Abstract
Mammalian gonadotropin-releasing hormone (GnRH1) and nonmammalian immunoreactive GnRH subtypes were examined in transgenic rats carrying an enhanced GFP (EGFP) reporter gene driven by a rat GnRH1 promoter. Double-label immunocytochemistry was performed on EGFP+/GnRH1 brain sections by using antisera against GnRH1, GnRH2 (chicken II), GnRH3 (salmon), or seabream GnRH. EGFP+/GnRH1 neurons were in the septal–preoptic hypothalamus but not in the midbrain, consistent with GnRH1-immunopositive neurons in WT rats. Apparent coexpression of EGFP+/GnRH1 with other GnRH subtypes was observed. All EGFP+ neurons in the septal–preoptic hypothalamus were GnRH1-immunopositive. However, only ≈80% of GnRH1-immunopositive neurons were EGFP+, which awaits further elucidation. GnRH subtypes-immunopositive fibers and EGFP+/GnRH1 fibers were conspicuous in the organum vasculosum of the lamina terminalis, median eminence, and surrounding the ependymal walls of the third ventricle and the aqueduct in the midbrain. These results demonstrate that the expression of the EGFP–GnRH1 transgene is restricted to the bona fide GnRH1 population and provide clear morphological evidence supporting the existence of GnRH1 neuronal subpopulations in the septal–preoptic hypothalamus, which might be driven by different segments of the GnRH promoter. This genetic construct permits analyses of promoter usage in GnRH neurons, and our histochemical approaches open questions about functional relations among isoforms of this peptide, which regulates reproductive physiology in its behavioral and endocrine aspects.
Keywords: luteinizing hormone-releasing hormone, double-label immunocytochemistry, green fluorescent protein, hypothalamus, preoptic area
Until recently, the widely held view was that mammalian gonadotropin-releasing hormone (GnRH1), a decapeptide essential for reproduction and reproductive behavior in vertebrates, is the sole GnRH in the forebrain of mammals (1). However, from morphological, physiological, and evolutionary perspectives there is compelling evidence of distinct subpopulations of GnRH neurons in the mammalian forebrain. For example, retrograde tracer studies (2–6) and computerized 3D reconstruction (7) reveal distinct GnRH subsets within the forebrain GnRH neuronal population in rodents. Furthermore, new populations of GnRH neurons have been reported in the forebrain of rodents and primates whose developmental origin is different (8, 9) from the well documented placodal origin of GnRH1 neurons (10). Also, under certain physiological circumstances only a subpopulation of preoptic GnRH neurons expresses c-Fos protein, receptors for N-methyl-d-aspartate, galanin, or steroid hormones (11, 12). Furthermore, from an evolutionary perspective, it has become increasingly clear that some nonmammalian and mammalian vertebrates possess two or more GnRH subtypes in the brain (13–15), which include in addition to GnRH1, chicken II GnRH2 and salmon GnRH3 in rodents (16, 17). The cloning of GnRH receptor subtypes from the brains of several vertebrate species has further increased the likelihood that more than one molecular form of GnRH is present in the mammalian brain and that GnRH subtypes have multiple functions in addition to stimulating the release of gonadotropins (15, 18) and promoting sexual behavior (19, 20). Taken together, these studies emphasize the possibility that in the forebrain of mammals there exist GnRH populations functionally distinct from the originally discovered GnRH1.
Over the last decade, 16 structurally distinct GnRH forms have been isolated from different vertebrate species (15), and antisera against each GnRH subtype are readily available, which prompted us to investigate the possibility that more than one GnRH subtype can be expressed in different clonal neuronal populations in the rat brain. Transgenic rats carrying an EGFP reporter gene driven by a rat GnRH1 promoter were generated by Masakatsu Kato in our laboratory at Nippon Medical School (21), which we used for double-labeling studies with a variety of GnRH antisera. We compared these with the brain of WT rats immunoreacted with a battery of GnRH antisera specific for GnRH subtypes (mammalian GnRH1, chicken II GnRH2, salmon GnRH3, and seabream GnRH; see Materials and Methods).
Materials and Methods
WT and Transgenic Animals. Transgenic Wistar rats carrying an EGFP (Clontech) reporter gene, driven by 3.0 kb of rat GnRH1 promoter, were generated in our laboratory at Nippon Medical School (for details see ref. 21). The generation and use of transgenic rats were in accordance with the guidelines and approval of the Nippon Medical School Institutional Animal Care and Use Committee. WT and transgenic rats were housed under controlled conditions of temperature (24–26°C) and illumination (lights on 0800–2000 hours) with access to food and water ad libitium.
GnRH Antibodies. Several different GnRH antisera raised by different laboratories were used in the present study. Polyclonal rabbit antibodies specific for mammalian GnRH1 (635.5, gift from L. Jennes, University of Kentucky, Lexington; AB 1567, Chemicon); and monoclonal mouse anti-GnRH1 (LRH13, gift from K. Wakabayashi, Gunma University, Maebashi, Japan) were used. Polyclonal rabbit antibodies specific for GnRH2 (chicken II GnRH,1458, gift from J. King, University of Cape Town, Cape Town, South Africa; aCII6, gift from K. Okuzawa, National Research Institute of Aquaculture, Mie, Japan; ISP-II, supplied by I.S.P.); GnRH3 (salmon GnRH, lot 2, a gift from K. Aida, University of Tokyo, Tokyo; GF 6, a gift from N. Sherwood, University of Victoria, Victoria, Canada), and seabream GnRH (ISP 1, supplied by I.S.P.) also were used. The crossreactivities of GnRH1, GnRH2, GnRH3, and seabream GnRH antisera with various synthetic peptides have been determined by RIA and expressed as percentage crossreactivity with mammalian GnRH1 (for details of crossreactivities see refs. 8 and 22–24; Table 1).
Table 1. Distribution of GnRH subtype immunopositive neurons in the brain of WT and EGFP+/GnRH1 and double-label GnRH neurons in transgenic rats.
| GnRH antibody
|
Antibody
|
%CR with GnRH1
|
Soma/fibers
|
Septal-preoptic hypothalamus
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Concentration | Code | TT-SHi | MS-DB | MPOA | LS | OVLT | ME | Pe | SO | VMHvl | Am | HB | PVP | CA1 | MM | CG | IP | IR cells | IF cells | EGFP+ cells | DL cells | % DL | ||
| GnRH1-EGFP+ | — | — | — | +/+ | +/+ | +/+ | -/+ | -/+ | -/+ | +/+ | +/+ | +/+ | -/+ | +/+ | -/+ | -/+ | +/+ | -/+ | -/+ | — | — | 901 ± 25 | — | — |
| GnRH1 | 1:2,500 | LRH13 | 100 | +/+ | +/+ | +/+ | -/- | -/+ | -/+ | +/+ | +/+ | +/+ | -/+ | -/+ | -/+ | -/+ | -/+ | -/+ | -/+ | 962 ± 26 | 1044 ± 51 | 908 ± 49 | 860 ± 42 | 82 ± 1 |
| GnRH1 | 1:8,000 | 635.5 | NA | +/+ | +/+ | +/+ | -/- | -/+ | -/+ | -/+ | +/+ | -/+ | -/+ | -/+ | -/+ | -/+ | -/+ | -/+ | -/+ | 964 ± 88 | ||||
| GnRH1 | 1:2,000 | AB1567 | 100 | -/+ | +/+ | +/+ | -/- | -/+ | -/+ | -/- | -/+ | -/- | -/+ | -/+ | -/+ | -/+ | -/+ | -/+ | -/+ | 124 ± 28 | ||||
| GnRH2 | 1:4,000 | aCII6 | 0.01 | -/- | +/+ | +/+ | -/+ | -/+ | -/+ | -/+ | -/+ | -/- | +/+ | -/+ | -/+ | -/- | -/+ | -/+ | -/+ | 456 ± 84 | 413 ± 74 | 861 ± 41 | 288 ± 82 | 70 ± 2 |
| GnRH2 | 1:3,000 | 1458 | 2.5 | +/+ | +/+ | +/+ | -/- | -/+ | -/+ | +/+ | +/+ | -/+ | -/+ | -/+ | -/+ | -/+ | -/- | -/+ | -/+ | 472 ± 8 | ||||
| GnRH2 | 1:3,000 | ISP II | NA | -/- | -/- | +/+ | -/- | -/+ | -/+ | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | 84 ± 20 | ||||
| GnRH3 | 1:11,500 | Lot. 2 | 0.01 | +/+ | +/+ | +/+ | -/- | -/+ | -/+ | -/+ | +/+ | +/+ | -/+ | -/+ | -/+ | -/- | -/+ | -/+ | -/+ | 516 ± 80 | 670 ± 76 | 972 ± 52 | 406 ± 24 | 60 ± 7 |
| GnRH3 | 1:3,500 | GF6 | 100 | +/+ | +/+ | +/+ | -/- | -/+ | -/+ | +/+ | +/+ | +/+ | -/+ | -/+ | -/+ | -/- | -/+ | -/+ | -/- | 616 ± 18 | ||||
| Seabream | 1:5,000 | ISP1 | 0.01 | -/- | +/+ | +/+ | -/- | -/+ | -/+ | -/- | -/+ | -/+ | -/+ | -/+ | -/- | -/- | -/+ | -/+ | -/- | 45 ± 8 | ||||
CR, cross reactivity; Shi, septohippocampal nucleus; MS-DB, medial septum/diagonal band of Broca; LS, lateral septum; Pe, periventricular hypothalamic nucleus; SO, supraoptic nucleus; VMHvl, ventrolateral part of ventromedial hypothalamus; Am, medial amygdaloid nucleus; PVP, periventricular thalamic nucleus; CA1, CA1 field of the hippocampus; CG, midbrain central gray; IP, interpeduncular nucleus; IR, peroxidase immunoreactivity; IF, immunofluorescence; DL, double-label; NA, not available. % DL, DL/IF × 100. +/- indicates presence/absence of cell soma (Left) and fibers (Right). GnRH immunoreactive and EGFP+ cell numbers are mean ± SEM from three to four animals per antibody and four EGFP+ animals. The sources (represented by code) of the antisera are given in Materials and Methods.
Peroxidase-Based Immunocytochemistry in WT Rats. Wistar rats (adult females, n = 12; adult males, n = 3) weighing 150–200 g were anesthetized with sodium pentobarbital (Nembutal, 35 mg/kg of body weight; Abbott). A single injection of colchicine (10 μg/5 μl) was placed stereotaxically into the lateral ventricle. The next day, the animals were anesthetized with an overdose of Nembutal and transcardially perfused with 200 ml of ice-cold PBS followed by 200 ml of 4% paraformaldehyde dissolved in 0.01 M phosphate buffer (PB; pH 7.5). The brains were removed, postfixed overnight in the same fixative, and cryoprotected in 20% sucrose in PB at 4°C overnight.
Coronal brain cryostat sections (30 μm thick) from Bregma +1.70 to –6.30 (25) were placed sequentially into one of three vessels, incubated in one of the polyclonal primary antiserum against GnRH subtypes (see Table 1), and processed for free-floating immunocytochemistry following a protocol modified from Parhar et al. (26). In brief, after incubation in primary antiserum, sections were incubated in biotinylated anti-rabbit IgG or anti-mouse IgG and avidin-biotinylated horseradish peroxidase complex (Vectastain ABC Elite kit, Vector Laboratories) and reacted with 0.05% 3,3′-diaminobenzidine tetrahydrochloride (Sigma) used as chromogen. Sections were then mounted onto slides, dehydrated, and cleared in xylene, and coverslips were applied with Permount (Fisher). Immunoreactivity was observed with the aid of an Olympus microscope.
Controls for Immunocytochemistry. Wistar rats (adult females, n = 2; adult males, n = 5) weighing 200–250 g were used as controls. To demonstrate the specificity of the primary antisera to GnRH, adjacent brain sections were incubated with primary antiserum preabsorbed with heterologous or its homologous GnRH peptide at concentrations ranging from 1 to 8 μg/ml of the primary antiserum at its working dilution overnight before use (see Table l). BSA was added to preabsorb anti-BSA. Additional negative controls included omission of one of the primary antisera from the immunostaining protocol, to further eliminate possible nonspecific reaction.
Identification of Transgenic Rats. Transgenic rats (n = 9, 8–12 weeks old) carrying the EGFP–GnRH1 reporter gene were identified by PCR analysis of genomic DNA isolated from ear biopsies by using a Puregene DNA isolation kit (Gentra Systems). PCR was performed by using a thermal cycler (PerkinElmer GeneAmp PCR system 9700, Applied Biosystems). The final amplification mix (Applied Biosystems) included the sense primer (F2: 3′-TAC TAT GGT CTA CGC TGC ACT-5′, rGnRH1 promoter base pairs 3017–3037), antisense primer (ERl: 5′-ACT TGA AGA AGT CGT GCT GCT-3′, pEGFP-1 base pairs 335–355) specific for EGFP-coding sequences, and 20 ng of genomic DNA to amplify a 1,015-bp fragment of the GFP gene. Following the standard PCR conditions, each PCR product was analyzed on 1% agarose gel containing ethidium bromide and photographed with a gel visualization system (Electronic UV Transilluminator, Ultra-Lum, Claremont, CA) (Fig. 1).
Fig. 1.
Gel showing expression of EGFP amplicons. EGFP+/GnRH1 rats show a 1,057-bp PCR amplicon in lanes 1, 3, 5, 8, 9, and 10, and EGFP– littermates in lanes 2, 4, 6, and 7. DNA size marker (M) in base pairs, is given.
Double-Label Immunofluorescence in Transgenic Rats. Under Nembutal anesthesia (35 mg/kg of body weight), EGFP+/GnRH1 rats (males, n = 2; females, n = 7) received a single stereotaxic injection of colchicine (10 μl/5 μl) into the lateral ventricle. The next day, animals were killed, and the brains were processed as in peroxidase-based immunocytochemistry (see above).
Coronal brain cryostat sections (30 μm thick) from Bregma +1.70 to –6.30 (25) were visualized for EGFP+/GnRH1 neurons, and then incubated overnight at room temperature with antisera to GnRH1 (LRH13), GnRH2 (aCI16), or GnRH3 (lot 2). These antisera were used at a dilution of 1,500–2,000 with 0.01 M PBS (pH 7.6). After washes, sections were placed in Cy3 goat-anti-rabbit IgG (1:400 in PBS) or anti-mouse IgG for 2 h at room temperature and then mounted onto slides, and coverslips were applied with Vectashield (Vector Laboratories). Sections were viewed under a fluorescent microscope (DM RXA2, Leica Microsystems, Wetzlar, Germany) by using Texas red filter to reveal GnRH cells labeled with Cy3 (red fluorescence; Research Organics, Cleveland) and fluorescein-isothiocyanate filter to reveal the EGFP+/GnRH1 neurons (green fluorescence). Digital images were captured on an Image Analysis System (Q5501W, Leica Microsystems) and superimposed for the observation of double-labeled cells. With photoshop 4.0 (Adobe Systems, Seattle) the images were arranged into plates.
Cell Counts. In each brain three different GnRH antisera were applied to every third 30-μm section made from Bregma +1.70 to –6.30. The total number of GnRH-immunoreactive neurons in WT, EGFP+/GnRH1, and double-labeled neurons in transgenic animals was counted in every third section spanning the septal–preoptic hypothalamus (Bregma +1.70 to –1.80). These counts were averaged across animals to determine mean ± SEM values for GnRH-immunopositive, EGFP+/GnRH1, and double-labeled cell populations per individual brain (Table 1). All GnRH cells cut through the plane of the nucleus were counted in each section. Because the diameter of the GnRH cell nucleus is considerably smaller than the thickness of each cryostat section, no correction for double counting of cells was made.
Results
EGFP+/GnRH1 neurons were observed in the tenia tecta (TT), septal–preoptic hypothalamus (septohippocampal nucleus, diagonal band of Broca, medial septum, medial preoptic area, retrochiasmatic supraoptic nucleus, ventrolateral part of the ventromedial hypothalamus), consistent with GnRH1-immunopositive cells in WT rats (Table 1). GnRH expression was not observed in the tectum of our adult rats, which is reported to be transiently expressed only during early development (9, 27).
Peroxidase-Based Immunocytochemistry in WT Rats. In the septal–preoptic hypothalamus, cells immunopositive for GnRH1 were more numerous than GnRH3 > GnRH2 > seabream GnRH (Table 1); these cells were fusiform in shape. GnRH cells were not segregated into nuclear clusters, instead they appeared as a loose continuum, which stretched from the TT to the septal–preoptic hypothalamus (Table 1).
Fibers immunoreactive to GnRH1, GnRH2, and GnRH3 were detected in the medial regions of the anterior olfactory nucleus, TT, septal–preoptic area, bed nucleus of the stria terminalis, periventricular hypothalamic nucleus, medial amygdaloid nucleus, and medial mammillary body (MM), and along the ventral border of the interpeduncular nucleus (Table 1). GnRH fibers were consistently seen to course beneath the ependymal walls of the third ventricle and the aqueduct (Aq) in the midbrain central gray. However, the most conspicuous GnRH fibers were seen in the organum vasculosum of the lamina terminalis (OVLT) and the median eminence (ME) (Table 1).
No differences were detected between males and females when using any of the GnRH antisera. Immunostaining with seabream GnRH antisera was very weak. In control experiments, in which the GnRH antisera were preabsorbed with their respective homologous antigen or omitted from the procedure, no staining was evident in any of the sections. GnRH antisera preabsorbed with heterologous peptides did not abolish immunostaining.
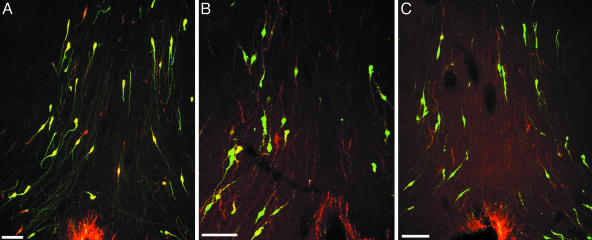
Double-Label Immunofluorescence in Transgenic Rats. EGFP+/GnRH1 cells were distributed as a loose continuum, which stretched from the TT to the septal–preoptic hypothalamus (Table 1 and Figs. 2 and 3). In the septal–preoptic hypothalamus, the number of EGFP+/GnRH1 cells immunoreactive to antisera against GnRH subtypes varied in the following order: GnRH1 > GnRH3 > GnRH2. Double-label immunofluorescence, undertaken on brain sections from transgenic rats, revealed that ≈80% of GnRH1-immunopositive cells were EGFP+ (Table 1 and Fig. 3A). Despite colocalization, cells immunoreactive to various GnRH antisera were seen scattered among EGFP+/GnRH1 cells in the septal–preoptic hypothalamus (Fig. 3). EGFP+/GnRH1 cells observed in the habenula (HB) and the MM were not immunoreactive to antisera against any of the GnRH subtypes (Fig. 2 B and D), whereas cells in the red nucleus were autofluorescent (data not shown).
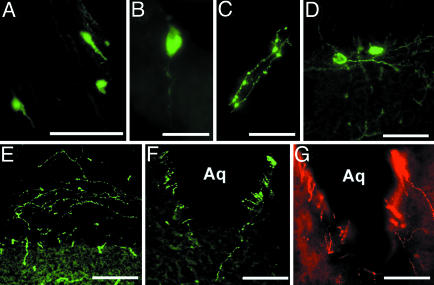
Fig. 2.
Distribution of GnRH soma and fibers in the brain of EGFP+/GnRH1 rats is shown. (A) TT. (B and C) Soma (B) and fibers (C) in the HB. (D and E) Soma (D) and fibers (E) in the MM. (F and G) EGFP+/GnRH1 fibers (F) and Cy3-labeled GnRH2 fibers (G) surrounding the Aq in the midbrain. (Scale bars: A and D–G, 100 μm; B and C, 50 μm.)
Fig. 3.
Coronal sections through the caudal region of the septal–preoptic area of EGFP+/GnRH1 rats immunoreactive to GnRH subtypes are shown. Cy3-labeled soma and fibers (red) reveal GnRH1 (LRH13) (A), GnRH3 (lot 2) (B), and GnRH2 (aCII6) (C). Note the typical fusiform shape of GnRH neurons and the abundance of EGFP+ cells immunoreactive to GnRH1 in A; EGFP+ soma and fibers coexpressing each GnRH subtype appear in yellow. Cy3-labeled nonmammalian GnRH soma and fibers (red) intermingled with EGFP+/GnRH1 cells (green). (Scale bars, 50 μm.)
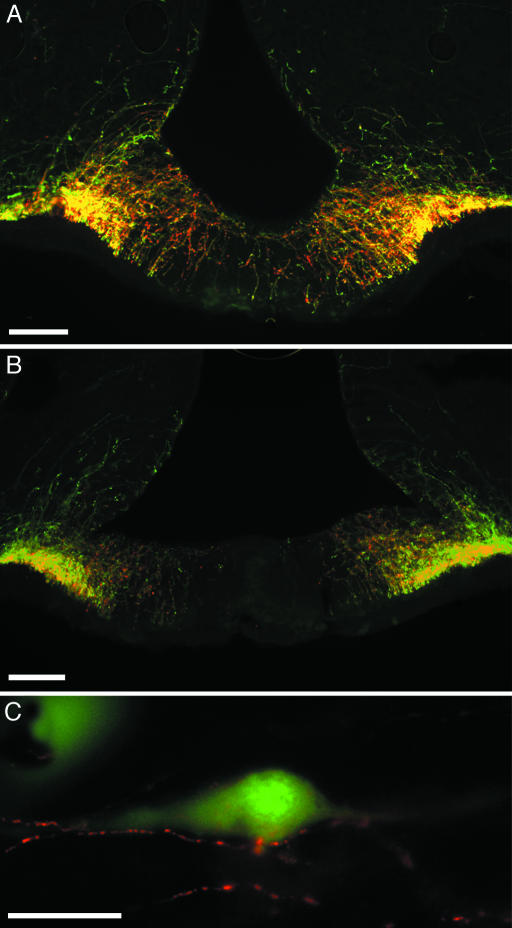
EGFP+/GnRH1 fibers were seen in the TT, septal–preoptic hypothalamus, HB, and the medial MM (Table 1 and Fig. 2 C and E). Fibers were consistently observed in the OVLT and ME, beneath the ependymal walls of the third ventricle, and in the Aq in the midbrain central gray (Figs. 2 F and G, 3, and 4 A and B). EGFP+/GnRH1-, GnRH1-, GnRH2-, and GnRH3-immunopositive fibers were seen in close apposition or double-labeled in the lateral regions of the ME (Fig. 4 A and B) and in close apposition to EGFP+/GnRH1 cell soma and fibers in the septal–preoptic hypothalamus (Figs. 3 and 4C).
Fig. 4.
Coronal sections through the medial region of the ME of EGFP+/GnRH1 rats immunoreactive to GnRH subtypes are shown. (A and B) Cy3-labeled (red) fibers immunoreactive to GnRH1 (LRH 13) (A) or GnRH2 (aCII6) (B). EGFP+ fibers coexpressing either nonmammalian GnRH appear in yellow. (C) Cy3-labeled GnRH2 (aCII6) fibers (red) in close contact with EGFP+/GnRH1 soma (green) in the septal–preoptic area are shown. (Scale bars: A and B, 100 μm; C, 50 μm.)
Discussion
EGFP+ neurons were detected in the TT, septal–preoptic hypothalamus, OVLT, and the ME, regions known to contain immunoreactive GnRH neurons and fibers in the rat brain (1) and in the TT of the Syrian hamster (28) and the musk shrew (29). Double-label studies confirmed that all EGFP+ soma and terminal segments of axons were GnRH1-immunopositive (94%) but not all GnRH1-immunopositive cells were EGFP+ (82%), which suggests the existence of GnRH neuronal subpopulations. In addition, it shows GnRH1 antiserum (LRH13) recognizes the precursor and the mature peptide indiscriminately (22), but differences in the availability of the absolute concentration of antigen along the axon preterminals, caused by rates of transport and/or sites of processing of the precursor, results in differences in double-labeling in preterminals versus terminal segments of axons in the OVLT and ME. Our results also demonstrate that the EGFP reporter gene driven by a rat GnRH1 promoter is more efficient for targeted expression of the reporter gene to GnRH-expressing neurons of the septal–preoptic hypothalamus compared with other reporter genes (e.g., luc and lacZ) driven by the human or murine GnRH promoters, which fail to provide GnRH neuron-restricted expression in transgenic mice (30).
Septal–Preoptic Hypothalamus EGFP+/GnRH Neurons. The present results show that the EGFP+/GnRH1 neurons in the septal–preoptic hypothalamus are the bona fide GnRH neurons; they synthesize the authentic GnRH1 and are detected by GnRH1 antisera. Therefore, the expression of EGFP–GnRH1 transgene in the septal–preoptic hypothalamus is restricted to the GnRH1 population described in the rat brain (1). The role of GnRH1 is evolutionarily conserved. GnRH1 functions as a hypophysiotropic hormone for the control of gonadotropin release and is crucial for reproduction throughout the vertebrate species (1, 15).
In the extrahypothalamic area, EGFP+/GnRH1 soma and fibers were seen in the medial HB similar to GnRH2-like immunoreactivity in the musk shrew (31). The medial HB has been implicated in the regulation of female sexual receptivity in rodents (32) and courtship behavior with associated increase in GnRH-containing mast cells in ringdoves and rats (1, 33).
Septal–Preoptic Hypothalamus EGFP–/GnRH Neurons. In the septal–preoptic hypothalamus, ≈80% of neurons immunoreactive to GnRH1 were EGFP+, but the remaining 20% of GnRH1 neurons were EGFP–. Whether EGFP– neurons represent a second population of septal–preoptic hypothalamic GnRH neurons, which synthesize a distinct GnRH gene product as in advance teleost and in the guinea pig (15, 34), or contain a posttranscriptionally modified hydroxyprolinated GnRH molecule (35) or GnRH fragments rather than fully mature GnRH1 (8) whose spatiotemporal origin might be different from the placodal origin of GnRH1 neurons (8–10, 15) remains to be determined because a genomewide search failed to detect homologies to GnRH2, GnRH3, and lamprey III GnRH peptide or their ORFs in the rat and mouse databases (36). Therefore, GnRH2, GnRH3, and lamprey III GnRH immunoreactivity in the brain of rodents (16, 17, 37) is questionable, and the role of lamprey III GnRH in the control of follicle-stimulating hormone is debatable (37, 38).
We speculate that the EGFP–GnRH transgene is incapable of exhaustive targeting of the total GnRH1 neuronal population or EGFP– neurons might express lower levels of the fusion protein, which do not produce visible levels of fluorescence and, therefore, remain undetectable. Alternatively, it is also possible that the synthesis of GnRH in EGFP– neurons in the forebrain is directed by different segments of the GnRH promoter. The existence of two independent promoter regions directing tissue-specific expression of human GnRH gene has been characterized (39). These two GnRH promoters have differential usage: two nonoverlapping 5′ control elements, each containing only one of the two transcriptional start sites are capable of directing reporter gene expression in tumor cells derived from reproductive tissues or hypothalamic neurons (39). Similarly, in rats, two regulatory regions in the GnRH 5′ flanking DNA have been identified as essential for cell type-specific expression in hypothalamic neurons: a 300-bp enhancer and a 173-bp proximal promoter (40). It is, therefore, conceivable that different promoter segments can direct GnRH synthesis in EGFP+ and EGFP– neuronal subpopulations in the rat.
Midbrain Central Gray. Up to now, the most accepted view recognizes GnRH2 as phylogenetically the conserved GnRH isoform, synthesized by neurons localized exclusively in the midbrain (13–15, 18). Biochemical assays have shown the presence of GnRH2 in the brain of all vertebrate species studied to date, but neurons expressing GnRH2 mRNA and peptide have been successfully localized in the midbrain of only teleost and primitive placental mammals (15, 41). The presence of GnRH2 neurons in the midbrain of higher vertebrates including rodents remains an enigma. So far, two reports (42, 43) have shown GnRH2 neurons in the midbrain of rodents, which appear to be false positive based on the following reasons. First, although using the same GnRH2 antiserum (aCII6), the two reports show different cell populations in the rodent midbrain as GnRH2-immunopositive. According to the reports, the cells are either scattered along the Aq in the midbrain (42) or reside in the oculomotor and red nuclei (43). Second, the GnRH2 neurons shown in the midbrain (42) morphologically do not appear as typical neurons. Third, the GnRH2 antiserum (aCII6) used in the two studies has been conjugated to BSA, which those authors did not attempt to immunoneutralize (42, 43), causing possible false-positive immunoreaction. Using three different GnRH2 antisera (ISP-II, 1458, and aCII6), we did not find GnRH2-immunopositive soma in the midbrain in our study. Nonspecific GnRH2 immunoreactivity was detected in the oculomotor and red nuclei of rats and in the oculomotor nucleus of teleosts (unpublished observation). Some might argue that the lack of immunopositive GnRH2 cells in the midbrain is caused by technical limitation to detect low levels of GnRH2 peptide or rapid turnover of the peptide. However, the immunocytochemical approach used in this study is well established in our laboratory at Nippon Medical School and has been used to demonstrate GnRH2-immunopositive soma and fibers in teleosts along with successful immunoneutralization (26). In addition, increasing the antiserum concentration or colchicine should have blocked axonal transport of GnRH2; if there were any GnRH2-synthesizing neurons in the midbrain, they would be visible. However, they were not. Thus, in rodents, GnRH2 peptide is probably not transcribed in the midbrain.
Administration of GnRH1 into the midbrain can enhance sexual behavior in female rats (20). Cells in the midbrain could be targets of GnRH1, which is supported by the presence of GnRH1-immunopositive and EGFP+ fiber terminals beneath the ependymal walls of the Aq in the midbrain. Because high doses of GnRH1 were needed to induce lordosis (20), GnRH1 receptors in the midbrain may be low in abundance or exogenous GnRH1 might have acted via the type II GnRH receptors, which have been hypothesized to be involved in behavioral regulation (18). Indeed, GnRH2 is more potent than GnRH1 in stimulating reproductive behaviors in many vertebrates (15, 18, 44–46).
GnRH Fibers. The most conspicuous EGFP+/GnRH1 and GnRH subtype-immunopositive fibers were seen in the OVLT and the caudal ME, suggesting the classical role of GnRH as a hypophysiotropic hormone for the regulation of gonadotropin release. The distribution of EGFP+/GnRH1 and GnRH subtype-immunopositive fibers throughout the septal–preoptic hypothalamus, beneath the ependymal walls of the third ventricle, and surrounding the Aq of the midbrain is consistent with the proposed role of GnRH as a neuromodulator (18). Axosomatic and axodendritic synapses between GnRH1 elements have been reported in the rat and rhesus monkey (1, 47). We now show GnRH fiber contacts between EGFP+ and EGFP– neurons, which may assist in the coordination of neuroendocrine and neuromodulatory actions of GnRH1.
Acknowledgments
We thank Dr. M. Kato for providing the transgenic rats. Generous gifts of antibodies from many colleagues (listed in Materials and Methods) are also acknowledged. This work was supported in part by Ministry of Education, Culture, Sports, Science, and Technology Grants 14580777 (to I.S.P.), 14370025 and 16086210 (to Y.S.), and Ministry of Health, Labour and Welfare Grant H16-Kagaku-002 (to I.S.P.). The Leica Image Analysis System was purchased with a grant from Japanese Ministry of Education and Science for independent colleges and universities (2000).
Author contributions: I.S.P. designed research; T.S. and Satoshi Ogawa performed research; I.S.P. and Y.S. contributed new reagents/analytic tools; I.S.P., T.S., and Satoshi Ogawa analyzed data; I.S.P. wrote the paper; and Sonoko Ogawa, Y.S., and D.W.P. provided suggestions and edited the manuscript.
Abbreviations: Aq, aqueduct; GnRH, gonadotropin-releasing hormone; HB, habenula; ME, median eminence; MM, mammillary body; MPOA, medial and median preoptic area; OVLT, organum vasculosum of the lamina terminalis; TT, tenia tecta.
References
- 1.Silverman, A. J., Livne, I. & Witkin, J. W. (1994) in The Physiology of Reproduction, eds. Knobil, E. & Neill, J. D. (Raven, New York), 2nd Ed., pp. 1683–1709.
- 2.Jennes, L. & Stumpf, W. E. (1986) Neuroscience 18, 403–416. [DOI] [PubMed] [Google Scholar]
- 3.Silverman, A. J., Jhamandas, J. H. & Renaud, L. P. (1987) J. Neurosci. 7, 2312–2319. [PMC free article] [PubMed] [Google Scholar]
- 4.Merchenthaler, I., Setalo, G., Csontos, C., Petrusz, P., Flerko, B. & Negro-Vilar, A. (1989) Endocrinology 125, 2812–2821. [DOI] [PubMed] [Google Scholar]
- 5.Witkin, J. W. (1990) Neuroscience 37, 501–506. [DOI] [PubMed] [Google Scholar]
- 6.Rajendren, G. (2001) Brain Res. 918, 74–79. [DOI] [PubMed] [Google Scholar]
- 7.Hiatt, E. S., Brunetta, P. G., Seiler, G. R., Barney, S. A., Selles, W. D., Wooledge, K. H. & King, J. C. (1992) Endocrinology 130, 1030–1043. [DOI] [PubMed] [Google Scholar]
- 8.Terasawa, E., Busser, B. W., Luchansky, L. L., Sherwood, N. M., Jennes, L., Millar, R. P., Glucksman, M. J. & Roberts, J. L. (2001) J. Comp. Neurol. 439, 491–504. [DOI] [PubMed] [Google Scholar]
- 9.Skynner, M. J., Slater, R., Sim, J. A., Allen, N. D. & Herbison, A. E. (1999) J. Neurosci. 19, 5955–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwanzel-Fukuda, M. & Pfaff, D. W. (1989) Nature 338, 161–164. [DOI] [PubMed] [Google Scholar]
- 11.Herbison, A. E. & Pape, J. R. (2001) Front. Neuroendocrinol. 22, 292–308. [DOI] [PubMed] [Google Scholar]
- 12.Gore, A. C. (2002) GnRH: The Master Molecule of Reproduction (Kluwer, Dordrecht, The Netherlands).
- 13.Sherwood, N. M., Schalburg, K. V. & Lescheid, D. W. (1997) in GnRH Neurons: Gene to Behavior, eds. Parhar, I. S. & Sakuma, Y. (Brain Shuppan, Tokyo), pp. 3–25.
- 14.King, J. A. & Millar, R. P. (1997) in GnRH Neurons: Gene to Behavior, eds. Parhar, I. S. & Sakuma, Y. (Brain Shuppan, Tokyo), pp. 51–77.
- 15.Parhar, I. S. (2002) Prog. Brain Res. 141, 3–17. [DOI] [PubMed] [Google Scholar]
- 16.Yahalom, D., Chen, A., Ben-Aroya, N., Rahimipour, S., Kaganovsky, E., Okon, E., Fridkin, M. & Koch, Y. (1999) FEBS Lett. 463, 289–294. [DOI] [PubMed] [Google Scholar]
- 17.Montaner, A. D., Affanni, J. M., King, J. A., Bianchini, J. J., Tonarelli, G. & Somoza, G. M. (1999) Cell. Mol. Neurobiol. 19, 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millar, R. P. (2003) Trends Endocrinol. Metab. 14, 35–43. [DOI] [PubMed] [Google Scholar]
- 19.Pfaff, D. W. (1973) Science 182, 1148–1149. [DOI] [PubMed] [Google Scholar]
- 20.Sakuma, Y. & Pfaff, D. W. (1980) Nature 283, 566–567. [DOI] [PubMed] [Google Scholar]
- 21.Kato, M., Ui-Tei, K., Watanabe, M. & Sakuma, Y. (2003) Endocrinology 144, 5118–5125. [DOI] [PubMed] [Google Scholar]
- 22.Park, M. K. & Wakabayashi, K. (1986) Endocrinol. Jpn. 33, 257–272. [DOI] [PubMed] [Google Scholar]
- 23.Blähser, S., King, J. A. & Kuenzel, W. J. (1989) Histochemistry 93, 39–48. [DOI] [PubMed] [Google Scholar]
- 24.Senthilkumaran, B., Okuzawa, K., Gen, K., Ookura, T. & Kagawa, H. (1999) J. Neuroendocrinol. 11, 181–186. [DOI] [PubMed] [Google Scholar]
- 25.Paxinos, G. & Watson, C. (1997) The Rat Brain in Stereotaxic Coordinates (Academic, San Diego), 3rd Ed.
- 26.Parhar, I. S., Pfaff, D. W. & Schwanzel-Fukuda, M. (1996) Brain Res. Mol. Brain Res. 41, 216–227. [DOI] [PubMed] [Google Scholar]
- 27.Wu, T. J., Gibson, M. J. & Silverman, A. J. (1995) J. Neuroendocrinol. 7, 899–902. [DOI] [PubMed] [Google Scholar]
- 28.Richardson, H. N., Parfitt, D. B., Thompson, R. C. & Sisk, C. L. (2002) J. Neuroendocrinol. 14, 375–383. [DOI] [PubMed] [Google Scholar]
- 29.Dellovade, T. L., Hunter, E. & Rissman, E. F. (1995) Neuroendocrinology 62, 385–395. [DOI] [PubMed] [Google Scholar]
- 30.Spergel, D. J., Kruth, U., Shimshek, D. R., Sprengel, R. & Seeburg, P. H. (2001) Prog. Neurobiol. 63, 673–686. [DOI] [PubMed] [Google Scholar]
- 31.Rissman, E. F. (1996) Biol. Reprod. 54, 413–419. [DOI] [PubMed] [Google Scholar]
- 32.Swanson, L. W. & Sawchenko, P. E. (1983) Annu. Rev. Neurosci. 6, 269–324. [DOI] [PubMed] [Google Scholar]
- 33.Khalil, M. H., Silverman, A. J. & Silver, R. (2003) J. Neurobiol. 56, 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jimenez-Linan, M., Rubin, B. S. & King, J. C. (1997) Endocrinology 138, 4123–4130. [DOI] [PubMed] [Google Scholar]
- 35.Gautron, J. P., Pattou, E., Bauer, K. & Kordon, C. (1991) Neurochem. Int. 18, 221–235. [DOI] [PubMed] [Google Scholar]
- 36.Morgan, K. & Millar, R. P. (2004) Gen. Comp. Endocrinol. 139, 191–197. [DOI] [PubMed] [Google Scholar]
- 37.McCann, S. M., Karanth, S., Mastronardi, C. A., Dees, W. L., Childs, G., Miller, B., Sower, S. & Yu, W. H. (2002) Prog. Brain Res. 141, 151–164. [DOI] [PubMed] [Google Scholar]
- 38.Kovacs, M., Seprodi, J., Koppan, M., Horvath, J. E., Vincze, B., Teplan, I. & Flerko, B. (2002) J. Neuroendocrinol. 14, 647–655. [DOI] [PubMed] [Google Scholar]
- 39.Dong, K.-W., Yu, K.-L., Chen, Z.-G., Chen, Y.-D. & Roberts, J. L. (1997) Endocrinology 138, 2754–2762. [DOI] [PubMed] [Google Scholar]
- 40.Lawson, M. A., Macconell, L. A., Kim, J., Powl, B. T., Nelson, S. B. & Mellon, P. L. (2002) Endocrinology 143, 1404–1412. [DOI] [PubMed] [Google Scholar]
- 41.Rissman, E. F., Alones, V. E., Craig-Veit, C. B. & Millam, J. R. (1995) J. Comp. Neurol. 357, 524–531. [DOI] [PubMed] [Google Scholar]
- 42.Gestrin, E. D., White, R. B. & Fernald, R. D. (1999) FEBS Lett. 448, 289–291. [DOI] [PubMed] [Google Scholar]
- 43.Chen, A., Yahalom, D., Ben-Aroya, N., Kaganovsky, E., Okon, E. & Koch, Y. (1998) FEBS Lett. 435, 199–203. [DOI] [PubMed] [Google Scholar]
- 44.Volkoff, H. & Peter, R. E. (1999) Gen. Comp. Endocrinol. 116, 347–355. [DOI] [PubMed] [Google Scholar]
- 45.Maney, D. L., Richardson, R. D. & Wingfield, J. C. (1997) Horm. Behav. 32, 11–18. [DOI] [PubMed] [Google Scholar]
- 46.Kauffman, A. S. & Rissman, E. F. (2004) Endocrinology 145, 3639–3646. [DOI] [PubMed] [Google Scholar]
- 47.Witkin, J. W. (1999) Microsc. Res. Tech. 44, 11–18. [DOI] [PubMed] [Google Scholar]