Main conclusion
Epigenetic memory affects the timing of bud burst phenology and the expression of bud burst-related genes in genetically identical Norway spruce epitypes in a manner usually associated with ecotypes.
In Norway spruce, a temperature-dependent epigenetic memory established during embryogenesis affects the timing of bud burst and bud set in a reproducible and predictable manner. We hypothesize that the clinal variation in these phenological traits, which is associated with adaptation to growth under frost-free conditions, has an epigenetic component. In Norway spruce, dehydrins (DHNs) have been associated with extreme frost tolerance. DHN transcript levels decrease gradually prior to flushing, a time when trees are highly sensitive to frost. Furthermore, EARLY BUD BREAK 1 genes (EBB1) and the FT-TFL1-LIKE 2-gene (PaFTL2) were previously suggested to be implied in control of bud phenology. Here we report an analysis of transcript levels of 12 DHNs, 3 EBB1 genes and FTL2 in epitypes of the same genotype generated at different epitype-inducing temperatures, before and during spring bud burst. Earlier flushing of epitypes originating from embryos developed at 18 °C as compared to 28 °C, was associated with differential expression of these genes between epitypes and between buds and last year’s needles. The majority of these genes showed significantly different expressions between epitypes in at least one time point. The general trend in DHN expression pattern in buds showed the expected reduction in transcript levels when approaching flushing, whereas, surprisingly, transcript levels peaked later in needles, mainly at the moment of bud burst. Collectively, our results demonstrate that the epigenetic memory of temperature during embryogenesis affects bud burst phenology and expression of the bud burst-related DHN, EBB1 and FTL2 genes in genetically identical Norway spruce epitypes.
Electronic supplementary material
The online version of this article (doi:10.1007/s00425-017-2713-9) contains supplementary material, which is available to authorized users.
Keywords: Bud phenology, Dehydrins, EBB1 genes, Epigenetic memory, FTL2, Picea abies
Introduction
Changes in global climate have been considered to represent a significant challenge for sufficiently rapid adaptation of species subjected to strong seasonal environmental stresses. Plants can cope with these stressful conditions and become more resistant to future exposures through an epigenetic memory mechanism. Epigenetic mechanisms that generate or remove epigenetic marks play an important role in plasticity responses to the environment and contribute to stress memory and adaptation in plants (Sahu et al. 2013; Thellier and Lüttge 2013; Baulcombe and Dean 2014; Avramova 2015; Crisp et al. 2016). Epigenetic mechanisms involve covalent modifications of DNA and histones of the chromatin, affecting transcriptional activity by altering the regulation of gene expression. Therefore, epigenetic mechanisms could modulate the development, morphology and physiology of an organism, contributing to an adaptive capacity of species such as forest trees (Bräutigam et al. 2013; Pikaard and Mittelsten Scheid 2014). Stress-induced epigenetic modifications are reversible but can be mitotically and meiotically transmitted in the form of heritable epialleles (Iwasaki and Paszkowski 2014).
The existence of an epigenetic memory that regulates bud phenology and cold acclimation in Norway spruce is well documented in studies of plants resulting from zygotic embryogenesis indoors in greenhouses compared to outdoors (Bjørnstad 1981; Johnsen 1989a, b; Skrøppa et al. 2007, 2010; Yakovlev et al. 2010, 2012). Furthermore, the temperature and photoperiod conditions during zygotic and somatic embryogenesis epigenetically shift the growth cycle program of the embryos, giving rise to different epitypes from the same genotype (Yakovlev et al. 2014). Depending on the temperature sum experienced by the developing embryo, phenological events such as bud burst or bud set can be advanced or delayed in time. A warmer embryogenesis environment delays their onset compared to colder conditions (Johnsen et al. 1996; Hänninen et al. 2007; Kvaalen and Johnsen 2008). During embryogenesis 329 out of 735 genes encoding putative epigenetic regulators were shown to be differentially expressed (DEG) in different epitype-inducing temperatures. The majority of these epigenetic regulator DEGs are related to DNA and histone methylation, the sRNA pathway and putative thermosensing and signaling genes (Yakovlev et al. 2016). These genes could be the main epigenetic regulators impacting chromatin status during formation of the epigenetic memory. In plants from epitype seeds the expression of siRNA pathways genes, miRNAs and phytochromes show significant DEG between epitypes (Johnsen et al. 2005; Yakovlev et al. 2010, 2011), but other transcriptional differences between such epitypes has not been surveyed.
Despite the increasing evidence for the epigenetic impact on bud burst phenology (Yakovlev et al. 2011; Bräutigam et al. 2013; Yordanov et al. 2014), generally relatively little is known about the control of dormancy release, bud burst and re-initiation of growth in trees. Studies on transcript profiling and gene expression during dormancy release and bud burst have been carried out in woody species such as Picea (Yakovlev et al. 2006; El Kayal et al. 2011; Busov et al. 2016), Populus (Rohde et al. 2007; Yordanov et al. 2014), Quercus (Derory et al. 2006; Uneo et al. 2013), Prunus (Basset et al. 2006; Yamane et al. 2008), Malus (Wisniewski et al. 2015) and Pyrus (Tuan et al. 2016). During dormancy there is a temporary suspension of morphogenetic activity. After dormancy release, bud burst marks the onset of the growing season, a time when trees are susceptible to environmental stressors such as late spring/early summer frosts. Onset of growth is water demanding for rehydration of meristems, cell expansion and metabolic pathway recovery, thus water stress-related proteins are likely important in the bud burst processes. Dehydrins (DHNs) are hydrophilic members of the late embryogenesis abundant class of proteins and are described as putative dehydration protective proteins. In trees, high levels of DHNs have been associated with tolerance to freezing temperatures, winter dormancy and protection against water stress (Basset et al. 2006; Yakovlev et al. 2008; Perdiguero et al. 2012; Eldhuset et al. 2013; Strimbeck et al. 2015). Due to their physicochemical properties, DHNs are considered to play a protective role as stabilizers of nuclear or cytoplasmic macromolecules and membranes under conditions of low water availability (Campbell and Close 1997; Danyluk et al. 1998; Koag et al. 2003). Rinne et al. (1999) suggested that DHNs might create local pools of water molecules that sustain the metabolic processes crucial during stress and re-growth. Water-binding capacity of DHNs may account for their role in protecting enzymes during cold stress (Rinne et al. 1999; Graether and Boddington 2014) and decreasing the damages created by ice crystal formation (Wisniewski et al. 1999). Their water-binding ability may also promote vitrification or act to prevent membrane–membrane interactions to stop or counteract the loss of water (Strimbeck et al. 2015).
Besides DHNs, other genes have been described to affect bud phenology. In woody perennials, a putative APETALA2/Ethylene responsive transcription factor named EARLY BUD-BREAK 1 (EBB1) was suggested to regulate the re-initiation of shoot growth after winter dormancy, since transcript levels increase prior to and during bud burst (Yordanov et al. 2014; Wisniewski et al. 2015; Busov et al. 2016; Tuan et al. 2016). EBB1 has been suggested to be involved in the shoot apical meristem activation via stimulation of cell proliferation (Yordanov et al. 2014).
Modulation of tree growth in response to seasonal changes in temperature, day-length and light quality is controlled genetically, sharing common traits with control of flowering (Gyllestrand et al. 2007; Olsen 2010; Asante et al. 2011; Olsen and Lee 2011). In Populus, short day-induced bud formation is closely linked to a decreased expression of FLOWERING LOCUS T (FT) (Böhlenius et al. 2006). Norway spruce lacks such an FT gene (Nystedt et al. 2013), but contains an FT-like gene (FTL2) with similarity also to TERMINAL FLOWER 1 (TFL1) in Arabidopsis thaliana. FTL2 shows substantially increased expression in short days and light quality conditions resulting in bud set, indicating a critical involvement in inhibiting growth and induction of bud set, as verified in Norway spruce plants overexpressing the gene (Gyllestrand et al. 2007; Asante et al. 2011; Karlgren et al. 2011, 2013; Opseth et al. 2015).
The aim of the study was to examine if the epigenetic memory of temperature during embryogenesis impacts on the bud burst-related DHNs, EBB1 and FTL2 genes in Norway spruce by examining their differential expression profiles before and during bud burst in spring in buds and last year’s needles in different epitypes.
Materials and methods
Plant materials and sample collection
Samples for studying gene expression during bud burst initiation were collected from a single genotype (ID#A2K) of the full-sib family of Picea abies (L.) Karst. arising from cross ♀#2650 × ♂#2707. This genotype was generated by somatic embryogenesis, where the embryogenic cultures were subjected to temperature treatments known to induce the formation of different epitypes from the same genotype in a predictable and reproducible manner (Kvaalen and Johnsen 2008). Two epitypes were used, originating from “cold” embryogenesis environment at 18 °C (C-epitype; CE) and “warm” embryogenesis environment at 28 °C (W-epitype; WE), respectively. For each epitype, terminal branches (nearly 15 cm long) with the terminal bud and needles of the previous year were collected at the Norwegian Institute for Bioeconomy Research field trial (Hoxmark, Ås-Norway; 59º40′07,5N/10º43′07,7E) and immediately frozen in liquid nitrogen and stored at −80 °C until use. Four biological replicates from each of the different clones (four individual trees per clone) were collected per epitype. Samples were collected weekly from the same trees in 2011 at six time points from April 20th until May 25th, covering the bud burst time point for both epitypes. The plants were also inspected weekly for their bud burst status.
RNA extraction
To study the dynamics of gene expression at different time points in the different epitypes a total of 96 samples, i.e. 48 bud and 48 needle samples, were processed separately. The rationale for studying needles in addition to buds was that leaves like shoot tips perceive environmental signals and that signaling from needles to buds in dormancy-related processes cannot be excluded (Wareing 1954; Thomas and Vince-Prue 1997). Also, to survive the winter, like shoot tips, needles have to be cold hardened and will de-harden in the spring before bud burst occur. The needles surrounding the terminal buds were removed to avoid interference with needles of the previous year. For tissue disruption a tissue lyser (RETCH MM300) bead mill was used, and RNA was purified using Epicentre MasterPure™ Plant RNA Purification Kit (Epicentre, Madison, WI, USA, #MPR09100) according to the manufacturer’s instructions. Contaminating DNA was removed from the total RNA samples using the above-mentioned kit, according to the supplier’s protocol. The total RNA preparations were stored at −80 °C until gene expression analyses. The quantity of total RNA was assessed by a NanoDrop 2000 Spectrophotometer (Thermo Scientific).
Genes searching and sequence analysis
DHNs were selected from a screened and annotated set of ESTs in a Norway spruce database from suppressive subtraction hybridization (SSH) cDNA libraries, and the FTL2 gene used has accession number EF633467 (previously named TFL1) (Yakovlev et al. 2008; Asante et al. 2011). For the expression study of the three spruce EBB1 orthologs we used the http://www.congenie.org functional genome resource for various tissues. These orthologs were shown to be highly expressed in vegetative buds (Busov et al. 2016).
Reverse transcription quantitative real-time PCR (RT-qPCR) analysis
RNA extracted from the four independent biological replicates of terminal buds and needles was employed for cDNA synthesis and subsequent RT-qPCR analysis. First-strand cDNA was synthesized from 375 ng of total RNA in 50 μl reaction volume using TaqMan® Reverse Transcription Reagents (Applied Biosystems, Carlsbad, CA, USA, #N8080234) according to the manufacturer’s procedure. RT-qPCR amplification was performed in a 10 μl reaction volume, using 2 μl of cDNA solution as template, 5 μl of 2X Fast® SYBR Green Master Mix and 200 nM of each primer. Gene specific primers for 12 Norway spruce DHNs, 3 orthologs of EBB1 genes (PaEBB1.1, PaEBB1.2 and PaEBB1.3) and PaFTL2 were designed with the Primer 3 software (Rozen and Skaletsky 2000) with default parameters and amendments according to the following criteria: melting temperature around 70 °C and product size between 80 and 150 bp (Table 1). Gene expression analyses were performed using the ViiA 7 Real-time PCR system (Applied Biosystems) with standard cycling parameters. For data analysis, the arithmetic mean of four different biological replicates was calculated and a no-template control was run for each primer pair. Target gene expression was normalized to the average of transcript levels of the spruce ACTIN (PaACTIN), TRANSLATION INITIATION FACTOR-5-ALPHA (PaelF5α) and α-TUBULIN (Paα-TUB). Quantification was performed using the ViiA 7 Software (Applied Biosystems).
Table 1.
Primer sequences used for RT-qPCR analyses of twelve dehydrins, three EARLY BUD-BREAK 1 orthologs and the FLOWERING LOCUS T-LIKE 2 transcripts and three reference genes. Sequences are listed in the 5′–3′ direction
| Gene IDs | Accession no.a | Forward primer | Reverse primer | Product length (bp) |
|---|---|---|---|---|
| PaDHN 1 | MA_95995g0010 | GCGGCCTATGCGGCAAGAA | TCGACGAGCCCCGCCTTCTG | 95 |
| PaDHN 2.2 | MA_187114g0010 | CGCGGGCTGTTCGGTTTGTT | CAGCGGACGCAAGCAGAGGA | 115 |
| PaDHN 4.3 | MA_8320994g0010 | GCGGCGGACAGCATTCTTCG | TTGTTATCGTGGCCCGGAAGC | 100 |
| PaDHN 6 | MA_747559g0010 | TCCCGGAGGCCGGAACAAGT | CGAAAGCGACATGGAGAGGTAGCC | 102 |
| PaDHN 9 | MA_2408574g0010 | TCACGGTCAGCAGGGGCAAG | AACCGGAGCCGGAGCCATGT | 101 |
| PaDHN 13 | MA_205576g0010 | CCAGCTCAGAAGGCGGGGTTT | TGGCATCCAGGCAGCATCTC | 116 |
| PaDHN 23 | MA_144878g0010 | GACTACCAGGACCGCAGCCACA | GAGTCTGGCCTCCGGGAATCA | 103 |
| PaDHN 24 | MA_12179g0010 | CCCGGCTGTCTGGAATGCTC | CCGCCAAAACCCCTAGCAGAACA | 90 |
| PaDHN 35 | MA_10428426g0010 | GGACGAAGGAACGCAGGATGA | CTGGCATCCGGGCAGCTTCT | 154 |
| PaDHN 39 | MA_86965g0010 | CGAGGAGGATAAGGGCGGGAAT | TGCGTGGGTTGTAGCAGGTG | 115 |
| PaDHN 40 | MA_10257300g0010 | CGTGGCAGGAGCAGGCATCA | GAGCCGGAGCGCAAAGACCA | 102 |
| PaDHN 41 | MA_10434136g0010 | CCGCGAGAAGCCCGTCCATAC | CACCAGCAAGAACACCGGCTGA | 98 |
| PaEBB1.1 | MA_27642g0010 | TGGCTTCGACACAACTGATCCTACCA | TGTGGTTGATCTTGTGGCTGCTGT | 114 |
| PaEBB1.2 | MA_30120g0010 | CGCTCCTTCTTACTTTGGCTTCGAC | CGTGTTGCTGCTGCTGTAATTCTGG | 119 |
| PaEBB1.3 | MA_77420g0010 | CCACCTCGGGTTGTGGTGTTTGTC | GTCATAAGGCCAGTTGAGCCAATGC | 88 |
|
PaFTL2
(Previously TFL1) |
EF633467 | ATGTTGGAGGAGACGACTTG | GTGTTGGATCGCTTGGACTA | 81 |
| Pa ACTIN | AY961918/MA_10427661g0030 | TGAGCTCCCTGATGGGCAGGTGA | TGGATACCAGCAGCTTCCATCCCAAT | 105 |
| Pa αTub |
X57980/ MA_93486g0010 |
GGCATACCGGCAGCTCTTC | AAGTTGTTGGCGGCGTCTT | 66 |
| Pa elF5α |
AY961932/ MA_103714g0010 |
GCCGATGCGGGAGCTTCCAA | TGCAGGGCCTGGCCTTAATGACG | 88 |
aAccession no. based on Norway spruce genome sequence v.1. (http://congenie.org/)
Statistical analysis
One-way ANOVA and Tukey’s test were done for buds and needles separately using IBM SPSS Statistics for Windows, Version 22.0 (Armonk, NY: IBM Corp). (Supplemental ANOVA File 1). Gene expression values were also analyzed with a principal component analysis (PCA) in R Statistical Environment (R Core Team 2017) core functions plus the package FactoMineR (Lê et al. 2008) after correcting the missing values with the package missMDA (Josse and Husson 2016).
Results
Timing of bud burst differs between genetically identical Norway spruce epitypes
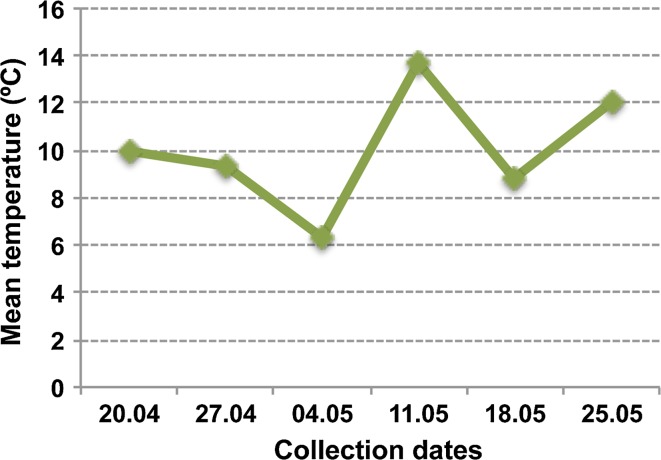
The epitypes generated from one single genotype examined in this work, were generated by somatic embryogenesis at two epitype-inducing temperatures (Fig. 1) as shown by Kvaalen and Johnsen (2008). The epitypes showed reproducible and predictable phenotypic differences related to the timing of bud burst (Fig. 1c) and bud set (Kvaalen and Johnsen 2008). Plants originating from embryos developed at 18 °C (CE) showed advanced bud burst with up to 2 weeks compared to plants originating from a warm embryogenic environment at 28 °C (WE) (Fig. 1c). For CE and WE bud burst occurred on May 11th and 25th (2011), respectively, which correlated with the first and second higher mean temperature periods recorded that spring (Fig. 2).
Fig. 1.
Epitypes of Picea abies a Somatic embryos cultured at 18 °C. Bar 1 mm. b Forest plantation of epitype trees generated from somatic embryos exposed to epitype-inducing temperature conditions. c Epitypes under identical spring conditions showing marked phenotypic differences in timing of bud burst-related to epigenetic memory of cold (CE; 18 °C) and warm (WE; 28 °C) temperature conditions during embryogenesis
Fig. 2.
Mean temperatures registered at the different time points in spring in year 2011. CE and WE showed bud burst on May 11th and May 25th, respectively. Data were obtained from the Sørås Field Station for Agroclimatic studies, Norwegian University of Life Sciences, Ås, Norway
Gene expression profiles differ between epitypes and plant parts
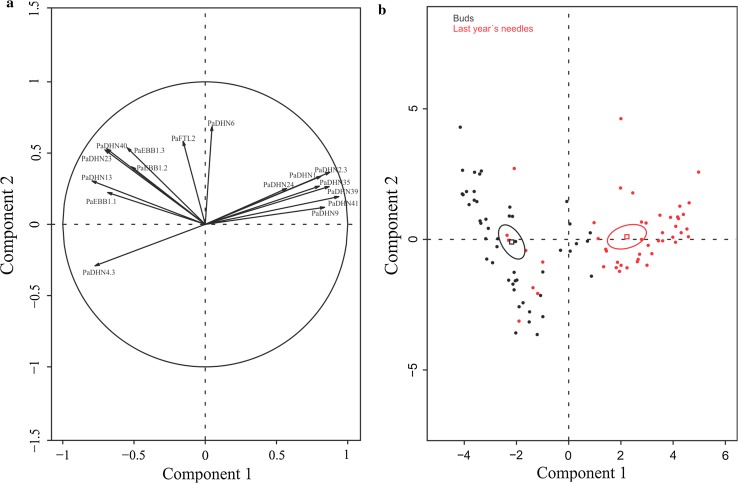
To study the overall pattern of gene expression and plant parts (buds and last year’s needles), PCA analyses were performed. In the analysis including both plant parts, the first component explained half of the variability and was significantly associated with plant part (R 2 = 0.6, probability value = 1.16e−20) (Fig. 3a, b). For this component, 14 out of the 16 genes studied showed significant correlation (Supplemental Table S1). The second component explained 15.94% of the variation with a weak but significant association with epitype (R 2 = 0.13, P value = 2.22e−4). No significant association was found among any of the components shown and sampling date (result not shown). Overall, two clear groups of DHNs could be identified: on one hand, PaDHN 40, PaDHN 23, PaDHN 4.3 and PaDHN 13, and on the other hand, PaDHN 41, PaDHN 39, PaDHN 35, PaDHN 2.2, PaDHN 24, PaDHN 1 and PaDHN 9 (Fig. 3a).
Fig. 3.
PCA factor maps for buds and last year’s needles from CE and WE epitypes of Picea abies. CE and WE trees originate from embryos formed under cold and warm epitype-inducing temperatures, respectively. Gene expression distribution (a) and sample distribution according to tissue type showing means (squares) and confidence ellipses (b). Arrows represent contribution intensity and direction of contribution
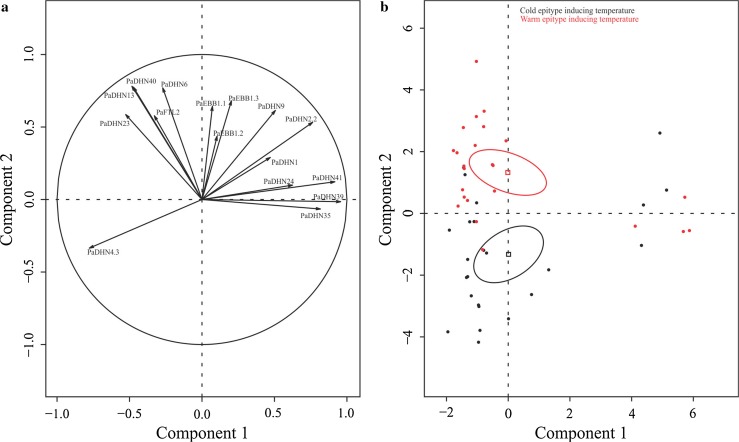
Given the clear separation between buds and last year’s needles, PCA analyses were performed for each of the plant parts (Figs. 4 and 5) in order to get insight into the effects of the epitype-inducing temperature and the sampling date on gene expression. The DHNs maintained a similar grouping as described above (Figs. 4a, 5a). In the PCA for buds, the first component explained 34.05% of the variability and was mainly associated with the sampling date (R 2 = 0.93, P value = 1.45e−23; data not shown), with the 1st collection date being significantly different from the others (data not shown). The second component explained 27.69% of the variability and was significantly associated with the epitype-inducing temperature (Fig. 4b), (R 2 = 0.39, P value = 1.59e−06).
Fig. 4.
PCA factor maps for the buds of the CE and WE epitypes of Picea abies. CE and WE trees originate from embryos formed under cold and warm epitype-inducing temperatures, respectively. Gene expression distribution (a) and sample distribution showing means (squares) and confidence ellipses (b). Arrows represent contribution intensity and direction of contribution
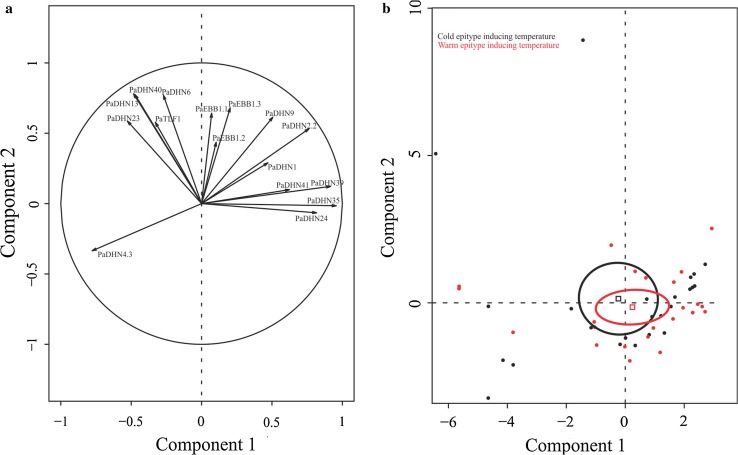
Fig. 5.
PCA factor maps for last year’s needles from CE and WE epitypes of Picea abies. CE and WE trees originate from embryos formed under cold and warm epitype-inducing temperatures, respectively. Gene expression distribution (a) and sample distribution according to tissue type showing means (squares) and confidence ellipses (b). Arrows represent contribution intensity and direction of contribution
For last year’s needles, component one explained 39.7% of the variability and the second 22.19% (Fig. 5a). In contrast to buds, there was no significant association of epitype with any of the components (Fig. 5b). Nevertheless, the majority of the genes showed significant correlation with both components (Supplemental Table S2). The sampling date was significantly associated with component one (R 2 = 0.64, P value 1.40e−08), and the variability for the 3rd and 4th collection dates was explained by the positive part of the component, while the variability for the 1st collection date was explained by the negative part. Moreover, sampling date was also significantly associated with component two (R 2 = 0.22, P value 0.04) and the 5th collection date was partially explained by the positive part.
Differences in gene expression profiles between epitypes
To examine if the timing of the expression of 12 DHNs, 3 EBB1 genes and PaFTL2 differed between epitypes, RT-qPCR analyses of expression pattern of these genes were carried out separately in buds and last year’s needles.
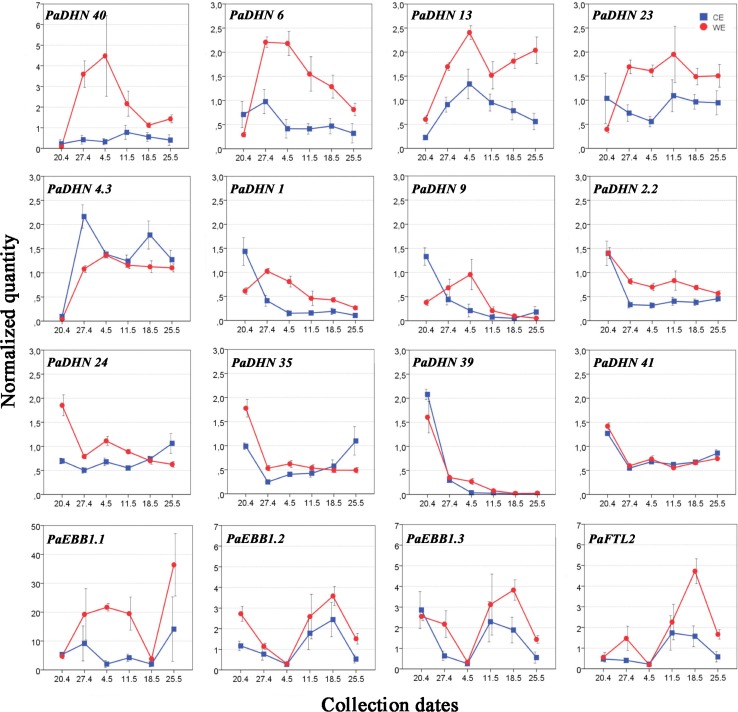
In buds, the majority of the DHN genes (PaDHN 40, PaDHN 6, PaDHN 13, PaDHN 23, PaDHN 1, PaDHN 2.2, PaDHN 24, PaDHN 35 and PaDHN 39) showed differences in expression between the epitypes at several time points (mainly at 2nd and 3rd collection dates: 27th of April and 4th of May). Overall, the expression of these PaDHNs was significantly higher in the WE, consistent with its delayed bud burst compared to CE. Furthermore, DHNs could be grouped according to their expression patterns within CE or WE. For WE, the DHN genes PaDHN 40, PaDHN 6 and PaDHN 1 showed an increasing expression pattern up to 2nd and/or 3rd collection dates (27th of April and 4th of May), followed by a decreasing transcript level (Fig. 6). Even though no significant differences were observed for PaDHN 9 when comparing 1st and 2nd collection dates, it showed a similar expression pattern. PaDHN 40 showed the highest transcript level for this epitype 3 weeks before bud burst (4th of May), reaching levels twofold higher than PaDHN 6 at this time point. In WE, PaDHN 13 had the same pattern as described above but at bud burst (25th of May) it showed increased expression. In this epitype, PaDHN 23 and PaDHN 4.3, showed increasing expression up to four and 3 weeks before bud burst, respectively (27th of April and 4th of May), and then was maintained at quite stable levels during the sampling period (Fig. 6). In WE, PaDHN 2.2, PaDHN 24, PaDHN 35, PaDHN 39 and PaDHN 41 followed a decreasing trend towards bud burst as compared to the first sampling point (Fig. 6). Furthermore, in CE, an increasing expression of PaDHN 13 and PaDHN 4.3 was detected up to 1 or 2 weeks before bud burst (4th of May and 27th of April), followed by a decreasing transcript level (Fig. 6). Within the period studied there seemed to be a small increase in transcript levels for PaDHN 24 and PaDHN 35 after bud burst. In CE, PaDHN 1, PaDHN 9, PaDHN 2.2, PaDHN 39 and PaDHN 41 showed decreasing expression pattern towards bud burst (Fig. 6). Finally, several DHNs such as PaDHN 40, PaDHN 6 and PaDHN 23 showed more stable transcript levels in CE than in WE, as no significant differences in transcript levels were detected along the sampling period.
Fig. 6.
Expression profiles of the Picea abies dehydrins, the EBB1 orthologs and the FTL2 gene in terminal buds (or shoot tips after bud burst occurred) in CE and WE. For CE and WE, bud burst occurred on May 11th and 25th (2011), respectively. Data represent the arithmetic mean ± standard error of four different biological replicates at each sampling point. Quantified transcript level was normalized to the average of the spruce reference genes PaACTIN, PaelF5α and Paα-TUB
EBB1 genes and PaFTL2 showed differences in expression between the epitypes at several time points with overall significant higher expression in the WE epitype. For this epitype differences were observed for PaEBB1.1 three and 2 weeks before bud burst (4th and 11th of May), and for PaEBB1.2, PaEBB1.3 and PaFTL2 at bud burst (25th of May) (Fig. 6). The highest transcript level was detected for PaEBB1.1, with a significant increment at WE flushing compared to earlier time points. CE and WE showed similar expression patterns with ups and downs for PaEBB1.2, PaEBB1.3 and PaFTL2. No differential expression between CE and WE was detected for these genes at the time point corresponding to 1 week before bud burst for CE and 3 weeks before bud burst for WE (4th of May). Thereafter, their expression increased for the next 2 weeks and decreased drastically 2 weeks after flushing for CE and at bud burst for WE (25th of May) (Fig. 6).
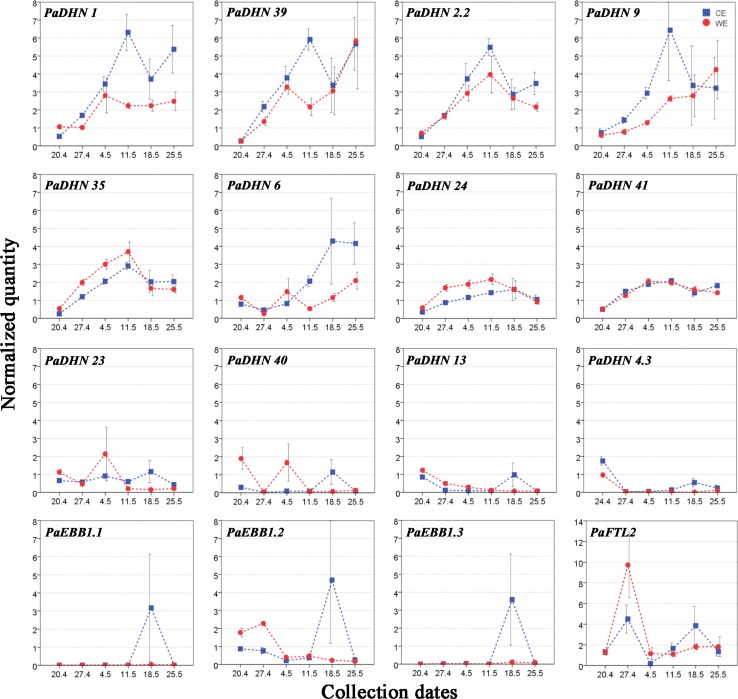
In last year’s needles, the majority of the DHNs showed differences in expression between the epitypes at several time points, except for PaDHN 2.2 where no significant differences were detected. For CE, PaDHN 1, PaDHN 39 and PaDHN 2.2 showed increasing gene expression up to bud burst with the highest transcript level at this time point (11th of May). This was followed by a decrease the week thereafter before the levels of these transcripts again increased (Fig. 7). For this epitype, PaDHN 9 and PaDHN 35 showed similar patterns but did not increase towards the final time point (Fig. 7). Despite that these DHNs showed the highest transcript level at bud burst, the transcript level of PaDHN 35 showed less increase compared to PaDHN 1, PaDHN 39, PaDHN 2.2 and PaDHN 9, which increased about twice as much. Also, in CE, PaDHN 6 showed a clear increasing trend during the sampling period (Fig. 7). However, for this epitype very low or barely detectable expression was observed for PaDHN 23, PaDHN 40, PaDHN 13 and PaDHN 4.3 although slightly higher expression was detected a week after bud burst (18th of May) (Fig. 7). For the WE epitype, PaDHN 1, PaDHN 39, PaDHN 9 and PaDHN 6 in last year’s needles showed overall increasing transcript levels towards bud burst (Fig. 7). In this epitype, an increasing transcript level was observed for PaDHN 2.2 and PaDHN 35 with the highest expression 2 weeks before bud burst (11th of May) followed by decreased expression (Fig. 7). A similar trend but with lower amplitude was detected for PaDHN 24 and PaDHN 41 for both epitypes. For WE, PaDHN 13 and PaDHN 4.3 showed low transcripts level at first collection date (20th of April) followed by decrease to very low or barely detectable levels. Even though no significant differences were found for PaDHN 23 and PaDHN 40, they showed a similar trend but with slightly higher transcript levels 4 weeks before bud burst (4th of May) (Fig. 7).
Fig. 7.
Expression profiles of the Picea abies dehydrins, the EBB1 orthologs and the FTL2 gene in last year’s needles of CE and WE. For CE and WE, bud burst occurred on May 11th and 25th (2011), respectively. Data represent the arithmetic mean ± standard error of four different biological replicates at each sampling point. Quantified transcript level was normalized to the average of the spruce reference genes PaACTIN, PaelF5α and Paα-TUB
In last year’s needles a significant difference was observed between epitypes for PaEBB1.2 (three first collection dates). For the early-flushing CE, the EBB1 genes showed no significant differences between collection dates, only a trend of higher expression the week after bud burst (Fig. 7). For the late-flushing WE, decreasing expression towards bud burst was observed for PaEBB1.2 whereas PaEBB1.1 and PaEBB1.3 showed no differential expression (Fig. 7). The epitypes showed similar behavior for PaFTL2 with the highest transcript levels at 27th of April (Fig. 7).
Differences in gene expression profiles between plant parts within epitypes
When buds and last year’s needles were compared, the expression pattern of the studied genes showed similar behavior within each epitype (Supplemental Figs. S1 and S2). For PaDHN 2.2, PaDHN 35, PaDHN 39, PaDHN 9, PaDHN 1, PaDHN 41 and PaDHN 24 transcript levels were significantly higher in needles compared to terminal buds for CE as well as WE. PaDHN 6 and PaFTL2 also showed this difference for CE. On the contrary, PaDHN 4.3, PaDHN 13 and PaEBB1.1 transcript levels were significantly higher for buds in both epitypes. PaEBB1.2 and PaEBB1.3 were significantly higher in buds than needles for WE.
In relation to bud burst, independently of the epitype, an opposite expression pattern was observed between buds and needles. Approaching bud burst, expression of DHNs decreased in buds (e.g. PaDHN 2.2, PaDHN 39, PaDHN 9, PaDHN 1, PaDHN 41, PaDHN 24, PaDHN 6, PaDHN 40). However, in needles, transcript levels of PaDHN 39, PaDHN 9, PaDHN 1, and PaDHN 6 were low at early spring (Supplemental Figs. S1 and S2), showing the highest expression level mainly at bud burst. It is noteworthy that some DHNs (including PaDHN 4.3, PaDHN 13, PaDHN 23 and PaDHN 40), EBB1 genes and PaFTL2, in CE needles showed a peak of expression at one specific time point (18th of May) corresponding to the week after bud burst (Supplemental Figs. S1 and S2).
Discussion
The main aim of this work was to investigate how the epigenetic memory of temperature during embryogenesis regulates bud burst in genetically identical epitypes (CE and WE) of Norway spruce. The studied eight year-old epitypes showed differences in the timing of bud burst. Consistent with the previously reported marked phenotypic differences (i.e. differences in timing of bud set) related to the memory of temperature during somatic embryo development (Kvaalen and Johnsen 2008), the CE showed flushing of terminal buds about 2 weeks earlier than WE. It is worth to mention that the first higher mean temperature recorded that spring, corresponded to the moment when CE exhibited bud burst (May 11th 2011), and the second higher temperature period corresponded to WE bud burst (May 25th 2011) (Fig. 2). The results confirm the existence of an epigenetic memory mechanism in Norway spruce that operates during embryo development and adjusts the timing of bud burst in the progeny in accordance with the temperature conditions during embryogenesis. At the time of this study, the epitype trees were 8 years old, which reflects that the epigenetic memory effect on bud burst is long-lasting, having implications for long-term growth under field conditions as observed by Skrøppa et al. (2007).
Here we report for the first time that the epigenetic memory affects expression of DHN genes, EBB1 genes and PaFTL2 in Norway spruce epitypes in relation to the timing of bud burst. The patterns of DHN gene expression between the two epitypes were noticeably different. Out of the 12 DHNs selected, transcript levels of 9 of them were significantly higher in buds of the late-flushing WE (Fig. 6) as compared to the early-flushing CE. These results resembles previous results in Norway spruce family materials where transcript levels of DHNs remained considerably higher in late-flushing compared to early-flushing families in the spring (Yakovlev et al. 2008). It is noteworthy that the regulation of DHNs is under tight control in this species as variation in transcript levels of DHNs within each epitype in our study was relatively low, allowing detection of significant differences between the epitypes under natural field conditions. Differences between the two epitypes were also observed for EBB1 genes and PaFTL2 in buds with the highest transcript levels for the WE. PCA for buds including all genes confirmed the significant effect of the epitype-inducing temperature (Fig. 4; Supplemental Table S3). In last year’s needles, the expression differed significantly in epitypes at several time points for 11 out of the 12 DHNs and for PaEBB1.2 (Fig. 7). Early-flushing CE showed higher transcript levels for seven of the DHNs. However, transcript levels were higher for late-flushing WE for four of the DHNs and for PaEBB1.2. These differences show that the expression of these genes is affected by the epigenetic memory in buds as well as in last year’s needles. However, in spite of these specific significant differences (ANOVA) for the last year’s needles, and in contrast to the situation for buds, the PCA for the needles, did not show any clear association between epitype and gene expression (Fig. 5; Supplemental Table S2). These results for the needles demonstrate absence of a differential global expression pattern for the analyzed genes between epitypes. This may suggest a lack of a clear role of last year’s needles in the differential timing of bud burst in the epitypes.
It is likely that the differential expression patterns between epitypes in response to the increasing spring temperature are due to specific chromatin modifications established during embryogenesis. A diverse range of environmental stresses can alter such epigenetic marks (Eichten et al. 2014). Of these, vernalization (defined as the acquisition of flowering competence by prolonged cold exposure) remains the best-understood environmentally responsive process impacted by epigenetic mechanisms. In this respect, epigenetic memory enables plants to remember their experience of winter conditions to flower the following spring. The floral repressor gene FLOWERING LOCUS C (FLC) in Arabidopsis thaliana is transcriptionally repressed by cold exposure (Baulcombe and Dean 2014) and repression is epigenetically maintained during subsequent development in warmer temperatures. Physiologically, bud burst is the final stage of a series of processes related to dormancy release and cold deacclimation. Like vernalization, dormancy release requires or is promoted by long-term exposure to low temperatures, and chilling restores the ability to grow but does not promote growth (Rohde and Bhalerao 2007). The physiological similarities between vernalization and dormancy release lead to the hypothesis that trees such as Norway spruce might employ a molecular mechanism analogous to vernalization regarding the establishment of an epigenetic memory, as Norway spruce epitypes remember the prolonged cold winter to bud burst the next spring.
An opposite behavior of the buds and last year’s needles was particularly clear for the genes selected, as also confirmed by the overall PCA analysis including both plant parts (Fig. 3; Supplemental Figs. S1 and S2). It was unexpected that the level of most DHNs was kept so high in last year’s needles (Supplemental Figs. S1 and S2). Differences between buds and needles were also observed for EBB1 genes, showing higher transcript levels in buds in most cases. On the contrary, for PaFTL2 needles generally had the highest transcript levels (See Figs. S1 and S2, in Supplemental). The expression pattern of most of the DHNs in buds was significantly decreased as bud burst was approached, whereas in needles transcript levels were low at early spring, when plants supposedly were still frost resistant, showing an increment of transcripts over time.
In Norway spruce, the timing of bud burst was shown to be associated with a high rate of net photosynthesis followed by decrease in amount of sugar and increase in starch compounds (Egger et al. 1996). Studies on metabolite profiling have been performed in Norway spruce during bud development (Lee et al. 2014; Dhuli et al. 2014). These authors assessed changes in metabolite profiles not only in sugars but also in other solutes such as ABA, antioxidants, flavonoids, terpenoids, amino acids and lipids, which were accumulated in cells during short days and frost, corroborating their cryoprotective properties. Dhuli et al. (2014) examined metabolite changes in buds and needles of Norway spruce and European silver fir during artificial forcing, observing higher levels of distinct carbohydrates in needles compared to buds. In evergreen conifers, carbohydrates and photosynthates from the previous year’s needles support shoot growth until new needles develop (Hansen and Beck 1994). Major changes in sugar metabolism occur in conifer needles during acclimation and deacclimation (Larcher 2003; Angelcheva et al. 2014). The differences in carbohydrate metabolism between buds and needles might possibly explain the differences in the regulation of DHNs, EBB1 genes and PaFTL2. On the other hand, DHNs play a role in cold tolerance of plants by maintaining low local water content, protecting the tissues against frost damage (Wisniewski et al. 1999). In Norway spruce, transcript levels of DHNs decrease gradually during the period immediately preceding bud burst (Yakovlev et al. 2008; Asante et al. 2009) which could be explained by the stage of development of buds at this period. The internal vegetative bud development preceding bud burst in Norway spruce during the spring includes the growth of the primordial shoot and the swelling of buds (Sutinen et al. 2009, 2012; Viherä-Aarnio et al. 2014). Simultaneously, the water content of the buds increases as a result of the growth of vascular tissue and lower concentration of sugar, reducing the frost hardiness significantly (de Fäy et al. 2000; Luoranen et al. 2010). Mature needles are highly protected from freezing and the dehydrin levels in last year’s needles are in general higher and not down-regulated during the studied period such as in buds, while flushed buds have no tolerance to frost at all. It seems that down-regulation of the DHNs needed for protection from freezing is a prerequisite for start of cell division and growth in Norway spruce (Yakovlev et al. 2008; Asante et al. 2009). Thus, since last year’s needles are fully-grown and do not continue to elongate they do not need to down-regulate the DHNs and can maintain high levels of these protective proteins during the spring.
EBB1 family genes seem to play an essential role in a conserved mechanism controlling bud break in perennial plants (Busov et al. 2016). In poplar, EBB1 has been suggested to regulate the re-initiation of shoot growth after winter dormancy, and transcript levels are undetectable in buds during the majority of the dormancy period but appear prior to and during bud break (Yordanov et al. 2014). EBB1 homologs are found to be associated with the timing of bud burst in apple, grape, spruce and pear (Wisniewski et al. 2015; Busov et al. 2016; Tuan et al. 2016). The dynamics of EBB1 gene expression in buds in Norway spruce appears more complex than its angiosperm trees. In angiosperms, EBB1 orthologs are down-regulated in dormant and up-regulated in actively growing apices. In our work, PaEBB1.2 and PaEBB1.3 transcript levels increased at bud burst in CE and towards bud burst in WE (Fig. 6) and reverted to low levels after bud burst in CE and at bud burst in WE. Differences in expression pattern between angiosperms and gymnosperms could be related to the difference in the biology of dormancy between these two groups.
A similar expression pattern to PaEBB1.2 and PaEBB1.3 was observed for PaFTL2. Our results show a peak of expression for this gene at bud burst for CE and 1 week before bud burst for the WE. FTL2 was shown to be up-regulated under short days in Norway spruce, indicating a critical involvement in inhibiting growth and induction of bud set (Gyllestrand et al. 2007; Asante et al. 2011; Karlgren et al. 2013; Opseth et al. 2015). Our study did not include bud set, but demonstrates relatively high expression also around bud burst. According to Gyllestrand et al. (2007), buds from adult trees in natural stands confirmed low expression levels for PaFT4, renamed by Karlgren et al. (2011) as PaFTL2, at early stages of bud burst in spring but a small increase was observed when buds had completely burst. Furthermore, it has been described that high expression of PaFTL2 is retained in needles during bud set until spring, when it drops to an intermediate level concurrent with increasing day-length and temperature (Gyllestrand et al. 2007; Karlgren et al. 2013). This observation is consistent with our results for both CE and WE, as low expression levels of this gene were detected in last year’s needles in late spring.
In conclusion, we have revealed important differences in bud phenology in epitypes formed in response to cold and warm epitype-inducing temperatures as a result of an epigenetic memory of temperature during embryogenesis. Of the 12 DHNs analyzed, 9 were found to have significantly different expression patterns in buds related to epitype-inducing temperatures during embryogenesis. Also the expression of EBB1-genes and PaFTL2 is clearly affected by the epigenetic memory. The epigenetic memory mechanism apparently provides plasticity in climatic adaptation of Norway spruce that impacts the expression of these genes.
Author contribution statement
CGF, IY and EC conceived and designed the research. CGF, IY and EC conducted the experiments. EC ran RT-qPCRs and performed ANOVAs and Tukey’s tests. MV performed PCA analysis. EC and CGF wrote the manuscript with assistance from IY, MV and JEO. All authors read and approved the final manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Inger Heldal and Anne E. Nilsen (Norwegian Institute for Bioeconomy Research) for valuable technical assistance. This work was financially supported by the EU FP7 Project ProCoGen and the NFR-FRIMEDBIO Grant 240766/F20. Elena Carneros was supported by a Grant from Iceland, Liechtenstein and Norway through the EEA Financial Mechanism. Operated by Universidad Complutense de Madrid.
Abbreviations
- CE
Cold embryogenesis environment (18 °C)
- DHNs
Dehydrins
- EBB1
EARLY BUD BREAK 1-genes
- FTL2
FT-TFL1-LIKE 2-gene
- PCA
Principal component analysis
- WE
Warm embryogenesis environment (28 °C)
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s00425-017-2713-9) contains supplementary material, which is available to authorized users.
References
- Angelcheva L, Mishra Y, Antti H, Kjellsen TD, Funk C, Strimbeck GR, Schröder WP. Metabolomic analysis of extreme freezing tolerance in Siberian spruce (Picea obovata) New Phytol. 2014;204:545–555. doi: 10.1111/nph.12950. [DOI] [PubMed] [Google Scholar]
- Asante DKA, Yakovlev IA, Fossdal CG, Timmerhaus G, Partanen J, Johnsen Ø. Effect of bud burst forcing on transcript expression of selected genes in needles of Norway spruce during autumn. Plant Physiol Biochem. 2009;47:681–689. doi: 10.1016/j.plaphy.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Asante DKA, Yakovlev IA, Fossdal CG, Holefors A, Opseth L, Olsen JE, Junttila O, Johnsen Ø. Gene expression changes during short day induced terminal bud formation in Norway spruce. Plant Cell Envrion. 2011;34:332–346. doi: 10.1111/j.1365-3040.2010.02247.x. [DOI] [PubMed] [Google Scholar]
- Avramova Z. Transcriptional ‘memory’ of a stress: transient chromatin and memory (epigenetic) marks at stress-response genes. Plant J. 2015;83(1):149–159. doi: 10.1111/tpj.12832. [DOI] [PubMed] [Google Scholar]
- Basset CL, Wisniewski ME, Timothy SA, Norelli L, Renaut J, Farrel RE. Global analysis of genes regulated by low temperature and photoperiod in peach bark. J Amer Soc Hort Sci. 2006;131(4):551–563. [Google Scholar]
- Baulcombe DC, Dean C. Epigenetic regulation in plant responses to the environment. Cold Spring Harb Perspect Biol. 2014;6(9):a019471. doi: 10.1101/cshperspect.a019471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørstand Å (1981) Photoperiodical after-effect of parent plant environment in Norway spruce (Picea abies (L.) Karst.) seedlings. Meddelelser fra Norsk Institutt for Skogforsking 36.6, 30s
- Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1039–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- Bräutigam K, Vining KJ, Lafon-Placette C, Fossdal CG, Mirouze M, Marcos JG, Fluch S, Fraga MF, Guevara MÁ, Abarca D, Johnsen Ø, Maury S, Strauss SH, Campbell MM, Rohde A, Díaz-Sala C, Cervera M-T. Epigenetic regulation of adaptive responses of forest tree species to the environment. Ecol Evol. 2013;3(2):399–415. doi: 10.1002/ece3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busov V, Carneros E, Yakovlev I. EARLY BUD-BREAK1 (EBB1) defines a conserved mechanism for control of bud-break in woody perennials. Plant Signal Behav. 2016;11(2):e1073873. doi: 10.1080/15592324.2015.1073873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell SA, Close TJ. Dehydrins: genes, proteins and associations with phenotypic traits. New Phytol. 1997;137:61–74. doi: 10.1046/j.1469-8137.1997.00831.x. [DOI] [Google Scholar]
- Crisp PA, Ganguly D, Eichten SR, Borevitz JO, Pogson BJ. Reconsidering plant memory: intersections between stress recovery. RNA turnover and epigenetics. Sci Adv. 2016;2:e1501340. doi: 10.1126/sciadv.1501340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danyluk J, Perron A, Houde M, Limin A, Fowler B, Benhamou N, Sarhan F. Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell. 1998;10:623–638. doi: 10.1105/tpc.10.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Fäy E, Vacher V, Humbert F. Water-related phenomena in winter buds and twigs of Picea abies L. (Karst.) until bud-burst: a biological, histological and NMR study. Ann Bot. 2000;86:1097–1107. doi: 10.1006/anbo.2000.1276. [DOI] [Google Scholar]
- Derory J, Leger P, García V, Schaerffer J, Hauser MT, Salin F, Lusching C, Plomion C, Glossl Kremmer A. Transcriptome analysis of bud burst in sessile oak (Quercus petraea) New Phytol. 2006;170:723–738. doi: 10.1111/j.1469-8137.2006.01721.x. [DOI] [PubMed] [Google Scholar]
- Dhuli P, Rohloff J, Strimbeck R. Metabolite changes in conifer buds and needles during forced bud break in Norway spruce (Picea abies) and European silver fir (Abies alba) Front Plant Sci. 2014;5:706. doi: 10.3389/fpls.2014.00706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Einig W, Sclereth A, Wallenda T, Magel E, Loewe A, Hampp R. Carbohydrate metabolism in one- and two-year-old spruce needles, and stem carbohydrates from three months before until three months after bud break. Physiol Plant. 1996;96:91–100. doi: 10.1111/j.1399-3054.1996.tb00188.x. [DOI] [Google Scholar]
- Eichten SR, Schmitz RJ, Springer NM. Epigenetics: beyond chromatin modifications and complex genetic regulation. Plant Physiol. 2014;165:933–947. doi: 10.1104/pp.113.234211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kayal W, Allen CCG, Ju CJT, Adams E, King-Jones S, Zaharia LI, Abrams SR, Cooke JEK. Molecular events of apical bud formation in white spruce, Picea glauca. Plant Cell Environ. 2011;34:480–500. doi: 10.1111/j.1365-3040.2010.02257.x. [DOI] [PubMed] [Google Scholar]
- Eldhuset TD, Nagy NE, Volařík D, Børja I, Gebauer R, Yakovlev I, Krokene P. Drought affects tracheid structure, dehydrin expression, and above- and belowground growth in 5-year-old Norway spruce. Plant Soil. 2013;366:305–320. doi: 10.1007/s11104-012-1432-z. [DOI] [Google Scholar]
- Graether SP, Boddington KF. Disorder and function: a review of the dehydrin protein family. Front Plant Sci. 2014;5(576):1–12. doi: 10.3389/fpls.2014.00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllestrand N, Clapham D, Källman T, Lagercranzt U. A Norway spruce FLOWERING LOCUS T homolog is implicated in control of growth rhythm in conifers. Plant Physiol. 2007;144:248–257. doi: 10.1104/pp.107.095802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänninen H, Slaney M, Linder S. Dormancy release of Norway spruce under climatic warming: testing ecophysiological models of bud burst with a whole-tree chamber experiment. Tree Physiol. 2007;27:291–300. doi: 10.1093/treephys/27.2.291. [DOI] [PubMed] [Google Scholar]
- Hansen J, Beck E. Seasonal changes in the utilization and turnover of assimilation products in 8-year-old Scots pine (Pinus sylvestris L.) trees. Trees. 1994;8:172–182. doi: 10.1007/BF00196844. [DOI] [Google Scholar]
- Iwasaki M, Paszkowski J. Epigenetic memory in plants. EMBO J. 2014;33(18):1987–1998. doi: 10.15252/embj.201488883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen Ø. Phenotypic changes in progenies of northern clones of Picea abies (L.) Karst. grown in a southern seed orchard. I. Frost hardiness in a phytotron experiment. Scan J For Res. 1989;4:331–341. doi: 10.1080/02827588909382570. [DOI] [Google Scholar]
- Johnsen Ø. Phenotypic changes in progenies of northern clones of Picea abies (L.) Karst. grown in a southern seed orchard. II. Seasonal growth rhythm and height in field trials. Scand J For Res. 1989;4:317–330. doi: 10.1080/02827588909382569. [DOI] [Google Scholar]
- Johnsen Ø, Skrøppa T, Junttila O, Dæhlen OG. Influence of female flowering environment on autumn frost-hardiness of Picea abies progenies. Theor Appl Genet. 1996;92:797–802. doi: 10.1007/BF00221890. [DOI] [PubMed] [Google Scholar]
- Johnsen Ø, Fossdal CG, Nagy N, Molmann J, Dælen OG, Skrøppa T. Climatic adaptation in Picea abies progenies is affected by the temperature during zygotic embryogenesis and seed maturation. Plant Cell Environ. 2005;28:1090–1102. doi: 10.1111/j.1365-3040.2005.01356.x. [DOI] [Google Scholar]
- Josse J, Husson F. missMDA: a package for handling missing values in multivariate data analysis. J Stat Softw. 2016;70(1):1–31. doi: 10.18637/jss.v070.i01. [DOI] [Google Scholar]
- Karlgren A, Gyllenstrand N, Kallman T, Sundstrom JF, Moore D, Lascoux M, Lagercranzt U. Evolution of the PEBP gene family in plants: functional diversification in seed plant evolution. Plant Phyisiol. 2011;156:1967–1977. doi: 10.1104/pp.111.176206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlgren A, Gyllenstrand N, Clapham D, Lagercrantz U. FLOWERING LOCUS T/TERMINAL FLOWER1-Like genes affect growth rhythm and bud set in Norway spruce. Plant Physiol. 2013;163:792–803. doi: 10.1104/pp.113.224139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavaalen H, Johnsen Ø. Timing of bud set in Picea abies is regulated by a memory of temperature during zygotic and somatic embryogenesis. New Phytol. 2008;177:49–59. doi: 10.1111/j.1469-8137.2007.02222.x. [DOI] [PubMed] [Google Scholar]
- Koag MC, Fenton RD, Wilkens S, Close TJ. The binding of maize DHN1 to lipid vesicles. Gain of structure and lipid specificity. Plant Physiol. 2003;131:309–316. doi: 10.1104/pp.011171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larcher W. Physiological plant ecology—ecophysiology and stress physiology of functional groups. Berlin: Springer; 2003. [Google Scholar]
- Lê S, Josse J, Husson F. FactoMineR: an R package for multivariate analysis. J Stat Softw. 2008;25:1–18. doi: 10.18637/jss.v025.i01. [DOI] [Google Scholar]
- Lee YK, Alexdander D, Wulff J, Olsen JE. Changes in metabolite profiles in Norway spruce shoot tips during short-day induced winter bud development and long-day induced bud flush. Metabolomics. 2014;10:842–858. doi: 10.1007/s11306-014-0646-x. [DOI] [Google Scholar]
- Luoranen J, Sutinen S, Rikala R. Predicting spring frost sensitivity by bud development and temperature sum in Norway spruce seedlings. Trees. 2010;23:683–700. [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, Vicedomini R, Sahlin K, Sherwood E, Elfstrand M, Gramzow L, Holmberg K, Hallman J, Keech O, Klasson L, Koriabine M, Kucukoglu M, Kaller M, Luthman J, Lysholm F, Niittyla T, Olson A, Rilakovic N, Ritland C, Rossello JA, Sena J, Svensson T, Talavera-Lopez C, Theiszen G, Tuominen H, Vanneste K, Wu ZQ, Zhang B, Zerbe P, Arvestad L, Bhalerao R, Bohlmann J, Bousquet J, Garcia Gil R, Hvidsten TR, de Jong P, MacKay J, Morgante M, Ritland K, Sundberg B, Lee Thompson S, Van de Peer Y, Andersson B, Nilsson O, Ingvarsson PK, Lundeberg J, Jansson S. The Norway spruce genome sequence and conifer genome evolution. Nature. 2013;497:579–584. doi: 10.1038/nature12211. [DOI] [PubMed] [Google Scholar]
- Olsen JE. Light and temperature sensing and signalling in induction of bud dormancy in woody plants. Plant Mol Biol. 2010;73:37–47. doi: 10.1007/s11103-010-9620-9. [DOI] [PubMed] [Google Scholar]
- Olsen JE, Lee YK. Trees and boreal forests. In: Storey K, Tanino K, editors. Temperature adaptation in a changing climate: Nature at risk. CABI Climate Change Series 3. Wallington: CABI; 2011. pp. 160–178. [Google Scholar]
- Opseth L, Holefors A, Rosnes AKR, Lee YK, Olsen JE. FTL2 expression preceding bud set corresponds with timing of bud set in Norway spruce under different light quality treatments. Environ Exp Bot. 2015;121:121–131. doi: 10.1016/j.envexpbot.2015.05.016. [DOI] [Google Scholar]
- Perdiguero P, Barbero MC, Cervera MT, Soto A, Collada C. Novel conserved segments are associated with differential expression pattern for Pinaceae dehydrins. Planta. 2012;236:1863–1874. doi: 10.1007/s00425-012-1737-4. [DOI] [PubMed] [Google Scholar]
- Pikaard CS, Mittelsten Scheid O. Epigenetic regulation in plants. Cold Spring Harb Perspect Biol. 2014;6:a019315. doi: 10.1101/cshperspect.a019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2017) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/. Accessed 26 Apr 2017
- Rinne PL, Kaikuranta PL, van der Plas LH, van der Schoot C. Dehydrins in cold-acclimated apices of birch (Betula pubescens Ehrh.): production, localization and potential role in rescuing enzyme function during dehydration. Planta. 1999;209:377–388. doi: 10.1007/s004250050740. [DOI] [PubMed] [Google Scholar]
- Rohde A, Bhalerao RP. Plant dormancy in the perennial context. Trends Plant Sci. 2007;12:217–223. doi: 10.1016/j.tplants.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Rohde A, Ruttink T, Hostyn V, Sterck L, Van Driessche K, Boerjan W. Gene expression during the induction, maintenance and release of dormancy in apical buds of poplar. J Exp Bot. 2007;58:4047–4060. doi: 10.1093/jxb/erm261. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: Methods in molecular biology. Totowa: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sahu P, Pandey G, Sharma N, Puranik S, Muthamilarasan M, Prasad M. Epigenetic mechanisms of plant stress responses and adaptation. Plant Cell Rep. 2013;32:1151–1159. doi: 10.1007/s00299-013-1462-x. [DOI] [PubMed] [Google Scholar]
- Skrøppa T, Kohmann K, Johnsen Ø, Steffenrem A, Edvardsen ØM. Field performance and early test results of offspring from two Norway spruce seed orchards containing clones transferred to warmer climates. Can J For Res. 2007;37:515–522. doi: 10.1139/X06-253. [DOI] [Google Scholar]
- Skrøppa T, Tollesfrud MM, Sperisen C, Johnsen Ø. Rapid change in adaptive performance from one generation to the next in Picea abies—Central European trees in a Nordic environment. Trees Genet Genom. 2010;6:93–99. doi: 10.1007/s11295-009-0231-z. [DOI] [Google Scholar]
- Strimbeck GR, Schaberg PG, Fossdal CG, Schröder WP, Kjellsen TD. Extreme low temperature tolerance in woody plants. Front Plant Sci. 2015;6:884. doi: 10.3389/fpls.2015.00884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutinen S, Partanen J, Viherä-Aarnio A, Häkkinen R. Anatomy and morphology in developing vegetative buds on detached Norway spruce branches in controlled conditions before bud burst. Tree Physiol. 2009;29:1457–1465. doi: 10.1093/treephys/tpp078. [DOI] [PubMed] [Google Scholar]
- Sutinen S, Partanen J, Viherä-Aarnio A, Häkkinen R. Development and growth of primordial shoots in Norway spruce buds before visible bud burst in relation to time and temperature in the field. Tree Physiol. 2012;32:987–997. doi: 10.1093/treephys/tps063. [DOI] [PubMed] [Google Scholar]
- Thellier M, Lüttge U. Plant memory: a tentative model. Plant Biol. 2013;15:1–12. doi: 10.1111/j.1438-8677.2012.00674.x. [DOI] [PubMed] [Google Scholar]
- Thomas B, Vince-Prue D. Photoperiodism in plants. London: Academic Press; 1997. [Google Scholar]
- Tuan P, Bai S, Saito T, Imai T, Ito A, Moriguchi T. Involvement of EARLY BUD-BREAK, an AP2/ERF transcription factor gene, in bud break in Japanese pear (Pyrus pyrifolia Nakai) lateral flower buds: expression, histone modifications, and possible target genes. Plant Cell Physiol. 2016;57:1038–1047. doi: 10.1093/pcp/pcw041. [DOI] [PubMed] [Google Scholar]
- Uneo S, Klopp C, Leplé JC, Derory J, Noirot C, Léger V, Prince E, Kremer A, Plomion C, Le Provost G. Transcriptional profiling of bud dormancy induction and release in oak by next-generation sequencing. BMC Genomics. 2013;14:236–250. doi: 10.1186/1471-2164-14-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viherä-Aarnio A, Sutinen S, Partanen J, Häkkinen R. Internal development of vegetative buds of Norway spruce trees in relation to accumulated chilling and forcing temperatures. Tree Physiol. 2014;34:547–556. doi: 10.1093/treephys/tpu038. [DOI] [PubMed] [Google Scholar]
- Wareing PF. Growth studies in woody species. Physiol Plant. 1954;7:261–277. doi: 10.1111/j.1399-3054.1954.tb07575.x. [DOI] [Google Scholar]
- Wisniewski ME, Webb R, Balsamo R, Close TJ, Yu-Xiao M, Griffith M. Purification, immunolocalization, cryoprotective, and antifreeze activity of PCA60: a dehydrin from peach (Prunus persica) Physiol Plant. 1999;105:600–608. doi: 10.1034/j.1399-3054.1999.105402.x. [DOI] [Google Scholar]
- Wisniewski M, Norelli J, Artlip T. Overexpression of a peach CBF gene in apple: a model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front Plant Sci. 2015;6:85. doi: 10.3389/fpls.2015.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlev IA, Fossdal CG, Johnsen Ø, Junttila O, Skrøppa T. Analysis of gene expression during bud burst initiation in Norway spruce via ESTs from subtracted cDNA libraries. Tree Genet Genom. 2006;2:39–52. doi: 10.1007/s11295-005-0031-z. [DOI] [Google Scholar]
- Yakovlev IA, Asante DKA, Fossdal CG, Partanen J, Junttila O, Johnsen Ø. Dehydrins expression related to timing of bud burst in Norway spruce. Planta. 2008;228:459–472. doi: 10.1007/s00425-008-0750-0. [DOI] [PubMed] [Google Scholar]
- Yakovlev I, Fossdal CG, Johnsen Ø. MicroRNAs, the epigenetic memory and climatic adaptation in Norway spruce. New Phytol. 2010;187:1154–1169. doi: 10.1111/j.1469-8137.2010.03341.x. [DOI] [PubMed] [Google Scholar]
- Yakovlev I, Asante D, Fossdal CG, Junttila O, Johnsen Ø. Differential gene expression related to an epigenetic memory affecting climatic adaptation in Norway spruce. Plant Sci. 2011;180:132–139. doi: 10.1016/j.plantsci.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Yakovlev I, Fossdal CG, Skrøppa T, Olsen JE, Jahren AH, Johnsen Ø. An adaptive epigenetic memory in conifers with important implications for seed production. Seed Sci Res. 2012;22(2):63–76. doi: 10.1017/S0960258511000535. [DOI] [Google Scholar]
- Yakovlev I, Lee YY, Rotter B, Olsen JE, Skrøppa T, Johnsen Ø, Fossdal CG. Temperature-dependent differential transcriptomes during formation of an epigenetic memory in Norway spruce embryogenesis. Tree Genet Genom. 2014;10:355–366. doi: 10.1007/s11295-013-0691-z. [DOI] [Google Scholar]
- Yakovlev I, Carneros E, Lee YY, Olsen JE, Fossdal CG. Transcriptional profiling of epigenetic regulators in somatic embryos during temperature induced formation of an epigenetic memory in Norway spruce. Planta. 2016;243(5):1237–1249. doi: 10.1007/s00425-016-2484-8. [DOI] [PubMed] [Google Scholar]
- Yamane H, Kashiwa Y, Ooka T, Tao R, Yonemori K. Suppression subtractive hybridization and differential screening reveals endodormancy-associated expression of an SVP/AGL24-type MADS-box gene in lateral vegetative buds of Japanese apricot. J Amer Soc Hort Sci. 2008;133(5):708–716. [Google Scholar]
- Yordanov YS, Ma C, Strauss SH, Busov V. EARLY BUD-BREAK 1 (EBB1) is a regulator of release of seasonal dormancy in poplar trees. Proc Natl Acad Sci USA. 2014;111:10001–10006. doi: 10.1073/pnas.1405621111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.