Abstract
Cis- and trans-regulatory mutations are important contributors to transcriptome evolution. Quantifying their relative contributions to intraspecific variation in gene expression is essential for understanding the population genetic processes that underlie evolutionary changes in gene expression. Here, we have examined this issue by quantifying genome-wide, allele-specific expression (ASE) variation using a crossing scheme that produces F1 hybrids between 18 different Drosophila melanogaster strains sampled from the Drosophila Genetic Reference Panel and a reference strain from another population. Head and body samples from F1 adult females were subjected to RNA sequencing and the subsequent ASE quantification. Cis- and trans-regulatory effects on expression variation were estimated from these data. A higher proportion of genes showed significant cis-regulatory variation (∼28%) than those that showed significant trans-regulatory variation (∼9%). The sizes of cis-regulatory effects on expression variation were 1.98 and 1.88 times larger than trans-regulatory effects in heads and bodies, respectively. A generalized linear model analysis revealed that both cis- and trans-regulated expression variation was strongly associated with nonsynonymous nucleotide diversity and tissue specificity. Interestingly, trans-regulated variation showed a negative correlation with local recombination rate. Also, our analysis on proximal transposable element (TE) insertions suggested that they affect transcription levels of ovary-expressed genes more pronouncedly than genes not expressed in the ovary, possibly due to defense mechanisms against TE mobility in the germline. Collectively, our detailed quantification of ASE variations from a natural population has revealed a number of new relationships between genomic factors and the effects of cis- and trans-regulatory factors on expression variation.
Keywords: allele-specific expression, cis-regulatory variation, nucleotide diversity, trans-regulatory variation, transposable element
EVOLUTIONARY changes in the patterns of gene expression have been suggested to have a substantial impact on organismal phenotypic evolution (Zuckerkandl and Pauling 1965). Therefore, mutations that change gene expression at either the cis- or trans-regulatory level are essential sources of species diversification. Cis-regulatory mutations in diploid organisms can be defined as those that change gene expression in an allele-specific manner; while trans-regulatory mutations influence gene expression in a diffusible manner, such as mutations in transcription factors (Emerson and Li 2010). Recent advances in large-scale quantification methods for examining gene expression have enabled us to quantify the relative contributions of cis- and trans-regulatory mutations to the evolution of gene expression. In Drosophila, comparisons of expression levels using chromosomal substitution lines (Hughes et al. 2006; Osada et al. 2006; Genissel et al. 2008; Lemos et al. 2008; Wang et al. 2008) have revealed a high estimated proportion (∼30–70%) of genes with significant intraspecific cis-regulatory variation. Conversely, parent-hybrid methods, which compare the allele-specific expression (ASE) level of the hybrid to the expression levels in its parental strains (Wittkopp et al. 2008; Suvorov et al. 2013; Coolon et al. 2014; Graze et al. 2014), have suggested smaller estimated proportions (∼5–50%).
One of the disadvantages of the previous experimental designs is that comparisons were made using a limited number of genotypes or strains. In addition, these studies typically relied on inbred strains or homozygous genotypes. As the effect size of trans-regulatory mutations is shown to be sensitive to masking in heterozygous genotypes (Lemos et al. 2008), some transcripts from highly inbred strains might be spuriously affected by rare recessive alleles. The Drosophila Genetics Reference Panel (DGRP) provides inbred Drosophila melanogaster strains derived from a natural population, with all genotypes publicly accessible (Ayroles et al. 2009; Mackay et al. 2012). Using these resources, we designed a crossing scheme to quantify the abundance of cis- and trans-regulatory expression variation within the population. This crossing scheme generates F1 hybrids from crosses between 18 different strains sampled from the DGRP and a reference strain from another population. Our method has advantages in that it does not rely on any particular pair of strains or require information on homozygous parental gene expression patterns (Gruber and Long 2009; Bickel et al. 2011; Miyagi et al. 2015). It also allowed us to compare different genomic parameters to cis- as well as trans-regulatory variation, which was difficult to unravel using prior experimental designs.
The genetic variation that underlies cis- and trans-regulated expression variation comprises SNPs and indels, which include transposable element (TE) insertions. Indeed, the presence or absence of TE insertions constitutes a considerable portion of the genetic variation observed between Drosophila genomes (Fontanillas et al. 2007; Kofler et al. 2012, 2015; Linheiro and Bergman 2012; Cridland et al. 2013). More than TE insertions have been identified in the DGRP (Linheiro and Bergman 2012; Mackay et al. 2012; Cridland et al. 2013, 2015), with the majority being unique (Cridland et al. 2013). TEs could potentially disrupt regulatory structures (Dunn and Laurie 1995; Lerman et al. 2003) or, in rare occasions, serve as novel enhancers (Chung et al. 2007). Most insertion events are likely to be deleterious for the host genome and therefore defense mechanisms against TE mobility have evolved (reviewed in Kavi et al. 2005 and Slotkin and Martienssen 2007). These include transcriptional and post-transcriptional silencing by the well-characterized Piwi-interacting RNA (piRNA) system present in germline cells (reviewed in Senti and Brennecke 2010 and Iwasaki et al. 2015). However, there are conflicting opinions as to whether transcriptional silencing through the modification of local chromatin states can spread to neighboring genes (Sienski et al. 2012; Le Thomas et al. 2013; Lee 2015). One prediction that has not yet been tested is that transcriptional perturbation by proximal TE insertion is limited to ovarian tissues, as TE silencing by the Piwi system is limited to germline cells and some somatic cells of the ovary. A detailed comparison of transcript-level changes due to proximal TE insertions in ovarian and nonovarian tissues may clarify this issue.
In this study, we have employed an outcrossing experimental design using a reference strain to extract within-population cis- and trans-regulatory variation and analyze genomic factors that associate with their effect sizes. We have also examined the effects of proximal TE insertions on transcriptional perturbation by comparing published TE-insertion panels to our precise ASE data.
Materials and Methods
Fly strains
The following inbred strains of D. melanogaster from the DGRP (Ayroles et al. 2009; Mackay et al. 2012; Massouras et al. 2012) were used in this study: RAL-208, RAL-324, RAL-335, RAL-358, RAL-360, RAL-365, RAL-375, RAL-379, RAL-380, RAL-486, RAL-517, RAL-555, RAL-707, RAL-774, RAL-786, RAL-799, RAL-820, and RAL-852. These strains were chosen arbitrarily from the 40 strains with initial microarray expression data (Ayroles et al. 2009), which were not used in this study. In addition, we used an inbred strain of D. melanogaster, Mel6 (G59), which originated from Benin, West Africa (Takahashi and Takano-Shimizu 2005), as a reference strain. Flies were kept at 25° and on a 12-hr light–dark cycle with standard cornmeal fly medium.
Genome sequences and single nucleotide variant calling
Genomic DNA was extracted from 100 female Mel6 flies using QIAGEN (Valencia, CA) Genomic-tip 100/G. Paired-end sequencing with a 100-bp read length was performed using an Illumina HiSeq2000. The genotype of Mel6 was determined with ∼160-fold coverage of resequencing data. Initial mapping was performed using Bowtie2 software (Langmead and Salzberg 2012) with the default parameter settings. In total, 99.999% of euchromatic regions had at least onefold coverage with high quality bases [Phred quality value (QV) ≥ 30], and 99.570% had 10-fold coverage with the same quality. Single nucleotide variant (SNV) calling was performed using the SAMtools mpileup command (version 0.1.17) with a genotype quality cut-off score of 40 (Li et al. 2009). Only homozygous SNVs (794,305) in protein-coding genes were considered in subsequent analysis. We also used the GATK SNV caller (UnifiedGenotyper) (McKenna et al. 2010) for SNV calling, applying the Best Practices workflow (DePristo et al. 2011; Van der Auwera et al. 2013). GATK is less stringent than SAMtools and reported more potential SNVs. However, almost all homozygous SNVs called by SAMtools also overlapped with SNVs called by GATK. As it was critical to avoid sequencing errors that could introduce strong bias for ASE, we chose to use only SAMtools-identified SNVs for analysis. The genome sequences of DGRP strains were downloaded from the project database (http://dgrp.gnets.ncsu.edu/). When constructed genome sequences were not available, paired-end short sequences were downloaded and SNVs were called using the same pipeline as described above.
RNA sequencing
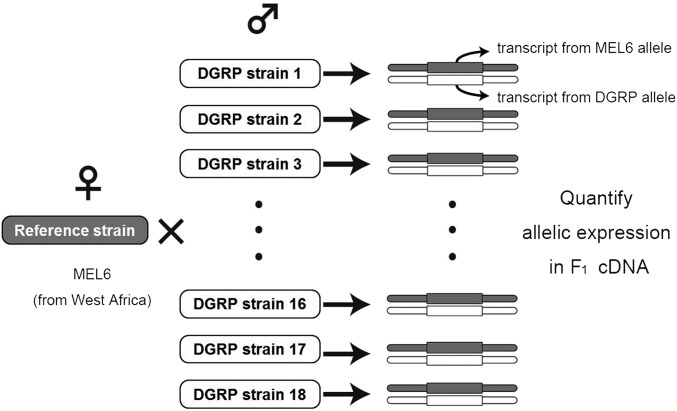
In total, males of 18 DGRP strains were crossed to females of the reference strain (Mel6). F1 hybrids were then subjected to RNA sequencing (RNA-seq) analyses (Figure 1). F1 virgin females and males from crosses were collected within 8 hr of eclosion and kept separately on regular food media. After 4–7 days, 100 flies per sample were flash frozen in liquid nitrogen within 1–2 hr after the lights were turned on, and kept in −80°. Heads were separated from bodies on dry ice by forcing frozen samples through a stainless mesh (opening: 710 μm) using a paintbrush. Appendages from both head and body that came off during this process fell through the second mesh (opening: 425 μm) and were discarded. The total RNA from head and headless body samples were extracted using a TRIzol Plus RNA Purification Kit (Life Technologies). The extracted total RNA was quantified using a Nanodrop 2000c (Thermo Fisher Scientific) and quality was assessed using a Bioanalyzer 2100 (Agilent Technologies). For RNA-seq, 250 ng of total RNA was used for library construction. Libraries were constructed using an Illumina TruSeq RNA-seq Sample Prep Kit. For sequencing, six samples were bar-code indexed and pooled on each lane. Single-end reads of 100 bp were obtained using an Illumina Hiseq2000 and two biological replicates with randomized bar-code indices were obtained for the female samples.
Figure 1.
Schematic figure describing the crossing design used in this study. Males (♂) from 18 DGRP strains were crossed with Mel6 females (♀). Transcripts from DGRP and Mel6 alleles in F1 females were quantified using RNA-seq. Since both alleles share the same trans-regulatory environment in the F1 nucleus, the differences in allele-specific transcript quantities can be attributed to cis-regulatory differences. Also, because the reference allele is present in all F1 samples, its allele-specific transcript quantities can be used to estimate trans-regulatory variation.
Estimation of ASE level
To accurately quantify allele-specific transcript abundance, the following procedures were employed. Initially, to reduce the mapping bias of reads to genomes with allelic differences (Degner et al. 2009; Satya et al. 2012), RNA-seq reads from each sample were mapped simultaneously onto the Mel6 and the corresponding DGRP genomes, using the TopHat2 program (Kim et al. 2013). Because these reconstructed genomes contained N’s in ambiguously defined regions, the reads were also mapped to the reference D. melanogaster genome (version dmel5.2) to minimize mapping error and to use the read-count information from those regions. Mapping information was subsequently merged into a single alignment file (details in Supplemental Material, Figure S1). The reference genome sequence was used to cover regions with ambiguously defined sequences, potentially causing mapping errors, in the Mel6 and DGRP genomes. The normalized expression level, measured in fragments per kilobase of transcript per million mapped reads (FPKM), for each gene was then estimated using Cufflinks and the upper-quartile normalization method (Trapnell et al. 2012).
Tag SNVs, which are the SNVs detected in both the genome sequence and the RNA-seq reads, were identified using RNA-seq reads and the genome sequences of the reference (Mel6) and DGRP strains. Several filtering criteria were applied to reduce mapping biases and errors. First, low quality bases (QV < 15) were filtered out. In addition, because we focused on relatively minor gene expression changes due to cis- and trans-regulatory mutations, genes that showed strong allelic expression bias [represented by low minor frequency SNVs (<0.05) due to sequencing errors] were also filtered. Tag SNVs within the 100-bp regions on either side of indels were also filtered out. After these filtering processes to reduce mapping biases (see Figure S2), genes that had a coverage >50 after summing up all tag SNVs within a gene were selected for further analysis. The level of ASE was estimated by dividing the FPKM values according to the ratio of tag SNVs. FPKM was log2 transformed (FPKMlog) before being used in ANOVA.
Estimation of cis- and trans-regulated gene expression variances
A type-II ANOVA was conducted to estimate the variances due to cis- and trans-regulatory effects for each gene using female ASE data with a biological replicate. Cis- and trans-regulatory effects were estimated separately using the following generalized linear model (GLM): ASE = μ + cis + trans + ϵ. Category assignments for cis and trans effects are shown in Figure S3. Because the experimental design was not orthogonal, cis-by-trans interaction could not be estimated, i.e., the cis effect was estimated with the given trans effect, and the trans effect was estimated with the given cis effect. The type-II ANOVA was performed using the CAR package in R (R Core Team 2016).
GLM approach using genomic factors
A GLM was formulated to analyze genomic factors that associated with the cis- and trans-regulatory effects on expression variation, Vcis and Vtrans, respectively. The FPKM values used for the analysis were calculated using FPKMlog means across F1 females from the 18 parental strain combinations (Figure 1). Tissue specificity index τ (Yanai et al. 2005) was calculated for each gene using tissue-specific expression level data from the FlyAtlas database (Chintapalli et al. 2007). For the calculation, 22 nonoverlapping tissues were chosen for the analyses (Table S1). Presence/absence calls and expression levels were estimated using the MAS5 and RMA algorithms implemented in the Affymetrix Expression Console, respectively. Genes were called as present when more than two out of four biological replicates showed statistically significant expression. The expression status of each gene in the ovary was also determined using this data set. Female bias in expression was calculated as the logit of female FPKM/(female FPKM + male FPKM) after male and female FPKMs were normalized by adjusting the median. Nucleotide diversities at synonymous sites and nonsynonymous sites were calculated using sequence data from the 18 DGRP genomes. Gene density was calculated as a proportion of the exonic regions per 100 kb. Recombination rates were obtained from Comeron et al. (2012). Enrichment of origin recognition complex (ORC) binding sites within 10 kb of the annotated gene region was calculated from the MA2C score obtained by chromatin immunoprecipitation with dORC2 antibody in asynchronous Kc167 cells (Gene Expression Omnibus accession: GSE17282; MacAlpine et al. 2010). A generalized liner regression analysis with a gamma distribution and a log link was performed using the glm function implemented in R (R Core Team 2016).
Analysis of the effect of TE insertions
TE insertion calls were obtained from Cridland et al. (2015). TE insertion was counted when the upstream or downstream breakpoint was within various distances from the annotated gene region. A simulation to generate Spearman’s correlation coefficients (ρ’s) between log-transformed ASEs of the removed and the remaining samples after random removal of strain(s) was performed using a custom script in R (R Core Team 2016).
Data availability
All raw sequence data were deposited in the DNA Data Bank of Japan Sequence Read Archive database (http://trace.ddbj.nig.ac.jp/dra/) with accession number DRA002265. The R code and raw data for conducting type-II ANOVA are in File S1 and those for generating Spearman’s correlation coefficients between ASEs of the removed and the remaining samples after random removal of strain(s) are in File S2.
Results
Quantification of cis- and trans-regulated gene expression variation
We designed a unique crossing experiment to estimate cis- and trans-regulatory variation within a population at the genome-wide level. Males from 18 inbred DGRP strains were crossed to females from the reference Mel6 strain. F1 females from these crosses were subjected to RNA-seq analyses (Figure 1). We limited our analysis to 3213 genes in the head and 3919 genes in the body that had a reliable number of read counts and SNV information to discriminate alleles (see Materials and Methods). To quantitatively evaluate the influence of cis- and trans-regulatory effects, we estimated the ASE levels in units of FPKM, rather than using the ratios of specifically mapped RNA-seq reads to one of the parental chromosomes. Because FPKM values are known to have a log-normal distribution (Bengtsson et al. 2005), an allele-specific FPKM was treated as a normal variate after log transformation. Log-transformed ASE levels for each gene were then subjected to a type-II ANOVA to estimate the variances due to cis-regulatory (Vcis) and trans-regulatory effects (Vtrans) (Figure S3). Calculated Vcis, and Vtrans values are listed in Table S2 and Table S3. These estimates showed that a large fraction of the variance in differentially expressed genes between genotypes is explained by cis-regulatory changes. The ratios averaged across genes were ∼1.98 and ∼1.88 for heads and bodies, respectively.
GLM approach on cis- and trans-regulated gene expression variation
To investigate potential factors that associate with the sizes of cis- and trans-regulatory effects on gene expression, multivariate analyses using a GLM were conducted. or (dependent variables) and eight independent variables were considered. These include expression properties (FPKM, τ, female bias), nucleotide diversities and regional properties (gene density, recombination rate, and ORC enrichment) (Table S2 and Table S3). The summaries of the head and body anal0yses are shown in Table 1. The variables with the most significant effects on cis-regulatory contribution (Vcis) were tissue specificity index (τ) and local nucleotide diversity of nonsynonymous sites in both head and body tissues. A negative correlation between and gene density was observed in both head and body tissues. strongly correlated with τ and also moderately with Interestingly, associated with the local recombination rate in both head and body tissues, which indicated that genes in regions with high recombination had slightly smaller trans-regulatory contributions.
Table 1. Effects of expression properties, genetic diversities, and regional properties on Vcis and Vtrans by GLM analysis.
| Variables | Head (N = 3044) | Body (N = 3681) | ||
|---|---|---|---|---|
| Sign of coefficient | P-value | Sign of coefficient | P-value | |
| Expression properties | ||||
| FPKM | + | 0.533 | − | 0.498 |
| τa | + | <10−7*** | + | <10−6*** |
| Female bias | + | 0.210 | − | <10−3*** |
| Nucleotide diversity | ||||
| + | 0.486 | + | 0.046* | |
| + | <10−5*** | + | <10−4*** | |
| Regional properties | ||||
| Gene densityb | − | 0.045* | − | 0.002** |
| Recombination ratec | − | 0.026* | − | 0.180 |
| ORC enrichmentd | − | 0.859 | + | 0.625 |
| Expression properties | ||||
| FPKM | − | 0.222 | − | <10−3*** |
| τa | + | <10−13*** | + | <10−15*** |
| Female bias | + | 0.159 | + | 0.428 |
| Nucleotide diversity | ||||
| − | 0.944 | + | 0.084 | |
| + | 0.001** | + | 0.011* | |
| Regional properties | ||||
| Gene densityb | − | 0.393 | − | 0.076 |
| Recombination ratec | − | 0.028* | − | <10−3*** |
| ORC enrichmentd | + | 0.936 | − | 0.026* |
P < 0.05, ** P < 0.01, *** P < 0.001.
Tissue-specific index (Yanai et al. 2005).
Gene density (proportion of exonic regions) per 100 kb.
Local recombination rate estimate from Comeron et al. (2012).
Enrichment of ORC binding site within 10 kb (MacAlpine et al. 2010).
TE insertion and cis-regulatory variation
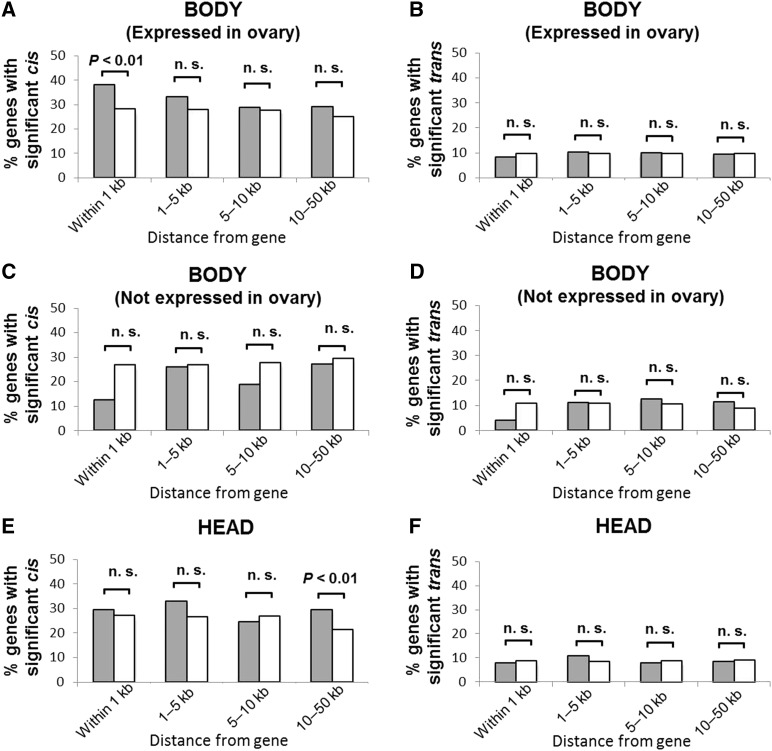
TE-insertion sites have been mapped in the genomes of DGRP flies (Linheiro and Bergman 2012; Mackay et al. 2012; Cridland et al. 2013, 2015), and detailed investigations by Cridland et al. (2015) has shown that TE insertions profoundly affect the expression of closely located genes. Their analysis was conducted using microarray data and inbred strains. To further investigate the effects of TE insertions on cis- and trans-regulatory variations, the proportions of genes with significant cis- and trans-regulatory variations were compared between genes with TEs inserted within various distances and those with no TE insertion (Figure 2). Genes expressed in the ovary were analyzed separately from genes not expressed in this tissue to investigate the effect of TE silencing mechanisms present in germline cells. Ovary-expressed genes were defined as those that showed statistically significant expression in the ovary in two out of four biological replicates in Chintapalli et al. (2007) (see Materials and Methods). A higher proportion of genes with a significant cis-regulatory contribution to expression variation was found in ovary-expressed genes that had a TE insertion within 1 kb (in ≥1 strain), compared to genes with no TE insertion (absent in all strains) within 1 kb (Figure 2A). There was no difference in the proportion of genes with significant cis-regulatory variation between TE-inserted and TE-absent genes within any distance for genes not expressed in ovary (Figure 2C). This was also demonstrated in head genes, except for genes with a TE insertion within 10–50 kb (Figure 2E). This distant effect may not be a direct influence of the inserted TE, but is likely linked to genomic properties associated with regional TE abundances. No difference in the proportion of genes with significant trans-regulatory variation was detected between TE-inserted and TE-absent genes within any distance (Figure 2, B, D, and F). These results suggested that TE insertion within 1 kb of the gene is associated with a larger cis-regulatory contribution to expression variation, but only in ovary-expressed genes.
Figure 2.
Proportion of genes that showed significant cis- or trans-regulatory contribution to expression variation. The proportion of genes with (shaded bar) or without (open bar) TE insertions within various distances that showed significant cis-regulatory variation in (A) body genes expressed in the ovary (N = 3312), (C) body genes not expressed in the ovary (N = 412), and (E) head genes (N = 3213). Additionally, genes that showed significant trans-regulatory variation in (B) body genes expressed in the ovary, (D) body genes not expressed in the ovary, and (F) head genes. P-values after Bonferroni correction for multiple (four) tests using χ2-tests are shown. n.s., nonsignificant pairs.
We next investigated the direct effects of TE insertions on ASEDGRP (DGRP allele-specific FPKMlog). Our data did not indicate any significant directional changes in ASEDGRP that associated with TE insertions within 1 kb (body genes expressed in ovary: paired t-test, t = −1.01, d.f. = 251, P = 0.31; body genes not expressed in ovary: paired t-test, t = 0.14, d.f. = 48, P = 0.89; head genes: paired t-test, t = −0.95, d.f. = 344, P = 0.34). On average, the ASEDGRP of the samples with TE insertions was reduced by 9.0% SD in ovary-expressed body genes and by 0.4% SD in body genes not expressed in the ovary. In head genes, this was reduced by 4.8% SD. The slight reduction in ASEDGRP for samples with TE insertions supports the finding reported by Cridland et al. (2015) that the general effect of TE insertion on nearby genes is to reduce their expression levels.
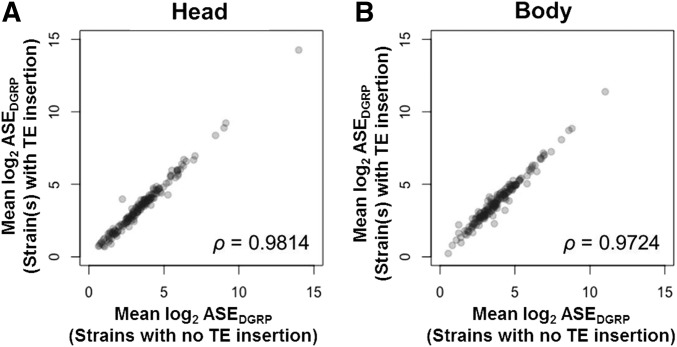
The comparison of ASEDGRP among ovary-expressed genes commonly expressed and analyzed in both heads and bodies (n = 174) showed that Spearman’s correlation coefficient (ρ) between ASEs of the strain(s) with and without TE insertions were slightly lower in bodies (ρ = 0.9724, Figure 3B) relative to heads (ρ = 0.9814, Figure 3A). To investigate if this was due to differential effects caused by TE insertions on ASEDGRP between heads and bodies, we conducted a simulation by randomly removing the same number of sample(s) as those of TE-inserted strain(s) (within 1 kb) for each gene. Spearman’s ρ-values calculated between ASEDGRP of the removed and the remaining samples after the random removals were obtained from 10,000 iterations and compared to the observed values. In heads, the probability for observing ρ < 0.9814 was 0.0819; whereas in bodies, the probability for observing ρ < 0.9724 was 0.0056. This indicated that the effects of TE insertions within 1 kb on the ASEDGRP of ovary-expressed genes were significant in the body but not in the head. Therefore, TE-associated elevation of cis-regulatory expression variation is likely linked to changes in transcription level in the ovary.
Figure 3.
Effect of removing strains with TE insertions on ASE. Comparisons of mean log-transformed ASE of DGRP alleles (ASEDGRP) between strains with no TE insertion within 1 kb (x-axis) and strain(s) with a TE insertion within 1 kb (y-axis), using 174 genes commonly found in heads and bodies. (A) ASE in heads. Spearman’s ρ = 0.9814, P < (B) ASE in bodies. Spearman’s ρ = 0.9724, P <
Discussion
Our study has quantified the segregating cis- and trans-regulatory effects on expression variation in a natural population of Drosophila using a unique strategy. Our method has several advantages over other approaches. First, while many studies have used a small number of strains or genotypes, our design quantified variation among 18 strains originally sampled from a natural population. Therefore, our experimental design is suitable for capturing and characterizing naturally segregating regulatory variation. Second, it employs an outbred crossing scheme and expression levels are compared in a heterozygous state, more closely resembling individual conditions in a natural population. Many studies using Drosophila rely on measurements of expression differences between inbred strains or homozygous genotypes that could be biased by the effects of rare recessive alleles that are normally masked. Indeed in Drosophila, a large proportion of gene expression differences between homozygous inbred strains can be concealed by heterozygous individuals (Lemos et al. 2008). We should note, however, that the effect of recessive alleles is a less compelling issue in yeast studies, because outcrossing frequency is generally low in Saccharomyces (Johnson et al. 2004; Aa et al. 2006; Ruderfer et al. 2006).
There are also some caveats when interpreting outcomes from our study. First, cis-regulatory effects on expression variance analyzed by a type-II ANOVA were calculated from the differences among 18 DGRP genomes plus the reference genome from an African population (Figure S3). If interpopulation expression differences are considerably large, our estimates may inflate the variances to some extent. Second, our model does not take into account the effect of epistatic cis-by-trans interactions. Particularly, compensatory interactions (cis- and trans-regulatory changes with opposite effect on gene expression) are often detected in the analyses using closely related species and their hybrids in yeast (Tirosh et al. 2009; Metzger et al. 2017) as well as in Drosophila (Landry et al. 2005; McManus et al. 2010). Information on the prevalence of such regulatory interactions within species is still limited, but their effect on expression variation may not be negligible in some genes (Genissel et al. 2008; Suvorov et al. 2013; Miyagi et al. 2015). Finally, a shortcoming of using the outbred crossing design is that this method is likely to miss recessive regulatory mutations, which also have influences on the dynamics of transcriptome evolution (Lemos et al. 2008; Gruber et al. 2012; Metzger et al. 2017). Expression quantity data from all the parental inbred strains used in this study would allow us to directly compare and accurately determine the differences between our method and the more commonly used methods based on parent-hybrid comparisons. Conducting such comparisons in the future would help elucidate precise genetic architectures underlying regulatory variations.
Using our method, we have shown that expression variation due to cis-regulatory effects (Vcis) was about twice as large as variation due to trans-regulatory effects (Vtrans). In addition, a higher proportion of genes showed significant cis-regulatory variation (∼28%) than trans-regulatory variation (∼9%), although these results are sensitive to the gene set we have analyzed and ASE variances between replicates, which were relatively large in our samples. Enrichment of the cis- regulatory effects on expression variation was consistent with previous studies that employed different experimental designs using Drosophila (Osada et al. 2006; Genissel et al. 2008; Lemos et al. 2008; Wittkopp et al. 2008; Graze et al. 2014), but see Wayne et al. (2004), Wang et al. (2008), and Suvorov et al. (2013). Our estimate was also similar to the proportion of cis-expression QTL (eQTL)-associated transcripts detected in the DGRP, which was 26% of the 7889 genes tested at a false discovery rate of <10% (Massouras et al. 2012).
Regarding trans-regulatory contribution, our analysis using the outbred crossing design may have provided smaller estimates compared to studies using pure inbred strains. This may be because the effect size of trans-regulatory mutations is particularly sensitive to masking in heterozygous genotypes (Lemos et al. 2008). In yeast, possibly due to low outcrossing rate (thus a small masking effect), contribution of trans-regulatory effect on intraspecific expression variation is estimated to be more extensive compared to cis-regulatory effect in studies using eQTL (Brem et al. 2002; Yvert et al. 2003) and ASE (Wang et al. 2007; Sung et al. 2009; Emerson et al. 2010). Interestingly, when a different timescale is considered, cis-regulatory changes play a larger role in shaping expression divergence between species than expression variation within species in yeast (Emerson et al. 2010; Metzger et al. 2017) as well as in Drosophila (Wittkopp et al. 2008; Coolon et al. 2014).
A positive correlation between local nucleotide diversity and expression variation has been shown repeatedly in various organisms, including Saccharomyces cerevisiae (Ronald et al. 2005), D. simulans (Lawniczak et al. 2008), and Arabidopsis thaliana (Kliebenstein et al. 2006). By separately analyzing the cis- and trans-regulatory contributions, our GLM analysis (Table 1) has depicted a strong association between the level of nonsynonymous site nucleotide diversity and both cis- and trans-regulatory variation (Vcis and Vtrans, respectively). However, there was no significant association detected between and nor between and Because and strongly correlate with each other (heads: Spearman’s ρ = 0.36, P < 10–15; bodies: Spearman’s ρ = 0.37, P < 10–15), both do correlate with and using univariate regression, but in our GLM model, is more strongly associated with and than This relationship parallels reports indicating a positive correlation between rates of protein divergence and expression divergence in Drosophila (Nuzhdin et al. 2004; Lemos et al. 2005; Good et al. 2006; but see Larracuente et al. 2008). Our data have also shown strong positive correlations between the tissue specificity index (τ) and both and (Table 1). This is consistent with the picture obtained from protein divergence analysis that demonstrated that broadly expressed genes (with small τ) tend to be under stronger purifying selection (Larracuente et al. 2008). It is noteworthy that not only the expression variation due to cis-regulatory effects but that due to trans-regulatory effects is also coupled to the overall constraint on amino acid sequences.
The negative correlation observed between recombination rate and (Table 1) is contradictory to the well-described positive correlation that exists between recombination rate and nucleotide polymorphism in Drosophila (Begun and Aquadro 1992; Andolfatto and Przeworski 2001; Presgraves 2005; Shapiro et al. 2007; Comeron et al. 2012; Campos et al. 2014). There was also no clear relationship between recombination rate and These results suggest that the degree of expression variation is largely uncoupled to linked targets of natural selection shaping nucleotide polymorphism patterns along recombining chromosomes. Factors that determine recombination rates are not fully understood in Drosophila, but an association between recombination rate and active transcription during early meiosis has been reported (Adrian and Comeron 2013). Although the exact causal relationship underlying the negative correlation observed between recombination rate and is not clear at this point, recombination rate may be associated with an unknown property (e.g., chromatin accessibility) that affects transcriptional activity controlled by trans-acting factors.
Our results also suggest that TE insertions within 1 kb of a gene associate with a greater contribution of cis-regulation to expression variation in ovary-expressed genes (Figure 2). A comparison of the direct effects of TE insertions on expression levels between heads and bodies using the same set of ovary-expressed genes clearly showed that proximal TE insertion within 1 kb perturbs transcription in bodies but not detectably in heads (Figure 3). Transcripts from the body are from a heterogeneous set of tissues, but should sufficiently reflect transcripts from ovarian tissues. Therefore, these results indicate that proximal TE insertions are likely to affect transcription in the ovary but not as strongly in other tissues at a detectable level. TE insertions can potentially affect transcription by physically disrupting the regulatory sequences of nearby genes, but in this case, insertions should affect all tissues equally and there is no reason to expect that this effect should be stronger in ovary-expressed genes.
The ovary-specific effect of TE insertions on transcription may arise from their mobility in germline cells. Due to the deleterious effects of TE mobility, ovarian tissues of flies have acquired defense mechanisms against it (reviewed in Kavi et al. 2005 and Slotkin and Martienssen 2007). A well-described mechanism in Drosophila germline cells is the piRNA system, which enforces transposition repression by affecting local chromatin states (Klenov et al. 2007; Malone and Hannon 2009; Siomi et al. 2011; Wang and Elgin 2011; Sienski et al. 2012; Le Thomas et al. 2013). Therefore, there is a possibility that epigenetic modifications in the regions surrounding TE insertion sites may perturb the transcription levels of proximal genes in the ovary.
Despite evidence for TE silencing through modifications of local chromatin states, whether repressive states at TE insertion sites can spread to the surrounding DNA has remained unknown. It has been shown using ovarian cell culture that the repressive chromatin structure induced by piRNA-mediated TE silencing can potentially spread from silenced TE sequences to adjacent genes (Sienski et al. 2012), but this effect was not detected in the ovaries of adult flies (Le Thomas et al. 2013). Nevertheless, Lee (2015) has used TE panels of DGRP flies and compared them to the modENCODE H3K9me (Nègre et al. 2011) and published piRNA data (Shpiz et al. 2014). This has revealed that the repressive chromatin mark is elevated in sequences adjacent to TE insertions and also that the heterochromatic state of the gene depends on whether the nearest TE is targeted by piRNA. Therefore, at least from these genome-wide comparative analyses (Lee 2015), it can be concluded that repressive chromatin-marked regions are capable of spreading and affecting the transcription of adjacent genes. However, the relative effect sizes of transcription perturbation in adjacent genes, due to chromatin modification and physical disruption of the regulatory sequences, are yet to be fully investigated. Our data indicated a stronger association between TE insertions and the degree of transcription perturbation in ovary tissue, supporting the view that adjacent gene transcription is affected by defense-associated chromatin modification.
In summary, dichotomizing expression variation into cis- and trans-regulatory effects using our outcrossing design has revealed that (1) cis-regulatory variation is more prominent than trans-regulatory variation; (2) the degree of purifying selection on coding sequences is reflected in the size of cis-regulatory variation and also, to a smaller but considerable extent, in the size of trans-regulatory variation; and (3) unlike nucleotide diversity, expression variation is largely uncoupled to the polymorphism landscape positively correlated with local recombination rate. Furthermore, our precise quantification of transcript levels suggested that TE insertions, even those that are present in a natural population, may affect the expression levels of proximal genes through TE silencing mechanisms in the ovary.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.201459/-/DC1.
Acknowledgments
We thank Y. Hiromi for providing fly food, the Bloomington Stock Center for the fly stocks, and Hokkaido System Science for running the HiSeq2000. This work was supported by Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (no. 23113008) and by the Sumitomo Foundation (no. 160999).
Footnotes
Communicating editor: D. A. Barbash
Literature Cited
- Aa E., Townsend J. P., Adams R. I., Nielsen K. M., Taylor J. W., 2006. Population structure and gene evolution in Saccharomyces cerevisiae. FEMS Yeast Res. 6: 702–715. [DOI] [PubMed] [Google Scholar]
- Adrian A. B., Comeron J. M., 2013. The Drosophila early ovarian transcriptome provides insight to the molecular causes of recombination rate variation across genomes. BMC Genomics 14: 794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P., Przeworski M., 2001. Regions of lower crossing over harbor more rare variants in African populations of Drosophila melanogaster. Genetics 158: 657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles J. F., Carbone M. A., Stone E. A., Jordan K. W., Lyman R. F., et al. , 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 41: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun D. J., Aquadro C. F., 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356: 519–520. [DOI] [PubMed] [Google Scholar]
- Bengtsson M., Ståhlberg A., Rorsman P., Kubista M., 2005. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 15: 1388–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel R. D., Kopp A., Nuzhdin S. V., 2011. Composite effects of polymorphisms near multiple regulatory elements create a major-effect QTL. PLoS Genet. 7: e1001275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R. B., Yvert G., Clinton R., Kruglyak L., 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755. [DOI] [PubMed] [Google Scholar]
- Campos J. L., Halligan D. L., Haddrill P. R., Charlesworth B., 2014. The relation between recombination rate and patterns of molecular evolution and variation in Drosophila melanogaster. Mol. Biol. Evol. 31: 1010–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J., Dow J. A. T., 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39: 715–720. [DOI] [PubMed] [Google Scholar]
- Chung H., Bogwitz M. R., McCart C., Andrianopoulos A., Ffrench-Constant R. H., et al. , 2007. Cis-regulatory elements in the Accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics 175: 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolon J. D., McManus C. J., Stevenson K. R., Graveley B. R., Wittkopp P. J., 2014. Tempo and mode of regulatory evolution in Drosophila. Genome Res. 24: 797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland J. M., Macdonald S. J., Long A. D., Thornton K. R., 2013. Abundance and distribution of transposable elements in two Drosophila QTL mapping resources. Mol. Biol. Evol. 30: 2311–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland J. M., Thornton K. R., Long A. D., 2015. Gene expression variation in Drosophila melanogaster due to rare transposable element insertion alleles of large effect. Genetics 199: 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner J. F., Marioni J. C., Pai A. A., Pickrell J. K., Nkadori E., et al. , 2009. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics 25: 3207–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M., Banks E., Poplin R., Garimella K., Maguire J., et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn R. C., Laurie C. C., 1995. Effects of a transposable element insertion on alcohol dehydrogenase expression in Drosophila melanogaster. Genetics 140: 667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson J. J., Li W.-H., 2010. The genetic basis of evolutionary change in gene expression levels. Phil. Trans. R. Soc. B. 365: 2581–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson J. J., Hsieh L. C., Sung H. M., Wang T. Y., Huang C. J., et al. , 2010. Natural selection on cis and trans regulation in yeasts. Genome Res. 20: 826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanillas P., Hartl D. L., Reuter M., 2007. Genome organization and gene expression shape the transposable element distribution in the Drosophila melanogaster euchromatin. PLoS Genet. 3: e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genissel A., McIntyre L. M., Wayne M. L., Nuzhdin S. V., 2008. Cis and trans regulatory effects contribute to natural variation in transcriptome of Drosophila melanogaster. Mol. Biol. Evol. 25: 101–110. [DOI] [PubMed] [Google Scholar]
- Good J. M., Hayden C. A., Wheeler T. J., 2006. Adaptive protein evolution and regulatory divergence in Drosophila. Mol. Biol. Evol. 23: 1101–1103. [DOI] [PubMed] [Google Scholar]
- Graze R. M., McIntyre L. M., Morse A. M., Boyd B. M., Nuzhdin S. V., et al. , 2014. What the X has to do with it: differences in regulatory variability between the sexes in Drosophila simulans. Genome Biol. Evol. 6: 818–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber J. D., Long A. D., 2009. Cis-regulatory variation is typically polyallelic in Drosophila. Genetics 181: 661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber J. D., Vogel K., Kalay G., Wittkopp P. J., 2012. Contrasting properties of gene-specific regulatory, coding, and copy number mutations in Saccharomyces cerevisiae: frequency, effects, and dominance. PLoS Genet. 8: e1002497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K. A., Ayroles J. F., Reedy M. M., Drnevich J. M., Rowe K. C., et al. , 2006. Segregating variation in the transcriptome: Cis regulation and additivity of effects. Genetics 173: 1347–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki Y. W., Siomi M. C., Siomi H., 2015. PIWI-interacting RNA: its biogenesis and functions. Annu. Rev. Biochem. 84: 405–433. [DOI] [PubMed] [Google Scholar]
- Johnson L. J., Koufopanou V., Goddard M. R., Hetherington R., Schäfer S. M., et al. , 2004. Population genetics of the wild yeast Saccharomyces paradoxus. Genetics 166: 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavi H. H., Fernandez H. R., Xie W., Birchler J. A., 2005. RNA silencing in Drosophila. FEBS Lett. 579: 5940–5949. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., et al. , 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov M. S., Lavrov S. A., Stolyarenko A. D., Ryazansky S. S., Aravin A. A., et al. , 2007. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 35: 5430–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D. J., West M. A., van Leeuwen H., Kim K., Doerge R. W., et al. , 2006. Genomic survey of gene expression diversity in Arabidopsis thaliana. Genetics 172: 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R., Betancourt A. J., Schlötterer C., 2012. Sequencing of pooled DNA samples (Pool-Seq) uncovers complex dynamics of transposable element insertions in Drosophila melanogaster. PLoS Genet. 8: e1002487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R., Nolte V., Schlötterer C., 2015. Tempo and mode of transposable element activity in Drosophila. PLoS Genet. 11: e1005406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry C. R., Wittkopp P. J., Taubes C. H., Ranz J. M., Clark A. G., et al. , 2005. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics 171: 1813–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente A. M., Sackton T. B., Greenberg A. J., Wong A., Singh N. D., et al. , 2008. Evolution of protein-coding genes in Drosophila. Trends Genet. 24: 114–123. [DOI] [PubMed] [Google Scholar]
- Lawniczak M. K., Holloway A. K., Begun D. J., Jones C. D., 2008. Genomic analysis of the relationship between gene expression variation and DNA polymorphism in Drosophila simulans. Genome Biol. 9: R125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. C., 2015. The role of piRNA-mediated epigenetic silencing in the population dynamics of transposable elements in Drosophila melanogaster. PLoS Genet. 11: e1005269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B., Bettencourt B. R., Meiklejohn C. D., Hartl D. L., 2005. Evolution of proteins and gene expression levels are coupled in Drosophila and are independently associated with mRNA abundance, protein length, and number of protein-protein interactions. Mol. Biol. Evol. 22: 1345–1354. [DOI] [PubMed] [Google Scholar]
- Lemos B., Araripe L. O., Fontanillas P., Hartl D. L., 2008. Dominance and the evolutionary accumulation of cis- and trans-effects on gene expression. Proc. Natl. Acad. Sci. USA 105: 14471–14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman D. N., Michalak P., Helin A. B., Bettencourt B. R., Feder M. E., 2003. Modification of heat-shock gene expression in Drosophila melanogaster populations via transposable elements. Mol. Biol. Evol. 20: 135–144. [DOI] [PubMed] [Google Scholar]
- Le Thomas A., Rogers A. K., Webster A., Marinov G. K., Liao S. E., et al. , 2013. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 27: 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linheiro R. S., Bergman C. M., 2012. Whole genome resequencing reveals natural target site preferences of transposable elements in Drosophila melanogaster. PLoS One 7: e30008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine H. K., Gordan R., Powell S. K., Hartemink A. J., MacAlpine D. M., 2010. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 20: 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster genetic reference panel. Nature 482: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C. D., Hannon G. J., 2009. Small RNAs as guardians of the genome. Cell 136: 656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massouras A., Waszak S. M., Albarca-Aguilera M., Hens K., Holcombe W., et al. , 2012. Genomic variation and its impact on gene expression in Drosophila melanogaster. PLoS Genet. 8: e1003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus C. J., Coolon J. D., Duff M. O., Eipper-Mains J., Graveley B. R., et al. , 2010. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 6: 816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger B. P. H., Wittkopp P. J., Coolon J. D., 2017. Evolutionary dynamics of regulatory changes underlying gene expression divergence among Saccharomyces species. Genome Biol. Evol. 9: 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi R., Akiyama N., Osada N., Takahashi A., 2015. Complex patterns of cis-regulatory polymorphisms in ebony underlie standing pigmentation variation in Drosophila melanogaster. Mol. Ecol. 24: 5829–5841. [DOI] [PubMed] [Google Scholar]
- Nègre N., Brown C. D., Ma L., Aaron Bristow C., Miller S. W., et al. , 2011. A cis-regulatory map of the Drosophila genome. Nature 471: 527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin S. V., Wayne M. L., Harmon K. L., McIntyre L. M., 2004. Common pattern of evolution of gene expression level and protein sequence in Drosophila. Mol. Biol. Evol. 21: 1308–1317. [DOI] [PubMed] [Google Scholar]
- Osada N., Kohn M. H., Wu C.-I., 2006. Genomic inferences of the cis-regulatory nucleotide polymorphisms underlying gene expression differences between Drosophila melanogaster mating races. Mol. Biol. Evol. 23: 1585–1591. [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., 2005. Recombination enhances protein adaptation in Drosophila melanogaster. Curr. Biol. 15: 1651–1656. [DOI] [PubMed] [Google Scholar]
- R Core Team, 2016 R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna, Austria. Available at: https://www.R-project.org/.
- Ronald J., Brem R. B., Whittle J., Kruglyak L., 2005. Local regulatory variation in Saccharomyces cerevisiae. PLoS Genet. 1: e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderfer D. M., Pratt S. C., Seidel H. S., Kruglyak L., 2006. Population genomic analysis of outcrossing and recombination in yeast. Nat. Genet. 38: 1077–1081. [DOI] [PubMed] [Google Scholar]
- Satya R. V., Zavaljevski N., Reifman J., 2012. A new strategy to reduce allelic bias in RNA-Seq readmapping. Nucleic Acids Res. 40: e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti K. A., Brennecke J., 2010. The piRNA pathway: a fly’s perspective on the guardian of the genome. Trends Genet. 26: 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A., Huang W., Zhang C., Hubisz M. J., Lu J., et al. , 2007. Adaptive genic evolution in the Drosophila genomes. Proc. Natl. Acad. Sci. USA 104: 2271–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S., Ryazansky S., Olovnikov I., Abramov Y., Kalmykova A., 2014. Euchromatic transposon insertions trigger production of novel pi- and endo-siRNAs at the target sites in the Drosophila germline. PLoS Genet. 10: e1004138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G., Donertas D., Brennecke J., 2012. Transcriptional silencing of transposons by Piwi and Maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi M. C., Sato K., Pezic D., Aravin A. A., 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12: 246–258. [DOI] [PubMed] [Google Scholar]
- Slotkin R. K., Martienssen R., 2007. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8: 272–285. [DOI] [PubMed] [Google Scholar]
- Sung H. M., Wang T. Y., Wang D., Huang Y. S., Wu J. P., et al. , 2009. Roles of trans and cis variation in yeast intraspecies evolution of gene expression. Mol. Biol. Evol. 26: 2533–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorov A., Nolte V., Pandey R. V., Franssen S. U., Futschik A., et al. , 2013. Intra-specific regulatory variation in Drosophila pseudoobscura. PLoS One 8: e83547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A., Takano-Shimizu T., 2005. A high-frequency null mutant of an odorant-binding protein gene, Obp57e, in Drosophila melanogaster. Genetics 170: 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I., Reikhav S., Levy A. A., Barkai N., 2009. A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324: 659–662. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera G. A., Carneiro M., Hartl C., Poplin R., del Angel G., et al. , 2013. From FastQ data to high-confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43: 11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Sung H. M., Wang T. Y., Huang C. J., Yang P., et al. , 2007. Expression evolution in yeast genes of single-input modules is mainly due to changes in trans-acting factors. Genome Res. 17: 1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.-Y., Fu Y., McPeek M. S., Lu X., Nuzhdin S., et al. , 2008. Complex genetic interactions underlying expression differences between Drosophila races: analysis of chromosome substitutions. Proc. Natl. Acad. Sci. USA 105: 6362–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. H., Elgin S. C. R., 2011. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc. Natl. Acad. Sci. USA 108: 21164–21169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne M. L., Pan Y. J., Nuzhdin S. V., McIntyre L. M., 2004. Additivity and trans-acting effects on gene expression in male Drosophila simulans. Genetics 168: 1413–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp P. J., Haerum B. K., Clark A. G., 2008. Regulatory changes underlying expression differences within and between Drosophila species. Nat. Genet. 40: 346–350. [DOI] [PubMed] [Google Scholar]
- Yanai I., Benjamin H., Shmoish M., Chalifa-Caspi V., Shklar M., et al. , 2005. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21: 650–659. [DOI] [PubMed] [Google Scholar]
- Yvert G., Brem R. B., Whittle J., Akey J. M., Foss E., et al. , 2003. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat. Genet. 35: 57–64. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L., 1965. Evolutionary divergence and convergence in proteins, pp. 97–166 in Evolving Genes and Proteins, edited by Bryson V., Vogel H. J. Academic Press, New York. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequence data were deposited in the DNA Data Bank of Japan Sequence Read Archive database (http://trace.ddbj.nig.ac.jp/dra/) with accession number DRA002265. The R code and raw data for conducting type-II ANOVA are in File S1 and those for generating Spearman’s correlation coefficients between ASEs of the removed and the remaining samples after random removal of strain(s) are in File S2.