ABSTRACT
Urease, a nickel-containing metalloenzyme, was the first enzyme to be crystallized and has a prominent position in the history of biochemistry. In the present study, we identified a nickel urease gene cluster, ureABCEFGDH, in Bacillus paralicheniformis ATCC 9945a and characterized it in Escherichia coli. Enzymatic assays demonstrate that this oxygen-stable urease is also an iron-containing acid urease. Heterologous expression assays of UreH suggest that this accessory protein is involved in the transmembrane transportation of nickel and iron ions. Moreover, this iron-containing acid urease has a potential application in the degradation of urea in rice wine. The present study not only enhances our understanding of the mechanism of activation of urease but also provides insight into the evolution of metalloenzymes.
IMPORTANCE An iron-containing, oxygen-stable acid urease from B. paralicheniformis ATCC 9945a with good enzymatic properties was characterized. This acid urease shows activities toward both urea and ethyl carbamate. After digestion with 6 U/ml urease, approximately 92% of the urea in rice wine was removed, suggesting that this urease has great potential in the food industry.
KEYWORDS: iron-containing urease, acid urease, UreH, metal transporter, urea, ethyl carbamate
INTRODUCTION
Ureases, enzymes that hydrolyze urea to ammonia and carbamic acid (H2N-CO-NH2 + H2O → H2N-COOH + NH3 and H2N-COOH + H2O → H2CO3 + NH3), have been found in plants and many microorganisms (1–5). Because of their multifaceted contributions to the environmental cycling of nitrogen and localized neutralization of pH (1, 6, 7), ureases have always attracted intensive attention. In fact, jack bean urease (Canavalia ensiformis) was identified as the first metalloenzyme to contain a nickel ion (8) and was the first enzyme to be crystallized (9). Thus, ureases possess a prominent place in the history of biochemistry. To understand the activation mechanisms of ureases and the roles of accessory proteins in the posttranslational modifications of ureases, many different microbial nickel-containing ureases have been identified and characterized. Although the structural subunits and enzyme active sites have been well studied (3–5, 10–15), the function of each accessory protein and the urease maturation mechanism are still poorly understood (3, 4, 16–20).
In most of the microbial ureases identified, the accessory proteins (UreE, UreD, UreG, and UreF) are essential for active-site metalation and urease maturation (3, 4, 20). UreE, a metallochaperone, delivers nickel to urease apoprotein via the chaperone complex (UreD-UreF-UreG)3, in which UreD functions as a scaffold for the recruitment of other accessory subunits, UreF guarantees metallocenter fidelity, and UreG is a GTPase for the incorporation of nickel ions (3, 4, 19–21). Although the ureD gene in Helicobacter species (18) was renamed ureH, its function is consistent with that of other UreD accessory proteins. However, in some Bacillus species, the ureD and ureH genes are both conserved (3), and the function of this heterogeneous UreH is still unclear. In addition, enzymatic elimination of urea by acid ureases has been considered to be the most efficient method of decreasing the concentration of the carcinogenic compound ethyl carbamate (EC; also known as urethane) (22–26) in traditional fermented foods (27–30). Thus, the identification of a novel microbial acid urease not only contributes to our understanding of the assembly process but also provides an alternative way of eliminating urea in rice wine.
RESULTS
Cloning and sequence alignment of the urease gene cluster from Bacillus paralicheniformis ATCC 9945a.
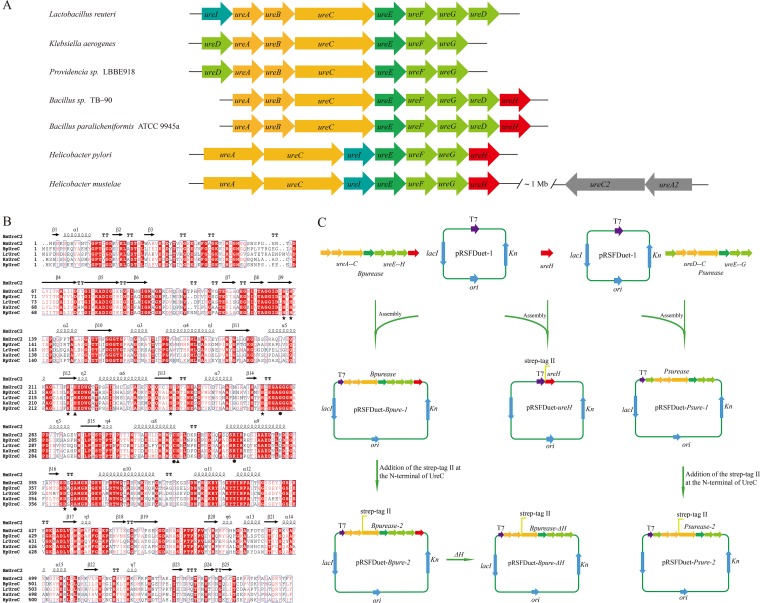
The urease gene cluster (ureABCEFGDH) was cloned from the genome of B. paralicheniformis ATCC 9945a. As shown in Fig. 1A, the B. paralicheniformis ATCC 9945a urease (BpUrease) gene cluster consists of three structural genes (ureA, ureB, and ureC) and five accessory genes (ureE, ureF, ureG, ureD, and ureH). The structural genes ureA, ureB, and ureC and the accessory genes ureE, ureF, ureG, and ureD are highly conserved in all microorganisms except Helicobacter species, in which the structural genes ureA and ureA2 are fusions of ureA and ureB. In comparison, other accessory genes, for instance, the urea transporter gene ureI, are conserved only in Lactobacillus reuteri and Helicobacter species, and an accessory gene, ureH, with unknown function was present in the BpUrease cluster. Notably, the recently identified oxygen-labile, iron-containing Helicobacter mustelae urease (HmUrease) cluster contains an extra copy each of the structural genes ureA and ureC (31). Remarkably, sequence analysis showed that BpUreC shares 61.1% identity with HmUreC2. In addition, a multiple-sequence alignment of BpUreC with other UreC proteins, including the iron-containing HmUreC2 (Fig. 1B), confirmed that all the amino acids involved in nickel binding (His137, His139, Lys220, His247, His275, and Asp363) and catalysis (His222 and His323) are highly conserved (3, 4, 31, 32). The Gly280, Cys322, Arg339, and Ala366 residues, which are thought to be important for the catalytic mechanism, but not for metal binding, are also strictly conserved (32).
FIG 1.
Characterization of urease gene clusters and construction of plasmids. (A) Structures of representative urease gene clusters in different microorganisms. The structural genes (ureA, ureB, and ureC), ureE, the urease accessory genes (ureF, ureG, ureD), ureI, ureH, and ureC2A2 are color-coded orange, dark green, light green, light blue, red, and gray, respectively. (B) Alignment of the structural C subunits of the ureases. HmUreC2, BpUreC, LrUreC, KaUreC, and HpUreC represent the C subunits of the ureases from H. mustelae (UreC2), B. paralicheniformis ATCC 9945a, L. reuteri, K. aerogenes, and H. pylori, respectively. Metal ligands are labeled with stars; two functional histidine residues are labeled with filled triangles; four residues thought to be important in the catalytic mechanism are labeled with circles. (C) Construction of the recombinant plasmid for the expression of urease and ureH in E. coli. The Strep tag (WSHPQFEK) was fused at the N termini of both UreC and UreH for protein purification.
Analysis of the specificity of BpUrease for metal ions.
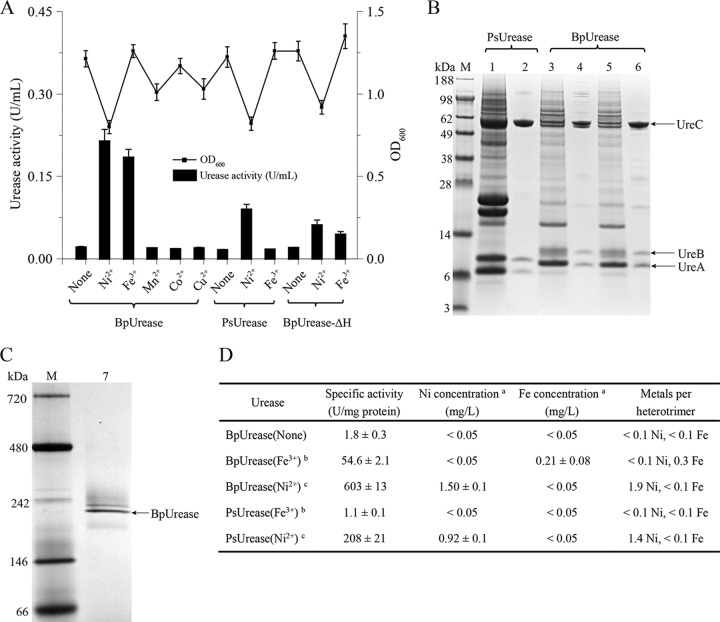
After amplification and sequence analysis, the BpUrease gene cluster was subcloned into expression plasmid pRSFDuet-1 (Fig. 1C), and the resulting plasmid, pRSFDuet-Bpure-2, was transformed into Escherichia coli BL21(DE3) for investigation. To eliminate uncertainty, modified Riesenberg medium (minimal medium) was used for investigating the specificity of BpUrease for metal ions. Different metal ion salts (NiSO4, FeCl3, MnCl2, CuSO4, and CoSO4) were added during cultivation in order to examine their effects on BpUrease activity. Measurements of BpUrease activity were carried out with extracted enzyme. As shown in Fig. 2A, only the addition of NiSO4 or FeCl3 resulted in an obvious accumulation of urease activity, at 0.21 U/ml or 0.18 U/ml, respectively, suggesting that only Ni2+ and Fe3+ can be coordinated to yield an active urease. In contrast, a Providencia sp. urease (PsUrease), encoded by the ureDABCEFG gene cluster, exhibited urease activity (0.09 U/ml) only with the addition of NiSO4. These results suggested that BpUrease can be actively expressed in the presence of Ni2+ or Fe3+.
FIG 2.
Identification of the iron-containing urease. (A) Effects of medium supplementation with metal ion salts and knockout of ureH from the urease gene cluster on recombinant E. coli urease activity. (B) SDS-PAGE analysis of purified recombinant urease. Lane M, protein molecular mass standard; lane 1, crude supernatant of PsUrease, which was supplemented with NiSO4 during expression; lane 2, purified PsUrease; lane 3, crude supernatant of BpUrease (Fe3+), which was supplemented with FeCl3 during expression; lane 4, purified BpUrease (Fe3+); lane 5, crude supernatant of BpUrease (Ni2+), which was supplemented with NiSO4 during expression; lane 6, purified BpUrease (Ni2+). (C) Native PAGE analysis of purified BpUrease. Lanes: M, markers; 7, purified BpUrease (Fe3+). (D) Metal contents of the purified proteins. a, the metal content in 1,000 mg/liter urease was determined by atomic absorption spectrometry; b, urease was expressed in the presence of 1 mM FeCl3; c, urease was expressed in the presence of 1 mM NiSO4.
BpUrease is an iron-containing urease.
The recombinant PsUrease and BpUrease (supplemented with Fe3+ or Ni2+) were fused with a Strep tag (WSHPQFEK) (Fig. 1C) and were purified with StrepTrap HP affinity columns (Fig. 2B). To confirm the composition of the active BpUrease, the purified urease was also investigated by native polyacrylamide gel electrophoresis (PAGE) analysis. As shown in Fig. 2C, the molecular mass of BpUrease was approximately 220 kDa, suggesting that BpUrease forms a (UreABC)3 structure that is consistent with those of most other microbial ureases. To confirm that BpUrease is an iron-containing urease, the metal ion (Fe3+ or Ni2+) contents of purified PsUrease and BpUrease were determined with an atomic absorption spectrometer (Agilent, USA). As shown in Fig. 2D, BpUrease supplemented with FeCl3 [BpUrease (Fe3+)] and BpUrease supplemented with NiSO4 [BpUrease (Ni2+)] contained much higher concentrations of Fe3+ (0.21 ± 0.08 mg/liter, corresponding to 0.3 equivalent of Fe3+ per heterotrimer) and Ni2+ (1.5 ± 0.1 mg/liter, corresponding to 2 equivalents of Ni2+ per heterotrimer), respectively, than the control BpUrease [BpUrease (None)]. In contrast, no detectable Fe3+ or Ni2+ was detected in BpUrease (Ni2+) or BpUrease (Fe3+), respectively. Meanwhile, PsUrease (Fe3+) failed to integrate Fe3+, resulting in a specific activity much lower than that of PsUrease (Ni2+). Taken together, these results demonstrate that the BpUrease identified is an iron-containing urease.
Functional identification of the accessory protein BpUreH.
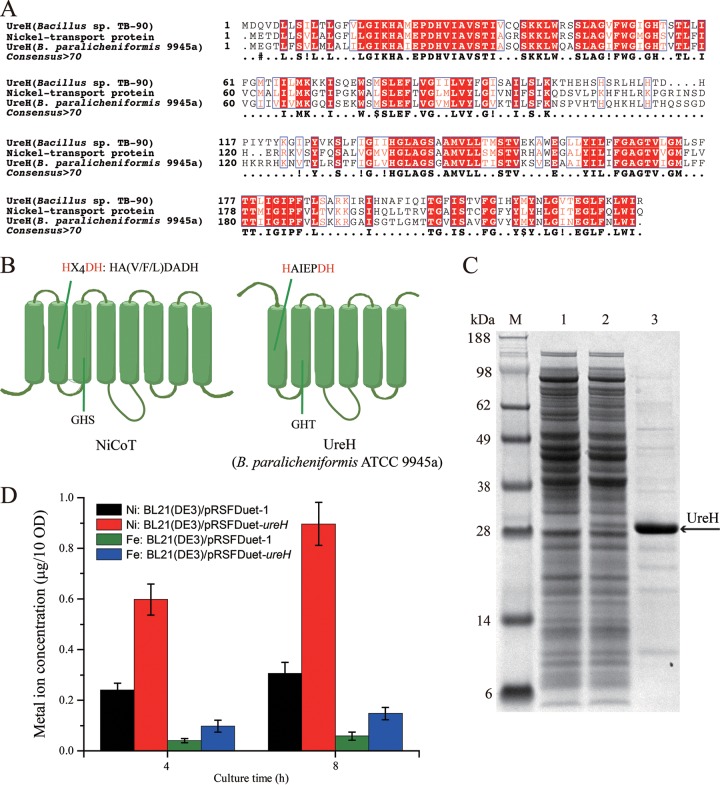
To explore the function of the accessory protein BpUreH, multiple-sequence alignment analysis was carried out. As shown in Fig. 3A, BpUreH shares 60% identity with the nickel transport protein from Lihuaxuella thermophila and with Bacillus sp. strain TB-90 UreH (33), but it has <16.1% amino acid identity with H. pylori UreH (18), indicating that BpUreH is a nickel transporter. Thus, the topological model of BpUreH was predicted by the TMHMM server and was compared with that of a well-studied nickel/cobalt transporter (NiCoT) (Fig. 3B). BpUreH contains six transmembrane domains (TMDs) and the conserved signature sequence (HX4DH) of the NiCoTs (34). To experimentally reveal the function of BpUreH, the effects of UreH inactivation on urease activity were first investigated (Fig. 1C). As shown in Fig. 2A, deletion of UreH dramatically decreased urease activity, suggesting the critical role of UreH in urease activity. Then BpUreH was subcloned into pRSFDuet-1 (Fig. 1C) and was overexpressed in E. coli BL21(DE3) (Fig. 3C), and the intracellular concentrations of Ni2+ and Fe3+ were comparatively investigated. As shown in Fig. 3D, overexpression of BpUreH resulted in significant increases in both Ni2+ and Fe3+ concentrations. In conclusion, the results demonstrate that in contrast to H. pylori UreH, BpUreH is likely a transmembrane protein capable of transferring nickel and iron ions for active urease assembly.
FIG 3.
Functional analysis of UreH. (A) Alignment of UreH from B. paralicheniformis ATCC 9945a with the nickel transport protein from L. thermophila and with Bacillus sp. strain TB-90 UreH. (B) Topology of nickel/cobalt transporters (NiCoTs) and UreH from B. paralicheniformis ATCC 9945a. The signature sequence HX4DH in TMD II of the NiCoT and the corresponding sequence HAIEPDH in UreH are shown above the diagrams. (C) SDS-PAGE analysis of UreH expression. Lane M, protein molecular weight standard; lane 1, BL21(DE3)/pRSFDuet-1 cell lysis supernatant; lane 2, BL21(DE3)/pRSFDuet-ureH cell lysis supernatant; lane 3, purified UreH. (D) Intracellular metal ion contents of recombinant strains. After induction for 4 or 8 h, strains were washed and lysed, and the soluble extracts were used for the analysis of metal ion contents by atomic absorption spectrometry.
Biochemical characterization of BpUrease.
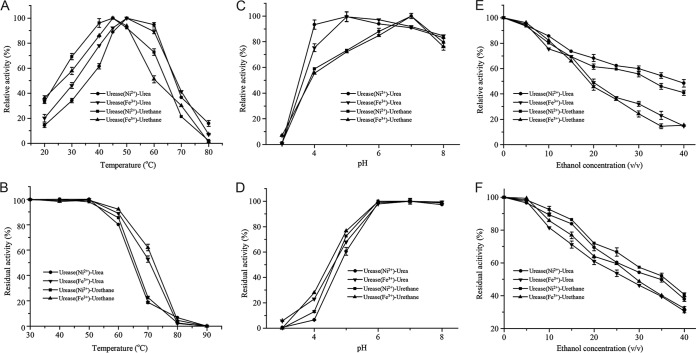
To evaluate the potential applications of BpUrease, its kinetic parameters and biochemical properties were examined. The specific activity and catalytic efficiency (kcat/Km) of BpUrease (Ni2+) (603 U/mg and 1.2E+05 s−1 M−1, respectively) were much higher than those of BpUrease (Fe3+) (54.6 U/mg and 9,557 s−1 M−1, respectively) (Table 1), indicating that Ni2+ was preferred over Fe3+. However, the optimal temperature and pH were 50°C and 5.0, respectively, for both BpUrease (Fe3+) and BpUrease (Ni2+) (Fig. 4A and C), suggesting that BpUrease is an acid urease. Moreover, both forms of BpUrease, that with Fe3+ and that with Ni2+, were stable at temperatures lower than 50°C (Fig. 4B) and in the pH range of 6.0 to 8.0 (Fig. 4D). BpUrease (Fe3+) showed higher thermostability at 60°C than BpUrease (Ni2+) (Fig. 4B) and higher acid resistance at pH 4.0. (Fig. 4D). Both BpUrease (Fe3+) and BpUrease (Ni2+) showed high ethanol tolerance. More than 70% of their maximal activity was detected in the presence of 15% ethanol (Fig. 4E) or after treatment with 5% ethanol for 2 h (Fig. 4F), indicating the potential application of this acid urease for eliminating urea from a model rice wine with a pH of 4.5 and an ethanol concentration of 15%. Additionally, this urease showed enzymatic activity toward EC (Table 1; Fig. 4).
TABLE 1.
Kinetic parameters of BpUrease
| Substrate and enzyme | Sp act (U · mg−1) | Km (mM) | Vmax (μmol · mg−1 · min−1) | kcat (s−1) | kcat/Km (s−1 · M−1) |
|---|---|---|---|---|---|
| Urethane | |||||
| BpUrease (Ni2+) | 23.1 ± 0.4 | 1,018 ± 58 | 118 ± 5 | 433 ± 22 | 425 ± 19 |
| BpUrease (Fe3+) | 2.5 ± 0.2 | 958 ± 52 | 15.2 ± 1.1 | 56 ± 3 | 58 ± 3.9 |
| Urea | |||||
| BpUrease (Ni2+) | 603 ± 13 | 23.7 ± 1.5 | 769 ± 39 | 2,820 ± 135 | (1.2 ± 0.1)E + 05 |
| BpUrease (Fe3+) | 54.6 ± 2.1 | 28.2 ± 1.7 | 73.5 ± 5.3 | 270 ± 14 | 9,557 ± 478 |
FIG 4.
Effects of temperature, pH, and ethanol on enzyme activity. (A and B) Effects of temperature on enzyme activity (A) and stability (B). (C and D) Effects of pH on enzyme activity (C) and stability (D). (E and F) Effects of ethanol on enzyme activity (E) and stability (F).
Based on its chemical characterization, the ability of the purified urease to eliminate urea and EC from rice wine (pH 4.5; 15% ethanol; 50 mg/liter urea; 450 μg/liter EC) was evaluated (Fig. 5). After digestion with 2 U/ml urease at 37°C for 50 h, approximately 56.4% of the urea was removed by BpUrease (Fe3+). When urease was added to 6 U/ml, approximately 92% of the urea was removed, suggesting the potential application of this urease for the degradation of urea in rice wine. In contrast, no hydrolysis of EC was observed (Fig. 5), which could be ascribed to the low affinity (Km) and catalytic efficiency (kcat/Km) of this urease toward EC (Table 1; see also Fig. S1 in the supplemental material).
FIG 5.
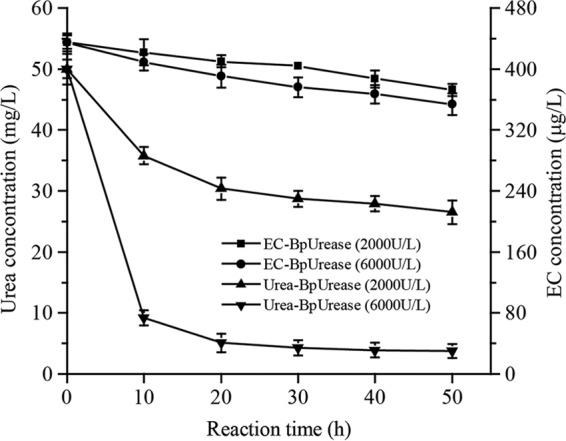
Evaluation of urea and urethane degradation by BpUrease (Fe3+) in rice wine The purified BpUrease (Fe3+) (at a final titer of 2 or 6 U/ml) was added to a wine sample (pH 4.5; 15% [vol/vol] ethanol), and the reaction system was incubated at 37°C for 2 days.
DISCUSSION
Although ureases are ancient metalloenzymes, the identification and characterization of ureases, and the investigation of their maturation machinery, are still of great significance because of their special position in the history of biochemistry and their complicated assembly process (3). In the present study, we identified an iron-containing acid urease with activities toward urea and EC. The biochemical characterization results showed that this urease assimilates only nickel and iron ions during the formation of its active site. Moreover, our results demonstrate that the accessory protein BpUreH is involved in the transmembrane transportation of nickel and iron ions for the assembly of active urease.
Recently, Carter et al. investigated and characterized an iron-dependent urease (UreA2B2) in H. mustelae (31). In contrast to other reported archetypical nickel-containing ureases, this neutral urease is oxygen labile and Fe2+ dependent, and its maturation does not require the typical accessory proteins. Moreover, the transcription of ureA2B2 is upregulated by iron and downregulated by nickel (35). Contrary to the iron-containing urease from H. mustelae, BpUrease identified in this study is an oxygen-stable, nickel- or iron-dependent acid urease, and it requires accessory proteins for its maturation, which are similar to those of other nickel ureases. Additionally, a multiple-sequence alignment of BpUreC with other UreC proteins (nickel or iron containing) (Fig. 1B) showed that all the amino acids involved in nickel binding (His137, His139, Lys220, His247, His275, and Asp363) and catalysis (His222 and His323) were highly conserved. This suggests that the compatibility of BpUrease with both nickel and iron ions, in contrast to the iron dependence of the H. mustelae urease (31), should be ascribed mainly to the coordinated function of the accessory proteins UreG, UreF, and UreD.
In addition to the structural subunits UreABC, the functions of four accessory subunits, UreEFGD, especially those from Klebsiella aerogenes, have been well characterized (3, 4, 21). Although a UreH subunit was identified in the ureases from Helicobacter species (18), its function has been demonstrated to be consistent with those of other UreD accessory proteins. Interestingly, only BpUrease and the urease from Bacillus sp. strain TB-90 (BsUrease) preserve both UreH and UreD (Fig. 1A), although the function of the UreH subunit is still unclear. In this study, our results confirmed that BpUreH is involved in the transmembrane transportation of nickel and iron ions for the assembly of active urease. In fact, BpUrease showed higher specific activity than PsUrease and contained more nickel ions per heterotrimer (Fig. 2D), which also indirectly demonstrates the contribution of BpUreH to the formation of active sites by providing more nickel or iron ions than PsUrease, which was expressed in the UreH-lacking strains. With regard to the elimination of urea from rice wine (27), many ureases from different species, for instance, Lactobacillus fermentum (28) and L. reuteri (30), have been purified and evaluated for potential applications. Especially, the acid ureases from L. fermentum and Arthrobacter mobilis have already been made commercially available. Although these nickel-dependent acid ureases can efficiently decompose the urea in alcoholic beverages, the existence of nickel ions always gives rise to a safety concern. The iron-dependent acid urease identified in this study also exhibits good properties (Fig. 4 and 5; Table 1) for the degradation of urea in rice wine. Hence, achievement of its high-level expression with the food-grade Bacillus subtilis expression system (36) should be an attractive means of promoting practical applications.
In conclusion, an iron-containing, oxygen-stable acid urease from B. paralicheniformis ATCC 9945a was identified and characterized. Analysis of results from the deletion and overexpression of UreH confirmed that the accessory protein UreH is involved in the transmembrane transportation of nickel and iron ions for the formation of active sites. Moreover, this iron-containing acid urease showed good properties for urea decomposition in rice wine. The present results not only enhance our understanding of the activation mechanism of urease but also provide an alternative method for eliminating urea in rice wine.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 2, and the primers are listed in Table 3. Escherichia coli strains were grown in Luria-Bertani (LB) medium supplemented with appropriate antibiotics (50 mg/liter kanamycin) when necessary and in Terrific Broth (TB) or modified Riesenberg medium (37) supplemented with 2 mM NiSO4 or 2 mM FeCl3 for protein expression. The modified Riesenberg medium contains glycerol at 8.0 g/liter, (NH4)2HPO4 at 6.0 g/liter, KH2PO4 at 10.5 g/liter, citric acid at 1.7 g/liter, MgSO4 at 1.7 g/liter, thiamine HCl at 0.005 g/liter, and a trace metal solution at 10 ml/liter (pH 6.5). The trace metal solution contains ZnSO4·7H2O at 5.25 g/liter, CuSO4·5H2O at 3.0 g/liter, MnSO4·4H2O at 0.5 g/liter, Na2B4O7·10H2O at 0.23 g/liter, CaCl2 at 2.0 g/liter, and (NH4)6Mo7O2 at 0.1 g/liter (37). All chemicals were of reagent grade and were purchased from Sigma-Aldrich (St. Louis, MO, USA).
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source |
|---|---|---|
| Strains | ||
| Bacillus paralicheniformis ATCC 9945a | Used for cloning the iron-containing urease | CGMCC |
| Providencia sp. LBBE 918 | Used for cloning the nickel-containing urease | CCTCC |
| Escherichia coli | Used for plasmid screening and propagation | Invitrogen |
| JM109 | ||
| BL21(DE3) | Used for heterologous protein expression | Novagen |
| BL21(DE3)/pRSFDuet-Bpure-1 | BL21(DE3) derivative harboring plasmid pRSFDuet-Bpure-1 | This work |
| BL21(DE3)/pRSFDuet-Bpure-2 | BL21(DE3) derivative harboring plasmid pRSFDuet-Bpure-2 | This work |
| BL21(DE3)/pRSFDuet-Bpure-ΔH | BL21(DE3) derivative harboring plasmid pRSFDuet-Bpure-ΔH | This work |
| BL21(DE3)/pRSFDuet-ureH | BL21(DE3) derivative harboring plasmid pRSFDuet-ureH | This work |
| BL21(DE3)/pRSFDuet-Psure-1 | BL21(DE3) derivative harboring plasmid pRSFDuet-Psure-1 | This work |
| BL21(DE3)/pRSFDuet-Psure-2 | BL21(DE3) derivative harboring plasmid pRSFDuet-Psure-2 | This work |
| Plasmids | ||
| pRSFDuet-1 | Kanr; T7 promoter | Novagen |
| pRSFDuet-Bpure-1 | Overexpression of BpUrease gene cluster in E. coli | This work |
| pRSFDuet-Bpure-2 | Overexpression of BpUrease gene cluster in which a Strep tag was added to the amino terminus of the C subunit in E. coli | This work |
| pRSFDuet-Bpure-ΔH | Overexpression of BpUrease gene cluster in which ureH was knocked out from the gene cluster in E. coli | This work |
| pRSFDuet-ureH | Overexpression of ureH in E. coli | This work |
| pRSFDuet-Psure-1 | Overexpression of PsUrease gene cluster in E. coli | This work |
| pRSFDuet-Psure-2 | Overexpression of PsUrease gene cluster in which a Strep tag was added to the amino terminus of the C subunit in E. coli | This work |
TABLE 3.
Primers used in this study

Construction of the plasmids and strains.
The urease gene clusters from Providencia sp. strain LBBE (encoding PsUrease; 5.0 kb, containing ureDABCEFG; GenBank accession no. MF099656) and B. paralicheniformis 9945A (encoding BpUrease; 5.7 kb, containing ureABCEFGDH; GenBank accession no. CP005965.1) were amplified with primers Psure-F1 and Psure-R1 and primers Bpure-F1 and Bpure-R1, respectively (Fig. 1A; Table 3). Then the PCR products were cloned into expression vector pRSFDuet-1 with the ClonExpress II one-step cloning kit (Vazyme, Nanjing, China) to generate pRSFDuet-Psure-1 and pRSFDuet-Bpure-1. Specifically, pRSFDuet-1 was linearized by primers pRSFDuet-F and pRSFDuet-R. Then pRSFDuet-Psure-1 and pRSFDuet-Bpure-1 were amplified with primers Psure-F2 and Psure-R2 and primers Bpure-F2 and ure-R2, respectively, and the PCR fragments were phosphorylated and ligated with a blunting kination ligation (BKL) kit (TaKaRa, Japan) to yield plasmids pRSFDuet-Psure-2 and pRSFDuet-Bpure-2. pRSFDuet-Bpure-2 was first amplified using primers Bpure-F3 and Bpure-R3 and then phosphorylated and ligated to construct plasmid pRSFDuet-Bpure-ΔH. Eventually, the recombinant plasmids were transformed into E. coli BL21(DE3) to construct the recombinant E. coli strains BL21(DE3)/pRSFDuet-Psure-1, BL21(DE3)/pRSFDuet-Psure-2, BL21(DE3)/pRSFDuet-Bpure-1, BL21(DE3)/pRSFDuet-Bpure-2, and BL21(DE3)/pRSFDuet-Bpure-ΔH.
The ureH gene from B. paralicheniformis 9945A was amplified with primers BpureH-F and BpureH-R. After purification, the PCR products were assembled with the linear plasmid pRSFDuet-1 to generate pRSFDuet-ureH. Then pRSFDuet-ureH was transformed into E. coli BL21(DE3) to generate BL21(DE3)/pRSFDuet-ureH.
Sequence analysis.
Multiple alignments of protein sequences were conducted using the Clustal Omega program (http://www.ebi.ac.uk/Tools/msa/clustalo/) and Basic Local Alignment Search Tool programs (http://www.ncbi.nlm.nih.gov/BLAST/). The transmembrane helices in proteins were predicted by the TMHMM server, version 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/). The ESPript 3.0 (38) Web server (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) was used to predict the secondary structures of the urease proteins shown in Fig. 1B, using the structure of the iron-containing urease from H. mustelae as a template (PDB code 3QGA).
Cultivation of recombinant E. coli strains for urease expression.
Recombinant E. coli BL21(DE3)/pRSFDuet-Bpure-2 and BL21(DE3)/pRSFDuet-Psure-2 cells were inoculated into LB medium, supplemented separately with 50 mg/liter kanamycin, and incubated overnight at 37°C with shaking at 220 rpm. Then 4% (vol/vol) of the culture, washed three times with sterilized physiological saline, was transferred into 25 ml of modified Riesenberg medium containing 50 mg/liter kanamycin in 250-ml Erlenmeyer flasks. At the same time, 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to induce urease expression. Metal ion salts (NiSO4, FeCl3, MnCl2, CuSO4, or CoSO4) at 1 mM were also added to the medium, and the transformants were grown for 12 h at 30°C and 220 rpm. The cells were harvested by centrifugation at 10,000 × g for 8 min, washed three times with 50 mM phosphate buffer (pH 4.5), and resuspended. Then the cell suspension was disrupted by sonicating 99 times for 3-s periods at 5-s intervals at 70 W. The soluble cell extract that was obtained following centrifugation at 10,000 × g for 20 min at 4°C was used for urease activity measurements.
Determination of urease activity.
Enzyme activity was determined by measuring the rate of release of ammonia from urea (or EC), as assessed by the formation of indophenol at 625 nm (39). Briefly, 50 μl of diluted enzyme solution was incubated in citrate phosphate buffer (50 mM; pH 4.5) containing 350 μl of 500 mM urea (or 340 mM EC) at 37°C for 20 min. Then 200 μl of 10% trichloroacetic acid was added to terminate the reaction. Subsequently, 200 μl of chromogenic reagent I (15 g phenol and 0.625 g sodium nitroprusside in 250 ml of ultrapure water) and 200 μl of chromogenic reagent II (13.125 g NaOH and 7.5 ml NaClO in 250 ml of ultrapure water) (40) were added to the mixture, which was incubated for another 20 min. Then the absorbance was measured at 625 nm. One unit of enzyme activity was defined as the amount of enzyme (molar extinction coefficient [ε] = 138.8 mM−1 cm−1) required to hydrolyze 1 μmol urea (or EC) per min at 37°C and pH 4.5 under standard conditions.
Purification of recombinant urease.
The recombinant urease was purified using an ÄKTA Explorer system (GE Healthcare Life Sciences, Piscataway, NJ, USA). The cell extract was applied to a StrepTrap HP affinity column (GE Healthcare Life Sciences), which had been equilibrated with binding buffer (20 mM sodium phosphate, 280 mM NaCl, and 6 mM potassium chloride [pH 7.4]). Subsequently, the target protein was purified by eluting with elution buffer (20 mM sodium phosphate, 280 mM NaCl, 6 mM potassium chloride, and 2.5 mM desthiobiotin [pH 7.4]). Then the column were regenerated with regeneration buffer (0.5 M NaOH), before the next purification was started. The eluates were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and native PAGE. The protein concentration was determined by the modified Bradford protein assay kit (Sangon, Shanghai, China).
Characterization of recombinant ureases.
The optimal temperature of the recombinant urease was determined by quantifying its activity after incubation at different temperatures (20 to 80°C). Thermal stability was investigated by quantifying the residual activity at 37°C after incubation of the purified enzyme at various temperatures (indicated in Fig. 4B) for 30 min. The optimal pH was determined by quantifying the activity over a pH range of 3.0 to 8.0 using citrate phosphate buffer (50 mM). pH stability was investigated by quantifying the residual activity after incubation of the purified enzyme at various pHs (indicated in Fig. 4D) at 4°C for 6 h. The effect of alcohol on enzyme activity was determined by measuring the enzyme activity at different alcohol concentrations. Alcohol stability was analyzed by quantifying the residual activity after incubation of the purified enzyme with different concentrations of alcohol at 37°C for 2 h. A Lineweaver-Burk plot was used to obtain the kinetic parameters of the recombinant urease, and the initial rates were determined with substrate concentrations of 2 to 80 mM for urea and 100 to 1,800 mM for urethane in citrate phosphate buffer (50 mM; pH 4.5). All the experiments were performed under standard assay conditions.
Overexpression of UreH and metal ion determination.
Recombinant E. coli BL21(DE3)/pRSFDuet-ureH cells were inoculated into LB medium supplemented with 50 mg/liter kanamycin and were incubated overnight at 37°C and 220 rpm. Then 1% (vol/vol) of the culture was transferred into 25 ml of TB medium containing 50 mg/liter kanamycin in 250-ml Erlenmeyer flasks. When the optical density at 600 nm (OD600) reached 0.6, 1 mM IPTG was added to induce UreH expression. Specifically, 1 mM metal ion salts (NiSO4 or FeCl3) were also added to the medium at the same time. After induction for 4 or 8 h, the same number of cells was harvested by centrifugation at 10,000 × g for 8 min, washed five times, and resuspended in 20 ml of 50 mM phosphate buffer (pH 4.5). Then the cell suspension was disrupted by sonicating 99 times for 3-s periods at 5-s intervals at 70 W. An atomic absorption spectrometer (Agilent, USA) was used for detecting the nickel and iron concentrations in the soluble cell extracts (10,000 × g for 20 min at 4°C) and purified protein (1,000 mg/liter).
Elimination of urea and EC from rice wine.
To eliminate the urea and EC from rice wine, purified BpUrease (Fe3+) (at a final concentration of 2 or 6 U/ml) was added to a wine sample (pH 4.5; 15% [vol/vol] ethanol), and the reaction system was incubated at 37°C for 2 days. Diacetyl monoxime reactions (41) were used to quantify the residual concentration of urea in the reaction system. The EC concentration was determined by gas chromatography–mass spectrometry as described previously (42).
Accession number(s).
The urease gene cluster ureABCEFGDH identified from Providencia sp. LBBE in this study was deposited in GenBank and assigned accession number MF099656.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the Fundamental Research Funds for the Central Universities (JUSRP51707A), the Program for Changjiang Scholars and Innovative Research Team in University (grant IRT_15R26), and the 111 Project.
We declare that there are no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01258-17.
REFERENCES
- 1.Mobley HL, Hausinger RP. 1989. Microbial ureases: significance, regulation, and molecular characterization. Microbiol Rev 53:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krajewska B. 2009. Ureases. I. Functional, catalytic and kinetic properties: a review. J Mol Catal B Enzym 59:9–21. doi: 10.1016/j.molcatb.2009.01.003. [DOI] [Google Scholar]
- 3.Carter EL, Flugga N, Boer JL, Mulrooney SB, Hausinger RP. 2009. Interplay of metal ions and urease. Metallomics 1:207–221. doi: 10.1039/b903311d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farrugia MA, Macomber L, Hausinger RP. 2013. Biosynthesis of the urease metallocenter. J Biol Chem 288:13178–13185. doi: 10.1074/jbc.R112.446526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balasubramanian A, Ponnuraj K. 2010. Crystal structure of the first plant urease from jack bean: 83 years of journey from its first crystal to molecular structure. J Mol Biol 400:274–283. doi: 10.1016/j.jmb.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Scott DR, Marcus EA, Weeks DL, Sachs G. 2002. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology 123:187–195. doi: 10.1053/gast.2002.34218. [DOI] [PubMed] [Google Scholar]
- 7.Witte C-P. 2011. Urea metabolism in plants. Plant Sci 180:431–438. doi: 10.1016/j.plantsci.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Dixon NE, Gazzola C, Blakeley RL, Zerner B. 1975. Jack bean urease (EC 3.5.1.5). Metalloenzyme. Simple biological role for nickel. J Am Chem Soc 97:4131–4133. [DOI] [PubMed] [Google Scholar]
- 9.Sumner JB. 1926. The isolation and crystallization of the enzyme urease: preliminary paper. J Biol Chem 69:435–441. [Google Scholar]
- 10.Park I, Hausinger R. 1995. Requirement of carbon dioxide for in vitro assembly of the urease nickel metallocenter. Science 267:1156–1158. doi: 10.1126/science.7855593. [DOI] [PubMed] [Google Scholar]
- 11.Jabri E, Carr M, Hausinger R, Karplus P. 1995. The crystal structure of urease from Klebsiella aerogenes. Science 268:998–1004. doi: 10.1126/science.7754395. [DOI] [PubMed] [Google Scholar]
- 12.Volkmer D, Hommerich B, Griesar K, Haase W, Krebs B. 1996. Dinuclear nickel(II) complexes as models for the active site of urease. Inorg Chem 35:3792–3803. doi: 10.1021/ic951567x. [DOI] [PubMed] [Google Scholar]
- 13.Benini S, Rypniewski WR, Wilson KS, Miletti S, Ciurli S, Mangani S. 1999. A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels. Structure 7:205–216. doi: 10.1016/S0969-2126(99)80026-4. [DOI] [PubMed] [Google Scholar]
- 14.Zambelli B, Musiani F, Benini S, Ciurli S. 2011. Chemistry of Ni2+ in urease: sensing, trafficking, and catalysis. Acc Chem Res 44:520–530. doi: 10.1021/ar200041k. [DOI] [PubMed] [Google Scholar]
- 15.Mazzei L, Cianci M, Benini S, Bertini L, Musiani F, Ciurli S. 2016. Kinetic and structural studies reveal a unique binding mode of sulfite to the nickel center in urease. J Inorg Biochem 154:42–49. doi: 10.1016/j.jinorgbio.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Carter EL, Hausinger RP. 2010. Characterization of the Klebsiella aerogenes urease accessory protein UreD in fusion with the maltose binding protein. J Bacteriol 192:2294–2304. doi: 10.1128/JB.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fong YH, Wong HC, Chuck CP, Chen YW, Sun H, Wong K-B. 2011. Assembly of preactivation complex for urease maturation in Helicobacter pylori: crystal structure of UreF-UreH protein complex. J Biol Chem 286:43241–43249. doi: 10.1074/jbc.M111.296830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fong YH, Wong HC, Yuen MH, Lau PH, Chen YW, Wong K-B. 2013. Structure of UreG/UreF/UreH complex reveals how urease accessory proteins facilitate maturation of Helicobacter pylori urease. PLoS Biol 11:e1001678. doi: 10.1371/journal.pbio.1001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merloni A, Dobrovolska O, Zambelli B, Agostini F, Bazzani M, Musiani F, Ciurli S. 2014. Molecular landscape of the interaction between the urease accessory proteins UreE and UreG. Biochim Biophys Acta 1844:1662–1674. doi: 10.1016/j.bbapap.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 20.Farrugia MA, Wang B, Feig M, Hausinger RP. 2015. Mutational and computational evidence that a nickel-transfer tunnel in UreD is used for activation of Klebsiella aerogenes urease. Biochemistry 54:6392–6401. doi: 10.1021/acs.biochem.5b00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrugia MA, Han L, Zhong Y, Boer JL, Ruotolo BT, Hausinger RP. 2013. Analysis of a soluble (UreD:UreF:UreG)2 accessory protein complex and its interactions with Klebsiella aerogenes urease by mass spectrometry. J Am Soc Mass Spectrom 24:1328–1337. doi: 10.1007/s13361-013-0677-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hübner P, Groux PM, Weibel B, Sengstag C, Horlbeck J, Leong-Morgenthaler P-M, Lüthy J. 1997. Genotoxicity of ethyl carbamate (urethane) in Salmonella, yeast and human lymphoblastoid cells. Mutat Res 390:11–19. doi: 10.1016/S0165-1218(96)00160-7. [DOI] [PubMed] [Google Scholar]
- 23.Forkert P-G. 2010. Mechanisms of lung tumorigenesis by ethyl carbamate and vinyl carbamate. Drug Metab Rev 42:355–378. doi: 10.3109/03602531003611915. [DOI] [PubMed] [Google Scholar]
- 24.Zhao X, Du G, Zou H, Fu J, Zhou J, Chen J. 2013. Progress in preventing the accumulation of ethyl carbamate in alcoholic beverages. Trends Food Sci Technol 32:97–107. doi: 10.1016/j.tifs.2013.05.009. [DOI] [Google Scholar]
- 25.Ough CS. 1976. Ethylcarbamate in fermented beverages and foods. I. Naturally occurring ethylcarbamate. J Agric Food Chem 24:323–328. doi: 10.1021/jf60204a033. [DOI] [PubMed] [Google Scholar]
- 26.Schehl B, Senn T, Lachenmeier DW, Rodicio R, Heinisch JJ. 2007. Contribution of the fermenting yeast strain to ethyl carbamate generation in stone fruit spirits. Appl Microbiol Biotechnol 74:843–850. doi: 10.1007/s00253-006-0736-4. [DOI] [PubMed] [Google Scholar]
- 27.Cerreti M, Fidaleo M, Benucci I, Liburdi K, Tamborra P, Moresi M. 2016. Assessing the potential content of ethyl carbamate in white, red, and rosé wines as a key factor for pursuing urea degradation by purified acid urease. J Food Sci 81:C1603–C1612. doi: 10.1111/1750-3841.13344. [DOI] [PubMed] [Google Scholar]
- 28.Fidaleo M, Esti M, Moresi M. 2006. Assessment of urea degradation rate in model wine solutions by acid urease from Lactobacillus fermentum. J Agric Food Chem 54:6226–6235. doi: 10.1021/jf060934s. [DOI] [PubMed] [Google Scholar]
- 29.Miyagawa K, Sumida M, Nakao M, Harada M, Yamamoto H, Kusumi T, Yoshizawa K, Amachi T, Nakayama T. 1999. Purification, characterization, and application of an acid urease from Arthrobacter mobilis. J Biotechnol 68:227–236. doi: 10.1016/S0168-1656(98)00210-7. [DOI] [PubMed] [Google Scholar]
- 30.Yang Y, Kang Z, Zhou J, Chen J, Du G. 2015. High-level expression and characterization of recombinant acid urease for enzymatic degradation of urea in rice wine. Appl Microbiol Biotechnol 99:301–308. doi: 10.1007/s00253-014-5916-z. [DOI] [PubMed] [Google Scholar]
- 31.Carter EL, Tronrud DE, Taber SR, Karplus PA, Hausinger RP. 2011. Iron-containing urease in a pathogenic bacterium. Proc Natl Acad Sci U S A 108:13095–13099. doi: 10.1073/pnas.1106915108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maroney MJ, Ciurli S. 2014. Nonredox nickel enzymes. Chem Rev 114:4206–4228. doi: 10.1021/cr4004488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maeda M, Hidaka M, Nakamura A, Masaki H, Uozumi T. 1994. Cloning, sequencing, and expression of thermophilic Bacillus sp. strain TB-90 urease gene complex in Escherichia coli. J Bacteriol 176:432–442. doi: 10.1128/jb.176.2.432-442.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eitinger T, Suhr J, Moore L, Smith JAC. 2005. Secondary transporters for nickel and cobalt ions: theme and variations. Biometals 18:399–405. doi: 10.1007/s10534-005-3714-x. [DOI] [PubMed] [Google Scholar]
- 35.Stoof J, Breijer S, Pot RGJ, van der Neut D, Kuipers EJ, Kusters JG, van Vliet AHM. 2008. Inverse nickel-responsive regulation of two urease enzymes in the gastric pathogen Helicobacter mustelae. Environ Microbiol 10:2586–2597. doi: 10.1111/j.1462-2920.2008.01681.x. [DOI] [PubMed] [Google Scholar]
- 36.Yang S, Kang Z, Cao W, Du G, Chen J. 2016. Construction of a novel, stable, food-grade expression system by engineering the endogenous toxin-antitoxin system in Bacillus subtilis. J Biotechnol 219:40–47. doi: 10.1016/j.jbiotec.2015.12.029. [DOI] [PubMed] [Google Scholar]
- 37.Cheng J, Wu D, Chen S, Chen J, Wu J. 2011. High-level extracellular production of α-cyclodextrin glycosyltransferase with recombinant Escherichia coli BL21(DE3). J Agric Food Chem 59:3797–3802. doi: 10.1021/jf200033m. [DOI] [PubMed] [Google Scholar]
- 38.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weatherburn MW. 1967. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem 39:971–974. doi: 10.1021/ac60252a045. [DOI] [Google Scholar]
- 40.Zhou ND, Gu XL, Tian YP. 2013. Isolation and characterization of urethanase from Penicillium variabile and its application to reduce ethyl carbamate contamination in Chinese rice wine. Appl Biochem Biotechnol 170:718–728. doi: 10.1007/s12010-013-0178-2. [DOI] [PubMed] [Google Scholar]
- 41.Yang L, Wang S, Tian Y. 2010. Purification, properties, and application of a novel acid urease from Enterobacter sp. Appl Biochem Biotechnol 160:303–313. doi: 10.1007/s12010-008-8159-6. [DOI] [PubMed] [Google Scholar]
- 42.Zhao X, Zou H, Fu J, Zhou J, Du G, Chen J. 2014. Metabolic engineering of the regulators in nitrogen catabolite repression to reduce the production of ethyl carbamate in a model rice wine system. Appl Environ Microbiol 80:392–398. doi: 10.1128/AEM.03055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.