Abstract
Anthranilate, one of tryptophan degradation products has been reported to interfere with biofilm formation by Pseudomonas aeruginosa. Here, we investigated the effects of anthranilate on biofilm formation by various bacteria and the mechanisms responsible. Anthranilate commonly inhibited biofilm formation by P. aeruginosa, Vibrio vulnificus, Bacillus subtilis, Salmonella enterica serovar Typhimurium, and Staphylococcus aureus, and disrupted biofilms preformed by these bacteria. Because anthranilate reduced intracellular c-di-GMP and enhanced swimming and swarming motilities in P. aeruginosa, V. vulnificus, B. subtilis, and S. enterica, it is likely that anthranilate disrupts biofilms by inducing the dispersion of these bacteria. On the other hand, in S. aureus, a non-flagellate bacterium that has no c-di-GMP signaling, anthranilate probably inhibits biofilm formation by reducing slime production. These results suggest that anthranilate has multiple ways for biofilm inhibition. Furthermore, because of its good biofilm inhibitory effects and lack of cytotoxicity to human cells even at high concentration, anthranilate appears to be a promising agent for inhibiting biofilm formation by a broad range of bacteria.
Introduction
Biofilms are surface-attached microbial communities encapsulated in extracellular polymeric substances (EPS)1, 2. Biofilms cause serious infections and help make infections chronic. One study showed biofilms were detected in 60% of chronic wound specimens, and according to researchers at the Centers for Disease Control and Prevention (CDC), nearly 80% of chronic infections are mediated by the microbial biofilms3, 4. Moreover, bacterial cells within biofilms are nearly 1000 times more resistant to antibiotics than their planktonic counterparts5, 6.
Many biofilm inhibitors, such as anti-microbial peptides, metal chelators, quorum sensing inhibitors, and amino acids have been discovered for decades4, 7–11. The inhibition of biofilm formation by D-amino acids has been reported in the gram-positive species, Bacillus subtilis and Staphylococcus aureus 12, 13. D-Tryptophan inhibits B. subtilis biofilm formation by incorporated into peptidoglycan synthesis and decreasing the expressions of the matrix-producing operons, epsA and tapA 14. Tryptophan has been reported to inhibit biofilm formation by the gram-negative pathogens Escherichia coli and Pseudomonas aeruginosa 15, 16. Furthermore, tryptophan-coated biological dressing has been reported to have a beneficial effect on wound healing and closure17–19.
Indole is a tryptophan metabolite and is produced by a wide range of bacterial species that harbor tnaA gene encoding tryptophanase20. In E. coli, indole has been shown to reduce biofilm formation by interacting with SidA-mediated transcription21. Its derivatives, 5-hydroxyindole and 7-hydroxyindole, have also been shown to reduce biofilm formation by E.coli 22. In recent studies, indole was found to enhance biofilm formation by P. aeruginosa in a quorum sensing (QS)-independent manner21, 23.
Some other bacteria, such as, P. aeruginosa, metabolize tryptophan to anthranilate through a kynurenine pathway using kynBAU genes24. Recently, it was reported in P. aeruginosa that anthranilate degradation is related to biofilm formation. P. aeruginosa mutants unable to degrade anthranilate formed less biofilm, and the expressions of genes in the anthranilate degradation pathway were enhanced during biofilm formation without activation of the synthetic pathway for Pseudomonas quinolone signal (PQS; 2-heptyl-3-hydroxy-4-quinolone). Since anthranilate is a precursor of PQS, this result suggests anthranilate is exhausted by degradation via the TCA cycle when biofilms are formed25. Later, it was found that anthranilate can disrupt biofilm structures and induce the detachment of preformed P. aeruginosa biofilms by reducing intracellular c-di-GMP levels and modulating the expressions of genes involved in EPS production23.
However, it remains unclear whether anthranilate affects biofilm formation by other microorganisms, despite its potent biofilm-inhibiting effects. In this study, we investigated the effects of anthranilate on biofilm formation by various bacteria, that is, P. aeruginosa, Vibrio vulnificus, B. subtilis, S. aureus, and Salmonella enterica serovar Typhimurium. We found that anthranilate disrupted biofilm formation by a wide range of bacteria and that it did so via multiple mechanisms. Since we confirmed that anthranilate has no cytotoxicity to human cells even at high concentration, we suggest that anthranilate is a promising anti-biofilm agent to inhibit biofilm formation of broad range of bacteria.
Materials and Methods
Bacterial strains and culture conditions
The bacterial strains and plasmids used in this study are listed in Table S1. P. aeruginosa, S. aureus, B. subtilis, and S. enterica strains were grown in Luria-Bertani (LB) medium at 37 °C with constant shaking (170 rpm), but the NaCl concentration was increased to 2.0% (wt/vol) for V. vulnificus (this medium is indicated as LBS). Anthranilate was dissolved at 1 M in dimethyl sulfoxide (DMSO) as a stock solution and diluted into media to a final concentration of 0.1 mM. Cell growth was determined by measuring optical density at 600 nm (OD600). To select plasmid-harboring strains, carbenicillin (Car) or erythromycin (Erm) were used at 150 μg/ml for P. aeruginosa strains harboring pSKcdrA or at 10 μg/ml for S. aureus AH1121, respectively. To induce GFP (green fluorescence protein) in S. aureus AH1121 strain, anhydrotetracycline (aTet) was added at 0.4 μg/ml.
Biofilm formation and assay
Biofilm analyses were performed in the following media; modified M63 minimal medium (15 mM (NH4)2SO4, 100 mM KH2PO4, 1.8 μM FeSO4·7H2O, 0.2% glycerol, and 1 mM MgSO4) for P. aeruginosa 16, modified VFMG-CF (50 mM Tris-HCl, pH 7.2, 50 mM MgSO4, 300 mM NaCl, 10 mM KCl, 0.33 mM K2HPO4, 18.5 mM NH4Cl, and 32.6 mM glycerol, 10 mM CaCl2) for V. vulnificus 26, TSB medium (30 g/l of BactoTM Tryptic Soy Broth, BD) supplemented with 0.5% glucose for S. aureus and B. subtilis, and LB without NaCl but supplemented with 2% glucose for S. enterica 27. For flow cell biofilm analyses, differently diluted TSBs were used for S. aureus, S. enterica (0.6 g/l supplemented with 0.2% glucose) and B. subtilis (3 g/l)28.
The static biofilm assay was performed as previously described16, 26. Briefly, overnight cultures of bacterial strains were diluted 1:100 (vol/vol) in fresh medium, put into individual wells (200 μl) in 96-well polystyrene plates, and incubated for 12–16 hours at 37 °C without shaking. After measuring OD600, planktonic cells were poured out, and plates were washed with water and dried for 10 minutes. Then, 200 μl of crystal violet (0.1%, wt/vol) was added to each well and incubated for 10 minutes to stain biofilms attached to well surfaces. After a brief wash, the biofilm-staining crystal violet was dissolved in 200 μl of 30% acetic acid, and staining levels were assessed by measuring absorbance at 595 nm (A595). The data were normalized by cell growth (OD600). For biofilm dispersion assays, biofilms were formed in this static system for 12 hours without anthranilate. Spent medium was then discarded and planktonic cells were completely removed by washing 3 times with distilled water. Wells were then filled with 200 μl of fresh PBS containing 100 μM anthranilate. After incubation at 37 °C for 12 hours, wells were washed with distilled water, and remaining biofilms were stained with crystal violet and quantitated as described above.
Flow cell biofilm analysis was carried out as previously described23, 26, 28, 29. Briefly, overnight cultures of bacteria were diluted to OD600 of 0.01. 300 μl of the diluted cultures was then injected into each flow cell chamber, incubated for 1 hour without flow to allow attachment, and then fresh medium was pumped through the chamber using a peristaltic pump. Biofilms in flow cells were grown for 3–5 days at room temperature under constant flow, and biofilms in flow cells were visualized using GFP or by staining with 0.1% SYTO 9 for 15 minutes and analyzed by fluorescence microscopy, as described below. For the biofilm dispersion analysis, biofilms were allowed to form in a flow cell system for 2–3 days without anthranilate, and then these preformed biofilms were then treated with 0.1 mM anthranilate for 24 hours.
Biofilm imaging and quantification
Biofilms in flow cells were imaged by confocal laser scanning microscopy (CLSM) (Olympus; FV10i) or by fluorescence microscopy (Zeiss; Axioskop FL). Three-dimensional images of biofilms were reconstructed from plane images using Bitplane Imaris 6.3.1 image analysis software. Fluorescence intensities of flow cell biofilms were quantified using ImageJ software.
β-Galactosidase activity assay
β-Galactosidase activity was measured using the Galacto-Light Plus™ kit (Applied Biosystems) as previously described30. Briefly, OD600 values were measured and then 100 μl aliquots of cultures were mixed with 10 μl of chloroform. After vigorous vortexing and incubation for 15 minutes at room temperature, 10 μl aliquot of supernatants were transferred to new tubes and substrate solution of the Galacto-Light Plus™ kit was added. After 50-minute incubation in the dark for 50 minutes at room temperature, 150 μl of light emission solution (Accelerator II) of the kit was added and luminescence was promptly measured using a multi-well plate reader (Tristar LB941; Berthold). Measured luminescence was normalized by OD600 of culture and β-galactosidase activities are presented as luminescence/OD600.
Motility assay
Swimming, swarming, twitching (for P. aeruginosa, V. vulnificus, B. subtilis, and S. enterica), and spreading motilities (for S. aureus) were measured as previously described31–36. Swimming was assayed on the following media; LB and LBS media solidified with 0.3% (wt/vol) agar were used for P. aeruginosa and V. vulnificus, respectively. Nutrient broth (NB) supplemented with 0.5% glucose and solidified with 0.3% agar was used for S. enterica, and tryptone medium (1% tryptone, 0.5% NaCl) solidified with 0.3% agar was used for B. subtilis. Overnight cultures (3 μl) were inoculated into the middle of agar and incubated at 37 °C for 24 hours. Degrees of swimming motility are presented as the average of the largest and smallest diameters of swimming areas. Swarming was assayed on NB medium supplemented with 0.5% glucose and solidified with 0.5% agar for P. aeruginosa, B. subtilis, and S. enterica, or on Brain Heart Infusion (BHI) medium containing 2% NaCl and solidified with 0.3% agar for the V. vulnificus. To assess swarming motilities, 3 μl of overnight culture was dropped on the surfaces of agar plates and incubated at 37 °C for 24 hours. Degrees of swarming motility are presented in the same manner as swimming motility. Twitching was assayed on LB or LBS (for V. vulnificus) medium solidified with 1% agar. The strains were stab-inoculated with a sharp toothpick to the bottoms of plates and incubated for 24 hours at 37 °C. Twitching zones at plate-agar interfaces were visualized by staining with 1% crystal violet and their sizes were measured as described above. Spreading motility (for S. aureus) was assayed on tryptone medium solidified with 0.3% agar. Briefly, 5 μl of overnight cultures were dropped onto the surfaces of agar plates and incubated at 37 °C for 24 hours. Degrees of spreading were measured as described for swimming motility.
Measurement of intracellular c-di-GMP
Intracellular c-di-GMP levels were measured as described elsewhere37, 38. Bacterial cells were cultured in LB or LBS media containing 100 mM anthranilate at 37 °C with agitation for 8 hours. Cells were harvested and lysed by sonication in a buffer (10 mM Tris-HCl, 100 mM NaCl, pH 8.0). Protein concentrations of lysates were determined using Bio-Rad Protein Assay Dye Reagent (Bio-Rad). Cellular macromolecules were precipitated by adding perchloric acid (final concentration, 12%) on ice for 10 minutes, and then neutralized by adding a neutralizing buffer (3 M KOH, 0.4 M Tris, 2 M NaCl). After centrifugation, the soluble fraction was filtered through a 0.2 μm filter and 3 kD exclusion column. Thiazole orange (TO) was then added (final concentration, 30 μM) and incubated at 4 °C for 12 hours. Fluorescence was measured using a Tristar LB941 unit (Berthold; excitation 510 nm and emission 580 nm). Results were validated by comparing with independent measurements of intracellular c-di-GMP using cdrA-lacZ reporter in P. aeruginosa, as previously described23.
In vitro cytotoxicity
Cytotoxicity of anthranilate was measured using the human HepG2 hepatocyte cell line, as previously described39. Cells were cultured in high glucose Dulbecco’s Modified Eagle Medium (DMEM, Nissui) supplemented with 100 mg/ml streptomycin, 2.5 μg/ml amphotericin B, and 10% heat-treated fetal bovine serum (FBS) at 37 °C in a 5% CO2 humidified incubator. Cells were seeded in a 96-well microtiter tissue culture plate and allowed to adhere overnight, and then treated with 2 μl anthranilate for 24 hours at various concentrations. Control cells were treated with 2 μl DMSO. To measure cell viability, 10 μl of water soluble tetrazolium reagent (EZ-CyTox, Daeil Lab Service) was added to each well and incubated at 37 °C for 1 hour. Absorbances were measured using a iMark Microplate Absorbance Reader (Bio-Rad) at a wavelength of 450 nm.
Slime production assay
Slime production by S. aureus was assayed on Congo red agar plates (BHI medium containing 3.6% sucrose, 0.8 g/L Congo red, and 1% agar), as previously described40. Aliquots (3 μl) of overnight culture dropped on plates and incubated at 37 °C for 24 hours. Since slime is stained by Congo red, slime production by S. aureus appears dark brown around colonies on Congo red agar plates.
Statistical analysis
The student’s t-test (two-sample assuming equal variances) in MS Office Excel (Microsoft) was used to determine the significances of differences. P-values of less than 0.05 were considered significant. At least, all experiments were repeated twice in triplicate.
Results
Anthranilate inhibits biofilm formation by a wide range of bacteria
For this study, we selected five bacterial species, that is, P. aeruginosa, V. vulnificus, B. subtilis, S. enterica serovar Typhimurium, and S. aureus, because these strains represent a variety of bacteria in terms of their genetic classification, whether or not they produce anthranilates, type of motility, and their importance (Table S2). We investigated the effect of anthranilate on biofilm formation in a static biofilm formation system. Biofilm formations of all bacterial species were significantly inhibited by 36.2% to 48.6% (Fig. 1). We note that in our previous study23, anthranilate was demonstrated to increase the initial attachment of P. aeruginosa, although it eventually decreased the biofilm formation. In the previous study, citrate and casamino acids were used for carbon source of culture medium, but in this study, we used glycerol medium in which the biofilm formation by P. aeruginosa was inhibited by anthranilate throughout growth.
Figure 1.
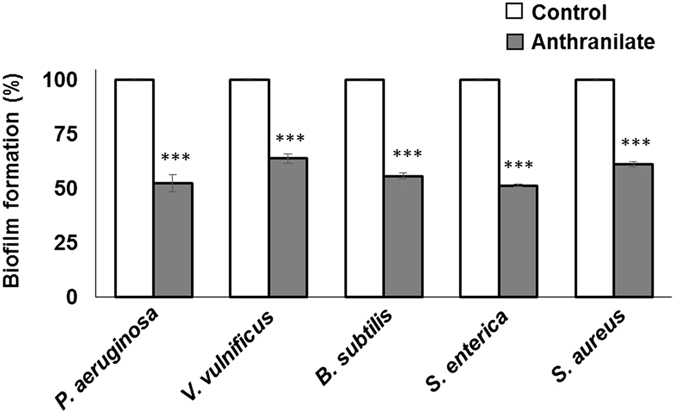
Inhibition of biofilm formation by anthranilate Static biofilm analysis was carried out with various bacteria. Biofilms were formed for 12–16 hours in a static biofilm system in the presence of 0.1 mM anthranilate. Since anthranilate was added to media from 1 M stock made up in dimethyl sulfoxide (DMSO), the same amount of DMSO was added to controls. Biofilm amounts were quantified by crystal violet staining as described in Materials and Methods and are presented as percentages of controls. ***p < 0.005.
When we examined the minimum effective concentration of anthranilate on the biofilm formation, P. aeruginosa showed the highest sensitivity to anthranilate (lower than 7.5 μM) (Fig. 2). The minimum effective concentrations for other bacteria were as follows; V. vulnificus (15 μM), B. subtilis (15 μM), S. enterica (100 μM), and S. aureus (100 μM) (Fig. 2). Furthermore, we investigated biofilm inhibition at high concentration of anthranilate and the results showed that higher concentrations of anthranilate had greater inhibitory effects (Fig. 3).
Figure 2.

Effective biofilm-inhibitory concentration of anthranilate P. aeruginosa (A), V. vulnificus (B), B. subtilis (C), S. enterica (D), and S. aureus (E) were treated with different concentrations of anthranilate and biofilm formations were quantified by static biofilm analysis. Results are presented as percentages of the control. *p < 0.05; ***p < 0.005.
Figure 3.
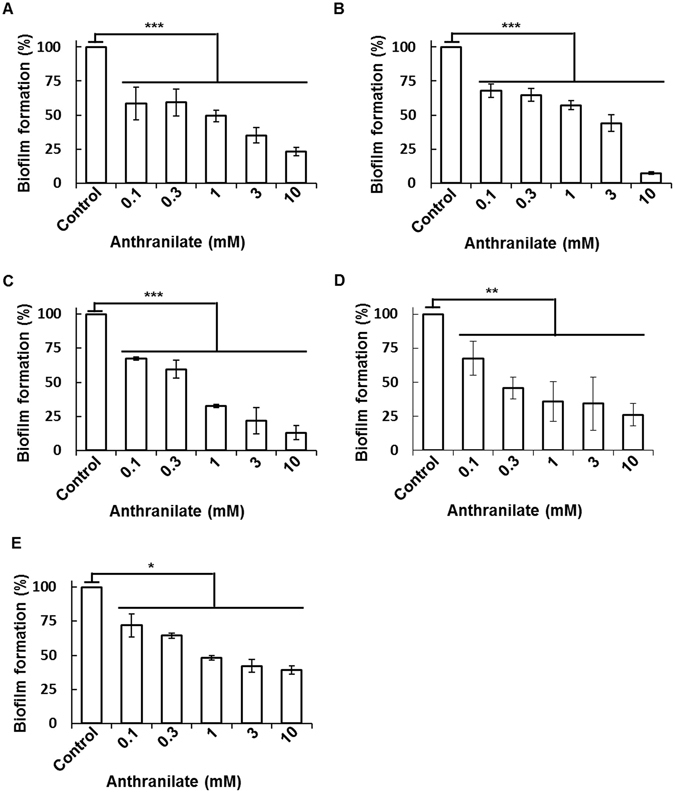
Strong inhibition of biofilms by high concentration of anthranilate P. aeruginosa (A), V. vulnificus (B), B. subtilis (C), S. enterica (D), and S. aureus (E) were treated with high concentration of anthranilate and biofilm formations were quantified by static biofilm analysis. Results are presented as percentages of the control. *p < 0.05; **p < 0.01; ***p < 0.005.
Anthranilate has been reported to inhibit biofilm formation by P. aeruginosa in a flow cell system23. Similarly, the biofilm inhibition by anthranilate was tested in the flow cell system for other four bacterial species and anthranilate was able to prevent biofilm formation by all bacterial strains in the flow cell system (Fig. 4). Taken together, we conclude that biofilm inhibition by anthranilate is common in a variety of bacteria.
Figure 4.
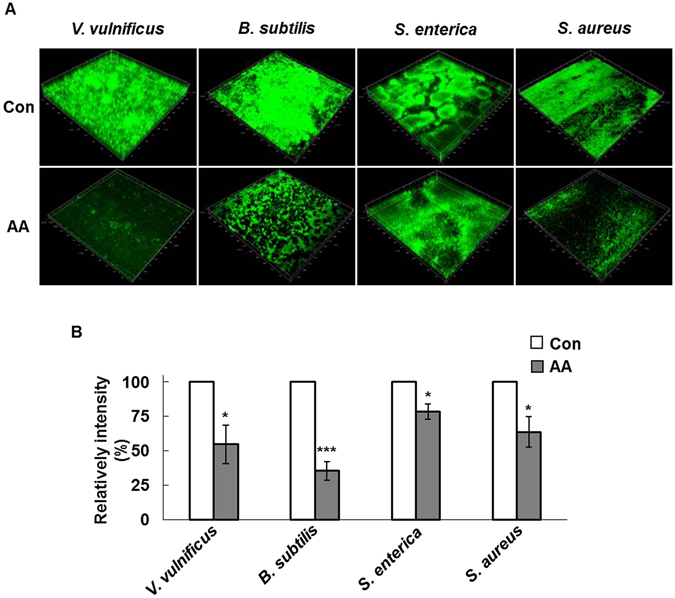
Biofilm inhibition by anthranilate in the presence of shear All strains were grown in flow cell chambers in the presence of 0.1 mM anthranilate for 3–5 days to form biofilms. V. vulnificus, B. subtilis and S. enterica biofilms were stained with 0.1% SYTO 9 for 15 minutes before microscopy, but S. aureus biofilms were detected directly because S. aureus harbors a GFP-expressing plasmid (pAH13). All biofilms were imaged by CLSM (A). Fluorescence intensities were quantified and graphed (B). Con, control; AA, 0.1 mM anthranilate. *p < 0.05; ***p < 0.005.
Anthranilate can disrupt preformed biofilms and induce detachment
Anthranilate has been reported to induce the detachment of preformed P. aeruginosa biofilms23. We first investigated biofilm detachment with other bacteria in the static biofilm system. Biofilms were preformed in the absence of anthranilate for 2 days and then anthranilate was applied for 12 hours. Crystal violet staining showed that biofilm masses of V. vulnificus, B. subtilis, S. enterica, and S. aureus were significantly decreased by anthranilate treatment (Fig. 5A).
Figure 5.
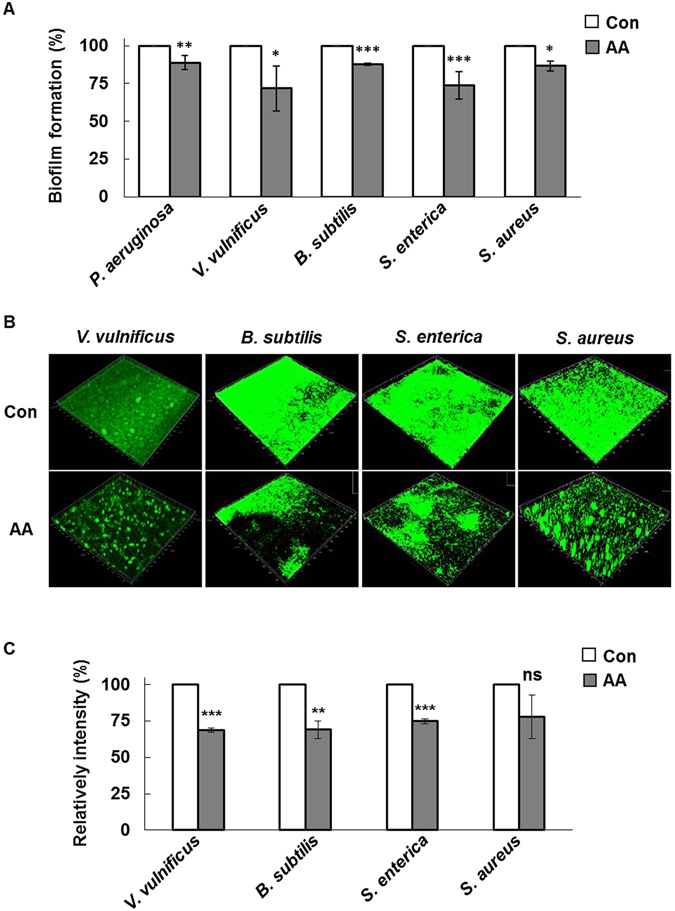
Detachment of preformed biofilms by anthranilate (A), bacterial biofilms were formed in the absence of anthranilate for 12 hours in the static biofilm system and then 0.1 mM anthranilate was added. After 12 hours of incubation, residual biofilms were quantified by crystal violet staining. (B) Biofilms were formed in the absence of anthranilate for 2–3 days in the flow cell system, and then treated with 0.1 mM anthranilate for 24 hours. A preformed biofilm was incubated without anthranilate under the same conditions as a control. All biofilms were imaged by CLSM (B) and fluorescence intensities were quantified (C). Con, control; AA, 0.1 mM anthranilate. *p < 0.05; **p < 0.01; ***p < 0.005; ns, no significant difference (p > 0.05).
The biofilm-detaching effect of anthranilate appeared weak in the static biofilm system. However, biofilm detachment is generally facilitated by shear force in the presence of flow. Therefore, we investigated biofilm detachment in the flow cell system. Preformed biofilm masses of V. vulnificus, B. subtilis, and S. enterica were significantly reduced by anthranilate treatment (Fig. 5B,C). Since preformed biofilms are generally firmly adherent to surfaces, this reduction in the biofilm mass by anthranilate treatment can be attributed to the biofilm detachment.
Interestingly, the mass of S. aureus preformed biofilm was not significantly reduced by anthranilate (Fig. 5C), although they tended to decrease. This result implies that the biofilm-detaching effect of anthranilate is not effective in S. aureus. The reason the ineffectiveness of anthranilate in S. aureus is discussed below.
Anthranilate reduces intracellular c-di-GMP levels
There are several distinct fashions in biofilm detachment. Biofilm dispersion is an active detachment process triggered by several specific biological signals, the most important of which is a decrease in intracellular c-di-GMP (3′,5′-cyclic diguanylate) level41. Briefly, this decrease facilitates biofilm dispersion and a planktonic existence, whereas an increase in intracellular c-di-GMP facilitates biofilm formation and a sessile existence42. This c-di-GMP-mediated regulation of biofilms has been reported in a variety of microorganisms, and notably, anthranilate has been found to decrease intracellular c-di-GMP levels in P. aeruginosa 23. Accordingly, we investigated whether anthranilate affects biofilm formation by modulating intracellular c-di-GMP levels of V. vulnificus, B. subtilis, and S. enterica; intracellular c-di-GMP levels were assessed using a thiazole orange (TO)-based fluorescence assay (TO produces fluorescence when bound to c-di-GMP). This method was validated by comparing result with that of a cdrA-lacZ reporter-based measurement of c-di-GMP, which has been used in P. aeruginosa 23 (Fig. S1). In P. aeruginosa, intracellular c-di-GMP levels were reduced by 18% in the presence of anthranilate (Fig. 6, Fig. S1) and this reduction resulted in the dispersion of P. aeruginosa biofilms23. In V. vulnificus, B. subtilis, and S. enterica, anthranilate significantly reduced intracellular levels of c-di-GMP by 27%, 18%, and 5%, respectively (Fig. 6), which are comparable with that observed for P. aeruginosa. Thus, our results suggest that anthranilate induces biofilm dispersion by reducing intracellular c-di-GMP levels in P. aeruginosa, V. vulnificus, B. subtilis, and S. enterica.
Figure 6.
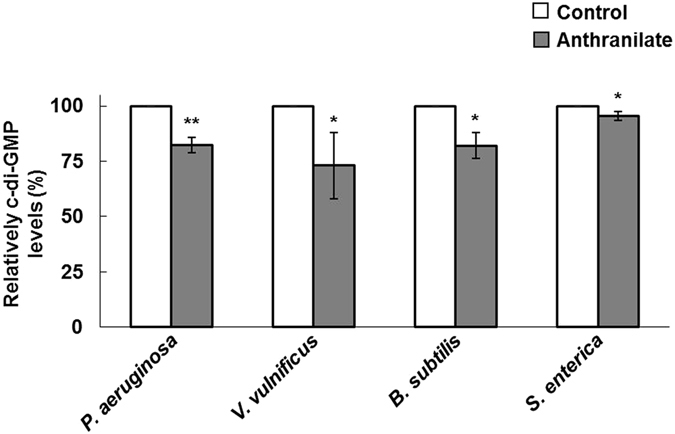
Reduction of intracellular c-di-GMP by anthranilate Intracellular c-di-GMP levels in various bacteria were measured after treatment with 0.1 mM anthranilate using a Thiazole orange (TO)-based fluorescence assay. Results are presented as percentages of the control. *p < 0.05; **p < 0.01.
Intracellular c-di-GMP levels were not measured in S. aureus, because intracellular c-di-GMP signaling does not exist in this species, despite the fact that S. aureus has a conserved GGDEF domain protein43, 44.
Anthranilate enhances swimming and swarming motilities
Bacterial motility is regarded as a phenotype that should be repressed for irreversible attachment to surface at early stage of biofilm formation, but should be induced for biofilm dispersion41. Therefore, enhanced motility can accelerate biofilm dispersion45–48. We found anthranilate significantly enhanced the swimming and swarming motilities of P. aeruginosa, V. vulnificus, B. subtilis, and S. enterica (Fig. 7). These results are consistent with our biofilm dispersion results and indicate anthranilate facilitates biofilm dispersion by promoting swimming and swarming motilities and by decreasing intracellular c-di-GMP levels.
Figure 7.
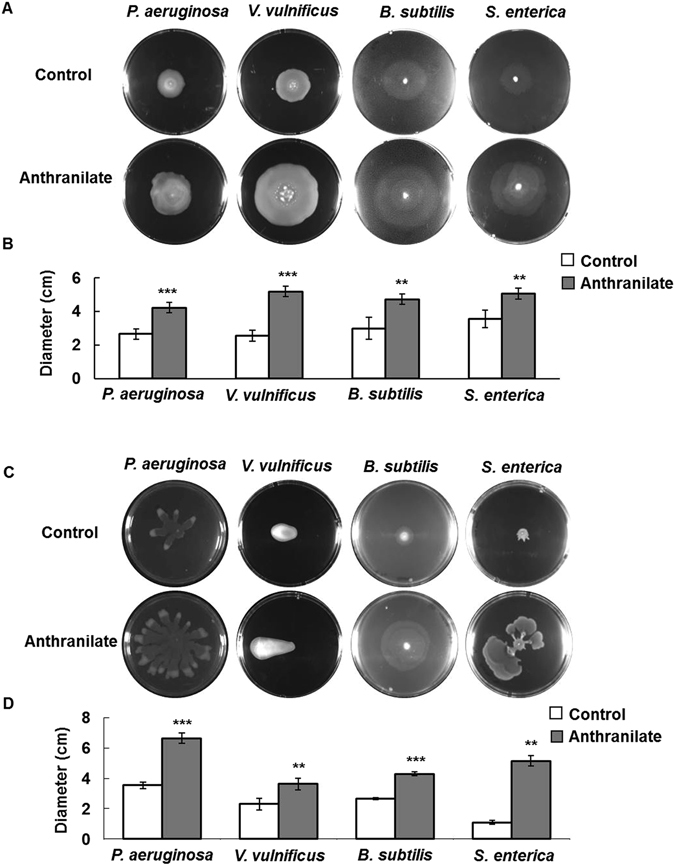
Effect of anthranilate on bacterial motility Swimming motilities of the bacterial strains were examined on semi-solid agar plates in the presence of 0.1 mM anthranilate (A). Diameters of swimming zones were graphed (B). Swarming motilities were also measured on solid agar plate containing 0.1 mM anthranilate (C) and diameters of swarming zones were graphed (D). **p < 0.01; ***p < 0.005.
Swimming and swarming are flagella-dependent motilities whereas twitching is pilus-dependent. Interestingly, the twitching motilities of P. aeruginosa, V. vulnificus, B. subtilis, and S. enterica were not affected by anthranilate (Fig. S2), which suggests anthranilate targets flagellar function.
Biofilm formation by S. aureus is inhibited by anthranilate in a different manner
Because S. aureus is a non-flagellated bacterium, we expected anthranilate would not affect its motility. S. aureus has a different type of motility, that is, colony spreading. In fact, anthranilate had no effect on the spreading motility of S. aureus (Fig. S3), which also reconfirmed anthranilate affects only flagella-dependent motilities. If so, the S. aureus biofilm reduction by anthranilate should have other reasons, because S. aureus does not have c-di-GMP signaling.
In general, the EPS production is very important for biofilm maturation and in the case of S. aureus, staphylococcal slime is associated with biofilm formation as a major EPS44. When we tested slime production by S. aureus, we found it was significantly reduced by anthranilate treatment (Fig. 8). This result suggests that in S. aureus, anthranilate inhibits the biofilm maturation by interfering with the EPS production, rather than induces the biofilm dispersion by affecting c-di-GMP signaling or motility. This suggestion explains why the biofilm-detaching effect of anthranilate was not effective in S. aureus (Fig. 5C). Thus, it appears anthranilate inhibits bacterial biofilm formation in several multiple ways, that is, by affecting c-di-GMP signaling or motility to induce biofilm dispersion (in P. aeruginosa, V. vulnificus, B. subtilis, and S. enterica), or by suppressing EPS production to repress biofilm maturation (in S. aureus).
Figure 8.
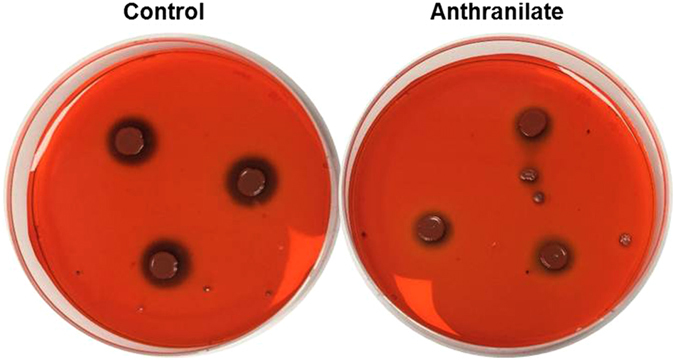
Inhibition of slime production by S. aureus by anthranilate Slime production by S. aureus was assessed by Congo red staining. S. aureus overnight culture (3 μl) was inoculated on a CRA plate containing 0.1 mM anthranilate and incubated at 37 °C for 24 hours. Slime production was visualized as a dark red around colonies.
Anthranilate is more effective at inhibiting biofilms than L-tryptophan
L-Tryptophan at concentrations >1.6 mM has been shown to repress biofilm formation by P. aeruginosa ATCC 27853 strain by 50% and to enhance swimming motility16. Since this tryptophan effect on the biofilm is similar to the anthranilate effect observed in this study, we compared the anti-biofilm effects of anthranilate and tryptophan. In our comparison with P. aeruginosa PAO1 strain, L-tryptophan showed an inhibitory effect at 10 mM, whereas anthranilate exhibited a similar effect at 0.1 mM (Fig. 9). Similarly, about 10 mM L-tryptophan was required for half-level inhibition of the V. vulnificus biofilm, whereas anthranilate had a similar inhibitory effect at 0.1 mM (Fig. 9). L-Tryptophan showed a slight inhibitory effect on S. enterica biofilm, and no effect on B. subtilis and S. aureus biofilms even at 10 mM, whereas anthranilate at 0.1 mM had significant effects on all bacterial strains examined (Fig. 9). This result suggests that anthranilate is more effective in biofilm inhibition and has broader application range.
Figure 9.
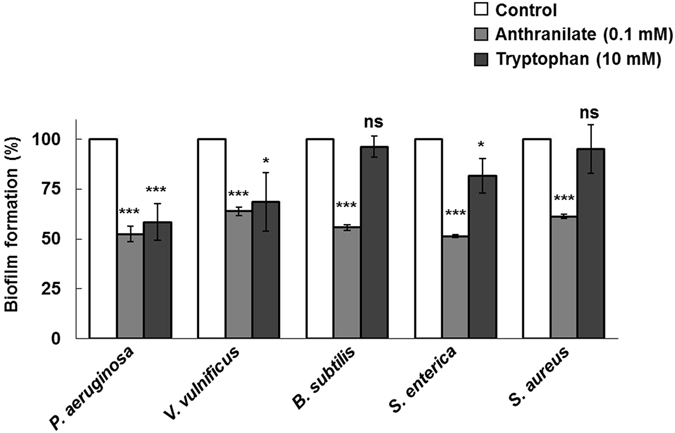
Comparison of the anti-biofilm effects of anthranilate and L-tryptophan The anti-biofilm effects of L-tryptophan (10 mM) and anthranilate (0.1 mM) were compared by static biofilm analysis. Results are presented as percentages of controls. *p < 0.05; ***p < 0.005; ns, no significant difference (p > 0.05).
Even if it has a good effect, it should be non-toxic to develop into an anti-biofilm substance. So, we examined the toxicity of anthranilate to human cells. When HepG2 cells (an immortalized human hepatocyte cell line) were exposed to anthranilate for 12 hours, anthranilate did not cause cell death or lysis even at a concentration of 10 mM (Fig. S4). This result demonstrates that anthranilate has no direct cytotoxic effect.
Discussion
Our results show the following: 1) anthranilate exerts biofilm-inhibiting effects on a wide range of bacteria; 2) in P. aeruginosa, V. vulnificus, B. subtilis, and S. enterica, anthranilate reduces intracellular c-di-GMP levels and enhances swarming and swimming motilities to induce biofilm dispersion; 3) in S. aureus, anthranilate reduces slime production to inhibit biofilm formation. In our previous work, we found anthranilate can disrupt the biofilm structure of P. aeruginosa and finally making a flat biofilm23. In this study, we further investigated the effects of anthranilate on biofilm formation by various bacterial species and found that anthranilate had antibiofilm effects on all bacterial species tested. Since the strains used in this study represented various bacterial physiology and characteristics (Table S2), these results indicate anthranilate has antibiofilm effects on a wide range of bacterial species.
Then, how does anthranilate deteriorate the biofilm? Developmental cycle of biofilm includes multiple stages; initial attachment of planktonic cells, microcolony formation, biofilm maturation in EPS matrix, and biofilm detachment49, 50, and thus, biofilm formation can be prevented by inhibiting cell attachment at early stage or interfering with EPS production at middle stage, or inducing biofilm detachment at late stage51–53. Anthranilate was found to have two effects on P. aeruginosa, V. vulnificus, B. subtilis, and S. enterica, that is, it reduced intracellular c-di-GMP levels and enhanced motility, which are related to biofilm detachment at late stage. Biofilm detachment is the most general term to indicate the reduction of biofilm mass and can be classified as: 1) sloughing, 2) erosion, and 3) seeding dispersal41. Sloughing and erosion are passive processes induced by shear forces, whereas dispersal is active process by biological activity41. We suggest anthranilate probably induces dispersal, because of its effects on intracellular levels of c-di-GMP and motility.
Studies have demonstrated negative relation between motility and biofilm formation, as the inhibition of motility has been shown to enhance biofilm formation by a wide range of microorganisms54. In B. subtilis, the eps operon that synthesizes EPS also encodes a bifunctional protein EpsE that inhibits flagella motility by blocking flagella rotor during biofilm formation55, 56. In P. aeruginosa and V. cholera, motility inhibition caused by repressing flagella gene expression is achieved by intracellular c-di-GMP up-regulation during biofilm formation57, 58. In E. coli, both the activation of capsular biosynthesis and suppression of flagella biogenesis occur by Rcs phosphorelay59, 60. According to these studies, the inhibition and enhancement of motility are requirements of biofilm formation and dispersal, respectively, and our observations that anthranilate inhibited P. aeruginosa, V. vulnificus, B. subtilis, and S. enterica biofilm formation by enhancing swarming and swimming motilities are consistent with the findings of these previous studies. Furthermore, it would seem anthranilate only enhances a flagella-mediated motility, as non-flagellar motilities, such as the twitching of flagellate bacteria and the spreading of S. aureus, a non-flagellate bacterium were not affected by anthranilate (Fig. S2, Fig. S3).
However, anthranilate seems to have effects other than the induction of dispersion. We previously reported anthranilate disrupted P. aeruginosa biofilm formation by reducing intracellular c-di-GMP levels and modulating EPS expression. In that study, we have found that the anthranilate modulates the expressions of the synthetic genes for Psl, Pel, and alginate which are major components of EPS23. This finding indicates anthranilate can influence biofilm maturation as well as dispersion. Furthermore, in this study, anthranilate reduced slime production by S. aureus (Fig. 8), which is probably responsible for the anti-biofilm effect of anthranilate on S. aureus, because this bacterium does not possess a c-di-GMP signaling system and its motility was unaffected by anthranilate.
Anthranilate is a natural product, has a simple molecular structure, is cheap and widely used in industry. When we compared the effectivenesses of L-tryptophan and anthranilate on the biofilm inhibition, anthranilate was found to be more potent than L-tryptophan (Fig. 9). Our cytotoxicity study showed anthranilate had no cytotoxic effect (Fig. S4). Anthranilate is naturally produced in man and animals by the kynurenine pathway, which is the major route for the metabolism of L-tryptophan61, 62. So, we suggest that anthranilate is a promising substance for the development of anti-biofilm agent.
Electronic supplementary material
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (NRF-2016R1A2B4014963). For Soo-Kyoung Kim, this work was also supported by the NRF grant funded by the Korea government (MSIP) (2016R1C1B1006671).
Author Contributions
Xi-Hui Li performed most of the experiments and wrote the manuscript. Soo-Kyoung Kim first discovered the anthranilate effect and gave helpful discussion. Joon-Hee Lee designed the study and wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-06540-1
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wei Q, Ma LZ. Biofilm matrix and its regulation in Pseudomonas aeruginosa. International journal of molecular sciences. 2013;14:20983–21005. doi: 10.3390/ijms141020983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiology and Molecular Biology Reviews. 2009;73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.James GA, et al. Biofilms in chronic wounds. Wound Repair and regeneration. 2008;16:37–44. doi: 10.1111/j.1524-475X.2007.00321.x. [DOI] [PubMed] [Google Scholar]
- 4.Akers KS, et al. Biofilms and persistent wound infections in United States military trauma patients: a case–control analysis. BMC infectious diseases. 2014;14:1. doi: 10.1186/1471-2334-14-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fux C, Costerton J, Stewart P, Stoodley P. Survival strategies of infectious biofilms. Trends in microbiology. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Hengzhuang W, Wu H, Ciofu O, Song Z, Høiby N. Pharmacokinetics/pharmacodynamics of colistin and imipenem on mucoid and nonmucoid Pseudomonas aeruginosa biofilms. Antimicrobial agents and chemotherapy. 2011;55:4469–4474. doi: 10.1128/AAC.00126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Favre-Bonté S, Köhler T, Van Delden C. Biofilm formation by Pseudomonas aeruginosa: role of the C4-HSL cell-to-cell signal and inhibition by azithromycin. Journal of antimicrobial chemotherapy. 2003;52:598–604. doi: 10.1093/jac/dkg397. [DOI] [PubMed] [Google Scholar]
- 8.Bala A, Kumar R, Harjai K. Inhibition of quorum sensing in Pseudomonas aeruginosa by azithromycin and its effectiveness in urinary tract infections. Journal of medical microbiology. 2011;60:300–306. doi: 10.1099/jmm.0.025387-0. [DOI] [PubMed] [Google Scholar]
- 9.Singh PK, Parsek MR, Greenberg EP, Welsh MJ. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- 10.Mulcahy H, Lewenza S. Magnesium limitation is an environmental trigger of the Pseudomonas aeruginosa biofilm lifestyle. PLoS One. 2011;6:e23307. doi: 10.1371/journal.pone.0023307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez CJ, et al. Effects of local delivery of D-amino acids from biofilm-dispersive scaffolds on infection in contaminated rat segmental defects. Biomaterials. 2013;34:7533–7543. doi: 10.1016/j.biomaterials.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 12.Kolodkin-Gal I, et al. D-amino acids trigger biofilm disassembly. Science. 2010;328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hochbaum AI, et al. Inhibitory effects of D-amino acids on Staphylococcus aureus biofilm development. Journal of bacteriology. 2011;193:5616–5622. doi: 10.1128/JB.05534-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leiman SA, et al. D-amino acids indirectly inhibit biofilm formation in Bacillus subtilis by interfering with protein synthesis. Journal of bacteriology. 2013;195:5391–5395. doi: 10.1128/JB.00975-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimazaki J, Furukawa S, Ogihara H, Morinaga Y. L-Tryptophan prevents Escherichia coli biofilm formation and triggers biofilm degradation. Biochemical and biophysical research communications. 2012;419:715–718. doi: 10.1016/j.bbrc.2012.02.085. [DOI] [PubMed] [Google Scholar]
- 16.Brandenburg KS, et al. Tryptophan inhibits biofilm formation by Pseudomonas aeruginosa. Antimicrobial agents and chemotherapy. 2013;57:1921–1925. doi: 10.1128/AAC.00007-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bandeira LG, Bortolot BS, Cecatto MJ, Monte-Alto-Costa A, Romana-Souza B. Exogenous Tryptophan Promotes Cutaneous Wound Healing of Chronically Stressed Mice through Inhibition of TNF-α and IDO Activation. PloS one. 2015;10:e0128439. doi: 10.1371/journal.pone.0128439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barouti N, Mainetti C, Fontao L, Sorg O. L-Tryptophan as a novel potential pharmacological treatment for wound healing via aryl hydrocarbon receptor activation. Dermatology. 2015;230:332–339. doi: 10.1159/000371876. [DOI] [PubMed] [Google Scholar]
- 19.Brandenburg KS, et al. Inhibition of Pseudomonas aeruginosa biofilm formation on wound dressings. Wound Repair and Regeneration. 2015;23:842–854. doi: 10.1111/wrr.12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J-H, Wood TK, Lee J. Roles of indole as an interspecies and interkingdom signaling molecule. Trends in microbiology. 2015;23:707–718. doi: 10.1016/j.tim.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Lee J, Jayaraman A, Wood TK. Indole is an inter-species biofilm signal mediated by SdiA. BMC microbiology. 2007;7:1. doi: 10.1186/1471-2180-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J, Bansal T, Jayaraman A, Bentley WE, Wood TK. Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Applied and environmental microbiology. 2007;73:4100–4109. doi: 10.1128/AEM.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SK, Park HY, Lee JH. Anthranilate deteriorates the structure of Pseudomonas aeruginosa biofilms and antagonizes the biofilm-enhancing indole effect. Applied and environmental microbiology. 2015;81:2328–2338. doi: 10.1128/AEM.03551-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurnasov O, et al. Aerobic tryptophan degradation pathway in bacteria: novel kynurenine formamidase. FEMS microbiology letters. 2003;227:219–227. doi: 10.1016/S0378-1097(03)00684-0. [DOI] [PubMed] [Google Scholar]
- 25.Costaglioli P, et al. Evidence for the involvement of the anthranilate degradation pathway in Pseudomonas aeruginosa biofilm formation. Microbiologyopen. 2012;1:326–339. doi: 10.1002/mbo3.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park JH, et al. The cabABC Operon Essential for Biofilm and Rugose Colony Development in Vibrio vulnificus. PLoS Pathog. 2015;11:e1005192. doi: 10.1371/journal.ppat.1005192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mireles JR, II, Toguchi A, Harshey RM. Salmonella enterica serovar typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. Journal of bacteriology. 2001;183:5848–5854. doi: 10.1128/JB.183.20.5848-5854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boles BR, Thoendel M, Roth AJ, Horswill AR. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PloS one. 2010;5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grantcharova N, Peters V, Monteiro C, Zakikhany K, Römling U. Bistable expression of CsgD in biofilm development of Salmonella enterica serovar typhimurium. Journal of bacteriology. 2010;192:456–466. doi: 10.1128/JB.01826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim SK, Im SJ, Yeom DH, Lee JH. AntR-mediated bidirectional activation of antA and antR, anthranilate degradative genes in Pseudomonas aeruginosa. Gene. 2012;505:146–152. doi: 10.1016/j.gene.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Rashid MH, Kornberg A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences. 2000;97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim HS, Park SJ, Lee KH. Role of NtrC‐regulated exopolysaccharides in the biofilm formation and pathogenic interaction of Vibrio vulnificus. Molecular microbiology. 2009;74:436–453. doi: 10.1111/j.1365-2958.2009.06875.x. [DOI] [PubMed] [Google Scholar]
- 33.Kim C-M, et al. Vibrio vulnificus metalloprotease VvpE is essentially required for swarming. FEMS microbiology letters. 2007;269:170–179. doi: 10.1111/j.1574-6968.2006.00622.x. [DOI] [PubMed] [Google Scholar]
- 34.Singh A, Gupta R, Pandey R. Rice Seed Priming with Picomolar Rutin Enhances Rhizospheric Bacillus subtilis CIM Colonization and Plant Growth. PloS one. 2016;11:e0146013. doi: 10.1371/journal.pone.0146013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim W, Killam T, Sood V, Surette MG. Swarm-cell differentiation in Salmonella enterica serovar Typhimurium results in elevated resistance to multiple antibiotics. Journal of bacteriology. 2003;185:3111–3117. doi: 10.1128/JB.185.10.3111-3117.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borges, A., Simões, L. C., Serra, C., Saavedra, M. J. & Simões, M. Activity of allylisothiocyanate and 2-phenylethylisothiocyanate on motility and biofilm prevention of pathogenic bacteria. Worldwide Research Efforts in the Fighting against Microbial Pathogens: From Basic Research to Technological Developments, 8–12 (2013).
- 37.Nakayama S, et al. Thiazole orange-induced c-di-GMP quadruplex formation facilitates a simple fluorescent detection of this ubiquitous biofilm regulating molecule. Journal of the American Chemical Society. 2011;133:4856–4864. doi: 10.1021/ja1091062. [DOI] [PubMed] [Google Scholar]
- 38.Chen G, et al. Oligoribonuclease is required for the type III secretion system and pathogenesis of Pseudomonas aeruginosa. Microbiological research. 2016;188:90–96. doi: 10.1016/j.micres.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Su M, et al. The In Vitro and In Vivo Anti-Inflammatory Effects of a Phthalimide PPAR-γ Agonist. Marine Drugs. 2017;15:7. doi: 10.3390/md15010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Podbielska A, Galkowska H, Stelmach E, Mlynarczyk G, Olszewski WL. Slime production by Staphylococcus aureus and Staphylococcus epidermidis strains isolated from patients with diabetic foot ulcers. Arch Immunol Ther Exp (Warsz) 2010;58:321–324. doi: 10.1007/s00005-010-0079-9. [DOI] [PubMed] [Google Scholar]
- 41.Kim SK, Lee JH. Biofilm dispersion in Pseudomonas aeruginosa. J Microbiol. 2016;54:71–85. doi: 10.1007/s12275-016-5528-7. [DOI] [PubMed] [Google Scholar]
- 42.Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiology and Molecular Biology Reviews. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Grundling A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS pathogens. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holland LM, et al. A staphylococcal GGDEF domain protein regulates biofilm formation independently of cyclic dimeric GMP. Journal of bacteriology. 2008;190:5178–5189. doi: 10.1128/JB.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernier SP, Ha D-G, Khan W, Merritt JH, O’Toole GA. Modulation of Pseudomonas aeruginosa surface-associated group behaviors by individual amino acids through c-di-GMP signaling. Research in microbiology. 2011;162:680–688. doi: 10.1016/j.resmic.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conrad JC, et al. Flagella and pili-mediated near-surface single-cell motility mechanisms in P. aeruginosa. Biophysical journal. 2011;100:1608–1616. doi: 10.1016/j.bpj.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Molecular microbiology. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 48.Ruer S, Stender S, Filloux A, de Bentzmann S. Assembly of fimbrial structures in Pseudomonas aeruginosa: functionality and specificity of chaperone-usher machineries. Journal of bacteriology. 2007;189:3547–3555. doi: 10.1128/JB.00093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costerton JW, et al. Bacterial biofilms in nature and disease. Annual Reviews in Microbiology. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 50.Harmsen M, Yang L, Pamp SJ, Tolker-Nielsen T. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunology & Medical Microbiology. 2010;59:253–268. doi: 10.1111/j.1574-695X.2010.00690.x. [DOI] [PubMed] [Google Scholar]
- 51.Cheng G, Zhang Z, Chen S, Bryers JD, Jiang S. Inhibition of bacterial adhesion and biofilm formation on zwitterionic surfaces. Biomaterials. 2007;28:4192–4199. doi: 10.1016/j.biomaterials.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boles BR, Thoendel M, Singh PK. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Molecular microbiology. 2005;57:1210–1223. doi: 10.1111/j.1365-2958.2005.04743.x. [DOI] [PubMed] [Google Scholar]
- 53.Ueda A, Wood TK. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885) PLoS Pathog. 2009;5:e1000483. doi: 10.1371/journal.ppat.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guttenplan SB, Kearns DB. Regulation of flagellar motility during biofilm formation. FEMS microbiology reviews. 2013;37:849–871. doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. A molecular clutch disables flagella in the Bacillus subtilis biofilm. science. 2008;320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- 56.Guttenplan SB, Blair KM, Kearns DB. The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet. 2010;6:e1001243. doi: 10.1371/journal.pgen.1001243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krasteva PV, et al. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baraquet C, Murakami K, Parsek MR, Harwood CS. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 2012;40:7207–7218. doi: 10.1093/nar/gks384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Francez‐Charlot A, et al. RcsCDB His‐Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Molecular microbiology. 2003;49:823–832. doi: 10.1046/j.1365-2958.2003.03601.x. [DOI] [PubMed] [Google Scholar]
- 60.Fredericks CE, Shibata S, Aizawa SI, Reimann SA, Wolfe AJ. Acetyl phosphate‐sensitive regulation of flagellar biogenesis and capsular biosynthesis depends on the Rcs phosphorelay. Molecular microbiology. 2006;61:734–747. doi: 10.1111/j.1365-2958.2006.05260.x. [DOI] [PubMed] [Google Scholar]
- 61.Jones SP, et al. Expression of the kynurenine pathway in human peripheral blood mononuclear cells: implications for inflammatory and neurodegenerative disease. PloS one. 2015;10:e0131389. doi: 10.1371/journal.pone.0131389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao, K. et al. Tryptophan metabolism in animals: important roles in nutrition and health. Front Biosci (Schol Ed.) 3, 286–297 (2011). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


