Abstract
Inherited retinal dystrophies are characterized by progressive retina degeneration and mutations in at least 250 genes have been associated as disease-causing. CRB1 is one of many genes analyzed in molecular diagnosis for inherited retinal dystrophy. Crumbs homolog-1 protein encoded by CRB1 is important for cell-to-cell contact, polarization of epithelial cells and the morphogenesis of photoreceptors. Pathogenic variants in CRB1 lead to a huge variety of phenotypes ranging from milder forms of inherited retinal dystrophy, such as retinitis pigmentosa to more severe phenotypes such as Leber congenital amaurosis. In this study, seven novel likely-pathogenic variants were identified: four missense variants (p.Leu479Pro, p.Ala921Pro, p.Cys948Arg and p.Asp1031Asn), two frameshift deletions (c.2536_2542del7 and c.3460_3461delTG) and one frameshift indel variant (c.276_294delinsTGAACACTGTAC). Furthermore, two patients with cone-rod dystrophy due to mutations in CRB1 were reported, supporting previous data, in which mutations in CRB1 can also cause cone-rod dystrophy. Finally, our data suggested there was a direct relation between phenotype severity and the mutation effect on protein functionality in 15 Brazilian CRB1 patients.
Introduction
The CRB1 gene is associated with some inherited retinal dystrophies (IRD). In humans, it is located on chromosome 1q31.3, composed of 12 exons and encodes a protein with 1406 amino acids, called Crumbs homolog-1. This protein participates in a conserved protein network involved in the morphogenesis of photoreceptors and the establishment and maintenance of apico-basal polarization and adherent junctions of epithelial cells1–3.
Crumbs homolog-1 is in a subapical region of photoreceptors, it has a large extracellular part composed of 19 epidermal growth factor (EGF)-like domains and 3 laminin A globular (AG)-like domains, one transmembrane segment and a small cytoplasmic domain. The intracellular domain has a juxtamembrane FERM-binding motif and a carboxy-terminal PDZ-binding motif, by means of which CRB1 interacts with other proteins forming a complex that participates in adherent junction formation and links to cytoskeletons3, 4.
Mutations in CRB1 lead to retinal abnormalities such as thickening, coarse lamination patterns and loss of photoreceptor signalling1. Currently, more than 200 mutations in CRB1 have been cited in the Human Gene Mutation Database - HGMD5. The main diseases caused by mutations in CRB1 are: retinitis pigmentosa (RP) either with or without paraarteriolar preservation of retinal pigment epithelium (PPRPE), Leber congenital amaurosis (LCA) and pigmented paravenous chorioretinal atrophy6, 7.
In this study, a large number of medical records of IRD Brazilian patients were reviewed, where 15 patients with CRB1 mutations were selected, and two of them presented cone-rod dystrophy (CRD). Seven new disease-causing variants were reported and a direct relation between phenotype severity and the impact on protein functionality caused by mutation was observed.
Results
Among the 230 medical records of IRD patients analyzed, 15 cases of unrelated patients with CRB1 variants were selected, where 13 of them had conclusive molecular diagnosis, whereas in the other two, only one variant was found, presenting a non-conclusive molecular diagnosis. All 15 patients received clinical diagnoses, wherein eight of them were diagnosed as LCA, three as RP, two as CRD and two as early-onset retinal dystrophy (EORD).
Clinical findings
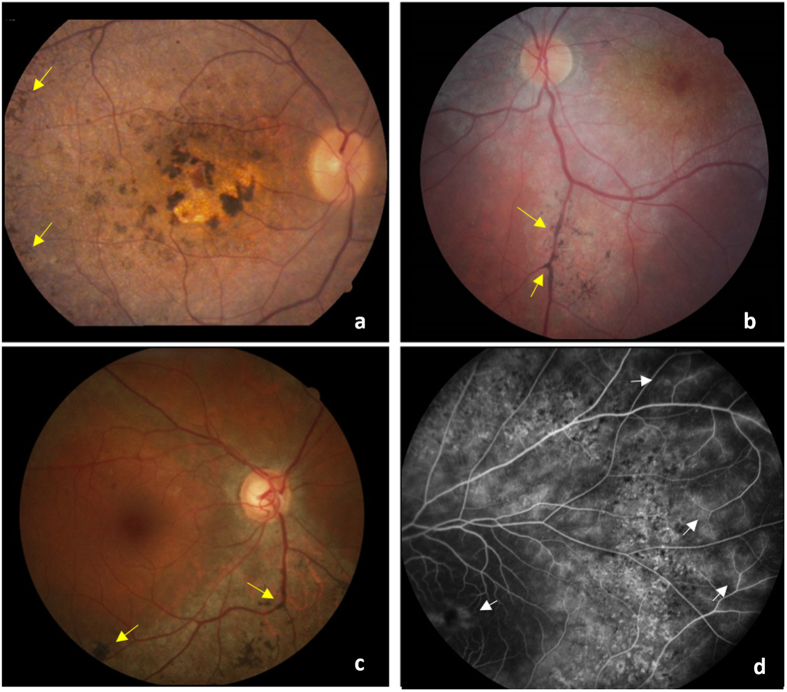
All eight patients with LCA exhibited the initial symptoms before the first year after birth. Low vision and nystagmus were the most striking features of this group. Only patient 5 did not present nystagmus. Visual acuity of LCA patients ranged from: reduced vision (patient 1) to severe visual loss (patient 4) (Table 1). The typical nummular pigmentation and macular atrophy could be observed from the fundus photographs. In some of them, there was a yellow deposit present in the macular area and widespread white dots in the retinal pigment epithelium (RPE) (Fig. 1a and Supplementary Figure S1).
Table 1.
Clinical Data of CRB1 patients.
| Patient | Signs and Symptoms | Onset of First Symptoms | Age at time of Diagnosis | Visual Acuities (OD; OS) | Clinical Diagnosis |
|---|---|---|---|---|---|
| 1 | Nystagmus; Reduced visual acuity improved with the development of patient. | first year of life | 27 | 20/60; 20/100 | LCA |
| 2 | Nystagmus | since birth | 6 months | good fix and follow behavior | LCA |
| 3 | Nystagmus; Deep reduced visual acuity; mild enophthalmos. | since birth | 27 | 20/1600; 20/1600 | LCA |
| 4 | Nystagmus; Severe visual loss; Minimum residual temporal visual field in the right eye; Divergent strabismus in the left eye. | first year of life | 20 | Counting fingers | LCA |
| 5 | Non-Nystagmus; Reduced visual acuity; Intermittent exotropia | since birth | 7 | 20/200; 20/200 | LCA |
| 6 | Nystagmus | 3 months of life | 3 | hand movements perception | LCA |
| 7 | Nystagmus; Sub-normal vision | 2 months of life | 2 | 20/200; 20/200 | LCA |
| 8 | Nystagmus; Progressive reduced visual acuity | first year of life | 16 | 20/80; 20/50 | LCA |
| 9 | Non-Nystagmus; Tubular visual field; Strabismus | 5 years old | 10 | 20/60; 20/60 | EORD |
| 10 | Non-Nystagmus; Reduced visual acuity; Nyctalopia | 6 years old | 12 | 20/400; 20/400 | EORD |
| 11 | Non-Nystagmus in the beginning; Nyctalopia | 9 years old | 9 | temporal perception of light and light movement | CRD |
| 12 | Non-Nystagmus; Reduced central visual acuity. | 7 years old | 24 | 20/200; 20/400 | CRD |
| 13 | Non-Nystagmus; Reduced visual acuity even with glasses; Tubular visual field; Nyctalopia | adolescence | 18 | 20/80; 20/80 | RP |
| 14 | Non-Nystagmus; Convergent strabismus; Hearing loss; Myopia; Glaucoma; Tubular visual field; Nyctalopia | adolescence | 47 | 20/40; 20/25 | RP |
| 15 | Non-Nystagmus; Tubular visual field; Nyctalopia | adulthood | 59 | 20/20; 20/30 | RP |
Figure 1.
Fundus appearance from CRB1 patients. (a) Color fundus photograph of LCA patient showing the nummular pigmentation and macular atrophy. (b) Color fundus photograph of CRD patient showing macular atrophy. (c) Color fundus photograph of RP patient showing RPE atrophy and macular area perverted. (d) Fluorescein Angiography with fluorescein leakage in peripheral vessels and at the macula. Yellow arrows indicate bone spicules and white arrows indicate leakage of fluorescein.
Four patients had showed the first signs and symptoms since their childhood. No nystagmus was present in any of them. Patient 9 with EORD had peripheral vision impairment (tunnel vision) with nummular pigmentation in the RPE and, patient 10 had a central vision impairment, midperiphery with bone spicules and granular pigmentation in the RPE (Supplementary Figure S2). On the other hand, patients diagnosed with CRD (11 and 12) had a more severe impairment of central vision (Table 1) and the fundus examination showed bone spicules with perivascular pattern, as well as macular atrophy characteristics of CRD (Fig. 1b and Supplementary Figure S2). In addition, patient 12 presented an atypical fundus pattern for cone-rod dystrophies, with a well-delimited hyperfluorescent area (Supplementary Figure S3).
As expected for the RP group, the first signs and symptoms appeared either during adolescence or later, and the absence of nystagmus was common in all, with visual acuity 20/80 or less (Table 1). Fundus analysis showed macular preservation compatible with their visual acuity, granular pigments in the RPE and the peripheral presence of bone spicules (Fig. 1c and Supplementary Figure S2). Only patient 13 had RP with PPRPE.
In relation to vascular aspects, patients 2, 4, 6 and 9 showed increased vascular tortuosity. The increased vascular permeability compatible with Coats-like disease onset was noted in two patients with LCA (patient 1 and 8), two with RP (patients 13 and 15), one with EORD (patient 10) and another with CRD (patient 11). Leakage of fluid and blood in Coats-like diseases usually occurs in peripheral vessels, but it may also occur in the macula, causing cystoid macular edema, as observed in patient 15 (Fig. 1d).
Genetic findings
Table 2 shows the genotypes of patients in this study. All presented variants are classified as pathogenic according to HGMD5, except the new variants, highlighted in bold. Patients 14 and 15 did not have a conclusive molecular result because the second pathogenic CRB1 variant was not found. Ten patients of the 13 genetically concluded cases are compound heterozygotes, whereas the remaining three are homozygotes (patients 3, 8 and 11). In addition, patient 11 is descended from a consanguineous marriage.
Table 2.
Genotypes of patients with CRB1 variants.
| Patient | Allele 1 | Allele 2 | Clinical Diagnosis | ||
|---|---|---|---|---|---|
| Nucleotide Change | Protein Change | Nucleotide Change | Protein Change | ||
| 1 | c.2843 G > A | p.Cys948Tyr | c.3676 G > T | p.Gly1226* | LCA |
| 2 | c.2536_2542del7 | p.Gly846Serfs*8 | c.2843 G > A | p.Cys948Tyr | LCA |
| 3 | c.984 G > A | p.Trp328* | c.984 G > A | p.Trp328* | LCA |
| 4 | c.2536_2542del7 | p.Gly846Serfs*8 | c.2843 G > A | p.Cys948Tyr | LCA |
| 5 | c.984 G > A | p.Trp328* | c.2843 G > A | p.Cys948Tyr | LCA |
| 6 | c.2842 T > C | p.Cys948Arg | c.2843 G > A | p.Cys948Tyr | LCA |
| 7 | c.2842 T > C | p.Cys948Arg | c.3460_3461delTG | p.Cys1154* | LCA |
| 8 | c.2843 G > A | p.Cys948Tyr | c.2843 G > A | p.Cys948Tyr | LCA |
| 9 | c.2291 G > A | p.Arg764His | c.4168 C > T | p.Arg1390* | EORD |
| 10 | c.276_294delinsTGAACACTGTAC | p.Arg92Serfs*54 | c.2506 C > A | p.Pro836Thr | EORD |
| 11 | c.1436 T > C | p.Leu479Pro | c.1436 T > C | p.Leu479Pro | CRD |
| 12 | c.2761 G > C | p.Ala921Pro | c.3091 G > A | p.Asp1031Asn | CRD |
| 13 | c.2506 C > A | p.Pro836Thr | c.3320 T > G | p.Leu1107Arg | RP |
| 14 | c.498_506del9 | p.Ile167_Gly169del | not found | not found | RP |
| 15 | c.614 T > C | p. Ile205Thr | not found | not found | RP |
The novel variants are indicated in bold.
Interestingly, all LCA subjects have more severe pathogenic variants in both alleles. These variants cause premature termination or structural change of the protein due to loss of cysteines involved in disulfide bond formation. In our study, two cysteines forming disulfide bonds are affected: Cys948 and Cys1154. On the other hand, RP and CRD patients have mostly missense variants that do not affect cysteines involved in disulfide bonds. The only exception was patient 14, who has one in-frame deletion with loss of three amino acids (Ile-Asp-Gly), which does not induce formation of a premature stop codon.
Sixteen different variants in CRB1 were found in these subjects. All of them are located in protein domains (Laminin(AG)-like or EGF-like domains) in crumbs homolog-1 extracellular segment, the only exception was p.Arg1390* variant that is in the cytoplasmic domain (Fig. 2). The variants found in this study are preferably located in exons 2, 7 and 9 (three variants at exon 2 and 7 and five variants at exon 9). p.Cys948Tyr was the most frequent in this study (7 alleles in 30 analyzed) (Table 3).
Figure 2.

Distribution of CRB1 variants in the protein.
Table 3.
Variants Data.
| CRB1 Variant | Exon | Protein Region | Protein Domain | Reported phenotype in HGMD (accession) | Allele Frequency† |
|---|---|---|---|---|---|
| c.276_294delinsTGAACACTGTAC (p.Arg92Serfs*54) | 2 | Extracellular | EGF 2 | not reported | 1/30 |
| c.498_506del9 (p.Ile167_ Gly169del) | 2 | Extracellular | EGF 4 | LCA, RP, Stargardt (CD061397) | 1/30 |
| c.614 T > C (p. Ile205Thr) | 2 | Extracellular | EGF 5 | RP, LCA (CM033359) | 1/30 |
| c.984 G > A (p.Trp328*) | 4 | Extracellular | EGF 8 | LCA (CM1310165) | 3/30 |
| c.1436 T > C (p.Leu479Pro) | 6 | Extracellular | EGF 11 | not reported | 2/30 |
| c.2291 G > A (p.Arg764His) | 7 | Extracellular | Laminin AG 2 | RP (CM130791) | 1/30 |
| c.2506 C > A (p.Pro836Thr) | 7 | Extracellular | Laminin AG 2 | RP (CM043271) | 2/30 |
| c.2536_2542del7 (p.Gly846Serfs*8) | 7 | Extracellular | Laminin AG 2 | not reported | 2/30 |
| c.2761 G > C (p.Ala921Pro) | 8 | Extracellular | EGF 13 | not reported | 1/30 |
| c.2842 T > C (p.Cys948Arg) | 8 | Extracellular | EGF 14 | not reported | 2/30 |
| c.2843 G > A (p.Cys948Tyr) | 9 | Extracellular | EGF 14 | RP, LCA, EORD (CM992152) | 7/30 |
| c.3091 G > A (p.Asp1031Asn) | 9 | Extracellular | Laminin AG 3 | not reported | 1/30 |
| c.3320 T > G (p.Leu1107Arg) | 9 | Extracellular | Laminin AG 3 | LCA (CM057656) | 1/30 |
| c.3460_3461delTG (p.Cys1154*) | 9 | Extracellular | EGF 15 | not reported | 1/30 |
| c.3676 G > T (p.Gly1226*) | 9 | Extracellular | EGF 17 | LCA (CM113150) | 1/30 |
| c.4168 C > T (p.Arg1390*) | 12 | Cytoplasmic | none | RP (CM130803) | 1/30 |
†Allele frequency in this study.
Seven subjects presented new changes in the CRB1 gene, wherein four novel missense variants were found (p.Leu479Pro, p.Ala921Pro, p.Cys948Arg and p.Asp1031Asn), two frameshift deletions (c.2536_2542del7 and c.3460_3461delTG) and one frameshift indel variant (c.276_294delinsTGAACACTGTAC) (see Table 4). None of them were present in the ClinVar8, ESP9, ExAC10 and 1000 Genomes Project11 databases. All new frameshift variants occur in the extracellular domain, leading to premature termination of the protein with the loss of the transmembrane region, contrasting with data found in patient 9 (p.Arg1390*) where protein truncation caused the loss of the PDZ-binding motif but the transmembrane domain was preserved.
Table 4.
Novel likely-pathogenic variants in CRB1 gene identified in this study.
| Nucleotide Change | Protein Change | Effect | in silico Analysis | Pathogenicity | ||
|---|---|---|---|---|---|---|
| Poly-Phen2† | PROVEAN | SIFT | ||||
| c.276_294delinsTGAACACTGTAC | p.Arg92Serfs*54 | Frameshift/protein truncation | — | — | — | Likely pathogenic |
| c.1436 T > C | p.Leu479Pro | Change of highly conserved residue | D | D | T | Likely pathogenic |
| c.2536_2542del7 | p.Gly846Serfs*8 | Frameshift/protein truncation | — | — | — | Likely pathogenic |
| c.2761 G > C | p.Ala921Pro | Change of highly conserved residue | D | D | T | Likely pathogenic |
| c.2842 T > C | p.Cys948Arg | Change of highly conserved residue | D | D | D | Likely pathogenic |
| c.3091 G > A | p.Asp1031Asn | Change of highly conserved residue | D | D | D | Likely pathogenic |
| c.3460_3461delTG | p.Cys1154* | Frameshift/protein truncation | — | — | — | Likely pathogenic |
†Poly-Phen2 HumVar; D – Probably Damaging; T – Tolerated.
In silico analysis of new missense variants showed that, they were classified as likely pathogenic for at least two predictors (Table 4). In comparison with other species, it is noted that amino acids changed are highly conserved (Fig. 3), mainly among primates (first 12 species of Fig. 3).
Figure 3.

Amino acid conservation analysis of novel missense variants identified in this study.
All novel variants found in this study can be classified as likely pathogenic according to the criteria of effect in the protein structure, amino acid conservation, frequency in population and classification by pathogenic predictors (Table 4).
Discussion
Mutations in the CRB1 gene lead to visual impairment and even complete blindness in individuals with many different clinical IRD phenotypes, including LCA, EORD and RP6, 12–14. Despite the large phenotypic heterogeneity among CRB1 patients, some signs, symptoms and ophthalmologic findings can be observed with more frequency12. Overall, the subjects in this study presented many of typical characteristics including: nummular pigmentation, macular atrophy, bone spicules, nystagmus and poor central vision in patients with LCA and macular preservation, peripheral bone spicules, pigmentation changes of the RPE, nyctalopia and tunnel vision in typical RP patients.
In two cases (patients 14 and 15), the molecular result was not conclusive because only one pathogenic variant was found. As the inheritance pattern of IRD caused by CRB1 mutations is autosomal recessive, then the presence of pathogenic variants in both alleles are required for the molecular test to be conclusive. While advances in the molecular diagnosis of IRD are moving fast, the next-generation sequencing still do not solve 35–45% of IRD cases15–18.
In the past, negative or inconclusive diagnoses have occurred through the screening of known mutations, such as in patient 15, tested by DNA microarray (APEX). The Sanger sequencing technique subsequently allowed for an improved analysis of specific genes and identification of known and new variants, such was the case of patients 1, 3, 4, 13 and 14.
Sanger sequencing of patient 14 identified, in addition to the pathogenic variant (c.498_506del9 - p.Ile167_Gly169del), a further three heterozygous variants: c.849-35 T > C (rs1337167), c.989-53 G > T (rs2786098) and c.*28 T > C (rs41302107). They are not rare in population databases10, 11 and considered likely benign8, 19. Conte and coworkers (2015) showed that retinal dystrophy could be caused by mutations in seed regions of miRNA20. Therefore, the 3′UTR variant was analyzed in PolymiRTS Database 3.021 and TargetScan v.7.022 to identify whether it could cause changes in miRNAs or in those target regions. The c.*28 T > C variant possibly changed some miRNA binding sites, as shown in Supplementary Figure S4. Changes in miRNA binding sites may affect CRB1 expression and contribute to the patient’s phenotype. However, the real effect of this change requires further investigation.
Two CRD patients showed likely pathogenic variants in CRB1 gene. CRD is caused by mutation in many genes such as ABCA4, ADAM9, C8orf37, CDHR1, CRX, DRAM2, GUCA1A, GUCY2D, PITPNM3, POC1B, PROM1, RAB28, RAX2, RIMS1, RPGRIP1, SEMA4A, and TTLL5 6. Genetic reference databases, including OMIM6 and RetNet7, do not indicate an association between CRB1 and CRD. Up to now, only three studies have found this causal relationship, with one describing three unrelated subjects23, another, a consanguineous nuclear family24 and the third, one proband with a novel splice-site mutation25. Our data supports the hypothesis that CRB1 can also cause CRD and thus the CRB1 gene might be included in target list for CRD genetic testing.
CRB1 mutations are considered a risk factor in the development of Coats-like vasculopathy13, 26, and, because of this, retinal vascular characteristics should be always evaluated in CRB1 patients. Among 15 patients analyzed in this study, approximately 67% showed vascular abnormalities such as: vascular tortuosity, arteriolar sclerosis, increased vascular permeability and leakage of fluid and blood, which may mean the beginning of a Coats-like disease. An interesting aspect to note is that mutations in CRB1 can cause osteoclast deposition on top of vessels or in the paravenous region (Fig. 1). It was not possible to associate these vascular phenotypes with a specific retinal dystrophy or specific mutation in these subjects, corroborating data in the literature which states that Coats-like vasculopathy does not develop solely in RP patients13, 23, 27.
Bujakowska and coworkers (2012) published an extensive review of CRB1 cases, showing that exons 7 and 9 have the highest concentration of pathogenic variants, and p.Cys948Tyr is the most frequent of them13. Our findings are similar to these, approximately 69% of variants found in this study are located in the exons 2, 7 and 9 and p.Cys948Tyr is also the most frequent in our samples - it was present in 23% of alleles analyzed.
Interestingly, patient 6 has two different missense variants in the same codon (p.Cys948Tyr and p.Cys948Arg). p.Cys948Arg is not described in literature and also it was found in patient 7. Cysteines have an important role in structure and function of proteins, and variations in this residue are highly likely to cause deleterious phenotypes, especially if the mutated cysteine is part of disulfide bonds28, as well as in Cys948 in crumbs homolog-1 protein. Mutations at codon 948 affect the correct folding of 14th EGF-like domain29. A large number of exonic variants, missense or synonymous, have already been shown to possess disease-causing effects by disrupting the pre-mRNA’s editing process, causing aberrant splicing30–32. Both variants were analyzed in Human Splicing Finder33, which indicated them as potential alterations of splicing. Perhaps the greatest deleterious effect caused by changes in 948th codon is not due to the exchange of a conserved cysteine but to mRNA processing problems.
Our data shows a striking pattern between mutation type and the patient’s phenotype. Individuals with severe retinal dystrophy, such as LCA, have two variants affecting the protein function or structure most severely (e.g. frameshift changes, premature stop codon formation, aberrant splicing and lack of disulfide bond, due to mutated cysteine). On the other hand, patients with milder IRD have missense variants or in-frame deletions. Patient 9 and 10 with EORD have an intermediate phenotype and genotype, i.e. a missense variant (p.Arg764His and p.Pro836Thr respectively) and a premature truncation (p.Arg1390* and p.Arg92Serfs*54 respectively). Despite the fact that a genotype-phenotype relationship has not been clearly established in previous studies, they also noted that patients with more severe phenotypes, for example macular atrophy, tend to have protein truncation (nonsense or frameshift deletions) and/or p.Cys948Tyr variants13, 23, 26.
To establish a genotype-phenotype correlation in CRB1 patients is not an easy task. There is a substantial phenotypic overlap and variability between CRB1-related diseases and a small number of patients with mutations in this gene. Moreover, the phenotypic modulation possibly occurs due to environmental factors and other genetic factors12, 13, 26, 34, 35, such as unknown genes, silent variants causing aberrant splicing36, deep intronic mutations36–38, copy number variations39, complex genomic rearrangements40, 41, multigenic inheritance patterns, genetic modifiers42–44 and regulators of gene expression20, 45. In addition, technical limitations, such as uncovered or low-depth regions in NGS analysis, may hinder the correct molecular diagnosis46.
Nowadays, molecular diagnoses are strongly orienting clinical practice in cases of IRD, where there is high genotype and phenotype heterogeneity. The more that new mutations are described and new genotypic-phenotypic associations are made, the greater the knowledge regarding these diseases. Our study highlighted a direct relation between phenotype severity and the mutation effect on protein functionality in CRB1 Brazilian patients, contributing to current knowledge about disease-causing variants and supporting the association between the CRB1 gene and cone-rod dystrophy.
Methods
This retrospective study reviewed 230 medical records of Brazilian patients with IRD assisted at the Universidade Federal de São Paulo and Instituto de Genética Ocular in São Paulo, Brazil between January 2006 and February 2017. The condicio sine qua non to include patients was that they must have already performed at least one genetic test for IRD. Table 5 shows the commercial genetic tests performed on each CRB1 patient.
Table 5.
Type of Genetic Test performed on CRB1 patients.
| Patient | Genetic Test | Number of Genes Analyzed | Test Date |
|---|---|---|---|
| 1 | Sanger Sequencing Panel | 10 | 2009 |
| 2 | Next-Generation Sequencing Panel | 19 | 2012 |
| 3 | Sanger Sequencing Panel | 17 | 2011 |
| 4 | Sanger Sequencing | 1 | 2011 |
| 5 | Next-Generation Sequencing Panel | 19 | 2015 |
| 6 | Whole Exome Sequencing | 2015 | |
| 7 | Next-Generation Sequencing Panel | 226 | 2017 |
| 8 | Next-Generation Sequencing Panel | 226 | 2017 |
| 9 | Next-Generation Sequencing Panel | 19 | 2013 |
| 10 | Next-Generation Sequencing Panel | 226 | 2017 |
| 11 | Next-Generation Sequencing Panel | 131 | 2014 |
| 12 | Next-Generation Sequencing Panel | 131 | 2014 |
| 13 | Sanger Sequencing Panel | 3 | 2011 |
| 14 | Sanger Sequencing | 1 | 2014 |
| 15 | Arrayed Primer Extension (APEX) | 18 (585 mutations/SNPs tested) | 2009 |
In addition to the genetic data, medical history and eye exams were collected. The clinical hypothesis for patient classification of patients was created based on their signs and symptoms, age of onset and fundus features.
The classification of new variants according to pathogenicity was based on the following criteria: variants with the most potential to cause disease are those that result in truncated protein production (premature stop codon and frameshift changes) or missense changes in highly conserved amino acids, as well as a rare frequency variation in genetic population databases and classified as likely damaging by the pathogenicity predictors. The databases consulted were: HGMD5, ExAC10, 1000 Genomes Project11, Exome Sequencing Project (ESP)9 and ClinVar8. The pathogenicity predictor softwares consulted were: Poly-Phen247, SIFT48 and PROVEAN49. Combined Annotation Dependent Depletion (CADD) software19 was used to evaluate changes in non-coding regions of the CRB1 gene. The Human Splicing Finder33 was used to check possible aberrant splicing. The bioinformatics tools PolymiRTS Database 3.021 and TargetScan v.7.022 were used to evaluate changes in miRNAs or miRNA binding sites.
For amino acid conservation analysis, CRB1 gene of 38 species was compared. A multiple sequence alignment was built using PRALINE online toolkit50, where all previous selected sequences were submitted to multiple alignment using default parameters. The alignment file was open in Clustal X51 in order to build alignment figures. The amino acids were classified as: highly conserved (changed in a maximum of three species), moderately conserved (changed in four to six species) and weakly conserved (changed in more than six species).
Nucleotide numbering is based on reference sequence NM_201253, where A of initiation codon (ATG) is the number 1.
The Ethics Committee in Research of Federal University of São Paulo approved this study (CEP: 0415/2016). Written informed consent for the use of personal medical data for scientific purposes and publication was obtained from all patients and/or their legal guardians. In addition, this study was performed in accordance with the ethical standards of the 1964 Declaration of Helsinki and its subsequent amendments.
Electronic supplementary material
Acknowledgements
The authors are grateful for fellowship support by CAPES (FLM, MVS and KAC), CNPq (RFS) and FAPESP (RPM).
Author Contributions
F.L.M. revised the medical records, collected data and drafted the manuscript. F.L.M. and J.M.F.S. analyzed and interpreted the data. M.V.S. provided clinical support. K.A.C. provided technical support. R.F.S. and R.P.M. prepared Figures 2 and 3 and provided bioinformatics support. J.M.F.S., M.V.S. and R.P.M. revised the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-09035-1
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jacobson SG, et al. Crumbs homolog 1 (CRB1) mutations result in a thick human retina with abnormal lamination. Hum. Mol. Genet. 2003;12:1073–8. doi: 10.1093/hmg/ddg117. [DOI] [PubMed] [Google Scholar]
- 2.Richard, M. et al. Towards understanding CRUMBS function in retinal dystrophies. Hum. Mol. Genet. R235-43. doi:10.1093/hmg/ddl195 (2006) [DOI] [PubMed]
- 3.Pocha SM, Knust E. Complexities of Crumbs function and regulation in tissue morphogenesis. Curr. Biol. 2013;23:R289–93. doi: 10.1016/j.cub.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Gosens I, den Hollander AI, Cremers FPM, Roepman R. Composition and function of the Crumbs protein complex in the mammalian retina. Exp. Eye Res. 2008;86:713–726. doi: 10.1016/j.exer.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Stenson PD, et al. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1 doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamosh A, Scott AF, Amberger J, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM) Hum. Mutat. 2000;15:57–61. doi: 10.1002/(SICI)1098-1004(200001)15:1<57::AID-HUMU12>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 7.Daiger S, Rossiter B, Greenberg J, Christoffels A, Hide W. Data services and software for identifying genes and mutations causing retinal degeneration. Investig. Ophthalmol. Vis. Sci. 1998;39 [Google Scholar]
- 8.Landrum MJ, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–8. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Exome Variant Server. NHLBI GO Exome Sequencing Project (ESP). http://evs.gs.washington.edu/EVS/ (accessed 5 Jan 2017).
- 10.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature526, 68–74 (2015). [DOI] [PMC free article] [PubMed]
- 12.Ehrenberg M, Pierce EA, Cox GF, Fulton AB. CRB1: One Gene, Many Phenotypes. Semin. Ophthalmol. 2013;28:397–405. doi: 10.3109/08820538.2013.825277. [DOI] [PubMed] [Google Scholar]
- 13.Bujakowska K, et al. CRB1 mutations in inherited retinal dystrophies. Hum. Mutat. 2012;33:306–315. doi: 10.1002/humu.21653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kousal B, et al. Phenotypic features of CRB1-associated early-onset severe retinal dystrophy and the different molecular approaches to identifying the disease-causing variants. Graefe’s Arch. Clin. Exp. Ophthalmol. 2016;254:1833–1839. doi: 10.1007/s00417-016-3358-2. [DOI] [PubMed] [Google Scholar]
- 15.Audo I, et al. Development and application of a next-generation-sequencing (NGS) approach to detect known and novel gene defects underlying retinal diseases. Orphanet J. Rare Dis. 2012;7 doi: 10.1186/1750-1172-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, et al. Comprehensive molecular diagnosis of 179 Leber congenital amaurosis and juvenile retinitis pigmentosa patients by targeted next generation sequencing. J. Med. Genet. 2013;50:674–88. doi: 10.1136/jmedgenet-2013-101558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiang JP, Trzupek K. The current status of molecular diagnosis of inherited retinal dystrophies. Curr. Opin. Ophthalmol. 2015;26:346–51. doi: 10.1097/ICU.0000000000000185. [DOI] [PubMed] [Google Scholar]
- 18.Saudi Mendeliome Group Comprehensive gene panels provide advantages over clinical exome sequencing for Mendelian diseases. Genome Biol. 2015;16 doi: 10.1186/s13059-015-0693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conte I, et al. MiR-204 is responsible for inherited retinal dystrophy associated with ocular coloboma. Proc. Natl. Acad. Sci. USA. 2015;112:E3236–45. doi: 10.1073/pnas.1401464112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhattacharya A, Ziebarth JD, Cui Y. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014;42:D86–91. doi: 10.1093/nar/gkt1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agarwal V, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:101–112. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henderson RH, et al. Phenotypic variability in patients with retinal dystrophies due to mutations in CRB1. Br. J. Ophthalmol. 2011;95:811–817. doi: 10.1136/bjo.2010.186882. [DOI] [PubMed] [Google Scholar]
- 24.Khan AO, Aldahmesh MA, Abu-Safieh L, Alkuraya FS. Childhood cone-rod dystrophy with macular cystic degeneration from recessive CRB1 mutation. Ophthalmic Genet. 2014;35:1–8. doi: 10.3109/13816810.2014.926942. [DOI] [PubMed] [Google Scholar]
- 25.Oishi M, et al. Next-generation sequencing-based comprehensive molecular analysis of 43 Japanese patients with cone and cone-rod dystrophies. Mol. Vis. 2016;22:150–60. [PMC free article] [PubMed] [Google Scholar]
- 26.den Hollander AI, et al. Leber congenital amaurosis and retinitis pigmentosa with Coats-like exudative vasculopathy are associated with mutations in the crumbs homologue 1 (CRB1) gene. Am. J. Hum. Genet. 2001;69:198–203. doi: 10.1086/321263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasan SM, Azmeh A, Mostafa O, Megarbane A. Coat’s like vasculopathy in leber congenital amaurosis secondary to homozygous mutations in CRB1: a case report and discussion of the management options. BMC Res. Notes. 2016;9 doi: 10.1186/s13104-016-1917-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raimondi, D., Orlando, G., Messens, J. & Vranken, W. F. Investigating the Molecular Mechanisms Behind Uncharacterized Cysteine Losses from Prediction of Their Oxidation State. Hum. Mutat. doi:10.1002/humu.23129 (2016) [DOI] [PubMed]
- 29.Cremers, F. P. M., Maugeri, A., den Hollander, A. I. & Hoyng, C. B. The expanding roles of ABCA4 and CRB1 in inherited blindness. Novartis Found. Symp. 255, 68-79-84, 177–8 (2004). [DOI] [PubMed]
- 30.Ward AJ, Cooper TA. The pathobiology of splicing. J. Pathol. 2010;220:152–63. doi: 10.1002/path.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 2011;12:683–91. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 32.Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 33.Desmet F-O, et al. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37 doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathijssen, I. B. et al. Long-Term Follow-Up Of Patients With Retinitis Pigmentosa Type 12 Caused By Crb1 MutatiONS: A Severe Phenotype With Considerable Interindividual Variability. Retina. doi:10.1097/IAE.0000000000001127 (2016) [DOI] [PubMed]
- 35.den Hollander AI, et al. CRB1 mutation spectrum in inherited retinal dystrophies. Hum. Mutat. 2004;24:355–369. doi: 10.1002/humu.20093. [DOI] [PubMed] [Google Scholar]
- 36.Braun TA, et al. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Hum. Mol. Genet. 2013;22:5136–45. doi: 10.1093/hmg/ddt367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.den Hollander AI, et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liquori A, et al. Whole USH2A Gene Sequencing Identifies Several New Deep Intronic Mutations. Hum. Mutat. 2016;37:184–93. doi: 10.1002/humu.22926. [DOI] [PubMed] [Google Scholar]
- 39.Bujakowska, K. M. et al. Copy-number variation is an important contributor to the genetic causality of inherited retinal degenerations. Genet. Med. doi:10.1038/gim.2016.158 (2016) [DOI] [PMC free article] [PubMed]
- 40.Nishiguchi KM, et al. Whole genome sequencing in patients with retinitis pigmentosa reveals pathogenic DNA structural changes and NEK2 as a new disease gene. Proc. Natl. Acad. Sci. USA. 2013;110:16139–44. doi: 10.1073/pnas.1308243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sullivan LS, et al. Genomic rearrangements of the PRPF31 gene account for 2.5% of autosomal dominant retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 2006;47:4579–88. doi: 10.1167/iovs.06-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ebermann I, et al. PDZD7 is a modifier of retinal disease and a contributor to digenic Usher syndrome. J. Clin. Invest. 2010;120:1812–23. doi: 10.1172/JCI39715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiang JP-W, et al. Progress and prospects of next-generation sequencing testing for inherited retinal dystrophy. Expert Rev. Mol. Diagn. 2015;15:1269–75. doi: 10.1586/14737159.2015.1081057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alves CH, Pellissier LP, Wijnholds J. The CRB1 and adherens junction complex proteins in retinal development and maintenance. Prog. Retin. Eye Res. 2014;40:35–52. doi: 10.1016/j.preteyeres.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 45.Chung SH, et al. Profiling of microRNAs involved in retinal degeneration caused by selective Müller cell ablation. PLoS One. 2015;10 doi: 10.1371/journal.pone.0118949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang X-F, Wu J, Lv J-N, Zhang X, Jin Z-B. Identification of false-negative mutations missed by next-generation sequencing in retinitis pigmentosa patients: a complementary approach to clinical genetic diagnostic testing. Genet. Med. 2015;17:307–11. doi: 10.1038/gim.2014.193. [DOI] [PubMed] [Google Scholar]
- 47.Adzhubei I, Jordan DM, Sunyaev SR. Predicting Functional Effect of Human Missense Mutations Using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013;72020:1–741. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 49.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–7. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bawono P, Heringa J. PRALINE: a versatile multiple sequence alignment toolkit. Methods Mol. Biol. 2014;1079:245–62. doi: 10.1007/978-1-62703-646-7_16. [DOI] [PubMed] [Google Scholar]
- 51.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.