Abstract
Background
Ventral incisional hernia is the most common long-term complication after abdominal surgery. Among newly-diagnosed colorectal cancer patients, we screened the pre-surgical plasma proteome to explore predictive markers for the development of an incisional hernia.
Methods
We utilized pre-operative plasma samples of 72 newly diagnosed colorectal cancer patients who underwent midline incision for tumor resection between 2010 and 2013. 21 patients with incisional hernia occurrence were matched with 51 patients with at least 18 months follow-up without an incisional hernia by gender, age, and BMI. To assess predictive markers of incisional hernia risk we screened the plasma proteome for >2,000 distinct proteins using a well-validated antibody microarray test. Paired t-tests were used to compare protein levels between cases and controls. A gene-set-enrichment analysis (Gene Ontology and KEGG) was applied to test for differences in signaling pathways between the two groups.
Results
The proteome screen identified 25 proteins that showed elevated or reduced plasma levels in the hernia group compared to the control group (nominal p-values <0.05). Several proteins were in pathways associated with wound healing (CCL21, SHBG, BRF2) or cell adhesion (PCDH15, CDH3, EPCAM).
Conclusion
Our study shows that there are multiple individual and groups of plasma proteins that could feasibly predict the personal hernia risk prior to undergoing surgery. Further investigations in larger independent sample sets are warranted to replicate findings and validate clinical utility of potential biomarkers. After validation, such a biomarker could be incorporated into a multifactorial risk model to guide clinical decision making.
Introduction
Ventral incisional hernia is the most common long-term complication after laparotomy and is a continuing problem for the abdominal surgeon. By definition, hernias develop due to a defect in an aponeurotic tissue layer, e.g. at the incisional site following abdominal surgery, leading to the protrusion of an organ out of its natural cavity.1 Incisional hernias occur more frequently after midline incisions compared with a transverse incisional approach.2 Incisional hernia incidence after elective midline incision, e.g., for the oncologic resection of colorectal cancer, is high with reported 10–40%.3, 4 Most incisional hernias develop within 2 years after surgery5 and can cause severe health and cosmetic problems. Each year, over 340,000 hernia repairs are performed in the United States, causing health care costs of at least $3.2 billion.6 These figures illustrate the tremendous economic burden associated with the incidence and repair of incisional hernia, and, consequently, demonstrate the importance of a multidirectional approach in the prevention of this complication. Preventive measures are available, such as prophylactic mesh implantation, leading to a reduction of an incisional hernia within 2 years after surgery.7 However, after analysis of the existing evidence, the European Hernia Society concludes “Although the data are favorable and consistent for prophylactic mesh augmentation, the Guidelines Development Group decided that larger trials are needed to make a strong recommendation to perform prophylactic mesh augmentation for all patients within certain risk groups.”8
Not all patients experience the same risk of developing an incisional hernia, and the pathogenic mechanism of incisional hernia development is not fully understood. The genesis of an incisional hernia is multifactorial: Generally, conditions that negatively affect wound healing make patients susceptible to incisional hernia. Contributing factors may be divided into patient- and surgeon-related factors, which are to some extent directly controllable.1, 9–15 Surgery-related factors include suture material, poor technique, and the need for emergency surgery.16 Furthermore, postoperative wound infection at the surgical site has been identified as a major contributing factor, with up to 25 percent of patients developing an incisional hernia.17, 18 Patient-related factors that contribute to the development of incisional hernia include, among others, age, obesity, smoking, malnutrition, immunosuppressive therapy, and connective tissue disorders.19 Moreover, a positive family history of incisional hernia contributes to an elevated risk.20 These findings support the hypothesis that certain individual biological factors may significantly increase the risk for incisional hernia development. Alterations in connective tissue metabolism such as changes in the extracellular matrix have been reported, including thinner collagen fibrils, imbalance between type I and type III collagen, and increased activity of collagen degrading matrix metalloproteinases (MMPs).21, 22 A decreased ratio of type I to III collagen and MMP-1/MMP-2 ratios in fascia tissue23 and a systemic alteration of collagen metabolism24 have been reported in hernia patients that might explain biological activities of key elements in the development of incisional hernia.
A predictive tool to precisely identify patients that are at high-risk for an incisional hernia is an unmet clinical need. A validated risk model shows predictive value for abdominal wound dehiscence16 and a predictive model for estimating the risk of early incisional hernia has been developed.25 However, no presurgical biomarker exists to date that predicts incisional hernia development prior to planned surgery. Such a biomarker, alone or incorporated into a multifactorial model, could guide decision making for early preventive measures (e.g., mesh implantation at primary surgery) and personalized recommendations for patients at risk.
Thus, we tested an array-based strategy for the discovery of predictive biomarker candidates among newly-diagnosed colorectal cancer patients undergoing median laparotomy using presurgical plasma samples while accounting for known clinical risk factors.
Patients and methods
Patients
This pilot case-control study is nested in the Heidelberg (Germany) site of the ColoCare Consortium. ColoCare is an international, multicenter, prospective cohort, with additional US sites at the Fred Hutchinson Cancer Research Center, Seattle (Washington), H. Lee Moffitt Cancer Center and Research Institute, Tampa (Florida) and Huntsman Cancer Institute, Salt Lake City (Utah), recruiting newly-diagnosed colorectal cancer patients (ICD-10 C18–C20) of all stages with the goal to investigate predictors of cancer recurrence, survival, treatment toxicities and health-related quality of life (ClinicalTrials.gov: NCT02328677).26–28
All ColoCare patients included in this study underwent clinically indicated midline incision along the linea alba for colorectal cancer removal at a single institution (Department of General, Visceral and Transplantation Surgery, University Hospital of Heidelberg) between October 2010 and January 2013.
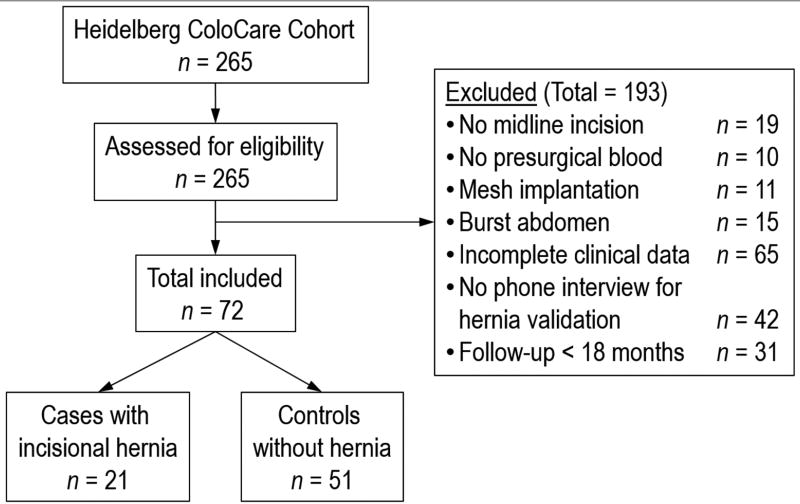
Out of n=265 patients enrolled in ColoCare, n=72 patients were included in the study who met the following inclusion criteria: midline incision, presurgical blood, no mesh implantation at the primary surgery, no burst abdomen in the control group, complete clinical data on risk factors, follow-up >18 months, and a phone interview to confirm presence or absence of incisional hernia. A study flow diagram is presented in Figure 1. Characteristics of the n=193 patients excluded from the study were similar to the patients included in the study, except that the included patients were an average of 5 years younger (see Supplementary Table 1). Of the n=72 included patients, n=21 had developed an incisional hernia (=cases) and were matched to the n=51 patients without hernia (=controls). A step-wise matching procedure was performed. In a first matching step we included all variables of interest: gender, age, BMI, smoking, adjuvant chemotherapy, impaired wound healing and diabetes. In the proceeding matching steps variables were dropped consecutively. Patients were matched by at least gender, age and BMI. Table 1 shows patient characteristics abstracted from patient’s medical records.
Figure 1.
Study flow diagram.
Table 1.
Patient characteristics of incisional hernia cases and controls.
| Variable | Cases with incisional hernia (n = 21) |
Controls withou hernia (n = 51) |
|---|---|---|
| Age, mean ± SD | 59.8 ± 11.4 | 58.9 ± 11.7 |
| Gender, n (%) | ||
| Male | 16 (76) | 30 (59) |
| Female | 5 (24) | 21 (41) |
| BMI (kg/m2), mean ± SD | 28.0 ± 4.6 | 26.3 ± 3.6 |
| Tumor location, n (%) | ||
| C18/C19 Colon or rectosigmoid | 10 (48) | 23 (45) |
| C20 Rectum | 11 (52) | 28 (55) |
| Stage, n (%) | ||
| 0/I/II | 15 (71) | 32 (63) |
| III/IV | 6 (29) | 19 (37) |
| Adjuvant therapy, n (%) | 10 (48) | 26 (51) |
| Neoadjuvant therapy, n (%) | 6 (29) | 19 (37) |
| Smoker, n (%) | ||
| Current smoker | 2 (9) | 10 (20) |
| Ex-Smoker | 1 (5) | 4 (8) |
| Never smoker | 18 (86) | 37 (72) |
| Postoperative wound disorder, n (%) | 5 (24) | 4 (8) |
| Previous surgeries, n (%) | ||
| Appendectomy | 4 (19) | 8 (16) |
| Hysterectomy | 0 (0) | 1 (2) |
| Ostomy | 1 (5) | 2 (4) |
| Cholecystectomy | 2 (10) | 2 (4) |
| Sterilization (male) | 0 (0) | 1 (2) |
| Sterilization (female) | 1 (5) | 0 (0) |
| Prostatectomy | 1 (5) | 0 (0) |
| Nephrectomy | 0 (0) | 1 (2) |
| Umbilical herniotomy | 1 (5) | 0 (0) |
| Caesarean section | 0 (0) | 2 (4) |
| Suture technique, n (%) | ||
| Continuous | 16 (76) | 39 (76) |
| Interrupted | 0 (0) | 1 (2) |
| Combined | 4 (19) | 11 (22) |
| Unknown | 1 (5) | 0 (0) |
| Suture material, n (%) | ||
| Maxon | 11 (52) | 22 (43) |
| PDS | 4 (19) | 11 (22) |
| Monoplus | 0 (0) | 1 (2) |
| unknown | 6 (29) | 17 (33) |
| Medication, n (%) | ||
| ACE-Inhibitors | 1 (2) | 7 (14) |
| Steroids | 0 (0) | 2 (4) |
| NSAR | 3 (6) | 8 (16) |
| Co-morbidities, n (%) | ||
| Diabetes | 3 (14) | 3 (6) |
| Chronic lung disease | 2 (10) | 0 (0) |
| Aneurysm | 1 (5) | 0 (0) |
| Collagen diseases | 0 (0) | 0 (0) |
| Previous hernia (inguinal or umbilical) | 4 (19) | 5 (10) |
| Chronic renal disease | 0 (0) | 1 (2) |
| Anemia | 0 (0) | 5 (10) |
| Albumin pre-operative (g/l), mean ± SD | 44.2 ± 2.9 | 45.2 ± 2.9 |
| Time between surgery and incisional hernia development, n (%) | ||
| <6 months | 8 (38) | - |
| 6–12 months | 5 (24) | - |
| >12 months | 7 (33) | - |
| unknown | 1 (5) | - |
| Follow-up (months), mean ± SD | 22.0 ± 6.4 | 26.2 ± 5.7 |
n: absolute number; SD: standard deviation
The study was approved by the Institutional Review Board of the Medical Faculty at the University of Heidelberg and all participants provided written informed consent.
Definition of incisional hernia cases and controls
The incidence of an incisional hernia at the midline incision site was assessed by a dual assessment of questionnaires and phone interviews. As a first step, we queried all patients for incisional hernia occurrences (along with a lay explanation and graphical illustration) in questionnaires at 6, 12, and 24 months post-surgery (see Supplementary Table 2). Second, we performed a comprehensive phone interview with the patient to verify whether an incisional hernia had occurred or not (see Supplementary Table 3). If a patient was not available by phone or the presence of incisional hernia remained unclear, the interview was conducted with the corresponding general practitioner. All incisional hernia cases reported the confirmation of hernia as part of a study-unrelated physical examination (e.g., with their general practitioner) or a surgical hernia repair. In addition, some cases reported confirmation by sonography (n=6) and internal CT scans were evaluated that confirmed hernia formation in n=3 cases. Patients were defined as controls if no incisional hernia had occurred within at least 18 months post-surgery, which was the time of the latest case of incisional hernia reported in the case group.
Plasma processing
Blood samples (EDTA) were collected from patients prior to surgery. Preparation of plasma was performed within 4 hours after blood-draw by retaining the supernatant after centrifugation (2500 g; 15 min) and storing in aliquots at −80 °C until analysis. 50 µl of each patient’s plasma were shipped on dry ice to the Fred Hutchinson Cancer Research Center (FHCRC, Seattle, Washington) for the antibody microarray experiments.
Antibody Microarray Experiments
Antibody microarray experiments were performed in the laboratory of Dr. Paul Lampe at FHCRC according to previously established methods.29–31 Briefly, antibodies were printed in triplicates on Nexterion H hydrogel slides (Schott, Mainz, Germany) in 48 blocks with a 15×15 block format for a total of 3,600 unique features at a final concentration of 275 µg/ml. After depletion of albumin and IgG, the remaining plasma proteins (200 µg) were labeled with Cy5 (cases and controls) or Cy3 (reference) (GE Health Biosciences, Pittsburgh, PA) following a “case/control versus reference” procedure to remove dye bias from the analysis. After plasma was incubated on arrays, arrays were washed and scanned using an Axon GenePix 4200A microarray scanner (Molecular Devices, Sunnyvale, CA). GenePix Pro 6.0 image analysis software was used to analyze the scanned array images.
Statistical analysis
Detailed descriptions of array statistical analysis have been reported previously.32 Briefly, for each antibody feature, the fold change of case and control signal (red channel) relative to the reference (green channel) was calculated as log2(Rc/Gc), where Rc is red corrected and Gc is green corrected applying a normexp background correction method.33 Paired t-test was performed to assess the difference between case and control signal for each antibody. The p-values calculated via the paired t-test represent the ability of the protein marker to distinguish cases from controls. Protein markers were then ranked based on the coefficient, also known as the odds ratio, which is a log2-based measurement of signal intensity and represents a 2-fold change. Markers with positive coefficients are greater in cases versus controls, and negative coefficients represent the converse.
Gene Set Enrichment Analysis (GSEA) was performed based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) gene sets that are available from the Molecular Signatures Database (MSigDB) (http://www.broadinstitute.org/gsea/msigdb/index.jsp). The 889 antibodies available for analysis correspond to 732 unique genes. Of the 990 GO gene sets analyzed, our array included at least 5 gene members in 835 of these gene sets. With respect to KEGG gene sets, our arrays included at least 5 proteins corresponding to gene members in 129 of the 131 gene sets available. We then tested whether the proteins corresponding to groups of genes in a given gene set had a higher statistical ranking than the proteins not in this gene set based on Wilcoxon testing.
R statistical computing software program was used for all statistical array analyses incorporating the “limma” package for microarray read-in and normalization.34
Matching of cases and controls and the analyses of the study population’s characteristics were performed using SAS 9.3 (SAS Institute, Cary, NC, USA). Comparison of quantitative variables was performed by parametric Student’s t-tests. Pearson χ2 tests were used to investigate differences between categorical variables.
Results
The case and control group were balanced with respect to age, gender, BMI, tumor location and stage. Somewhat higher proportions of postoperative wound disorder, diabetes, and a history of other type of hernia (e.g., inguinal hernia) were found in the incisional hernia case group. Most incisional hernias (65%) occurred within the first 12 months post-surgery. The study population’s demographic and clinical characteristics are shown in Table 1.
Of the 3,290 proteins assessed in the paired t-test, 25 (0.8%) showed statistically significant differences in signal between cases and controls at p<0.05 with a reasonable effect size and log2 ratio of >1 or <−1. Of these 25 proteins, 12 were lower and 13 higher in cases compared to controls (Table 2). The top half of proteins that were lower in cases than in controls included proteasome subunit beta type 5 (− 1.52 log2 ratio), sex hormone-binding protein (−1.49 log2 ratio), defensin alpha 1 (−1.34 log2 ratio), chemokine (C-C motif) ligand 21 (−1.3 log2 ratio), SRA (steroid receptor RNA activator) stem-loop interacting RNA binding protein (−1.22 log2 ratio), and epithelial cell adhesion molecule (−1.13 log2 ratio). The top half of proteins that were higher in cases than in controls included RNA polymerase III transcription initiation factor (1.78 log2 ratio), calreticulin 3 (1.36 log2 ratio), estrogen receptor 1 (1.32 log2 ratio), harvey rat sarcoma viral oncogene homolog (1.2 log2 ratio), and fibronectin 1 (1.18 log2 ratio). A lower effect size of >0.5 or <−0.5 with p-values <0.05 revealed an additional 62 proteins that were different between cases and controls (Table 3), including Collagen Type I Alpha 1 (0.82 log2 ratio), Interleukin 1 Beta (0.78 log2 ratio), Fibrillin 2 (0.77 log2 ratio), Granulin (−0.51 log2 ratio), Nidogen 1 (− 0.63 log2 ratio), and Collagen Type XXIV Alpha 1 (−0.90 log2 ratio).
Table 2.
Top ranked antibodies with p-values <0.05 and log2 ratios of > 1 or < −1. Log2 represents a 2-fold change.
| Antibody name | Gene name | log2 ratio | p-value |
|---|---|---|---|
| RNA polymerase III transcription initiation factor 50 kDa subunit | BRF2 | 1.78 | 0.001 |
| Calreticulin 3 | CALR3 | 1.36 | 0.017 |
| Estrogen receptor 1 | ESR1 | 1.32 | 0.021 |
| Harvey rat sarcoma viral oncogene homolog | HRAS | 1.20 | 0.003 |
| Fibronectin 1 | FN1 | 1.18 | 0.021 |
| Heparan sulfate proteoglycan 2 | HSPG2 | 1.17 | 0.001 |
| Interferon-induced protein with tetratricopeptide repeats 1 | IFIT1 | 1.13 | 0.042 |
| Cyclin-dependent kinase inhibitor 1B | CDKN1B | 1.12 | 0.017 |
| Filamin A, alpha | FLNA | 1.07 | 0.019 |
| Selectin E | SELE | 1.04 | 0.030 |
| TBC1 domain family, member 3 | TBC1D3 | 1.01 | 0.041 |
| Ras-related protein Rap-1A | RAP1A | 1.01 | 0.036 |
| Interleukin 12A | IL12A | 1.00 | 0.006 |
| Post-GPI Attachment To Proteins 3 | PERLD1 | −1.05 | 0.038 |
| Conserved helix-loop-helix ubiquitous kinase | CHUK | −1.05 | 0.010 |
| Coagulation factor II (thrombin) | F2 | −1.05 | 0.022 |
| Protocadherin-15 | PCDH15 | −1.05 | 0.018 |
| P-cadherin | CDH3 | −1.07 | 0.001 |
| Colon cancer secreted protein-1 | CCSP-1 | −1.12 | 0.016 |
| Epithelial cell adhesion molecule | EPCAM | −1.13 | 0.010 |
| SRA stem-loop-interacting RNA-binding protein | SLIRP | −1.22 | 0.022 |
| Chemokine (C-C motif) ligand 21 | CCL21 | −1.30 | 0.019 |
| Defensin alpha 1 | DEFA1 | −1.34 | 0.033 |
| Sex hormone-binding globulin | SHBG | −1.49 | 0.002 |
| Proteasome subunit beta type-5 | PSMB5 | −1.52 | 0.011 |
Table 3.
Antibodies with p-values <0.05 and log2 ratios between +/−0.5 and +/−1. Log2 represents a 2-fold change.
| Kallikrein Related Peptidase 5 | KLK5 | 0.98 | 0.042 |
| Phospholipid scramblase | PLSCR1 | 0.93 | 0.017 |
| Checkpoint Kinase 1 | CHEK1 | 0.92 | 0.01 |
| Thrombospondin 3 | THBS3 | 0.92 | 0.034 |
| Aryl Hydrocarbon Receptor Nuclear Translocator | arnT | 0.92 | 0.037 |
| YES Proto-Oncogene 1, Src Family Tyrosine Kinase | YES1 | 0.9 | 0.032 |
| Tumor Protein, Translationally-Controlled 1 | TPT1 | 0.84 | 0.005 |
| Lactalbumin Alpha | LALBA | 0.82 | 0.02 |
| Collagen Type I Alpha 1 | COL1A1 | 0.82 | 0.03 |
| Von Hippel-Lindau Tumor Suppressor | VHL | 0.78 | 0.01 |
| SMAD Family Member 2 | SMAD2 | 0.78 | 0.004 |
| Interleukin 1 Beta | IL1B | 0.78 | 0.039 |
| Cytochrome P450 Family 2 Subfamily C Member 8 | CYP2C8 | 0.77 | 0.036 |
| Fibrillin 2 | FBN2 | 0.77 | 0.022 |
| Enolase 1 | ENO1 | 0.77 | 0.039 |
| PH Domain And Leucine Rich Repeat Protein Phosphatase | PHLPPL | 0.73 | 0.037 |
| Deoxycytidine Kinase | DCK | 0.71 | 0.048 |
| Cut Like Homeobox 1 | CUX1 | 0.69 | 0.039 |
| Wingless-Type MMTV Integration Site Family, Member 7A | WNT7A | 0.68 | 0.042 |
| Membrane Spanning 4-Domains A7 | MS4A7 | 0.66 | 0.035 |
| Acyloxyacyl Hydrolase | AOAH | 0.66 | 0.026 |
| Oncostatin M | OSM | 0.65 | 0.045 |
| CTD Phosphatase Subunit 1 | CTDP1 | 0.64 | 0.037 |
| Forkhead Box A1 | foxa1 | 0.63 | 0.045 |
| MDM2 Proto-Oncogene | MDM2 | 0.61 | 0.043 |
| G Protein Subunit Alpha I3 | GNAI3 | 0.59 | 0.015 |
| Paraoxonase 1 | PON1 | 0.58 | 0.04 |
| Trefoil Factor 3 | TFF3 | 0.54 | 0.035 |
| GCSF, Colony Stimulating Factor 3 (Granulocyte) | CSF3 | 0.53 | 0.039 |
| Myotubularin Related Protein 11 | MTMR11 | 0.52 | 0.031 |
| Granulin | GRN | −0.51 | 0.048 |
| C-C Motif Chemokine Ligand 20 | CCL20 | −0.55 | 0.031 |
| Glucokinase | GCK | −0.58 | 0.046 |
| Stromal Cell Derived Factor 4 | SDF4 | −0.59 | 0.024 |
| Cyclin B1 | CCNB1 | −0.59 | 0.049 |
| Phosphoglycerate Mutase 1 | PGAM1 | −0.61 | 0.028 |
| NCK Adaptor Protein 1 | NCK1 | −0.62 | 0.007 |
| Neuropeptide Y | NPY | −0.62 | 0.024 |
| Prostaglandin-Endoperoxide Synthase 1 | PTGS1 | −0.62 | 0.03 |
| Nidogen 1 | NID1 | −0.63 | 0.04 |
| Hypoxia Inducible Factor 1 Alpha Subunit | HIF1A | −0.66 | 0.039 |
| Deleted In Malignant Brain Tumors 1 | DMBT1 | −0.66 | 0.031 |
| Mitogen-Activated Protein Kinase-Activated Protein Kinase 3 | MAPKAPK3 | −0.67 | 0.04 |
| Ribosomal Protein | RPS | −0.69 | 0.024 |
| Epidermal Growth Factor Receptor | EGFR | −0.69 | 0.05 |
| Minichromosome Maintenance 8 Homologous RecombinationRepair Factor | MCM8UV | −0.71 | 0.041 |
| Parkinsonism Associated Deglycase | PARK7 | −0.71 | 0.045 |
| Protein Phosphatase 2 Regulatory Subunit B, Alpha | PPP2R2AUV | −0.72 | 0.036 |
| Prohibitin | PHB_SDI | −0.77 | 0.028 |
| Peptidylprolyl Isomerase Like 3 | PPIL3UV | −0.77 | 0.035 |
| Homeobox D13 | HOXD13 | −0.78 | 0.011 |
| C-X-C Motif Chemokine Ligand 12 | CXCL12 | −0.81 | 0.031 |
| Early Endosome Antigen 1 | EEA1 | −0.81 | 0.023 |
| Orosomucoid 1 | ORM1 | −0.82 | 0.031 |
| Transforming Growth Factor Alpha | TGFA | −0.82 | 0.04 |
| Myocyte Enhancer Factor 2C | MEF2C | −0.88 | 0.034 |
| Keratin 23 | KRT23 | −0.89 | 0.002 |
| Collagen Type XXIV Alpha 1 | COL24A1 | −0.9 | 0.022 |
| Glyceraldehyde-3-Phosphate Dehydrogenase | GAPDH | −0.92 | 0.026 |
| Achaete-Scute Family BHLH Transcription Factor 1 | ASCL1 | −0.92 | 0.042 |
| T-Cell Leukemia Homeobox 2 | TLX2 | −0.94 | 0.04 |
In our gene set analysis, a total of 11 KEGG and a total of 123 GO gene sets had a p-value <0.05 (Table 4). These pathways include ECM receptor interaction (p=0.0002), intestinal immune network for IgA production (p=0.0024), progesterone mediated oocyte maturation (p=0.0062), melanoma (p=0.0073), bladder cancer (p=0.0084), complement and coagulation cascades (p=0.0112), ubiquitin mediated proteolysis (p=0.0175), focal adhesion (p=0.0224), regulation of actin cytoskeleton (p=0.0267), pathways in cancer (p=0.0272), and toll-like receptor signaling pathway (p=0.0005) (Table 4).
Table 4.
KEGG sets with p-values <0.05.
| Number of genes in set |
Number of unique genes observed |
AUC | p-value | |
|---|---|---|---|---|
| KEGG sets | ||||
| ECM receptor interaction | 84 | 4 | 0.60 | 0.0002 |
| Intestinal immune network for IgA production | 48 | 1 | 0.41 | 0.0024 |
| Progesterone mediated oocyte maturation | 86 | 3 | 0.60 | 0.0062 |
| Melanoma | 71 | 4 | 0.58 | 0.0073 |
| Bladder cancer | 42 | 3 | 0.58 | 0.0084 |
| Complement and coagulation cascades | 69 | 1 | 0.39 | 0.0112 |
| Ubiquitin mediated proteolysis | 138 | 2 | 0.62 | 0.0175 |
| Focal adhesion | 201 | 8 | 0.55 | 0.0224 |
| Regulation of actin cytoskeleton | 216 | 4 | 0.56 | 0.0267 |
| Pathways in cancer | 328 | 12 | 0.54 | 0.0272 |
| Toll like receptor signaling pathway | 102 | 3 | 0.75 | 0.0005 |
Results were consistent when removing n=9 patients (n=4 cases; n=5 controls) with a previous inguinal or umbilical hernia from the analysis.
Discussion
Given the tremendous economic and patient burden, precision prevention of incisional hernias is an unmet clinical need. A precise risk prediction would allow personalized surgical measures in high-risk patients (e.g., preventive mesh implantation during the primary surgery) and individual patient recommendations to prevent the development of an incisional hernia. Certain risk factors have been identified. However, to date, no predictive pre-surgery blood-based biomarker is available to evaluate a patient’s individual risk for an incisional hernia.
In presurgical plasma of colorectal cancer patients, we identified 25 proteins that were either nominally significantly elevated (13) or reduced (12) with a reasonable effect size (log2 ratio > 1 or < −1) in future incisional hernia patients. Most of these proteins are connected to cell adhesion, wound healing and inflammation. The cell adhesion molecules protocadherin-15, P-cadherin, and epithelial cell adhesion molecule (EPCAM) were decreased in future incisional hernia patients, and have previously been associated with wound healing and tissue integrity.35–37 Furthermore, we observed decreased plasma levels of CC-chemokine ligand 21, a chemokine involved in inflammatory processes, in hernia cases compared to controls. Intradermal injection of CC-chemokine ligand 21 has been shown to increase the migration of mesenchymal stem cells resulting in accelerated wound repair in mice.38 Moreover, sex hormone-binding globulin (SHBG) was decreased in future incisional hernia cases by a log2 ratio of −1.49 compared to controls, while estrogen receptor 1 was increased (log2 ratio of 1.32). Topical estrogen application has been shown to support wound healing in a randomized controlled trial39 and elevated estrogen receptor 1 plasma levels may reflect receptor upregulation due to decreased estrogen binding by SHBG. RNA polymerase III transcription initiation factor (BRF2) was elevated in cases compared to controls (log2 ratio of 1.32). Overexpression of BRF2 has been associated with abnormal expression of E-cadherin and N-cadherin, marker proteins of the epithelial-mesenchymal transition (EMT).40 Interestingly, proteasome subunit beta type-5 was decreased most (log2 ratio of −1.52) in future incisional hernia patients. As part of the 20S proteasome complex, it is responsible for recognizing damaged proteins for protein quality control and proteasome inhibition, and has been associated with decreased proliferation of lens epithelial cells.41 Finally, we discovered several proteins that differ between cases and control, for which no association with wound healing or connective tissue pathophysiology has been described in the literature thus far. GSEA analysis revealed sets or families of proteins that were changed in cases compared to controls. Based on this approach, pathways of interest included ECM (extracellular matrix) receptor interaction, focal adhesion, melanoma, bladder and pathways of cancer (Table 4).
One limitation of this study is that incisional hernias were assessed by a questionnaire and a phone interview, as physical examination by an experienced surgeon and imaging was not part of the ColoCare study design. Hence, small asymptomatic hernias may have been missed and misdiagnosis of a midline diastasis as a hernia may have occurred. Thus, our study reflects clinically apparent incisional hernias, which should be more important for clinical management. However, in future studies, we suggest to perform prospective study-related physical examination and respective imaging at pre-defined time points for the assessment of incisional hernia.
Another limitation is that our study population was restricted to colorectal cancer patients only. By our stringent inclusion criteria our case-control groups were very homogenous in their risk of developing an incisional hernia. However, we suggest in future directions that studies should not be limited to a specific disease group to identify a robust biomarker independent of clinical or demographic parameters.
A further limitation is that the medical chart reviews to collect data on clinical parameters were done retrospectively.
A limitation of the experimental array approach used is that we were only able to evaluate biomarkers for which antibodies were included on the array. Hence, no fully comprehensive assessment of the plasma proteome was performed and the potential of biomarkers not included on the array could not be assessed. This in particular limited our gene set analyses as we were limited by the candidates included on the array. However, the array covered a wide range of possible biomarker candidates yielding data on 3,290 proteins. The interpretation of the results is challenging given the study’s inherent pilot discovery nature. Nonetheless, this study shows proof-of-principle for array-based discovery of unique predictive biomarkers specific to incisional hernia and points to several proteins and pathways worth of further investigation.
In this pilot study, we applied, for the first time, an antibody microarray to compare the plasma proteome of future incisional hernia patients with matched clinical controls. We showed proof-of-principle for the discovery of novel plasma based biomarkers for incisional hernia occurrence by utilizing presurgical plasma samples. Our study shows that there are multiple individual and groups of plasma proteins that could predict the individual hernia risk prior to undergoing surgery. We were able to identify several protein biomarkers with a known association with wound healing or connective tissue pathophysiology. Promising biomarkers warrant further investigations in larger independent sample sets to further characterize and validate their potential clinical utility. After validation, such a biomarker could be incorporated into a multifactorial risk model to guide clinical decision making.
Supplementary Material
Acknowledgments
The authors thank all ColoCare study participants and the entire ColoCare study team, especially Susanne Jakob and Torsten Kölsch for patient recruitment and follow-up, and Dr. Werner Diehl for data management.
This work has been funded by the Lackas Foundation, the German Cancer Consortium (DKTK), by the Department of Preventive Oncology at the German Cancer Research Center, by the Huntsman Cancer Foundation and NIH grant number CA152746.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Park AE, Roth JS, Kavic SM. Abdominal wall hernia. Current problems in surgery. 2006;43(5):326–75. doi: 10.1067/j.cpsurg.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Le Huu Nho R, Mege D, Ouaissi M, Sielezneff I, Sastre B. Incidence and prevention of ventral incisional hernia. Journal of visceral surgery. 2012;149(5 Suppl):e3–14. doi: 10.1016/j.jviscsurg.2012.05.004. Epub 2012/11/13. [DOI] [PubMed] [Google Scholar]
- 3.Diener MK, Voss S, Jensen K, Buchler MW, Seiler CM. Elective midline laparotomy closure: the INLINE systematic review and meta-analysis. Annals of surgery. 2010;251(5):843–56. doi: 10.1097/SLA.0b013e3181d973e4. Epub 2010/04/17. [DOI] [PubMed] [Google Scholar]
- 4.Pereira JA, Pera M, Grande L. Incidence of incisional hernia after open and laparoscopic colorectal cancer resection. Cirugia espanola. 2013;91(1):44–9. doi: 10.1016/j.ciresp.2012.05.004. Epub 2012/07/10. Elevada incidencia de hernia incisional tras reseccion abierta y laparoscopica por cancer colorrectal. [DOI] [PubMed] [Google Scholar]
- 5.Skipworth JR, Khan Y, Motson RW, Arulampalam TH, Engledow AH. Incisional hernia rates following laparoscopic colorectal resection. International journal of surgery (London, England) 2010;8(6):470–3. doi: 10.1016/j.ijsu.2010.06.008. Epub 2010/07/07. [DOI] [PubMed] [Google Scholar]
- 6.Bower C, Roth JS. Economics of abdominal wall reconstruction. The Surgical clinics of North America. 2013;93(5):1241–53. doi: 10.1016/j.suc.2013.06.007. Epub 2013/09/17. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Urena MA, Lopez-Monclus J, Hernando LA, Montes DM, Valle de Lersundi AR, Pavon CC, et al. Randomized controlled trial of the use of a large-pore polypropylene mesh to prevent incisional hernia in colorectal surgery. Annals of surgery. 2015;261(5):876–81. doi: 10.1097/SLA.0000000000001116. Epub 2015/01/13. [DOI] [PubMed] [Google Scholar]
- 8.Muysoms FE, Antoniou SA, Bury K, Campanelli G, Conze J, Cuccurullo D, et al. European Hernia Society guidelines on the closure of abdominal wall incisions. Hernia : the journal of hernias and abdominal wall surgery. 2015;19(1):1–24. doi: 10.1007/s10029-014-1342-5. Epub 2015/01/27. [DOI] [PubMed] [Google Scholar]
- 9.Pares D, Shamali A, Stefan S, Flashman K, O'Leary D, Conti J, et al. Predictive factors for extraction site hernia after laparoscopic right colectomy. International journal of colorectal disease. 2016;31(7):1323–8. doi: 10.1007/s00384-016-2610-x. [DOI] [PubMed] [Google Scholar]
- 10.Brook AJ, Mansfield SD, Daniels IR, Smart NJ. Incisional hernia following closure of loop ileostomy: The main predictor is the patient, not the surgeon. Surgeon. 2016 doi: 10.1016/j.surge.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Nilsson JH, Strandberg Holka P, Sturesson C. Incisional hernia after open resections for colorectal liver metastases - incidence and risk factors. HPB (Oxford) 2016;18(5):436–41. doi: 10.1016/j.hpb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakayama M, Yoshimatsu K, Yokomizo H, Yano Y, Okayama S, Satake M, et al. Incidence and risk factors for incisional hernia after open surgery for colorectal cancer. Hepatogastroenterology. 2014;61(133):1220–3. [PubMed] [Google Scholar]
- 13.Aquina CT, Rickles AS, Probst CP, Kelly KN, Deeb AP, Monson JR, et al. Visceral obesity, not elevated BMI, is strongly associated with incisional hernia after colorectal surgery. Dis Colon Rectum. 2015;58(2):220–7. doi: 10.1097/DCR.0000000000000261. [DOI] [PubMed] [Google Scholar]
- 14.Yamada T, Okabayashi K, Hasegawa H, Tsuruta M, Abe Y, Ishida T, et al. Age, Preoperative Subcutaneous Fat Area, and Open Laparotomy are Risk Factors for Incisional Hernia following Colorectal Cancer Surgery. Ann Surg Oncol. 2016;23(Suppl 2):S236–41. doi: 10.1245/s10434-015-4462-y. [DOI] [PubMed] [Google Scholar]
- 15.Song IH, Ha HK, Choi SG, Jeon BG, Kim MJ, Park KJ. Analysis of risk factors for the development of incisional and parastomal hernias in patients after colorectal surgery. J Korean Soc Coloproctol. 2012;28(6):299–303. doi: 10.3393/jksc.2012.28.6.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Ramshorst GH, Nieuwenhuizen J, Hop WC, Arends P, Boom J, Jeekel J, et al. Abdominal wound dehiscence in adults: development and validation of a risk model. World journal of surgery. 2010;34(1):20–7. doi: 10.1007/s00268-009-0277-y. Epub 2009/11/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith CT, Katz MG, Foley D, Welch B, Leverson GE, Funk LM, et al. Incidence and risk factors of incisional hernia formation following abdominal organ transplantation. Surgical endoscopy. 2015;29(2):398–404. doi: 10.1007/s00464-014-3682-8. Epub 2014/08/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bucknall TE, Cox PJ, Ellis H. Burst abdomen and incisional hernia: a prospective study of 1129 major laparotomies. British medical journal (Clinical research ed) 1982;284(6320):931–3. doi: 10.1136/bmj.284.6320.931. Epub 1982/03/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.George CD, Ellis H. The results of incisional hernia repair: a twelve year review. Annals of the Royal College of Surgeons of England. 1986;68(4):185–7. Epub 1986/07/01. [PMC free article] [PubMed] [Google Scholar]
- 20.Zoller B, Ji J, Sundquist J, Sundquist K. Shared and nonshared familial susceptibility to surgically treated inguinal hernia, femoral hernia, incisional hernia, epigastric hernia, and umbilical hernia. Journal of the American College of Surgeons. 2013;217(2):289–99 e1. doi: 10.1016/j.jamcollsurg.2013.04.020. Epub 2013/07/23. [DOI] [PubMed] [Google Scholar]
- 21.Klinge U, Si ZY, Zheng H, Schumpelick V, Bhardwaj RS, Klosterhalfen B. Collagen I/III and matrix metalloproteinases (MMP) 1 and 13 in the fascia of patients with incisional hernias. Journal of investigative surgery : the official journal of the Academy of Surgical Research. 2001;14(1):47–54. doi: 10.1080/089419301750072202. Epub 2001/04/12. [DOI] [PubMed] [Google Scholar]
- 22.Guillen-Marti J, Diaz R, Quiles MT, Lopez-Cano M, Vilallonga R, Huguet P, et al. MMPs/TIMPs and inflammatory signalling de-regulation in human incisional hernia tissues. Journal of cellular and molecular medicine. 2009;13(11–12):4432–43. doi: 10.1111/j.1582-4934.2008.00637.x. Epub 2009/04/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salameh JR, Talbott LM, May W, Gosheh B, Vig PJ, McDaniel DO. Role of biomarkers in incisional hernias. The American surgeon. 2007;73(6):561–7. discussion 7–8. Epub 2007/07/31. [PubMed] [Google Scholar]
- 24.Henriksen NA, Mortensen JH, Sorensen LT, Bay-Jensen AC, Agren MS, Jorgensen LN, et al. The collagen turnover profile is altered in patients with inguinal and incisional hernia. Surgery. 2015;157(2):312–21. doi: 10.1016/j.surg.2014.09.006. Epub 2015/01/27. [DOI] [PubMed] [Google Scholar]
- 25.Veljkovic R, Protic M, Gluhovic A, Potic Z, Milosevic Z, Stojadinovic A. Prospective clinical trial of factors predicting the early development of incisional hernia after midline laparotomy. Journal of the American College of Surgeons. 2010;210(2):210–9. doi: 10.1016/j.jamcollsurg.2009.10.013. Epub 2010/02/02. [DOI] [PubMed] [Google Scholar]
- 26.Ristau J, Staffa J, Schrotz-King P, Gigic B, Makar KW, Hoffmeister M, et al. Suitability of circulating miRNAs as potential prognostic markers in colorectal cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2014;23(12):2632–7. doi: 10.1158/1055-9965.EPI-14-0556. Epub 2014/12/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skender S, Schrotz-King P, Boehm J, Abbenhardt C, Gigic B, Chang-Claude J, et al. Repeat physical activity measurement by accelerometry among colorectal cancer patients-feasibility and minimal number of days of monitoring. BMC Res Notes. 2015;8(222) doi: 10.1186/s13104-015-1168-y. Epub 2015/06/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liesenfeld D, Habermann N, Toth R, Owen R, Frei E, Böhm J, et al. Changes in urinary metabolic profiles of colorectal cancer patients enrolled in a prospective cohort study (ColoCare) Metabolomics. 2015:1–15. doi: 10.1007/s11306-014-0758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rho JH, Lampe PD. High-throughput screening for native autoantigen-autoantibody complexes using antibody microarrays. Journal of proteome research. 2013;12(5):2311–20. doi: 10.1021/pr4001674. Epub 2013/04/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buas MF, Rho JH, Chai X, Zhang Y, Lampe PD, Li CI. Candidate early detection protein biomarkers for ER+/PR+ invasive ductal breast carcinoma identified using pre-clinical plasma from the WHI observational study. Breast cancer research and treatment. 2015;153(2):445–54. doi: 10.1007/s10549-015-3554-5. Epub 2015/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rho JH, Mead JR, Wright WS, Brenner DE, Stave JW, Gildersleeve JC, et al. Discovery of sialyl Lewis A and Lewis X modified protein cancer biomarkers using high density antibody arrays. Journal of proteomics. 2014;96:291–9. doi: 10.1016/j.jprot.2013.10.030. Epub 2013/11/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mirus JE, Zhang Y, Hollingsworth MA, Solan JL, Lampe PD, Hingorani SR. Spatiotemporal proteomic analyses during pancreas cancer progression identifies serine/threonine stress kinase 4 (STK4) as a novel candidate biomarker for early stage disease. Molecular & cellular proteomics : MCP. 2014;13(12):3484–96. doi: 10.1074/mcp.M113.036517. Epub 2014/09/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods (San Diego, Calif) 2003;31(4):265–73. doi: 10.1016/s1046-2023(03)00155-5. Epub 2003/11/05. [DOI] [PubMed] [Google Scholar]
- 34.Smyth GK. limma: Linear Models for Microarray Data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York, NY: Springer New York; 2005. pp. 397–420. [Google Scholar]
- 35.Frank M, Kemler R. Protocadherins. Current opinion in cell biology. 2002;14(5):557–62. doi: 10.1016/s0955-0674(02)00365-4. Epub 2002/09/17. [DOI] [PubMed] [Google Scholar]
- 36.Litvinov SV, Velders MP, Bakker HA, Fleuren GJ, Warnaar SO. Ep-CAM: a human epithelial antigen is a homophilic cell-cell adhesion molecule. The Journal of cell biology. 1994;125(2):437–46. doi: 10.1083/jcb.125.2.437. Epub 1994/04/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Marck V, Stove C, Van Den Bossche K, Stove V, Paredes J, Vander Haeghen Y, et al. P-cadherin promotes cell-cell adhesion and counteracts invasion in human melanoma. Cancer research. 2005;65(19):8774–83. doi: 10.1158/0008-5472.CAN-04-4414. Epub 2005/10/06. [DOI] [PubMed] [Google Scholar]
- 38.Sasaki M, Abe R, Fujita Y, Ando S, Inokuma D, Shimizu H. Mesenchymal stem cells are recruited into wounded skin and contribute to wound repair by transdifferentiation into multiple skin cell type. Journal of immunology (Baltimore, Md: 1950) 2008;180(4):2581–7. doi: 10.4049/jimmunol.180.4.2581. Epub 2008/02/06. [DOI] [PubMed] [Google Scholar]
- 39.Ashcroft GS, Greenwell-Wild T, Horan MA, Wahl SM, Ferguson MW. Topical estrogen accelerates cutaneous wound healing in aged humans associated with an altered inflammatory response. The American journal of pathology. 1999;155(4):1137–46. doi: 10.1016/S0002-9440(10)65217-0. Epub 1999/10/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian Y, Lu M, Yue W, Li L, Li S, Gao C, et al. TFIIB-related factor 2 is associated with poor prognosis of nonsmall cell lung cancer patients through promoting tumor epithelial-mesenchymal transition. BioMed research international. 2014;2014:530786. doi: 10.1155/2014/530786. Epub 2014/04/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Awasthi N, Wagner BJ. Suppression of human lens epithelial cell proliferation by proteasome inhibition, a potential defense against posterior capsular opacification. Investigative ophthalmology & visual science. 2006;47(10):4482–9. doi: 10.1167/iovs.06-0139. Epub 2006/09/28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.