Abstract
Aeromonas sobria is opportunistic pathogen frequently found in environment and food. Interfering with its quorum sensing (QS) system could be a promising way to alleviate its virulence. In this study, curcumin liposomes were prepared and their characteristics like particle size, zeta potential, PDI (Polymey Disperse Index), encapsulation efficiency and loading capacity were measured. The quorum sensing inhibitory effect of curcumin liposomes under sub-MIC (Minimum Inhibitory Concentration) on siderophore production, swimming and swarming motility, extracellular proteases, biofilm formation and AHLs (N-acylhomoserine lactones) production of A. sobria were also determined. The results showed that, the curcumin liposomes with high encapsulation capacity (84.51 ± 0.58%) were stable and homogeneous. QS-regulated phenotypes of the pathogen were significantly inhibited by curcumin liposomes. The in silico analysis revealed that the QS system of A. sobria may be inhibited by released curcumin from curcumin liposomes through interacting with the built LuxI type protein and blocking the production of AHLs.
Introduction
Aeromonas sobria is a Gram-negative, rod-shaped, facultative anaerobic, motile, and flagellated bacterium of the Aeromonadaceae family1 . A. sobria usually thrives in fresh water, sea water, sewage, soil and food. As an opportunistic pathogen for animals and humans, it can cause extra-intestinal infections and gastrointestinal diseases2. It is also a causative bacterium of child diarrhea and soft-tissue infections in fish3. Currently, antibiotics and synthetic antibacterials are widely used to treat bacterial-caused diseases. However, improper use of antibiotics has created the problems of antibiotics resistance and environmental contamination.
Quorum sensing is a cell-to-cell communication mechanism, allowing bacteria to monitor their population density by secreting signaling molecules or autoinducers4. These chemical signals release from bacteria and accumulate in the environment, and once the threshold concentration reaches, they can bind to receptor proteins and trigger the expression of a series of QS targeting genes5. Most Gram-negative bacteria use N-acylhomoserine lactones (AHLs) as the major QS signal molecules, while Gram-positive bacteria use signal peptides to regulate physiological functions, such as bioluminescence, biosynthesis of antibiotic, release of virulence factors, biofilm formation, siderophore, extracellaluar proteases, swimming and swarming motility, etc.6, 7. Therefore, blocking the bacterial QS system has been considered as an alternative of traditional antibiotics, for it would not induce the resistance of bacteria due to its distinctive antibacterial mechanism8. It has been reported that A. sobria produces AHLs such as C4-HSL and C6-HSL to regulate its virulence factors9. Meanwhile, many natural products and synthetic materials such as iberin10, baicalein11, quercetin12, and 6-Gingerol13 have been proven to be quorum sensing inhibitors (QSIs). Among them, the brominated furanones such as furanone C-30 have been reported to inhibit QS-regulated biofilm formation and production of virulence factors14. However, their toxicity to mammalian cells has not been reported15. On the other hand, curcumin, the major ingredient from Curcuma longa has shown multiple pharmaceutical properties, such as antitumor16, 17, anti-HIV-118, antibacterial19, etc. Curcumin has also been shown to have inhibitory effect on the virulence, QS, and biofilm initiation of Pseudomonas aeruginosa PAO1 in the whole plant and animal models20. Similarly, curcumin also exhibited inhibitory effect on the QS system of uropathogens, including Escherichia coli, Proteus mirabilis, Pseudomonas aeruginosa PAO1 and Serratia marcescens 21. More importantly, curcumin has been applied as a dietary supplement and alternative medicine, and it is believed to be safe and efficacy when administered at very high doses clinically22. However, the bioavailability of curcumin was limited due to its poor water solubility and fast metabolism21, 23. Liposomes have been effective carriers for hydrophobic drugs24. Additionally, the inhibitory effect of curcumin liposomes acted as QSIs on the QS system of A. sobria has not been demonstrated. Herein, in the present study, the curcumin liposomes were prepared to study the inhibitory effect on quorum sensing system of A. sobria. The possible inhibitory mechanism was further analyzed by molecular docking techniques.
Materials and Methods
Materials and bacterial strains
Curcumin, cholesterol, lecithin, 2, 3-bis (2-methoxy-4-nitro-5-sulfo-phenyl)-2H-tetrazolium-5- carboxanilide (XTT) and menadione were purchased from Sigma-Aldrich (St. Louis, MO, USA). Standard AHLs including N-butanoyl-L-homoserine lactone (C4-HSL), N-hexanoyl-L-homoserine lactone (C6-HSL), N-octanoyl-L-homoserine lactone (C8-HSL), N-decanoyl-L- homoserine lactone (C10-HSL), N-dodecanoyl-L-homoserine lactone (C12-HSL) and N-tetradecanoyl-L-homoserine lactone (C14-HSL) were also purchased from Sigma-Aldrich. The AHLs C4-HSL, C6-HSL, C8-HSL, C10-HSL, C12-HSL and C14-HSL were dissolved by methanol (at 20 µM). The strains Chromobacterium violaceum CV026 and Agrobacterium tumefaciens A136 were kind gifts from Dr. Yang (Xinjiang Shihezi University, Xinjiang, China). C. violaceum CV026 is a mini-Tn5 mutant deficient in AHLs production, but it responds to exogenous AHLs and produces violacein in purple color25. Herein, it was used as an indicator bacterium for short chain AHLs detection. A. tumefaciens A136, which carries a lacZ fusion to traI, was used to detect long chain AHLs. It produces blue color in the presence of long chain AHLs and X-Gal (5-bromo-4- chloro-3-indoyl-β-D-galactopyranoside). C. violaceum CV026 was grown in 10 ml of Luria-Bertani (LB) (Beijing aoboxing bio-tech CO., LTD, China) medium containing 10 μl of kanamycin (20 μg/ml). A. tumefaciens A136 was grown in 10 ml of LB medium containing 10 μl of tetracycline (20 μg/ml) and 50 μl of spectinomycin (20 μg/ml). A. sobria was isolated and identified by our laboratory from deteriorated aquatic products. Curcumin was dissolved in 1% dimethyl sulfoxide (DMSO, dissolved in distilled water). Methanol was chromatographically pure and other chemicals were analytical reagents and all chemical reagents are commercially available.
Preparation of curcumin liposomes
The curcumin liposomes were prepared by using a conventional thin film-dispersed method with slight modification26. A total of 100 mg lecithin, 42 mg cholesterol and 5 mg curcumin were dissolved into 10 ml chloroform, respectively. And 1 ml of each solution was added to a rotary flask and mixed for 5 min (30 °C, 140 bar, 90 r/min) to obtain a thin film. A total of 1 ml 5% glucose (w/v) was added to the film and mixed by rotary evaporation under normal pressure (50 °C) for 2 hours. Then the solution was sonicated using an ultrasonic cell disruptor (DS-650Y, Doosy, Shanghai, China) for 100 cycles (2 s on, 2 s off, 400 W) until the solution was homogeneous. The temperature was controlled by an ice bath. The curcumin liposomes were obtained by filtration through a micro-pore filter (0.22 µm).
Determination of characteristics of curcumin liposomes
The transparent yellow solution of curcumin liposomes was obtained by sephadex gel column chromatography. The sample (0.1 ml) was added to 1.9 ml methanol for demulsification and the characteristics of curcumin liposomes were measured.
Measurement of encapsulation efficiency and loading capacity
The amount of entrapped curcumin was measured by high performance liquid chromatography (HPLC) at the detection wavelength of 425 nm, which was the maximum absorbance wavelength of curcumin. And it was analyzed quantitatively by comparing with a standard curve. The experiments were repeated three times. The loading capacity (LC %) and encapsulation efficiency (EE %) was derived by the equations below (1 and 2), respectively.
| 1 |
| 2 |
Measurement of particle size, size distribution as well as zeta potential
Particle size, zeta potential as well as size distribution of curcumin liposomes was tested by Malvern Zetasizer Nano ZS90 (Malvern Instruments Ltd., Malvern, UK). The particle size distribution was considered as the polydispersity index (PDI).
Morphological observation of curcumin liposomes
Curcumin liposomes (2.5 µl) were added to a piece of glass (5 mm × 5 mm × 0.2 mm) and dried slowly with nitrogen, in order to get a homogeneous film. The liposomes were coated with gold in a sputter coater for 4–5 min and the morphological observation of gold-sprayed liposomes was carried out by SEM (FESEM S-4800, JEOL, Japan). Images were obtained from random positions of curcumin liposomes on the glass. Each measurement was done in triplicate.
Curcumin release
The release of curcumin from the liposomes was determined using equilibrium dialysis in PBS (0.01 M, pH 5.8) containing 30% ethanol (w/v). A total of 5 ml curcumin liposomes solution was added into a dialysis bag and then exposed to 15 ml release medium at 37 °C. The sample (1 ml) was withdrawn from the dialysis bag at predetermined time intervals and replaced with fresh release medium. The content of curcumin was measured by HPLC and calculated as cumulative percent released. The curcumin was taken as control. The release experiments were done in triplicate.
QS inhibitory test
Detection of AHLs
The AHLs produced by A. sobria were detected by C. violaceum CV026 and A. tumefaciens A136. C. violaceum CV026 is the mini-Tn5 mutant of C. violaceum ATCC31532, which is unable to produce AHLs and violacein itself, but it is susceptible to different exogenous AHLs contained acyl side chains of 4–6 carbon atoms. A. tumefaciens A136, which carries a lacZ fusion to traI, is able to produce a blue color in the presence of X-Gal and long chain AHLs. C. violaceum CV026 was grown in 10 ml of LB medium containing 10 μl kanamycin (20 μg/ml) at 28 °C, shaking at 160 rpm to an OD600 of 1.0. A. tumefaciens A136 was grown in 10 ml of LB medium containing 10 μl of tetracycline (20 μg/ml) and 50 μl of spectinomycin (20 μg/ml) at 28 °C, shaking at 160 rpm to an OD600 of 1.0. C. violaceum CV026 (1 ml) and A. tumefaciens A136 (1 ml) were added to 100 ml of nutrient agar, respectively. After the temperature of the medium dropped to room temperature, A. sobria was streaked on the two plates respectively to detect the presence of quorum sensing molecules. The A. tumefaciens A136 plates were supplemented with 100 μl of X-Gal (20 mg/ml).
Determination of MIC of curcumin liposomes and free curcumin
The MIC of curcumin liposomes and free curcumin against C. violaceum CV026, A. tumefaciens A136 and A. sobria respectively was measured by broth macrodilution method27. It was performed to ensure that the curcumin liposomes and free curcumin interfered with the QS system of bacteria instead of inhibiting their growth. An overnight culture of C. violaceum CV026 (1.0 OD at 600 nm), A. tumefaciens A136 (1.0 OD at 600 nm) and A. sobria (1.0 OD at 600 nm) were diluted 1:100 in 10 ml LB medium, respectively. To determine the MIC of curcumin liposomes and free curcumin, the medium was supplemented with twofold diluted free curcumin and curcumin liposomes respectively and incubated at 28 °C for 24 h. The absorbance of the medium at 600 nm was measured before and after incubation by spectrophotometer. The MIC was recorded as the lowest concentration, which exhibited complete inhibition of visible growth of test bacteria. The following anti-quorum sensing assays (biofilm formation, siderophore, extracellular protease, swimming and swarming motility as well as AHLs production) of curcumin liposomes and free curcumin were performed only at sub-MIC concentrations. Cholesterol, lecithin, 1% DMSO as well as the sterile water were served as blank control.
Violacein inhibition assay and β-galactosidase assay
The violacein inhibitory assay and β-galactosidase assay were performed according to the method of Issac28. Violacein inhibitory assay was tested by using C. violaceum CV026. Violacein production is dependent on the external addition of 10 μl of C6-HSL (20 μg/ml). In A. tumefaciens A136, the gene lacZ expressed and broke down X-Gal to produce blue color due to the addition of 10 μl of C8-HSL (20 μg/ml). Overnight culture of C. violaceum CV026 (1.0 OD at 600 nm) and A. tumefaciens A136 (1.0 OD at 600 nm) were diluted 1:100 in 10 ml LB medium, respectively. The diluted C. violaceum CV026 (1 ml) was grown at 28 °C on LB plates (20 ml) containing 1.5% agar and 20 μg/ml C6-AHL (10 μl). A. tumefaciens A136 (1 ml) was grown at 28 °C on LB plates (20 ml) containing 1.5% agar and 20 μg/ml C8-AHL (10 μl) as well as 100 μl of X-Gal (20 mg/ml). After the medium solidified, curcumin liposomes (100 μl) at sub-MIC concentration were added to the plates. Cholesterol (50 μl) and lecithin (50 μl) were mixed and served as blank control. The plates were incubated at 28 °C for 24 h and the reduction in violacein production and blue color was observed.
Growth curve analysis
In order to ensure that the curcumin liposomes affected the quorum sensing system of bacteria instead of interfering with bacterial growth, growth curve was measured in the presence of different concertrations of curcumin liposomes and free curcumin under sub-MIC, respectively. The determination method of growth curve was performed according to the previous literature29 with slight modification. Overnight culture (1%) of A. sobria (adjusted to 0.1 OD at 600 nm) and the curcumin liposomes under MIC were added to 10 ml LB medium. As control, the growth curve was obtained in the absence of curcumin liposomes. The solution of curcumin liposomes with fresh LB medium was taken as blank control to measure background curcumin liposomes level. The tubes were put in an incubator shaker at 30 °C 160 r/min for 24 h and the biomass of the A. sobria was measured by enzymatic analyzer (Perkin Elmer V3, USA) at 600 nm every four hours. The growth curve of A. sobria in the presence of free curcumin was measured in the same method.
Preparation of cell-free supernatant
Overnight culture of A. sobria (1.0 OD at 600 nm) treated or untreated with curcumin liposomes and free curcumin respectively was centrifuged at 12000 r/min for 15 min. The bacterial cells were discarded and the supernatant was filtered by using 0.22 μm sterilized film. The supernatant was stored at −20 °C for extracellular protease assay and siderophore production assay.
Determination of biofilm formation, SEM (Scanning electron microscope) observation and confocal laser scanning microscope (CLSM) observation
The qualitative determination of biofilm formation was performed following the method of Gordon Ramage et al.30 with some modifications. This method was performed to measure the effect of curcumin liposomes and free curcumin on biofilm formation of A. sobria, respectively. Briefly, the assay was carried out in 96 well flat bottom plastic microplates (Sarstedt Inc, USA). An overnight culture of A. sobria (grown in LB medium, 1.0 OD at 600 nm) was diluted 1:100 in fresh sterile LB medium. The dilution of A. sobria in the presence or absence of curcumin liposomes under sub-MIC was added to each well. The dilution of A. sobria with sterile water in the absence of curcumin liposomes was used as control. Curcumin liposomes with fresh LB medium was served as blank control to measure background curcumin liposomes level. The microtiter plate was incubated in at 28 °C for 48 h. Subsequently, the biofilm was washed with phosphate buffered saline (PBS, 7 mM Na2HPO4, 3 mM Na2H2PO4, and 130 mM NaCl at pH 7.4) for three times to remove loosely adherent cells. XTT was dissolved in PBS to obtain a final concentration of 0.5 mg/ml. This solution was filtered through a 0.22 μm filter and then stored at −70 °C. Menadione was dissolved in acetone (10 mM). Before the determination of biofilm formation, menadione was added to XTT to attain the final concentration of 1 μM. XTT-menadione solution (200 μl) was added to the wells and incubated in the dark at 37 °C for 2 h, and the absorbance at 490 nm was determined with a microplate reader (Bio-tek, Vermont, USA). The values were the mean of four measurements. The effect of curcumin on biofilm formation of A. sobria was performed in the same method.
An overnight culture of A. sobria (grown in LB medium, 1.0 OD at 600 nm) was diluted 1:100 in LB medium. The diluted culture (20 ml) of A. sobria was added to a petri dish. Then the curcumin liposomes were added to the petri dish to make the concentration of 420 μg/ml. As a control, the diluted culture of A. sobria was grown in the absence of curcumin liposomes. A piece of polished zinc (5 mm × 5 mm × 0.2 mm) was immersed in the above culture to allow the attachment of biofilm. The microtiter plate was statically incubated at 28 °C for 48 h. After incubation, the zinc was taken out and washed gently with sterile water and air dried for 30 min. Then the zinc was immersed into 2–5% (v/v) glutaraldehyde (precooling at 4 °C) for 4 hours and then washed with 50%, 70%, 80% and 90% (v/v) ethanol for 10 min respectively and 100% ethanol for 15 min for twice. Lastly, the zinc was immersed in 25% isoamyl acetate for twice, 15 min each and air dried. The morphological observation of the biofilms on the piece of zinc was performed by SEM after the biofilms were gold coating by a sputter coater. Images were obtained from random positions of biofilms. The SEM observation of biofilm treated with free curcumin was performed in the same method. Each measurement was done in triplicate.
A piece of above zinc attached with biofilms was washed with PBS for three times. Then, it was stained with 0.01% acridine orange (w/v, dissolved in PBS) for 15 min in the dark. After staining, it was washed with sterile PBS for three times and dried for 30 min. Antifade mounting medium (10 μl) was added to the biofilm, and then the biofilms were observed by CLSM (ZEISS, German) (emission: 525 nm, excitation: 488 nm). The 3-dimensional images of biofilms were obtained by ZEN Lite 2012 software (Carl Zeiss Microscopy GmbH, Germany).
Determination of extracellular protease
The determination of extracellular protease was performed following the method of Vijayaraghavan31. It was determined to elucidate the effect of curcumin liposomes on the QS-regulated production of extracellular protease of A. sobria. A transparent zone around the hole indicated the presence of extracellular protease activities. A total of 10 ml 1.5% (w/v) milk (115 °C, 0.06 MPa, 30 mim) was added to 90 ml nutrient agar when the temperature dropped to 50 °C. The above filtered cell-free supernatant (section 2.4.5) in the presence of curcumin liposomes (100 μl) was added to the well and sterilized water (100 μl) was served as the blank control. The extracellular protease was determined by measuring the diameter of the transparent zone around the hole in the milk plates. The effect of free curcumin on production of extracellular protease was performed in the same method. Each measurement was done in triplicate.
Determination of swimming and swarming motility
The effect of curcumin liposomes and free curcumin on the QS-regulated swimming and swarming motility in A. sobria was determined, respectively. Swimming medium contained 1 g tryptone, 0.5 g sodium chloride, 0.3 g agar and 100 ml deionized water. Swarming medium contained 1 g peptone, 0.5 g sodium chloride, 0.3 g agar, 0.5 g D-fructose and 100 ml deionized water. The medium were sterilized at 121 °C for 15 min and solidified at room temperature. Every swimming and swarming plate contained 20 ml medium. A total of 1.5 μl of overnight culture of A. sobria (grown in LB medium, 1.0 OD at 600 nm) supplemented with curcumin liposomes under sub-MIC was inoculated in the swimming and swarming plates, respectively. The control plates were prepared without curcumin liposomes. The plates were incubated at 28 °C for 24 h. The migration distance of A. sobria was measured. Every experiment was performed in triplicate.
Determination of AHLs production
Extraction of the AHLs. The AHLs produced by A. sobria in the presence and absence of curcumin liposomes and free curcumin was extracted, respectively. The overnight cultures of A. sobria treated with curcumin liposomes or free curcumin under sub-MIC were centrifugated at 12,000 g for 4 min respectively to obtain the cell-free supernatant. Then, the supernatant was extracted by ethyl acetate (containing 0.1% acetic acid) for twice. Then the supernatant was concentrated by rotary evaporation at 35 °C. The extract was re-dissolved by using methanol (1 ml) and filtered through 0.22 μm film for GC-MS (Gas-chromatography-mass-spectrometry) detection. As control, the overnight cultures of A. sobria were in the absence of curcumin liposomes and free curcumin, respectively.
The identification of the AHLs via GC-MS. Analyses of the AHLs were performed via a GC-MS Agilent 7890 N/5975 (Agilent, USA) following the method of Zhu et al.32 with slight modification. All sample injections were done in the split mode (50:1) into a HP-5 MS capillary column (30 m length × 0.25 mm internal inameter × 0.25 μm film thichness). Helium was used as the carrier gas at a flow rate of 1 ml/min. The GC injector temperature was 200 °C and the oven temperature was programmed as followed: 150 °C ramped at 10 °C/min to 220 °C, and ramped at 5 °C/min to 250 °C, then ramped at 0.5 °C/min to 252.5 °C. Mass spectrometry conditions were as followed: electron ionization source was set to 70 eV; MS Quad 150 °C, emission current 500 μA, MS Source 230 °C. Data were acquired by full-scan mode (m/z 35–800) and selected ion monitoring (SIM) mode (m/z 143).
Determination of siderophore production
The effect of curcumin liposomes and free curcumin on the QS-regulated siderophore production in A. sobria was determined on solid media using Chrome-Azurol-S CAS-agar, respectively. The color of the plates was blue due to the CAS (Chrome Azurol Sulphonate) complex, ferric iron and hexadecyltrimethylammonium (HDTMA). The color changed to orange when iron was removed by siderophores. Solution A: 0.07 g CAS was dissolved into 50 ml distilled water and 10 ml of 1 mmol/l FeCl3 was added (containing 10 mmol HCl); Solution B: 0.06 g HDTMA was dissolved into 40 ml distilled water; Solution C: Solution A was added to solution B slowly and mixed and the solution C was obtained. Then solution C was sterilized at 121 °C for 15 min. Subsequently, 0.2 ml of 1 mmol/l CaCl2, 0.2 ml of 1 mmol/l MgSO4·7H2O, 2 g agar and 6 ml of 10% (w/v) acid hydrolysis of casein (121 °C sterilized separately) together with Pipes (Sigma) (adjust pH 6.8–7.0) were diluted with water to 100 ml and sterilized at 121 °C for 15 min. Solution C (5 ml) was added to the above medium at 50 °C and mixed. Every plate contained 20 ml medium. The above mentioned filtered cell-free supernatant (section 2.4.5) in the presence of curcumin liposomes (100 μl) was added to CAS plates. The filtered cell-free supernatant without curcumin liposomes or free curcumin was served as control. The plates were incubated at 28 °C for 24 h. The relative amount of siderophores produced were determined by mesuring the diameters of orange halos.
In silico analysis
Protein structure file and ligands
All calculations were performed on a LENOVO Z460 PC equipped with a double processor with the SYBYL-X 2.1 (Triops, Triops Inc., USA). The amino acids sequences of acyl-homoserine-lactone synthase and transcriptional regulators of A. sobria were obtained from the National Centre for Biotechnology Infromation (NCBI) (acyl-homoserine-lactone synthase, WP_042019484; transcriptional regulator, WP_042019486). The models of acyl-homoserine-lactone synthase and transcriptional regulators of A. sobria were built and assessed by using SWISS-MODEL33, 34 (https://swissmodel.expasy.org) and RAMPAGE (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php), respectively. The most homologous sequence was used as template for modeling. A total of 50 models were built by SWISS-MODEL and the best model with the highest similarity and GMQE (Global Model Quality Estimation) score was selected. The models of acyl-homoserine-lactone synthase and transcriptional regulators of A. sobria were subjected for energy minimization by using the SYBYL-X 2.1 program. The 3D structure of curcumin, furfuran C-30 and C6-HSL were obtained from ZINC (http://zinc.docking.org/browse/subsets/) and fully geometry-optimized by using the standard Tripos molecular mechanics force field. The convergence criterion was set to be 0.005 kcal/(Å mol). Partial atomic charges were calculated through the Gasteiger-Hückel method. The structure of curcumin, furfuran C-30 and C6-HSL were converted into mol2 files for molecular docking.
Model assessment and molecular docking
The proteins models of acyl-homoserine-lactone synthase and transcriptional regulators i.e. LuxI and LuxR type proteins of A. sobria were assessed by using SWISS-MODEL Protein Structure & Model Assessment Tools and RAMPAGE. The docking analysis was performed to describe the interactions between the ligands and the modeled LuxI and LuxR type proteins of A. sobria by surflex-Dock in SYBYL-X 2.1 program. The two built proteins were analyzed and prepared by SYBYL-X 2.1 and the water molecules in the models were removed. The docking boxes of two built proteins of A. sobria were chosen to be a radius of 5 Å around the ligands, and the residues which were not in the active site were hided. The mol2 files of curcumin, furfuran C-30 and C6-HSL were docked with the built models and the best conformation of each ligand could be obtained.
Statistical analysis
The results were expressed as means standard deviation (SD). Analysis of variance was conducted and differences between variables were tested for significance by one-way ANOVA with Tukey test using the SPSS Statistics 20.0, Origin Pro 9.0; Differences at P < 0.05 were considered statistically significant. All the experimental data represented the mean of triplicate values.
Data availability statement
All data are fully available without restriction.
Results and Discussion
SEM analysis and characteristics of curcumin liposomes
The SEM image of curcumin liposomes was shown in Fig. 1(A), the curcumin liposomes were in spherical or ellipsoidal shapes and distributed uniformly. The characteristics including particle size, zeta potential, PDI value, encapsulation efficiency and loading capacity were then measured. As listed in Table 1, the curcumin liposomes were similar in size (see Supplemental Fig. S1) and the encapsulation efficiency was high (84.51 ± 0.58%). The liposomes were further stored in 4 °C and their particle size and encapsulation efficiency were measured every two days for two weeks. The results showed that, the particle size and encapsulation efficiency varied very little, confirming that the curcumin liposomes were very stable. Chen et al. had prepared different curcumin-loaded liposomes by using soybean phospholipids, hydrogenated soybean phospholipids and egg yolk phospholipids. The highest encapsulation efficiency of these liposomes was 82.32 ± 3.91%, which was lower than that of our study17. The results indicated that liposomes were excellent carriers for curcumin. Basak et al. also used liposomes to pack curcumin-difluorinated to treat head and neck squamous cell carcinoma35. Narayanan et al. used the liposome to encapsulate curcumin and resveratrol and they found that, the two drugs in combination had the ability to reduce prostate cancer incidence in PTEN knockout mice36. Similarly, Agashe and coworkers used liposomes to package EF24-HPβCD and they found that, the drugs had an anti-proliferative effect in lung and prostate cancer cell lines37. Therefore, it was practicable to use liposomes to package curcumin to improve the its bioavailability.
Figure 1.
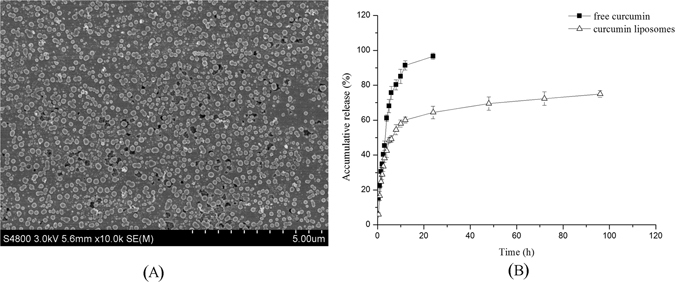
SEM image (A) and curcumin release (B) of prepared curcumin liposomes.
Table 1.
Characteristics of curcumin liposomes.
| Characteristics | Particle size (nm) | Zeta potential (mV) | PDI | Encapsulation efficiency (%) | Loading capacity (%) |
|---|---|---|---|---|---|
| Value | 159.3 ± 6.8 | −39 ± 2.70 | 0.156 ± 0.003 | 84.51 ± 0.58 | 2.87 ± 0.02 |
The release of curcumin from the curcumin liposomes was then examined. As shown in Fig. 1(B), the curcumin liposomes revealed a sustained-release trend. 34.65% of curcumin was released from the release medium, but only 28.87% of curcumin was released from curcumin liposomes within the first 2 h. 96.65% of the curcumin was released in medium, while only 64.39% of curcumin was released from curcumin liposomes after 24 h. The figure showed that, curcumin released quickly from curcumin liposomes at first and then given a slower release, resulting in a higher drug concentration at first, and then slow release of the remaining curcumin maintained efficacy for a longer time.
MIC of curcumin liposomes and free curcumin
Since curcumin has shown better antimicrobial activities, MIC should be measured to ensure that the inhibitory effect was due to interfering with the QS system of bacteria rather than inhibiting their growth. MIC of curcumin liposomes or free curcumin was the lowest concentration that exhibited complete inhibition of visible growth of C. violaceum CV026, A. tumefaciens A136 and A. sobria. The MIC of curcumin liposomes against C. violaceum CV026, A. tumefaciens A136 and A. sobria were 400 µg/ml, 460 µg/ml and 420 µg/ml, respectively. The MIC of free curcumin against C. violaceum CV026, A. tumefaciens A136 and A. sobria were 200 µg/ml, 370 µg/ml, and 280 µg/ml, respectively. All further studies were carried out at sub-MIC concentrations.
Violacein inhibition test and β-galactosidase assay
The detection of QSIs was largely facilitated by the application of biosensors, which allow rapid screening of substances with QS inhibitory activities. As shown in Fig. 2(A,B), AHLs produced by A. sobria were detected by C. violaceum CV026 and A. tumefaciens A136. Result showed that, A. sobria produced both short and long chain AHLs. In Fig. 2(C), the color of control was purple, and this was because of the expression of CviR (a luxR homologue) by acceptance of C6-HSL in the plate38. While there was loss of purple pigment instead of transparent and clear zone around the hole treated with curcumin liposomes. In Fig. 2(D), there was halo zone around the hole in the presence of curcumin liposomes, while the color of the control remains blue. The result indicated that, the curcumin liposomes had exhibited inhibitory effect on the QS system of bacteria rather than affecting the bacterial growth.
Figure 2.

Detection of AHLs produced by A. sobria via indicator bacteria of C. violaceum CV026 (A) and A. tumefaciens A136 (B) as well as violacein inhibition assay (C) and β-galactosidase assay (D) of curcumin liposomes.
Effects of curcumin liposomes on swimming and swarming motility, siderophore as well as extracellaluar proteases of A. sobria
Bacteria mainly relied on flagella in their motility, which was closely related to biofilm formation. Swarming was a group action, while swimming was individual behavior. Both the two bacterial motilities were controlled by QS system, by which bacteria could evade antibacterial agents and body’s immune system39. Many bacteria have shown swimming and swarming motility like Proteusbacillus vulgaris, Vibrio spp., Bacillus spp., Salmonella spp. and Serratia marcescens. As could be seen from Fig. 3 (A–F), the strain of A. sobria migrated from inoculation zone and formed round colonies in swimming and swarming plates. Curcumin liposomes and free curcumin showed a considerable reduction in the motility of A. sobria when compared with control, indicating that the QS-controlled swimming and swarming motility of A. sobria was significantly inhibited. Additionally, curcumin liposomes were more effective than free curcumin in inhibiting swimming and swarming motility of A. sobria. Similarly, Kazemian and coworkers have found that, the extract of Chamaemelum nobile has shown inhibitory effect on the QS-regulated swarming motility and biofilm formation of Pseudomonas aeruginosa 40.
Figure 3.
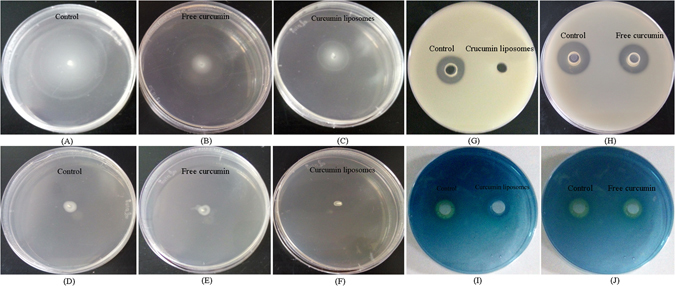
Inhibitory effect of curcumin liposomes and free curcumin (420 µg/ml of liposomes and 280 µg/ml of free curcumin) on swimming (A–C) and swarming motility (D–F), extracellaluar proteases (G,H) as well as siderophore (I,J) of A. sobria.
Spoilage microorganisms decompose proteins in food into small molecular peptides and amino acids by secreting extracellular protease, and ultimately produce volatile and stimulating nitrogen- and sulfur-containing compounds via a series of bacterial metabolisms, resulting in the spoilage of food. Extracellular protease was also one of the most important QS-regulated virulence factors in many bacteria. Christensen et al. studied the expression of QS-controlled extracellular protease in Serratia proteamaculans and they found that, the AHL (3-oxo-C6-HSL) had a very large impact on exoenzymes activies41. In the skimmed milk plates, there was a large and transparent zone around the hole which was in the absence of curcumin liposomes, while the size of transparent zone around the hole treated with free curcumin was smaller than that of control. Further, there was no significant change around the hole in the present of curcumin liposomes (Fig. 3(G,H)). The results indicated that, the inhibitory effect of curcumin liposomes in inhibiting the QS-regulated secretion of extracellular protease was better than that of free curcumin.
Siderophore generally refers to a series of small molecular chelates (1–2 kDa) with strong and specific affinity to Fe3+, which is synthesized by microorganisms through non-ribosome pathway in low iron condition. The CAS plate was blue due to the ternary complex (CAS, HDTMA and Fe3+), but a faint yellow halo appeared around the hole when there were siderophore produced by bacteria. As shown in Fig. 3(I,J), there was no faint yellow halo around the hole in the presence of curcumin liposomes, which was contrary to that of control, suggesting that the QS-regulated siderophore production in A. sobria was inhibited by curcumin liposomes. The inhibitory effect of curcumin in siderophore production in A. sobria was not significant.
Effects of curcumin liposomes on the growth curve and biofilm formation of A. sobria
Biofilms are aggregation of one or several species of bacteria, which are embedded in a extracellular polymeric substance (EPS)13, 40. EPS consists of extracellular DNA, polysaccharides and proteins, etc. Biofilms can attach to various surfaces, including food, medical devices, and host tissues42, 43. And once it forms, it is difficult to be removed even use modern technologies, causing great damages to human health and safety44, 45. Recent studies have found that the formation of biofilm was regulated by QS system42. Chan et al. have found that Aeromonas veronii biovar sobria strain 159 showed QS activities mediated by N-acyl homoserine lactone46. Suntharalingam et al. have found that the biofilm formation of Streptococci was associated with their QS system47. Similarly, Birgit Huber and coworkers48 found that, the biofilm formation and swarming motility of Burkholderia cepacia were regulated by its QS system. Thus, it is a feasible and potential way to hinder the bacterial biofilm formation by blocking their quorum sensing pathways.
In qualitative analysis, the biofilm formation of A. sobria was inhibited by free curcumin and curcumin liposomes under sub-MIC in a concentration-dependent manner. The biofilm has fallen by 51.90% when treated with free curcumin at 280 µg/ml, whereas, curcumin liposomes inhibited biofilm formation up to 93.35% when compared to control (p < 0.05) at 420 µg/ml. More importantly, the growth curve of A. sobria was typical microbial growth curve (Fig. 4), indicating that the curcumin liposomes did not affect the growth of A. sobria but interfered with their QS system.
Figure 4.
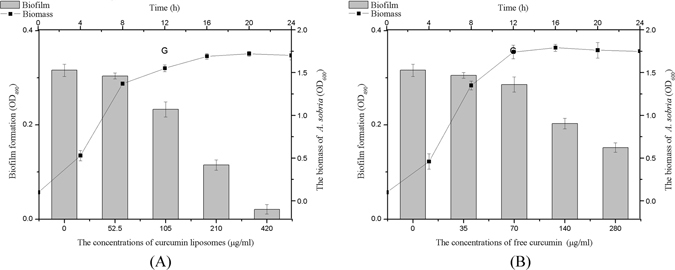
Effect of different concentrations of curcumin liposomes (1) or free curcumin (2) on growth and biofilm formation of A. sobria. Each bar and line represents mean of three replicates and standard deviations.
The biofilm produced by A. sobria could be observed clearly by SEM. The bacterial cells were rod-shaped, aggregated, and formed dense biofilm. Conversely, the biofilm was looser and bacterial cells were scattered when treated with free curcumin and curcumin liposomes. Little biofilms were formed when treated with curcumin liposomes (Fig. 5(A–C)). What’s more, the morphology of A. sobria was normal. The result also suggested that, the curcumin liposomes had inhibitory effect on the biofilm formation of A. sobria and they did not interfere with normal growth of bacteria. Qu Lin et al. got the similar conclusion that norspermidine could prevent and eradicate the biofilm formation of P. aeruginosa. And they also observed the initial attachment of biofilm by SEM44. QS-regulated production of EPS contributes the mature of biofilm49. From SEM images, EPS of biofilms treated with curcumin liposomes decreased significantly when compared with that of free curcumin and control. The reason may be that, the curcumin released from curcumin liposomes penetrated into bacterial cells due to the reduction of EPS and interfered with the QS system of bacteria. Junli Zhu et al. also found that, green tea polyphenols had inhibitory effect on the EPS production of Shewanella balitica 50. Interestingly, some of the curcumin liposomes were attached to the surface of bacterial cells. CLSM images (Fig. 5(D–F)) showed that, the color of control was very green and the biofilm was thick, dense, and compact, whereas the biofilm treated with free curcumin was dispersed and the intensity of green color was lower. The biofilm treated with curcumin liposomes was the thinnest and the green color was the lowest. The results revealed that curcumin liposomes significantly decreased the biofilm formation in A. sobria. The result was corresponded to the qualitative analysis.
Figure 5.
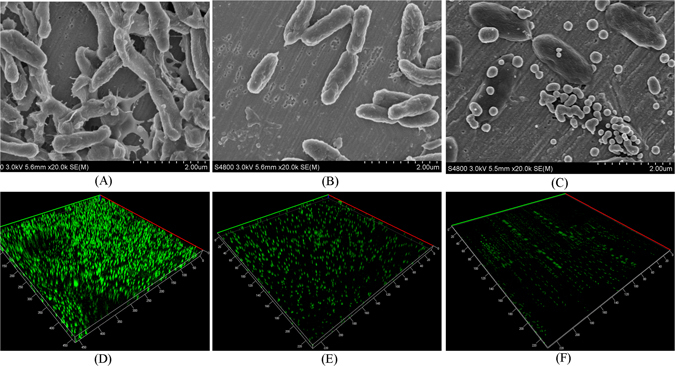
SEM analysis (A–C) and CLSM observation (D–F) of biofilm of A. sobria treated with curcumin liposomes and free curcumin. (A,D): biofilm of A. sobria; (B,E) biofilm of A. sobria treated with free curcumin; (C,F): biofilm of A. sobria treated with curcumin liposomes.
Effects of curcumin liposomes on AHL production of A. sobria
AHLs were the signal molecules used by most Gram-negative bacteria, and the AHLs might be different in N-acyl chain and carbon chain produced by various bacterial species51. Numerous studies have shown that AHLs-mediated quorum sensing was closely associated with bacterial pathogenicity25, 45.
It has been more accurate to detect and analyze AHLs quantitatively by using GC-MS (see Supplemental Fig. S2). Table 2 was the changes in AHLs production of A. sobria in the presence and absence of free curcumin and curcumin liposomes. A. sobria produced C4-HSL, C6-HSL, C10-HSL, C12-HSL and C14-HSL, while it produced C4-HSL, C6-HSL, C10-HSL and C14-HSL when treated with free curcumin. Only C4-HSL and C14-HSL produced when treated with curcumin liposomes under sub-MIC. C4-HSL, C6-HSL, C10-HSL and C14-HSL treated with free curcumin decreased by 64.65%, 37.67%, 83.60% and 73.00%, respectively. While C4-HSL and C4-HSL decreased by 71.36% and 93.55% respectively when treated with curcumin liposomes. The result showed that, the inhibitory effect of curcumin liposomes on AHLs production of A. sobria was more significant than that of free curcumin. This study was consistent with Luciardi et al. that the volatile metabolites of Merremia dissecta creeper had the ability to hinder the QS system of Pseudomonas aeruginosa. And the AHLs content decreased 63–75% by GC-MS detection52.
Table 2.
Effect of curcumin liposomes and free curcumin on AHLs production of A. sobria.
| Signal molecules | Peak time | Peak Area | ||
|---|---|---|---|---|
| Control | Treated with free curcumin | Treated with curcumin liposomes | ||
| C4-HSL | 4.431 | 4,766 | 1,685 | 1,365 |
| C6- HSL | 6.286 | 4,959 | 3,091 | — |
| C8- HSL | 8.333 | — | — | — |
| C10- HSL | 10.720 | 76,520 | 12,550 | — |
| C12- HSL | 13.422 | 37,501 | — | — |
| C14- HSL | 16.939 | 99,515 | 26,862 | 6,419 |
–: Not detected.
Sequence alignment and homology modeling
For a variety of difficulties, the structures of certain proteins could not be known, therefore, plenty methods were utilized to solve this problem including homology modeling. Computer homology modeling techniques are generally used to build three dimensional (3D) proteins with unknown structures. The 3D structures of LuxI and LuxR type proteins of A. sobria were built via sequence alignment and homology modeling by SWISS-MODEL. The models were built through Blast and HHBlits search to match the target amino acid sequence in the SWISS-MODEL template library. Plenty sequences were searched by SWISS-MODEL and the templates which similarity above 30% were selected to build models. Three and five models for LuxI and LuxR type protein respectively were selected based on the sequence similarities and GMQE score.
The sequence alignment results were shown in Fig. 6(A,B). The sequence alignment of the target protein with template proteins exhibited that, the residues in the active site were strictly conserved. There were three matches for LuxI type protein (acyl-homoserinelactone synthase EsaI, autoinducer synthesis protein lasI and AHL synthase) and five matches for LuxR type protein (transcriptional regulator of ftsQAZ gene cluster, regulatory protein SdiA, probable transcriptional activator protein traR, CviR transcriptional regulator and quorum-sensing control repressor) of A. sobria to build models (Table 3).
Figure 6.
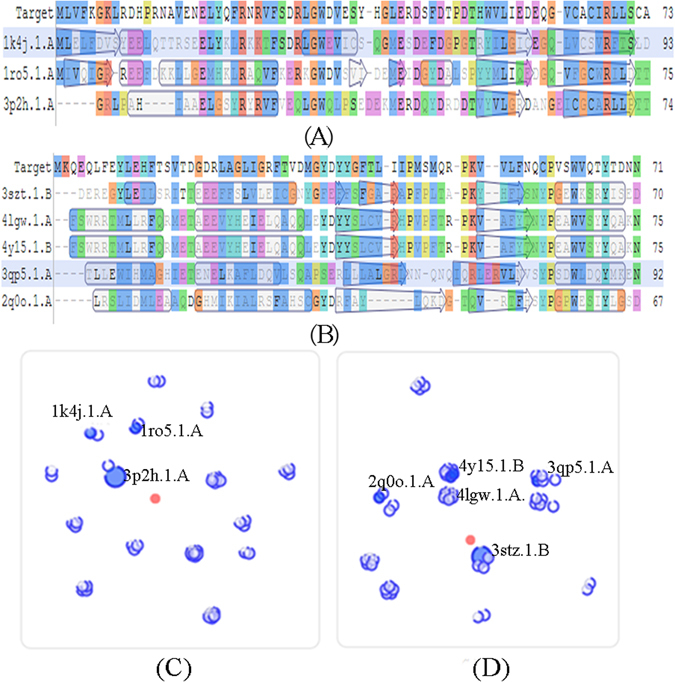
Part of sequence alignment results of LuxI (1) and LuxR type (2) proteins of A. sobria and their plots of scores (3: LuxI; 4: LuxR).
Table 3.
Structures and scores of LuxI and LuxR type proteins models of A. sobria.
| Proteins | Template | Description | GMQE | QMEAN | Model |
|---|---|---|---|---|---|
| LuxI type | 1k4j.1.A | acyl-homoserinelactone synthase EsaI | 0.54 | −4.55 |
|
| 1ro5.1.A | autoinducer synthesis protein lasI | 0.55 | −2.28 |
|
|
| 3p2h.1.A | AHL synthase | 0.61 | −3.14 |
|
|
| LuxR type | 4y15.1.B | transcriptional regulator of ftsQAZ gene cluster | 0.63 | −3.36 |
|
| 4lgw.1.A | regulatory protein SdiA | 0.64 | −2.91 |

|
|
| 2q0o.1.A | probable transcriptional activator protein traR | 0.60 | −3.88 |
|
|
| 3qp5.1.A | CviR transcriptional regulator | 0.62 | −4.85 |
|
|
| 3stz.1.B | quorum-sensing control repressor | 0.63 | −2.54 |
|
The 3D models of LuxI and LuxR type proteins were built from the most homologous sequence of each protein. The AHL synthase (3p2h.1.A) and quorum-sensing control repressor (3stz.1.B) were selected as the templates for LuxI and LuxR type proteins modeling respectively based on the GMQE and QMEAN score53. The plots of scores of the proteins (Fig. 6(C,D)) also showed that, the 3p2h.1.A and 3stz.1.B for LuxI and LuxR type proteins respectively were nearest to the center points (target sequence), suggesting that the two models were the most suitable templates for builting LuxI and LuxR type proteins of A. sobria. The similarity of the templates was all above 30%, indicating that the build models could make a good prediction of the target proteins.
Model quality assessment
The quality of build models were evaluated by using the SWISS-MODEL and RAMPAGE. QMEAN is a composite scoring function for both the estimation of the global quality of the entire model as well as for the local per-residue analysis of different regions within a model53. The QMEAN score and Z-score were 0.68 and −0.88 for LuxI type protein, and 0.76 and −0.10 for LuxR type protein of A. sobria (Fig. 7(A–F)). The results indicated that the models are valid, and the built models could make a good prediction for the 3D structures of LuxI and LuxR type proteins. The Ramachandran plot analysis (Fig. 7(G,H)) showed that, in LuxI type models, 89.1% residues were in favoured region, 9.8% in allowed region, 1.1% in the outlier region; whereas the corresponding values for the Lux R type protein were 91.7%, 7.0%, 1.3%, respectively. This ensured that the quality of generated model was good. Similarly, Al-Khayyat et al. have built the LuxI protein model, which was similar to the closely-related known structures by homology modeling method. And model 4 with the QMEAN6 score of 0.732 had better quality among the five built models and they selected ten compounds for docking analysis with the built models54.
Figure 7.
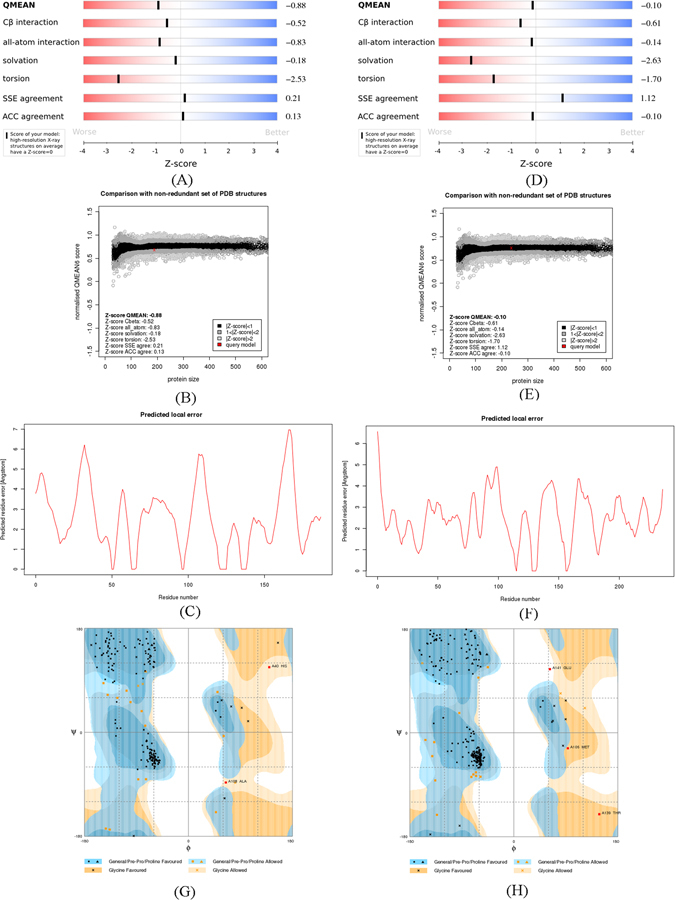
Assessment of the quality of LuxI (1-3) and LuxR (4–6) type proteins of A. sobria by SWISS-MODEL and RAMPAGE (7: Ramachandran Plot Analysis of LuxI type protein; 8: Ramachandran Plot Analysis of LuxR type protein).
Docking analysis
Molecular docking analysis was performed to elucidate the mechanism of curcumin acted on the QS system of A. sobria. In order to clarify the interactions between curcumin and the two built proteins of A. sobria, curcumin was docked with the active sites of built LuxI and LuxR type proteins. Futher, C6-HSL was used as native ligand when docked with LuxR type protein. Furanone C-30 and its derivate i.e. furanone-56 were known inhibitors of QS system, which could interfere with the combination of AHLs with their receptor proteins. It has been reported to inhibit the production of biofilm and virulence factors as well as other physiological activities of Pseudomonas aeruginosa 55. Thus, furanone C-30 was used as positive control to be docked with LuxR type protein of A. sobria.
Docking results of LuxI and LuxR proteins with their ligands were listed in Table 4. The results showed that, curcumin docked with the active site of LuxI and LuxR type proteins of A. sobria with total scores of 8.0610 and 6.4729, respectively. In addition, C6-HSL and furanone C-30 were also docked with Lux R type protein of A. sobria with total scores of 4.5625 and 1.2519, respectively. Molecular docking studies revealed that, curcumin showed higher in silico binding affinity to the LuxI type protein of A. sobria. The docking calculations with 20 conformations for each ligand (Fig. 8(E–H)) showed that, the conformations distribution of curcumin with LuxI type protein were more centralized than that of others. The above results suggested that the inhibitory mechanism of curcumin on QS system of A. sobria might be that, the curcumin combined with LuxI type protein and blocked the production of AHLs in A. sobria.
Table 4.
Docking results of LuxI and LuxR type proteins of A. sobria with curcumin, C6-HSL and furanone C-30.
| Protein | Ligand | Total score | Crash | Polar | Compound structure |
|---|---|---|---|---|---|
| LuxI type | curcumin | 8.0610 | −1.5280 | 3.7959 |
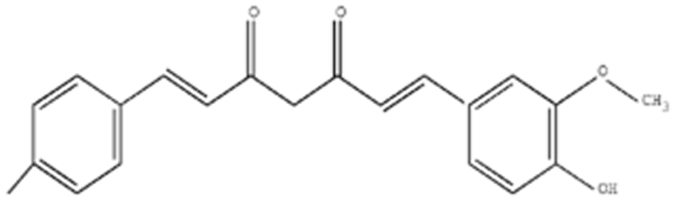
|
| LuxR type | curcumin | 6.4729 | −1.7836 | 2.0901 | |
| C6-HSL | 4.5625 | −0.8459 | 1.9111 |
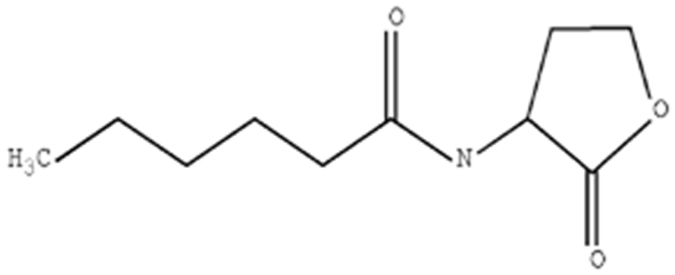
|
|
| furanone C-30 | 1.2519 | −0.1880 | 0.0000 |
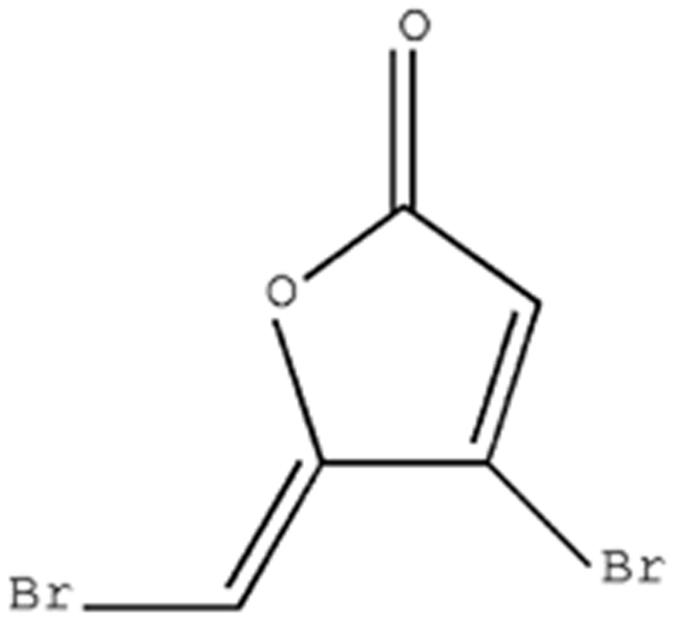
|
Figure 8.
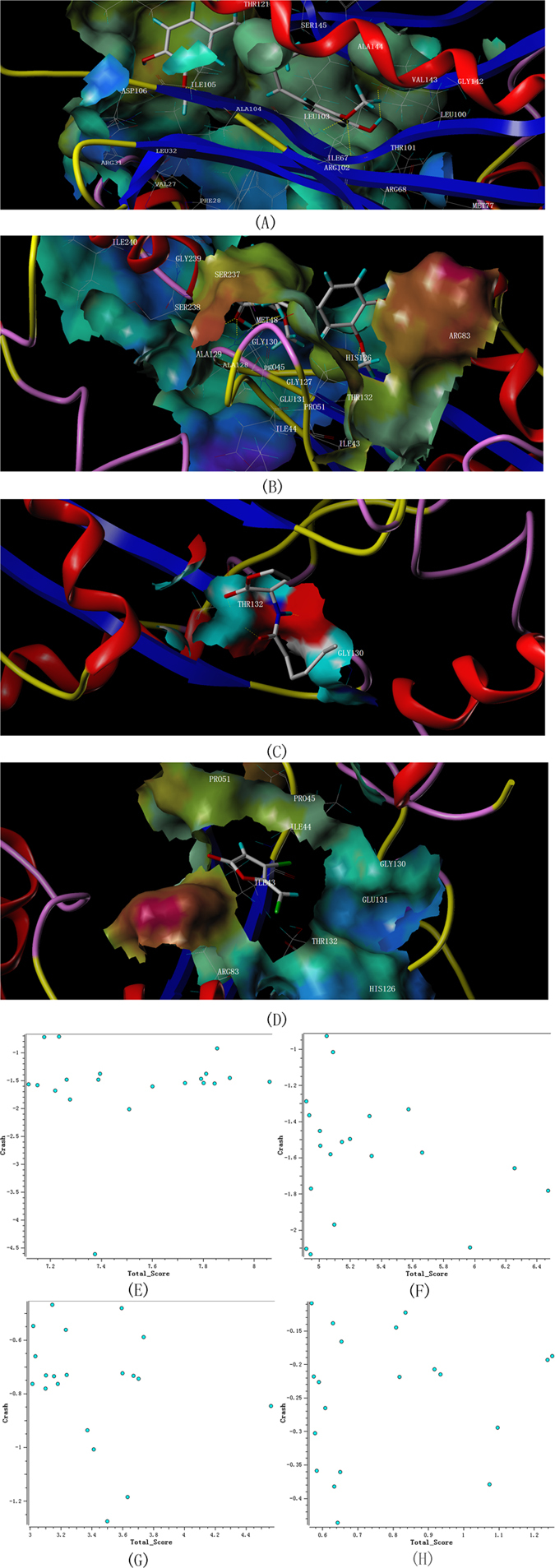
kGraphical representation of LuxI and LuxR type proteins of A. sobria docked with curcumin, C6-HSL and furanone C-30 as well as the conformations of the ligands. Yellow dashed line represents the Hydrogen-bonds. Residues in the active site were labeled. (1) Curcumin docked with modeled LuxI type protein (2) Curcumin docked with modeled LuxR type protein (3) C6-HSL docked with modeled LuxR type protein (4) Furanone C-30 docked with modeled LuxR type protein (5) Conformations of curcumin docked with LuxI type protein (6) Conformations of curcumin docked with LuxR type protein (7) Conformations of C6-HSL docked with LuxR type protein (8) Conformations of furanone C-30 docked with LuxR type protein.
The interactions between the ligands and the two built models of A. sobria were analyzed by SYBYL-X 2.1. As shown in Fig. 8(A–D), curcumin interacted with key residues (VAL143, ARG102, LEU103, SER145 and ILE105) of LuxI type protein via hydrogen-bonding interactions. Similarly, the residues of LuxR type protein of A. sobria interacted with curcumin were found to be THR132, GLY130, ALA129 and GLY127. For the native ligand, C6-HSL has formed hydrogen bonds with GLY130 and THR132 residues in LuxR type protein, while there were no hydrogen bonds formed between furanone C-30 and LuxR type protein of A. sobria. Overall, the above results had demonstrated that, the QS system inhibited by curcumin liposomes was largely because that, the curcumin released from curcumin liposomes interacted with LuxI type protein by forming hydrogen bonds in A. sobria.
Conclusion
This study exhibited that the prepared curcumin liposomes had significantly inhibited the QS-controlled siderophore production, swimming and swarming motility, extracellular proteases, biofilm formation and AHLs production of A. sobria. In silico molecular docking analysis revealed that, the QS system inhibited by curcumin liposomes was largely because that, the curcumin released from curcumin liposomes interacted with LuxI type protein by forming hydrogen bonds, which hindered the production of AHLs in A. sobria. The built LuxI and LuxR proteins might be useful and potential targets to design novel quorum sensing inhibitors.
Electronic supplementary material
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (No. 31471639, No. 31301572), China Postdoctoral Science Foundation (No. 2014M552302).
Author Contributions
Ting Ding carried out the experiments and wrote the manuscript. Tingting Li, Jianrong Li and Zhi Wang revised the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Ting Ding and Tingting Li contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-08986-9
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Spadaro S, et al. Aeromonas sobria necrotizing fasciitis and sepsis in an immunocompromised patient: a case report and review of the literature. J Med Case Rep. 2014;8 doi: 10.1186/1752-1947-8-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang H, Hung YS, Shie SS, Lin TL. Fulminant necrotizing fasciitis caused by Aeromonas sobria in neutropenic patients. Intern Med. 2012;51:3287–3290. doi: 10.2169/internalmedicine.51.6281. [DOI] [PubMed] [Google Scholar]
- 3.Litwinowicz A, Blaszkowska J. Preventing infective complications following leech therapy: elimination of symbiotic Aeromonas spp. from the intestine of Hirudo verbana using antibiotic feeding. Surg Infect (Larchmt) 2014;15:757–762. doi: 10.1089/sur.2014.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bucio-Cano A, et al. Targeting quorum sensing by designing azoline derivatives to inhibit the N-hexanoyl homoserine lactone-receptor CviR: Synthesis as well as biological and theoretical evaluations. Bioorg Med Chem. 2015;23:7565–7577. doi: 10.1016/j.bmc.2015.10.046. [DOI] [PubMed] [Google Scholar]
- 5.Boyle KE, Monaco H, van Ditmarsch D, Deforet M, Xavier JB. Integration of Metabolic and Quorum Sensing Signals Governing the Decision to Cooperate in a Bacterial Social Trait. PLoS computational biology. 2015;11 doi: 10.1371/journal.pcbi.1004279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alasil SM, Omar R, Ismail S, Yusof MY. Inhibition of Quorum Sensing-Controlled Virulence Factors and Biofilm Formation in Pseudomonas aeruginosa by Culture Extract from Novel Bacterial Species of Paenibacillus Using a Rat Model of Chronic Lung Infection. International journal of bacteriology. 2015;2015 doi: 10.1155/2015/671562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busetti A, et al. Marine-derived quorum-sensing inhibitory activities enhance the antibacterial efficacy of tobramycin against Pseudomonas aeruginosa. Marine drugs. 2015;13:1–28. doi: 10.3390/md13010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Priya K, Yin WF, Chan KG. Anti-quorum sensing activity of the traditional Chinese herb. Phyllanthus amarus. Sensors. 2013;13:14558–14569. doi: 10.3390/s131114558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan KG, et al. Quorum sensing in Aeromonas species isolated from patients in Malaysia. Curr Microbiol. 2011;62:167–172. doi: 10.1007/s00284-010-9689-z. [DOI] [PubMed] [Google Scholar]
- 10.Jakobsen TH, et al. Food as a source for quorum sensing inhibitors: iberin from horseradish revealed as a quorum sensing inhibitor of Pseudomonas aeruginosa. Applied and environmental microbiology. 2012;78:2410–2421. doi: 10.1128/AEM.05992-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo J, et al. Baicalein attenuates the quorum sensing-controlled virulence factors of Pseudomonas aeruginosa and relieves the inflammatory response in P. aeruginosa-infected macrophages by downregulating the MAPK and NFkappaB signal-transduction pathways. Drug design, development and therapy. 2016;10:183–203. doi: 10.2147/DDDT.S97221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gopu V, Meena CK, Shetty PH. Quercetin Influences Quorum Sensing in Food Borne Bacteria: In-Vitro and In-Silico Evidence. PLoS One. 2015;10 doi: 10.1371/journal.pone.0134684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HS, Lee SH, Byun Y, Park HD. 6-Gingerol reduces Pseudomonas aeruginosa biofilm formation and virulence via quorum sensing inhibition. Scientific reports. 2015;5 doi: 10.1038/srep08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssens JC, et al. Brominated furanones inhibit biofilm formation by Salmonella enterica serovar Typhimurium. Appl Environ Microbiol. 2008;74:6639–6648. doi: 10.1128/AEM.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang S, et al. Bicyclic brominated furanones: a new class of quorum sensing modulators that inhibit bacterial biofilm formation. Bioorg Med Chem. 2014;22:1313–1317. doi: 10.1016/j.bmc.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdel-Lateef E, et al. Bioactive chemical constituents of Curcuma longa L. rhizomes extract inhibit the growth of human hepatoma cell line (HepG2) Acta Pharm. 2016;66:387–398. doi: 10.1515/acph-2016-0028. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, et al. Preparation of curcumin-loaded liposomes and evaluation of their skin permeation and pharmacodynamics. Molecules. 2012;17:5972–5987. doi: 10.3390/molecules17055972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ali A, Banerjea AC. Curcumin inhibits HIV-1 by promoting Tat protein degradation. Sci Rep. 2016;6 doi: 10.1038/srep27539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kali A, Bhuvaneshwar D, Charles PM, Seetha KS. Antibacterial synergy of curcumin with antibiotics against biofilm producing clinical bacterial isolates. J Basic Clin Pharm. 2016;7:93–96. doi: 10.4103/0976-0105.183265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thimmaraju Rudrappa HPB. Curcumin, a known phenolic from Curcuma longa, attenuates the virulence of Pseudomonas aeruginosa PAO1 in whole plant and animal pathogenicity models. J. Agric. Food Chem. 2008;56:1955–1962. doi: 10.1021/jf072591j. [DOI] [PubMed] [Google Scholar]
- 21.Packiavathy IA, Priya S, Pandian SK, Ravi AV. Inhibition of biofilm development of uropathogens by curcumin - an anti-quorum sensing agent from Curcuma longa. Food Chem. 2014;148:453–460. doi: 10.1016/j.foodchem.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Cheng AL HC, et al. Phase I clinical trial of curcumin, a chemopreventive agent, in patients with high-risk or pre-malignant lesions. Anticancer Res. 2001;21:2895–2900. [PubMed] [Google Scholar]
- 23.Thimmaraju Rudrappa, H. P. B. Curcumin, a known phenolic from Curcuma longa, attenuates the virulence of Pseudomonas aeruginosa PAO1 in whole plant and animal pathogenicity models. J. Agric. Food Chem. 1955–1962 (2008). [DOI] [PubMed]
- 24.Takahashi M, et al. Characterization and bioavailability of liposomes containing a ukon extract. Biosci Biotechnol Biochem. 2008;72:1199–1205. doi: 10.1271/bbb.70659. [DOI] [PubMed] [Google Scholar]
- 25.Zhang C, Zhu S, Jatt AN, Zeng M. Characterization of N-acyl homoserine lactones (AHLs) producing bacteria isolated from vacuum-packaged refrigerated turbot (Scophthalmus maximus) and possible influence of exogenous AHLs on bacterial phenotype. J Gen Appl Microbiol. 2016;62:60–67. doi: 10.2323/jgam.62.60. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Y, et al. Preparation and in vitro evaluation of povidone-sodium cholate-phospholipid mixed micelles for the solubilization of poorly soluble drugs. Arch Pharm Res. 2010;33:911–917. doi: 10.1007/s12272-010-0614-6. [DOI] [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, approved standard (7th ed.). Clinical and Laboratory, Standards Institute documentM7-A7 (2006).
- 28.Issac Abraham Sybiya Vasantha Packiavathy PA, Khadar Syed Musthafa SK, Pandian AVR. Antibiofilm and quorum sensing inhibitory potential of Cuminum cyminum and its secondary metabolite methyl eugenol against Gram negative bacterial pathogens. Food Research International. 2012;45:85–92. doi: 10.1016/j.foodres.2011.10.022. [DOI] [Google Scholar]
- 29.Zhang J, et al. Polyphenolic extract from Rosa rugosa tea inhibits bacterial quorum sensing and biofilm formation. Food Control. 2014;42:125–131. doi: 10.1016/j.foodcont.2014.02.001. [DOI] [Google Scholar]
- 30.Ramage G, Saville SP, Wickes BL, Lopez-Ribot JL. Inhibition of Candida albicans Biofilm Formation by Farnesol, a Quorum-Sensing Molecule. Applied and environmental microbiology. 2002;68:5459–5463. doi: 10.1128/AEM.68.11.5459-5463.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vijayaraghavan P, Vincent SGP. A simple method for the detection of protease activity on agar plates using bromocresolgreen dye. Journal of Biochemical Technology. 2013;4:628–630. [Google Scholar]
- 32.Zhu J, Zhao A, Feng L, Gao H. Quorum sensing signals affect spoilage of refrigerated large yellow croaker (Pseudosciaena crocea) by Shewanella baltica. International journal of food microbiology. 2016;217:146–155. doi: 10.1016/j.ijfoodmicro.2015.10.020. [DOI] [PubMed] [Google Scholar]
- 33.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 35.Basak SK, et al. Liposome encapsulated curcumin-difluorinated (CDF) inhibits the growth of cisplatin resistant head and neck cancer stem cells. Oncotarget. 2015;6:18504–18517. doi: 10.18632/oncotarget.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narayanan NK, Nargi D, Randolph C, Narayanan BA. Liposome encapsulation of curcumin and resveratrol in combination reduces prostate cancer incidence in PTEN knockout mice. Int J Cancer. 2009;125:1–8. doi: 10.1002/ijc.24336. [DOI] [PubMed] [Google Scholar]
- 37.Agashe H, et al. Liposome-encapsulated EF24-HPbetaCD inclusion complex: a preformulation study and biodistribution in a rat model. J Nanopart Res. 2011;13:2609–2623. doi: 10.1007/s11051-010-0154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McClean KH, et al. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143(Pt 12):3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, et al. A novel regulator PA5022 (aefA) is involved in swimming motility, biofilm formation and elastase activity of Pseudomonas aeruginosa. Microbiological research. 2015;176:14–20. doi: 10.1016/j.micres.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Kazemian H, et al. Antibacterial, anti-swarming and anti-biofilm formation activities of Chamaemelum nobile against Pseudomonas aeruginosa. Revista da Sociedade Brasileira de Medicina Tropical. 2015;48:432–436. doi: 10.1590/0037-8682-0065-2015. [DOI] [PubMed] [Google Scholar]
- 41.Christensen AB, et al. Quorum-sensing-directed protein expression in Serratia proteamaculans B5a. Microbiology. 2003;149:471–483. doi: 10.1099/mic.0.25575-0. [DOI] [PubMed] [Google Scholar]
- 42.Rasamiravaka T, et al. Pseudomonas aeruginosa Biofilm Formation and Persistence, along with the Production of Quorum Sensing-Dependent Virulence Factors, Are Disrupted by a Triterpenoid Coumarate Ester Isolated from Dalbergia trichocarpa, a Tropical Legume. PloS one. 2015;10 doi: 10.1371/journal.pone.0132791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maurer S, et al. Tasting Pseudomonas aeruginosa Biofilms: Human Neutrophils Express the Bitter Receptor T2R38 as Sensor for the Quorum Sensing Molecule N-(3-Oxododecanoyl)-l-Homoserine Lactone. Frontiers in immunology. 2015;6 doi: 10.3389/fimmu.2015.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu L, et al. Effects of norspermidine on Pseudomonas aeruginosa biofilm formation and eradication. MicrobiologyOpen. 2016;5:402–412. doi: 10.1002/mbo3.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Husain FM, et al. Sub-MICs of Mentha piperita essential oil and menthol inhibits AHL mediated quorum sensing and biofilm of Gram-negative bacteria. Frontiers in microbiology. 2015;6 doi: 10.3389/fmicb.2015.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan XY, How KY, Yin WF, Chan KG. N-Acyl Homoserine Lactone-Mediated Quorum Sensing in Aeromonas veronii biovar sobria Strain 159: Identification of LuxRI Homologs. Front Cell Infect Microbiol. 2016;6 doi: 10.3389/fcimb.2016.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suntharalingam P, Cvitkovitch DG. Quorum sensing in streptococcal biofilm formation. Trends Microbiol. 2005;13:3–6. doi: 10.1016/j.tim.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 48.Birgit Huber, K. R. et al. Leo Eberl. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 2517–2528 (2001). [DOI] [PubMed]
- 49.Packiavathy IA, Sasikumar P, Pandian SK, Veera Ravi A. Prevention of quorum-sensing-mediated biofilm development and virulence factors production in Vibrio spp. by curcumin. Applied microbiology and biotechnology. 2013;97:10177–10187. doi: 10.1007/s00253-013-4704-5. [DOI] [PubMed] [Google Scholar]
- 50.Zhu J, Huang X, Zhang F, Feng L, Li J. Inhibition of quorum sensing, biofilm, and spoilage potential in Shewanella baltica by green tea polyphenols. Journal of microbiology. 2015;53:829–836. doi: 10.1007/s12275-015-5123-3. [DOI] [PubMed] [Google Scholar]
- 51.Kumar AS, Bryan JN, Kumar SR. Bacterial quorum sensing molecule N-3-oxo-dodecanoyl-L-homoserine lactone causes direct cytotoxicity and reduced cell motility in human pancreatic carcinoma cells. PloS one. 2014;9 doi: 10.1371/journal.pone.0106480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luciardi MC, et al. Volatiles from Subtropical Convolvulaceae That Interfere with Bacterial Cell-to-Cell Communication as Potential Antipathogenic Drugs. Evidence-based complementary and alternative medicine: eCAM. 2016;2016 doi: 10.1155/2016/7890260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Al-Khayyat MZ, Al-Dabbagh AG. In silico Prediction and Docking of Tertiary Structure of LuxI, an Inducer Synthase of Vibrio fischeri. Rep Biochem Mol Biol. 2016;4:66–75. [PMC free article] [PubMed] [Google Scholar]
- 55.Morten Hentzer KR, et al. Kjelleberg, Michael Givskov. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. 2002;148:87–102. doi: 10.1099/00221287-148-1-87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are fully available without restriction.


