Abstract
Glioblastoma multiforme (GBM) is the most common form of malignant glioma, characterized by genetic instability, intratumoral histopathological variability, and unpredictable clinical behavior. We investigated global gene expression in surgical samples of brain tumors. Gene expression profiling revealed large differences between normal brain samples and tumor tissues and between GBMs and lower-grade oligodendroglial tumors. Extensive differences in gene expression were found among GBMs, particularly in genes involved in angiogenesis, immune cell infiltration, and extracellular matrix remodeling. We found that the gene expression patterns in paired specimens from the same GBM invariably were more closely related to each other than to any other tumor, even when the paired specimens had strikingly divergent histologies. Survival analyses revealed a set of ≈70 genes more highly expressed in rapidly progressing tumors that stratified GBMs into two groups that differed by >4-fold in median duration of survival. We further investigated one gene from the group, FABP7, and confirmed its association with survival in two unrelated cohorts totaling 105 patients. Expression of FABP7 enhanced the motility of glioma-derived cells in vitro. Our analyses thus identify and validate a prognostic marker of both biologic and clinical significance and provide a series of putative markers for additional evaluation.
Keywords: brain, glioma, tumor, FABP7, prognosis
Glial tumors are the most common primary brain malignancies. Glioblastoma multiforme (GBM) (Grade IV astrocytoma) accounts for 80% of malignant astrocytomas and is marked by an extremely poor prognosis: half of all patients die within 1 year of diagnosis (1). Key histopathological features of GBM, such as necrosis and endothelial proliferation, distinguish these tumors from lower-grade astrocytic tumors that have a much better prognosis (2). The mechanisms that underlie the correlation between high grade and poor prognosis are unknown, because all grades of astrocytoma are characterized by inappropriate proliferation, invasion of normal brain tissue, and disruption of normal brain functions. As indicated in the name of the tumor, GBM is characterized by marked cytologic and histologic variation and displays extensive genetic and biological variability (3).
Genome-scale gene expression profiling using microarrays allows the molecular characterization of intertumor variability, revealing molecular signatures that reflect underlying pathogenic mechanisms and molecular features that may be associated with survival (4, 5). Recent studies of gliomas have documented patterns of gene expression associated with specific clinical grades of oligodendroglioma (ODG) (6) and astrocytoma (7–14), patterns associated with specific genetic alterations (15, 16), and patterns that differentiate neoplastic tissues and normal brain (17–19). In the case of medulloblastoma, a non-glial childhood brain tumor, similar analyses have led to the identification of putative prognostic markers (20, 21). However, the gene expression profiles associated with the heterogeneous clinical behavior of GBM have not been explored, and a reliable molecular marker that predicts survival of patients with this tumor is still lacking. In this study, we used cDNA microarrays to analyze gene expression patterns in a series of GBM specimens and identified markers that are likely to predict patient survival.
Materials and Methods
Tissue Samples. Frozen specimens and paraffin-embedded tissues were obtained from the Neurosurgery Tissue Bank at the University of California, San Francisco. These patients were on clinical protocols, and follow-up data were available. Additional samples were from the M. D. Anderson Cancer Center, University of Texas (Houston). Postmortem specimens from normal brains were from Stanford University Hospital. Each frozen tumor tissue was sectioned and histopathologically evaluated by a single neuropathologist (A.W.B.), and specimens containing less than ≈25% tumor cells were not included for study. All samples were obtained with informed consent after approval of the Human Research Committee at University of California, San Francisco, and M. D. Anderson Cancer Center.
Sample Preparation and Analysis. The microarray methods followed closely those of previous studies (4, 5). Briefly, total RNA was extracted by using TRIzol followed by mRNA purification using FastTrack (Invitrogen). mRNA was reverse-transcribed and labeled with Cy dyes (Amersham Pharmacia Biosciences) before hybridization. Detailed methods can be found at http://microarraypubs.stanford.edu/gbm.
Bioinformatic Analyses. Agglomerative hierarchical clustering was performed by using the cluster program (22). Stand-alone Perl scripts were written to facilitate the analyses. We used a two-step algorithm to identify survival-associated genes. First, Cox regression coefficients were calculated for all clustered genes, and a moving average of these values was calculated for consecutive genes in the hierarchically clustered list. Only one sample was used from each tumor for this calculation. By using 1,000 random permutations of the array labels, we identified the moving average Cox coefficients above and below which clusters were associated with survival at P < 0.01. To further limit genes on which to focus follow-up studies, we used an algorithm to identify “tumor-intrinsic” genes expressed most consistently in samples from different areas of the same tumor but that varied in samples from different tumors (5). Kaplan–Meier survival analysis was done by using winstat.
Immunohistochemistry. Antigen retrieval and immunostaining of FABP7 in paraffin samples were performed as described (23). Rabbit polyclonal antibodies against FABP7 were generous gifts of R. Godbout (University of Alberta, Alberta, ON, Canada) and N. Heintz (Rockefeller University, New York) (24, 25). Scoring was semiquantitative based on extent and intensity of nuclear staining by a single neuropathologist (K.D.A.).
Migration Assay. SF767MG cells were transfected with pcDNA3 or pcDNA3-FABP7 by using FuGENE (Roche Molecular Biochemicals). For migration assays, 1 × 104 cells were seeded into the upper chamber of TransWell FluoroBlok (Corning BD Biosciences), and 10% FBS in DMEM was used as chemoattractant. Migrated cells were counted by using standard methods (see Supporting Text, which is published as supporting information on the PNAS web site).
Results
Overview of Glioma Gene Expression Signatures. We used DNA microarrays consisting of ≈23,000 elements (representing ≈18,000 unique UniGene clusters) to survey gene expression in 31 patients with primary brain malignancies. The tumor specimens included 25 glioblastomas (four in which two distinct regions were analyzed) and six oligodendroglial tumors [including oligoastrocytomas (OACs) and ODGs]. For comparison, we also analyzed three postmortem samples from normal brains.
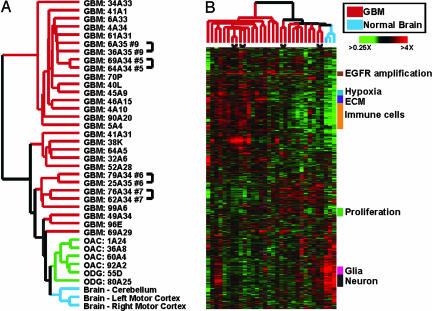
To explore relationships among the clinical specimens and the genes they expressed, we performed agglomerative hierarchical cluster analyses of both genes and samples using genes showing significant variation across these specimens. As can be seen in Fig. 1A, the normal brain and oligodendroglial neoplasms were more closely related to each other than to any GBM specimen. The gene expression patterns of GBMs showed high variability and separated the tumors into two groups, one of which was more closely related to OACs/ODGs and normal brain than to the other GBMs. The gene expression signatures of the OACs/ODGs included relatively elevated expression of the canonical oligodendrocyte markers OLIG1 and OLIG2. The GBMs that clustered with the oligodendroglial tumors also tended to express higher levels of OLIG1 and OLIG2 (see Fig. 6, which is published as supporting information on the PNAS web site), consistent with a previous report of their variable expression in GBM (26).
Fig. 1.
Gene expression in normal and malignant brain samples. (A) Unsupervised hierarchical clustering of 38 samples including GBMs, OACs, ODGs, and normal brain specimens. Array elements that varied at least 2-fold from the median on at least five microarrays were included (2,490 cDNA elements representing ≈2,000 genes). Samples from different regions of the same tumor are indicated by brackets. (B) Unsupervised hierarchical clustering of 32 samples including GBMs and normal brain. Array elements that varied at least 2-fold from the median on at least five microarrays were included (2,188 cDNA elements representing ≈1,800 genes). The data are displayed as a hierarchical cluster where rows represent genes (unique cDNA elements), and columns represent samples. Colored pixels capture the magnitude of the response for any gene, where shades of red and green represent induction and repression, respectively, relative to the median for each gene. Black pixels reflect no change from the median, and gray pixels represent missing data. Groups of genes with common functional or biological themes are indicated. A searchable version of this cluster is available upon request to the corresponding author.
We next sought to analyze how the extensive histological variation within a single GBM might be related to its patterns of gene expression. We therefore collected paired tumor specimens from four GBM patients. In each case, these tissues were obtained by stereotactically guided biopsies from regions of tumors whose radiographic appearances suggested they might vary histologically. Microscopic examination of these samples showed histological variations characteristic of GBM (e.g., variable necrosis, vascular/endothelial proliferation, etc.; see Fig. 7, which is published as supporting information on the PNAS web site). The hallmark histopathologic features present in the paired specimens from the same tumor were reflected in their expression patterns. For example, a significant difference in mitotic index (data not shown) between the two specimens from GBM no. 7 was paralleled by differential expression of genes involved in cellular proliferation. Similarly, genes regulated by hypoxia were more highly expressed in one sample of GBM no. 9, which showed extensive necrosis, than in the other less necrotic sample from the same tumor (see below and Fig. 7).
Although histological characteristics varied greatly between these paired samples, we found that the gene expression patterns of different specimens from the same tumor were always more closely related to each other than to any other tumor (Fig. 1). Thus, although the GBMs showed significant intratumor histological heterogeneity, differences between tumors were significantly greater than variations within a tumor, and tumor-specific gene expression signatures could readily be recognized. The observation that a tumor's global pattern of gene expression is highly specific despite intratumoral histological variability was consistent with data from other solid tumors (5, 27). These results suggest that histologic characteristics of GBMs are unlikely to account for or mark the molecular (28) or pathologic (29) heterogeneity that characterizes these tumors. This observation also implies that expression patterns might manifest the intrinsic differences of these tumors better than histological criteria.
An important caveat for these analyses is that the histological heterogeneity of GBMs could confound our stereotactic sample analysis, because even small local specimens might contain mixtures of histologies not appreciated in the sections that were microscopically examined.
To focus on gene expression variability among different GBMs, we further analyzed gene expression patterns in only the GBM and normal brain specimens. Fig. 1B depicts the complex variations in gene expression patterns found among these samples. As expected, genes involved in related biological processes or expressed in the same cell type clustered together, because their expression patterns were more closely correlated to each other than to functionally unrelated genes in the data set. Many genes were more highly expressed in GBMs than in normal brain, and most of these genes were variably expressed among the GBMs. Functionally, many of these genes fell into several broad categories, including genes related to immune cell infiltration, the extracellular matrix (ECM), hypoxia, and proliferation.
Normal Brain Signature. One of the most striking gene expression patterns consisted of genes with higher expression in normal brain compared with the tumors. These genes could be divided into two broad classes: those characteristically expressed in neurons and those characteristically expressed in glia. Some tumor samples displayed relatively high expression of both neural and glial genes, perhaps reflecting the invasion of normal brain by the tumors.
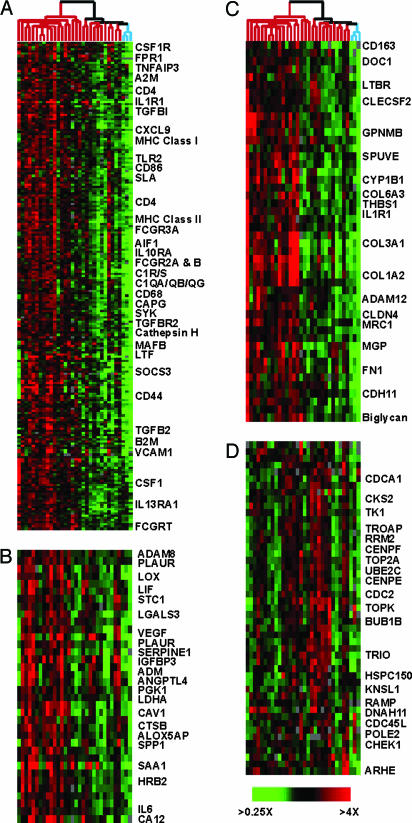
Immune Cell Signature. The largest cluster of differentially expressed genes was enriched for genes typically expressed in macrophages, microglia, and lymphocytes (e.g., MHC class II genes, CD4, CD53, and CD68) and genes associated with macrophage activation (e.g., CD44) (30) (Fig. 2A). This expression signature was consistent with the known infiltration of macrophages/microglia in GBMs; up to one-third of cells in some GBM specimens are microglia (31, 32). Furthermore, the covariation between the immune cell and hypoxia signatures (node mean correlation, R = 0.71) echoes the correlation between macrophage infiltration and vessel count in glioma (33) and may reflect the role macrophages and other immune cells can play during angiogenesis (34).
Fig. 2.
Expanded view of biologically distinct expression signatures among GBMs. Data were extracted from Fig. 1B and are displayed. Individual clusters depict genes associated with immune cells (A), hypoxia (B), ECM (C), and cellular proliferation (D). Many of the array elements encode uncharacterized genes, and only a subset of named genes is shown.
Hypoxia Signature. Microvascular proliferation and necrosis are two key histologic features of GBM. Hypoxia, an invariant characteristic of necrotic tumor regions, induces the expression of VEGF (35). A cluster of genes that included VEGF, ADM, PLAUR, and SERPINE1, which have previously been implicated in hypoxia and angiogenesis, was more highly expressed in most GBMs than in normal brain (Fig. 2B).
ECM Signature. A prominent cluster composed primarily of genes encoding ECM-related proteins was expressed at variably elevated levels in GBMs (Fig. 2C). This signature included multiple collagens and fibronectin 1, which are not highly expressed in the normal mammalian nervous system but are found in the CNS vasculature (36). Many of the proteins encoded by genes in this cluster are localized to the basement membrane of CNS blood vessels. Immunohistochemical analysis of GBM tissues confirmed the presence of type 1 collagen and fibronectin 1 in the thickened ECM of tumor vessels, including the ECM of proliferating microvasculature (data not shown). Notably, three of the genes in this cluster (COL1A2, COL3A1, and COL6A3) were also among a group of genes identified by serial analysis of gene expression (SAGE) as being elevated in colon tumor endothelium (37). The genes in this cluster are thus likely related to the formation of the tumor vasculature.
Proliferation Signature. This cluster contained genes involved in proliferation and progression through the cell cycle. Most of these genes have been shown to be periodically expressed during the cell cycle (Fig. 2D) (38). These genes may provide an opportunity to develop a sensitive molecular measure of the proliferation rate of tumor cells.
Survival Analysis Reveals Two Subgroups of GBM. Both the structure of the tumor dendrogram in Fig. 1 and the gene expression signatures described reflect the molecular heterogeneity of GBM and suggest the possibility of identifying molecularly distinguishable subtypes. We initially hypothesized that the group of GBMs most similar to OAC/ODG tumors and normal brain (Fig. 1) might differ clinically from the remaining tumors. However, none of the recorded clinical characteristics, such as patient age and survival from diagnosis, or pathologic measures, such as percentage of tumor cells, differed significantly between the groups.
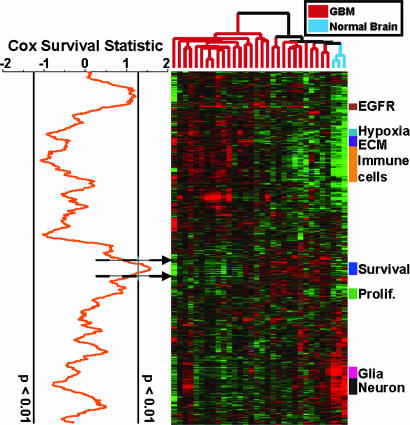
As an alternative approach, we searched deliberately for gene expression signatures that could subdivide GBMs into groups with differences in prognosis. Using the 20 GBMs for which we could obtain duration of survival and other clinical data, we plotted a local Cox survival statistic next to the hierarchically clustered data from Fig. 1B by using a moving average algorithm (Fig. 3). This approach combines the functional organization and noise reduction of hierarchical clustering with the supervised methodology of Cox survival analysis. Only gene expression clusters containing multiple genes strongly correlated with survival will show significant Cox score peaks; thus, the approach highlights large sets of coregulated genes whose expression is associated with patient survival. To assess the statistical significance of these peaks, we randomly permuted the patient labels 1,000 times and focused on Cox score peaks that were significant at P < 0.01. As can be seen in Fig. 3, the only significant peak was centered on a small cluster of coregulated genes. We did not observe a significant peak of negative Cox scores, implying that none of the gene expression clusters in our dataset were strong positive prognostic signatures. For comparison, we have performed a supervised Cox survival analysis for every gene included in Fig. 1B, and these data are available in Supporting Text.
Fig. 3.
Coregulated genes that correlate with patient survival. The Cox survival statistic was calculated for every gene by using the 20 GBMs for which we could obtain survival data. Genes correlating with poorer prognosis received positive Cox scores, whereas genes correlating with better prognosis received negative Cox scores. A moving average (window size, 71 elements) of these scores was plotted adjacent to the gene expression map from Fig. 1B. A permutation-based algorithm was used to calculate moving average scores that were significant at P < 0.01, and these are indicated by vertical lines.
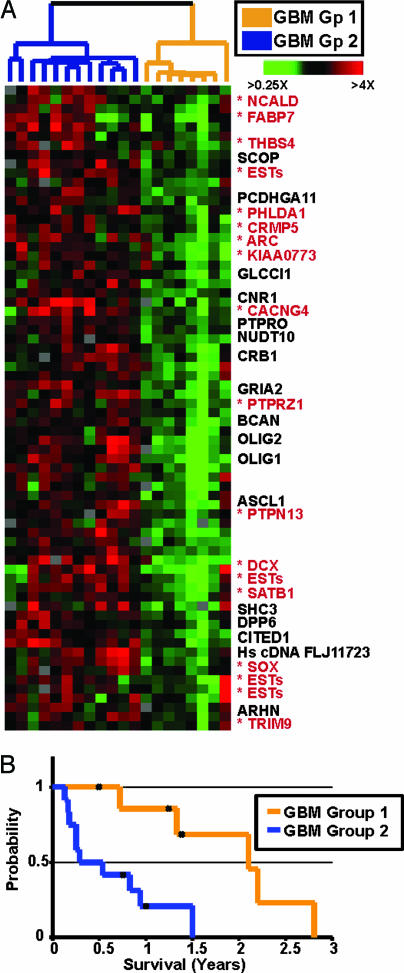
To further analyze the survival-associated gene expression signature, we hierarchically clustered these 20 GBMs using the genes identified by the largest peak shown in Fig. 3. This divided them into two groups of comparable size (Fig. 4A). Kaplan–Meier analysis revealed a significant difference in survival between the two groups, with median survival times of 25 months for Group 1 and 4 months for Group 2 (Fig. 4B). Three tumors in Group 1 and one in Group 2 are derived from recurrent cases, in which the survival time was defined as the time of diagnosis to death. Other clinical parameters were not significantly different between the groups (see Table 1, which is published as supporting information on the PNAS web site). Expression of the survival-associated genes was very different in the two groups of GBMs, with relatively low expression in the better prognostic group (Group 1). Several of these genes have previously been shown to be variably expressed in GBMs (including OLIG2 and FABP7) (25, 26). A subset of the genes in this cluster has been implicated in the migration of glial or neural progenitors (e.g., BCAN, FABP7, and CRMP5), suggesting that their expression in GBMs might be related to the invasive potential of the tumor cells.
Fig. 4.
Expression of survival-associated genes stratifies GBMs into two distinct subgroups. (A) Expanded view of the survival-associated expression signature identified in Fig. 3 among 20 GBMs with survival data. The tumors are classified as belonging to Group 1 (orange) or Group 2 (blue) based on the array dendrogram. Genes marked with asterisks were among 500 genes with the top “intrinsic” scores (see text). (B) Kaplan–Meier analysis of the two subgroups indicates they have significantly different median survivals.
We extracted Gene Ontology terms from the SOURCE database (39) and systematically compared their frequency among the survival-associated genes with their overall frequency among all of the genes from Fig. 1B. Among the most enriched “biological process” terms was neurogenesis (P < 0.01 by hypergeometric distribution), suggesting that at least a subset of the genes in this cluster may have important roles in CNS development.
We next identified the subset of survival-associated genes whose expression was most consistent among paired samples taken from the same tumor. We reasoned that these genes would more likely reflect “intrinsic” deterministic properties of the tumors, such as prognosis. Using an algorithm similar to one described in a previous study (5), we used the eight paired tumor samples to select 500 genes that showed the greatest variation among different tumors compared with that between paired samples from the same tumor. The survival-associated genes that were in this intrinsic gene set are marked in Fig. 4A.
Expression of FABP7 Is Correlated with Survival in Independent Groups of GBM. Based on the preceding analyses, the lack of known functional significance, and the availability of antibodies, we chose to investigate FABP7 expression as a prognostic marker for GBM. FABP7 mRNA and protein have previously been detected in human GBM specimens and cell lines (25). FABP7 is highly expressed in developing brain and retina, and its expression decreases significantly in the adult CNS (40). Based on in vitro results, it has been suggested that FABP7 is required for the establishment of the radial glial system of the developing brain (24).
To verify our array results, we analyzed protein expression of FABP7 by immunohistochemistry in a subset of the GBMs we had examined in Fig. 1. FABP7 protein was barely detectable in normal brain but showed moderate to strong nuclear and cytoplasmic expression in several GBMs. Tumor specimens with FABP7 mRNA expression similar to normal brain tissues had only a few scattered immunopositive cells. In contrast, numerous neoplastic cells with strong FABP7 immunoreactivity were found in tumors with high FABP7 mRNA expression (see Fig. 8, which is published as supporting information on the PNAS web site).
To test the hypothesis that FABP7 might be a useful prognostic marker, we examined FABP7 protein expression in an independent set of 61 GBM (Fig. 9, which is published as supporting information on the PNAS web site). This cadre of patients had a median survival of 0.9 years, and age was closely correlated to survival (P < 0.001; hazard ratio = 1.06; 95% of CI, 1.03–1.08), suggesting that this group was representative of GBM patients in general. Using Cox regression analysis, survival of these patients was loosely associated with nuclear immunoreactivity of FABP7 (P = 0.056). In a Cox model incorporating age and FABP7 nuclear staining, high FABP7 expression was negatively correlated with survival (P = 0.014; hazard ratio = 1.52; 95% of CI, 1.09–2.13) (data not shown). The significant inverse correlation of FABP7 expression with survival after correcting for age indicated that FABP7 carried prognostic information above and beyond age in this group of patients. Because the correlation of nuclear staining and patient survival became stronger after adjusting for age, we stratified the patients by age and reexamined the survival data. Nuclear FABP7 expression significantly correlated with survival in patients younger than the median age (<53 yr; n = 32; 18 were score 0; P = 0.017; hazard ratio = 1.71; 95% of CI, 1.1–2.7) but not in the older patients (>53 yr; n = 29; 17 were score 0; P = 0.75; hazard ratio = 1.09; 95% of CI, 0.63–1.91).
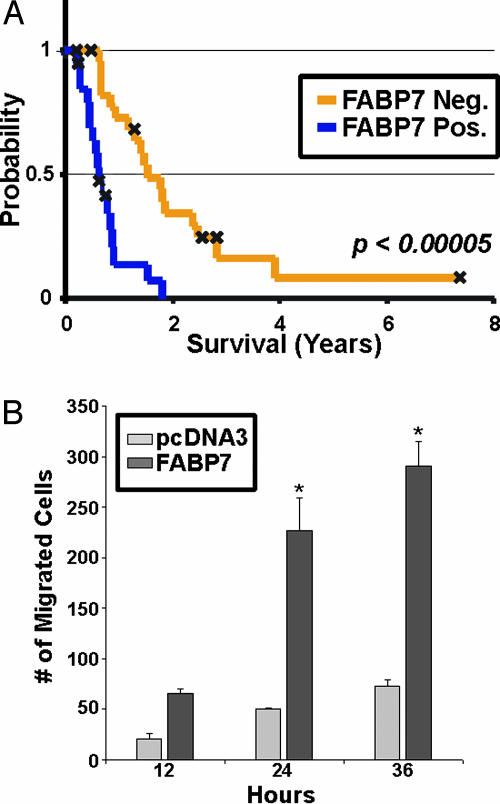
Despite the consistent inverse association between FABP7 expression and survival, the multiple analyses clouded the true prognostic significance of nuclear FABP7 immunoreactivity. We therefore specifically tested the hypothesis that nuclear FABP7 predicts shorter survival in another independent set of 44 GBMs whose median age was 49 years. The median survival of this group of patients was 0.8 years, and age significantly correlated with survival (P = 0.02; hazard ratio = 1.04; 95% of CI, 1.01–1.07). We found that nuclear FABP7 staining (22 were score 0) again had a strong negative correlation with survival (scores 1 or 2 vs. score 0; P < 0.001; hazard ratio = 4.68; 95% of CI, 2.151–10.172). Fig. 5A shows the Kaplan–Meier survival curve of this second set, stratified by FABP7 immunoreactivity. Thus, in GBM, nuclear FABP7, presumably representing the larger gene expression program we observed in our global gene expression analysis, is strongly predictive of poor prognosis, especially in younger patients.
Fig. 5.
FABP7 protein expression is associated with survival in independent cohorts of GBM patients and enhances glioma cell motility in vitro. (A) Kaplan–Meier analysis of nuclear FABP7 expression in an independent cohort of 44 GBM patients. FABP7 immunoreactivity (orange for score 0, blue for score of 1 or 2) is correlated with survival (P < 0.00005). (B) SF767MG cells were transfected and plated in the upper chamber of a transwell system (see text). Results represent average number of migrated cells from three separate transfections. Error bars represent standard error of the mean. *, P < 0.001.
FABP7 Enhances Glioma Cell Migration. We sought to identify a role for FABP7 in the pathogenesis of GBM. Transient expression experiments did not reveal an effect of expression on either cell cycle progression or the rate of spontaneous apoptosis (data not shown). Given the documented expression of FABP7 in radial glia, which extend processes and migrate through the developing nervous system, we investigated the effect of FABP7 expression on the motility of glioma cells. We transfected FABP7 cDNA into SF767 glioma cells, which express low levels of FABP7, and examined the motility of these cells. We found that FABP7-transfected cells displayed 5-fold greater migration than control cells (Fig. 5B). This suggests that the relatively poorer prognosis portended by overexpression of FABP7 may be due to increased migration and invasion of tumor cells into the surrounding brain parenchyma.
Discussion
This systematic characterization of gene expression patterns in GBM has given us insights into its biology and clinical behavior. The hallmark histopathologic features of these tumors were reflected in their gene expression profiles. Samples from different regions of the same tumor, which varied in histopathological features (e.g., mitotic index or necrosis), had corresponding differences in gene expression profiles. As revealed by hierarchical cluster analysis, the expression patterns in paired specimens from the same tumors were more similar to each other than to any other GBM specimen, indicating that the dominant features of gene expression profiles reflected the intrinsic characteristics of the tumors and did not depend on intratumoral histopathological variation. Gene expression profiles may thus more accurately reflect both biologic and clinical differences between GBMs than histological parameters.
Using a combination of unsupervised and supervised algorithms, we identified two molecularly distinct subgroups of GBM. Although these two groups expressed similar levels of many of the GBM-specific genes (such as those involved in angiogenesis, microglial/macrophage infiltration, and chromosomal amplifications), they showed a striking difference in expression of a group of genes correlated with survival but not obviously associated with known diagnostic characteristics. Using paired tumor specimens, we focused on an “intrinsic” subset of genes and confirmed the association of protein expression of one of these (FABP7) with survival in two independent groups of patients.
Understanding the functional roles of the genes whose expression predicts survival could enhance our understanding of the pathogenesis of GBM. Many of these genes are more highly expressed during CNS development than in mature brains, and some are specifically expressed in developing glial and oligodendroglial cells (FABP7, OLIG1, OLIG2, and PTPRZ1). This expression signature may reflect the pretransformation precursor of Group 2 GBMs. Because tumors commonly “inherit” important aspects of their behavior from their normal progenitors, biological properties of these precursor cells may lead to GBMs that are more aggressive.
A significant proportion of survival-associated genes are involved in cell migration, including glial-guided neuronal migration and adhesion of glioma cells to various ECM proteins (e.g., BCAN, PTPRZ1, CRMP5, and FABP7) (41–44). Evidence for a role of brevican in glioma invasion has been demonstrated in animal models (45). We found that glioma cells overexpressing FABP7 demonstrated increased migration rates in vitro. Thus, Group 2 tumors may be more infiltrative and aggressive, resulting in shorter survival times.
An important extension of these results will be to refine our understanding of the prognostic value of the gene expression patterns we have identified and to develop a small panel of well characterized markers that can be rapidly analyzed in clinical laboratories. Although our analysis does not validate a specific signature of multiple genes associated with survival, our examination of FABP7 in two independent sets of tumors provides strong evidence that the gene expression signature associated with survival will contain other genes of prognostic import. We suggest that stratification of GBMs, based on expression of FABP7 and other markers that identify poor-prognosis GBMs, may allow more accurate prognostic predictions and the development of therapies optimized for each subtype.
Supplementary Material
Acknowledgments
We thank the members of the P.O.B., D.B., and M.A.I. laboratories for helpful advice and discussions. The University of California, San Francisco, is a National Cancer Institute-designated Specialized Program of Research Excellence for Brain Tumors. This work was supported by National Institutes of Health Grant CA85129-04 (to P.O.B. and D.B.), National Institute of General Medical Sciences Training Grant GM07365 (to M.D.), the Theodora B. Betz Foundation and the Jordan and Kyra Memorial Fund (to M.A.I.), and a University of California, San Francisco, Cancer Center Core Grant (to K.R.L.). P.O.B. is an associate investigator of the Howard Hughes Medical Institute.
Author contributions: Y.L., M.D., A.W.B., K.D.A., M.K.N., K.R.L., D.B., P.O.B., and M.A.I. designed research; Y.L., M.D., A.W.B., K.D.A., and M.S.B. performed research; Y.L., M.D., A.W.B., K.D.A., K.R.L., P.O.B., and M.A.I. analyzed data; and Y.L., M.D., K.R.L., P.O.B., and M.A.I. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GBM, glioblastoma multiforme; OAC, oligoastrocytoma; ODG, oligodendroglioma; ECM, extracellular matrix.
Data deposition: The raw array data have been deposited in the Stanford Microarray Database, smd.stanford.edu.
References
- 1.Lacroix, M., Abi-Said, D., Fourney, D. R., Gokaslan, Z. L., Shi, W., DeMonte, F., Lang, F. F., McCutcheon, I. E., Hassenbusch, S. J., Holland, E., et al. (2001) J. Neurosurg. 95, 190–198. [DOI] [PubMed] [Google Scholar]
- 2.Kleihues, P., Louis, D. N., Scheithauer, B. W., Rorke, L. B., Reifenberger, G., Burger, P. C. & Cavenee, W. K. (2002) J. Neuropathol. Exp. Neurol. 61, 215–225; discussion 226–229. [DOI] [PubMed] [Google Scholar]
- 3.Nagane, M., Huang, H. J. & Cavenee, W. K. (1997) Curr. Opin. Oncol. 9, 215–222. [DOI] [PubMed] [Google Scholar]
- 4.Alizadeh, A. A., Eisen, M. B., Davis, R. E., Ma, C., Lossos, I. S., Rosenwald, A., Boldrick, J. C., Sabet, H., Tran, T., Yu, X., et al. (2000) Nature 403, 503–511. [DOI] [PubMed] [Google Scholar]
- 5.Perou, C. M., Sorlie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., Rees, C. A., Pollack, J. R., Ross, D. T., Johnsen, H., Akslen, L. A., et al. (2000) Nature 406, 747–752. [DOI] [PubMed] [Google Scholar]
- 6.Watson, M. A., Perry, A., Budhjara, V., Hicks, C., Shannon, W. D. & Rich, K. M. (2001) Cancer Res. 61, 1825–1829. [PubMed] [Google Scholar]
- 7.Fuller, G. N., Rhee, C. H., Hess, K. R., Caskey, L. S., Wang, R., Bruner, J. M., Yung, W. K. & Zhang, W. (1999) Cancer Res. 59, 4228–4232. [PubMed] [Google Scholar]
- 8.Sallinen, S. L., Sallinen, P. K., Haapasalo, H. K., Helin, H. J., Helen, P. T., Schraml, P., Kallioniemi, O. P. & Kononen, J. (2000) Cancer Res. 60, 6617–6622. [PubMed] [Google Scholar]
- 9.Rickman, D. S., Bobek, M. P., Misek, D. E., Kuick, R., Blaivas, M., Kurnit, D. M., Taylor, J. & Hanash, S. M. (2001) Cancer Res. 61, 6885–6891. [PubMed] [Google Scholar]
- 10.Hunter, S., Young, A., Olson, J., Brat, D. J., Bowers, G., Wilcox, J. N., Jaye, D., Mendrinos, S. & Neish, A. (2002) J. Neuropathol. Exp. Neurol. 61, 275–281. [DOI] [PubMed] [Google Scholar]
- 11.Shai, R., Shi, T., Kremen, T. J., Horvath, S., Liau, L M., Cloughesy, T. F., Mischel, P. S. & Nelson, S. F. (2003) Oncogene 22, 4918–4923. [DOI] [PubMed] [Google Scholar]
- 12.Godard, S., Getz, G., Delorenzi, M., Farmer, P., Kobayashi, H., Desbaillets, I., Nozaki, M., Diserens, A. C., Hamou, M. F., Dietrich, P. Y., et al. (2003) Cancer Res. 63, 6613–6625. [PubMed] [Google Scholar]
- 13.van den Boom, J., Wolter, M., Kuick, R., Misek, D. E., Youkilis, A. S., Wechsler, D. S., Sommer, C., Reifenberger, G. & Hanash, S. M. (2003) Am. J. Pathol. 163, 1033–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nutt, C. L., Mani, D. R., Betensky, R. A., Tamayo, P., Cairncross, J. G., Ladd, C., Pohl, U., Hartmann, C., McLaughlin, M. E., Batchelor, T. T., et al. (2003) Cancer Res. 63, 1602–1607. [PubMed] [Google Scholar]
- 15.Mukasa, A., Ueki, K., Matsumoto, S., Tsutsumi, S., Nishikawa, R., Fujimaki, T., Asai, A., Kirino, T. & Aburatani, H. (2002) Oncogene 21, 3961–3968. [DOI] [PubMed] [Google Scholar]
- 16.Mischel, P. S., Shai, R., Shi, T., Horvath, S., Lu, K. V., Choe, G., Seligson, D., Kremen, T. J., Palotie, A., Liau, L. M., et al. (2003) Oncogene 22, 2361–2373. [DOI] [PubMed] [Google Scholar]
- 17.Huang, H., Colella, S., Kurrer, M., Yonekawa, Y., Kleihues, P. & Ohgaki, H. (2000) Cancer Res. 60, 6868–6874. [PubMed] [Google Scholar]
- 18.Ljubimova, J. Y., Lakhter, A. J., Loksh, A., Yong, W. H., Riedinger, M. S., Miner, J. H., Sorokin, L. M., Ljubimov, A. V. & Black, K. L. (2001) Cancer Res. 61, 5601–5610. [PubMed] [Google Scholar]
- 19.Markert, J. M., Fuller, C. M., Gillespie, G. Y., Bubien, J. K., McLean, L. A., Hong, R. L., Lee, K., Gullans, S. R., Mapstone, T. B. & Benos, D. J. (2001) Physiol. Genom. 5, 21–33. [DOI] [PubMed] [Google Scholar]
- 20.MacDonald, T. J., Brown, K. M., LaFleur, B., Peterson, K., Lawlor, C., Chen, Y., Packer, R.J., Cogen, P. & Stephan, D.A. (2001) Nat. Genet. 29, 143–152. [DOI] [PubMed] [Google Scholar]
- 21.Pomeroy, S. L., Tamayo, P., Gaasenbeek, M., Sturla, L. M., Angelo, M., McLaughlin, M. E., Kim, J. Y., Goumnerova, L. C., Black, P. M., Lau, C., et al. (2002) Nature 415, 436–442. [DOI] [PubMed] [Google Scholar]
- 22.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simmons, M. L., Lamborn, K. R., Takahashi, M., Chen, P., Israel, M. A., Berger, M. S., Godfrey, T., Nigro, J., Prados, M., Chang, S., et al. (2001) Cancer Res. 61, 1122–1128. [PubMed] [Google Scholar]
- 24.Feng, L., Hatten, M. E. & Heintz, N. (1994) Neuron 12, 895–908. [DOI] [PubMed] [Google Scholar]
- 25.Godbout, R., Bisgrove, D. A., Shkolny, D. & Day, R. S., III (1998) Oncogene 16, 1955–1962. [DOI] [PubMed] [Google Scholar]
- 26.Lu, Q. R., Park, J. K., Noll, E., Chan, J. A., Alberta, J., Yuk, D., Alzamora, M. G., Louis, D. N., Stiles, C. D., Rowitch, D. H., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 10851–10856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen, X., Cheung, S. T., So, S., Fan, S. T., Barry, C., Higgins, J., Lai, K. M., Ji, J., Dudoit, S., Ng, I. O., et al. (2002) Mol. Biol. Cell 13, 1929–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung, V., Romeike, B. F., Henn, W., Feiden, W., Moringlane, J. R., Zang, K. D. & Urbschat, S. (1999) J. Neuropathol. Exp. Neurol. 58, 993–999. [DOI] [PubMed] [Google Scholar]
- 29.Burger, P. C. & Kleihues, P. (1989) Cancer 63, 2014–2023. [DOI] [PubMed] [Google Scholar]
- 30.Ricciardi-Castagnoli, P. & Granucci, F. (2002) Nat. Rev. Immunol. 2, 881–889. [DOI] [PubMed] [Google Scholar]
- 31.Graeber, M. B., Scheithauer, B. W. & Kreutzberg, G. W. (2002) Glia 40, 252–259. [DOI] [PubMed] [Google Scholar]
- 32.Roggendorf, W., Strupp, S. & Paulus, W. (1996) Acta Neuropathol. 92, 288–293. [DOI] [PubMed] [Google Scholar]
- 33.Nishie, A., Ono, M., Shono, T., Fukushi, J., Otsubo, M., Onoue, H., Ito, Y., Inamura, T., Ikezaki, K., Fukui, M., et al. (1999) Clin. Cancer Res. 5, 1107–1113. [PubMed] [Google Scholar]
- 34.Crowther, M., Brown, N. J., Bishop, E. T. & Lewis, C. E. (2001) J. Leukocyte Biol. 70, 478–490. [PubMed] [Google Scholar]
- 35.Ziemer, L. S., Koch, C. J., Maity, A., Magarelli, D. P., Horan, A. M. & Evans, S. M. (2001) Neoplasia 3, 500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chintala, S. K., Sawaya, R., Gokaslan, Z. L., Fuller, G. & Rao, J. S. (1996) Cancer Lett. 101, 107–114. [DOI] [PubMed] [Google Scholar]
- 37.St Croix, B., Rago, C., Velculescu, V., Traverso, G., Romans, K. E., Montgomery, E., Lal, A., Riggins, G. J., Lengauer, C., Vogelstein, B., et al. (2000) Science 289, 1197–1202. [DOI] [PubMed] [Google Scholar]
- 38.Whitfield, M. L., Sherlock, G., Saldanha, A. J., Murray, J. I., Ball, C. A., Alexander, K. E., Matese, J. C., Perou, C. M., Hurt, M. M., Brown, P. O., et al. (2002) Mol. Biol. Cell 13, 1977–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diehn, M., Sherlock, G., Binkley, G., Jin, H., Matese, J. C., Hernandez-Boussard, T., Rees, C. A., Cherry, J. M., Botstein, D., Brown, P. O., et al. (2003) Nucleic Acids Res. 31, 219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimizu, F., Watanabe, T. K., Shinomiya, H., Nakamura, Y. & Fujiwara, T. (1997) Biochim. Biophys. Acta 1354, 24–28. [DOI] [PubMed] [Google Scholar]
- 41.Grumet, M., Friedlander, D. R. & Sakurai, T. (1996) Perspect. Dev. Neurobiol. 319–342. [PubMed]
- 42.Nutt, C. L., Matthews, R. T. & Hockfield, S. (2001) Neuroscientist 7, 113–122. [DOI] [PubMed] [Google Scholar]
- 43.Wang, L. H. & Strittmatter, S. M. (1996) J. Neurosci. 16, 6197–6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng, C., Heintz, N. & Hatten, M. E. (1996) Science 272, 417–419. [DOI] [PubMed] [Google Scholar]
- 45.Nutt, C. L., Zerillo, C. A., Kelly, G. M. & Hockfield, S. (2001) Cancer Res. 61, 7056–7059. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.