National Center for Biotechnology Information (NCBI), Phytozome, and MaizeGDB, and aligned using MUSCLE (Edgar 2004). The evolutionary history was inferred by the maximum-likelihood method using MEGA6.0 (Tamura et al. 2013). The percentage of trees in which the associated taxa clustered together is shown next to the branches. The analysis included proteins encoded by the following 33 gene models: RTE/GRMZM2G166159/KP751214, RTE2/GRMZM2G082203 (NCBI KY053531), RTE3/GRMZM2G051753 (corrected gene model; NCBI KY129660), RTE4/GRMZM2G374989, RTE5/GRMZM2G302559, and RTE6/GRMZM2G454327 (corrected gene model; NCBI KY129661) (maize); OsBOR1/AK070617, OsBOR2/DQ421408, OsBOR3/AK072421, and OsBOR4/DQ421409 (rice); Sb08g018440, Sb09g005350, and Sb03g004180 (sorghum); TaBOR1.1/BAO98796, TaBOR1.2/BAO98797, TaBOR1.3/BAO98798, TaBOR2/ABX26206, TaBot-D5b/AHY28551, and TaBot-B5b/AHY28552.1 (wheat); ABX26122 (Hordeum vulgare); AtBOR1/AT2G47160, AtBOR2/AT3G62270, AtBOR3/AT3G06450, AtBOR4/AT1G15460, AtBOR5/AT1G74810, AtBOR6/AT5G25430, and AtBOR7/AT4G32510 (A. thaliana); S1g057770, S6g071500, S3g120020, and S8g066960 (Solanum lycopersicum); and ScBOR1, NP_014124 (Saccharomyces cerevisiae). ScBOR1 was used as an outgroup.
The comparison of colinearity within genomic regions of different RTE-like genes was performed using CoGe SynMap (Lyons et al. 2008; https://genomevolution.org/coge).
Expression analysis
Total RNA was extracted from different tissues obtained from pools of three or more B73 plants using TRIzol reagents (ThermoFisher Scientific) as per manufacturer’s instructions. Samples used for analysis included embryos and endosperm at 10 days after pollination, mature leaf blades, seedling shoots and roots, pollen, and 1-cm tassel and ear primordia. Complementary DNA (cDNA) was obtained using the qScript cDNA Synthesis Kit and amplified with PerfeCTa SYBR Green FastMix (Quanta Biosciences). For tissue-specific expression, quantitative real-time reverse-transcription PCR (qRT-PCR) was performed using gene-specific primers (Table S3 in File S1). UBIQUITIN was used as internal control. The cycle threshold (CT) values for all genes in different RNA samples were normalized to the CT value of the internal control gene. Relative messenger RNA (mRNA) levels of each gene in different tissue samples were calculated using the 2−ΔΔCt method (Livak and Schmittgen 2001). Two biological replicates with at least three technical replicates per each pooled sample were used in the analysis.
Tissue-specific RNA-sequencing (RNA-seq) data were taken from Walley et al. (2016). Normalized average values of fragments per kilobase of transcript per million mapped reads from three replicates were converted to log10 + 1 and plotted using the aheatmap function in the R package NMF (Gaujoux and Seoighe 2010).
For expression analysis of RTE2 in Arabidopsis, RNA was extracted from rosette leaves of T1 plants and treated as described above. For analysis of RTE2 expression in insertion lines, RNA from homozygous plants and wild-type siblings for all three alleles was extracted from seedling shoots in two separate biological replicates. RT-PCR products were gel purified and sequenced to verify the identity of the amplified fragment. Primers used for qRT-PCR and RT-PCR are listed in Table S3 in File S1.
For in situ hybridizations, the 5′ and 3′ UTR of RTE and RTE2 were cloned into the pGEM-T Easy Vector System (Promega, Madison, WI) using primers RTE-UTR and RTE2-UTR (Table S3 in File S1). The 5′ UTR of RTE shares 78% identity with RTE2, and the 3′ UTR of RTE shares 79% identity with RTE2. Purified PCR products obtained using primers M13F and M13R were used as templates for synthesizing sense and antisense probes using SP6 and T7 RNA polymerases (Promega), respectively. For each experiment, the 5′ UTR and 3′ UTR RNA probes were mixed in equal ratios and used for hybridizations. The full-length RTE2 antisense probe was subjected to carbonate hydrolysis prior to its use. Seedling roots and young inflorescences were fixed in a 4% paraformaldehyde solution, dehydrated in ethanol, and embedded in paraplast. Hybridizations were conducted at 59° overnight. After several washes and treatment with anti-DIG antibody, signals of DIG-labeled probes were detected using NBT/BCIP (Promega), and images were acquired using a Leica DM5500B microscope.
Transient expression in tobacco
To create the 35S::RTE2:YFP construct, the RTE2 coding sequence (without stop codon) was PCR amplified and cloned in pBJ36+2x35Spro-YFP. The resulting plasmid was digested with NotI and the 2x35S-RTE2-YFP cassette was subcloned into the NotI sites of pMLBART. Expression plasmids for 35S-RTE-YFP and 35S-YFP were described previously (Chatterjee et al. 2014). Plasmids were transformed into Agrobacterium and used for transient expression in tobacco as described previously (Chatterjee et al. 2014). An mCHERRY-labeled nuclear marker containing the maize BAF1 transcription factor (Gallavotti et al. 2011) was cloned into pEarlyGate104-mCHERRY (Gutierrez et al. 2009) and co-injected into tobacco. Leaf disks were imaged on a Leica SP5 confocal microscope using 514-nm excitation and 520- to 575-nm emission for YFP, and 594-nm excitation and 610- to 640-nm emission for mCHERRY. Image processing was performed with ImageJ.
Genomic sequences of RTE4 and RTE5
RTE4 and RTE5 genes shared 98% identity with ambiguous ORFs, possibly due to genomic sequence assembly errors. To determine the correct genomic sequences of RTE4 and RTE5, we amplified RTE4 from DNA isolated from the OMA 9.41 line that contained chromosome 9 of the maize B73 inbred line (Rines et al. 2009). The RTE5 locus was instead isolated using B73 genomic DNA. The primers used to amplify both genomic sequences are listed in Table S3 in File S1.
cDNA cloning
The ORFs of RTE and RTE2 were PCR amplified from ear cDNA. RTE3 (KY129660) was isolated from leaf cDNA pools, while RTE6 (KY129661) was isolated from pollen cDNA pools. Protein sequence analysis was performed using the following databases: http://aramemnon.botanik.uni-koeln.de, www.ebi.ac.uk/interpro, and http://ExPASy.org.
RTE2 complementation test
To complement the Arabidopsis bor1-3 mutant (Kasai et al. 2011), the B73 RTE2 coding sequence was amplified with Phusion DNA polymerase (NEB) using primers RTE2 EcoRI-F1 and RTE2 HindIII-R1 (Table S2 in File S1). The amplicon was subsequently cloned into pBJ36+2x35S vector. The 35Spro:RTE2 cassette was isolated using NotI digestion and cloned into the transfer DNA binary vector pMLBART. Homozygous bor1-3 mutants were propagated by supplementation with 100 μM boric acid and transformed by the floral dip method. Primary transformants containing the 35Spro:RTE2 construct were selected on soil with Basta and assessed for the phenotypic rescue of bor1-3 defects. Complementation of the bor1-3 mutant with RTE was previously described (Chatterjee et al. 2014).
Histology and microscopy
Sections of maize shoot apical meristems and young tassels were stained with Toluidine blue (2 min, 0.1% solution in 0.6% boric acid) and Safranin O-Alcian Blue (20 min, 0.02% in 0.1 M Na acetate, pH 5.0). Following a rinse in deionized water, slides were mounted using Permount (Fisher Scientific, Pittsburgh, PA) and visualized using a Leica DM5500B microscope.
Data availability
All materials generated in this study are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Identification of RTE-like genes in maize
To identify additional members of the boron efflux transporter family that may play a role in transport and distribution of the microelement, we discovered five predicted boron transporter genes in the maize genome. Based on sequence similarity to the RTE gene, these genes were named RTE2 (GRMZM2G082203), RTE3 (GRMZM2G051753), RTE4 (GRMZM2G374989), RTE5 (GRMZM2G302559), and RTE6 (GRMZM2G454327).
To better understand the evolutionary relationship between the maize boron transporter genes and previously characterized boron transporters, a phylogenetic analysis of the predicted protein sequences of 33 boron transporters from different species was performed. Similarly to what we previously reported (Chatterjee et al. 2014), the phylogenetic tree clearly separated the boron transporters into two classes (Figure S1A in File S1) that coincide with differences in gene function. Class I contained AtBOR1, AtBOR2, and OsBOR1-like boron transporters, which are essential for efficient xylem loading under boron-deficient conditions (Miwa et al. 2006, 2013; Nakagawa et al. 2007). Class II, on the other hand, contained members like AtBOR4, TaBOR2, and HvBOR2 which are responsible for tolerance to boron toxicity (Miwa et al. 2007; Reid 2007; Sutton et al. 2007). RTE and RTE2 belonged to class I, while RTE3, RTE4, RTE5, and RTE6 were found in the class II clade. RTE4, RTE5, and RTE6 were in the same clade and showed a high percentage of identity with OsBOR4, a pollen-specific boron transporter required for pollen germination and tube elongation (Tanaka et al. 2013).
Among all family members, RTE is most similar to its paralog RTE2. Both genes share an identical gene structure and exhibit conserved intron–exon boundaries (Figure 1A) with an overall 94% nucleotide sequence identity in the coding sequence, and 95% identity at the amino acid level (Figure S2 in File S1). RTE3 showed lower similarity with both RTE/RTE2 and RTE4/RTE5/RTE6 groups. RTE4, RTE5, and RTE6 showed significant similarity among themselves at the nucleotide level in the coding region (>90%). To confirm the predicted coding sequences of these genes, we isolated full-length cDNAs of RTE2, RTE3, and RTE6. The ORFs of RTE2, RTE3, and RTE6 are 2109, 2028, and 2022 bp in length and encode proteins of 702, 675, and 673 aa, respectively (Figure S2 in File S1). RTE4, RTE5, and RTE6 reside on extensively duplicated regions on chromosomes 9, 3, and 8, respectively (Figure S3B in File S1). Gene annotation of RTE4 and RTE5 in the maize B73v3 genome was ambiguous. To rule out any improper assembly of the highly similar genomic regions of both genes, we first sequenced the entire RTE4 locus from an oat-maize addition line carrying maize B73 chromosome 9 (Rines et al. 2009), and the RTE5 locus directly from B73 genomic DNA. This confirmed that both loci are present in the maize genome on chromosomes 9 and 3, respectively. However, after several attempts we were not able to isolate full-length ORFs of either gene. Based on sequence comparison with the experimentally determined RTE6 coding sequence, we noticed that RTE4 had a single base pair deletion in the predicted fourth exon (position +501). Similarly, when compared with RTE6, we determined that a stop codon was present in the predicted fifth exon of RTE5 (position +586). This suggests that both RTE4 and RTE5 are likely pseudogenes, and we removed them from our subsequent analysis. All RTE protein family members are predicted antiporters and share a similar structure containing 12 transmembrane domains (Takano et al. 2002; Chatterjee et al. 2014) (Figure S2 in File S1).
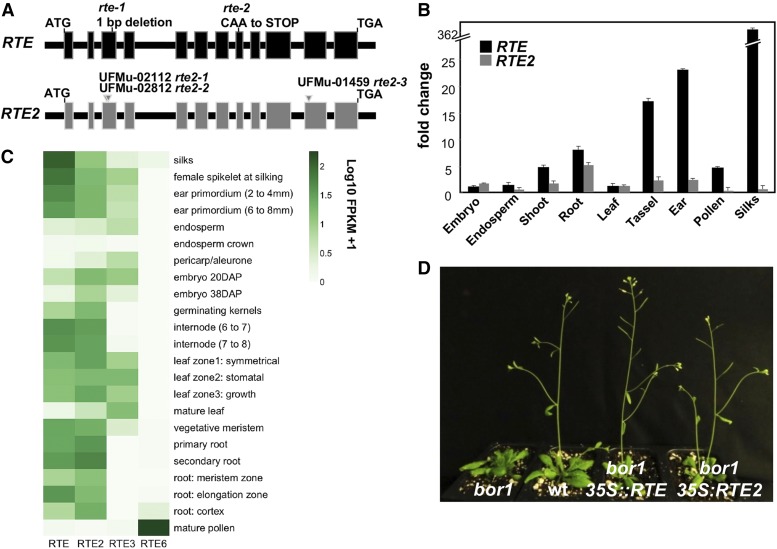
Figure 1.
RTE and RTE2 are duplicated genes. (A) RTE and RTE2 mutant alleles used in this study. (B) qRT-PCR analysis of RTE and RTE2 expression in different tissues relative to leaf. (C) Tissue-specific expression of RTE, RTE2, RTE3, and RTE6 based on RNA-seq data sets from Walley et al. (2016). (D) Both RTE and RTE2 rescue the Arabidopsis bor1 mutant when overexpressed. FPKM, fragments per kilobase of transcript per million mapped reads.
qRT-PCR was performed to assess the tissue-specific expression pattern of all RTE family members. The transcript abundance of the RTE family members differed in terms of tissue specificity (Figure 1, B and C, and Figure S1C in File S1). RTE mRNA was present in all the tissues examined including leaf, root, ear, tassel, and pollen, but was most abundant in ears and silks. Interestingly, RTE2 showed the highest expression in roots among all tissue tested. On the other hand, RTE3 was expressed at highest levels in leaves while RTE6 transcripts were predominantly present in pollen, in accordance with the phylogenetic grouping. We verified the relative expression of the RTE family genes in RNA-seq data sets from published sources (Figure 1C) and those agreed well with our qRT-PCR data (Walley et al. 2016).
RTE2 is a functional ortholog of the Arabidopsis boron transporter BOR1
RTE2 encodes a boron transporter highly similar to RTE. Analysis of the genomic regions of RTE and RTE2 showed strong colinearity and evidence that the two genes resided in duplicated regions of chromosome 1 and 3, respectively (Figure 1 and Figure S3A in File S1). We therefore wondered if RTE2 played a similar role to RTE in maize development. To investigate the function of RTE2 in maize, we identified three independent transposon insertions, two in the 3rd exon (UFMu-02112 and UFMu-02812) and one in the 11th exon of RTE2 (UFMu-01459; Figure 1A). We renamed these insertions as rte2-1, rte2-2, and rte2-3, respectively. Each insertion was predicted to completely disrupt the function of RTE2, yet none of the three insertion lines showed any vegetative or reproductive developmental defects in the shoot, even when grown in boron-deficient conditions. We checked if RTE2 was still expressed in homozygous insertion lines. While we could recover full-length RTE2 transcripts in all siblings without insertions, we failed to do so for all rte2 alleles after several attempts (data not shown). RTE2 expression is still detectable in these insertion lines but we only recovered aberrantly spliced transcripts and transcripts containing the transposon (Figure S4 in File S1). These results suggest that all rte2 alleles are likely functional null.
We previously demonstrated that RTE is a functional ortholog of AtBOR1 (Chatterjee et al. 2014). To assess whether RTE2 also shares a similar role during development and if any of the amino acids that differed with RTE could impair its function, we transformed the maize RTE2 gene under the control of the 35S promoter into the Arabidopsis bor1-3 mutant (Kasai et al. 2011). All vegetative and reproductive defects of bor1-3 plants such as loss of apical dominance and reduced fertility were completely rescued in ∼40% of the 38 independent lines overexpressing RTE2 (Figure 1D and Table S4 in File S1). We checked the expression levels of RTE2 in a few representative Arabidopsis lines, and the level of phenotypic rescue correlated with higher expression levels of the transgene (Figure S4B in File S1). We also measured boron concentration in a subset of fully and partially rescued lines and observed >100% higher levels in leaves when compared to the Arabidopsis bor1 mutant in both cases (Table S1 in File S1). Altogether these results indicated that RTE2 encodes a functional boron transporter and can complement the developmental and fertility defects of the Arabidopsis bor1-3 mutant.
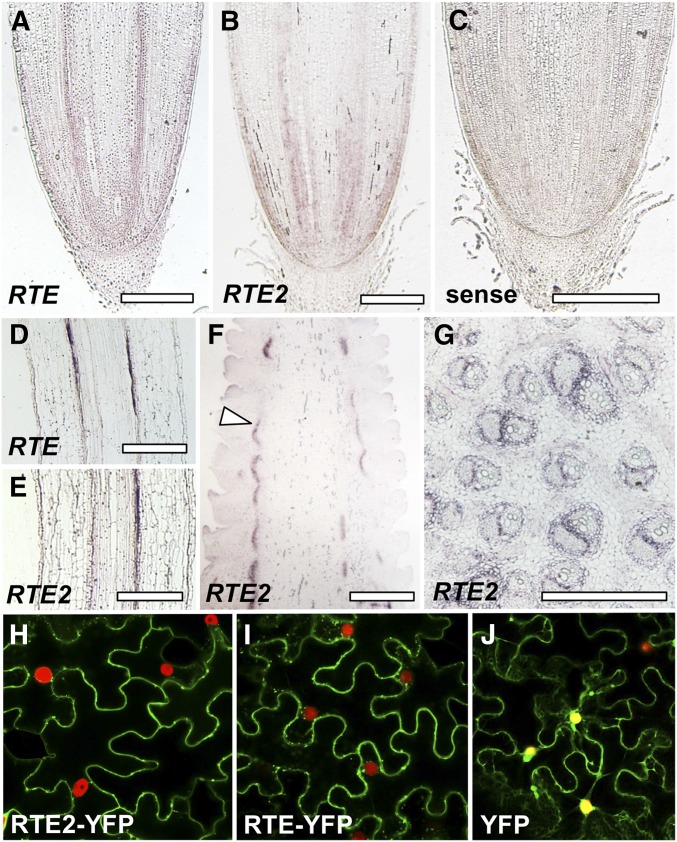
To further understand if expression differences may explain the lack of a mutant phenotype in rte2 single mutants we performed in situ hybridizations in root and inflorescence tissues using antisense probes specific to the 5′ and 3′ UTR regions, as well as full-length RTE2 probe. Longitudinal sections of roots from 5-day-old seedlings showed identical expression of RTE and RTE2 in the vasculature and root tips (Figure 2, A–E). Similarly, in longitudinal sections of developing inflorescences, RTE2 transcripts localized to vasculature-surrounding regions (Figure 2, F and G) in an identical fashion to what we previously observed with RTE (Chatterjee et al. 2014).
Figure 2.
Expression and subcellular localization analysis of RTE and RTE2. In situ hybridizations of longitudinal sections of seedling roots showing RTE and RTE2 expression in (A and B) root tips and (D and E) vasculature. (C) RTE2 sense control. (F) Longitudinal section of an immature ear showing RTE2 expression in vasculature (arrowhead). (G) Stem cross section. Bar, 500 µm. (H–J) Confocal images of tobacco epidermal cells expressing RTE2-YFP, RTE-YFP, YFP control, and the nuclear marker BAF1-mCHERRY (Gallavotti et al. 2011).
We also checked RTE2 subcellular localization. RTE2-YFP was predominantly localized to the plasma membrane by confocal imaging of tobacco leaves in transient expression assays (Figure 2, H–J). The observed localization was identical to what we previously reported for RTE (Chatterjee et al. 2014). Altogether, these results indicate that RTE2 and RTE share similar functions in maize development.
RTE and RTE2 are required for maize growth in boron-deficient conditions
In rte plants, tassels fail to produce branches and spikelets while ears remain small and show widespread cell death (Chatterjee et al. 2014). We therefore generated rte;rte2 double mutant plants which showed a strong enhancement of the rte single mutant phenotype in soils with poor boron content (0.35 ppm, Rutgers field; Table S2 in File S1). rte;rte2 double mutant plants showed stunted growth (Figure 3, A and B), with leaves displaying narrow white stripes along their length which subsequently widened and became papery and translucent. rte;rte2 plants also showed rudimentary undeveloped ear-like structures (Figure 3C) and did not develop beyond 10–15 cm, eventually dying off after producing ∼7–8 leaves (Figure 3, F and G). These plants showed a 50% decrease in leaf boron content when compared to wild type (Table S1 in File S1). However, when grown in soil with adequate boron content; such as greenhouse soil or in the fields of Molokai, Hawaii (0.20 mg/liter and 2.35 ppm, respectively; Table S2 in File S1); rte;rte2 double mutants did not show any vegetative defects, and resembled single rte mutants grown under the same conditions (Figure S5 in File S1).
However, Rutgers and Molokai soils differ for various parameters, not only for boron concentration (Table S2 in File S1). To unequivocally show that the rte;rte2 double mutant defects were caused by inadequate boron levels in the soil, we grew double mutant plants in pots containing soil from Rutgers, and watered them with 200 μM boric acid, a concentration that we previously showed being sufficient to fully rescue the vegetative and reproductive defects of rte mutants without causing toxicity symptoms in normal plants (Chatterjee et al. 2014). Double mutant plants watered with boric acid did not show any of the vegetative phenotypes observed in control plants. While control rte;rte2 plants already showed severe defects such as broad necrotic lesions and rolled up leaves 40 days after planting, those defects were not visible in rte;rte2 plants supplemented with boric acid and those plants eventually produced fully formed and fertile tassels (Figure 4; n = 6). We then measured boron levels in treated and control plants. In rte;rte2-treated plants, the amount of boron was >200% higher than in untreated plants (Figure 4D). We also quantified the level of boron in both control and treated soils, and we determined that in pots watered with boric acid the concentration of boron was more than four times higher compared to control pots after treatment (Table S2 in File S1). These results unambiguously show that the defects observed in rte;rte2 mutants are due to lack of adequate boron supply to growing tissues.
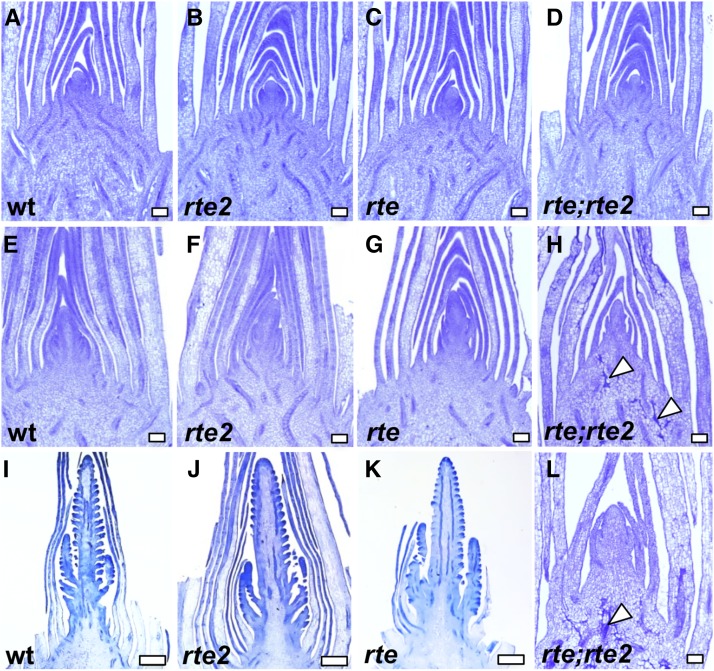
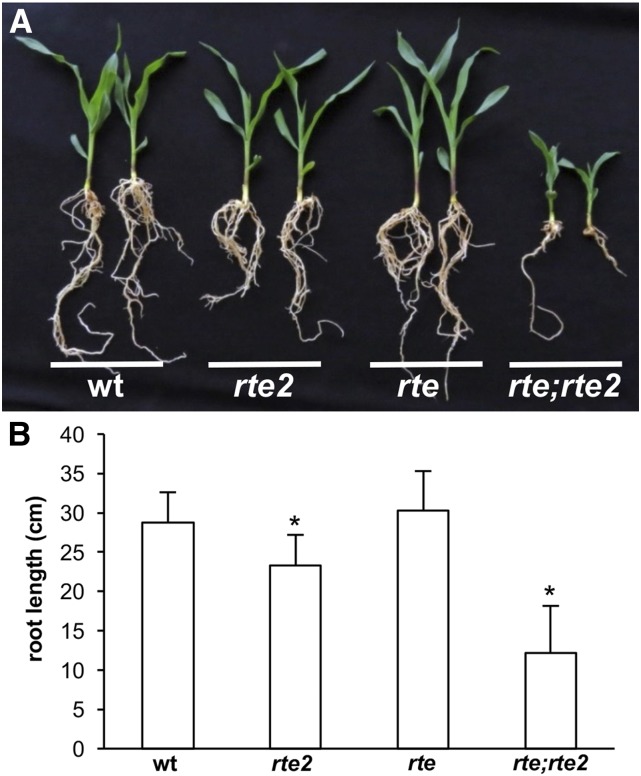
To understand how shoots of single rte or rte;rte2 double mutants were affected in boron-deficient conditions, we analyzed different developmental stages. Shoot tips of wild type, rte, rte2, and rte;rte2 were collected from 3- to 5-week-old plants grown in Rutgers fields. At 3 weeks, wild-type, rte, rte2, and rte;rte2 plants showed a normal developing shoot apical meristem and leaf primordia (Figure 5, A–D). As development progressed, wild-type, rte, and rte2 plants transitioned normally to reproductive development. However, rte;rte2 doubled mutants began showing several lesions in the stem ground tissue (Figure 5, E–H and L). Staining with Safranin O-Alcian Blue suggested that those lesions corresponded to patches of lignified tissues (Figure 4E and Figure 5, H and L). At 5 weeks, wild type, rte, and rte2 mutants showed initiation of several axillary meristems in immature tassels. However, the shoot apical meristem of rte;rte2 double mutants appeared arrested (Figure 5, I–L, and Figure S5 in File S1). Eventually, wild-type and rte2 plants produced fully fertile inflorescences, while rte mutants showed characteristic small, sterile ears with brown tips (Chatterjee et al. 2014). Although single rte2 mutants did not show any vegetative or reproductive phenotype in the shoots, a subtle phenotype in primary root development was evident in young seedlings (Figure 6). The primary roots of rte2 seedlings were significantly shorter than wild-type or rte single mutant seedlings. This phenotype was more extreme in roots of rte;rte2 double mutants which appeared significantly shorter and generated fewer lateral and seminal roots (Figure 6).
Figure 5.
Longitudinal sections of shoot apical meristems and inflorescence meristems at different stages of development. (A–D) 3-week-old shoot tip showing shoot apical meristem and developing leaf primordia, (E–H) 4-week-old shoot tips at transition stage, and (I–L) 5-week-old shoot tips showing developing tassels in wild type, rte, and rte2. rte;rte2 double mutant failed to grow. Arrowheads point to ground tissue lesions. (A–H and L) Bar, 100 µm. (I–K) Bar, 1000 µm. wt, wild type.
Figure 6.
(A) Root phenotype of 3-week-old wild type, rte, rte2, and rte;rte2 grown in boron-deficient soil. (B) Average length of primary roots (t-test between wild type, rte2, and rte;rte2, * P < 0.001). Error bars represent SD. wt, wild type.
Overall, these results demonstrate that the disruption of RTE2 strongly enhanced the phenotype of single rte mutants, suggesting that RTE and RTE2 act synergistically to sustain maize growth in boron-deficient conditions.
Discussion
We previously demonstrated that RTE encodes a functional coortholog of the Arabidopsis efflux transporter BOR1 (Chatterjee et al. 2014). In this study, we identified three additional boron transporter genes in the maize genome (RTE2, RTE3, and RTE6) that likely contribute to boron transport and distribution during maize development in different tissues.
Boron transporter genes vary in number, function, as well as expression in different plant species. Among the different family members, some genes have been reported to provide tolerance to boron deficiency, whereas others are known to prevent toxicity, and those genes belong to class I and class II, respectively (Miwa et al. 2006, 2007; Sutton et al. 2007; Pallotta et al. 2014). In several instances, copy number variation in boron transporters has been associated with tolerance to high levels of boron in soils. Some barley varieties have tandem copies for BOR1-like (Bot1) genes (Sutton et al. 2007), whereas different landraces in wheat show variation in Bot-B5/D5 alleles due to insertions of repetitive sequences in promoter regions or deletions in exons (Pallotta et al. 2014). These genes all belong to class II and are phylogenetically related to RTE3, suggesting that increasing the number of RTE3 copies may produce maize with tolerance to high boron soils.
Homologous boron transporters in various species have also been reported to show cell type-specific expression patterns. For example, OsBOR4 expression is mainly restricted to pollen, as is the expression of Arabidopsis BOR6 and BOR7 (Becker et al. 2003; Bock et al. 2006). In addition to differences in transcriptional regulation, boron transporters are also differentially regulated at the post-transcriptional level (Takano et al. 2005, 2010; Nakagawa et al. 2007; Leaungthitikanchana et al. 2013). BOR1 abundance at the plasma membrane is regulated via the endosomal/vacuolar recycling pathway whereby its activity at the plasma membrane is decreased or increased in high or low boron conditions, respectively (Takano et al. 2010; Kasai et al. 2011; Yoshinari et al. 2012). BOR2 was also reported to be regulated in the same fashion (Miwa et al. 2013). In rice, expression of OsBOR1 changes according to fluctuations of boron supply in the medium (Nakagawa et al. 2007). Maize RTE-like genes showed notable tissue-specific expression differences (Figure 1). RTE, RTE2, and RTE3 were expressed in all tissues examined, however each gene was preferentially expressed in certain tissue types. Expression of RTE was most abundant in ears and silks (Chatterjee et al. 2014), RTE2 in roots, and RTE3 in leaves. RTE6 was expressed at very low levels in most tissues but was extremely abundant in pollen, consistent with it belonging to the same clade as OsBOR4. Overall, the phylogenetic grouping of each member is consistent with expression specificity in various species, suggesting that family members belonging to the same clades carry out specific functions in different tissues.
RTE2 encodes a protein similar to its paralog RTE and can fully complement the Arabidopsis bor1 mutant (Figure 1). In situ hybridizations of both RTE and RTE2 in maize roots and inflorescences showed an essentially identical expression pattern (Figure 2). Surprisingly, transposon insertions in RTE2 did not show any vegetative or inflorescence defects. However, in young seedlings, single rte2 mutants showed a slight reduction in root length when grown in boron-deficient soil (Figure 6). This suggested a subtle difference in the function of RTE and RTE2 and, together with slightly higher expression levels of RTE2 in roots compared to other tissue, indicates that RTE2 main function may reside in roots. A comparable situation has been described for Arabidopsis bor2 mutants, whose root growth is affected under low boron conditions. BOR2 is a close paralog of BOR1 and both share 90% amino acid sequence identity (Miwa et al. 2013). By using fluorescent marker lines, BOR2 and BOR1 expression was reported to differ in roots, with stronger expression of BOR2 observed in lateral root caps and epidermis; while BOR1 was predominantly expressed in the root meristem, transition, and elongation zones, but not in lateral root caps. Double bor1;bor2 mutants show more severe growth defects than either single mutant, suggesting that BOR1 and BOR2 have partially overlapping roles in shoot and root growth when grown in boron-limiting conditions. This is similar to the synergistic interaction we observed in rte;rte2 mutants. rte;rte2 double mutants remained undeveloped, produced only a few leaves, and died after 4–6 weeks (Figure 3). This phenotype, visible in boron-poor soils (Rutgers) but not in nutrient-rich soils (Molokai), could be fully rescued by applications of boric acid.
The duplication events originating paralogous genes in each species are independent of each other (Figure S1 in File S1). RTE and RTE2 belong to maize 1 and 2 subgenomes, respectively, from the most recent maize whole-genome duplication that happened 5–12 MYA (Schnable et al. 2011; Hughes et al. 2014); while BOR1 and BOR2 are located on chromosomes 2 and 3, respectively, in duplicated regions possibly arisen from a more ancient whole-genome duplication event (Arabidopsis Genome Initiative 2000; Miwa et al. 2013). This different evolutionary history is reflected by a significantly higher identity between the paralogous proteins in maize than in Arabidopsis (95% vs. 90%). It is intriguing that a similar fate of paralogous genes is observed in two distantly related species for corresponding orthologs. While the expression differences of BOR1 and BOR2 suggest a case of subfunctionalization following a whole-genome duplication event (Miwa et al. 2013), we could not detect significant differences between RTE and RTE2 expression by in situ hybridizations. However, rte2 mutants have a subtle-root phenotype and it is therefore possible that RTE2 is a subfunctionalized gene similarly to BOR2 (Hughes et al. 2014). RTE and RTE2 protein localization may indeed reveal subtle differences that could explain the root rte2 phenotype. Another possible scenario is that RTE2 will eventually become an entirely nonfunctional gene, given that RTE in rte2 mutants is sufficient for normal development.
Altogether, our current and previous results suggest that, in normal conditions, RTE is the main boron transporter in maize and the loss of its function severely impairs maize fertility in conditions of both adequate and low boron availability (Chatterjee et al. 2014). RTE2 function, on the other hand, can be lost without significant repercussion on development and reproduction even in low boron conditions, presumably due to RTE function. When RTE function is lost, RTE2, which is highly expressed in roots, can supply enough boron to shoot tissues to allow plants to grow and form inflorescences, albeit severely compromised. However, when both gene functions are lost, diffusion and channel proteins cannot supply enough boron to sustain rapidly growing tissues in conditions of low boron availability. Indeed, in the current model of boron transport, BOR1-like proteins are required for the export of negatively charged borate from root endodermal cells to supply boron to xylem elements (Takano et al. 2008; Miwa and Fujiwara 2010). The severity of the rte;rte2 double mutant phenotype is very similar to that reported for single tls1 mutants grown in poor soils, which supports a two-step process for boron transport in maize whereby both RTE and RTE2 function downstream of TLS1 in boron uptake and loading in the root xylem (Miwa and Fujiwara 2010; Durbak et al. 2014). It is important to point out though that rte;rte2 double mutants are fully complemented by boric acid watering. Three possibilities could explain this result: rte2 mutants may not be completely null, additional not-yet-identified boron transporters may be expressed in roots of the genetic background used in our experiments, or diffusion and facilitated transport mechanisms may be able to overcome active boron transport deficiencies. In summary, our results show that under boron-deficient conditions RTE and RTE2 work synergistically to provide boron to support maize growth during vegetative and reproductive development.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.198275/-/DC1.
Acknowledgments
We are grateful to Paula McSteen, Michaela Matthes, Kimble Ashten, and the Missouri University Plant and Soil Analysis Facility for support with boron measurements; Nelson Garcia for providing oat-maize addition lines; Marc Probasco for greenhouse and field care; Zara Tabi for Figure 2, F and G; and the Maize Genetics Cooperation Stock Center for Uniform-Mu seeds. Q.L. was supported by the China Scholarship Council. This research was supported by the National Science Foundation (IOS-1114484 and IOS-1546873) to A.G.
Footnotes
Communicating editor: K. Bomblies
Literature Cited
- Agarwala S. C., Sharma P. N., Chatterjee C., Sharma C. P., 1981. Development and enzymatic changes during pollen development in boron deficient maize plants. J. Plant Nutr. 3: 329–336. [Google Scholar]
- Arabidopsis Genome Initiative , 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Becker J. D., Boavida L. C., Carneiro J., Haury M., Feijo J. A., 2003. Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol. 133: 713–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock K. W., Honys D., Ward J. M., Padmanaban S., Nawrocki E. P., et al. , 2006. Integrating membrane transport with male gametophyte development and function through transcriptomics. Plant Physiol. 140: 1151–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P. H., Bellaloui N., Wimmer M. A., Bassil E. S., Ruiz J., et al. , 2002. Boron in plant biology. Plant Biol. 4: 205–223. [Google Scholar]
- Canon P., Aquea F., de la Guardia A. R. H., Arce-Johnson P., 2013. Functional characterization of Citrus macrophylla BOR1 as a boron transporter. Physiol. Plant 149: 329–339. [DOI] [PubMed] [Google Scholar]
- Chatterjee M., Tabi Z., Galli M., Malcomber S., Buck A., et al. , 2014. The boron efflux transporter ROTTEN EAR is required for maize inflorescence development and fertility. Plant Cell 26: 2962–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dordas C., Brown P. H., 2001. Permeability and the mechanism of transport of boric acid across the plasma membrane of Xenopus laevis oocytes. Biol. Trace Elem. Res. 81: 127–139. [DOI] [PubMed] [Google Scholar]
- Durbak A. R., Phillips K. A., Pike S., O’Neill M. A., Mares J., et al. , 2014. Transport of boron by the tassel-less1 aquaporin is critical for vegetative and reproductive development in maize. Plant Cell 26: 2978–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A., Malcomber S., Gaines C., Stanfield S., Whipple C., et al. , 2011. BARREN STALK FASTIGIATE1 is an AT-hook protein required for the formation of maize ears. Plant Cell 23: 1756–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaujoux R., Seoighe C., 2010. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 11: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta U. C., Jame Y. W., Campbell C. A., Leyshon A. J., Nicholaichuk W., 1985. Boron toxicity and deficiency: a review. Can. J. Soil Sci. 65: 381–409. [Google Scholar]
- Gutierrez R., Lindeboom J. J., Paredez A. R., Emons A. M., Ehrhardt D. W., 2009. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11: 797–806. [DOI] [PubMed] [Google Scholar]
- Hayes J. E., Reid R. J., 2004. Boron tolerance in barley is mediated by efflux of boron from the roots. Plant Physiol. 136: 3376–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J. E., Pallotta M., Garcia M., Oz M. T., Rongala J., et al. , 2015. Diversity in boron toxicity tolerance of Australian barley (Hordeum vulgare L.) genotypes. BMC Plant Biol. 15: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H. N., Brown P. H., Labavitch J. M., 1996. Species variability in boron requirement is correlated with cell wall pectin. J. Exp. Bot. 47: 227–232. [Google Scholar]
- Hughes T. E., Langdale J. A., Kelly S., 2014. The impact of widespread regulatory neofunctionalization on homeolog gene evolution following whole-genome duplication in maize. Genome Res. 24: 1348–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai K., Takano J., Miwa K., Toyoda A., Fujiwara T., 2011. High boron-induced Ubiquitination regulates vacuolar sorting of the BOR1 borate transporter in Arabidopsis thaliana. J. Biol. Chem. 286: 6175–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Matoh T., Azuma J., 1996. Two chains of rhamnogalacturonan II are cross-linked by borate-diol ester bonds in higher plant cell walls. Plant Physiol. 110: 1017–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaungthitikanchana S., Fujibe T., Tanaka M., Wang S. L., Sotta N., et al. , 2013. Differential expression of three BOR1 genes corresponding to different genomes in response to boron conditions in hexaploid wheat (Triticum aestivum L.). Plant Cell Physiol. 54: 1056–1063. [DOI] [PubMed] [Google Scholar]
- Leonard A., Holloway B., Guo M., Rupe M., Yu G. X., et al. , 2014. Tassel-less1 encodes a boron channel protein required for inflorescence development in maize. Plant Cell Physiol. 55: 1044–1054. [DOI] [PubMed] [Google Scholar]
- Liu K., Liu L. L., Ren Y. L., Wang Z. Q., Zhou K. N., et al. , 2015. Dwarf and tiller-enhancing 1 regulates growth and development by influencing boron uptake in boron limited conditions in rice. Plant Sci. 236: 18–28. [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lordkaew S., Dell B., Jamjod S., Rerkasem B., 2011. Boron deficiency in maize. Plant Soil 342: 207–220. [Google Scholar]
- Lyons E., Pedersen B., Kane J., Alam M., Ming R., et al. , 2008. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 148: 1772–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelbart M. V., Hasegawa P. M., Bailey-Serres J., 2015. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 16: 237–251. [DOI] [PubMed] [Google Scholar]
- Miwa K., Fujiwara T., 2010. Boron transport in plants: co-ordinated regulation of transporters. Ann. Bot. 105: 1103–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa K., Takano J., Fujiwara T., 2006. Improvement of seed yields under boron-limiting conditions through overexpression of BOR1, a boron transporter for xylem loading, in Arabidopsis thaliana. Plant J. 46: 1084–1091. [DOI] [PubMed] [Google Scholar]
- Miwa K., Takano J., Omori H., Seki M., Shinozaki K., et al. , 2007. Plants tolerant of high boron levels. Science 318: 1417. [DOI] [PubMed] [Google Scholar]
- Miwa K., Wakuta S., Takada S., Ide K., Takano J., et al. , 2013. Roles of BOR2, a boron exporter, in cross linking of rhamnogalacturonan II and root elongation under boron limitation in Arabidopsis. Plant Physiol. 163: 1699–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y., Hanaoka H., Kobayashi M., Miyoshi K., Miwa K., et al. , 2007. Cell-type specificity of the expression of Os BOR1, a rice efflux boron transporter gene, is regulated in response to boron availability for efficient boron uptake and xylem loading. Plant Cell 19: 2624–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi K., Yasumori M., Imai T., Naito S., Matsunaga T., et al. , 1997. bor1–1, an Arabidopsis thaliana mutant that requires a high level of boron. Plant Physiol. 115: 901–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill M. A., Warrenfeltz D., Kates K., Pellerin P., Doco T., et al. , 1996. Rhamnogalacturonan-II, a pectic polysaccharide in the walls of growing plant cell, forms a dimer that is covalently cross-linked by a borate ester. In vitro conditions for the formation and hydrolysis of the dimer. J. Biol. Chem. 271: 22923–22930. [DOI] [PubMed] [Google Scholar]
- O’Neill M. A., Eberhard S., Albersheim P., Darvill A. G., 2001. Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294: 846–849. [DOI] [PubMed] [Google Scholar]
- Pallotta M., Schnurbusch T., Hayes J., Hay A., Baumann U., et al. , 2014. Molecular basis of adaptation to high soil boron in wheat landraces and elite cultivars. Nature 514: 88–91. [DOI] [PubMed] [Google Scholar]
- Perez-Castro R., Kasai K., Gainza-Cortes F., Ruiz-Lara S., Casaretto J. A., et al. , 2012. VvBOR1, the grapevine ortholog of AtBOR1, encodes an efflux boron transporter that is differentially expressed throughout reproductive development of Vitis vinifera L. Plant Cell Physiol. 53: 485–494. [DOI] [PubMed] [Google Scholar]
- Raven J. A., 1980. Short-distance and long-distance transport of boric-acid in plants. New Phytol. 84: 231–249. [Google Scholar]
- Reid R., 2007. Identification of boron transporter genes likely to be responsible for tolerance to boron toxicity in wheat and barley. Plant Cell Physiol. 48: 1673–1678. [DOI] [PubMed] [Google Scholar]
- Rines H. W., Phillips R. L., Kynast R. G., Okagaki R. J., Galatowitsch M. W., et al. , 2009. Addition of individual chromosomes of maize inbreds B73 and Mo17 to oat cultivars Starter and Sun II: maize chromosome retention, transmission, and plant phenotype. Theor. Appl. Genet. 119: 1255–1264. [DOI] [PubMed] [Google Scholar]
- Schnable J. C., Springer N. M., Freeling M., 2011. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 108: 4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnurbusch T., Hayes J., Sutton T., 2010. Boron toxicity tolerance in wheat and barley: Australian perspectives. Breed. Sci. 60: 297–304. [Google Scholar]
- Settles A. M., Holding D. R., Tan B. C., Latshaw S. P., Liu J., et al. , 2007. Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics 8: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelp B. J., Marentes E., Kitheka A. M., Vivekanandan P., 1995. Boron mobility in plants. Physiol. Plant. 94: 356–361. [Google Scholar]
- Shorrocks V. M., 1997. The occurrence and correction of boron deficiency. Plant Soil 193: 121–148. [Google Scholar]
- Stangoulis J. C. R., Brown P. H., Bellaloui N., Reid R. J., Graham R. D., 2001. The efficiency of boron utilisation in canola. Aust. J. Plant Physiol. 28: 1109–1114. [Google Scholar]
- Sun J. H., Shi L., Zhang C. Y., Xu F. S., 2012. Cloning and characterization of boron transporters in Brassica napus. Mol. Biol. Rep. 39: 1963–1973. [DOI] [PubMed] [Google Scholar]
- Sutton T., Baumann U., Hayes J., Collins N. C., Shi B. J., et al. , 2007. Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science 318: 1446–1449. [DOI] [PubMed] [Google Scholar]
- Takano J., Noguchi K., Yasumori M., Kobayashi M., Gajdos Z., et al. , 2002. Arabidopsis boron transporter for xylem loading. Nature 420: 337–340. [DOI] [PubMed] [Google Scholar]
- Takano J., Miwa K., Yuan L., von Wiren N., Fujiwara T., 2005. Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl. Acad. Sci. USA 102: 12276–12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J., Wada M., Ludewig U., Schaaf G., von Wiren N., et al. , 2006. The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 18: 1498–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J., Miwa K., Fujiwara T., 2008. Boron transport mechanisms: collaboration of channels and transporters. Trends Plant Sci. 13: 451–457. [DOI] [PubMed] [Google Scholar]
- Takano J., Tanaka M., Toyoda A., Miwa K., Kasai K., et al. , 2010. Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc. Natl. Acad. Sci. USA 107: 5220–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Wallace I. S., Takano J., Roberts D. M., Fujiwara T., 2008. NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell 20: 2860–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N., Uraguchi S., Saito A., Kajikawa M., Kasai K., et al. , 2013. Roles of pollen-specific boron efflux transporter, OsBOR4, in the rice fertilization process. Plant Cell Physiol. 54: 2011–2019. [DOI] [PubMed] [Google Scholar]
- Walley J. W., Sartor R. C., Shen Z., Schmitz R. J., Wu K. J., et al. , 2016. Integration of omic networks in a developmental atlas of maize. Science 353: 814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari A., Kasai K., Fujiwara T., Naito S., Takano J., 2012. Polar localization and endocytic degradation of a boron transporter, BOR1, is dependent on specific tyrosine residues. Plant Signal. Behav. 7: 46–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All materials generated in this study are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.