Abstract
Geographical origin determination of white rice has become the major issue of food industry. However, there is still lack of a high-throughput method for rapidly and reproducibly differentiating the geographical origins of commercial white rice. In this study, we developed a method that employed lipidomics and deep learning to discriminate white rice from Korea to China. A total of 126 white rice of 30 cultivars from different regions were utilized for the method development and validation. By using direct infusion-mass spectrometry-based targeted lipidomics, 17 lysoglycerophospholipids were simultaneously characterized within minutes per sample. Unsupervised data exploration showed a noticeable overlap of white rice between two countries. In addition, lysophosphatidylcholines (lysoPCs) were prominent in white rice from Korea while lysophosphatidylethanolamines (lysoPEs) were enriched in white rice from China. A deep learning prediction model was built using 2014 white rice and validated using two different batches of 2015 white rice. The model accurately discriminated white rice from two countries. Among 10 selected predictors, lysoPC(18:2), lysoPC(14:0), and lysoPE(16:0) were the three most important features. Random forest and gradient boosting machine models also worked well in this circumstance. In conclusion, this study provides an architecture for high-throughput classification of white rice from different geographical origins.
Introduction
The abiotic stress has a large impact on the constituents of plant sources, such as food additives, pharmaceuticals, flavors, and industrially important biochemicals1. In recent years, the demand for high-quality food products with geographical indications has substantially increased2. Adulteration practice, especially the falsification of food origins, is prejudicial to consumers as well as authorized producers and distributors2, 3. Therefore, the geographical origin determination and authenticity of food products have become the major issues of food industry. White rice, a main staple food of many countries in Asia and Africa, has been a potential target to adulteration regarding their similar physical properties4. Better authentication methods to detect the geographical origin are, indeed, required.
Trace elements and stable isotope ratios have been widely used to discriminate the geographical origins of rice5–8. When search for other potential chemical compositions that are capable to predict the geographical origins of commercial white rice, we found that phospholipids (PLs) are the attractive targets. Environmental factors, which are essentially different from countries to countries, greatly affect the concentrations of PLs in white rice. In addition, the deterioration of some PL species during storage contributes to the degradation of white rice9. In a previous preliminary experiment, we demonstrated that the differences of lysoglycerophospholipids (lysoGPLs) might be proper to differentiate white rice originated from different countries10.
There are many analytical methods for the determination of white rice geographical origins based on their chemical compositions2. In addition, chemometric-based classification techniques, especially partial least squares discriminant analysis (PLS-DA), have been formally applied for the authenticity of food products and herbal medicines, including white rice11–14. Interestingly, a recent survey provided a background about the statistical methods the researchers have used in metabolomics-related studies15. Univariate statistic has been a common practice, especially Student t-test (91%) and analysis of variance (89%). Other methods include Mann–Whitney U test (54%), Benjamini–Hochberg false discovery rate correction (50%), and Kruskal Wallis (44%). In multivariate analysis, principal component analysis (PCA) (96%) and PLS-DA (73%) are the two most widely used methods. However, random forest (RF) was employed in only 27%. It is worth mentioning that overoptimistic and overfitting results are the common problems of the PLS-DA and the abovementioned methods, except RF, are not the preferred options for the classification study16. Besides these well-known statistical and chemometric methods, the application of sophisticated machine learning techniques in the geographical classification has also emerged in recent years17. Supervised machine learning algorithms are very powerful and they can additionally be applied to get better insights into the alteration patterns of the biological targets under specific conditions18. Maione et al. successfully employed machine learning to classify the origins of rice of different regions within a country19. The experiment was executed using 20 trace elements and the origins of the samples was predicted by support vector machines, RF, and neural network20–22. The applied models were validated using repeated 10-time 10-fold cross-validation. Although the sample size was relatively small and there was no independent validation sample, the results demonstrated the great potential of the supervised learning techniques in geographical classification of white rice. Additionally, deep learning is an advanced machine learning approach and has recently become the cutting-edge algorithm because of its extraordinary performance of the prediction accuracy in many fields23–28. The good profile and advancement of deep learning encourage us to utilize this approach for the geographical classification of commercial white rice.
In the current paper, we developed a method for rapid, accurate, and reproducible discrimination of the geographical origins of white rice from different countries. Since the generalization of the results is crucially important in class prediction study, we have collected a large number of white rice samples belonging to 30 different cultivars (11 from Korea and 19 from China). In addition, white rice cultivated in two different years, 2014 and 2015, were collected in three different time points. Sixty representative samples of white rice cultivated from 2014 was collected in 2015. White rice cultivated from 2015 was collected in April (40 representative samples) and August 2016 (26 representative samples). Moreover, our recent developed method for simultaneous profiling of 17 prominant lysoGPLs in white rice using direct infusion-electrospray ionization-multiple reaction monitoring-mass spectrometry (DI-ESI-MRM-MS) was applied in this study10. This significantly reduced the time required to analyze data for the classification down to few minutes compared to the conventional chromatography coupled with MS methods. lysoGPL data were further processed, visualized, and analyzed using a wide range of techniques for data exploration and machine learning-based classification. Finally, the proposed prediction model from white rice cultivated in 2014 was implemented to predict the origins of the samples from two different batches of white rice cultivated in 2015. Our results indicate that the combination of DI-MRM-MS-based targeted lipidomics with the cutting-edge deep learning algorithm provides an effective framework for the authenticity and geographical origin determination of white rice.
Results and Discussion
Summary of 2014 white rice, 2015-early white rice and 2015-late white rice
A total of 126 samples belonging to 30 different cultivars were purchased in April-2015 (2014 white rice, batch 1), April-2016 (2015 white rice, batch 2), and August-2016 (2015 white rice, batch 3) at local markets. There were 60, 40, and 26 samples in batch 1, batch 2, and batch 3, respectively. The detailed information can be found in Table 1.
Table 1.
The geographical origins and the cultivars of white rice from Korea and China.
| Group | Korea | China | ||||
|---|---|---|---|---|---|---|
| Label* | Origin | Cultivar | Label* | Origin | Cultivar | |
| Batch 1 Training set | KR1 | Gyeonggi | Choochung | CN1 | Heilongjiang | Jinjingdao |
| KR2 | Gyeonggi | Samgwang | CN2 | Heilongjiang | Youjida | |
| KR3 | Gangwon | Ode | CN3 | Liaoning | Dongbeida | |
| KR4 | Jeonnam | Hopyeong | CN4 | Shandong | Dongbeida | |
| KR5 | Jeonnam | Ode | CN5 | Heilongjiang | Wuchangxiang | |
| KR6 | Jeonbuk | Shindongjin | CN6 | Liaoning | Zhenzhu | |
| KR7 | Jeonnam | Ode | CN7 | Jilin | Daohuaxiang | |
| KR8 | Gangwon | Ode | CN8 | Liaoning | Daohuaxiang | |
| KR9 | Jeonnam | Ilmi | CN9 | Heilongjiang | Zhanglixiang | |
| KR10 | Jeonnam | Ode | CN10 | Jilin | Baijinxiang | |
| KR11 | Gyeongbuk | Ilmi | CN11 | Liaoning | Zhenzhu | |
| KR12 | Jeonnam | Samgwang | CN12 | Heilongjiang | Fuxiangdao | |
| KR13 | Chungnam | Samgwang | CN13 | Liaoning | Yalujiang 7 xi | |
| KR14 | Gyeongbuk | Ilpum | CN14 | Jilin | Youjida | |
| KR15 | Chungnam | Samgwang | CN15 | Liaoning | Yalujiang 3 xi | |
| KR16 | Gyeongbuk | Senoori | CN16 | Shandong | Dongbeida | |
| KR17 | Gyeonggi | Choochung | CN17 | Heilongjiang | Zhanglixiang | |
| KR18 | Gangwon | Ode | CN18 | Heilongjiang | Yatian | |
| KR19 | Gyeonggi | Choochung | CN19 | Heilongjiang | Xuejingdao | |
| KR20 | Gangwon | Choochung | CN20 | Heilongjiang | Zhonghuahe | |
| KR21 | Jeonbuk | Shindongjin | CN21 | Heilongjiang | Wuchangda | |
| KR22 | Gyeongnam | Samgwang | CN22 | Jilin | Youjida | |
| KR23 | Chungbuk | Choochung | CN23 | Jilin | Daohuaxiang | |
| KR24 | Jeonnam | Hitomebore | CN24 | Shandong | Zhanglixiang | |
| KR25 | Jeonnam | Ilmi | CN25 | Heilongjiang | Youjida | |
| KR26 | Gyeonggi | Samgwang | CN26 | Liaoning | Daohuaxiang | |
| KR27 | Gyeonggi | Koshihikari | CN27 | Heilongjiang | Shengtaidao | |
| KR28 | Gyeongbuk | Ilmi | CN28 | Jilin | Luseda | |
| KR29 | Gyeonggi | Shindongjin | CN29 | Liaoning | Dongbeida | |
| KR30 | Gyeonggi | Jinsang | CN30 | Heilongjiang | Yueguangdaoxi | |
| Batch 2 Test set 1 | KR1 | Chungnam | Samgwang | CN1 | Jilin | Youjida |
| KR2 | Gyeongbuk | Ilpum | CN2 | Heilongjiang | Shengtaidao | |
| KR3 | Gyeonggi | Shindongjin | CN3 | Heilongjiang | Yatian | |
| KR4 | Gyeongbuk | Ilmi | CN4 | Jilin | Luseda | |
| KR5 | Gangwon | Choochung | CN5 | Jilin | Daohuaxiang | |
| KR6 | Jeonnam | Ode | CN6 | Heilongjiang | Fuxiangdao | |
| KR7 | Gyeonggi | Choochung | CN7 | Liaoning | Daohuaxiang | |
| KR8 | Gyeongbuk | Ilmi | CN8 | Heilongjiang | Xuejingdao | |
| KR9 | Gangwon | Ode | CN9 | Liaoning | Zhenzhu | |
| KR10 | Gyeonggi | Choochung | CN10 | Jilin | Daohuaxiang | |
| KR11 | Gyeongnam | Samgwang | CN11 | Jilin | Youjida | |
| KR12 | Jeonnam | Ilmi | CN12 | Shandong | Zhanglixiang | |
| KR13 | Gangwon | Ode | CN13 | Heilongjiang | Zhanglixiang | |
| KR14 | Gyeonggi | Jinsang | CN14 | Liaoning | Zhenzhu | |
| KR15 | Jeonnam | Hopyeong | CN15 | Liaoning | Yalujiang 7 xi | |
| KR16 | Jeonnam | Hitomebore | CN16 | Heilongjiang | Wuchangda | |
| KR17 | Chungnam | Samgwang | CN17 | Liaoning | Dongbeida | |
| KR18 | Chungbuk | Choochung | CN18 | Heilongjiang | Zhanglixiang | |
| KR19 | Jeonbuk | Shindongjin | CN19 | Liaoning | Dongbeida | |
| KR20 | Gyeonggi | Koshihikari | CN20 | Jilin | Baijinxiang | |
| Batch 3 Test set 2 | KR1 | Gyeonggi | Jinsang | CN1 | Heilongjiang | Fuxiangdao |
| KR2 | Gyeonggi | Choochung | CN2 | Liaoning | Yalujiang 7 xi | |
| KR3 | Gyeonggi | Koshihikari | CN3 | Heilongjiang | Dongbeida | |
| KR4 | Gyeongnam | Samgwang | CN4 | Jilin | Luseda | |
| KR5 | Chungnam | Samgwang | CN5 | Liaoning | Shengtai | |
| KR6 | Jeonbuk | Shindongjin | CN6 | Jilin | Baijinxiang | |
| KR7 | Gangwon | Ode | CN7 | Heilongjiang | Daohuaxiang | |
| KR8 | Gangwon | Ode | CN8 | Heilongjiang | Fuxiangdao | |
| KR9 | Gyeonggi | Choochung | CN9 | Jilin | Baijinxiang | |
| KR10 | Jeonbuk | Shindongjin | CN10 | Heilongjiang | Dongbeida | |
| KR11 | Gyeonggi | Choochung | CN11 | Jilin | Yatian | |
| KR12 | Gyeongbuk | Samgwang | CN12 | Heilongjiang | Daohuaxiang | |
| KR13 | Jeonnam | Ilmi | CN13 | Liaoning | Zhenzhu | |
Some cultivars were purchased at the same province. However, they were cultivated in different areas and processed by different companies.
In general, the geographical classification of white rice from different countries is difficult because there are many factors such as water, temperature, light, ion, nutrient, and reactive oxygen species that greatly affect the reproducibility of the results29. The cultivation and harvest time (within-year or different years), the diversity of white rice cultivars (genetically modified or not), and storage conditions are also particularly significant. From the practice aspect, the influence of the quality of the sample preparation and data gathering methods are remarkable. In this study, we developed an experimental design that aimed to partially overcome the abovementioned difficulties and to achieve the results with generalization. Indeed, we collected white rice that was cultivated in different years (2014 white rice and 2015 white rice), white rice that was cultivated in the same year but the farming season and storage period were different (early 2015 white rice and late 2015 white rice). The sample collection was performed with the intention to maximize the heterogeneity of the samples by sampling many cultivars or white rice with different within-country origins. Finally, it is also worth pointing out that lysoGPLs profiling of white rice were conducted in three different periods.
Characterization of lysoGPLs in white rice
Although the quantity of PLs is much lower than other compounds in white rice, nutritional impact of PLs has been regconized30. Furthermore, lysoGPLs, a member of PLs, has an important role in determining rice quality. lysoPCs and lysoPEs are two major types of lysoGPLs in white rice and lysoPEs are particularly vulnerable to environmental changes. lysoPGs, however, just occupy a very small quantity in rice endosperm9. The existent of other lysoGPLs such as lysophosphatidylinositol (lysoPIs), lysophosphatidylserine (lysoPSs), and lysophosphatidic acid (lysoPAs) are as-yet unknown. Our investigation aimed to characterize six classes of lysoGPLs in commercial white rice, including lysoPCs, lysoPEs, lysoPGs, lysoPIs, lysoPSs, and lysoPAs. However, only 17 lysoGPLs of lysoPC (6 species), lysoPE (7 species), and lysoPG (4 species) were capable to be detected10. Moreover, the divergence of the lysoGPLs in white rice samples originating from different countries was described. The study implemented DI-MRM-MS, which substantially reduces the quantity of samples and the analysis time yet yields valuable data. Therefore, 17 lysoGPLs were initially profiled in this study in search for an effective classification model to discriminate white rice between Korea and China.
lysoGPLs variation of white rice from different countries: data exploration and visualization
The density plots in Fig. 1 show the distribution of the intensities of 17 lysoGPLs in white rice originated from Korea and China of batch 1. The density plots of two batches of 2015 white rice are provided in Figure S1. In general, the relative differences in terms of the concentrations of 17 species among samples between two countries were small. Among three batches of samples, the concentrations of lysoPCs were higher in white rice from Korea. In contrary, the concentrations of lysoPEs were elevated in white rice from China. lysoPGs were likely enriched in Korean group, however, the results were not consistent. The fold change, P-value, and FDR of 17 lysoGPLs among three batches of samples can be found in Table 2. In 2014 white rice, the concentrations of 14 species were statistically significant differences, except lysoPC(14:0) and lysoPG(14:0), and lysoPG(18:2). Similarly, the concentrations of 12 species were statistically significant differences, except lysoPC(16:1), lysoPE(14:0), lysoPG(14:0), lysoPG(18:1), and lysoPG(18:2) in 2015-early white rice. Finally, the concentrations of 13 species were statistically significant differences, except lysoPC(16:0), lysoPC(16:1), lysoPC(18:2), and lysoPE(18:0), in 2015-late white rice. Noticeably, the values of fold changes were relative small and there was no big difference between two groups (with the criterion of 2). Collectively, these results suggested a slight deviation in terms of the lysoGPLs concentrations of white rice and this is likely results from the heterogeneity of many affecting factors, such as cultivation year and storage conditions.
Figure 1.
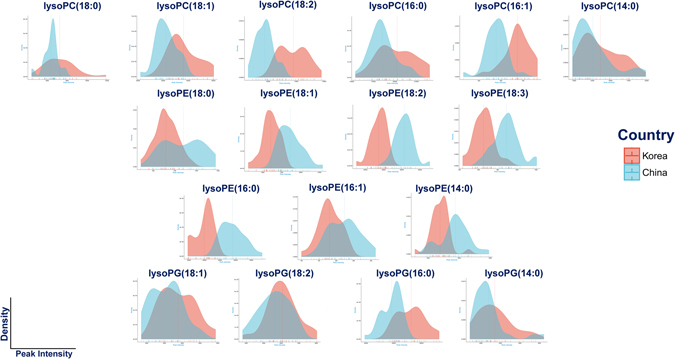
Density plots of 17 lysGPLs of 2014 white rice from Korea and China. lysoPCs are enriched in white rice from Korea while lysoPEs are prominent in white rice from China.
Table 2.
The fold change, P-value, and FDR of 17 lysoGPLs among three different batches of samples.
| Group | Compound | Fold change (K/C) | P-value | FDR |
|---|---|---|---|---|
| Batch 1 | lysoPC(14:0) | 1.08 | 8.78E-2 | 8.78E-2 |
| lysoPC(16:0) | 1.08 | 1.44E-3 | 2.04E-3 | |
| lysoPC(16:1) | 1.17 | 8.68E-8 | 2.46E-7 | |
| lysoPC(18:0) | 1.14 | 1.91E-2 | 2.32E-2 | |
| lysoPC(18:1) | 1.22 | 2.91E-6 | 6.19E-6 | |
| lysoPC(18:2) | 1.17 | 2.49E-13 | 1.41E-12 | |
| lysoPE(14:0) | 0.74 | 1.17E-5 | 2.20E-5 | |
| lysoPE(16:0) | 0.69 | 1.18E-16 | 2.01E-15 | |
| lysoPE(16:1) | 0.81 | 1.58E-4 | 2.68E-4 | |
| lysoPE(18:0) | 0.76 | 9.26E-4 | 1.43E-3 | |
| lysoPE(18:1) | 0.70 | 3.09E-10 | 1.31E-9 | |
| lysoPE(18:2) | 0.66 | 3.21E-16 | 2.73E-15 | |
| lysoPE(18:3) | 0.65 | 1.43E-8 | 4.86E-8 | |
| lysoPG(14:0) | 1.07 | 8.10E-2 | 8.61E-2 | |
| lysoPG(16:0) | 1.24 | 4.22E-7 | 1.02E-6 | |
| lysoPG(18:1) | 1.13 | 8.68E-3 | 1.13E-2 | |
| lysoPG(18:2) | 1.08 | 6.68E-2 | 7.56E-2 | |
| Batch 2 | lysoPC(14:0) | 1.50 | 2.32E-8 | 9.85E-8 |
| lysoPC(16:0) | 1.09 | 2.88E-6 | 5.45E-6 | |
| lysoPC(16:1) | 1.08 | 4.82E-1 | 5.12E-1 | |
| lysoPC(18:0) | 1.27 | 4.07E-7 | 1.10E-6 | |
| lysoPC(18:1) | 1.30 | 7.37E-9 | 4.18E-8 | |
| lysoPC(18:2) | 1.17 | 2.02E-9 | 1.71E-8 | |
| lysoPE(14:0) | 0.91 | 2.91E-1 | 3.30E-1 | |
| lysoPE(16:0) | 0.71 | 4.53E-7 | 1.10E-6 | |
| lysoPE(16:1) | 0.89 | 4.52E-2 | 6.41E-2 | |
| lysoPE(18:0) | 0.79 | 3.75E-4 | 6.37E-4 | |
| lysoPE(18:1) | 0.75 | 9.25E-7 | 1.97E-6 | |
| lysoPE(18:2) | 0.69 | 1.45E-11 | 2.47E-10 | |
| lysoPE(18:3) | 0.71 | 2.96E-7 | 1.01E-6 | |
| lysoPG(14:0) | 1.09 | 6.34E-2 | 8.30E-2 | |
| lysoPG(16:0) | 1.08 | 1.23E-2 | 1.91E-2 | |
| lysoPG(18:1) | 0.98 | 6.65E-1 | 6.65E-1 | |
| lysoPG(18:2) | 0.93 | 7.18E-2 | 8.72E-2 | |
| Batch 3 | lysoPC(14:0) | 1.13 | 1.02E-2 | 1.44E-2 |
| lysoPC(16:0) | 1.07 | 8.11E-2 | 9.20E-2 | |
| lysoPC(16:1) | 1.04 | 4.88E-1 | 5.19E-1 | |
| lysoPC(18:0) | 1.38 | 2.67E-5 | 9.09E-5 | |
| lysoPC(18:1) | 1.23 | 1.30E-4 | 3.69E-4 | |
| lysoPC(18:2) | 1.00 | 9.60E-1 | 9.60E-1 | |
| lysoPE(14:0) | 0.62 | 4.93E-4 | 1.05E-3 | |
| lysoPE(16:0) | 0.66 | 1.92E-7 | 1.09E-6 | |
| lysoPE(16:1) | 0.79 | 1.96E-2 | 2.56E-2 | |
| lysoPE(18:0) | 0.86 | 7.26E-2 | 8.82E-2 | |
| lysoPE(18:1) | 0.78 | 6.28E-4 | 1.19E-3 | |
| lysoPE(18:2) | 0.63 | 1.92E-7 | 1.09E-6 | |
| lysoPE(18:3) | 0.62 | 1.25E-3 | 2.08E-3 | |
| lysoPG(14:0) | 1.31 | 1.35E-3 | 2.08E-3 | |
| lysoPG(16:0) | 1.52 | 1.92E-7 | 1.09E-6 | |
| lysoPG(18:1) | 1.55 | 5.77E-6 | 2.45E-5 | |
| lysoPG(18:2) | 1.32 | 4.93E-4 | 1.05E-3 |
Univariate analysis does not consider the correlations among features, thus, we further conducted unsupervised multivariate exploratory data analyses to get better insights into our data sets31. PAM cluster analysis was first applied to observe the dissimilarity of the samples of three data sets. This algorithm is preffered because it is robust to outliers32. Unexpectedly, many samples that belonged to Korean group were clustered together with Chinese group (Fig. 2a) in 2014 white rice. In other two batches of samples from 2015 white rice, this unsupervised analysis showed a similar clustered tendency, however, with a lower degree since some samples of Korean group were clustered together with the samples from Chinese group (Fig. 2b and c). PCA, a data reduction unsupervised method, was conducted to explore the patterns of difference between white rice from Korea and China. As shown in Fig. 2d, a partly overlap (95% confident interval (CI)) between two groups was observed (PC1 + PC2 = 60.2%). Significantly, lysoPCs were shown to be important in Korean group while lysoPEs were prominent in Chinese group. Similar trends were also observed in two batches of 2015 white rice (Fig. 2e and f). Heatmap was also applied to get the intuitive visualization of our data sets. As shown in Fig. 3, the stronger colors focused on the lysoPEs and lysoPCs of Chinese groups and Korean groups, respectively. In general, there was no feature with unusually extremely colors in the three data sets. Collectively, the univariate analysis and multivariate unsupervised data exploration revealed that there was an overlap in some degree of white rice originated from two countries and cultivated in different years. The observation also implied that the geographical classification of white rice might be difficult for conventional methods. Consequenly, sophicated classification algorithms are more proper for this task.
Figure 2.
PAM and PCA analyses for data exploration. (a–c) Show two clusters of PAM of 2014 white rice, 2015-early white rice, and 2015-late white rice, respectively. (d–f) Show PCA biplots of 2014 white rice, 2015-early white rice, and 2015-late white rice, respectively. (a) 1–30: white rice from Korea, 31–60: white rice from China. (b) 1–20: white rice from Korea, 21:40: white rice from China. (c) 1–13: white rice from Korea, 14–26: white rice from China.
Figure 3.
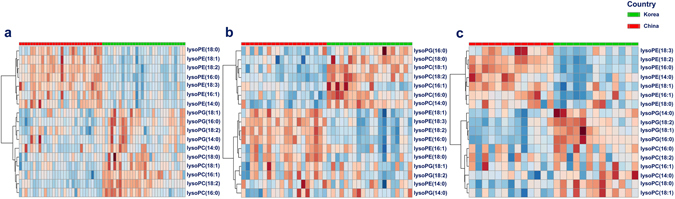
Heatmaps show the relative difference of concentrations of 17 lysoGPLs of (a) 2014 white rice, (b) 2015-early white rice, and (c) 2015-late white rice, respectively.
Development and validation of white rice geographical classification
Highly correlated variables, which include lysoPG(14:0), lysoPE(18:1), lysoPC(18:1), lysoPE(18:0), lysoPG(18:2), lysoPE(16:1), and lysoPG(18:1) were removed from the data sets. The correlation matrix can be seen in Figure S2. The 10 remaining predictors with a two-class label of 2014 white rice data set was finally used to train the deep learning model for geographical classification of white rice. The model was trained with an input layer, four hidden layers (200 neurons/layer), and an output layer. The iteration (epochs) of 10 was set. A five-fold cross-validation was applied to estimate the prediction performance of the model in the training set. We used the adaptive learning rate algorithm, as recommended by H2O. There are several regularization method options. Among them, dropout is currently the method of choice to prevent overfitting33. When select dropout regularization, random neurons in hidden layers will be excluded during the training process to prohibit the dependencies that might occur34. Thus, the rectified activation function with dropout (the dropout ratio – 0.5) was selected in this study. Early stopping was applied with the stopping metric – log loss, stopping tolerance – 0.001, and stopping rounds – 5. The variable importance was extracted from the prediction model. A seed number was set to get the reproducible results. Other parameters were kept as default.
The trained prediction model was then applied to predict the class of unseen samples from two batches of 2015 white rice. The two batches are different in terms of the collection time (April and August 2016). The results were surprisingly encouraging. As shown in Table 3, the RMSE and log loss values of three different classification analyses were small. For instane, the RMSE of the training set, test set 1, and test set 2 were 0.45, 0.54, and 0.46, respectively. Similarly, log los values of the training set, test set 1, and test set 2 were 0.55, 0.83, and 0.59, respectively. There was no class error so the MCE of classification analyses was 0 in three data sets. Furthermore, AUC, Gini, accuracy, sen, spec, TPR, and TNR were as the highest level (1.00). Look at the variable importance (Fig. 4), 10 predictors contribute significantly to the deep learning model. However, lysoPC(16:0) tended to be the least important predictor. The top three predictors were lysoPC(18:2), lysoPC(14:0), and lysoPE(16:0). Last, but not least, we were aware of the architecture of the above settings, which might be more complicated than needed. For example, the number of the layers could be decreased down to two, each with 200 neurons. Of note, we are free to tune the model using the training set as long as the tuned model is capable to predict the origins of the samples correctly. Nevertheless, the act of “training on the test set” should be avoided. The three data sets and corresponding R commands for deep learning classification are provided in Spreadsheet S1.
Table 3.
The performance of the deep learning prediction model on training and test sets.
| Data set | Total samples | RMSE | log loss | MCE | AUC | Gini | Accuracy | Sensitivity | Specitivity | TPV | TNV |
|---|---|---|---|---|---|---|---|---|---|---|---|
| White rice 2014 (Training set) | 60 | 0.45 | 0.55 | 0.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| White rice 2015 (A) (Test set 1) | 40 | 0.54 | 0.83 | 0.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| White rice 2015 (B) (Test set 2) | 26 | 0.46 | 0.59 | 0.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
RMSE: Root mean squared error.
LogLoss: Logarithmic loss.
MCE: Mean per-class error.
AUC: Area under the ROC curve.
Gini: Gini coefficient.
TPR: True positive rate.
TNR: True negative rate.
Figure 4.
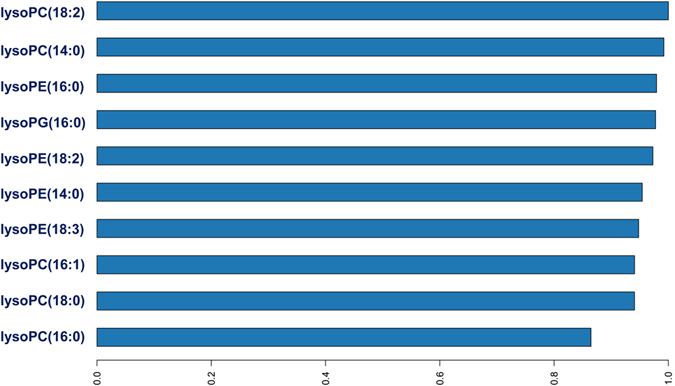
Variable importance plot of the optimal deep learning model. Top three predictors are lysoPC(18:2), lysoPC(14:0), and lysoPE(16:0).
Next, we examined the geographical classification of the RF model with the settings of followings: the number of tree in the forest (ntrees) – 1000, five-fold cross-validation, and other parameters were set at default. In addition, the parameters of the GBM for geographical classification were: ntrees – 100, five-fold cross-validation, learn rate – 0.1, stopping metric – log loss, stopping round – 5, stopping tolerance – 0.0001, score tree interval – 10. Other parameters were set as default. The results of both RF and GBM were convincing since there was only one sample from Chinese group (RF) and one sample from Korean group (GBM) of the 2015 white rice of the test set 1 were misclassified. The information of RMSE, log loss, MCE, AUC, Gini, and variable importance of the RF and GBM optimal models can be found in Figure S3. In the RF model, lysoPE(18:2), lysoPE(16:0), and lysoPC(18:2) turned out to be the top three important predictors whilst the role of lysoPC(18:0), lysoPC(14:0), and lysoPC(16:0) were insignificant. However, in the GBM model, lysoPE(18:2) and lysoPE(16:0) were the two most important features and the role of others appeared to be negligible.
Our study has several limitations. First, the sample size was relatively small due to the practical reasons. This might increase the overfitting of the classification models. However, we applied dropout and early stopping as well as external validation method using within-year and between-year samples to guarantee the regularization of the results. The sample size issue may also be solved when new white rice samples are available in the market. Second, the intended mixing ingradients of the samples between two countries were not investigated. Finally, the scope of the study was limitted to commercial white rice of Korea and China. Further investigations, therefore, are warranted to extend the utility of this approach to the real-world applications.
Conclusion
lysoGPLs can be considered as the potential features for geographical authenticity of white rice. In fact, our findings demonstrate the combination of simultaneous lysoGPL profiling method and advanced supervised learning algorithms can effectively predict the origins of the white rice. In addition to deep learning, random forest and gradient boosting machine techniques have proven to be the probable methods. In conclusion, this study suggests that machine learning algorithms possibly improve the geographical discrimination of white rice as well as other food products. Owing to the great potential of this approach, prospective studies are needed to broaden its application to a larger scale either in the coverage of geographical origins or the geographical authencity of other food products.
Materials and Methods
Materials and reagents
One hundred twenty-six white rice samples were randomly collected from local markets in Korea and China. After collection, the samples were immediately stored at −70 °C until further processed. The solvents (analytical grade), including methanol, acetonitrile, and isopropanol, were purchased from J. T. Baker (Avantor, Phillipsburg, NJ, USA). Caffeine was obtained from Sigma-Aldrich (St Louis, MO, USA). Polytetrafluoroethylene (PTFE) syringe filter (0.20 µm) was purchased from Advantec (Tokyo, Japan).
Sample preparation
White rice was freeze-dried and finely grinded to powder. The powder was then strained using two sieves with different sizes (250 µm and 125 µm) and extracted using a previously described protocol30. Concisely, 1 mg caffeine was added to 150 mg of powder samples. The mixture was extracted using 6 mL of 75% isopropanol in a water bath at 90 °C for 2 h and centrifuged at 16,000 g for 5 min. Thereafter, 1 mL of supernatant filtered by a PTFE syringe filter was transferred to a Agilent 1.5 mL screw vial (Agilent, CA, USA) for the analysis.
DI-MRM-MS analysis conditions
A triple-quadrupole mass spectrometry system (6460 QqQ LC-ESI-MS/MS, Agilent, CA, USA) was exploit to perform every experiment in order to ascertain the practical instrumental conditions. The following settings were adopted from our previously developed method10. The analysis of lysoPCs was conducted in positive ion mode. lysoPEs and lysoPGs, on the other hand, were characterized in negative ion mode. The contamination of ion source by sample injection was minimized using a constant flow of 50% acetonitrile (0.2 mL/min). The sample sequences of every experiment were set randomly to avoid possible technical bias. The mass spectrometer was following the acquisition settings: scan time −200 scans/sec, cell accelerator voltage −7 V, fragmentor voltage −135 V, nebulizer pressure −40 psi, dry gas temperature −325 °C, dry gas flow −11 L/min, and capillary −4 kV. Nitrogen was used as the collision, nebulizing, and drying gas. The system was operated at a collision energy of 20 eV for positive and negative ion modes. MRM transitions of each compound were set in accordance to the mass per charge ratios (m/z) of the highest intensity fragments of product ions. The experiment was tightly controlled and a variation criterion of 10% of relative standard deviations (RSD) in quality control (QC) samples was used to consider the quality of the analysis of targeted lipid species. Lastly, the lipid identification was confirmed using our in-house library.
Data preprocessing and univariate statistical analysis
DI-MRM-MS data were processed using Agilent Mass Hunter Workstation software version B.06.00. The peak intensities of 17 lysGPLs were normalized using peak intensities of caffeine. There were no near zero-variance and missing values in the three data sets. Density plots were used to visualize the intensity distributions of samples between two countries and Wilcoxon rank-sum test was performed to detect differentially expressed features. A P-value of <0.05 and a false discovery rate (FDR) for multiple testing of <0.1 were considered to be the level of statistical significance. The univariate analysis was performed using Metaboanalyst 3.0 and the density plot was illustrated using ggpubr 0.1.2 in R language 3.3.335–37.
Data visualization and multivariate Data Analysis
Since multivariate analysis does take the correlations among variables into account, it is considered particularly suitable for analyzing high-dimensional omics data31. In this study, partitioning around medoids clustering analysis (PAM), PCA, and heatmap analysis were applied to visualize the data and explore the tendency of separation among samples. Except heatmap analysis that was performed using metaboanalyst 3.0, other analysis and visualization techniques were performed using FactoMineR version 1.35, factoextra 1.0.4, and ggplot 2 2.2.1 in R language version 3.3.337–40.
Highly correlated predictor removal
Highly correlated predictors might affect the performance of the prediction models. Therefore, we removed all the predictors with absolute correlations of 0.70 or higher. The process was conducted using caret package 6.0–73. Correlation matrix was visualized using corrplot 0.77 package41, 42.
Deep learning classification
In this study, a feedforward deep neural network model for class prediction was established using 60 white rice samples cultivated in 2014. A five-fold cross-validation was utilized during training process as a model validation technique. The performance of the model was further validated using two independent batches of white rice cultivated in 2015. The training and testing processes were carried out using H2O package 3.10.3.6 in R language version 3.3.3. H2O provides cutting-edge machine learning algorithms and well-known regularization tools for big data analysis43. Although deep learning includes unsupervised and supervised settings, H2O provides a purely supervised learning protocol together with many innovative features that help getting the optimal prediction models in a short period. In addition, RF and gradient boosting machine (GBM), two major machine learning techniques, were additionally employed to build classification models37, 44, 45. The metrics to evaluate the model included root mean squared error (RMSE), cross-entropy loss function (log loss), mean per-class error (MCE), the area under the receiver operating characteristic (ROC) curve (AUC), and Gini coefficient (Gini) along with the prediction accuracy, sensitivity (sen), specificity (spec), true positive value (TPV), and true negative value (TNV).
Electronic supplementary material
Acknowledgements
This work was supported by the Rural Development Administration of Korea (PJ01164601), Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2016R1D1A1A02937257), and the BK21 Plus Program in 2016. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
Author Contributions
N.P.L. and S.W.K. conceived and designed the research. N.P.L. and D.K.L. carried out lipidomics experiments, from sampling to data processing. D.K.L., N.P.L., C.M., and G.K. undertook data collection and statistical analysis. N.P.L., D.K.L., and S.W.K. wrote the manuscript. All authors participated in the manuscript revision and approved the final version of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Nguyen Phuoc Long and Dong Kyu Lim contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-08892-0
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Akula R, Ravishankar GA. Influence of abiotic stress signals on secondary metabolites in plants. Plant signaling & behavior. 2011;6:1720–1731. doi: 10.4161/psb.6.11.17613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luykx DM, Van Ruth SM. An overview of analytical methods for determining the geographical origin of food products. Food Chemistry. 2008;107:897–911. doi: 10.1016/j.foodchem.2007.09.038. [DOI] [Google Scholar]
- 3.Nguyen HT, et al. A 1 H NMR-based metabolomics approach to evaluate the geographical authenticity of herbal medicine and its application in building a model effectively assessing the mixing proportion of intentional admixtures: A case study of Panax ginseng: Metabolomics for the authenticity of herbal medicine. Journal of pharmaceutical and biomedical analysis. 2016;124:120–128. doi: 10.1016/j.jpba.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 4.Vlachos A, Arvanitoyannis IS. A review of rice authenticity/adulteration methods and results. Critical reviews in food science and nutrition. 2008;48:553–598. doi: 10.1080/10408390701558175. [DOI] [PubMed] [Google Scholar]
- 5.Cheajesadagul P, Arnaudguilhem C, Shiowatana J, Siripinyanond A, Szpunar J. Discrimination of geographical origin of rice based on multi-element fingerprinting by high resolution inductively coupled plasma mass spectrometry. Food chemistry. 2013;141:3504–3509. doi: 10.1016/j.foodchem.2013.06.060. [DOI] [PubMed] [Google Scholar]
- 6.Li G, et al. Profiling the ionome of rice and its use in discriminating geographical origins at the regional scale, China. Journal of Environmental Sciences. 2013;25:144–154. doi: 10.1016/S1001-0742(12)60007-2. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalvez A, Armenta S, De La Guardia M. Trace-element composition and stable-isotope ratio for discrimination of foods with Protected Designation of Origin. TrAC Trends in Analytical Chemistry. 2009;28:1295–1311. doi: 10.1016/j.trac.2009.08.001. [DOI] [Google Scholar]
- 8.Suzuki Y, Chikaraishi Y, Ogawa NO, Ohkouchi N, Korenaga T. Geographical origin of polished rice based on multiple element and stable isotope analyses. Food Chemistry. 2008;109:470–475. doi: 10.1016/j.foodchem.2007.12.063. [DOI] [PubMed] [Google Scholar]
- 9.Liu L, Waters DL, Rose TJ, Bao J, King GJ. Phospholipids in rice: significance in grain quality and health benefits: a review. Food chemistry. 2013;139:1133–1145. doi: 10.1016/j.foodchem.2012.12.046. [DOI] [PubMed] [Google Scholar]
- 10.Lim DK, Mo C, Nguyen Phuoc L, Kim G, Kwon SW. Simultaneous profiling of lysoglycerophospholipids in rice (Oryza sativa L.) using direct infusion-tandem mass spectrometry with multiple reaction monitoring. Journal of Agricultural and Food Chemistry. 2017;65:2628–2634. doi: 10.1021/acs.jafc.7b00148. [DOI] [PubMed] [Google Scholar]
- 11.Barbosa RM, et al. A simple and practical control of the authenticity of organic sugarcane samples based on the use of machine-learning algorithms and trace elements determination by inductively coupled plasma mass spectrometry. Food chemistry. 2015;184:154–159. doi: 10.1016/j.foodchem.2015.02.146. [DOI] [PubMed] [Google Scholar]
- 12.Tahri K, Tiebe C, El Bari N, Hübert T, Bouchikhi B. Geographical provenience differentiation and adulteration detection of cumin by means of electronic sensing systems and SPME-GC-MS in combination with different chemometric approaches. Analytical Methods. 2016;8:7638–7649. doi: 10.1039/C6AY01906D. [DOI] [Google Scholar]
- 13.Kim N, et al. Metabolomic approach for age discrimination of Panax ginseng using UPLC-Q-Tof MS. Journal of agricultural and food chemistry. 2011;59:10435–10441. doi: 10.1021/jf201718r. [DOI] [PubMed] [Google Scholar]
- 14.Lo Feudo G, Naccarato A, Sindona G, Tagarelli A. Investigating the origin of tomatoes and triple concentrated tomato pastes through multielement determination by inductively coupled plasma mass spectrometry and statistical analysis. Journal of agricultural and food chemistry. 2010;58:3801–3807. doi: 10.1021/jf903868j. [DOI] [PubMed] [Google Scholar]
- 15.Weber RJ, et al. Computational tools and workflows in metabolomics: An international survey highlights the opportunity for harmonisation through Galaxy. Metabolomics. 2017;13 doi: 10.1007/s11306-016-1147-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gromski PS, et al. A tutorial review: Metabolomics and partial least squares-discriminant analysis–a marriage of convenience or a shotgun wedding. Analytica chimica acta. 2015;879:10–23. doi: 10.1016/j.aca.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 17.Chung I-M, Kim J-K, Lee J-K, Kim S-H. Discrimination of geographical origin of rice (Oryza sativa L.) by multielement analysis using inductively coupled plasma atomic emission spectroscopy and multivariate analysis. Journal of Cereal Science. 2015;65:252–259. doi: 10.1016/j.jcs.2015.08.001. [DOI] [Google Scholar]
- 18.Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends in biotechnology. 2004;22:245–252. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Maione C, Batista BL, Campiglia AD, Barbosa F, Barbosa RM. Classification of geographic origin of rice by data mining and inductively coupled plasma mass spectrometry. Computers and Electronics in Agriculture. 2016;121:101–107. doi: 10.1016/j.compag.2015.11.009. [DOI] [Google Scholar]
- 20.Burges CJ. A tutorial on support vector machines for pattern recognition. Data mining and knowledge discovery. 1998;2:121–167. doi: 10.1023/A:1009715923555. [DOI] [Google Scholar]
- 21.Cutler DR, et al. Random forests for classification in ecology. Ecology. 2007;88:2783–2792. doi: 10.1890/07-0539.1. [DOI] [PubMed] [Google Scholar]
- 22.Zhang GP. Neural networks for classification: a survey. IEEE Transactions on Systems, Man, and Cybernetics, Part C (Applications and Reviews) 2000;30:451–462. doi: 10.1109/5326.897072. [DOI] [Google Scholar]
- 23.Min, S., Lee, B. & Yoon, S. Deep learning in bioinformatics. Briefings in Bioinformatics, bbw068 (2016). [DOI] [PubMed]
- 24.Schmidhuber J. Deep learning in neural networks: An overview. Neural networks. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Narasingarao M, Manda R, Sridhar G, Madhu K, Rao A. A clinical decision support system using multilayer perceptron neural network to assess well being in diabetes. Journal of the Association of Physicians of India. 2009;57:127–133. [PubMed] [Google Scholar]
- 26.Angermueller C, Pärnamaa T, Parts L, Stegle O. Deep learning for computational biology. Molecular systems biology. 2016;12 doi: 10.15252/msb.20156651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aliper A, et al. Deep learning applications for predicting pharmacological properties of drugs and drug repurposing using transcriptomic data. Molecular pharmaceutics. 2016;13 doi: 10.1021/acs.molpharmaceut.6b00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohanty, S. P., Hughes, D. P. & Salathé, M. Using Deep Learning for Image-Based Plant Disease Detection. Frontiers in Plant Science7 (2016). [DOI] [PMC free article] [PubMed]
- 29.Obata T, Fernie AR. The use of metabolomics to dissect plant responses to abiotic stresses. Cellular and Molecular Life Sciences. 2012;69:3225–3243. doi: 10.1007/s00018-012-1091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L, et al. Determination of starch lysophospholipids in rice using liquid chromatography–mass spectrometry (LC-MS) Journal of agricultural and food chemistry. 2014;62:6600–6607. doi: 10.1021/jf500585j. [DOI] [PubMed] [Google Scholar]
- 31.Xia, J. & Wishart, D. S. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Current Protocols in Bioinformatics, 14.10. 11–14.10. 91 (2016). [DOI] [PubMed]
- 32.Lee BS, et al. A clustering method to identify who benefits most from the treatment group in clinical trials. Health Psychology and Behavioral Medicine: an Open Access Journal. 2014;2:723–734. doi: 10.1080/21642850.2014.924857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srivastava N, Hinton GE, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. Journal of Machine Learning Research. 2014;15:1929–1958. [Google Scholar]
- 34.Candel, A., Parmar, V., LeDell, E. & Arara, A. Deep Learning with H2O (2017).
- 35.Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic acids research. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots. R package version 0.1.2.999 (2017).
- 37.Team R Core. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria (2017).
- 38.Lê S, Josse J, Husson F. FactoMineR: an R package for multivariate analysis. Journal of statistical software. 2008;25:1–18. doi: 10.18637/jss.v025.i01. [DOI] [Google Scholar]
- 39.Kassambara, A. & Mundt, F. Factoextra: extract and visualize the results of multivariate data analyses. R package version 1.0.3 (2015).
- 40.Wickham H. ggplot2: elegant graphics for data analysis. Springer New York. 2009;1 [Google Scholar]
- 41.Kuhn, M. et al. Caret: Classification and Regression Training. R package version 6.0-73 (2016).
- 42.Wei, T. & Simko, V. corrplot: Visualization of a Correlation Matrix. R package version 0.77 (2016).
- 43.The H2O.ai Team. H2O: R Interface for H2O. R package version 3.10.3.6 (2017).
- 44.Checa A, Bedia C, Jaumot J. Lipidomic data analysis: tutorial, practical guidelines and applications. Analytica chimica acta. 2015;885:1–16. doi: 10.1016/j.aca.2015.02.068. [DOI] [PubMed] [Google Scholar]
- 45.Click, C., Malohlava, M., Candel, A., Roark, H. & Parmar, V. Gradient Boosting Machine with H2O (2017).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



