Abstract
We report that herpes simplex virus 1 (HSV-1) infection can activate and exploit a cellular DNA damage response that aids viral replication in nonneuronal cells. Early in HSV-1 infection, several members of the cellular DNA damage-sensing machinery are activated and accumulate at sites of viral DNA replication. When this cellular response is abrogated, formation of HSV-1 replication centers is retarded, and viral production is compromised. In neurons, HSV-1 replication centers fail to mature, and the DNA damage response is not initiated. These data suggest that the failure of neurons to mount a DNA damage response to HSV-1 may contribute to the establishment of latency.
Keywords: replication, Mre11, damage, signaling, ataxia telangiectasia mutated
Mammalian cells are equipped with complex machinery to monitor and repair damaged DNA (1–3). Central players in this pathway include the Mre11 complex, consisting of Mre11, Rad50, and Nbs1 (4), and the phosphatidylinositol 3-kinase-like kinases ataxia telangiectasia mutated (ATM) and ATM-Rad3 related (ATR), which are the signal transducers of the DNA damage response (5, 6). An early event in response to DNA double-strand breaks is activation of ATM kinase activity by dimer dissociation and intermolecular autophosphorylation on the serine 1981 residue (7). Many proteins are substrates for ATM and ATR (5), including the checkpoint kinases Chk1 and Chk2 and the DNA repair proteins 53BP1, BRCA1, and p53. In addition to detecting cellular DNA damage, we have suggested that the repair apparatus of the host cell can recognize exogenous genetic material during viral infections (8–10). We have shown that the DNA damage machinery presents an obstacle to adenovirus replication and that adenovirus disarms the cellular response by targeting the Mre11 complex to prevent genome concatemerization (8, 9). It is possible that the cellular damage machinery does not present an obstacle for all viruses and that there may be examples of viruses that exploit the host DNA repair apparatus to aid their lifecycle.
Herpes simplex virus 1 (HSV-1) is a large double-stranded DNA virus with a genome of 152 kb (11). After HSV-1 infection of nonneuronal cells, replication and subsequent death of the host cell usually occurs (11). In contrast, when HSV-1 infects sensory neurons, replication is limited, and the virus can establish latency where it is maintained for the lifetime of the host. The events governing the establishment and reactivation from latency are poorly understood, but the known stimuli for reactivation include DNA damaging agents and stress (11).
Expression from the HSV-1 genome is organized in a temporal cascade, with expression of immediate early (IE), early (E), and late (L) gene products. The IE genes are the first to be transcribed, and their products are required to coordinate the expression of the E and L genes. After infection, the incoming genome is thought to circularize, and, because this process does not depend on de novo protein synthesis from the virus, it is assumed that the end-joining is facilitated by cellular proteins. Until recently, it was generally accepted that a circular DNA molecule provided the template for HSV replication (11). This premise has recently been challenged by the proposal that the IE gene infected cell polypeptide (ICP) 0 controls the configuration of the viral genome inside the host cell and that the preferred template for replication is actually a linear molecule (12). ICP0 is a promiscuous activator of viral transcription that is required for efficient initiation of the viral lytic cascade and reactivation from latency (13). ICP0 is also an E3 ubiquitin ligase that induces degradation of the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs) and other cellular proteins associated with promyelocytic leukemia bodies (14–16).
Formation of viral replication compartments is thought to proceed by an ordered series of events, with distinct stages defined by assembly of viral protein complexes (17, 18). Seven essential HSV-1 replication proteins are necessary and sufficient for genome amplification (11). Prereplicative sites form in infected nuclei, with accumulation of the origin binding protein (UL9), the single-stranded DNA binding protein (ICP8, the product of the UL29 gene), and the helicase–primase heterotrimer (UL5/UL8/UL52) (18). After recruitment of the viral polymerase holoenzyme (UL30/UL42), a subset of these prereplicative sites matures to form viral replication compartments. Cellular factors including p53, PCNA, Rb, RPA, Nbs1, and Rad51 accumulate at viral replication centers (17, 19, 20), but the significance of these observations has remained unclear.
In this study, we explored whether a cellular DNA damage response is triggered during HSV-1 infection and what effect the response would have on the virus lifecycle. We identified key cellular proteins that are activated by HSV-1 and then investigated the consequences of mutating these proteins. We observed that activation of the DNA damage response is beneficial for viral replication but that this cellular response is abrogated in neuronal cells. These results suggest that cellular DNA damage proteins may be involved in controlling viral latency.
Materials and Methods
Cell Lines. HeLa, Vero, and IMR90 cells were purchased from the American Type Culture Collection. Ataxia telangiectasia-like disorder (A-TLD1) cells were obtained from J. Petrini (Memorial Sloan–Kettering Cancer Center, New York). Immortalized A-TLD1 cells and their complemented counterpart and the cells expressing E1b55K proteins are described in ref. 8. The ataxia telangiectasia (A-T) cell lines AT02052B, ATGM5849, and ATGM09607B were obtained from the Coriell Institute (Camden, NJ). The matched pair of transformed A-T cells was obtained from Y. Shiloh (Tel Aviv University, Tel Aviv). The ATR-inducible cell lines were obtained from P. Nghiem (Massachusetts General Hospital, Charlestown) and were induced with 1 μg/ml doxycycline for 48 h before infection (8). Cells were maintained as monolayers in DMEM supplemented with 10% or 20% FBS at 37°C in a humidified atmosphere containing 5% CO2.
Human Embryonic Stem Cells (hESCs). Nondifferentiated Cyth25 cells (CyThera, San Diego) were cultured on mitotically inactivated mouse embryonic fibroblasts (Specialty Media, Lavellette, NJ) in DMEM/F12 glutamax (GIBCO)/20% KnockOut Serum Replacement (GIBCO)/0.1 mM nonessential amino acids (GIBCO)/0.1 mM 2-mercaptoethanol (GIBCO)/4 ng/ml basic fibroblast growth factor-2 (bFGF-2) (R & D Systems). Cells were induced for neuronal differentiation by plating on a feeder layer of the mouse stromal cell line PA6 (21) in differentiation media (as above but with 10% KnockOut Serum Replacement and no bFGF-2).
Viruses. HSV-1 strain 17syn+ was used as WT in all experiments (22). The GFP-expressing virus 17+GFP contains a CMV GFP cassette inserted into US5. The IE gene-deficient virus 1764 27–4-GFP contains an inactivation in the transactivation domain of VP16 (23), complete deletions of the coding regions for ICP27 and ICP4, and a reporter gene cassette inserted into the virion host shutoff gene (vhs) (24). These viruses and the cell lines for propagation were obtained from R. Coffin (University College London, London) (24, 25). The fHSVΔpac (26) and GFP bacterial artificial chromosomes (BAC) (27) were obtained from Y. Saeki (Ohio State University Medical Center, Columbus), and the GFP amplicon was prepared by using an ICP27-deleted helper BAC (28). The ICP0 mutant virus n212 was obtained from P. Schaffer (Beth Israel Deaconess Medical Center, Boston) (29). The E1-deleted recombinant adenoviruses expressing E4orf6 and E1b55K were obtained from P. Branton (McGill University, Montreal) and propagated in 293 cells. The E4-deleted adenovirus mutant dl1004 was obtained from G. Ketner (Johns Hopkins University, Baltimore) and propagated on an E4-complementing cell line (30).
Antibodies. Viral replication centers were visualized by staining for ICP8 with either mouse monoclonal antibody (D. Knipe, Harvard Medical School, Boston) or rabbit polyclonal antisera (W. Ruyechan, University at Buffalo, Buffalo, NY). Primary antibodies were purchased from Novus Biologicals (Littleton, CO) (Nbs1, Nbs1-S343-P, and ATM), Genetex (San Antonio, TX) (Mre11 and Rad50), Santa Cruz Biotechnology (Ku70, Ku86, and DNA-PKcs), Upstate Biotechnology (Lake Placid, NY) (Chk2), Cell Signaling Technology (Beverly, MA) (Chk2-T68 and p53-S15), Covance (Princeton, NJ) (neuronal β-tubulin Tuj1), Pharmingen (neuronal MAP2a+b), and Rockland (Gilbertsville, PA) (ATM S1981-P). Antibody to phosphorylated 53BP1 was from P. Carpenter (University of Texas, Houston). Antibody to ICP0 was from R. Everett (MRC Virology Unit, Glasgow, Scotland). All secondary antibodies were from The Jackson Laboratory.
Infections, Drug Treatments, and Growth Curves. Infections were performed on monolayers of cultured cells in serum-free media. After 1 h at 37°C, monolayers were overlaid with DMEM containing 10% (standard lines) or 20% (primary cells) serum. Phosphonoacetic acid (PAA) was used at a final concentration of 400 μg/ml (31). These drugs were added with the virus and left in for the duration of infection. Caffeine was used at a final concentration of 5 mM for 1 h before virus infection and throughout the infection. Growth curves were carried out at a multiplicity of infection (moi) of 0.1, and samples were harvested for viral yield determination by plaque assay on Vero cells.
Quantitative Real-Time PCR. To determine genomic titer of a stock of GFP-expressing virus, capsids were denatured with 2M NaOH at 65°C for 30 min and neutralized with 2M HCl (32). Viral DNA was quantified by using the GFP-specific primers 5′-ATGGCCGACAAGCAGAAGAA and 5′-GCTGCCGTCCTCGATGTT. Amplifying sequences were detected by using SYBR green reporter dye in a Prism 7700 sequence detection system (PerkinElmer/Applied Biosystems). For experiments to determine amplification of viral DNA over time (Fig. 4D), cells were infected with a replication-competent virus expressing GFP (17+ GFP), and total DNA was extracted at various times postinfection (DNeasy kit, Qiagen, Valencia, CA). Viral DNA was amplified by using the GFP-specific primers 5′-AGTCCGCCCTGAGCAAAGA and 5′-TCACGAACTCCAGCAGGACC and detected by using the tetramethylrhodamine (TAMRA)-conjugated GFP-specific probe 5′-CCCAACGAGAAGCGCGATCACA. To determine viral genomes per cell, cellular DNA was amplified by using the human telomerase reverse transcriptase (hTERT)-specific primers 5′-GAGCTGCTCAGGTCTTTCTTTTATG and 5′-TTTGCAACTTGCTCCAGACACT and detected by using the TAMRA-conjugated hTERT-specific probe 5′-CCACGGAGACCACGTTTCAAAAGAACAGG.
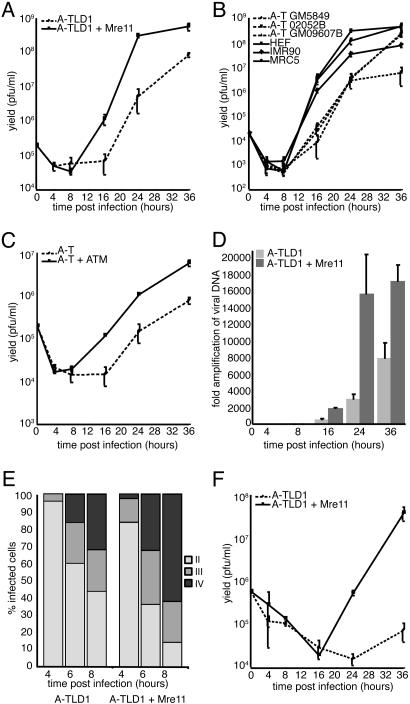
Fig. 4.
HSV-1 replication is impaired in the absence of a cellular DNA damage response. (A–C) Cells were infected at a moi of 0.1 and harvested at various times postinfection for determination of viral yield by plaque assay on Vero cells. All infections were carried out at least in duplicate. Growth curves were performed in mutant and complemented A-TLD1 (A), mutant A-T and WT (B), and mutant and complemented A-T (C) cells. (D) Effects of Mre11 on HSV-1 genome replication were assessed by using quantitative PCR (qPCR). A-TLD1 cells or A-TLD1 cells complemented with Mre11 were infected at a moi of 0.1, and DNA was extracted at various times postinfection for qPCR analysis to detect viral and cellular DNA. The number of viral genomes at 4 hpi indicated equal infection of the different cell lines (data not shown). The data were normalized to input and are presented as fold-amplification of viral genomes over time. (E) Formation of ICP8 foci was assessed by immunofluorescence in A-TLD1 cells. A-TLD1 cells or A-TLD1 cells complemented with Mre11 were infected with HSV-1 WT at a moi of 5. Cells were stained with an ICP8 antibody at 4, 6, or 8 hpi. The relative abundance of different stages of infection was quantified by counting 200 infected cells per condition and categorizing them as diffuse (stage II), punctate (stage III), or mature replication centers (stage IV), as described in the text and Fig. 10C. (F) Growth of an ICP0-null virus was assessed in A-TLD1 compared with Mre11-complemented cells.
Immunoblotting and Immunofluorescence. Infected cells were harvested after two washes in ice-cold PBS, and whole-cell lysates were prepared. Protein concentration was estimated by Lowry assay (Bio-Rad), and 30 μg of protein was loaded per well. Western blotting was carried out by standard protocols. For immunofluorescence, cells were grown on glass coverslips and infected at a moi of 1. Cells were washed with PBS and fixed at –20°C for 30 min with ice-cold methanol/acetone (1:1) or 3% paraformaldehyde for 20 min and extracted with 0.5% Triton X-100 in PBS for 10 min. For immunofluorescence with the Nbs1 antibody, the cells were washed and permeablized before paraformaldehyde fixation and extraction by a 7-min incubation in 0.5% Triton X-100/20 mM Hepes-KOH, pH 7.9/50 mM NaCl/3 mM MgCl2/300 mM sucrose. Immunoreactivity was visualized by using a Nikon Diaphot 300 or a Radiance 2100 confocal microscope (Bio-Rad). The confocal data were obtained by using a krypton/argon laser at the following excitation/emission wavelengths (in nm): FITC, 488 and 515/30; Cy3, 568 and 600/50; Cy5, 647 and 660/LP.
Results and Discussion
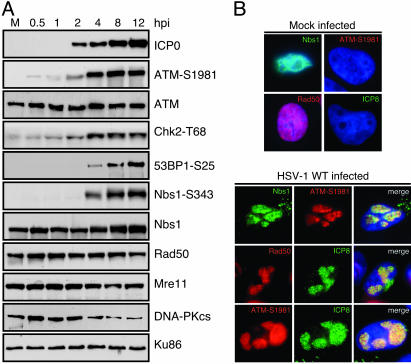
HSV-1 Infection Activates a Cellular DNA Damage Response. We investigated whether HSV-1 infection activated signaling cascades characteristic of the cellular DNA damage response. The effect of HSV-1 was examined by immunoblotting of lysates over a time course of infection using antibodies that recognize cellular proteins and specific phosphorylated residues known to be modified upon DNA damage (Fig. 1A). We observed activation of ATM as indicated by its autophosphorylation at S1981 (7), as well as phosphorylation of Chk2, 53BP1, and Nbs1. (Fig. 1 A). In contrast to adenovirus infection where the Mre11 complex is degraded (9), HSV-1 infection did not significantly alter the steady-state levels of the Mre11/Rad50/Nbs1 proteins or the ATM kinase. By 4 h postinfection (hpi), we detected down-regulation of DNA-PKcs (14, 15). Activation of ATM appeared to be an early event that preceded signaling to other proteins and expression of ICP0. Similar results were observed in a variety of cell types, including the primary IMR90 cell line (data not shown). Together, these results reveal that infection with WT HSV-1 results in ATM activation and signaling to proteins involved in the cellular DNA damage response.
Fig. 1.
HSV-1 infection activates a cellular DNA damage response. (A) HeLa cells were infected with HSV-1 at a moi of 5, and lysates were prepared for immunoblotting over a time course of infection. Autophosphorylation of ATM could be detected as early as 0.5 hpi. Signaling to downstream targets was observed by using the indicated phospho-specific antibodies. Degradation of DNA-PKcs was detected after 4 hpi. Ku86 served as a loading control. (B) Activation of ATM and accumulation of DNA repair proteins at HSV-1 replication centers was observed by immunofluorescence. Cells were infected at a moi of 1 and examined at 8 hpi.
We used immunofluorescence to examine the localization of DNA repair factors in infected cells (Fig. 1B). The Nbs1 and Rad50 proteins, as well as the autophosphorylated form of ATM, accumulated at sites of viral replication that costained with ICP8. We also observed accumulation of phosphorylated Nbs1 (data not shown). Reorganization of DNA damage proteins was an early event that could be seen as soon as the smallest ICP8 foci became detectable (Fig. 6, which is published as supporting information on the PNAS web site).
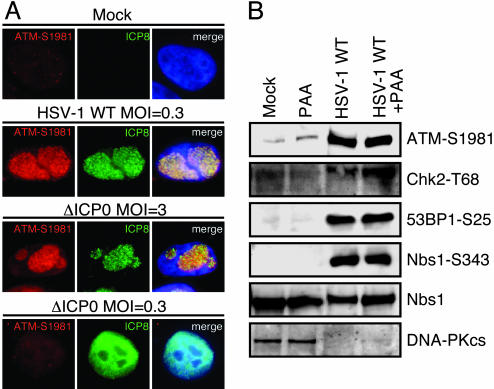
Requirements for the DNA Damage Response to HSV-1. We used mutant viruses and drug treatments to study the involvement of viral components or stages in the viral lifecycle in activation of the cellular damage response. No damage response was observed after infection with HSV-1 amplicon vectors that do not express any viral genes or with mutant virus unable to express significant amounts of any of the HSV-1 genes (Fig. 7, which is published as supporting information on the PNAS web site), indicating that neither the virion nor the incoming viral genome are sufficient to activate the response. In contrast, when cells were transfected with a bacterial artificial chromosome expressing all of the HSV-1 genes, activation of ATM was detected (Fig. 7), suggesting a requirement for expression of viral gene product(s). ICP0 is one of the most important IE genes expressed by HSV-1. Although ICP0 expression by plasmid transfection led to degradation of the promyelocytic leukemia protein, it did not appear to induce degradation or significant global redistribution of the Mre11 complex and did not activate the DNA damage response (Fig. 8, which is published as supporting information on the PNAS web site). ICP0-deficient mutant virus was still capable of causing ATM autophosphorylation (Fig. 2A), indicating that ICP0 is not required for the cellular response. The damage response to HSV-1 was also intact when degradation of DNA-PKcs was prevented by addition of proteasome inhibitors (data not shown). It is known that ICP0-null viruses display a moi dependency for replication (13). At high mois, they grow as WT, but at low mois, they resemble quiescent genomes in that replication is impaired and the viral genomes can be stably maintained until they are reactivated by exogenous ICP0. Therefore, it was interesting that infection with an ICP0-negative virus at low moi (<1 plaque-forming unit per cell) resulted in diffuse nuclear ICP8 staining and no ATM phosphorylation (Fig. 2 A). This finding raises the intriguing possibility that a damage response is not activated in situations that resemble latency.
Fig. 2.
The cellular response does not require ICP0 expression or replication by the viral polymerase. (A) HeLa cells were infected at the mois indicated with either WT or an ICP0-null virus (n212). At 16 hpi, the cells were fixed and assessed for ICP8 and phosphorylated ATM by immunofluorescence. (B) IMR90 cells were infected with WT virus at a moi of 10 in the presence of 400 μg/ml PAA where indicated. Samples were harvested at 16 hpi, and induction of a DNA damage response was assessed by immunoblotting of cell lysates.
The damage response to HSV-1 could still be detected in the presence of PAA, a specific inhibitor of the viral polymerase, suggesting that viral DNA replication mediated by the viral polymerase is not essential (Fig. 2B). However, we cannot rule out the possibility that replication of viral genetic material mediated by cellular DNA polymerases may be a contributing factor to the damage response we have observed. Interestingly, we observed a viral load dose dependence to the outcome of PAA addition, and at low moi, some effect of the drug was observed (Fig. 7). This observation suggests that there is a threshold below which the damage response is not activated or cannot be detected but that at a high moi, the input virus is sufficient to induce a response even in the absence of amplification by the viral polymerase. Infection with replication-incompetent viruses deleted individually for members of the viral helicase–primase complex results in diffuse ICP8 staining (18), and no ATM activation was observed with these mutants (data not shown). Together, these data support the hypothesis that activation of the cellular damage response to HSV-1 infection requires formation of prereplicative structures and recruitment of cellular repair factors without an absolute requirement for viral DNA replication by the viral polymerase.
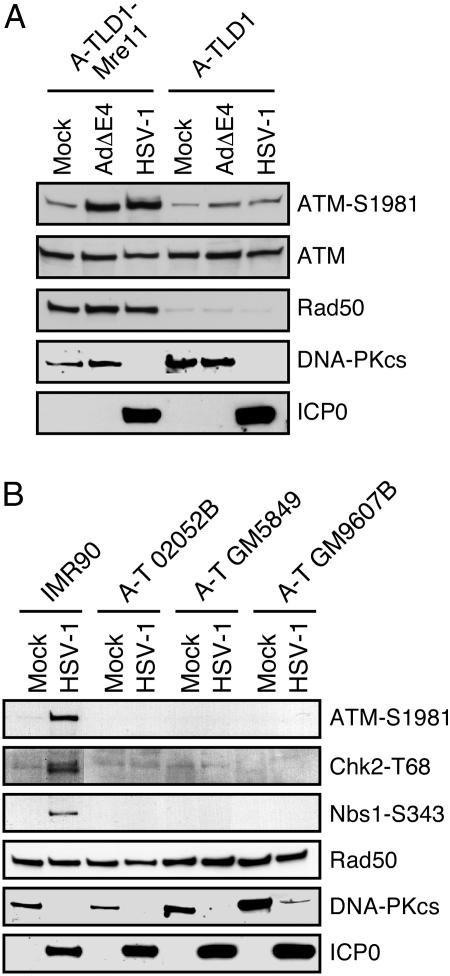
The cellular requirements for activation of the damage response were examined by using mutant cell lines. To determine whether the Mre11 complex was involved, we performed infections in the A-TLD1 patient line that expresses a truncated, hypomorphic form of the Mre11 protein (33). An E4-deleted adenovirus (which activates a damage response only in the presence of Mre11) was included as a control, and infections were compared with A-TLD1 cells that were complemented with a cDNA expressing the wild-type Mre11 protein (8). Autophosphorylation of ATM was minimal in A-TLD1 cells but was robust in the Mre11-complemented cells (Fig. 3A). Infections in three different cell lines from patients with A-T demonstrated that signaling to Chk2 and Nbs1 required the ATM kinase (Fig. 3B). We also found that ATM autophosphorylation and signaling to downstream targets was abrogated in cells in which degradation of the Mre11 complex was induced by expression of E1b55K/E4orf6 adenoviral proteins either by coinfections (Fig. 9A, which is published as supporting information on the PNAS web site) or in controlled cell lines that either can or cannot degrade the Mre11 complex (Fig. 9B). Together, these data demonstrate that the Mre11 complex is required for full activation of ATM in response to HSV-1 infection.
Fig. 3.
The DNA damage response to HSV-1 is mediated by Mre11 and ATM. Cells were infected at a moi of 5 and harvested at 16 hpi. (A) ATM activation and signaling were detected in A-TLD1 cells complemented with Mre11 but not in Mre11-deficient A-TLD1 cells. (B) ATM activation and signaling were detected in IMR90 cells but were absent in three different A-T cell lines.
Viral Replication and Growth Are Greater in the Presence of the DNA Damage Response. To investigate the biological consequence of activation of the cellular DNA damage signaling machinery by HSV-1, we assessed virus production in cell lines harboring mutations in Mre11 or ATM. Cells were infected with WT HSV-1, and viral yield was determined by plaque assay (Fig. 4). Comparing A-TLD1 cells with the Mre11-complemented cells revealed that the yield of virus at 24 hpi was reduced by almost 2 logs in cells deficient in Mre11 (Fig. 4A). Viral growth was also examined in three cell lines from patients with A-T and compared with three cell lines expressing ATM (Fig. 4B). Additionally, we compared a transformed A-T cell line to a matched line in which ATM had been restored (Fig. 4C). Again, we saw that virus production was decreased in the absence of ATM. HSV-1 infections in cells in which Mre11 was degraded by adenoviral proteins (8) also showed reduced viral yield at 36 h (Fig. 9C). Caffeine has been shown to inhibit the kinase activity of both ATM and ATR in vitro (34). When growth curves were performed in the presence of caffeine (Fig. 10A, which is published as supporting information on the PNAS web site), virus growth was reduced, which we believe was due to ATM inhibition, because viral yields were unaffected by overexpression of a transdominant kinase-dead ATR protein (Fig. 10B).
In addition to the reduced yield of virus, there was also an extended eclipse phase for virus growth in cells deficient for Mre11 and ATM (Figs. 4 A and C and 9), suggesting a delay in virus replication. To investigate this observation further, we performed quantitative PCR analysis of DNA extracted from infected cells and determined the degree of amplification of the viral genome over the time course of infection (Fig. 4D). Analysis of internalized viral genomes per cellular gene and expression of viral IE proteins at 4 hpi showed that the cells were infected with similar efficiencies (data not shown). A reduction was observed in accumulation of viral DNA in the A-TLD1 mutant cells, indicating that the effect on virus growth is due to a defect in HSV-1 replication. We also quantified the relative frequency of ICP8 patterns for HSV-1 infection in the A-TLD1 cells compared with the Mre11-complemented line. ICP8 showed diffuse nuclear staining, discrete punctate foci, and large globular replication compartments, which correspond to stages II, III, and IV of HSV-1 replication, respectively (17, 18). We found that in cells lacking functional Mre11, there was a delay in the formation of punctate ICP8 foci and mature replication compartments (Fig. 4E). Together, our results show a correlation between viral prereplication structures and induction of the cellular DNA damage signaling pathways. The exact viral protein components required to activate a response are unknown. A recent report showed that a polymerase-null mutant virus failed to cause a mobility shift in Nbs1 (17), raising the possibility that recruitment, but not activity, of the viral polymerase may be required. The Mre11 complex is found at cellular replication centers (35) and is required for stable replication fork formation in Xenopus (36). Our observations raise the possibility that Mre11 is required for efficient assembly or maintenance of viral replication structures.
The defect in HSV-1 growth in the absence of a DNA damage response was even more marked with an ICP0-deficient virus in A-TLD1 cells. We observed that the ICP0-null virus was almost incapable of replicating in the absence of functional Mre11 (Fig. 4F), suggesting that when a virus is already compromised for replication, the effects of Mre11 on viral production are amplified. This result is interesting because ICP0-negative viruses at low mois have been suggested to resemble quiescent genomes (especially in limited-passage human fibroblasts), and ICP0 expression is important for reactivation from latency (13).
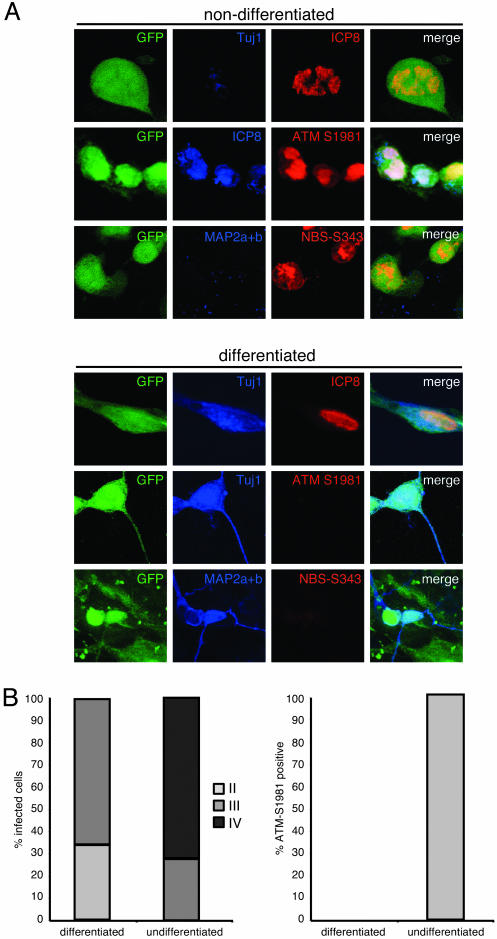
The DNA Damage Response to HSV-1 Is Abrogated in Neurons. Because neurons are the natural host cell for HSV-1 latency, we examined the damage response to HSV-1 in human neuronal cells. To examine this response in a controlled way, we used a recently developed system employing hESCs that can be induced to differentiate into authentic neurons in vitro and in vivo (ref. 37 and A.R.M., K. Nakashima, N. Toni, V. Sandler, and F.H.G., unpublished data). The hESCs were cocultured with a mouse stromal cell line, which induces their differentiation to a neuronal lineage (21), for 4 weeks until >95% of the colonies were neuronal, as assessed by the presence of extensive processes and positive staining for neuronal markers. Nondifferentiated hESCs were also plated as a control; both cell types were infected with HSV-1, and the damage response was assessed by staining for phosphorylated ATM and phosphorylated Nbs1 (Fig. 5A). We also stained neuronally differentiated and nondifferentiated hESCs infected with HSV-1 for ICP8 and determined the relative frequency of ICP8 patterns ranging from a diffuse nuclear pattern to mature replication compartments (Fig. 5B). We observed that HSV-1 infection of nondifferentiated hESCs resulted in formation of mature viral replication compartments that were stained positive for autophosphorylated ATM and phosphorylated Nbs1. In contrast, the neuronally differentiated hESCs showed predominantly diffuse ICP8 staining and were unable to mount a damage response to HSV-1 (Fig. 5B). In control experiments, we determined that these observations were not attributable to differences in cell division between the two cell lines (data not shown). These results suggest that in neuronal cells, replication compartment formation is impaired, and HSV-1 is unable to cause a DNA damage response. This finding is consistent with reports that neurons are inefficient at DNA repair (38, 39) and that ATM is specifically down-regulated when neuronal precursors are differentiated to a neuronal lineage (40). The cellular transcription factor Oct-1 is required for efficient assembly of HSV-1 replication compartments (41). Oct-1 expression is usually low in neurons (42), but, in several cell lines, it can be rapidly and transiently induced in response to a variety of DNA damaging agents or genotoxic stress (43), suggesting a potential further link between neurons, DNA damage, and HSV-1 latency.
Fig. 5.
Human neurons are unable to mount a damage response to HSV-1. (A) hESCs were plated on PA6 cells for 4–5 weeks to induce differentiation or were plated on mouse embryonic fibroblasts for at least 24 h as a nondifferentiated control. Cells were infected at a moi of 5 with 17+GFP. Activation of ATM, phosphorylation of Nbs1, and formation of HSV-1 replication centers was observed by immunofluorescence at 16 hpi. Tuj1 or MAP2a+b was used as a neuronal marker. (B) The relative abundance of different stages of the infection was quantified by counting 200 infected cells per condition and categorizing the ICP8 staining pattern as diffuse (stage II), punctate (stage III), or mature replication compartments (stage IV), as shown in Fig. 10C. The activation of ATM in neuronal and nonneuronal cells was quantified by counting 200 infected cells per condition and classifying them as positive or negative for ATM-S1981 staining.
In conclusion, we have shown that wild-type HSV-1 infection results in activation of the host cell's DNA damage machinery. This result is an interesting contrast to adenovirus, where the repair machinery represents an obstacle for productive infection (9). Our results have demonstrated that either specific DNA repair proteins or a DNA damage response environment in general are beneficial for productive HSV-1 infection. The formation or stability of concatemers and complex branched structures observed during HSV-1 infection (44) may require the Mre11 complex and the cellular DNA damage response machinery. In situations where the ability of HSV-1 to induce a DNA damage response was compromised (in cells with mutant Mre11 or ATM), we observed significant defects in viral replication. We propose that this situation is parallel to that occurring after HSV-1 infection of neurons, where we have shown that the virus is unable to elicit a DNA damage response. We suggest that the delay in viral growth in the absence of DNA repair proteins may be due to a reduced ability to form stable replication structures in a timely manner. In the absence of a damage response, the integrity of the replication fork or replication intermediates may be compromised, leading to deficiencies in viral replication and contributing to the establishment of latency.
Supplementary Material
Acknowledgments
We thank P. Branton, P. Carpenter, R. Coffin, R. Everett, G. Ketner, D. Knipe, P. Nghiem, J. Petrini, W. Ruyechan, Y. Saeki, P. Schaffer, and Y. Shiloh for gifts of reagents; the M.D.W. laboratory and colleagues at The Salk Institute for Biological Studies for critical reading of the manuscript; the Pendleton Microscopy Facility and Robert G. Summers for expert assistance with confocal microscopy; and the administration at The Salk Institute for the establishment of a non-National Institutes of Health core in the Stem Cell Facility (where the hESC work was performed by C.E.L. and A.R.M.) through support of the Lookout Fund and the Mathers Foundation. This work was supported by Wellcome Trust International Research Fellowship GR066559 (to C.E.L.), the Timken–Sturgis Foundation (C.T.C.), an ARCS scholarship (to C.T.C.), and the Pew Latin America Fellows (A.R.M.).
Author contributions: C.E.L. and M.D.W. designed research; C.E.L., C.T.C., and A.R.M. performed research; A.R.M. and F.H.G. contributed new reagents/analytic tools; C.E.L., F.H.G., and M.D.W. analyzed data; and C.E.L. and M.D.W. wrote the paper.
Abbreviations: HSV-1, herpes simplex virus 1; ATM, ataxia telangiectasia mutated; ATR, ATM-Rad3 related; DNA-PKcs, catalytic subunit of the DNA-dependent protein kinase; ICP, infected cell polypeptide; IE, immediate early; moi, multiplicity of infection; PAA, phosphonoacetic acid; hpi, h postinfection; A-TLD, ataxia telangiectasia-like disorder; A-T, ataxia telangiectasia.
References
- 1.Petrini, J. H. & Stracker, T. H. (2003) Trends Cell Biol. 13, 458–462. [DOI] [PubMed] [Google Scholar]
- 2.Zhou, B. B. & Elledge, S. J. (2000) Nature 408, 433–439. [DOI] [PubMed] [Google Scholar]
- 3.Jackson, S. P. (2002) Carcinogenesis 23, 687–696. [DOI] [PubMed] [Google Scholar]
- 4.D'Amours, D. & Jackson, S. P. (2002) Nat. Rev. Mol. Cell. Biol. 3, 317–327. [DOI] [PubMed] [Google Scholar]
- 5.Shiloh, Y. (2001) Curr. Opin. Genet. Dev. 11, 71–77. [DOI] [PubMed] [Google Scholar]
- 6.Kastan, M. B. & Lim, D. S. (2000) Nat. Rev. Mol. Cell. Biol. 1, 179–186. [DOI] [PubMed] [Google Scholar]
- 7.Bakkenist, C. J. & Kastan, M. B. (2003) Nature 421, 499–506. [DOI] [PubMed] [Google Scholar]
- 8.Carson, C. T., Schwartz, R. A., Stracker, T. H., Lilley, C. E., Lee, D. V. & Weitzman, M. D. (2003) EMBO J. 22, 6610–6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stracker, T. H., Carson, C. T. & Weitzman, M. D. (2002) Nature 418, 348–352. [DOI] [PubMed] [Google Scholar]
- 10.Weitzman, M. D., Carson, C. T., Schwartz, R. A. & Lilley, C. E. (2004) DNA Repair (Amst.) 3, 1165–1173. [DOI] [PubMed] [Google Scholar]
- 11.Roizman, B. & Knipe, D. M. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott Williams & Wilkins, Philadelphia), Vol. 2, pp. 2399–2460. [Google Scholar]
- 12.Jackson, S. A. & DeLuca, N. A. (2003) Proc. Natl. Acad. Sci. USA 100, 7871–7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett, R. D. (2000) Bioessays 22, 761–770. [DOI] [PubMed] [Google Scholar]
- 14.Everett, R. D., Freemont, P., Saitoh, H., Dasso, M., Orr, A., Kathoria, M. & Parkinson, J. (1998) J. Virol. 72, 6581–6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkinson, J., Lees-Miller, S. P. & Everett, R. D. (1999) J. Virol. 73, 650–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boutell, C., Sadis, S. & Everett, R. D. (2002) J. Virol. 76, 841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson, D. E. & Weller, S. K. (2004) J. Virol. 78, 4783–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukonis, C. J. & Weller, S. K. (1996) J. Virol. 70, 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilcock, D. & Lane, D. P. (1991) Nature 349, 429–431. [DOI] [PubMed] [Google Scholar]
- 20.de Bruyn Kops, A. & Knipe, D. M. (1988) Cell 55, 857–868. [DOI] [PubMed] [Google Scholar]
- 21.Kawasaki, H., Suemori, H., Mizuseki, K., Watanabe, K., Urano, F., Ichinose, H., Haruta, M., Takahashi, M., Yoshikawa, K., Nishikawa, S., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 1580–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown, S. M., Ritchie, D. A. & Subak-Sharpe, J. H. (1973) J. Gen. Virol. 18, 329–346. [DOI] [PubMed] [Google Scholar]
- 23.Ace, C. I., McKee, T. A., Ryan, J. M., Cameron, J. M. & Preston, C. M. (1989) J. Virol. 63, 2260–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lilley, C. E., Groutsi, F., Han, Z., Palmer, J. A., Anderson, P. N., Latchman, D. S. & Coffin, R. S. (2001) J. Virol. 75, 4343–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas, S. K., Lilley, C. E., Latchman, D. S. & Coffin, R. S. (1999) J. Virol. 73, 7399–7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saeki, Y., Ichikawa, T., Saeki, A., Chiocca, E. A., Tobler, K., Ackermann, M., Breakefield, X. O. & Fraefel, C. (1998) Hum. Gene Ther. 9, 2787–2794. [DOI] [PubMed] [Google Scholar]
- 27.Wade-Martins, R., Smith, E. R., Tyminski, E., Chiocca, E. A. & Saeki, Y. (2001) Nat. Biotechnol. 19, 1067–1070. [DOI] [PubMed] [Google Scholar]
- 28.Saeki, Y., Fraefel, C., Ichikawa, T., Breakefield, X. O. & Chiocca, E. A. (2001) Mol. Ther. 3, 591–601. [DOI] [PubMed] [Google Scholar]
- 29.Cai, W., Astor, T. L., Liptak, L. M., Cho, C., Coen, D. M. & Schaffer, P. A. (1993) J. Virol. 67, 7501–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bridge, E. & Ketner, G. (1990) Virology 174, 345–353. [DOI] [PubMed] [Google Scholar]
- 31.Schang, L. M., Phillips, J. & Schaffer, P. A. (1998) J. Virol. 72, 5626–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veldwijk, M. R., Topaly, J., Laufs, S., Hengge, U. R., Wenz, F., Zeller, W. J. & Fruehauf, S. (2002) Mol. Ther. 6, 272–278. [DOI] [PubMed] [Google Scholar]
- 33.Stewart, G. S., Maser, R. S., Stankovic, T., Bressan, D. A., Kaplan, M. I., Jaspers, N. G., Raams, A., Byrd, P. J., Petrini, J. H. & Taylor, A. M. (1999) Cell 99, 577–587. [DOI] [PubMed] [Google Scholar]
- 34.Sarkaria, J. N., Busby, E. C., Tibbetts, R. S., Roos, P., Taya, Y., Karnitz, L. M. & Abraham, R. T. (1999) Cancer Res. 59, 4375–4382. [PubMed] [Google Scholar]
- 35.Maser, R. S., Mirzoeva, O. K., Wells, J., Olivares, H., Williams, B. R., Zinkel, R. A., Farnham, P. J. & Petrini, J. H. (2001) Mol. Cell. Biol. 21, 6006–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Costanzo, V., Robertson, K., Bibikova, M., Kim, E., Grieco, D., Gottesman, M., Carroll, D. & Gautier, J. (2001) Mol. Cell 8, 137–147. [DOI] [PubMed] [Google Scholar]
- 37.Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S. & Jones, J. M. (1998) Science 282, 1145–1147. [DOI] [PubMed] [Google Scholar]
- 38.Nouspikel, T. & Hanawalt, P. C. (2000) Mol. Cell. Biol. 20, 1562–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gobbel, G. T., Bellinzona, M., Vogt, A. R., Gupta, N., Fike, J. R. & Chan, P. H. (1998) J. Neurosci. 18, 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allen, D. M., van Praag, H., Ray, J., Weaver, Z., Winrow, C. J., Carter, T. A., Braquet, R., Harrington, E., Ried, T., Brown, K. D., et al. (2001) Genes Dev. 15, 554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nogueira, M. L., Wang, V. E., Tantin, D., Sharp, P. A. & Kristie, T. M. (2004) Proc. Natl. Acad. Sci. USA 101, 1473–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He, X., Treacy, M. N., Simmons, D. M., Ingraham, H. A., Swanson, L. W. & Rosenfeld, M. G. (1989) Nature 340, 35–41. [DOI] [PubMed] [Google Scholar]
- 43.Zhao, H., Jin, S., Fan, F., Fan, W., Tong, T. & Zhan, Q. (2000) Cancer Res. 60, 6276–6280. [PubMed] [Google Scholar]
- 44.Wilkinson, D. E. & Weller, S. K. (2003) IUBMB Life 55, 451–458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.