Abstract
Genetic recombination occurs between homologous DNA molecules via a four-way (Holliday) junction intermediate. This ancient and ubiquitous process is important for the repair of double-stranded breaks, the restart of stalled replication forks, and the creation of genetic diversity. Once formed, the four-way junction alone can undergo the stepwise exchange of base pairs known as spontaneous branch migration. Conventional ensemble assays, useful for finding average migration rates over long sequences, have been unable to examine the affect of sequence and structure on the migration process. Here, we present a single-molecule spontaneous branch migration assay with single-base pair resolution in a study of individual DNA junctions that can undergo one step of migration. Junctions exhibit markedly different dynamics of exchange between stacking conformers depending on the point of strand exchange, allowing the moment at which branch migration occurs to be detected. The free energy landscape of spontaneous branch migration is found to be highly nonuniform and governed by two types of sequence-dependent barriers, with unmediated local migration being up to 10 times more rapid than the previously deduced average rate.
Keywords: FRET, single molecule, recombination, DNA structure
Homologous recombination is an essential process in maintaining genomic stability, and its defects can lead to serious human diseases, including cancer. To cope with DNA damage encountered during genome duplication, a four-way (Holliday) junction is formed by joining two nearly identical DNA molecules by strand exchange (1–3). To understand the mechanisms of cellular enzymes that recognize and process the Holliday junction, it is necessary to describe the static and dynamic structural properties of their natural substrates: homologous junctions. However, because there is no way to synchronize migration, structural properties have been determined only for junctions whose branch points are fixed, either by sequence (i.e., lack of homology) (4–13), the constraints of a crystal lattice (14–17), or other mechanical constructs (18). These studies have shown that in the absence of divalent metal ions the junction adopts an extended geometry in which the arms are directed toward the corners of a square with an open central region. But upon the addition of divalent metal ions, such as magnesium, the junction folds into the stacked X-structure, a pairwise, coaxial stacking of helices to form a right-handed, antiparallel cross. There are two possible conformers that differ in which pairs of helices are stacked against each other and a single junction can sample both stacking conformers, with continual conformational exchange between them (9–12, 19).
Although protein-mediated branch migration has been studied extensively for various enzymes (20–22) and recently at the single-molecule level for RuvAB (23–25), the mechanics of spontaneous migration remain largely unknown. Ensemble measurements of spontaneous branch migration in homologous junctions have detected a 1,000-fold decrease in the migration rate upon the addition of Mg2+ (26, 27). This finding suggests a relationship between structure and migratability, but without being able to watch both aspects simultaneously the nature of that relationship has remained unclear.
In this work, we map the kinetic and thermodynamic properties of junctions that can undergo one step of spontaneous branch migration by using single-molecule FRET. We observed stacking conformer transitions and single-step branch migration simultaneously from individual junctions and measured their transition rates for a variety of sequences and solution conditions. Multiple conformer transitions occur before a branch migration step is taken, such that conformational bias is effectively sampled at each branch point. Our data not only revealed a surprisingly rugged and highly sequence-dependent energy landscape of spontaneous branch migration, but also provided insight into its molecular mechanism, namely the individual stacking dynamics at each position of branch migration. These findings give rise to an intriguing possibility whereby local spontaneous branch migration can occur much more rapidly than the average rate, so that junctions can quickly find stable branch points if they are transiently free of protein in vivo.
Materials and Methods
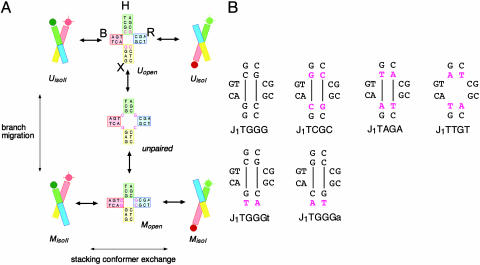
Junction Constructs. A series of four-way DNA (28) junctions were constructed by the hybridization of four 22-nt oligonucleotides. The central sequences were designed so that junctions could undergo a single step of branch migration between two species, one where all four arms were 11 bp in length (termed species U) and the other where two arms were 12 and two arms were 10 bp (species M, Fig. 1A). A series of such junctions were generated, with each differing only in the sequence around the point of strand exchange (see Fig. 1B for their central sequences). In each case, two helical arms (B and H) were terminally labeled with fluorophores, so that efficient energy transfer would occur when the conformation adopted brought arms B and H physically close as in the case of isoII and little energy transfer would occur when the arms were forced apart as in the case of isoI. Biotin was attached to a third arm (R) and was used to tether the junctions to a glass coverslip for extended observation with confocal microscopy.
Fig. 1.
Dynamics of structural interconversion in a junction capable of a single step of branch migration. (A) Structural interconversion possible in junction J1TGGG. The junction comprises four arms (B, H, R, and X) formed by annealing four ssDNA molecules of 22 nt each (named b, h, r, and x). The sequence allows one step of branch migration (vertical interconversion). In one species (U, Upper) the four arms are each 11 bp in length, whereas in the other form (M, Lower) the arms are 12 bp (B and R) or 10 bp (H and X). Each of these species can exist in either stacking conformer (horizontal interconversion) isoI (H on B stacking) or isoII (B on X stacking). Depicted intermediate species are not observed directly in our experiments. Other sequences varied either the migrating base pair (J1TCGC, J1TAGA, J1TTGT) or the base pair flanking the migrating base pair in the X arm (J1TGGGt, J1TGGGa) (see Materials and Methods for naming scheme). In all cases, the Cy3 donor, Cy5 acceptor, and biotin were attached to the 5′ termini of the h, b, and r strands, respectively. (B) The central sequences of the full set of junctions analyzed in these studies, with the sequence differences from J1TGGG highlighted in magenta. Lines indicate the base pairs that form in the alternative branch position.
The junctions were named according to the four nucleotides located immediately 5′ to the point of strand exchange, in the order b, h, r, and x. Lowercase letters follow a given nucleotide to designate further sequence in the 3′ direction if required.
Junction Preparation. Oligonucleotides were either purchased from IDT (Coralville, IA) or synthesized by using phosphoramidite chemistry implemented on ABI394 DNA synthesizers (Applied Biosystems). Strand sequences were constant for each junction with the exception of nucleotides 10–13 (denoted in the following by K) on each strand. The sequences were (all written 5′ to 3′): b, Cy5-CCCTAGCAA-K-CTGCTACGG; h, Cy3-CCGTAGCAGK-AGCGGTGGG; r, biotin-CCCACCGCT-K-TCAACTGGG; x, CCCAGTTGA-K-TTGCTAGGG. Each junction differed in the K sequence for each strand, i.e.: J1TGGG, b = GTCG, h = CGCG, r = CGCC, x = GGAC; J1TCGC, b = GTGG, h = CCCG, r = CGGC, x = GCAC; J1TAGA, b = GTTG, h = CACG, r = CGTC, x = GAAC; J1TTGT, b = GTAG, h = CTCG, r = CGAC, x = GTAC; J1TGGGt, b = GTCG, h = CGCG, r = CGCA, x = TGAC; J1TGGGa, b = GTCG, h = CGCG, r = CGCT, x = AGAC.
All strands were purified by PAGE under denaturing conditions. Junctions were annealed by mixing ≈10 μM of the b and x strands and ≈5 μM of the h and r strands in 10 mM Tris·HCl (pH 8.0) and 50 mM NaCl and slowly cooled from 85°C.
Single-Molecule Measurements. Details have been reported (11, 12). Streptavidin-coated glass surfaces were prepared by successive application of 1 mg·ml–1 biotinylated BSA (Sigma) and 0.2 mg·ml–1 streptavidin (Molecular Probes) in buffer A (10 mM Tris·HCl, pH 8.0/50 mM NaCl). Biotinylated junction molecules (10–50 pM in buffer A) were then added to the treated surface and immobilized. A 532-nm laser (Crystalaser, Reno, NV) was used to excite the donor, Cy3. Single-molecule data were obtained by using a confocal scanning microscope with two silicon avalanche photodiodes (PerkinElmer) by using a data integration time of 8 ms (29). For temperature regulation, a water-circulating bath (Neslab Instruments, Portsmouth, NH) was connected to the microscope objective (by a brass tubing collar) and used in conjunction with a Peltier cooling device (Tellurex, Traverse City, MI) applied to the sample area. The sample temperature was monitored directly at the objective/coverslip interface with a thermocouple. Unless otherwise specified, all measurements were made at 25°C in 10 mM Tris·HCl, pH 8.0/50 mM NaCl with an oxygen scavenging system [7% (wt/vol) glucose, 1% (vol/vol) 2-mercaptoethanol, 0.1 mg·ml–1 glucose oxidase, and 0.02 mg·ml–1 catalase] with specified amounts of MgCl2.
Data Analysis. To analyze traces we first filtered out blinking events (defined as a reversible transition of the acceptor to an inactive state, giving rise to an unquenched donor emission) and photobleaching of either fluorophore. We then performed a threshold analysis on the FRET efficiency time traces after partitioning them into their step position (U or M). FRET efficiency was calculated as the acceptor intensity divided by the sum of donor and acceptor intensities. The threshold varied, ranging from 0.35 to 0.5 dependingonMg2+ concentration. Transitions were not counted unless the state lasted for at least three data points, in an effort to remove the contribution of shorter lifetime photophysical effects. The dwell times of all low FRET states or all high FRET states from 20–50 molecules were plotted as a histogram and fitted to an exponential decay curve.
Hydroxyl Radical Probing of Junctions. The four strands of each junction were separately radioactively 5′-32P-labeled, and each was hybridized either to the remaining unlabeled strands of the junction or the complementary strand to form a perfect duplex. The cleavage reaction was carried out on the resulting eight species (30). In brief, 8 μl of a freshly prepared solution of the DNA junction was added to final concentrations of 0.1 mM Fe(II), 0.2 mM EDTA, 1 mM ascorbate, and 0.15% hydrogen peroxide. The solution was incubated at room temperature for 2 min, and the reaction was terminated by addition of thiourea to a final concentration of 0.1 M. Calf thymus DNA (10 ng/μl, final concentration) was added, and the DNA was precipitated with ethanol. The pellet was then resuspended in formamide, and the DNA was subjected to electrophoresis in a 12% polyacrylamide sequencing gel. Radioactivity was quantified by using phosphorimaging (Fuji BAS-1500).
Results
Single-Molecule Time Trajectories Reveal Four-State Behavior. Single-molecule fluorescence time traces of individual junction molecules exhibited anticorrelated fluctuations between donor and acceptor intensities (Fig. 2 and Figs. 5 and 6, which are published as supporting information on the PNAS web site) with average dwell times on the order of hundreds of milliseconds. This variation in FRET efficiency with similar lifetimes was previously observed in junctions that could not branch migrate and recognized as exchange between stacking conformers (11, 12). However, in this study it was observed that junctions switched between two distinct phases, each consisting of two-state fluctuations but with markedly different kinetics. The slow phase (S) displayed a strong bias toward the high FRET state, whereas the fast phase (F) was much less biased. Because such an alternation in conformer exchange kinetics was never observed in a number of junctions whose branch points were fixed, we conclude that the alternative phases represent the two possible branch migration species U and M. This conclusion is also in agreement with earlier studies that deduced that conformational exchange must occur multiple times between branch migration steps (11). Of the junctions studied, J1TGGG, J1TGGGa, J1TGGGt, and J1TTGT exhibited similar partitioning into species with distinct stacking conformer exchange rates. Two junctions, J1TCGC and J1TAGA, did not show evidence for partitioning and behaved similarly to junctions with fixed branch points.
Fig. 2.
Fluorescence time traces for single junction molecules. Solution conditions are as described in the text with a Mg2+ concentration of 50 mM. (A) A record of donor (green) and acceptor (red) fluorescence intensities (Upper) and calculated FRET efficiency (Lower) from a single molecule of J1TGGGa as a function of time. The molecule interconverts between high and low FRET efficiency and exhibits two distinct dynamic behaviors. Arrows denote a transition between these forms at 3 s and the reverse at 7 s. (B) FRET efficiency as a function of time for J1TGGG. (C) FRET efficiency as a function of time for J1TTGT.
Assigning Absolute Branch Points by Hydroxyl Radical Cleavage. Single-molecule data indicated that ≈95% of the population of J1TGGG exists at one of the branch points (S) at a given time. Although there was a significant difference in FRET efficiency between the two high-FRET states, SisoII and FisoII, the difference was too small to determine unambiguously whether the S state corresponded to the M or U species. We therefore used hydroxyl radical probing of this junction in free solution (Fig. 3 and Fig. 7, which is published as supporting information on the PNAS web site). Corresponding experiments using junctions incapable of spontaneous branch migration exhibit protection of the nucleotides flanking the point of strand exchange on the exchanging strands only (5). Using J1TGGG we observed significant protection at the centers of the b and r strands, clearly indicating that the major stacking conformer was that formed by coaxial stacking of B on X arms (isoII, where b and r are exchanging strands), in agreement with the single-molecule FRET data. By identifying which specific positions along the b and r strands were protected we could conclude that the dominant S species observed in single-molecule measurements was the U structure. The same bias was deduced for J1TGGGt. However hydroxyl radical cleavage experiments performed on junction J1TTGT generated a pattern of protection consistent with the M species as the predominant form (this time equating S with M and F with U), although as before the b and r strands were primarily protected, consistent with the isoII stacking conformer. J1TCGC and J1TAGA that showed only two-state behavior in single-molecule measurements did not reveal a clear preference for either M or U, but exhibited extended regions of protection on all four strands, consistent with the existence of significant populations of both branch positions and both stacking conformers.
Fig. 3.
Determination of the major conformational species in solution by hydroxyl radical probing. Four junctions were constructed by hybridization of four strands, one of which was radioactively 5′-32P-labeled. The same strand was also hybridized to its complement to make a perfect duplex for comparison. The junction and duplex species were subjected to hydroxyl radical attack, and the products were analyzed by gel electrophoresis under denaturing conditions and phosphorimaging. The radioactivity in each band was quantified from the phosphorimage files and presented in plotted form for the duplex (gray) and junction (red). Regions protected against radical attack are revealed by the lower intensity of the junction-derived peaks. (A) The profiles of radical attack for the four strands of junction J1TGGG. There are regions of protection apparent at the centers of the b and r strands (labeled with the corresponding nucleotides). (Insets) The ratios of intensities are also shown by the histograms, clearly showing the protection of the central nucleotides. The results show clearly that the major species in solution is in the isoII stacking conformation (where the exchanging b and r strands are protected) and has the U branch position (shown by the position of protection on these strands). The protected nucleotides are summarized in C.(B) Comparison of the radical attack profiles on the b strands of junctions J1TGGGt, J1TTGT, and J1TCGC. The positions of protection indicate that J1TGGGt is also in the U, isoII conformation, whereas J1TTGT is predominantly in the alternative branch position, i.e., M, isoII. By contrast, the protection of J1TCGC indicates that all of the stacking and branch positions are populated in this junction. The full data are shown in Figs. 9–12, which are published as supporting information on the PNAS web site, and the protected nucleotides are summarized in C. (C) Summary of the nucleotides protected against radical attack in the four junctions, shown by the yellow shading. The nucleotides highlighted in red can participate in branch migration. Junction J1TAGA (Fig. 12) is very similar to J1TCGC.
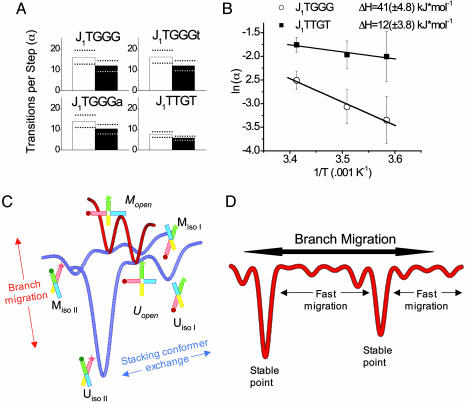
Large Variations in State Lifetimes. All junctions exhibiting two distinct phases were predominantly found in the more slowly oscillating phase. In other words, the branch point in which conformer exchange was slow was also more heavily populated. Conformer dwell time data exhibited extremely large sequence-dependent variations in the lifetimes of branch points and conformations (Table 1 and Fig. 8, which is published as supporting information on the PNAS web site), ranging from the heavily UisoII-dominated J1TGGGt (UisoII/UisoI/MisoII/MisoI of 100:47:4:1 and U/M of 32:1) to the slightly more homogenous J1TTGT (MisoII/MisoI/UisoII/UisoI of 24:2:1:1 and M/U of 14:1).
Table 1. Dynamics of branch migration and conformer exchange for single-step junctions as a function of nucleotide sequence.
| Junction | Uhigh lifetime, s | Ulow lifetime, s | Mhigh lifetime, s | Mlow lifetime, s | U lifetime, s | M lifetime, s | α, U | α, M |
|---|---|---|---|---|---|---|---|---|
| J1TGGG | 2.2 ± 0.05 | 0.317 ± 0.007 | 0.144 ± 0.004 | 0.044 ± 0.001 | 20 ± 5.14 | 1.2 ± 0.31 | 16 ± 4.13 | 12 ± 3.1 |
| J1TGGGt | 2.1 ± 0.01 | 0.92 ± 0.007 | 0.10 ± 0.005 | 0.022 ± 0.014 | 25 ± 8.2 | 0.78 ± 0.26 | 16 ± 5.4 | 10 ± 3.27 |
| J1TGGGa | 2.1 ± 0.005 | 0.63 ± 0.002 | 0.114 ± 0.002 | 0.061 ± 0.002 | 20 ± 4.57 | 1.1 ± 0.25 | 14 ± 3.09 | 10 ± 2.28 |
| J1TTGT | 0.097 ± 0.003 | 0.088 ± 0.002 | 2.3 ± 0.009 | 0.125 ± 0.0007 | 0.65 ± 0.11 | 8.9 ± 1.44 | 5.8 ± 0.94 | 7.6 ± 1.23 |
| J1TCGC | 2.26 ± 0.013 | 1.74 ± 0.006 | — | — | — | — | — | — |
| J1TAGA | 1.54 ± 0.03 | 0.55 ± 0.005 | — | — | — | — | — | — |
Dwell times and α values were measured in the presence of 50 mM Mg2+ ions. The U and M assignments were made by using hydroxyl radical footprinting.
Magnesium Ion Concentration Affects All Lifetimes Equivalently. At each branch position, the average dwell time of each conformer decreased with decreasing Mg2+ concentration, whereas the relative conformer populations remained constant (Table 2, which is published as supporting information on the PNAS web site). This behavior was also observed from junctions whose branch points were fixed (11), further supporting the interpretation that the transitions are the result of conformer exchanges. In addition, the average dwell time of each branch position (U and M) decreased as the Mg2+ concentration was lowered, in agreement with the acceleration of spontaneous branch migration at lower Mg2+ concentrations observed in ensemble studies (27). Although the Mg2+ concentrations used in our experiments were high (ranging from 10 to 50 mM) because of an inability to clearly discern M from U at lower concentrations, earlier single-molecule measurements on nonmigratable junctions (12) have shown that there is no qualitative change in junction structural properties from 10 to 0.5 mM other than a 10-fold acceleration in exchange rates. This behavior together with ensemble studies of branch migration lifetimes over a large concentration range (27) suggest that the trends exhibited for mono-migratable junctions over 10–50 mM Mg2+ will extend to the more physiologically relevant solution conditions.
Spontaneous Branch Migration Is Easier When It Involves A-T Base Pairs. Acceleration of both conformer exchange and spontaneous branch migration upon lowering Mg2+ concentration suggests that an unfolded form, which resembles the open structure of the junction adopted in the absence of added metal ions (4, 6, 14), must be visited before both transitions (11, 13, 26). To measure the relative degree of difficulty for an unfolded junction to migrate to an adjacent open state (migrate) compared with simply refolding without branch migration, we defined α as the average number of conformer transitions that took place in one branch position before a branch migration step was taken. For all sequences α values were very similar for the forward (M → U) and backward (U → M) steps. α remained relatively constant for the three sequences with G-C base pairs capable of branch migration (≈14 for J1TGGGt, J1TGGGa, and J1TGGG), but smaller by a factor of two for J1TTGT (≈7, Fig. 4A and Table 1) where the rearranged base pairs are A-T. This finding is consistent with base pair breaking as a major barrier in branch migration, given that A-T base pairs are weaker than G-C. To explore this property further we measured the transition rates and α values as a function of temperature to determine activation barriers. Although the rate of transition between adjacent open states, kOM, cannot be measured directly, α should be inversely proportional to kOM so that its temperature dependence was used to estimate the activation enthalpy for open-to-open transitions. The barriers against junction unfolding (stacked → open) for J1TTGT, J1TGGG, and fixed junctions previously studied (11) were all similar to each other. By contrast, the activation enthalpy associated with the open → open transitions was more than three times higher for J1TGGG compared with J1TTGT (42 kJ·mol–1 vs. 12 kJ·mol–1) (Fig. 4B), further indicating that the base pair breaking is rate limiting for spontaneous branch migration once the junction is unfolded.
Fig. 4.
Branch migration energy landscape. (A) The number of conformational transitions occurring before a migration step takes place (α) is approximately constant for G-C base pairs, but the value is halved for an A-T base pair. Empty bars represent the α value for slow-to-fast state transitions, and filled bars represent the α value for fast-to-slow transitions. (B) Arrhenius plot showing the dependence of the rate of branch migration on temperature. Note that the activation enthalpy for branch migration requiring the breakage of a G-C base pair (J1TGGG) is nearly three times that for an A-T pair (J1TTGT). (C) Illustrative scheme showing the important states and energy barriers for a single-step junction. Although open states are depicted, they are too short-lived for direct experimental observation in the presence of Mg2+ ions. Note that the open state is a common intermediate for both conformer exchange and branch migration. (D) Long-range branch migration will be a very uneven process, reflecting a rugged energy landscape. The data from junctions of different sequence indicate that the lifetimes of individual stacking conformer lifetimes vary markedly, thereby trapping junctions in particularly stable step positions while moving relatively rapidly through stretches of less stable positions.
Parallel Forms Are Undetectable. A version of junction J1TGGG that was labeled with donor and acceptor on the H and X arms (termed the HX vector) so that it would only exhibit high FRET efficiency if it adopted a parallel conformation was also examined. Similar experiments performed earlier on a fixed junction failed to detect any degree of the parallel form (12). Equivalent results were obtained for the single-step junction by using our 4- to 8-ms time resolution; junction molecules exhibited a constant FRET value of ≈0.2 before photobleaching (data not shown).
Discussion
We have observed and analyzed the dynamics of Holliday junction spontaneous branch migration of a single base pair and found that it exhibits a very rugged landscape dictated almost entirely by stacking conformer dynamics. At least a 100-fold variation in the stabilities of junction conformations was observed for a given sequence, far exceeding the 4-fold variations observed from junctions with fixed branch points. This finding is in marked contrast to the earlier suggestion that conformer bias should decrease for increased homology (31). We also observed >30-fold variations in the relative stability of branch points, nearly an order of magnitude greater than anything found when using physically constrained junctions (18).
The Dynamic Behavior of Single-Step Junctions Is Likely to Reflect That of Longer Homologous Sequences. We compared our findings with previous ensemble spontaneous branch migration studies (26) on highly homologous junctions. This comparison required a conversion factor because of different experimental conditions used. Our measurements of conformer lifetimes and ensemble measurements of stepping time show a 10-fold decrease every 14°C. Ensemble stepping times decreased 8-fold between 50 and 1 mM Mg2+. Because our junctions can migrate in only one direction, the migration rate in a highly homologous junction would be twice as high. The presence of 50 mM NaCl in our experiments is known to accelerate conformer exchange by a factor of two (11, 12). Altogether these differences require a correction factor of ≈100 between our conditions (50 mM Mg2+, 50 mM NaCl, and 20°C) and those used in ensemble measurements (1 mM Mg2+, no NaCl, and 37°C). The branch point dwell time averaged over J1TGGG, J1TGGGt, J1TGGGa, and J1TTGT is 9.7 s. Using the correction factor of ≈100 we calculate a 97-ms lifetime in the ensemble conditions, very close to the estimate of 61 ms. Such good agreement indicates that conclusions drawn from junctions that can undergo single-step migration are likely to be applicable to fully homologous junctions.
The Properties of Junctions Are Not Changed Fundamentally by the Ability to Undergo Branch Migration. Our data show that there is no fundamental change in the structural properties of junctions when their sequence permits spontaneous branch migration to occur. They continue to display stacking conformer transitions between antiparallel forms, with no evidence of parallel forms. Transition rates strongly depend on ion concentration, whereas conformer bias is still independent of ion concentration. Even the activation barrier for conformer exchange is almost identical between fixed junctions and those that can undergo single-step branch migration. Therefore, all junctions (irrespective of their ability to branch migrate) exist primarily as stacked structures with frequent but short-lived formation of the open form. If the sequence permits (i.e., the junction is homologous), this opening can lead to occasional migration events between adjacent branch points.
A Rugged Energy Landscape. Step-to-step transitions must be preceded by a stacked-to-unstacked, or opening transition, which explains why the lifetime at each branch position varies directly with the sum of the average lifetimes of the folded conformations at that branch position as Mg2+ concentration is varied or the sequence is modified (Table 2). If either stacked conformation is particularly stable at a given position because of the local DNA sequence, it takes longer to generate the open intermediate where there is an opportunity to jump to an open state at the adjacent branch point. Thus, the sequence-dependent barrier against junction unfolding is the largest obstacle to branch migration. Our comparison of junctions with A-T and G-C base pairs participating in the branch migration showed that base pair breaking is rate limiting for spontaneous branch migration once the junction has unfolded. These observations are summarized in Fig. 4C as a schematic 2D free energy landscape. Two kinds of energetic barrier exist, one to junction unfolding and the other to reorganization of base pairing. Much of the uneven character arises from the sequence-dependent stability of the folded forms, but in sequences capable of multistep migration the nature of the participating base pair (i.e., whether the breaking base pairs are G-C or A-T) will also contribute. If the profile is collapsed along the conformer transition axis and extrapolated to junctions that can migrate more extensively, the energy landscape along the branch migration axis would appear very rugged with sequence-dependent barriers and local energy minima as illustrated schematically in Fig. 4D.
Spontaneous Branch Migration in Vivo May Be Easier than Previously Supposed. Such large variations in branch point lifetimes in free junctions prompt us to reevaluate the potential role of spontaneous branch migration in vivo. Earlier ensemble studies suggested that unassisted branch migration is unimportant in vivo because it is very slow due to ion-induced folding, and because it is inhibited by even a single nonhomologous base pair (26). However, it subsequently became clear that a major role of homologous recombination is the restart of stalled replication fork in which the participating DNA molecules are highly homologous (32). Therefore spontaneous branch migration might be effective in such a context provided it is fast, at least locally. Although the average lifetimes of branch positions in our studies agree with that estimated from ensemble measurements, the individual lifetimes ranged from 245 ms in the case of the U state of J1TGGGt to 6.5 ms for the M state of J1TTGT when extrapolated to physiologically relevant conditions (1 mM Mg2+, 37°C). Such a large variation in branch point stability may explain why certain homologous junctions form crystals with a fixed branch position (17). Although clearly RuvAB and other branch migration enzymes are necessary for large-scale movement of the junction position (33, 34), our measurements suggest that local spontaneous branch migration in vivo can be 10 times faster than the average (26, 27) so that junctions can quickly find stable branch points if they are transiently free of protein in vivo. Whether structural constraints in the context of chromosomal junctions permit such behavior is yet to be determined.
The Central Role of the Stacked-X Structure. The general structural and dynamic properties of the Holliday junction are now clear. In free solution the dynamic character of four-way junctions and their ability to undergo spontaneous branch migration are almost entirely determined by the properties of the stacked X-form under conditions similar to those inside the cell. Conformer exchange and branch migration must share a common intermediate that is likely to resemble the open state deduced for free junctions in the absence of divalent metal ions. In light of this scheme it is possible to reevaluate the action of proteins that interact with Holliday junctions, many of which are structure-selective, but induce a marked alteration in the global structure of the junction (35). Because Holliday junctions created from homologous DNA sequences most closely resemble the stacked X-structure in the absence of any constraint, if some other structure is required for the biological function it must be imposed by the binding of the proteins. Indeed, many proteins that interact with Holliday junctions induce a marked alteration in the global structure of the junction (35). These could affect the properties described for the free junction.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grants PHY-0134916 and DBI-0215869 (to T.H.), National Institutes of Health Grant GM065367 (to T.H.), and Cancer Research UK (D.M.J.L). S.A.M. is a National Science Foundation Graduate Research Fellow and was also partially supported by National Institutes of Health Molecular Biophysics Training Grant T32GM008276.
Author contributions: S.A.M., D.M.L., and T.H. designed research; S.A.M. and A.D.J.F. performed research; A.D.J.F. contributed new reagents/analytic tools; S.A.M. and A.D.J.F. analyzed data; and S.A.M., D.M.L., and T.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Holliday, R. (1964) Genet. Res. 5, 282–304. [Google Scholar]
- 2.Potter, H. & Dressler, D. (1978) Proc. Natl. Acad. Sci. USA 75, 3698–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwacha, A. & Kleckner, N. (1995) Cell 83, 783–791. [DOI] [PubMed] [Google Scholar]
- 4.Duckett, D. R., Murchie, A. I., Diekmann, S., von Kitzing, E., Kemper, B. & Lilley, D. M. J. (1988) Cell 55, 79–89. [DOI] [PubMed] [Google Scholar]
- 5.Churchill, M. E., Tullius, T. D., Kallenbach, N. R. & Seeman, N. C. (1988) Proc. Natl. Acad. Sci. USA 85, 4653–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murchie, A. I. H., Clegg, R. M., von Kitzing, E., Duckett, D. R., Diekmann, S. & Lilley, D. M. J. (1989) Nature 341, 763–766. [DOI] [PubMed] [Google Scholar]
- 7.Cooper, J. P. & Hagerman, P. J. (1989) Proc. Natl. Acad. Sci. USA 86, 7336–7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlstrom, G. & Chazin, W. J. (1996) Biochemistry 35, 3534–3544. [DOI] [PubMed] [Google Scholar]
- 9.Overmars, F. J. & Altona, C. (1997) J. Mol. Biol. 273, 519–524. [DOI] [PubMed] [Google Scholar]
- 10.Grainger, R. J., Murchie, A. I. H. & Lilley, D. M. J. (1998) Biochemistry 37, 23–32. [DOI] [PubMed] [Google Scholar]
- 11.McKinney, S. A., Declais, A. C., Lilley, D. M. J. & Ha, T. (2003) Nat. Struct. Biol. 10, 93–97. [DOI] [PubMed] [Google Scholar]
- 12.Joo, C., McKinney, S. A., Lilley, D. M. J. & Ha, T. (2004) J. Mol. Biol. 341, 739–751. [DOI] [PubMed] [Google Scholar]
- 13.Lilley, D. M. J. (2000) Q. Rev. Biophys. 33, 109–159. [DOI] [PubMed] [Google Scholar]
- 14.Ortiz-Lombardia, M., Gonzalez, A., Eritja, R., Aymami, J., Azorin, F. & Coll, M. (1999) Nat. Struct. Biol. 6, 913–917. [DOI] [PubMed] [Google Scholar]
- 15.Nowakowski, J., Shim, P. J., Prasad, G. S., Stout, C. D. & Joyce, G. F. (1999) Nat. Struct. Biol. 6, 151–156. [DOI] [PubMed] [Google Scholar]
- 16.Thorpe, J. H., Gale, B. C., Teixeira, S. C. & Cardin, C. J. (2003) J. Mol. Biol. 327, 97–109. [DOI] [PubMed] [Google Scholar]
- 17.Eichman, B. F., Vargason, J. M., Mooers, B. H. & Ho, P. S. (2000) Proc. Natl. Acad. Sci. USA 97, 3971–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun, W., Mao, C., Liu, F. & Seeman, N. C. (1998) J. Mol. Biol. 282, 59–70. [DOI] [PubMed] [Google Scholar]
- 19.Miick, S. M., Fee, R. S., Millar, D. P. & Chazin, W. J. (1997) Proc. Natl. Acad. Sci. USA 94, 9080–9084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiom, K. & West, S. C. (1995) Cell 80, 787–793. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan, D. L. & O'Donnell, M. (2002) Mol. Cell 10, 647–657. [DOI] [PubMed] [Google Scholar]
- 22.Whitby, M. C. & Lloyd, R. G. (1995) EMBO J. 14, 3302–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dennis, C., Fedorov, A., Kas, E., Salome, L. & Grigoriev, M. (2004) EMBO J. 23, 2413–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amit, R., Gileadi, O. & Stavans, J. (2004) Proc. Natl. Acad. Sci. USA 101, 11605–11610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawid, A., Croquette, V., Grigoriev, M. & Heslot, F. (2004) Proc. Natl. Acad. Sci. USA 101, 11611–11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panyutin, I. G. & Hsieh, P. (1994) Proc. Natl. Acad. Sci. USA 91, 2021–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panyutin, I. G., Biswas, I. & Hsieh, P. (1995) EMBO J. 14, 1819–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lilley, D. M. J., Clegg, R. M., Diekmann, S., Seeman, N. C., von Kitzing, E. & Hagerman, P. (1995) Eur. J. Biochem. 230, 1–2. [DOI] [PubMed] [Google Scholar]
- 29.Ha, T. (2001) Methods 25, 78–86. [DOI] [PubMed] [Google Scholar]
- 30.Price, M. A. & Tullius, T. D. (1992) Methods Enzymol. 212, 194–219. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, S. & Seeman, N. C. (1994) J. Mol. Biol. 238, 658–668. [DOI] [PubMed] [Google Scholar]
- 32.Cox, M. M., Goodman, M. F., Kreuzer, K. N., Sherratt, D. J., Sandler, S. J. & Marians, K. J. (2000) Nature 404, 37–41. [DOI] [PubMed] [Google Scholar]
- 33.West, S. C. (1997) Annu. Rev. Genet. 31, 213–244. [DOI] [PubMed] [Google Scholar]
- 34.Briggs, G. S., Mahdi, A. A., Weller, G. R., Wen, Q. & Lloyd, R. G. (2004) Philos. Trans. R. Soc. London B 359, 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lilley, D. M. J. & White, M. F. (2001) Nat. Rev. Mol. Cell. Biol. 2, 433–443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.