Abstract
The pregnancy-associated glycoprotein-like family (PAG-L) is a large group of chorionic products, expressed in the pre-placental trophoblast and later in the post-implantational chorionic epithelium, and are involved in proper placenta development and embryo-maternal interaction in eutherians. This study describes identification of the PAG-L family in the genome of the Eurasian beaver (Castor fiber L.), named CfPAG-L. We identified 7657 bp of the CfPAG-L gDNA sequence (Acc. No. KX377932), encompassing nine exons (1–9) and eight introns (A–H). The length of the CfPAG-L exons (59–200 bp) was equivalently similar to the only known counterparts of bPAG1, bPAG2, and pPAG2. The length of the CfPAG-L introns ranged 288–1937 bp and was completely different from previously known PAG introns. The exonic CfPAG-L regions revealed 50.3–72.9% homology with equivalent segments of bPAG1 and pPAG2 structure. The intronic CfPAG-L regions alignments revealed a lack of homology. Within the entire CfPAG-L gene, 31 potential single nucleotide variants (SNV: 7 transversions and 24 transitions) were predicted. The identified exonic polymorphic loci did not affect the amino acid sequence of the CfPAG-L polypeptide precursor. This is the first report describing the CfPAG-L gene sequence, structural organization, and SNVs in the Eurasian beaver, one of the largest rodents.
Keywords: APs, Aspartic proteinases, Beaver, PAGs, SNVs, Exon-intron structure
Introduction
Since the Eurasian beaver is not a common subject of the scientific studies, the number of papers on this species is very limited. Within the Rodentia order, the Castoridae family is represented by only two still extant species, Castor canadensis in North America and Castor fiber (Cf) in Eurasia (http://www.iucnredlist.org/details/4007/0). Both species have a similar appearance and can be distinguished only by cytogenetic analyses indicating 40 or 48 chromosomes in the American and the Eurasian beaver, respectively (Lavrov and Orlov 1973). However, many aspects of the physiological knowledge of beavers still remains completely unknown, especially reproduction and pregnancy. Limited data originate from difficulties with tissue sampling of this taxon. Previous multi-gene studies have suggested Geomyoidea taxa to be the closest relatives of the beavers (Montgelard et al. 2008; Blanga-Kanfi et al. 2009). But, more recent molecular data strongly support the placement of Castor within a “mouse-related clade” with several families, including Pedetidae, Anomaluridae, Muridae, Dipodidae, Geomyidae, and Heteromyidae (Horn et al. 2011).
Genomic studies of most mammals to date have revealed a predominance of multi-gene families whose products are expressed in some reproductive organs. Within the placenta, the chorionic trophoblast constitutes the outer embryo-derived cells that form an essential interface between the maternal uterus and the embryo-originated placental membranes (Wallace et al. 2015).
Pregnancy-associated glycoproteins (PAGs) belong to the multigenic aspartic proteinase (AP) family, widely distributed in various taxa, which also includes pepsins (A, C and F), cathepsins (D and E), and various other enzymes as plasmepsins or napsins (see Szafranska et al. 2006). All members of the APs, possess a two-bilobe configuration with a cleft (with two Asp residues within two domains), capable of binding short peptides. Pepsins accomplish digestive functions outside the cell, whereas cathepsin D and E are typical intracellular zymogens, generally localized in the lysosomal compartment that provides the acidic environment necessary to accomplish their catalytic function (Kageyama 2002; Carginale et al. 2004). Interestingly, the PAG-L family products function as various chorionic signaling ligands interacting with gonadal and extra-gonadal gonadotropin receptors of cyclic pigs and cows (Szafranska et al. 2007) or early pregnant pigs (Panasiewicz et al. 2007).
To date, the entire exon-intron organization structures have been identified for only three genes: bovine PAG1 – bPAG1 (Xie et al. 1995), porcine PAG2 – pPAG2 (Szafranska et al. 2001), and bPAG2 (Telugu et al. 2009). The mammalian PAGs and many alternatively named PAG-Like (PAG-Ls) are the most closely related to the pepsin family. The identified PAGs possess a conserved structure that includes nine exons and eight introns (A–H), among which the intron F is the longest in pPAG2, bPAG1 and 2 (Wallace et al. 2015; Bieniek-Kobuszewska et al. 2016). However, the organizational structures of the PAGs have not been studied and, therefore, remain completely unknown in the genomes of the Rodentia taxa.
In our previous studies, we identified 1257 bp of the Castor fiber PAG-Like (CfPAG-L; KU245742) cDNA sequence, encoding 391 amino acid (aa) of entire polypeptide precursor, composed of 16 aa signal peptide, 46 aa pro-piece, and 329 aa of the mature protein, with one site of potential N-glycosylation and two Asp residues specific for the catalytic cleft of APs (A. Lipka et al. unpublished). In addition, among the diversified cellular and secretory CfPAG-L profiles, we identified dominant 58 kDa isoform, which was immuno-detected despite the fetus sex and the multiplicity of gestation. The CfPAG-L expression was localized within mono-nucleated and giant trophectodermal cells of the beaver discoidal placenta (A. Lipka et al. unpublished). The identified characteristics of the CfPAG-L family (placental cDNA encoding polypeptide precursor; also the cellular and secretory proteins) should be complemented by a genomic analysis. Thus, the aim of this study was identification and broad-based characterization of the CfPAG-L with its exon-intron structure and a potential polymorphism—single nucleotide variants (SNVs) in the genome of the Eurasian beaver, originating from the Polish population.
Materials and methods
Beavers were captured and euthanized with government permits from the Regional Directorate for Environmental Protection in Olsztyn (RDOS-28-OOP-6631-0007-638/09/10/pj), and the III Local Ethical Commission for Experiments on Animals at the Warsaw University of Life Sciences (11/2010), confirmed by the Local Ethical Commission for Experiments on Animals at the University of Warmia and Mazury in Olsztyn (UWM/111/2011/DTN).
Blood samples were harvested post mortem from the jugular veins of male and female beavers (N = 15). Collected samples were transported in ice to the laboratory and centrifuged (3.500×g) for 30 min at 4 °C. Plasma was discarded and the buffy coats of white cells were separated from red cells and immediately stored at −70 °C until genomic DNA isolation.
Identification of the PAG-L gene in the beaver genome
Leukocyte samples of 15 beavers: 5 females (pregnant); 5 males (potential fathers of the offspring) and 5 fetuses; were used as a source of DNA. Genomic DNA (gDNA) templates of Cf were isolated from leukocytes with the use of a commercial available kit (Sherlock AX, A&A Biotechnology, Poland). Only high-quality gDNA templates were used for PCR amplifications (700 ng) of the exonic and/or intronic CfPAG-L fragments. In order to identify the initial and partial nucleotide sequence of the CfPAG-L, the gDNA amplicons were produced with homological primers (Table 1), designed on the CfPAG-L cDNA sequence identified previously (KU245742). For effective multiple PCRs, JumpStart™ Taq ReadyMix™ (Sigma-Aldrich) was used for amplifications under the following conditions: initial activation (95 °C/2 min), then 40 cycles (95 °C/1 min for the denaturation of gDNA templates; 60 °C/1 min for primer annealing; and 72 °C/4,5 min for amplicon synthesis). Obtained amplicons: CfPAG-L gDNA, porcine PAG10 cDNA—used as a positive control, and negative control (without templates)—were separated in 1% agarose gels, parallel to a marker (100–3000 bp; Thermo Fisher Scientific, USA), UV-visualized using Midori Green Nucleic Acid Staining Solution (NIPPON Genetics Europe GmbH, Germany) and then archived (G:Box, Syngene, UK).
Table 1.
Specific primers applied for the CfPAG-L amplifications with gDNA templates
| Forward primers | Reverse primers | ||
|---|---|---|---|
| Name | Sequence (5′–3′) | Name | Sequence (5′–3′) |
| ATG_F | ATGAAGTGGATAGTGGTGGCC | ||
| Exon2_F | TTCCTGAAGAMSCACAA | Exon2_R | TCATGGAGAACACTGAAGTC |
| Exon3_F | GCTCCTCCAACCTGTGGGT | Exon3_R | ACCCACAGGTTGGAGGAGCC |
| Exon4_F | ACCTACCACACCGATAAGAAGA | Exon4_R | TCTTCTTATCGGTGTGGTAGGT |
| Exon5_F | GATGGCATCMTGGGSCTGGCCTA | Exon5_R | TAGGCCAGSCCCAKGATGCCATC |
| Exon6_F | GTGTGGACAAGCGCCTGTACAA | Exon6_R | CTCCCCCATCCTTAGAGCCTTC |
| Exon7_F | TTGTKGACACMGGCACCTCTCTG | Exon7_R | CAGAGAGGTGCCKGTGTCMACAA |
| Exon8_F | CTGCCCACACTCGACTTCATCA | Exon8_R | TGATGAAGTCGAGTGTGGGCAG |
| Exon9_F | ACTGCATGGTGGGAATCCAG | Exon9_R | GAAGACATCWCCMAGGATCCAA |
| IntronA_F | GCAGCCCTTGAATGACAGGTAT | IntronA_R | CCTCCTTTTTCAGTATGAGTGCAA |
| IntronB_F | CTTGTGAGCTAACAGCCTGCC | IntronA_R2 | TGGCTCTCACTTACAATCTCCATG |
| IntronD_F | GCAAGGTTGAAGAATGAGCCTAG | IntronA_R3 | TTCCTCCATGACACCAACAAAGAC |
| IntronF_F | CCACAGGTGTCCTCTGCATGA | IntronE_R | TTCGGGTTTCATGTAGAGTGAG |
| IntronG_F | GGGTTAGAGCAAAATTACTGGAAC | 3’utr_R | CCAGAGAAGAGGCACAGATAGA |
Electrophoresed CfPAG-L gDNA amplicons were gel-out purified and used as templates for capillary sequencing (3130 Genetic Analyzer, Applied Biosystems, USA) in sense and anti-sense directions. The gDNA amplicons labeling was performed with a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA), but with some modifications to the original manufacturer procedure. Briefly, the labeling conditions were: initial denaturation (at 96 °C for 1 min), and then 30 cycles (96 °C/10 s, 50 °C/5 s, 60 °C/4 min). Each labeling mix (20 μl) contained: 12 μl (5–10 ng) of amplicon templates, 1.2 μl Ready Reaction Mix, 4 μl BigDye Terminator v1.1/3.1 Sequencing buffer (5×), 2 μl of primer and 0.8 μl H2O. The labeled gDNA amplicons were purified with the BigDye X Terminator Purification Kit and separated in capillaries filled with POP-7™ polymer (Applied Biosystems, USA). The obtained CfPAG-L sequence data were analyzed by Geneious R7 software (http://www.geneious.com, Kearse et al. 2012). Exon-intron structure was predicted (http://www.cbs.dtu.dk/services/NetGene2/) and then confirmed by comparison of the entire CfPAG-L gDNA sequence with previously identified CfPAG-L cDNA (KU245742).
Results
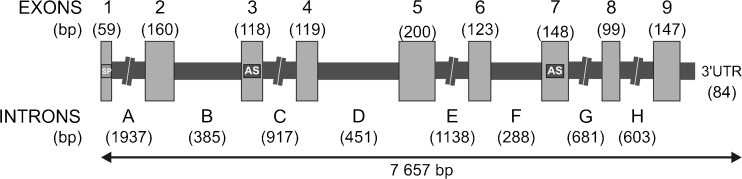
Homological primers used for PCR amplification of the gDNA templates produced numerous amplicons of the CfPAG-L fragments. Among the 750 electrophoresed, gel-out purified and sequenced CfPAG-L gDNA amplicons, 441 clear chromatograms (HQ range: 20–98.2%) were subjected for analyses with GENEIOUS R7 software, which was used to identify 7657 bp of the entire CfPAG-L gDNA sequence that have been deposited in the GenBank database (Acc. No. KX377932). Among the identified CfPAG-L gDNA sequence, nine exons and eight introns (named A–H) were identified (Fig. 1), as well as donor and acceptor site in exon-intron junctions (Table 2). The sequences at the 5′ donor and 3′ acceptor sites of all introns conformed to the GT-AG rules, and splice junctions were not restricted to any particular phase of a codon (Table 2). Two Asp residues within two domains specific for catalytic cleft of many AP members, predicted previously in CfPAG-L cDNA (A. Lipka et al. unpublished), were presently localized within exons 3 and 7 of CfPAG-L gDNA (Table 2, Fig. 1).
Fig. 1.
Exon-intron structure of the identified genomic CfPAG-L sequence. Abbreviations: SP signal peptide, AS active site sequences coding domains 1 and 2 of the catalytic cleft
Table 2.
Characteristics of exon-intron junctions of the CfPAG-L gDNA sequence
| Donor splice sites | Acceptor splice sites | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| exon | 5′- > 3’ | Phase | Intron | 5′- > 3’ | Intron | 5′- > 3’ | Phase | Exon | 5′- > 3’ |
| 1 | CAATATCCAG | 0 | A | GTGAGCTTGG | A | TGACTTACAG | 1 | 2 | TGACTTACAG |
| 2 | TTACATGGAT | 0 | B | GTGAGTCCTG | B | CTGCCCCCAG | 0 | 3 | GCTACTTACT |
| 3 | AGCGGCTGCT | 1 | C | GTGAGTGCGG | C | CCCTCTGCAG | 1 | 4 | CCACACATCC |
| 4 | CACTCTGACC | 0 | D | GTAAGTGCAG | D | TCCCCTCCAG | 1 | 5 | GTCAGAACAT |
| 5 | ACCTTGGCAG | 2 | E | GTGAGCATCT | E | TCTCATGCAG | 2 | 6 | GAAAGAAGGC |
| 6 | AAATCGAGGA | 2 | F | GTGAGTTTGT | F | TTCCATTCAG | 2 | 7 | GTTCCTCGTT |
| 7 | AGGAGAACCT | 2 | G | GTTTGGAGAG | G | CTCGCTGCAG | 0 | 8 | TATTTTGTGA |
| 8 | CATCATTAGT | 0 | H | GTAAGTCCTG | H | TGTTGAGCAG | 0 | 9 | GACGATGGCT |
Generally, the determined lengths of the CfPAG-L exons (1–9) were similar to exonic lengths of bPAG1, bPAG2, and pPAG2. However, CfPAG-L introns (A–H) completely differ compared to previously known PAG introns (Table 3). A megablast of the entire CfPAG-L gDNA sequence showed some homology only with various BAC clones and pepsinogen C of different species, but 1–5% query cover of these Blast Hits preclude regarding the results as significant. Also, a direct megablast of the CfPAG-L with bPAG1 (Acc. No. AH003454.1) or pPAG2 (Acc. Nos.: U39198–9; U41421–4; U39762–3; KF471015.1; KF492695.1; KF500427.1; KF527576.1; KF537535.1) revealed no significant similarities. Alignments performed separately for each CfPAG-L exon and intron with equivalent segments of bPAG1 and pPAG2 structure revealed a lack of homology in the case of intronic regions and 50.3–72.9% homology to exonic regions (Table 4). In silico analyses of the identified CfPAG-L-enabled prediction of 31 SNVs (single nucleotide variant), in which 7 were transversions and 24 transitions (Table 5). Among them, 5 identified SNVs were localized within exons and the remaining 26 SNVs within introns. Changes of nucleotide sequence within exons did not affect the amino acid sequence of the CfPAG-L polypeptide precursor, and all identified SNVs were synonymous.
Table 3.
Nucleotide lengths of the exons and introns of the CfPAG-L, bPAG1, bPAG2, and pPAG2 gDNA templates
| Segment name | CfPAG-L [bp] | bPAG1 [bp] | bPAG2 [bp] | pPAG2 [bp] |
|---|---|---|---|---|
| Exon 1 | 59 | 53 | 53 | 53 |
| Intron A | 1937 | 1100 | 1300 | 1350 |
| Exon 2 | 160 | 151 | 151 | 166 |
| Intron B | 385 | 1000 | 1000 | 1200 |
| Exon 3 | 118 | 118 | 118 | 118 |
| Intron C | 917 | 100 | 100 | 90 |
| Exon 4 | 119 | 119 | 119 | 119 |
| Intron D | 451 | 1200 | 1200 | 1200 |
| Exon 5 | 200 | 194 | 194 | 200 |
| Intron E | 1138 | 900 | 1100 | 850 |
| Exon 6 | 123 | 117 | 117 | 117 |
| Intron F | 288 | 1900 | 1700 | 1800 |
| Exon 7 | 148 | 142 | 142 | 136 |
| Intron G | 681 | 100 | 100 | 85 |
| Exon 8 | 99 | 99 | 99 | 99 |
| Intron H | 603 | 1700 | 1700 | 292 |
| Exon 9 | 147 | 150 | 150 | 156 |
Table 4.
Homology of the CfPAG-L exons with equivalents of bPAG1 and pPAG2 gDNA
| CfPAG-L | bPAG1 [%] | pPAG2 [%] |
|---|---|---|
| Exon 1 | 60.7 | 64.3 |
| Exon 2 | 50.3 | 60.4 |
| Exon 3 | 66.1 | 72.9 |
| Exon 4 | 56.9 | 59.2 |
| Exon 5 | 54.8 | 60.4 |
| Exon 6 | 56.9 | 62.2 |
| Exon 7 | 55.8 | 54.2 |
| Exon 8 | 57 | 57.4 |
| Exon 9 | 53.5 | 56.7 |
Table 5.
Identified SNVs, genotypes and allele frequency (px) within the CfPAG-L sequence
| Type of SNV | Position | Segment | Homozygous | Heterozygous | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nt | n | px | nt | n | px | nt | n | px | ||||
| 1 | Transition | 109 | Intron A | gg | 4 | 0.27 | aa | 0 | 0 | ga | 11 | 0.73 |
| 2 | Transversion | 112 | Intron A | tt | 4 | 0.27 | gg | 0 | 0 | tg | 11 | 0.73 |
| 3 | Transition | 2634 | Exon 3 | gg | 0 | 0 | aa | 0 | 0 | ga | 15 | 1.0 |
| 4 | Transition | 2655 | Exon 3 | cc | 0 | 0 | tt | 0 | 0 | ct | 15 | 1.0 |
| 5 | Transition | 2668 | Intron C | cc | 0 | 0 | tt | 0 | 0 | ct | 15 | 1.0 |
| 6 | Transition | 2671 | Intron C | gg | 0 | 0 | aa | 0 | 0 | ga | 15 | 1.0 |
| 7 | Transition | 2676 | Intron C | gg | 0 | 0 | aa | 0 | 0 | ga | 15 | 1.0 |
| 8 | Transition | 2702 | Intron C | gg | 0 | 0 | aa | 0 | 0 | ga | 15 | 1.0 |
| 9 | Transition | 2721 | Intron C | gg | 0 | 0 | aa | 0 | 0 | ga | 15 | 1.0 |
| 10 | Transition | 2738 | Intron C | gg | 0 | 0 | aa | 0 | 0 | ga | 15 | 1.0 |
| 11 | Transition | 3803 | Intron D | tt | 0 | 0 | cc | 0 | 0 | tc | 15 | 1.0 |
| 12 | Transition | 3822 | Intron D | gg | 0 | 0 | aa | 0 | 0 | ga | 15 | 1.0 |
| 13 | Transition | 3890 | Intron D | gg | 0 | 0 | aa | 0 | 0 | ga | 15 | 1.0 |
| 14 | Transition | 3912 | Intron D | gg | 2 | 0.14 | aa | 0 | 0 | ga | 13 | 0.86 |
| 15 | Transversion | 3936 | Intron D | gg | 4 | 0.27 | cc | 0 | 0 | gc | 11 | 0.73 |
| 16 | Transversion | 4098 | Intron D | gg | 0 | 0 | tt | 1 | 0.07 | gt | 14 | 0.93 |
| 17 | Transversion | 4124 | Intron D | aa | 1 | 0.07 | cc | 1 | 0.07 | ac | 13 | 0.86 |
| 18 | Transition | 4131 | Intron D | cc | 0 | 0 | tt | 0 | 0 | ct | 15 | 1.0 |
| 19 | Transition | 4141 | Intron D | cc | 0 | 0 | tt | 0 | 0 | ct | 15 | 1.0 |
| 20 | Transversion | 4221 | Exon 5 | aa | 0 | 0 | cc | 0 | 0 | ac | 15 | 1.0 |
| 21 | Transversion | 6101 | Intron G | tt | 0 | 0 | gg | 0 | 0 | tg | 15 | 1.0 |
| 22 | Transition | 6152 | Intron G | cc | 0 | 0 | tt | 0 | 0 | ct | 15 | 1.0 |
| 23 | Transition | 6948 | Intron H | cc | 0 | 0 | tt | 0 | 0 | ct | 15 | 1.0 |
| 24 | Transition | 6981 | Intron H | aa | 0 | 0 | gg | 0 | 0 | ag | 15 | 1.0 |
| 25 | Transition | 6993 | Intron H | cc | 12 | 0.8 | tt | 0 | 0 | ct | 3 | 0.2 |
| 26 | Transversion | 7138 | Intron H | aa | 0 | 0 | tt | 0 | 0 | at | 15 | 1.0 |
| 27 | Transition | 7140 | Intron H | tt | 0 | 0 | cc | 0 | 0 | tc | 15 | 1.0 |
| 28 | Transition | 7345 | Intron H | cc | 0 | 0 | tt | 0 | 0 | ct | 15 | 1.0 |
| 29 | Transition | 7406 | Intron H | cc | 0 | 0 | tt | 0 | 0 | ct | 15 | 1.0 |
| 30 | Transition | 7537 | Exon 9 | cc | 0 | 0 | tt | 0 | 0 | ct | 15 | 1.0 |
| 31 | Transition | 7549 | Exon 9 | cc | 0 | 0 | tt | 0 | 0 | ct | 15 | 1.0 |
Discussion
This is the first study identifying the PAGs in the genome of the Eurasian beaver (CfPAG-L). Among the identified 7657 bp of the CfPAG-L gDNA sequence, nine exons/eight introns (A–H) and 31 SNVs were found. Generally, the length of the CfPAG-L exons (1–9) is similar to exon lengths of bPAG1, bPAG2, and pPAG2. However, the length of the CfPAG-L introns (A–H) completely differ from previously known PAG introns. The localization of the two Asp residues (D), specific for catalytic cleft of APs, is also conserved in exons 3 and 7 of CfPAG-L. Despite the very strong resemblance of the coding region of CfPAG-L gene to pepsinogen C (A. Lipka et al. unpublished), there is no such similarity for intronic regions. Additionally, the identified intronic regions of the CfPAG-L did not exhibit significant homology to any sequences deposited in GenBank database.
Phylogenetic studies show that the PAG-L family arise as a result of a progene duplication or fusion, causing various reproductive capability that may be the result of positive selection of these genes during evolution (Hughes et al. 2003). Lately, limited studies of gDNA revealed the number-diversified presence of the PAG-L family in the genomes of some eutherian species, e.g., the elk, yak, wildebeest, impala, and several other antelopes; the pig, goat, horse, cow, sheep, deer, and wild boar and bison (see Szafranska et al. 2006); as well as the alpaca, dromedary, and Bactrian (Majewska et al. 2009). However, unknown exon-intron structure of the PAGs has not been studied in the aforementioned and other eutherian species. Previously, based on both pPAG1 and pPAG2 cDNA identification (Szafranska et al. 1995), the positions of the exonic and intronic boundaries have been established for the pPAG2 only (Szafranska et al. 2001). The pPAG2 represents the first member of the pPAG2-Like subfamily (pPAG2-L: pPAG4, 6, 8 and 10) encoding catalytically active APs, although potentially inactive members of the pPAG1-L (pPAG1-L: pPAG3 and 5) have also been identified (Panasiewicz et al. 2004). The pPAG2 structure (Szafranska et al. 2001), encompasses nine exons (99–200 bp) and eight introns (A–H; 85–1.8 kbp). The shorter introns have been fully sequenced (C, G and H), although the lengths of the longer introns were estimated after PCR amplification and electrophoretic analysis. Recently, nucleotide sequences of the remaining pPAG2 introns (A, B, D, E and F) have been sequenced and deposited in the GenBank database (KF471015.1; KF492695.1; KF500427.1; KF527576.1; KF537535.1; M. Bieniek-Kobuszewska et al. unpublished). The identified lengths of the introns A, B, D, E, and F are relatively comparable with the predicted ones, according to the pPAG2 structure (Szafranska et al. 2001). The final length of the pPAG2 with promoter region is equal to 8755 bp (Bieniek-Kobuszewska et al. 2016). The second known exon-intron organization concerns the bPAG1 gene (8095 bp) which is similar to other APs and whose intron sizes vary from 87 bp to 1.8 kbp and exon-intron boundaries conform to the standard GT-AG rule for 5′ donor and 3′ acceptor sites (Xie et al. 1995). Moreover, 18 full-length bPAG genes with the conserved 9-exon structure of various PAG-Ls are represented and properly annotated in the genome assembly (Telugu et al. 2009). Thus, our results are consistent with the exon-intron structures of three known PAG genes: bPAG1 (Xie et al. 1995), pPAG2 (Szafranska et al. 2001) and bPAG2 (Telugu et al. 2009).
The obtained results regarding SNVs within CfPAG-L gene may constitute a basis for further genome-wide association (GWA) studies (Appels et al. 2013; Akpinar et al. 2017). The relationship between a specific genotype and a phenotype can be used to predict genes that may correlate with observable traits in various animals. Presently, since our SNV data of the CfPAG-L cannot be compared in the beavers because such data are unavailable, we will therefore discuss the data in relation to the PAGs in other species. Previously, 32 SNPs/InDel and 42 SNPs/InDels have been identified within proximal and flanking distal regions of the pPAG2-L promoter in cross-breed pigs and in the Duroc breed, respectively (Bieniek-Kobuszewska et al. 2016). Many of those SNPs have been identified within transcription factor binding sites, which suggests the importance of allelic diversity and the significant influence on regulation of the pPAG2-L expression. Other studies concerning polymorphism in pPAG2-L gene indicate that the SNPs identified within the exon 6 and the intron F are associated with reproductive traits, i.e., the number of the born alive and weaned piglets (G. Panasiewicz et al. unpublished). Thus, in the case of our results it cannot be excluded that predicted SNVs may be involved in positive or negative regulation of placenta development, which may seriously affect pregnancy outcome, as it was previously suggested in human (Majewska et al. 2017). To date, GWA studies have been mainly focused on the selection of domestic animals with the best traits for breeding (Hering et al. 2014). In the future, our results may be useful to establish a genetic marker for the selection of unrelated representatives of various endangered animals for reconstruction or reintroduction programs.
Finally, since the beaver is not a common object of research it is extremely difficult to compare and discuss the obtained genetic results. However, considering the direct impact of this species on the environment and its significant influence on all other organisms inhabiting that environment, we are confident that the beaver will become a more common object of biological and economic interest.
This study provided pioneering data on the CfPAG-L family in the genome of the beaver, the largest rodent in Europe. Our data will presumably have an influence on further explanation of proper genetic regulation, efficient implantation and pregnancy maintenance in this species. Our results extend present knowledge about the beaver genome, and in the future, will help to improve the possibility of biodiversity conservation and genetic resource protection in Poland and other countries.
Acknowledgements
The authors thank Dr. Grzegorz Belzecki (The Kielanowski Institute of Animal Physiology and Nutrition of Polish Academy of Sciences) and Prof. Zygmunt Gizejewski (Institute of Animal Reproduction and Food Research of Polish Academy of Sciences) for enabling collection of the research material.
Compliance with ethical standards
The studies presented in this manuscript have been accomplished in accordance with the established standards for the use/care of animals and in full agreement with local ethical authorities: the Regional Directorate for Environmental Protection in Olsztyn (RDOS-28-OOP-6631-0007-638/09/10/pj), the III Local Ethical Commission for Experiments on Animals at the Warsaw University of Life Sciences (11/2010), and the Local Ethical Commission for Experiments on Animals at the University of Warmia and Mazury in Olsztyn (UWM/111/2011/DTN).
Funding
This study was supported by the National Science Centre (2012/07/N/NZ9/02050) and the University of Warmia and Mazury in Olsztyn (528–0206-881). The first author was a beneficiary of the scholarship programs co-financed by the European Social Fund: “DrINNO3 Scholarships for PhD students” (DrINNO3/85/2013) and “RIM WiM – Regional Investment in Young Scientists of Warmia and Mazury” (CIiTT/RIM WiM/2014/65).
Conflict of interest
The authors declare that they have no competing interests.
References
- Akpinar BA, Lucas S, Budak H. A large-scale chromosome-specific SNP discovery guideline. Funct Integr Genomics. 2017;17(1):97–105. doi: 10.1007/s10142-016-0536-6. [DOI] [PubMed] [Google Scholar]
- Appels R, Barrero R, Bellgard M. Advances in biotechnology and informatics to link variation in the genome to phenotypes in plants and animals. Funct Integr Genomics. 2013;13(1):1–9. doi: 10.1007/s10142-013-0319-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniek-Kobuszewska M, Panasiewicz G, Lipka A, Majewska M, Szafranska B. Novel SNPs and InDels discovered in two promoter regions of porcine pregnancy-associated glycoprotein 2-like subfamily (pPAG2-ls) in cross-breed pigs. Funct Integ Genomics. 2016;16(6):705–715. doi: 10.1007/s10142-016-0522-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanga-Kanfi S, Miranda H, Penn O, Pupko T, DeBry RW, Huchon D. Rodent phylogeny revised: analysis of six nuclear genes from all major rodent clades. BMC Evol Biol. 2009;9:71–82. doi: 10.1186/1471-2148-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carginale V, Trinchella F, Capasso C, Scudiero R, Riggio M, Parisi E. Adaptive evolution and functional divergence of pepsin gene family. Gene. 2004;333:81–90. doi: 10.1016/j.gene.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Hering DM, Oleński K, Ruść A, Kaminski S. Genome-wide association study for semen volume and total number of sperm in Holstein-Friesian bulls. Anim Reprod Sci. 2014;151:126–130. doi: 10.1016/j.anireprosci.2014.10.022. [DOI] [PubMed] [Google Scholar]
- Horn S, Durka W, Wolf R, Ermala A, Stubbe A, Stubbe M, Hofreiter M. Mitochondrial genomes reveal slow rates of molecular evolution and the timing of speciation in beavers (Castor), one of the largest rodent species. PLoS One. 2011;1:e14622. doi: 10.1371/journal.pone.0014622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Green JA, Piontkivska H, Roberts RM. Aspartic proteinase phylogeny and the origin of pregnancy-associated glycoproteins. Mol Biol Evol. 2003;20:1940–1945. doi: 10.1093/molbev/msg217. [DOI] [PubMed] [Google Scholar]
- Kageyama T. Pepsinogens, progastricsins, and prochymosins: structure, function, evolution, and development. Cell Mol Life Sci. 2002;59:288–306. doi: 10.1007/s00018-002-8423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentjies P, Drummond A. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;12:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrov LS, Orlov VN (1973) Karyotypes and taxonomy of modern beavers (Castor, Castoridae, Mammalia) Zoologicheskii Zhurnal. 52: 734–742, (In Russian; English abstract)
- Majewska M, Panasiewicz G, Louis KK, Olivera VM, Mamani JM, Abd-Elnaeim MM, Szafranska B. Pregnancy-associated glycoprotein (PAG) family: transcripts and gene amplicons in camelids. Reprod Biol. 2009;9:127–150. doi: 10.1016/S1642-431X(12)60022-9. [DOI] [PubMed] [Google Scholar]
- Majewska M, Lipka A, Paukszto L, Jastrzebski JP, Myszczynski K, Gowkielewicz M, Jozwik M, Majewski MK. Transcriptome profile of the human placenta. Funct Integr Genomics. 2017 doi: 10.1007/s10142-017-0555-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgelard C, Forty E, Arnal V, Matthee CA. Suprafamilial relationships among Rodentia and the phylogenetic effect of removing fast-evolving nucleotides in mitochondrial, exon and intron fragments. BMC Evol Biol. 2008;8:321. doi: 10.1186/1471-2148-8-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasiewicz G, Majewska M, Szafranska B. Trophoblastic cDNA cloning of porcine pregnancy-associated glycoprotein genes (pPAG) and in silico analysis of coded polypeptide precursors. Reprod Biol. 2004;4:131–141. [PubMed] [Google Scholar]
- Panasiewicz G, Majewska M, Romanowska A, Dajnowiec J, Szafranska B. Radiocompetition of secretory pregnancy-associated glycoproteins as chorionic ligands with luteal and uterine gonadotrophin receptors of pregnant pigs. Anim Reprod Sci. 2007;99:285–298. doi: 10.1016/j.anireprosci.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Szafranska B, Xie S, Green J, Roberts RM. Porcine pregnancy-associated glycoproteins: new members of the aspartic proteinase gene family expressed in trophectoderm. Biol Reprod. 1995;53:21–28. doi: 10.1095/biolreprod53.1.21. [DOI] [PubMed] [Google Scholar]
- Szafranska B, Miura R, Ghosh D, Ezashi T, Xie S, Roberts RM, Green JA. Gene for porcine pregnancy-associated glycoprotein 2 (poPAG2): its structural organization and analysis of its promoter. Mol Reprod Dev. 2001;60:137–146. doi: 10.1002/mrd.1070. [DOI] [PubMed] [Google Scholar]
- Szafranska B, Panasiewicz G, Majewska M. Biodiversity of multiple pregnancy-associated glycoprotein (PAG) family: gene cloning and chorionic protein purification in domestic and wild eutherians (Placentalia)—a review. Reprod Nutr Dev. 2006;5:481–502. doi: 10.1051/rnd:2006034. [DOI] [PubMed] [Google Scholar]
- Szafranska B, Panasiewicz G, Majewska M, Romanowska A, Dajnowiec J. Pregnancy-associated glycoprotein family (PAG)—as chorionic signaling ligands for gonadotropin receptors of cyclic animals. Anim Reprod Sci. 2007;99:269–284. doi: 10.1016/j.anireprosci.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Telugu BP, Walker AM, Green JA. Characterization of the bovine pregnancy-associated glycoprotein gene family—analysis of gene sequences, regulatory regions within the promoter and expression of selected genes. BMC Genomics. 2009;24(10):185. doi: 10.1186/1471-2164-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RM, Pohler KG, Smith MF, Green JA. Placental PAGs: gene origins, expression patterns, and use as markers of pregnancy. Reproduction. 2015;149:R115–R126. doi: 10.1530/REP-14-0485. [DOI] [PubMed] [Google Scholar]
- Xie S, Green J, Beckers JF, Roberts RM. The gene encoding bovine pregnancy-associated glycoprotein-1, an inactive member of the aspartic proteinase family. Gene. 1995;159:193–197. doi: 10.1016/0378-1119(94)00928-L. [DOI] [PubMed] [Google Scholar]