Abstract
Hedgehog (Hh) regulates the Cubitus interruptus (Ci) transcription factor in Drosophila melanogaster by activating full-length Ci-155 and blocking processing to the Ci-75 repressor. However, the interplay between the regulation of Ci-155 levels and activity, as well as processing-independent mechanisms that affect Ci-155 levels, have not been explored extensively. Here, we identified Mago Nashi (Mago) and Y14 core Exon Junction Complex (EJC) proteins, as well as the Srp54 splicing factor, as modifiers of Hh pathway activity under sensitized conditions. Mago inhibition reduced Hh pathway activity by altering the splicing pattern of ci to reduce Ci-155 levels. Srp54 inhibition also affected pathway activity by reducing ci RNA levels but additionally altered Ci-155 levels and activity independently of ci splicing. Further tests using ci transgenes and ci mutations confirmed evidence from studying the effects of Mago and Srp54 that relatively small changes in the level of Ci-155 primary translation product alter Hh pathway activity under a variety of sensitized conditions. We additionally used ci transgenes lacking intron sequences or the presumed translation initiation codon for an alternatively spliced ci RNA to provide further evidence that Mago acts principally by modulating the levels of the major ci RNA encoding Ci-155, and to show that ci introns are necessary to support the production of sufficient Ci-155 for robust Hh signaling and may also be important mediators of regulatory inputs.
Keywords: Hedgehog signaling, exon junction complex, cubitus interruptus, splicing, Drosophila
HEDGEHOG (Hh) signaling plays many roles in development and tissue maintenance from Drosophila to humans. Accordingly, genetic mutations that alter Hh signaling are associated with a wide range of birth defects and cancers, some of which are being treated with drugs that inhibit Hh signaling (Anderson et al. 2012; Petrova and Joyner 2014; Pak and Segal 2016). In Drosophila and in mammals, cells respond to Hh primarily by altering the activity of Gli-family transcription factors (Hui and Angers 2011; Xiong et al. 2015; Pak and Segal 2016). In the absence of Hh, the primary translation products of Drosophila Cubitus interruptus (Ci), as well as mammalian Gli-2 and Gli-3 orthologs, are proteolytically processed to C-terminally truncated forms that readily enter the nucleus and repress Hh target genes, while unprocessed full-length proteins remain largely cytoplasmic and inactive. When Hh binds to Patched (Ptc) and its coreceptors, the seven-transmembrane domain Smoothened (Smo) protein is activated with two key consequences; inhibition of Ci/Gli-2/3 processing and activation of full-length Ci/Gli transcriptional activators, including Gli-1, which is not subject to processing.
Ci/Gli processing involves phosphorylation by Protein Kinase A (PKA) and other protein kinases, scaffolded by a kinesin-like protein (Cos2/Kif7), to create a Cul1-dependent E3 ubiquitin ligase binding site; Hh is thought to inhibit Ci-155 processing by promoting dissociation of these phosphorylation complexes (Hui and Angers 2011; Xiong et al. 2015; Pak and Segal 2016). Inhibition of Ci-155 processing reduces or eliminates Ci-75 repressor but also increases Ci-155 levels. Ci-75 repressor maintains some key Hh target genes silent outside Hh signaling territory, but the relative importance of eliminating the repressor and increasing the levels of Ci-155 in cells responding to Hh has not been satisfactorily determined. High levels of Hh also promote Ci-155 degradation via a Cul3-dependent E3 ubiquitin ligase that has been considered as a possible negative feedback mechanism for limiting Ci-155 activity (Ohlmeyer and Kalderon 1998; Kent et al. 2006; Zhang et al. 2006). Altogether, the contribution of changes in Ci-155 levels to Hh signaling are complex and have been hard to assess.
The mechanism of full-length Ci/Gli activation is understood only in outline. It is thought principally to involve relief of inhibition by Suppressor of fused [Su(fu)], which binds directly to Ci/Gli proteins and, in Drosophila, it depends on Hh-activated Fused (Fu) protein kinase activity (Humke et al. 2010; Tukachinsky et al. 2010; Zhou and Kalderon 2011; Han et al. 2015; Oh et al. 2015; Zhang et al. 2016). It is also unclear to what degree regulation of Ci-155 activation, Ci-155, and Ci-75 levels must collaborate to produce graded Hh signaling. Dose-dependent signaling can be studied in wing discs by the quantitative induction of the universal Hh target gene, ptc, and the induction of target genes induced by low levels (decapentaplegic; dpp), intermediate levels (collier; col), or only by high levels (engrailed; en) of signaling (Vervoort 2000). There is some evidence of apparent redundancy, perhaps as a means to support robust signaling in a variety of settings. For example, Hh signaling is normal in the absence of Su(fu), suggesting that regulation of Ci-155 activation can be largely dispensable (Preat 1992; Ohlmeyer and Kalderon 1998). Conversely, synthetic activation of Fu kinase, which promotes Ci-155 activity, can suffice to induce strong Hh pathway activity without the strong inhibition of Ci-155 processing that normally accompanies Hh signaling (Zhou and Kalderon 2011).
To uncover new insights into the regulation of Ci-155 levels and activity we conducted a genetic screen in a sensitized background of diminished Hh signaling lacking any regulatory input from Fu kinase activity. Surprisingly, the screen revealed proteins involved in RNA processing, including core components of the Exon Junction Complex (EJC), which has recently been implicated in regulating RNA splicing (Roignant and Treisman 2010; Ashton-Beaucage and Therrien 2011; Hayashi et al. 2014; Malone et al. 2014; Le Hir et al. 2016), and the serine-arginine rich (SR) protein, Srp54. We provide evidence that the EJC and Srp54 target ci splicing to influence Hh signaling by altering the levels of Ci-155 primary translation product. We also explore the role of an alternative ci RNA and its presumed translation product.
Materials and Methods
Drosophila stocks
Drosophila stocks were maintained on standard cornmeal/molasses/agar medium at room temperature. For the modifier screen, yw hs-flp fumH63; FRT42D P[Fu+, w+] P[y+]/Cyo; Su(fu)LP C765-GAL4 ptc-lacZ/TM6B females were crossed to males with second or third chromosome deficiencies over balancer chromosomes from the Bloomington deletion library (Ryder et al. 2007) to produce male progeny lacking Fu kinase activity and heterozygous for both Su(fu) and the tested deficiency. The same females were crossed to UAS-RNAi (upstream activating sequence-RNA interference) stocks with or without gCi transgenes (16-kb genomic segments constructed as described in the section below) or ci94/Dp(1;4)1021, y+, svspa-pol to examine wing discs from male y (fu mutant) larval progeny. To analyze adult fu mutant male progeny with straight wings, the CyO balancer was replaced with Sp in the parental females.
yw hs-flp; Sp/Cyo; C765-GAL4 ptc-lacZ/TM6B females were crossed to UAS-RNAi transgenes (with or without gCi transgenes or UAS-Srp54 or UAS-CG3605 transgenes) to examine wing discs of progeny with normal Fu and Su(fu) activities. C765-GAL4 is expressed throughout developing wing discs. UAS-RNAi lines for CG3605 (GD-26250) and Srp54 (GD-51088) were recombined with UAS-Diap1 on the third chromosome (BL-6657); second chromosome UAS-mago RNAi and UAS-Y14 RNAi were provided by J. E. Treisman (Roignant and Treisman 2010) and other RNAi lines tested were from the Bloomington Drosophila Stock Center or from the Vienna Drosophila Resource Center (prefaced by GD or KK) (Dietzl et al. 2007), including those for eIF4AIII (KK-108580) and btz (GD-38722).
Clones in a Minute background were made by crossing yw hs-flp; FRT42D M(2)53[1] P[hs-GFP, y+]; ptc-lacZ/TM6B females to FRT42D Y14Δ18 dark/CyO males from J. E. Treisman (who also provided UAS-MAPK stocks) (Roignant and Treisman 2010).
To generate MARCM (mosaic analysis with a repressible clone marker) clones with activated Fu (and loss of smo activity) with or without UAS-RNAi expression (UAS-Diap1 was used as a control whenever testing UAS-Srp54 RNAi plus UAS-Diap1), yw hs-flp UAS-GFP; smo2 FRT42D P[smo+] tub-Gal80; C765 ptc-lacZ/TM6B females were crossed to yw; smo2 FRT42D (UAS-mago RNAi) UAS-GAP-Fu; (UAS-Diap1) (UAS- Srp54 RNAi) males. For smo cos2 clones and ptc clones, males had cos22 or ptcS2, respectively, in place of UAS-GAP-Fu.
For pka clones, yw hs-flp UAS-GFP; tub-Gal80 FRT40A; C765 ptc-lacZ/TM6B females were crossed to yw hs-flp; pka-C1H2 FRT40A (UAS-mago RNAi)/Cyo; (UAS-Diap1) (UAS-Srp54 RNAi) males. In all cases, clones were induced by a 1-hr heat-shock at 37° of first and second instar larvae.
To test replacement of ci with gCi or UAS-Ci transgenes in MARCM clones, yw hs-flp UAS-GFP; FRT42D P[ci+, w+] tub-GAL80/CyO; C765-GAL4 ptc-lacZ/TM6B; ci94/Dp(1;4)1021, y+, svspa-pol females were crossed to yw; FRT42D; gCi or UAS-Ci; ci94/Dp(1;4)1021, y+, svspa-pol males.
To test rescue of ci94 null animals, ptc-lacZ; ci94/Dp(1;4)1021, y+, svspa-pol females were crossed to gCi (or derivatives); ci94/Dp(1;4)1021, y+, svspa-pol males. Rescue in the presence of ciCe−2 was tested using males with this allele replacing ci94.
To test RNAi effects in the presence of only gCi or SV-1 in whole discs, yw hs-flp; Sp/Cyo ; (gCi) (SV-1) C765 ptc-lacZ/TM6B; ci94/Dp(1;4)1021, y+, svspa-pol females were crossed to yw hs-flp: (UAS-mago RNAi) (Sp)/Cyo; (gCi) (SV-1) (UAS-Srp54 RNAi) (UAS-Diap1); ci94/Dp(1;4)1021, y+, svspa-pol males. For the analogous test in pka mutant MARCM clones, yw hs-flp UAS-GFP; tub-Gal80 FRT40A; (gCi or SV-1) C765 ptc-lacZ/TM6B; ci94/Dp(1;4)1021, y+, svspa-pol females were crossed to yw hs-flp; pka-C1H2 FRT40A/CyO; (gCi or SV-1) UAS-Diap1 (UAS-Srp54 RNAi); ci94/Dp(1;4)1021, y+, svspa-pol males and yw hs-flp UAS-GFP; tub-Gal80 FRT40A; Su(fu)LP C765 ptc-lacZ/TM6B; ci94/Dp(1;4)1021, y+, svspa-pol females were crossed to yw hs-flp; pka-C1H2 FRT40A UAS-mago RNAi/CyO; (gCi or SV-1); ci94/Dp(1;4)1021, y+, svspa-pol males.
All wings or stained wing discs shown were from male animals when mutant for fu; in all other cases, wing discs were dissected from larvae without sorting males from females.
Mutagenesis and cloning
Genomic transgenes were created by cloning the entire 16-kb genomic ci region from a Bluescript-SK (BSK) vector [provided by K. Basler (Methot and Basler 1999)] into an att-Pacman Expression vector (Drosophila Genomics Resource Center). To facilitate mutagenesis, the 16-kb fragment was first separated into two parts. The region including the promoter, first exon, and part of the first intron (“Ci fragment 2”) was cloned as a BamHI-NheI fragment into BSK cut with BamHI and XbaI to create BSK-CiF2. The complementary NheI-KpnI fragment containing all other exons and the 3′-UTR (“Ci Fragment 1”) was cloned into BSK cut with SpeI and KpnI to create BSK-CiF1. BSK-CiF2 was cut with NotI and Bsp1201 to clone the whole CiF2 fragment into the P[acman]-CmR vector cut with NotI, so that RsrII and PmeI vector sites were downstream of ci first intron sequences in RP-CiF2. CiF1 was amplified from BSK-CiF1 by long-range PCR using PfuUltraII Fusion HS DNA polymerase (Agilent Technologies), adding RsrII and PmeI at either end and cloning the product into a Zero Blunt Topo cloning vector (Invitrogen, Carlsbad, CA). The RsrII-PmeI fragment was then cloned into RP-CiF2 cut with the same enzyme to create the final Pacman vector containing the entire 16 kb genomic ci DNA. The 28 kb gCi AttPacman transgene was then inserted at the att ZH-86Fb landing site at cytological location 86F8 (Rainbow Transgenic Services). To create gCi ATG-A and gCi ATG-B, ATG codons were first altered in BSK-CiF2 and BSK-CiF2, respectively, using the QuikChange II site-directed mutagenesis kit (Agilent Technologies) (primers are listed in Supplemental Material, Table S2 in File S1). To create SV-1 and Ci-1, Ci coding sequences were amplified from a full-length ci cDNA using primers beginning 2- nt upstream of the initiation codon and at the stop codon but preceded by an added XbaI site. This amplicon was partially digested with AatII and XbaI to generate a 4.2-kb coding region fragment that was cloned between AatII (which cuts 23 nt upstream of the initiation codon for Ci-A) and XbaI sites of BSK-CiF2. The resulting DNA was cut with Bsp1201 and NotI to release an 11.5-kb fragment that was cloned into the NotI site of attB-P[acman]-ApR (producing att-BP[acman]-PCi). A 1-kb segment of DNA downstream of the stop codon was amplified by PCR from BSK-CiF1, adding NotI and PacI to either end, and cloned between the NotI site and Pac1 sites of att-BP[acman]-PCi to produce the Ci-1 transgene. Alternatively, the 3′-UTR from SV40 was amplified from pUAST-attB with addition of NotI and PacI sites and cloned similarly into att-BP[acman]-PCi to produce the SV-1 transgene. The UAS-Srp54 transgene was constructed by amplifying the coding region from the FBcI0164286 cDNA clone (DGRC) with addition of NotI and XhoI sites, followed by cloning into pUAST-attB and introduction into the 86F att landing site. An analogous method was used to make UAS-CG3605, starting from the FBcl10177075 cDNA clone (DGRC), adding EcoRI and KpnI sites to clone it into pUAST-attB.
Immunohistochemistry
Wing discs were dissected from late third instar larvae in PBS and fixed in 4% paraformaldehyde (in PBS) for 30 min, rinsed 3× with PBS, blocked with 10% normal goat serum (Jackson ImmunoResearch Laboratories) in PBST (0.1% Triton) for 1 hr, and stained with the following primary antibodies: rabbit anti-caspase 3 (1:100; D175, Cell Signaling), rabbit anti-β-galactosidase (1:10000; MP Biomedicals), mouse 4D9 anti-engrailed (1:5; Developmental Studies Hybridoma Bank), rat 2A1 anti-Ci (1:3; Developmental Studies Hybridoma Bank), and mouse anti-collier (1:10,000 from Alain Vincent, Toulouse University, France) overnight at 4°. Inverted larvae were then washed three times in PBST for 10 min each and incubated with Alexa Fluor 488, 546, 594, or 647 secondary antibodies (1:1000; Molecular Probes, Eugene, OR) for 1 hr at room temperature. Larvae were washed twice in PBST for 20 min each, once in PBS for 10 min, and mounted, with or without additional Hoechst staining (1:1000; Molecular Probes), in Aqua/Poly mount (Polysciences, Warrington, PA).
Quantitation from fluorescent images
Fluorescence images were captured using 20×, or 63×, 1.4 NA oil immersion lenses on a confocal microscope (LSM 700; Zeiss [Carl Zeiss], Thornwood, NY). Whole wing disc images of 512 × 512 pixels at 8-bit depth were captured with a 20× lens at a resolution of 2.52 pixels/µm. Three stacks per wing disc where acquired with xzy scaling of 645, 645, and 5.3 µm. Centering of the middle stack was done based on the highest levels of ptc-lacZ and Ci-155 at the dorsal/ventral (D/V) boundary. The range indicator was used to set the appropriate laser intensity per experiment for each fluorophore such that the signal was in the linear range. Rotation and z-projection of the stacks were performed with ImageJ software (National Institutes of Health, Bethesda, MD). To measure intensity profiles along the anterior/posterior (AP) axis, an elongated rectangle was drawn on a central region of the wing pouch, avoiding the DV border. The y-axis shows the average fluorescence intensity over the height of the rectangle at each point on the x-axis (AP axis) for ptc-lacZ expression or Ci-155 protein, measured using Image J. At least three wings discs per condition were measured and averaged for each plot, using the posterior edge of ptc-lacZ expression as a reference point for the AP border. The ratio of the peak fluorescence intensity at the AP border for an experimental genotype relative to the appropriate control processed in parallel was calculated for each experiment. These values (expressed as a percentage of controls) from at least three independent experiments were used to calculate a mean and C.I. For clones, the average fluorescent intensity over the area of the GFP-marked clone was measured using Image J. The average fluorescence intensity in anterior cells outside the clones in the same wing disc was also measured. From these measurements, the ratio of fluorescence intensities (inside/outside) was calculated for each clone. These values from multiple control and experimental clones were used to derive mean and SEM values. Representative images of clones were acquired with a 63×, 1.4 NA oil immersion lens at a resolution of 2.52 pixels/μm.
Adult wings
Adult wings were pulled off anesthetized flies and placed in 70% ethanol for 5 min, transferred to 100% ethanol, and then mounted in Aqua/Poly Mount (Polysciences). They were imaged with Transmitted Light on a Nikon Diaphot 300 microscope at 10× (Nikon, Garden City, NY).
RNA analysis
For analysis, 30–40 wing discs per genotype were dissected from third instar larvae in cold nuclease-free PBS. RNA was isolated from the wing discs using an RNeasy mini kit (QIAGEN, Valencia, CA) with DNase (QIAGEN) and converted to cDNA using the Maxima H Minus First Strand cDNA Synthesis Kit (Thermo Scientific) using random primers or oligodT. Quantitative RT-PCR was performed in the StepOnePlus Real-Time PCR System with Power SYBR Green Mastermix (Applied Biosystems, Foster City, CA). Primers for amplifying each PCR product are listed in Table S3 in File S1. The relative abundance of RNAs for experimental discs vs. controls was calculated using the 2−ΔΔCt method, normalizing to the housekeeping genes Rpl15 and α4-tubulin. The level of ci-B RNA relative to ci-A RNA was calculated from the Ct values for each.
Statistics
To compare experimental values to controls for fluorescence intensity (of ptc-lacZ expression or Ci-155 protein) measured at the AP border of wing discs, the experimental/control ratio was first calculated from individual trials using at least three wing discs of each genotype, as described earlier. These ratios from multiple trials were then used to calculate a mean value and 95% C.I. for each experimental genotype, expressed as a percentage of control values. Mean values and C.I.s of experimental samples expressed as a percentage of controls were calculated analogously for RNA measurements after converting raw quantitative (q)RT-PCR results into an experimental/control ratio for each trial. For measurements of fluorescent intensity in clones, the average clone intensity relative to surrounding anterior cells was first calculated for experimental and control samples, as described earlier. The set of values obtained from multiple clones of each genotype were examined by a Shapiro–Wilk test for a fit to a normal distribution. Mean values for experimental and control clones were then calculated and a P-value for the significance of differences between them was calculated using an unpaired Student’s t-test for unequal variances. For each experiment, the number of samples and trials are given in the figure legend. The number of samples or trials was not predetermined; we aimed to test as many samples as possible (some required genotypes were rare among progeny). No samples were excluded for reasons other than poor staining or physical damage during processing.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully in the article. Drosophila stocks and other reagents are available upon request.
Results
EJC and splicing factors identified in a screen for modifiers of Hh pathway activity
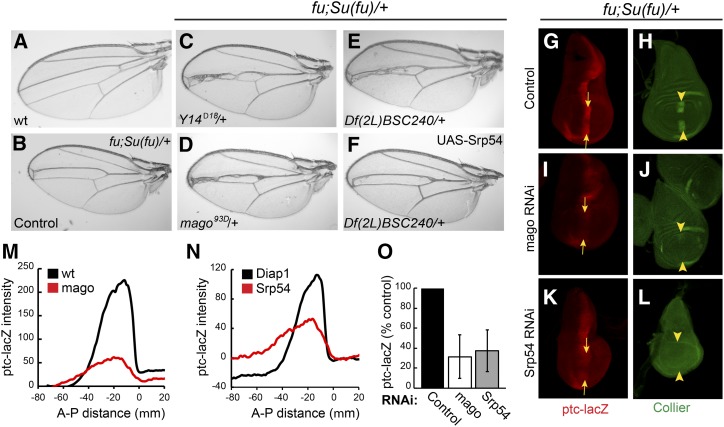
Hh is expressed in posterior compartment cells of the wing disc and is transported to a strip of neighboring anterior cells to induce transcription of several patterning genes at the AP border (Vervoort 2000; Xiong et al. 2015). These Hh target genes include ptc, conveniently reported by the level of ptc-lacZ expression, and col (Figure 1, G and H), which is essential for specifying the tissue between the central two veins of the mature wing. To identify new contributors to Hh signaling pathway activity, we tested a large set of heterozygous autosomal deficiencies for the ability to modify the wing phenotypes of flies heterozygous for Su(fu) and lacking Fu kinase activity; these flies have reduced intervein territory between veins 3 and 4 (v3–4, Figure 1, A and B) resulting from impaired Hh signaling. Only two deficiencies, including Su(fu) and cos2, respectively, increased v3–4 separation, consistent with enhanced Hh pathway activity (Table S1 in File S1). For each deficiency that narrowed the v3–4 interval, we tested several additional deletions to define a critical region. Ten intervals with at least two overlapping deficiencies narrowing v3–4 were identified from a total of ∼220 deficiencies successfully screened (Table S1 in File S1).
Figure 1.
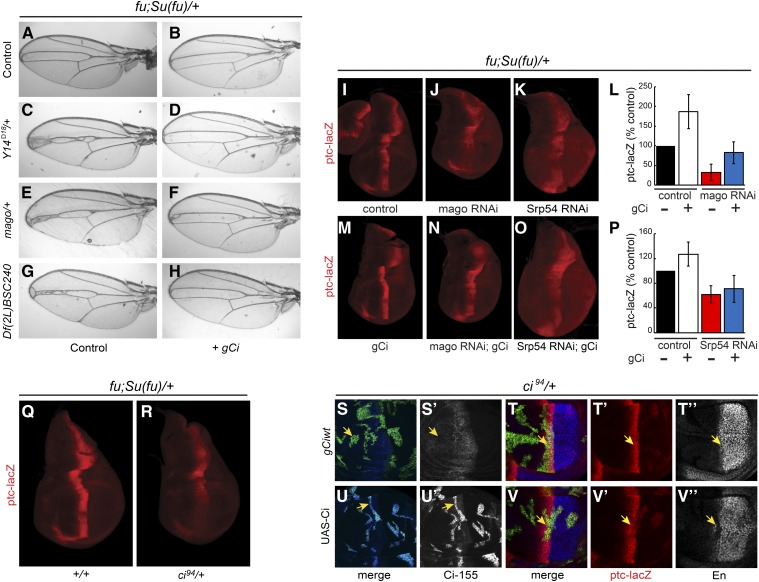
Screen for Hh pathway modifiers. (A–F) Wings from (A) wild-type males and (B–F) fu; Su(fu)/+ males. (B) Narrowing of the central veins was increased by heterozygosity for (C) Y14Δ18 and (D) mago93D. Narrowing due to (E) Df(2L) BSC240 was (F) largely suppressed by expression of UAS-Srp54 cDNA with C765-GAL4. (G and H) Wing discs from fu; Su(fu)/+ larvae have reduced expression at the AP border (yellow arrows) of (G) ptc-lacZ (red) and (H) Collier [green; restricted to the wing pouch (arrowheads)] relative to wild-type (data not shown). (I–L) Both ptc-lacZ and Collier were reduced further by C765-GAL4-driven expression of (I and J) UAS-mago RNAi or (K and L) UAS-Srp54 RNAi together with UAS-Diap1 (UAS-Diap1 was always used as a control for this genotype). Reduced ptc induction limits Ptc-induced endocytosis of Hh, so a weaker ptc-lacZ AP border stripe is also generally broader. (M and N) Average ptc-lacZ intensity along the AP axis (anterior to the left of zero) of the wing pouch for four wing discs in a single experiment, comparing (M) UAS-mago RNAi to control and (N) UAS-Srp54 RNAi to control (both also express UAS-Diap1). (O) Maximal ptc-lacZ intensity at the AP border (derived from profiles along the AP axis) as a percentage of controls for discs expressing mago and Srp54 RNAi, showing means and 95% C.I.s (n = 4 experiments for mago RNAi and n = 3 experiments for Srp54 RNAi). AP, anterior/posterior; Hh, Hedgehog; Ptc, Patched; RNAi, RNA interference; wt, wild-type.
The phenotype of one deficiency was reproduced by a point mutation in the tsunagi (tsu; aka Y14) gene (Figure 1C). Because Y14 and Mago nashi (Mago) are core members of the EJC, we also tested a null mutation of mago as a modifier and found that it also narrowed v3–4 spacing (Figure 1D). This spacing could theoretically be altered by reducing Hh pathway activity or by affecting downstream steps. The latter mechanism is likely for a set of overlapping deficiencies that included spalt and spalt-related, which have established roles in vein specification (Organista et al. 2015) (Table S1 in File S1). The MAPK rolled is also involved in vein formation and is a known target of Mago and Y14 (Blair 2007; Ashton-Beaucage et al. 2010; Roignant and Treisman 2010). However, the mago phenotype was not reversed by ectopic expression of a UAS-MAPK transgene shown previously to rescue MAPK function impaired by EJC depletion (Roignant and Treisman 2010) (Figure S1, D and E in File S1). Thus, Mago is not acting via MAPK and may instead be affecting the Hh pathway.
To identify the key gene within two additional narrowly defined deletion intervals, we tested the consequence of expressing all relevant available UAS-RNAi transgenes in a sensitized (fu; Su(fu)/+) background using C765-GAL4, which is expressed selectively in wing discs. RNAi directed toward the genes CG4602 (Srp54) and CG3605 did not permit eclosion of any adults but significantly reduced the activity of the universal Hh pathway reporter, ptc-lacZ, at the AP border of third instar larval wing discs (Figure S2C in File S1 and data not shown). However, both also variably reduced the size of wing discs, disrupted nuclear integrity, and induced activated caspase 3 staining, indicative of apoptosis (Figure S2C’ in File S1 and data not shown). Coexpression of the antiapoptotic Diap1 gene largely rescued these phenotypes for Srp54 RNAi (Figure S2, A’–D’ in File S1) but less completely for RNAi toward CG3605. In both cases, there was still a marked reduction in ptc-lacZ staining at the AP border of wild-type or fu; Su(fu)/+ wing discs (Figure 1, G, K, N, and O and Figures S1, F–M and S2, A–D in File S1). Expression of UAS-Srp54 using C765-GAL4 largely reversed the v3–4 narrowing of fu; Su(fu)/+ flies due to Df(2L)BSC240 (Figure 1, E and F), while UAS-CG3605 partially suppressed v3–4 narrowing due to Df(2L) Exel7014 (Figure S1, A–C in File S1), suggesting that Srp54 and CG3605 were critical modifier genes within the two deficiencies that affected Hh pathway activity. Srp54 belongs to the family of SR proteins, which contain RNA-binding and Arg/Ser-rich domains, and are generally involved in RNA splicing (Bradley et al. 2015). Both Srp54 and CG3605 have been identified in spliceosomal complexes and functionally implicated in RNA splicing (Kennedy et al. 1998; Park et al. 2004; Wu et al. 2006; Herold et al. 2009).
Collier is induced at the AP border of wing discs by moderately high levels of Hh signaling (Vervoort 2000) and consequently has lower expression in a fu; Su(fu)/+ background than in wild-type wing discs. Expression of Collier was eliminated by additional expression of Srp54 or CG3605 RNAi transgenes (Figure 1, H, J, and L and Figure S1, F’ and H’ in File S1). Likewise, mago RNAi eliminated Collier induction at the AP border of fu; Su(fu)/+ wing discs and strongly reduced ptc-lacZ expression (Figure 1, G–J, M, and O), directly implicating Mago in Hh signaling. Expression of mago RNAi did not induce cell death or other morphological phenotypes. Because the apoptotic phenotype of CG3605 inhibition was not fully suppressed by Diap1, we focused further studies on just Srp54 and Mago, and always expressed excess Diap1 together with Srp54 RNAi to suppress cell death.
Mago and Srp54 alter Ci-155 levels
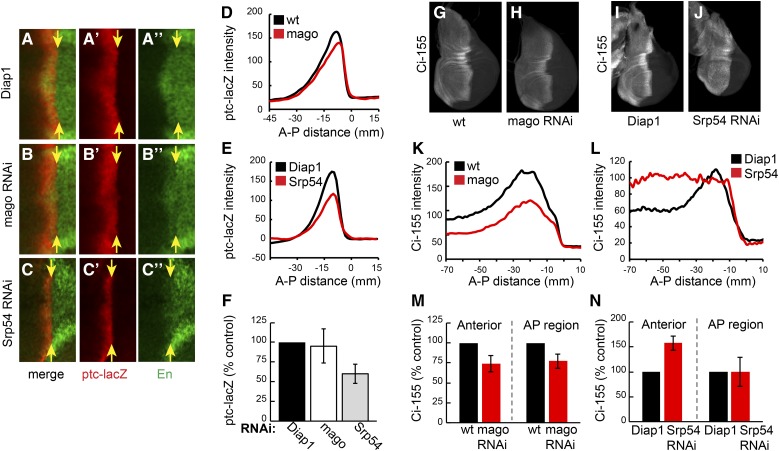
The effects of reducing Mago and Srp54 activity on Hh pathway activity and the levels of Ci-155 were measured in otherwise wild-type wing discs. Mago inhibition lowered Ci-155 levels at the AP border and in anterior regions by ∼30% (Figure 2, G, H, K, and M). RNAi transgenes directed to two other core pre-EJC components, Y14 and eIF4AIII, also reduced Ci-155 levels in wild-type wing discs and reduced ptc-lacZ expression in wing discs with impaired Hh signaling (fu; Su(fu)/+) (Figure S3 in File S1). Hh pathway activity, measured by expression of ptc-lacZ and the high-level Hh target gene, Engrailed (En) at the AP border, was not inhibited by RNAi directed to Mago, Y14, or eIF4AIII in wild-type wing discs (Figure 2, A, B, D, and F and data not shown). Inhibition of Barentsz (Btz), which is a largely cytoplasmic core EJC component (MLN51 in mammals), did not alter ptc-lacZ expression in fu, Su(fu)/+ wing discs or Ci-155 levels (Figure S3, D, E, J, and K). A nuclear function of Mago, Y14, and eIF4AIII that is not shared by Btz has been demonstrated for normal splicing of MAPK and piwi in Drosophila (Roignant and Treisman 2010; Ashton-Beaucage and Therrien 2011; Hayashi et al. 2014; Malone et al. 2014). Therefore, we hypothesized that Mago, Y14, and eIF4AIII core EJC components affect Hh pathway activity under sensitized conditions by acting directly on ci RNA splicing to reduce Ci-155 protein levels.
Figure 2.
Different effects of Mago and Srp54 inhibition on Ci-155 levels and activity. (A–C) Central region (around DV and AP borders) of wing discs expressing (A) UAS-Diap1, (B) UAS-mago RNAi, and (C) UAS-Srp54 RNAi plus UAS-Diap1 under the control of C765-GAL4, stained for ptc-lacZ (red) and Engrailed (En; green). Only Srp54 RNAi reduced ptc-lacZ expression and prevented Hh induction of anterior En (to the left of yellow arrows indicating the posterior edge of the AP border). (D and E) ptc-lacZ intensity profiles along the AP axis for (D) UAS-mago RNAi and (E) UAS-Srp54 RNAi compared to controls for a single experiment (average of 3–4 discs). (F) Maximal ptc-lacZ intensity as a percentage of controls for discs expressing mago and Srp54 RNAi, showing means and 95% C.I.s (n = 6 experiments for mago RNAi and n = 3 experiments for Srp54 RNAi). (G–J) Wing discs expressing (H) UAS-mago RNAi, (J) UAS-Srp54 RNAi, and (G and I) controls, stained for Ci-155 (white). (K–L) Ci-155 intensity profiles along the AP axis for single experiments (average of four wing discs), comparing (K) Mago and (L) Srp54 inhibition to controls. (M and N) Maximum Ci-155 intensity in anterior and AP border cells as a percentage of controls for discs expressing mago and Srp54 RNAi, showing means and 95% C.I.s (n = 5 experiments for mago RNAi and n = 4 experiments for Srp54 RNAi). AP, anterior/posterior; DV, ; Hh, Hedgehog; Ptc, Patched; RNAi, RNA interference; wt, wild-type.
Reduction of Srp54 activity also altered the profile of Ci-155 in wing discs but in a different manner. Ci-155 levels were not significantly altered at the AP border but were higher throughout the rest of the anterior compartment compared to wild-type discs (Figure 2, I, J, L, and N). Inhibition of Srp54 also reduced ptc-lacZ expression and almost eliminated anterior En expression at the AP border of wild-type wing discs (Figure 2, A, C, E, and F). The changes in Ci-155 levels and ptc-lacZ expression due to Srp54 RNAi were suppressed by expression of an Srp54 cDNA (Figure S2, E–H), confirming attribution to Srp54. The observed Ci-155 profile suggests that Srp54 RNAi may impair Ci-155 processing in anterior cells, so that Ci-155 levels cannot reliably report any additional changes in ci RNA.
Effects of Mago and Srp54 inhibition on ci RNA
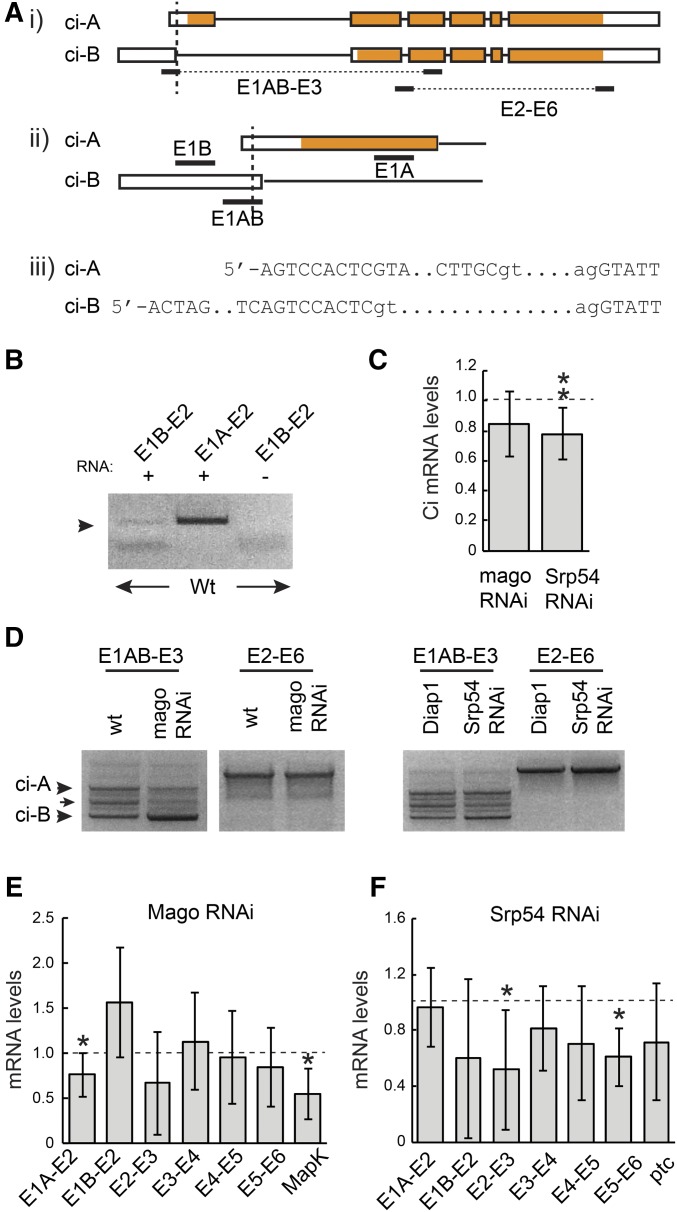
The most direct way that Mago might alter Ci-155 levels is by acting on ci RNA. The major documented ci RNA is denoted ci-A but a much less prevalent RNA, ci-B, has been deduced from a small number of cDNAs and RNA-seq data (FlyBase). Those data suggest that ci-B RNA initiates at a site upstream of ci-A and uses an alternative splice donor at the end of the first exon that is just 8 nt downstream of the ci-A 5′-end (Figure 3A). Using RNA from wild-type wing discs we were able to detect a product of the expected size using primers corresponding to exon 1B and exon 2, but at much lower levels than for an exon 1A to exon 2 primer pair (Figure 3B). These data confirm the presence of ci-B RNA in wing discs. The absence of a larger product corresponding to the E1A splice donor when using primer 1B supported the idea that RNAs initiating upstream of the E1B primer use only the E1B donor splice site. qRT-PCR experiments with the same primer pairs indicated that ci-A is roughly 200-fold more abundant than ci-B in wing discs (difference between Ct values was 7.5 ± 0.64). We then explored whether inhibition of Mago or Srp54 altered the amounts or splicing patterns of these ci RNAs.
Figure 3.
Mago and Srp54 regulate ci RNA. (A) Schematic representation of ci-A and ci-B RNAs, together with primer locations (exons are boxed with translated segments in orange). The TSS of B is 742 nt upstream of TSS-A. ci-B first exon donor splice site is 8 nt downstream of TSS-A. ci-A first exon is 469 nt and first intron is 3446 nt; ci-B first exon is 751 nt and first intron is 3906 nt. (B) RT-PCR products from wild-type wing disc RNA using the indicated primer pairs. (C) ci RNA levels as a fraction of controls (set at 1.0; dashed line) measured by qRT-PCR with primers from exon 3 and spanning the exon 2/3 junction for total RNA from wing discs expressing mago RNAi or Srp54 RNAi. (D) RT-PCR products obtained using the indicated primer pairs using RNA from wing discs with reduced Mago, Srp54, or their controls. Products of the expected sizes for ci-A and ci-B (arrowheads) were confirmed by sequencing. An intermediate band (arrow) revealed a clear product on one occasion, corresponding to splicing between the ci-B donor site and an acceptor 277 nt downstream, followed by splicing in the ci-A pattern (and therefore encoding the normal Ci-A protein product). (E and F) RNA levels as a fraction of controls (set at 1.0; dashed line) measured by qRT-PCR using primers spanning each exon junction of ci and exons 6–7 of MAPK for wing discs expressing (E) UAS-mago RNAi or (F) UAS-Srp54 RNAi compared to controls. (C, E, and F) Values in all qRT-PCR experiments were first normalized to Rp49 and Rpl45 levels. Means and 95% C.I.s are shown relative to controls (set at 1.0) for (C) n = 9 experiments and (E and F) n = 3 experiments for all except E1A–E2 for mago and Srp54 RNAi (n = 7), E1B–E2, E2–E3, E4–E5, and E5–E6 for mago RNAi (n = 4), MAPK (n = 5), and E5–E6 for Srp54 RNAi (n = 4). Significant differences to controls were also calculated by Student’s t-test (* P < 0.05, ** P < 0.01). qRT-PCR, quantitative RT-PCR; RNAi, RNA interference; TSS, transcription start site; wt, wild-type.
We observed reduced levels of ci RNA by qRT-PCR using primers that amplified sequences in exon 3 for both mago and Srp54 RNAi treatments of wing discs (Figure 3C). To compare ci-A and ci-B RNA levels, while selectively increasing detection sensitivity for ci-B RNA we used a primer (E1AB) that matched exon 1B throughout its 20 nt but matched exon 1A only at the last 9 nt. Using this primer together with an exon 3 primer for RT-PCR revealed three clear bands (Figure 3D). Two were sequenced and found to correspond to the expected sequences of ci-A and ci-B in this region. The ratio of these two bands was not discernibly altered in wing discs expressing Srp54 RNAi but Mago inhibition significantly increased the ci-B product relative to ci-A (Figure 3D). No novel RNA splicing patterns or included introns were detected in response to Mago or Srp54 inhibition for regions spanning exons 1–3 or 2–6 (Figure 3D). qRT-PCR measurements supported the inference that RNA with the splice characteristic of ci-B was increased, while RNA with the ci-A splice was reduced in response to Mago inhibition (Figure 3E).
RNA spliced across introns common to ci-A and ci-B was reduced by mago RNAi in some cases, but not consistently (Figure 3E). A known Mago target in the MAPK gene was used as a positive control (Roignant and Treisman 2010) and showed reduced spliced product, as expected. The alteration in the pattern of ci RNAs suggests that Mago may act directly on ci splicing. Loss of Mago may be affecting the choice of splice donor sites for the first exon to allow some E1B donor use for the major ci primary transcript. Alternatively, loss of Mago might be affecting the choice of transcription start sites, increasing initiation at the upstream site characteristic of ci-B and decreasing initiation from the major ci-A start site. In either case, the reduced levels of E1A–2 spliced RNA observed (Figure 3, D and E) would be expected to decrease Ci-155 protein levels, while increased ci-B RNA levels would only produce a very small increase in protein product because this RNA is present at only ∼1% of ci-A RNA levels.
Inhibition of Srp54 led to a reduction in spliced RNA across all main-body introns but, in contrast to Mago inhibition, it did not increase E1B–E2 spliced RNA or significantly reduce E1A–E2 spliced RNA levels (Figure 3, D and F). ptc RNA levels were reduced (Figure 3F), consistent with reduced ptc-lacZ expression at the AP border. The reduced amount of ci RNA measured across all constitutive splice junctions would be expected to result in reduced levels of Ci-155 primary translation product.
Cell autonomy of Mago and Srp54 effects on Hh pathway activity
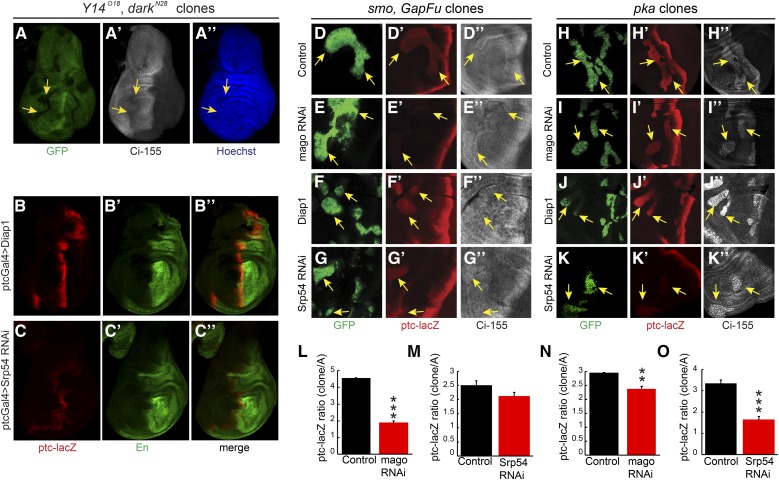
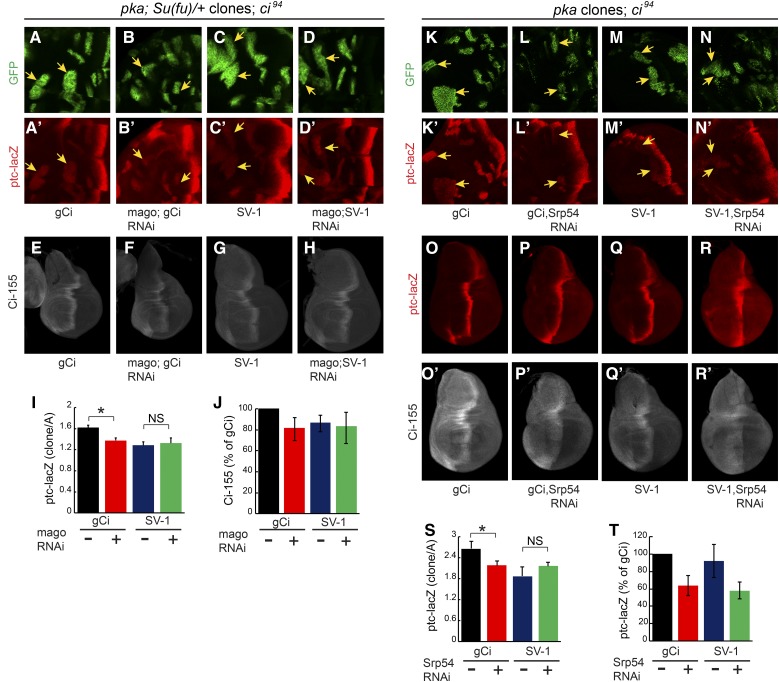
If EJC proteins or Srp54 affect Hh signaling by acting on ci RNA, we would expect them to act in cells responding to Hh, rather than in cells responsible for producing or transporting Hh. Strong alleles of Y14 or mago did not readily produced large homozygous mutant wing disc clones but we did observe a cell autonomous reduction of Ci-155 levels for Y14 clones when clone survival was enhanced by using a Minute background (Amoyel and Bach 2014) and a mutant allele of the proapoptotic dark gene (Roignant and Treisman 2010) (Figure 4A). We also observed a reduction in AP border ptc-lacZ and En expression when Srp54 RNAi expression was limited to anterior cells by using ptc-GAL4 as a driver (Figure 4, B and C). However, the most convincing evidence of Mago and Srp54 acting in Hh signal transduction was observed by creating MARCM clones (Lee and Luo 2001) that expressed the corresponding RNAi in anterior cells and had ectopic Hh pathway activity due to genetic alteration of PKA or Fu activity.
Figure 4.
Cell autonomous action of EJC members and Srp54 on Hh pathway activity. (A) Homozygous Y14Δ28 darkN28 clones (arrows) in a Minute background, marked by loss of GFP (green), had lower Ci-155 levels [white, (A’)] but no cell death or rearrangement evident from nuclear Hoechst staining [blue, (A”)]. (B–C) Expression of UAS-Srp54 RNAi with ptc-GAL4 driver reduced ptc-lacZ (red) and anterior En (green) expression relative to UAS-Diap1 controls. (D–G) Ectopic ptc-lacZ (red) was induced in anterior smo clones expressing UAS-GapFu (marked by GFP, green, arrows) but induction was reduced (E) greatly by UAS-mago RNAi and (G) slightly by UAS-Srp54 RNAi. (D”–G”) Changes in Ci-155 levels (white) were small. (H–K) Ectopic ptc-lacZ (red) was induced in anterior pka clones (marked by GFP, green, arrows) but induction was reduced (I) slightly by UAS-mago RNAi and (K) greatly by UAS-Srp54 RNAi. (H”–K”) The increase of Ci-155 (white) was clearly reduced by Srp54 RNAi in some pka clones. (L–O) ptc-lacZ intensity relative to controls within (L and M) smo GapFu clones or (N and O) pka clones, showing mean, SEM, and significant differences calculated by Student’s t-test (** P < 0.001, *** P < 0.0001), using (L) n = 4 control and n = 8 experimental clones, (M) n = 13 control and n = 21 experimental clones, (N) n = 21 control and n = 18 experimental clones, and (O) n = 15 control and n = 16 experimental clones. EJC, exon junction complex; En, engrailed; Hh, Hedgehog; Ptc, Patched; RNAi, RNA interference; wt, wild-type.
Reduced ptc-lacZ expression due to mago RNAi was clearest in clones expressing activated Fu elicited by expression of GAP-Fu, a membrane-tethered Fu fusion protein containing the myristoylation domain from Growth-Associated-Protein-43 (Claret et al. 2007; Zhou and Kalderon 2011) (reduced 60%, Figure 4, D, E, and L). Reduced ptc-lacZ expression was also evident in pka (Figure 4, H, I, and N) and cos2 (Figure S4, G, H, and K in File S1) mutant clones (reduced 20% in each case). A cell autonomous reduction of ptc-lacZ expression due to Srp54 inhibition was also seen for the same three types of clone. The effects of Srp54 inhibition were larger for pka mutant clones (reduced 50%, Figure 4, J, K, and O) than for cos2 mutant clones (reduced ∼20%, Figure S4, I, J, and L in File S1) or for clones with activated Fu (reduced 15%, Figure 4, F, G, and M). No significant reduction of ptc-lacZ due to Mago or Srp54 inhibition was observed in ptc mutant clones (Figure S4, A–F in File S1), which have very high levels of Hh pathway activity. These results are consistent with cell autonomous actions of Mago and Srp54, which contribute significantly to Hh pathway activity when the pathway is not maximally activated (as in pka, cos2, and GAP-Fu mutant clones and at the AP border of discs lacking Fu kinase activity).
We also observed that Ci-155 levels were reduced by Srp54 RNAi in pka mutant clones (Figure 4, J” and K”). There is no Ci-155 processing in the absence of PKA, so the reduced levels of Ci-155 likely represent a lower rate of Ci-155 production, consistent with the observation that Srp54 RNAi reduced ci RNA levels (Figure 3).
The additional effect of Srp54 RNAi reducing the rate of Ci-155 processing, inferred from elevated anterior Ci-155 in otherwise normal wing discs (Figure 2, J, L, and N), can explain the different impacts of Mago and Srp54 in different clones. Hh target gene activation appears to be more sensitive to reductions in Ci-155 levels in GAP-Fu clones than in pka clones based on the stronger effect of Mago inhibition in GAP-Fu clones. However, the reduction of Ci-155 levels expected from Srp54 RNAi lowering ci RNA is substantially offset in GAP-Fu clones (and not at all in pka clones) because Srp54 RNAi additionally reduces Ci-155 processing only in the GAP-Fu clones. Hence, Srp54 RNAi barely inhibits ptc-lacZ in GAP-Fu clones, whereas ptc-lacZ inhibition in pka clones is greater than for mago RNAi, probably because Srp54 RNAi causes a larger reduction in ci RNA (Figure 3). Altogether, clonal analyses are consistent with the hypotheses that both Mago and Srp54 inhibition reduce Hh pathway activity by reducing ci RNA levels, and that Srp54 inhibition also impairs Ci-155 processing.
Effects of Ci-155 levels on Hh pathway activity
We next explored whether reduced levels of Ci-155 primary translation product, inferred from studies of ci RNA and Ci-155 antibody staining, could plausibly explain the alterations in Hh pathway activity seen in response to inhibition of Mago and Srp54. In a fu; Su(fu)/+ background, heterozygosity for Srp54 (in Df(2L)BSC240), mago, or Y14 reduced v3–4 spacing. Normal spacing was restored by a single copy of a 16-kb ci genomic transgene (Methot and Basler 1999) inserted at an att site on chromosome 3 (“gCi”) (Figure 5, A–H). In the same genetic background (fu; Su(fu)/+), addition of the gCi transgene enhanced ptc-lacZ expression (Figure 5, I, L, and M), while heterozygosity for ci reduced ptc-lacZ expression (Figure 5, Q and R). Thus, reduced ci gene dosage can phenocopy the effects of reduced Mago or Srp54 activity and an extra ci transgene can suppress heterozygous Mago and Srp54 phenotypes, consistent with Mago and Srp54 acting through regulation of ci RNA levels.
Figure 5.
Dependence of Hh signaling on Ci-155 levels. (A–H) Narrowing of veins 3–4 in (A) fu; Su(fu)/+ controls is increased by heterozygosity for (C) Y14, (E) mago, or (G) Df(2L)BSC240, but these changes were (B, D, F, and H) suppressed by addition of a single copy of the gCi transgene. (I–P) In fu; Su(fu)/+ wing discs a single copy of the gCi transgene (I and M) increased ptc-lacZ expression and (J–O) suppressed inhibition of ptc-lacZ expression by (J and N) mago RNAi but (K and O) not by Srp54 RNAi. (L and P) Maximal ptc-lacZ intensity as a percentage of controls for fu; Su(fu)/+ wing discs expressing mago and Srp54 RNAi, with or without a gCi transgene, showing means and 95% C.I.s for n = 4 experiments. (Q and R) Loss of one copy of ci (R) reduced ptc-lacZ (red) expression in fu; Su(fu)/+ wing discs. (S–V) MARCM clones (marked by GFP, green) that lose a second chromosome gCi transgene in wing discs that are ci94/+ and either (S and T) include a third chromosome gCi transgene or (U and V) express UAS-Ci with C765-GAL4. (S and U) Ci-155 levels (white) were increased greatly by UAS-Ci expression but were not affected by exchange of gCi transgenes. (T and V) Only excess Ci-155 from UAS-Ci decreased ptc-lacZ (red) and anterior En (white) expression at the AP border (yellow arrows). AP, anterior/posterior; En, engrailed; Hh, Hedgehog; Ptc, Patched; RNAi, RNA interference; wt, wild-type.
When Mago activity was reduced more drastically using RNAi in fu; Su(fu)/+ discs, adding the gCi transgene substantially restored ptc-lacZ expression (Figure 5, J, L, and N), consistent with Mago acting solely by modifying ci RNA levels. However, analogous complementation was not observed for Srp54 RNAi (Figure 5, K, O, and P), suggesting that severe reduction in Srp54 activity does not act solely by reducing ci RNA.
The effect of excess Ci on Hh signaling was tested by expressing a UAS-Ci cDNA transgene using C765-GAL4 in MARCM clones at the AP border. Ci-155 levels were greatly elevated in clones and induced strong ptc-lacZ expression in posterior clones, as expected due to stimulation by Hh (Smelkinson et al. 2007), but did not induce ptc-lacZ in anterior clones (Figure 5, S–V). Surprisingly, clones at the AP border showed reduced expression of both ptc-lacZ and En (Figure 5V), whereas control clones had no such changes (Figure 5T), implying that excess Ci-155 can impair Hh signaling. Thus, small reductions in Ci-155 primary translation product can reduce Hh pathway activity under sensitized conditions, while excess Ci-155 can impair normal Hh signaling.
ci-A and ci-B RNA encode similar Ci activator functions
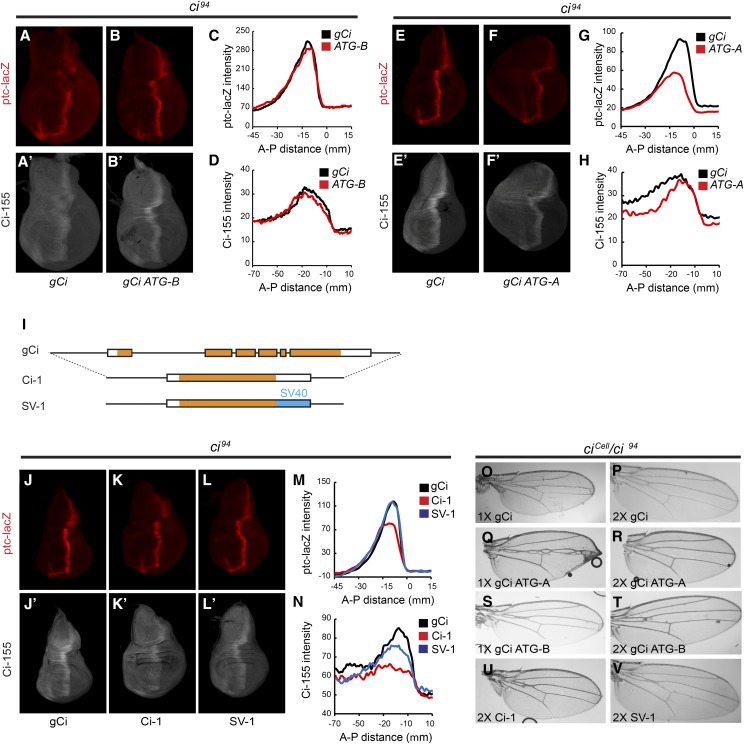
The confirmed existence of ci-B RNA in wing discs, albeit at much lower levels than ci-A RNA, as well as the potential for Mago to regulate the relative levels of ci-A and ci-B (Figure 3), led us to question the functional role of ci-B RNA. The ci-B RNA does not include the first coding exon of ci-A and it would be expected to encode a translation product that initiates at M119 of the Ci-A protein (Figure 3A). Therefore, we constructed two variants of the gCi transgene, in which the expected initiation codons for either Ci-A (gCi ATG-A) or Ci-B (gCi ATG-B) were altered to AAG Lys codons.
We found that gCi ATG-B (lacking Ci-B) rescued ci null (ci94) flies to adulthood with a similar efficiency to gCi WT (Table 1); the profile of ptc-lacZ expression and Ci-155 protein of rescued wing discs was also very similar (Figure 6, A–D), suggesting that Ci-B protein has no essential function under normal conditions. It remains possible that ci-B RNA has a function independent of protein products or that it encodes a protein initiating downstream of M119 that substitutes for Ci-B function when M119 is altered.
Table 1. Rescue to adulthood of ci94/ci94 null flies by gCi transgenes.
| transgene/+; ci94/ci94 (% transgene/+; ci94/+) | transgene/transgene; ci94/ ci94 (% transgene/+; ci94/+) | |
|---|---|---|
| gCi | 49 (n = 298) | 94 (n = 1052) |
| gCi ATG-B | 83 (n = 550) | 99 (n = 530) |
| gCi ATG-A | 0 (n > 300) | 0 (n > 300) |
| SV-1 | 23 (n = 699) | 75 (n = 350) |
| Ci-1 | 0 (n = 298) | 9 (n = 603) |
Figure 6.
Activities of Ci-A, Ci-B proteins, and intronless ci transgenes. (A, B, H, and F) Wing discs from ci null larvae with a gCi transgene that (A and E) is wild-type, or lacks the initiator codon for (B) Ci-B or (F) Ci-A, stained for ptc-lacZ (red) or Ci-155 (white). (C and G) ptc-lacZ and (D and H) Ci-155 intensity profiles along the AP axis for single experiments (n = 4 wing discs). (I) Diagram representing the structure of gCi, Ci-1, and SV-1 transgenes, in which introns are deleted (boxed regions are exons with coding sequence in orange). In SV-1, the 3′-UTR was replaced by SV40 3′-UTR sequences (blue). (J–L) ci null wing discs with one copy of (J) gCi, (K) Ci-1, or (L) SV-1, stained for ptc-lacZ (red) and Ci-155 (white). (M) ptc-lacZ and (N) Ci-155 intensity profiles along the AP axis for single experiments (n = 3 wing discs). (O–V) Wings from ciCell/ci94 flies with one copy or two copies of (O and P) gCi, (Q and R) gCi ATG-A, (S and T) gCi ATG-B, (U) Ci-1, or (V) SV-1. AP, anterior/posterior.
Neither one nor two copies of gCi ATG-A rescued any ci null animals to adulthood (Table 1), but rescue to third larval instar was observed. Wing discs from those animals showed reduced levels of AP border ptc-lacZ and Ci-155, together with enlarged anterior compartments (Figure 6, E–H). Ci-B (the sole expected product of gCi ATG-A) may have little or no Ci-75 repressor function based on the prior observation of loss of repressor activity for a Ci variant lacking residues 6–339 (Zhou and Kalderon 2010). Loss of Ci repressor leads to ectopic dpp induction in anterior cells and anterior expansion (Methot and Basler 1999). Consistent with the possibility that Ci-B defects stem largely from the failure to produce a repressor, gCi ATG-A was able to rescue adult flies transheterozygous for ci94 and the ciCe allele, which produces only a Ci repressor that is not regulated by Hh (Methot and Basler 1999)(Table 2). Rescue of ciCe/ci94 animals by a single copy of gCi ATG-A was less efficient than for gCi WT and resulted in more severe narrowing of the v3–4 interval, but both shortcomings were largely rectified by providing two copies of the transgene (Figure 6, O–V and Table 2). These properties suggest that gCi ATG-A generates a Ci activator that is regulated normally but present at slightly reduced levels, most likely because of less efficient use of the Ci-B translation initiation codon compared to Ci-A, or perhaps reduced stability of Ci-B protein compared to Ci-A.
Table 2. Rescue to adulthood of ciCell/ci94 flies by gCi transgenes.
| transgene/+; ciCe/ci94 (% transgene/+; ci94/+) | transgene/transgene; ciCe/ci94 (% transgene/+; ci94/+) | |
|---|---|---|
| gCi | 44 (n = 218) | 76 (n = 992) |
| gCi ATG-B | 20 (n = 208) | 92 (n = 286) |
| gCi ATG-A | 17 (n = 440) | 32 (n = 462) |
| SV-1 | 0 (n = 192) | 21 (n = 843) |
| Ci-1 | 0 (n = 546) | 0 (n = 941) |
Properties of intronless ci transgenes
It is possible that regulation of the efficiency of ci-A RNA splicing, or even a regulatory role of ci-B RNA independent of Ci-B protein production, are important for Hh signaling. To test these ideas, we constructed two gCi transgene variants, in which all intron sequences had been removed. One variant (Ci-1) retained the normal ci 3′-UTR sequences, while the other (SV-1) instead included 3′-UTR sequences from Simian Virus 40, with the expectation that this 3′-UTR, commonly employed for high-level gene expression, might enhance protein translation (Figure 6I).
Ci-1 rescued ci null adults only when present in two copies, and very inefficiently, while SV-1 had rescue activity intermediate between Ci-1 and gCi WT (Table 1). Ci-155 protein levels encoded by Ci-1 were much lower than for gCi WT, while SV-1 Ci-155 levels were only marginally lower (Figure 6, J’–L’, and N). We were not able to collect enough rescued wing discs to measure ci RNA to determine if intron removal in Ci-1 reduced Ci-155 protein because of reduced transcription, RNA stability, or translation. AP border expression of ptc-lacZ appeared normal in discs rescued by SV-1 but was slightly reduced in the very few wing discs rescued by Ci-1 (Figure 6, J–M).
Rescue of ciCe/ci94 animals showed a similar pattern. Ci-1 produced almost no adults, even when present in two copies, and did not support the normal v3–4 spacing endowed by two copies of gCi WT (Figure 6U and Table 2). SV-1 rescued no adults in one copy but in two copies rescue approached the efficiency of one copy of gCi WT, with a modest wing vein abnormality (Figure 6V and Table 2).
These results show that ci introns are important to support normal functional levels of Ci-155 and provide further evidence that relatively small reductions in Ci-155 levels compromise robust Hh signaling (measured by rescue efficiency and phenotypes) in a number of settings, including reduced ci gene dosage or the presence of constitutive Ci repressor.
The functional deficit of Ci-1 relative to SV-1 is likely due to reduced Ci-155 levels and SV-1 undoubtedly has considerable ability to support Hh signaling. However, the surprisingly poor rescue of ciCe animals by SV-1, given almost normal Ci-155 levels, suggests that intron removal may also compromise some regulatory input from Hh.
ci RNA as a direct target of Mago and Srp54 function in Hh signaling
The significant, though incomplete, rescue activity of the SV-1 transgene provided a way to test whether Mago or Srp54 acted directly on ci RNA splicing to impact Hh signaling. If so, we should see no effect of Mago or Srp54 inhibition on signaling through SV-1. We were able to test this most effectively in the context of pka mutant clones, where both mago RNAi and Srp54 RNAi reduced ptc-lacZ induction in otherwise wild-type wing discs (Figure 4, H–K, N, and O). When a single copy of the gCi WT transgene rescues a ci null, ectopic ptc-lacZ expression in pka mutant clones was much lower than in wild-type discs and not satisfactory for testing the effects of Mago and Srp54 inhibition (data not shown). However, ptc-lacZ expression could be increased to an intermediate level by using either a heterozygous Su(fu) background (Figure 7A) or two copies of the transgene (Figure 7K).
Figure 7.
An intronless ci transgene tests ci RNA splicing as a key target of Mago and Srp54. (A–D and K–N) Ectopic ptc-lacZ (red) induced in anterior pka clones (marked by GFP, green, arrows) in wing discs null for ci with (A–D) one copy of a ci transgene and heterozygous for Su(fu) or (K–N) two copies of a ci transgene, was measured in the presence or absence of (A–D) mago RNAi or (K–N) Srp54 RNAi. (A, C, K, and M) ptc-lacZ expression was higher for discs with gCi than for SV-1 and was reduced by mago RNAi and Srp54 RNAi (A, B, K, and L) in the presence of gCi but (C, D, M, and N) not in the presence of SV-1. (I and S) ptc-lacZ expression in pka clones in discs of the designated genotypes relative to discs with gCi (and no RNAi), showing mean, SEM, and significant differences by Student’s t-test (* P < 0.05) for (I) n = 3, n = 3, n = 4, and n = 3 clones, and (S) n = 11, n = 9, n = 7, and n = 12 clones in the order shown. (E–H) Ci-155 (white) in ci null wing discs with two copies of gCi or SV-1 in the presence or absence of mago RNAi expression. (J) Maximal Ci-155 intensity as a percentage of wing discs with gCi, showing means and 95% C.I.s from two independent experiments of n = 6 discs for each condition. Ci-155 levels were reduced by Mago inhibition for gCi but not for SV-1. (O–R and T) ptc-lacZ (red) and Ci-155 (white) in ci null wing discs expressing UAS-Diap1 with two copies of gCi or SV-1 in the presence or absence of Srp54 RNAi. (T) Maximal ptc-lacZ intensity at the AP border as a percentage of wing discs with gCi, showing means and 95% C.I.s from two independent experiments of n = 6 discs for each condition. AP, anterior/posterior; RNAi, RNA interference.
The level of ptc-lacZ induced in pka mutant clones in Su(fu)/+ flies with one copy of gCi WT (and otherwise ci null) was reduced ∼25% by expression of mago RNAi (Figure 7, A, B, and I). SV-1 supported a slightly lower level of ptc-lacZ induction than gCi WT, consistent with lower Ci-155 levels produced by SV-1, but mago RNAi produced no change (Figure 7, C, D, and I). Similarly, Ci-155 levels were reduced at the AP border of wing discs by mago RNAi for gCi but not SV-1 (Figure 7, E–H, and J). These results are consistent with mago RNAi normally reducing Hh pathway activity by reducing the levels of the major spliced ci RNA (ci-A) and primary Ci-155 translation product.
The level of ptc-lacZ induced in pka mutant clones by two copies of gCi WT (and otherwise ci null) was reduced almost 20% by expression of Srp54 RNAi (Figure 7, K, L, and S). SV-1 supported a lower level of ptc-lacZ induction and Srp54 RNAi produced no reduction in ptc-lacZ expression in the presence of only SV-1 (Figure 7, M, N, and S). This result is consistent with Srp54 RNAi normally reducing Hh pathway activity in pka mutant clones by reducing the levels of spliced ci RNA.
Srp54 RNAi, in contrast to mago RNAi, inhibited Hh signaling in wild-type wing discs, suggesting that this action may not be mediated by reducing ci RNA levels (Figure 2, A–F). Indeed, we found that Srp54 RNAi still decreased ptc-lacZ expression at the AP border of wing discs expressing only SV-1 (Figure 7, O–R and T). Elevated anterior Ci-155 in response to Srp54 RNAi was also evident in the presence of SV-1, confirming that this action of Srp54 is also not dependent on ci splicing (Figure 7, O’–R’). Thus, Srp54 affects ci RNA splicing slightly differently to Mago but with the same functional consequence of reducing ci-A RNA levels, leading to lowered rates of Ci-155 production and Hh pathway deficits in several settings, including pka mutant clones. Srp54 additionally, through currently uncharacterized mechanisms, appears to promote Ci-155 processing in anterior cells and Hh signaling at the AP border.
Discussion
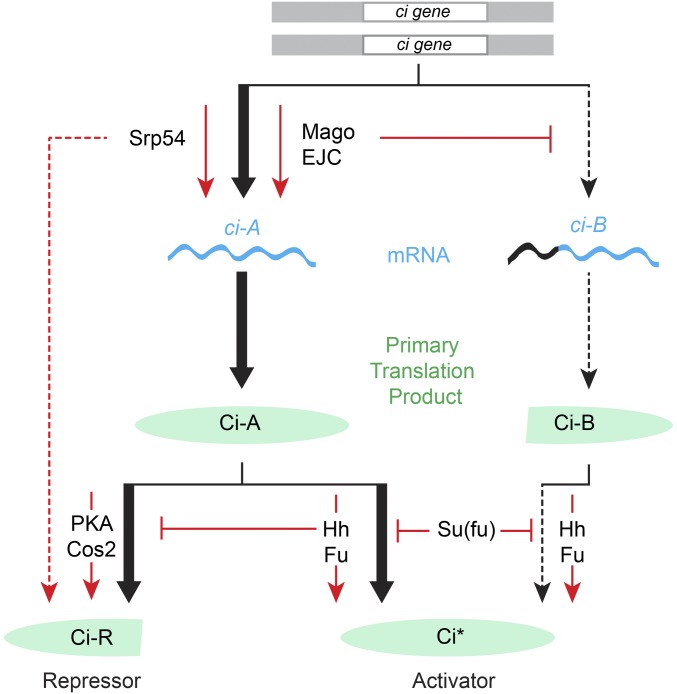
We used a genetic modifier screen to identify new components in Hh signal transduction. Surprisingly, the screen identified EJC components and an SR protein, all with established functions in regulating RNA. Further investigation revealed ci RNA as a direct target and clarified the roles of different ci RNAs and the levels of their primary translation products in Hh signaling (Figure 8).
Figure 8.
Role of the EJC, Srp54, and ci RNA production in Hh signal transduction. In the absence of Hh, full-length Ci-155 protein is largely inactive and processed slowly, with the participation of PKA and Cos2, into active Ci-75 repressor. Hh signal transduction via activation of Smo leads to inhibition of Ci-155 processing, leading to a loss of Ci-75 repressor and a greater accumulation of Ci-155 primary translation product. Hh also promotes activation of Ci-155 via Fu kinase and in opposition to Su(fu). Activated Ci-155 is subject to Cul3-mediated degradation (data not shown). The role of Ci-155 levels in contributing to Hh target gene activation is not well-studied. Here, we have found that reduced rates of Ci-155 production due to reduced ci gene dose, genetic removal of ci intronic sequences, or reduced production of the major ci RNA (ci-A) through inhibition of core nuclear EJC factors or Srp54, reduce Hh pathway activity under conditions of submaximal activation (by Hh in the absence of Fu kinase, by synthetic Fu activation, or by loss of PKA or Cos2). Loss of Su(fu), like reduced Ci-155 production, only affects Hh pathway activity under an overlapping set of conditions for submaximal pathway activation, illustrating the potential for partial redundancy in regulating Ci-155 levels and Ci-155 activity. The minor ci-B RNA is increased in the absence of Mago and encodes a protein with normally regulated activator function but appears not to be processed to a functional repressor. EJC, exon junction complex; Fu, Fused; Hh, Hedgehog; PKA, Protein kinase A; Smo, Smoothened; Su(fu), Suppressor of fused.
Direct actions of Mago and Srp54 that impact Hh signaling
Reducing the level of each of the core EJC components, Mago, Y14, and eIF4AIII, reduced Hh pathway activity under sensitized conditions, while inhibition of Btz did not affect Hh signaling. This pattern of EJC contributions has been observed previously for the archetypal studies showing EJC regulation of splicing of MAPK and piwi (Ashton-Beaucage et al. 2010; Roignant and Treisman 2010; Hayashi et al. 2014; Malone et al. 2014), and therefore suggested that RNA splicing may be the relevant EJC focus in Hh signaling. We found that loss of Mago reduced Hh pathway activity cell autonomously in pka mutant clones and in clones expressing activated Fu, indicating an effect on Hh signal transduction. Mago inhibition did not affect Hh signaling when mediated by a ci transgene (SV-1) that contained no introns, identifying ci RNA splicing as the key Mago target. We also observed changes in the pattern of ci RNAs when Mago was inhibited. Specifically, the level of a minor alternatively spliced product (ci-B) was increased while the level of the major splice form (ci-A) was decreased by an amount commensurate with observed reductions in Ci-155 protein levels and Hh target gene inhibition (Figure 8).
The evidence for Srp54 acting directly on ci RNA splicing is similar to that for Mago in terms of cell autonomous action and a failure to influence Hh pathway activity when the only source of Ci protein is an RNA (from the SV-1 transgene) that does not need to be spliced. However, unlike Mago depletion, inhibition of Srp54 reduced the levels of ci RNA measured across most introns but did not alter the ratio of ci-A and ci-B RNAs, so it is unlikely that Srp54 is acting on ci RNA in concert with EJC components, despite some evidence of association of Srp54 with EJCs in directing alternative splicing (Sakashita et al. 2004).
For Srp54, unlike Mago, we also found evidence for effects on the Hh pathway that are not mediated by alterations in ci RNA. First, Ci-155 protein was increased in anterior wing disc cells when Srp54 was inhibited (Figure 8). This most likely reflects impaired Ci-155 proteolytic processing to Ci-75 repressor because Srp54 inhibition actually lowered Ci-155 levels when Ci-155 processing was eliminated in pka mutant clones. Second, Srp54 inhibition reduced ptc-lacZ induction at the AP border of normal wing discs, whereas twofold reduction in ci dosage or Mago inhibition had no effect. Moreover, AP border ptc-lacZ inhibition was not rescued by providing excess ci and it was still observed when the only source of ci was an intronless transgene. Thus, Srp54 has additional uncharacterized actions that impinge on Hh signaling.
Ci-155 protein levels in Hh signaling
It has previously been recognized that Hh signaling alters both the amounts and activities of Ci proteins (Briscoe and Therond 2013). However, the impact of regulating Ci-155 levels has not yet been investigated carefully. In this study, we have altered the rate of production of the primary full-length Ci translation product through heterozygosity for ci, various ci transgenes, and Mago inhibition. We consistently observed that Hh pathway activity was altered by twofold or lesser changes in Ci-155 levels under conditions where normal Hh signaling was compromised (loss of Fu kinase) or only partially phenocopied (by loss of pka or synthetic Fu activation). The observed sensitivity to relatively small changes in Ci-155 makes it plausible that any mechanism that regulates Ci-155 levels, including regulation of ci splicing, contributes to normal Hh signaling (Figure 8).
Impact of alternative ci RNAs and ci splicing on Hh signal transduction
Our observation that Mago inhibition altered the proportion of ci-A and ci-B RNA prompted investigation of potentially distinctive roles of these RNAs. Both public data and our investigation of wing disc RNA showed ci-B to be much less abundant than ci-A. Hence, we first considered the hypothesis that ci-B might encode a hyperactive form of Ci activator, potentially induced as a feed-forward mechanism to achieve the highest levels of Hh signaling. Our investigations did not support that speculation. A ci transgene lacking the initiator codon for Ci-B supported normal Hh signaling, while another transgene lacking the initiation codon for Ci-A, and therefore expected to produce more Ci-B protein than normal, was not hyperactive. Instead, the gCi ATG-A transgene product appeared to be a normally regulated activator that does not generate an active processed repressor. At present, we cannot therefore assign a distinct function to the N-terminally truncated protein that is predicted to be encoded by ci-B RNA.
We additionally tested the properties of two ci transgenes lacking any intronic sequences. SV-1, which included an SV40 3′-UTR in place of ci sequences, produced significantly more Ci-155 protein than Ci-1 and had greater rescue activity in the absence of normal ci gene activity. Both were less active than a wild-type transgene but exhibited improved function in two doses. The dose-dependent and graded deficiencies of these transgenes provide further evidence of the importance of the levels of Ci-155 primary translation product for Hh signaling. We did not determine why intron removal reduced Ci-155 production, but two plausible possibilities are that the large first intron includes a transcriptional enhancer or that translation efficiency is compromised, as observed for some other genes following intron removal (Chorev and Carmel 2012). Although substantially reduced Ci-155 levels provide a sufficient explanation for major deficits of Ci-1, the rescue activity of SV-1, most notably in a ciCe background, was lower than expected from measurements of Ci-155 levels. This quantitative limitation of the activity of an intronless transgene suggests that the presence of introns in ci may also serve a regulatory role that is important for ci to transduce Hh signals robustly.
When assessing the impact of a regulatory process in Hh signaling, it is important to consider the potential for partially or fully redundant mechanisms (Figure 8). Su(fu) provides a notable example of a factor that is central to the mechanism of Hh signal transduction in Drosophila and mammals, but only discernibly affects Hh pathway activity in Drosophila under conditions that perturb normal Hh signaling (Preat 1992; Zhou and Kalderon 2011). In fact, Mago affects Hh pathway activity under exactly the same conditions as Su(fu) (e.g., in pka mutant clones or when Fu kinase is inactive) but not under normal conditions (Figure 8). Similarly, deficiencies of intronless ci transgenes are especially evident under sensitized conditions (reduced ci dosage, pka mutant clones, or in the presence of a constitutive repressor). To resolve the contribution of potentially regulated ci splicing, the EJC, Srp54, and translatable ci RNA levels to Hh signaling, it will be necessary to simultaneously eliminate regulation through Ci-155 activation, Ci-155 processing, and Ci-155 proteolysis individually, and perhaps in combination. At present, the studies described here have alerted us to the possibility that ci RNA transcription and processing, the EJC, and an SR protein splicing factor may all play a significant role in regulating the output of Hh signaling.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.202457/-/DC1.
Acknowledgments
We thank the Bloomington Drosophila Stock Center, the Vienna Drosophila Resource Center, the Developmental Studies Hybridoma Bank, Jessica Treisman, and Alain Vincent for fly stocks and antibodies; Sarah Finkelstein, Allyson Ray, Jessica Chan, Hana Littleford, and Maryam Mudasir for assistance with experiments; and Jessica Treisman, Jose F. de Celis, and Amy Reilein for discussions. This work was supported by the National Institutes of Health (grant GM-041815). The authors declare no competing or financial interests.
Author contributions: E.G.G., J.C.L., and D.K. were responsible for conceptualization and methodology; E.G.G., J.C.L., and D.K. for formal analysis and investigation; E.G.G. and D.K. for writing (original draft preparation); E.G.G., J.C.L., and D.K. for reviewing and editing the manuscript; E.G.G. and J.C.L. for visualization; and D.K. for funding acquisition.
Footnotes
Communicating editor: N. Perrimon
Literature Cited
- Amoyel M., Bach E. A., 2014. Cell competition: how to eliminate your neighbours. Development 141: 988–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E., Peluso S., Lettice L. A., Hill R. E., 2012. Human limb abnormalities caused by disruption of hedgehog signaling. Trends Genet. 28: 364–373. [DOI] [PubMed] [Google Scholar]
- Ashton-Beaucage D., Therrien M., 2011. The exon junction complex: a splicing factor for long intron containing transcripts? Fly (Austin) 5: 224–233. [DOI] [PubMed] [Google Scholar]
- Ashton-Beaucage D., Udell C. M., Lavoie H., Baril C., Lefrancois M., et al. , 2010. The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell 143: 251–262. [DOI] [PubMed] [Google Scholar]
- Blair S. S., 2007. Wing vein patterning in Drosophila and the analysis of intercellular signaling. Annu. Rev. Cell Dev. Biol. 23: 293–319. [DOI] [PubMed] [Google Scholar]
- Bradley T., Cook M. E., Blanchette M., 2015. SR proteins control a complex network of RNA-processing events. RNA 21: 75–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J., Therond P. P., 2013. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14: 416–429. [DOI] [PubMed] [Google Scholar]
- Chorev M., Carmel L., 2012. The function of introns. Front. Genet. 3: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claret S., Sanial M., Plessis A., 2007. Evidence for a novel feedback loop in the Hedgehog pathway involving smoothened and fused. Curr. Biol. 17: 1326–1333. [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. [DOI] [PubMed] [Google Scholar]
- Han Y., Shi Q., Jiang J., 2015. Multisite interaction with Sufu regulates Ci/Gli activity through distinct mechanisms in Hh signal transduction. Proc. Natl. Acad. Sci. USA 112: 6383–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi R., Handler D., Ish-Horowicz D., Brennecke J., 2014. The exon junction complex is required for definition and excision of neighboring introns in Drosophila. Genes Dev. 28: 1772–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold N., Will C. L., Wolf E., Kastner B., Urlaub H., et al. , 2009. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol. Cell. Biol. 29: 281–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui C. C., Angers S., 2011. Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 27: 513–537. [DOI] [PubMed] [Google Scholar]
- Humke E. W., Dorn K. V., Milenkovic L., Scott M. P., Rohatgi R., 2010. The output of Hedgehog signaling is controlled by the dynamic association between suppressor of fused and the Gli proteins. Genes Dev. 24: 670–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy C. F., Kramer A., Berget S. M., 1998. A role for SRp54 during intron bridging of small introns with pyrimidine tracts upstream of the branch point. Mol. Cell. Biol. 18: 5425–5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent D., Bush E. W., Hooper J. E., 2006. Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development 133: 2001–2010. [DOI] [PubMed] [Google Scholar]
- Le Hir H., Sauliere J., Wang Z., 2016. The exon junction complex as a node of post-transcriptional networks. Nat. Rev. Mol. Cell Biol. 17: 41–54. [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L., 2001. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24: 251–254 (erratum: Trends Neurosci. 24: 385). [DOI] [PubMed] [Google Scholar]
- Malone C. D., Mestdagh C., Akhtar J., Kreim N., Deinhard P., et al. , 2014. The exon junction complex controls transposable element activity by ensuring faithful splicing of the piwi transcript. Genes Dev. 28: 1786–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methot N., Basler K., 1999. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell 96: 819–831. [DOI] [PubMed] [Google Scholar]
- Oh S., Kato M., Zhang C., Guo Y., Beachy P. A., 2015. A comparison of Ci/Gli activity as regulated by Sufu in Drosophila and mammalian Hedgehog response. PLoS One 10: e0135804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmeyer J. T., Kalderon D., 1998. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature 396: 749–753. [DOI] [PubMed] [Google Scholar]
- Organista M. F., Martin M., de Celis J. M., Barrio R., Lopez-Varea A., et al. , 2015. The Spalt transcription factors generate the transcriptional landscape of the Drosophila melanogaster wing pouch central region. PLoS Genet. 11: e1005370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak E., Segal R. A., 2016. Hedgehog signal transduction: key players, oncogenic drivers, and cancer therapy. Dev. Cell 38: 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. W., Parisky K., Celotto A. M., Reenan R. A., Graveley B. R., 2004. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc. Natl. Acad. Sci. USA 101: 15974–15979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova R., Joyner A. L., 2014. Roles for Hedgehog signaling in adult organ homeostasis and repair. Development 141: 3445–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preat T., 1992. Characterization of suppressor of fused, a complete suppressor of the fused segment polarity gene of Drosophila melanogaster. Genetics 132: 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant J. Y., Treisman J. E., 2010. Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochromatic gene. Cell 143: 238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E., Ashburner M., Bautista-Llacer R., Drummond J., Webster J., et al. , 2007. The DrosDel deletion collection: a Drosophila genomewide chromosomal deficiency resource. Genetics 177: 615–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakashita E., Tatsumi S., Werner D., Endo H., Mayeda A., 2004. Human RNPS1 and its associated factors: a versatile alternative pre-mRNA splicing regulator in vivo. Mol. Cell. Biol. 24: 1174–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smelkinson M. G., Zhou Q., Kalderon D., 2007. Regulation of Ci-SCFSlimb binding, Ci proteolysis, and hedgehog pathway activity by Ci phosphorylation. Dev. Cell 13: 481–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukachinsky H., Lopez L. V., Salic A., 2010. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J. Cell Biol. 191: 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervoort M., 2000. Hedgehog and wing development in Drosophila: a morphogen at work? Bioessays 22: 460–468. [DOI] [PubMed] [Google Scholar]
- Wu J. Y., Kar A., Kuo D., Yu B., Havlioglu N., 2006. SRp54 (SFRS11), a regulator for tau exon 10 alternative splicing identified by an expression cloning strategy. Mol. Cell. Biol. 26: 6739–6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Liu C., Zhao Y., 2015. Decoding Ci: from partial degradation to inhibition. Dev. Growth Differ. 57: 98–108. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Zhang L., Wang B., Ou C. Y., Chien C. T., et al. , 2006. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev. Cell 10: 719–729. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Shen L., Law K., Zhang Z., Liu X., et al. , 2016. Suppressor of fused chaperones Gli proteins to generate transcriptional responses to Sonic Hedgehog signaling. Mol. Cell. Biol. 37: e00421-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Kalderon D., 2010. Costal 2 interactions with Cubitus interruptus (Ci) underlying Hedgehog-regulated Ci processing. Dev. Biol. 348: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Kalderon D., 2011. Hedgehog activates fused through phosphorylation to elicit a full spectrum of pathway responses. Dev. Cell 20: 802–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully in the article. Drosophila stocks and other reagents are available upon request.