Abstract
CircRNAs, a class of widespread circular RNAs produced from precursor mRNA back-splicing, have been implicated in regulation of gene expression in eukaryotes, but their biological functions in plants have not yet been elucidated. By deep sequencing of rRNA-removed and RNase R-digested RNA samples we have identified several thousands of putative back-splicing sites in tomato fruit (Solanum lycopersicum) and show that the abundance of some of these circRNAs derived from fruit pigment biosynthesis genes are regulated by fruit ripening. Herein, we overexpressed a circRNA derived from Phytoene Synthase 1 (PSY1) in tomato ‘Ailsa Craig’ and microTom. The PSY1 mRNA abundance, the lycopene and β-carotene accumulation were decreased significantly in the transgenic tomato fruits, likely due to the continuous highly expressed circRNAs and/or the low abundant linear RNAs generated from the overexpression vector. Besides, overexpression of a circRNA derived from Phytoene Desaturase (PDS) showed similar results. Our results provide biological insights into plant circRNAs.
Introduction
CircRNAs, a class of circular RNAs in eukaryotes, are derived from precursor mRNA back-splicing1. Although circRNAs have been identified more than 20 years ago2–4, they had been considered to be produced from aberrant splicing and their existence and functional potential were both underestimated. Nowadays, with the development of next-generation sequencing and bioinformatics, circRNAs have been identified in various eukaryotic species5–9. Most of the identified circRNAs are expressed at low levels, indicating the possibility that the majority of circRNAs might be splicing byproducts with little functional potential8, 10–12. However, many circRNAs are more abundant than their linear counterparts5–8, 13, suggesting the potential functional significance of these circular RNA molecules. Recent studies revealed that circRNAs are more stable than linear mRNAs, and most of them are cytoplasmic6, 7, 13. In addition, the circularization of circRNAs are conserved among species, and the expression of circRNAs are often cell, tissue and developmental stage-specific7, 8, 13–15. Recent studies have revealed that circRNAs may play roles in gene expression regulation, although the function of most circRNAs remain largely unknown16–18.
The biogenesis of circRNAs is considered to be regulated by both cis-elements and trans-acting factors16, 18. Complementary sequences or inverted repeats in the introns flanking the back-splice site could promote exon circularization by pairing to form hairpin structures10, 13, 19, 20, and multiple circRNAs may be produced from one single gene due to different sequence pairing, which is referred as to alternative circularization10. However, there are also circRNAs produced from exons without being bracketed by complementary sequences21–23, indicating that other cis-elements may account for the circularization, such as sequences recognized by RNA-binding proteins (RBPs)24–26. Exon circularization may occur during transcription20, 24 or post-transcriptionally19, 27, with trans-acting factors, such as the splicing factor Muscleblind (MBL)24 and Quaking (QKI)25. The circRNAs have been shown to regulate gene expression by acting as miRNA sponges7, 28, 29. However, miRNA inhibition may not be a general function of circRNAs since very few of them contain multiple binding sites for specific miRNAs12, 22, 30, 31. The biogenesis of circMbl, a circRNA in Drosophila, were demonstrated to compete with pre-mRNA splicing by binding and sequestering the splicing factor MBL indicating role of circRNAs in gene splicing24. A subtype of circRNAs with retained intron(s) (termed as exon-intron circRNAs or EIciRNAs), are mainly localized in the nucleus and revealed to promote the transcription of their parent genes providing evidence for direct regulation of transcription by circRNAs32. Moreover, recent studies demonstrated that circRNAs can be translated in vitro and in vivo 33–36.
In recent years, a large number of circRNAs have been identified and characterized in various eukaryotes, especially in animals, but much less has been known in plants, except the identification and validation of circRNAs in Arabidopsis, rice (Oryza sativa L.), tomato (Solanum lycopersicum L.), and barley (Hordeum vulgare L.)9, 22, 37–40. Tomato fruit has been served as the model system for studying fruit ripening, and has been extensively studied at the physiological, biochemical, molecular and genetic levels. The biosynthetic pathway of carotenoids in tomato fruit is well elucidated, but its regulation is complex and still needs to be clarified. Herein, by deep sequencing, we performed genome-wide identification of circRNAs in tomato fruits at different ripening stages, and analyzed the function significance of one special circRNA derived from Phytoene Synthase 1 (PSY1), a key gene in carotenoid biosynthesis.
Results
Bioinformatic detection and molecular validation of back-splice sites in ripening tomato fruits
To obtain circRNA sites in ripening fruits, we deep-sequenced rRNA-removed and RNase R-digested RNA samples of tomato fruits (Ailsa Craig) at mature green, breaker, and breaker +6 stages with two biological duplicates, and obtained a total of 266 million paired-end reads. The potential back-splice junctions were extracted with Segemehl (v 0.1.7) software41, and totally 9598 unique potential back-splice sites (≥2 reads) were identified. Although a large number of putative back-splice sites were obtained, the data showed low reproducibility, and only 223 putative back-splice sites were shared among samples (Table 1 and Fig. 1). We also analyzed the deep sequencing data with another tool CIRI (v 2.0.5)42, which filters out back-splice junctions without GU/AG splicing signals, and obtained 1018 unique putative circRNAs (with the default setting). The number of overlapped back-splice junctions between the two prediction algorithms is 796 (Fig. 1). The back-splice sites identified by both algorithms are listed in Supplementary Table 1. The circular RNAs within coding regions obtained by Segemehl prediction represented exonic circRNAs with back-splice sites derived from known splice sites (mostly the U2 splice signals (GU/AG), canonical sites)43, from unannotated splice sites (non-canonical sites), and intronic circular RNAs (ciRNAs). Validation of circRNAs derived from 16 genes associated with ripening by RNase R resistance and sequencing identified 16 canonical and 37 non-canonical circRNAs (Supplementary Table 2 and Fig. 2). We found that alternative circularization existed extensively in tomato fruits, including alternative back-splicing circularization (different back-splice sites originated from the same gene) and alternative splicing circularization (different transcripts with the same back-splice site) (Supplementary Fig. 1).
Table 1.
Yield of deep sequencing and putative back-splice sites detected.
| Samples | Yield | Segemehl (v 0.1.7, ≥2 reads) | CIRI (v 2.0.5, the default setting) | |||
|---|---|---|---|---|---|---|
| Mbases | #Reads | # circRNAs | Intersection | # circRNAs | Intersection | |
| Mature green-1 | 4424 | 43807106 | 1464 | 161 | 271 | 150 |
| Mature green-2 | 4701 | 46551038 | 1245 | 351 | ||
| Breaker-1 | 4770 | 47227328 | 1762 | 163 | 286 | 155 |
| Breaker-2 | 4446 | 44017308 | 1534 | 332 | ||
| Breaker +6-1 | 4241 | 41987128 | 2427 | 242 | 425 | 180 |
| Breaker +6-2 | 4291 | 42494150 | 2587 | 391 | ||
| Total | 26873 | 266084058 | 9598 | 1018 | ||
Figure 1.
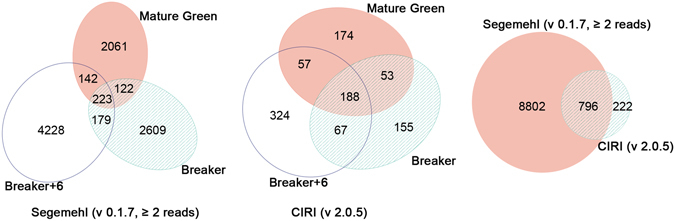
Distribution of circular RNAs among fruits at different ripening stages.
Figure 2.
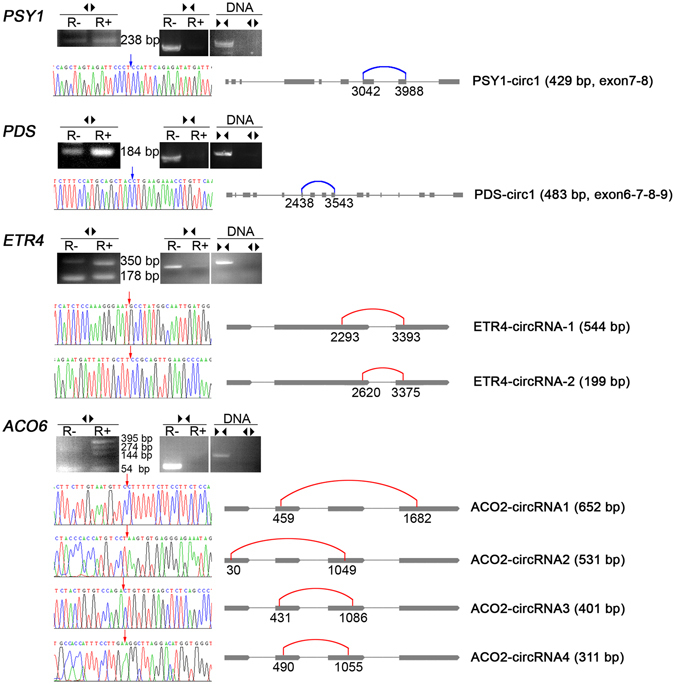
Canonical and non-canonical circRNAs validated by RNase R resistance and sequencing. RNase R resistance analysis. After DNase I treatment, total RNA from tomato fruit pericarp was digested with (R+, one hour at 37 °C) or without (R−, use water instead) RNase R. Both R+ and R− were reverse transcribed to cDNA using random primers. Divergent primers (◀▶) and convergent primers (▶◀) were used to identify circRNAs and linear RNAs respectively. Genomic DNA was also used as template to make certain that the site is not existent in genome. Sanger sequencing of divergent primers-amplified products were showed. The blue arrows and blue curves indicate canonical back-splice sites, while the red arrows and red curves indicate non-canonical back-splice sites.
Expression profiling of circRNAs in tomato fruits
The number of putative back-splice sites seems increased during ripening process (Table 1 and Fig. 1), suggesting potential relationship between circRNAs and fruit ripening. To validate this, we characterized the expression of a few canonical circRNAs derived from genes associated with fruit color, including those from PSY1, Phytoene Desaturase (PDS), 15-cis-zeta-carotene isomerase (ZISO) and DE-ETIOLATED 1 (DET1 or HP2), and found that most of them were up-regulated during fruit ripening and exhibited expression profiles similar to their parent genes (Fig. 3). The opposite expression trend of circRNAs derived from HP2 was likely due to alternative and competitive circularization. We also characterized the expression of these circRNAs in an ethylene-insensitive Nr (Never Ripe) mutant, and found that all these circRNAs were up-regulated during ripening, even though some of their parent genes were down-regulated at breaker +6 stage (Supplementary Fig. 2). These results suggest potential functional significance of some circRNAs in fruit ripening. Herein we focused on circRNAs generated from PSY1, a key gene involved in carotenoid biosynthesis. By RNA-seq, we detected all the three classes of circular RNAs (Supplementary Table 3), indicating that alternative circularization occurred on this gene. It seems that the abundance of PSY1 circRNAs increased during ripening (Supplementary Table 3), especially the three canonical circRNAs (Supplementary Fig. 3), suggesting that PSY1 circRNAs may be involved in fruit ripening.
Figure 3.
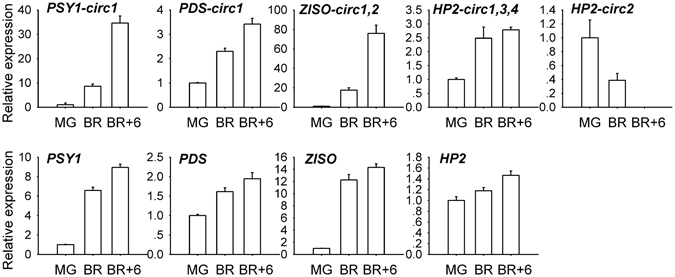
Expression profiling of a few circRNAs and their parent genes in Ailsa Craig fruits at different ripening stages. The data were normalized to Actin (NM_001321306.1) and presented as the means ± SD (Standard Deviation, n = 3). MG, mature green. BR, breaker.
Overexpression of PSY1-circ1 in tomato Ailsa Craig and microTom
To gain insight into the potential function of plant circRNAs in fruit ripening, we overexpressed a canonical circRNA derived from PSY1 gene (termed as PSY1-circ1) (Fig. 2) in tomato cvs. Ailsa Craig and microTom. We constructed the overexpression vector (using the CaMV 35S promoter) by introducing part of the flanking intron and inserting it into the other side of the exons in an opposite orientation (Supplementary Fig. 4)28, as it was shown that the pairing of complementary reverse sequence could promote circRNA biogenesis10. With quantitative real-time PCR and sequencing analysis, we found that PSY1-circ1 was highly expressed in the transgenic tomato fruits at different ripening stages (Fig. 4c and Supplementary Fig. 5c), indicating the high efficiency of circRNA overexpression system. To check whether the overexpression vector could generate undesired circular RNAs or large amounts of linear RNAs as shown in a previous report22, we used divergent and convergent primers to amplify the potential fragments, and the results indicate that the expression vector could produce the exact circRNA PSY1-circ1 with higher abundance, and also produce linear RNAs with much lower abundance (Supplementary Fig. 6 and Fig. 4c). In the overexpression vector, the endogenous non-complementary flanking sequences (281 bp upstream and 139 bp downstream) were retained (Supplementary Fig. 4), therefore the endogenous sequences flanking the back-splice site might contribute to the precision and high level of back-splicing in plants.
Figure 4.
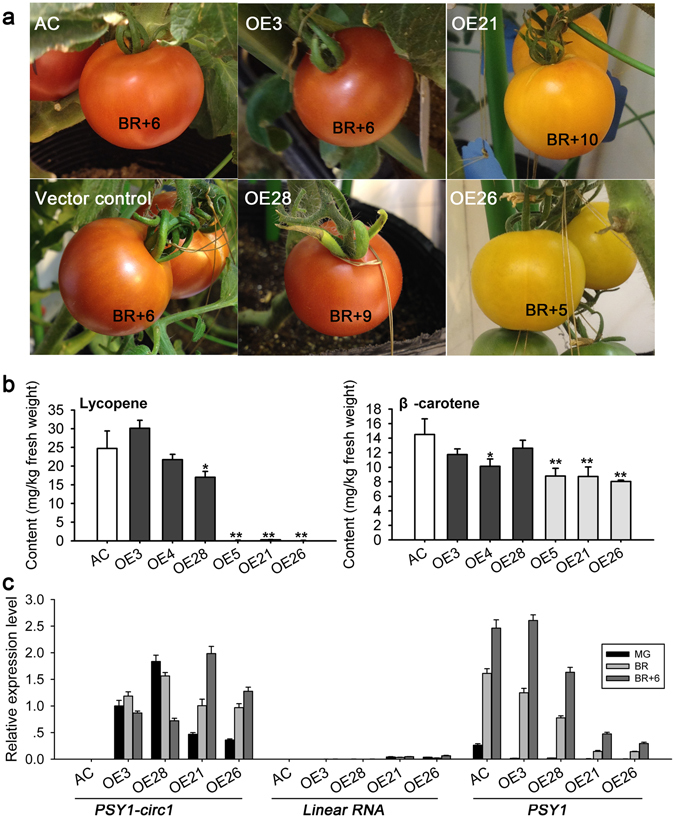
Overexpression of PSY1-circ1 in Ailsa Craig. (a) Fruits of different PSY1-circ1 overexpression tomato lines. OE3-OE28 stand for transgenic lines. Vector control stands for Ailsa Craig (AC) plants transformed with the empty vector pCAMBIA1301. (b) Lycopene and β-carotene contents in transgenic tomato fruits at breaker +10 stage. White, black and gray bars stand for control, transgenic lines with red fruits and yellow fruits respectively. The data with error bars were expressed as mean ± SEM (Standard Error of the Mean, n = 4), asterisks indicate statistically significant difference (with respect to control; *p value < 0.05; **p value < 0.01; two-way ANOVA followed by LSD (Least Significant Difference) test). (c) Expression profile of PSY1-circ1, linear RNAs generated by the overexpression vector, and PSY1 gene in transgenic tomato fruits at different ripening stages. The data are normalized to Actin and presented as means ± SD (n = 5).
Phenotype of PSY1-circ1 overexpressed tomato
The PSY1-circ1 overexpression Ailsa Craig plants showed different phenotypes, in which eight lines produced red fruits, while the other seven lines yielded yellow fruits (Fig. 4a and Supplementary Table 4). Similar to the transgenic Ailsa Craig plants, some transgenic microTom lines produced red fruits (24 lines), while the others produced yellow fruits (6 lines) (Supplementary Fig. 5a and Supplementary Table 4). In agreement with the fruit color, both the lycopene and β-carotene contents in the yellow transgenic fruits were decreased significantly, and their contents in some red transgenic fruits were slightly decreased (Fig. 4b and Supplementary Fig. 5b). The decrease of the β-carotene content was slighter than that of the lycopene content, and this result may be explained by the fact that β-carotene accumulates prior to ripening initiation44. Although with different fruit color, all the transgenic tomatoes had high expression levels of PSY1-circ1 (Fig. 4c and Supplementary Fig. 5c). We planted hygromycin-resistant T1 generation of a few transgenic microTom lines, and progenies from the yellow T0 lines produced mostly yellow fruits, and progenies from the red T0 lines produced mostly red fruits (Supplementary Table 4), indicating relative stable inheritance of the fruit color phenotype. Similar to T0 generation plants, both T1 generation plants with red and yellow fruits had high expression levels of PSY1-circ1 (Supplementary Fig. 7). In addition, the petals of some transgenic plants bearing yellow fruits (both Ailsa Craig and microTom) were white (Supplementary Fig. 8).
Expression of PSY1-circ1 and PSY1 in transgenic tomato fruits
In agreement with the fruit color phenotype, the expression of PSY1 in the ripening fruits of transgenic Ailsa Craig or microTom plants bearing yellow fruits, was down-regulated, and PSY1 expression in the red transgenic fruits were higher than in the yellow ones (Fig. 4c and Supplementary Fig. 5c), since PSY1 is a key gene in lycopene biogenesis. With regard to PSY1-circ1, we found that its expression showed different pattern between the red and yellow transgenic fruits. The expression of PSY1-circ1 in the yellow transgenic Ailsa Craig fruits showed an uptrend from the mature green to the breaker stage, then to the breaker +6 stage, and its expression in the yellow transgenic fruits was more abundant than that in the red ones at the breaker +6 stage. On the contrary, the PSY1-circ1 expression in the red transgenic fruits decreased from the breaker stage to the breaker +6 stage (Fig. 4c). Similar results were obtained in the PSY1-circ1 transgenic microTom fruits (Supplementary Fig. 5c). These results suggest that the continuous high expression of PSY-circ1 may play roles in inhibiting its parent mRNA accumulation. In addition, we found that the linear RNAs generated from the overexpression vector, though with low abundance, were more abundant in the yellow transgenic fruits than in the red ones (Fig. 4c), suggesting the possibility of co-suppression between the linear RNAs and the endogenous mRNAs.
Overexpression of PDS-circ1 in microTom
To examine whether other exonic canonical circRNAs play roles in fruit development and ripening, we overexpressed another circRNA derived from PDS, termed as PDS-circ1 (Fig. 2), in microTom in the same way as PSY1-circ1. Although most transgenic plants had no obvious phenotype, a few plants showed photobleached leaves, and a transgenic line (OE25) produced yellow fruits with photo-bleaching on the leaves, petals, and sepals (Fig. 5a). We found that the PDS expression level and the lycopene and β-carotene contents in the fruits of OE25 were decreased significantly (Fig. 5b,c), which is in agreement with the fruit color phenotype, since PDS is another critical gene in carotenoid biosynthesis. Besides, photo-bleaching is a typical phenotype of PDS silencing45. The results indicate that PDS-circ1 was highly expressed in all the transgenic tomato fruits, and its expression in OE25 fruits was increased dramatically after the breaker stage, and was even higher at the breaker +6 stage, while its expression in the fruits of the other transgenic lines, was much lower at the breaker +6 stage (Fig. 5c). Furthermore, the linear RNAs generated from the overexpression vector were expressed at low levels in the fruits of all the transgenic lines, and its expression in OE25 fruits was relatively higher than in the fruits of other transgenic lines (Fig. 5c). Therefore, these results suggest that tomato plants with the highest expression of PDS-circ1 showed inhibition of PDS expression in leaves and fruits.
Figure 5.
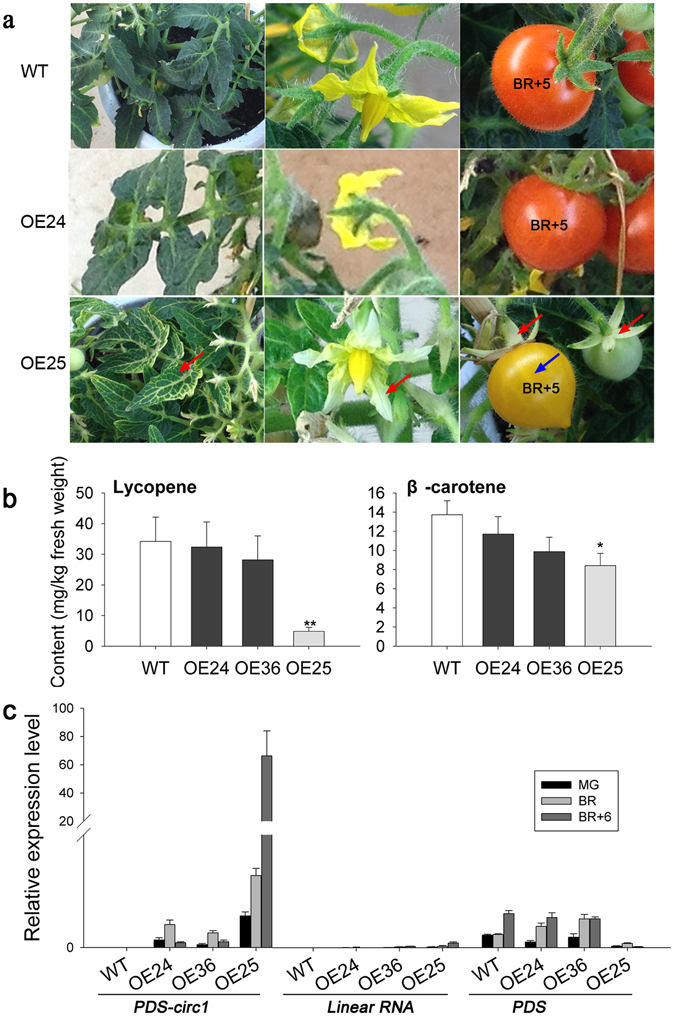
Overexpression of PDS-circ1 in microTom. (a) Leaves, flowers and fruits of different PDS-circ1 overexpression lines and the control (WT), red arrows indicate photo-bleaching of leaves, petals, and sepals, blue arrow indicates yellow fruit. (b) Lycopene and β-carotene contents in transgenic tomato fruits at breaker +10 stage. White, black and gray bars stand for control, transgenic lines with red fruits and yellow fruits respectively. The data with error bars are expressed as mean ± SEM (n = 5), asterisks indicate statistically significant difference (with respect to control; *p value < 0.05; **p value < 0.01; two-way ANOVA followed by LSD test). (c) Expression profile of PDS-circ1, linear RNAs generated by the overexpression vector, and PDS gene in transgenic tomato fruits at different ripening stages. The data were normalized to Actin and presented as means ± SD (n = 5). OE24-OE36 stand for different transgenic lines.
Discussion
Most of the putative circRNAs we identified were expressed at low levels, which is consistent with recent reports8, 10–12, and suggesting little functional potential of most circRNAs. It seems that there is potential relationship between circRNAs and fruit ripening since the total number of putative circRNAs was increased and a few circRNAs we validated were upregulated similar to their parent genes during ripening process. As reported in other plants37, the positive correlation of expression pattern between some circRNAs and their parent genes indicating that their biogenesis might share similar mechanism. However, in Nr mutant, PSY1-circ1 and PDS-circ1 were upregulated after the breaker stage while their parent genes were downregulated. Therefore, the relationship of the expression between the circRNAs and their parent genes in plants seems complicated, which merits further study. Despite that some circRNAs play roles by acting as miRNA sponges, very few of the identified circRNAs contain miRNA binding sites12, 22, 31. Herein by searching miRNA homologs within the database miRBase (http://www.mirbase.org/search.shtml), we found no miRNA targeting PSY1-circ1 (within miRNAs in Solanum lycopersicum, Solanum tuberosum, and Nicotiana tabacum). Therefore, there might be little functional potential of PSY1-circ1 action by miRNA inhibition.
As mentioned in another report28, we overexpressed circRNAs by introducing endogenous reverse complementary intronic sequences. The overexpression vector could generate the exact circRNAs with high abundance, however, it inevitably produced linear RNAs as well, although with much lower abundance. The linear RNA byproducts may interfere the function analysis of circRNAs. We found that both the circRNAs and the linear isoforms showed correlation with their parent mRNAs. It seems that the continuous high expression of PSY-circ1 or PDS-circ1 is related to their parent gene downregulation. However, the linear RNAs generated from the overexpression vectors may also contribute to the parent gene inhibition by co-suppression46.
In summary, we identified a large number of putative circRNAs, and characterized a few circRNAs derived from genes involved in the fruit pigment accumulation. PSY1 derived circRNA and/or its linear isoforms could negatively regulate the expression of their parent gene, and reduce lycopene and β-carotene accumulation in tomato fruits. These discoveries reveal the potential circRNA behavior in fruit ripening, and provide evidence for the biological implication of circRNAs in plants. Future work is needed to reveal the mechanism of down-regulation of parent mRNAs in the transgenic lines over-expression plant circRNAs. In addition, since circRNAs could be inhibited by siRNA approaches13, knockdown of PSY1-circ1 by introducing artificial miRNA (amiRNA) targeting the back-splice site might provide further evidence of the functional role of PSY1-circ1 during fruit ripening.
Methods
Plant materials and growth conditions
Tomato (Solanum lycopersicum L.) plants (cvs. Ailsa Craig and microTom) were grown under growth room conditions (24 °C, 75% relative humidity, 16/8-h light/dark cycle, 500 μmol m−2 s−1 photosynthetic photon flux density). The seeds were surface sterilized with 5% sodium hypochlorite (w/v) for 25 minutes, washed with sterilized water for six times, and then sown in pots containing nutritive soil.
Deep sequence of circular RNAs
Tomato (cv. Ailsa Craig) fruit pericarps at mature green, breaker, and breaker +6 stages were sampled for circular RNA detection (two independent biological replicates). Total RNA was extracted using Trizol reagent (Life Technologies, Carlsbad, CA). The rRNA was removed by a Ribo-Zero Magnetic kit (Epicentre, Madison, WI), and linear RNA was digested by RNase R (Epicentre, Madison, WI) at 37 °C for one hour. RNA-Seq library was prepared using TruSeq RNA LT Sample Prep kit v2 (Illumina, San Diego, CA) with insert length of 120–250 bp, and high-throughput sequencing was performed on HiSeq 2500 instrument with 100 bp paired-end reads (Genenergy Biotechnology Co., Ltd, Shanghai, China). The adaptors and low quality data were removed from the sequencing raw data using Trimmomatic (version 0.32) (parameters: ILLUMINACLIP: adaptor.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:50). The software Segemehl (v 0.1.7) and CIRI (v 2.0.5) were used for potential back-splice sites extraction41, 42.
RNase R resistance test
Total RNA was isolated by Trizol reagent (Life Technologies, Carlsbad, CA) followed by DNA residue depletion using DNase I (Fermentas, Glen Burnie, MD). After purification by RNA Clean & Concentrator kit (Zymo Research, Irvine, CA), the RNA samples were separated to two aliquots, one aliquot was digested with RNase R (Epicentre, Madison, WI) at 37 °C for one hour (R+), the other aliquot was mock-treated with water (R−). Both R+ and R− were reverse transcribed (M-MLV Reverse Transcriptase, Promega, Madison, WI) to cDNA using random primers and used as templates for circRNA identification. Convergent and divergent primers (Supplementary Table 5) were used for linear and circRNA amplification respectively. Genomic DNA was also used as template to make certain that the site is not existent in genome.
Quantitative real-time PCR analysis
Total RNA of tomato fruit pericarp was isolated using Trizol reagent followed by DNase I digestion at 37 °C for 30 minutes, then reverse transcribed to cDNA using random primers and used as templates for Real-time PCR. At least one of the divergent primers spanned the back-splice site, and the 3′ end of the primer that cross the junction was 3–6 bp in length. Real-time PCR was performed using the SYBR Select Master Mix on QuantStudio 6 Flex Real-time PCR System (Applied Biosystems, Foster City, CA). The primers used were listed in Supplementary Table 5.
Vector construction and genetic transformation
To construct a vector for PSY1-circ1 overexpression, a genomic fragment consisting of the exons to be cyclized and its flanking sequences (281 bp upstream and 681 downstream) was cloned, then part of the downstream sequence (517 bp) was amplified. The two fragments were introduced into the expression vector pCAMBIA1301 in the opposite orientation under CaMV 35 S promoter (Supplementary Fig. 4). The PDS-circ1 overexpression vector was constructed in the same way as PSY1-circ1, the upstream and downstream flanking sequences of the circularized exons are 465 bp and 660 bp respectively, and the reverse inserted fragment from the downstream flanking intron is 462 bp. The vectors were introduced into A. tumefaciens EHA105, which was subsequently used to transform tomato Ailsa Craig and microTom following punctured cotyledon transformation method47.
Lycopene and β-carotene content analysis
The fruit pericarp samples were excised, frozen with liquid nitrogen and stored at −80 °C. The frozen samples were ground to fine powder with liquid nitrogen, and approximately 0.2 g sample were extracted with 1.8 ml acetone-hexane (2:3) solvent for one hour. The optical density of the crude extract at 663 nm, 645 nm, 505 nm, and 453 nm were measured at the same time, using a UV-1800 spectrophotometer (Shimadzu, Kyoto, Japan). The contents of lycopene and β-carotene were estimated using the follow equations48: Lycopene (100 mg/100 mL) = −0.0458 A663 + 0.204 A645 + 0.372 A505 − 0.0806 A453, β-carotene (100 mg/100 mL) = 0.216 A663 − 1.22 A645 − 0.304 A505 + 0.452 A453.
Electronic supplementary material
Acknowledgements
We thank Dr. Avtar K. Handa (Purdue University) for critical reading the paper, and Jie Luo (Zhejiang Academy of Agricultural Sciences) for assistance with the data analysis. The study was financially supported by the Ministry of Science and Technology of China (973 program grant 2013CB127101 to Z.D.), and the National Natural Science Foundation of China (grant no. 31501766, 31401925 and 31571306 to J.T., Z.Z. and X.S. respectively).
Author Contributions
Z.D. and J.T. designed the experiments and wrote the paper. J.T., Z.Z., and Y.N. performed the experiments, J.T., X.S., and Z.D. analysed the data.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-08806-0
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nigro JM, et al. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-S. [DOI] [PubMed] [Google Scholar]
- 3.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 4.Pasman Z, Been MD, Garcia-Blanco MA. Exon circularization in mammalian nuclear extracts. RNA. 1996;2:603–610. [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen TB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 8.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang PL, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9 doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang XO, et al. Complementary Sequence-Mediated Exon Circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Bachmayr-Heyda A, et al. Correlation of circular RNA abundance with proliferation-exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5 doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeck WR, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szabo L, et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16 doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veno MT, et al. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16 doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salzman J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2016 doi: 10.1080/15476286.2016.1271524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 19.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramer MC, et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westholm JO, et al. Genome-wide Analysis of Drosophila Circular RNAs Reveals Their Structural and Sequence Properties and Age-Dependent Neural Accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu T, et al. Transcriptome-wide investigation of circular RNAs in rice. RNA. 2015;21:2076–2087. doi: 10.1261/rna.052282.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;9 doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashwal-Fluss R, et al. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol. Cell. 2014;56:55–56. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 25.Conn SJ, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, et al. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016;15:611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 28.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 29.Zheng Q, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nature communications. 2016;7 doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.You X, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 33.Abe N, et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci Rep. 2015;5 doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pamudurti NR, et al. Translation of CircRNAs. Mol. Cell. 2017;66:9–21. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Legnini I, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye C, Chen L, Liu C, Zhu Q, Fan L. Widespread noncoding circular RNAs in plants. New Phytol. 2015;208:88–95. doi: 10.1111/nph.13585. [DOI] [PubMed] [Google Scholar]
- 38.Sun X, et al. Integrative analysis of Arabidopsis thaliana transcriptomics reveals intuitive splicing mechanism for circular RNA. FEBS Lett. 2016;590:3510–3516. doi: 10.1002/1873-3468.12440. [DOI] [PubMed] [Google Scholar]
- 39.Zuo J, Wang Q, Zhu B, Luo Y, Gao L. Deciphering the roles of circRNAs on chilling injury in tomato. Biochem. Biophys. Res. Commun. 2016;479:132–138. doi: 10.1016/j.bbrc.2016.07.032. [DOI] [PubMed] [Google Scholar]
- 40.Darbani B, Noeparvar S, Borg S. Identification of Circular RNAs from the Parental Genes Involved in Multiple Aspects of Cellular Metabolism in Barley. Front. Plant Sci. 2016;7 doi: 10.3389/fpls.2016.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffmann S, et al. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing, and fusion detection. Genome Biol. 2014;15 doi: 10.1186/gb-2014-15-2-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16 doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nature Reviews Genetics. 2016;17:679–692. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ronen G, Cohen M, Zamir D, Hirschberg J. Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon‐cyclase is down‐regulated during ripening and is elevated in the mutantDelta. Plant J. 1999;17:341–351. doi: 10.1046/j.1365-313X.1999.00381.x. [DOI] [PubMed] [Google Scholar]
- 45.Kumagai M, et al. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc Natl Acad Sci USA. 1995;92:1679–1683. doi: 10.1073/pnas.92.5.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Napoli C, Lemieux C, Jorgensen R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rai GK, et al. Effects of explant age, germination medium, pre-culture parameters, inoculation medium, pH, washing medium, and selection regime on Agrobacterium-mediated transformation of tomato. In Vitro Cell. Dev. Biol. Plant. 2012;48:565–578. doi: 10.1007/s11627-012-9442-3. [DOI] [Google Scholar]
- 48.Nagata M, Yamashita I. Simple method for simultaneous determination of chlorophyll and carotenoids in tomato fruit. J. Japan. Soc. Food Sci. Technol. 1992;39:925–928. doi: 10.3136/nskkk1962.39.925. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


