ABSTRACT
Commensal bifidobacteria colonize the human gastrointestinal tract and catabolize glycans that are impervious to host digestion. Accordingly, Bifidobacterium longum typically secretes acetate and lactate as fermentative end products. This study tested the hypothesis that B. longum utilizes cranberry-derived xyloglucans in a strain-dependent manner. Interestingly, the B. longum strain that efficiently utilizes cranberry xyloglucans secretes 2.0 to 2.5 mol of acetate-lactate. The 1.5 acetate:lactate ratio theoretical yield obtained in hexose fermentations shifts during xyloglucan metabolism. Accordingly, this metabolic shift is characterized by increased acetate and formate production at the expense of lactate. α-l-Arabinofuranosidase, an arabinan endo-1,5-α-l-arabinosidase, and a β-xylosidase with a carbohydrate substrate-binding protein and carbohydrate ABC transporter membrane proteins are upregulated (>2-fold change), which suggests carbon flux through this catabolic pathway. Finally, syntrophic interactions occurred with strains that utilize carbohydrate products derived from initial degradation from heterologous bacteria.
IMPORTANCE This was a study of bacterial metabolism of complex cranberry carbohydrates termed xyloglucans that are likely not digested prior to reaching the colon. This is significant, as bifidobacteria interact with this dietary compound to potentially impact human host health through energy and metabolite production by utilizing these substrates. Specific bacterial strains utilize cranberry xyloglucans as a nutritive source, indicating unknown mechanisms that are not universal in bifidobacteria. In addition, xyloglucan metabolism proceeds by using an alternative pathway that could lead to further research to investigate mechanisms underlying this interaction. Finally, we observed cross-feeding between bacteria in which one strain degrades the cranberry xyloglucan to make it available to a second strain. Similar nutritive strategies are known to occur within the gut. In aggregate, this study may lead to novel foods or supplements used to impact human health through rational manipulation of the human microbiome.
KEYWORDS: bifidobacteria, food microbiology, prebiotics
INTRODUCTION
Microbial commensals colonize the mammalian gut and interact with their host through various interwoven metabolic networks. A well-characterized operation performed by microbiota is energy liberation from dietary polysaccharides. These complex carbohydrates are impervious to host digestion and are thus available for microbial populations to utilize. In turn, gut microorganisms produce metabolites sequestered by the host, including short-chain fatty acids (SCFAs) (1–4). Briefly, gut microorganisms compete for dietary molecules with metabolites secreted by one member often utilized by a secondary microbial population (5). This syntrophic metabolism is extended toward interactions with their host. For instance, microbial SCFAs (i.e., acetate, propionate, butyrate, and valerate) are substrates for specific host tissues. This includes butyrate, which provides a primary energy source for enterocytes (1, 2, 6).
Bifidobacterium longum is often dominant in the infant gut and colonizes adults at lower concentrations (7). Whereas B. longum subsp. infantis colonizes the infant gut, B. longum subsp. longum tends to populate adult microbiomes. As with all bifidobacteria, B. longum catabolizes carbohydrates via the fructose-6-phosphate phosphoketolase pathway, which is characteristic of the genus and thus termed the bifid shunt. This ATP-generating pathway results in acetate and lactate secretion to recycle cofactors required in substrate level phosphorylation (8, 9). The theoretical yield is an acetate:lactate ratio of 3:2 mol produced for every 2 mol of hexose that enters the bifid shunt. Bifidobacteria encode an assortment of glycosyl hydrolases (GHs) in order to utilize dietary glycans as fermentative substrates (10–12). Oligosaccharide utilization phenotypes are often consistent with the ecological niche that the bifidobacterial strain occupies (e.g., the adult versus the infant gut) (13). The B. longum subsp. longum genome, for example, encodes a large number of GHs dedicated to arabinose and xylose utilization (14). In contrast, the chromosome of the phylogenetic near neighbor and infant-colonizing bacterium B. longum subsp. infantis contains genes that enable human milk oligosaccharide utilization within the nursing infant gut (15–18). In general, B. longum subsp. infantis varies in its ability to utilize plant-derived carbohydrates (13).
Xyloglucans are cross-linking oligosaccharides found in type 1 plant cell walls (19) that exhibit a β(1→4)-glucan primary backbone with α(1→6)-linked xylosyl residues as substituents. Depending on the plant species and tissue of origin, xyloglucan branches may be extended by galactose, fucose, or arabinose residues (20, 21). The predominant cranberry xyloglucan structure was previously characterized as SSGG [S, β-d-glucose with α-l-Ara-(1, 2)-α-d-Xyl at the O-6 position; G, β(1→4)-glucan main chain] (22).
Oligosaccharides isolated from the cranberry (Vaccinium macrocarpon) cell wall prevent the adhesion of uropathogenic Escherichia coli and may limit biofilm production (23, 24). With regard to xyloglucan metabolism, certain gut microorganisms produce enzymes that degrade tamarind seed xyloglucans in their extracellular environment (25). However, the structure-function relationship between xyloglucans and specific populations of commensal bacteria remains unresolved. Thus, we evaluated B. longum strains for the ability to utilize cranberry-derived xyloglucans as a sole carbon source. To further understand in vitro metabolism, organic acids produced during fermentation were profiled. In addition, Lactobacillus strains that are recognized as probiotics (26) and have potential for use as synbiotics were subjected to growth on xyloglucans as their sole carbohydrate source.
RESULTS
The cranberry cell wall contains xyloglucans.
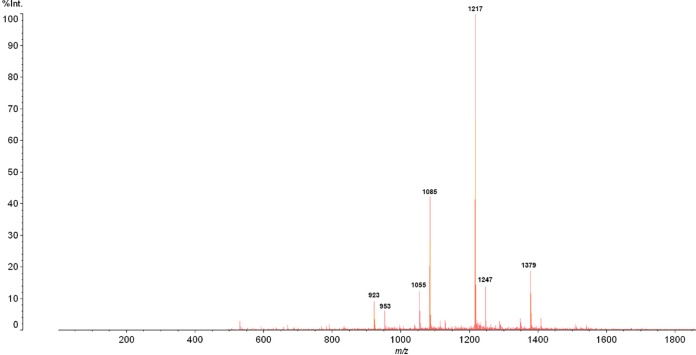
Oligosaccharides were purified from a cranberry derivative through reverse-phase C18 and size exclusion chromatography (see Fig. S1 in the supplemental material). The chemical properties of the purified oligosaccharides were subsequently assessed by matrix-assisted laser desorption ionization (MALDI-TOF) mass spectrometry (MS) (Fig. 1) and 1H nuclear magnetic resonance (NMR) spectroscopy (Fig. S2). Consistent with previous studies (22, 24), the purified oligosaccharides were identified as xyloglucans with degrees of polymerization (DPs) ranging from 6 to 9. MALDI-TOF MS analysis identified sodium adduct ions at 923, 953, 1,055, 1,085, 1,217, 1,247, and 1,379, which were interpreted as xyloglucan compositions of hexose3-pentose3 (H3P3), H4P2, H3P4, H4P3, H4P4, H5P3, and H5P4, respectively. Putative cranberry xyloglucan oligosaccharide structures were provisionally assigned as reported in Table S1.
FIG 1.
Purified cranberry xyloglucan putative structures. Shown is the positive reflectron mode MALDI-TOF MS spectrum of cranberry xyloglucan obtained from a HiPrep Sephacryl S-100 HR size exclusion column. The voltage was set at 80 kV, and 500 profiles were collected. Major peaks at m/z 923, 953, 1,055, 1,085, 1,217, 1,247, and 1,379 represent sodium adducts ([M + Na]+) of xyloglucan with DPs ranging from 6 to 9 and sugar compositions as follows: [H3P3+ Na]+, m/z 923; [H4P2+ Na]+, m/z 953; [H3P4+ Na]+, m/z 1,055; [H4P3+ Na]+, m/z 1,085; [H4P4+ Na]+, m/z 1,217; [H5P3+ Na]+, m/z 1,247; [H5P4+ Na]+, m/z 1,379.
B. longum xyloglucan utilization is strain dependent.
Among the bifidobacterial strains tested (Table 1), B. longum subsp. longum UCD401 exhibits the most growth on the purified xyloglucans, achieving a final optical density at 600 nm (OD600) of 0.15 ± 0.02 (Fig. 2). The growth rate of B. longum subsp. longum UCD401 while utilizing xyloglucans does not significantly differ from that during glucose utilization (P > 0.05) (Table 2). This indicates that B. longum subsp. longum UCD401 does not have a metabolic preference for glucose relative to xyloglucans. As expected, while fermenting xyloglucans, B. longum subsp. longum UCD401 achieves a modest biomass relative to that achieved on glucose (P < 0.05) and no growth was observed on the negative control. This indicates that UCD401 does not utilize these carbohydrates with similar efficiency. Bifidobacteria that utilize both plant and milk oligosaccharides have exhibited similar growth profiles (27, 28). Despite similar growth rates on glucose and xyloglucans, B. longum subsp. longum UCD401 ferments xyloglucans more slowly than glucose to achieve maximal growth (tc) (P < 0.05) (Table 2). In contrast to UCD401, B. longum subsp. infantis ATCC 15697 and JCM1260 did not metabolize cranberry xyloglucans.
TABLE 1.
Strains used in this study
| Straina | Species or subspecies | Origin |
|---|---|---|
| ATCC 15697 | B. longum subsp. infantis | Human infant feces |
| JCM 1260 | B. longum subsp. infantis | Human infant feces |
| JCM 1272 | B. longum subsp. infantis | Human infant feces |
| JCM 7007 | B. longum subsp. infantis | Human infant feces |
| JCM 11347 | B. longum subsp. longum | Human feces |
| ATCC 15708 | B. longum subsp. longum | Human feces |
| UCD401 | B. longum subsp. longum | Human feces |
| ATCC BAA-793 | L. plantarum | Human saliva |
| ATCC 33200 | L. johnsonii | Human blood |
UCD, University of California Davis Culture Collection; ATCC, American Type Culture Collection; JCM, Japanese Collection of Microorganisms.
FIG 2.
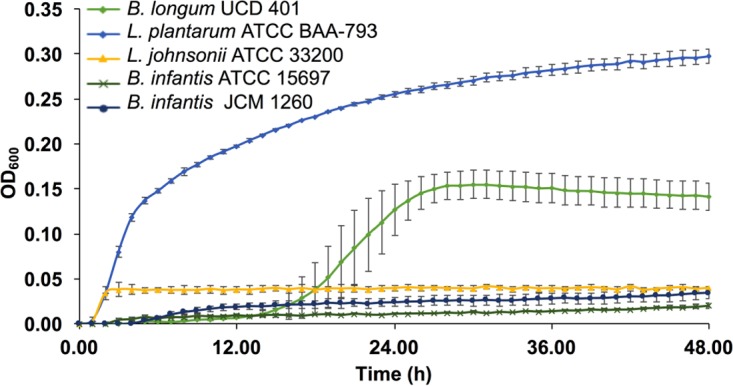
Bacterial growth while utilizing cranberry xyloglucans. Growth curves of B. longum subsp. longum UCD401, L. plantarum ATCC BAA-793, L. johnsonii ATCC 33200, and B. longum subsp. infantis ATCC 15697 and JCM 1260 grown on mMRS medium containing 2% (wt/vol) xyloglucans. The curves are drawn from an average of three independent experiments.
TABLE 2.
Analysis of bacterial growth kinetics calculated with Wolfram Mathematica 10.3
| Strain | 2% xyloglucans |
2% glucose |
||||
|---|---|---|---|---|---|---|
| k (h−1) | ΔODasym | tc (h) | k (h−1) | ΔODasym | tc (h) | |
| B. infantis JCM 1260 | NDa | ND | ND | ND | ND | ND |
| B. infantis ATCC 15697 | ND | ND | ND | ND | ND | ND |
| B. longum UCD401 | 0.555 ± 0.055 | 0.15 ± 0.02b | 19.7 ± 2.5d | 0.614 ± 0.048 | 1.25 ± 0.09 | 13.7 ± 0.6 |
| L. plantarum ATCC BAA-793 | 0.386 ± 0.049f | 0.29 ± 0.03c | 2.3 ± 0.60e | 0.569 ± 0.005 | 1.48 ± 0.02 | 5.3 ± 0.60 |
| L. johnsonii ATCC 33200 | ND | ND | ND | ND | ND | ND |
ND, not determined.
Significant difference in the asymptotic OD value of B. longum UCD401 compared to L. plantarum and to the positive control, glucose (P < 0.05).
Significant difference in the asymptotic OD value of L. plantarum compared to B. longum UCD401 and to the positive control, glucose (P < 0.05).
Significant difference in the inflection point (tc) of B. longum UCD401 compared to L. plantarum and to the positive control (P < 0.05).
Significant difference in the inflection point (tc) of L. plantarum compared to B. longum UCD401 (P < 0.05).
Significant difference in the growth rate (k) of L. plantarum on xyloglucans compared to that of other strains on xyloglucans and glucose (P < 0.05).
Lactobacillus strains were evaluated to test if other fermentative bacteria could harness cranberry xyloglucan carbon. Lactobacillus plantarum ATCC BAA-793 and Lactobacillus johnsonii ATCC 33200 were subjected to growth on xyloglucans as a sole carbohydrate source as depicted in Fig. 2 with growth kinetics displayed in Table 2. L. johnsonii ATCC 33200 did not utilize cranberry-derived oligosaccharides. In contrast, L. plantarum ATCC BAA-793 achieved a final OD600 of 0.29 ± 0.03, which is higher than that of all of the B. longum strains tested (P < 0.05) (Fig. 2). This Lactobacillus strain does prefer glucose, as it has a significantly lower growth rate while consuming xyloglucans (P < 0.05, Table 2). Its maximal growth (tc) did not vary between xyloglucan and glucose utilization (P > 0.05), despite the lower growth rate during xyloglucan utilization. Interestingly, L. plantarum has a lower growth rate than B. longum subsp. longum UCD401 while fermenting purified xyloglucans despite achieving a higher biomass (P < 0.05). The time required to achieve maximum growth (tc) while utilizing xyloglucan is significantly shorter for L. plantarum than for B. longum subsp. longum UCD401 (P < 0.05). This suggests that L. plantarum does not prefer these complex carbohydrates, despite having a higher utilization efficiency than the B. longum strain.
B. longum consumes xyloglucans from the growth medium.
The extent to which UCD401 utilizes xyloglucans was determined by profiling spent fermentation medium from growth conducted in microcentrifuge tubes. Interestingly, degraded glycans were not detected in spent medium compared with the control (i.e., preinoculated medium) (Fig. S3 and S4). The extracted ion counts for each xyloglucan were quantitated as percentages of the baseline control (t = 0) (Fig. S5 and S6). Of interest, detectable xyloglucans with DPs of 6 to 9 exhibited >120% of the extracted ion counts of the baseline (t = 0). Thus, there was an extensive accumulation of xyloglucans with all DPs following fermentation by B. longum subsp. longum UCD401. Significantly, xyloglucans with DPs of <7 did not accumulate to the same extent. This is indicative of an ability to utilize the short-chain oligosaccharides. Accordingly, we did not detect monosaccharide accumulation in the spent fermentation medium. A similar observation has been reported previously with bifidobacterial metabolism of human milk oligosaccharides (29, 30). As previously postulated, B. longum is able to transport only lower-molecular-weight oligosaccharides across its envelope. This suggests that a dynamic equilibrium has been achieved between the import of lower-DP molecules and the extracellular hydrolysis of higher-DP molecules. This is consistent with higher-DP oligosaccharide accumulation in the growth medium. A similar glycan distribution was observed in L. plantarum qualitatively, along with corresponding degradation phenotypes (Fig. S3). It is important to note that glycoprofiling was performed with bacteria grown in microcentrifuge tubes.
Bifidobacteria metabolize xyloglucans via the bifid shunt.
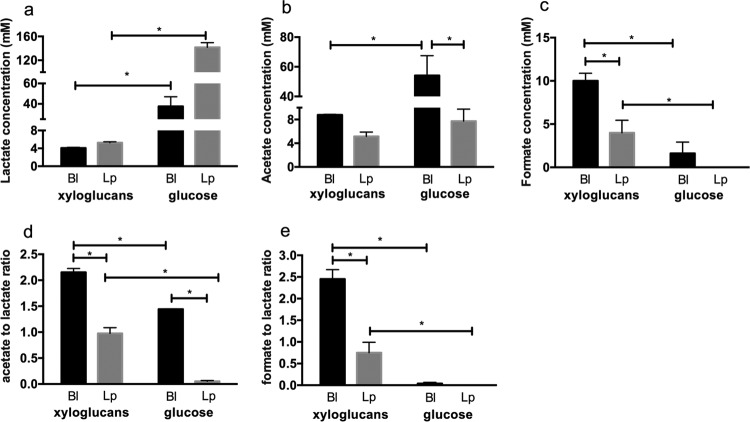
Metabolite end products secreted while utilizing xyloglucans were quantified by high-performance liquid chromatography (HPLC) of cell-free supernatants. The absolute concentrations of acetate, lactate, and formate secreted by B. longum subsp. longum UCD401 in the early stationary phase of xyloglucan or glucose utilization are depicted in Fig. 3. B. longum subsp. longum UCD401 secretes significantly lower lactate and acetate concentrations while utilizing xyloglucans (4.08 ± 0.16 and 8.78 ± 0.10 mM, respectively) than when subsisting on glucose (37.60 ± 9.41 and 54.11 ± 13.40 mM, respectively). This is expected and consistent with the lower biomass achieved (P < 0.05). In addition, less metabolic investment is required to catabolize glucose in the bifid shunt. The absolute concentrations were normalized with respect to the OD600 reached in microcentrifuge tubes. UCD401 exhibited higher acetate concentrations in xyloglucan metabolism (172.54 ± 10.52 mM) than in glucose metabolism (124.56 ± 22.70 mM), whereas the lactate concentrations were similar (80.31 ± 6.76 and 86.50 ± 15.98 mM). Interestingly, and despite a lower biomass, B. longum subsp. longum UCD401 secretes high concentrations of formate while metabolizing cranberry xyloglucans (10.01 ± 0.86 mM). This is significantly higher than during glucose utilization (1.62 ± 1.30 mM) (P < 0.05), indicating a shift in metabolism to harvest energy more efficiently from this substrate.
FIG 3.
Bacterial fermentative end products of cranberry xyloglucan utilization. Shown are lactate (a), acetate (b), and formate (c) production; acetate/lactate ratios (d); and formate/lactate ratios (e). Bl, B. longum subsp. longum UCD401; Lp, Lactobacillus plantarum ATCC BAA-793. Averages of independent biological triplicates are shown, and bars represent the standard deviation of the mean. Organic acid production is expressed in millimolar absolute concentrations. Asterisks represent significant differences determined by two-way ANOVA and Sidak's multiple-comparison test (P < 0.05).
The bifid shunt catabolizes hexose sugars to yield a theoretical acetate:lactate ratio of 1.5. As expected, UCD401 achieved this while utilizing glucose (1.44 ± 0.01); however, xyloglucan fermentation shifted the ratio significantly toward acetate at the expense of lactate (2.15 ± 0.08) (P < 0.05) (Fig. 3d). When normalizing the absolute concentrations with respect to biomass, more acetate production from xyloglucan metabolism than from glucose metabolism occurred. This has been observed in a previous study, as more ATP is produced by flux through acetate-producing pathways (31). Accordingly, formate secretion was significantly increased with a formate:lactate ratio of 2.50 ± 0.28 while fermenting xyloglucans, in contrast to 0.05 ± 0.01 during glucose metabolism (Fig. 3e).
Organic acids produced while lactobacilli ferment xyloglucans were subsequently profiled (Fig. 3). In addition to comparative physiology, this approach may inform future mixed-culture probiotics (i.e., lactobacilli and bifidobacteria), as well as synbiotic strategies that incorporate xyloglucans with one or more probiotic strain. Lactate was produced at significantly lower concentrations (5.27 ± 0.19 mM, P < 0.05) by L. plantarum ATCC BAA-793 than during glucose consumption. In addition, acetate secretion was observed with an acetate:lactate ratio of 1:1 when L. plantarum ATCC BAA-793 utilized cranberry xyloglucans (Fig. 3d). While fermenting glucose, L. plantarum exhibited an acetate:lactate ratio of 0.05 ± 0.01 (P < 0.05) (Fig. 3d). We expected L. plantarum, as a heterofermentative species, to secrete acetate. Interestingly, formate production was detected during xyloglucan consumption, although it was not detected following glucose fermentation. The secretion of acetate and formate has been previously observed when inefficient substrate utilization yielded less biomass (32, 33).
Bacterial commensals exhibit differential growth phenotypes while consuming oligosaccharide extracts containing secondary plant material.
In addition to purified xyloglucans, crude cranberry cell wall extracts were evaluated as a growth substrate. These fractions are more crudely enriched for oligosaccharides and thus more closely mimic what would be encountered by the microbiota following the ingestion of cranberries. Moreover, prebiotic formulations would likely use a crude cranberry extract for functional or processing reasons.
We tested two crude fractions, termed A2, which retains a pink color and is extracted from whole cranberries, and A6, which is derived from the primary A2 fraction. Both of these preparations have been previously reported (22). However, the precise distribution of glycans and noncarbohydrate molecules in A2 is not fully characterized. Accordingly, B. longum strains achieved more pronounced growth phenotypes while consuming A2 and A6. Cellular growth (Fig. 4) and kinetics varied by the particular strain tested (Table 3). A2 was most vigorously utilized by B. longum subsp. infantis JCM7007 (OD600 of 0.58 ± 0.34), B. longum subsp. infantis JCM1272 (OD600 of 0.38 ± 0.10), and B. longum subsp. longum UCD401 (0.37 ± 0.07) (Fig. 4a; Table 3). With the exception of UCD401, these strains did not grow on purified xyloglucans. Interestingly, there are two B. longum subsp. infantis strains that utilize A2 as a substrate. This is significant, as B. longum subsp. infantis is predicted not to utilize xyloglucans; thus, other components of the A2 fraction were fermented or cofermented. In addition, B. longum subsp. infantis JCM1272 achieved moderate growth on A2 (OD600 of 0.17 ± 0.08), whereas growth on purified xyloglucans was not observed.
FIG 4.
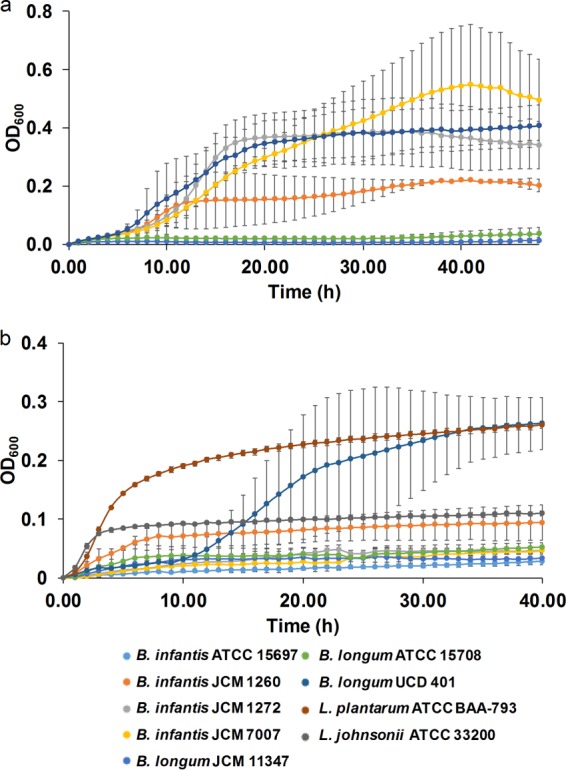
Bacterial growth on additional fractions of cranberry xyloglucans. Growth curves represent B. longum subsp. infantis ATCC 15697, JCM 1260, JCM 1272, and JCM 7007; B. longum subsp. longum JCM 11347, ATCC 15708, and UCD401; Lactobacillus plantarum ATCC BAA-793; and Lactobacillus johnsonii ATCC 33200 grown on mMRS medium containing 2% (wt/vol) xyloglucan fraction A2 (a) or 2% (wt/vol) xyloglucan fraction A6 (b). The curves are drawn from the average of at least three independent experiments, with the exception of strains JCM 1260, JCM 7007, JCM 11347, and JCM 15708 which are based on duplicates in panel a.
TABLE 3.
Kinetic analysis of bacterial growth on A2 and A6 fraction xyloglucans calculated with Wolfram Mathematica 10.3
| Strain | Xyloglucans from fraction: |
|||||
|---|---|---|---|---|---|---|
| A2 |
A6 |
|||||
| k (h−1) | ΔODasym | tc (h) | k (h−1) | ΔODasyma | tc (h) | |
| B. infantis JCM 1260 | 0.464 ± 0.280 | 0.17 ± 0.08 | 6.4 ± 3.3 | 1.188 ± 0.79 | 0.08 ± 0.02a | 3.3 ± 1.8 |
| B. infantis JCM 1272 | 0.427 ± 0.039 | 0.38 ± 0.10 | 12.4 ± 0.6 | NDc | ND | ND |
| B. infantis JCM 7007 | 0.215 ± 0.137 | 0.58 ± 0.34 | 17.6 ± 6.1 | ND | ND | ND |
| B. infantis ATCC 15697 | NAd | NA | NA | ND | ND | ND |
| B. longum JCM 11347 | ND | ND | ND | ND | ND | ND |
| B. longum ATCC 15708 | ND | ND | ND | ND | ND | ND |
| B. longum UCD401 | 0.438 ± 0.152 | 0.37 ± 0.08 | 11.3 ± 3.0 | 0.270 ± 0.084 | 0.27 ± 0.03b | 16.3 ± 0.6 |
| L. plantarum ATCC BAA-793 | NA | NA | NA | 0.513 ± 0.061 | 0.25 ± 0.02b | 2.9 ± 0.5 |
| L. johnsonii ATCC 33200 | NA | NA | NA | 0.944 ± 0.166 | 0.12 ± 0.01a | 1.4 ± 0.3 |
Significant difference in the asymptotic OD values for JCM 1260 and L. johnsonii compared to UCD401 and L. plantarum in A6 fraction treatment (P < 0.05).
Significant difference in the asymptotic OD values for UCD401 and L. plantarum compared to JCM 1260 and L. johnsonii in A6 fraction treatment (P < 0.05).
ND, not determined.
NA, not available.
In contrast, the more purified A6 fraction was utilized only by B. longum subsp. longum UCD401 and B. longum subsp. infantis JCM 1260, with final OD600s of 0.27 ± 0.03 and 0.08 ± 0.02, respectively (Fig. 4b). Clearly, the A6 fraction is not as efficiently utilized as the cruder and likely polyphenol-containing A2 fraction (Table 3; Fig. S7). UCD401 exhibited a gradual decline in growth efficiency while utilizing the crudest extract to the most purified form (Fig. S7b) (P < 0.05). This mirrors the general trend that the majority of bifidobacterial strains tested do not metabolize highly purified xyloglucans. This includes all B. longum subsp. infantis strains (Fig. S7c).
B. longum subsp. longum UCD401 utilizes A6, leading to an increase in the acetate:lactate ratio (1.90 ± 0.16) and absolute formate production (8.87 ± 2.63 mM) relative to those achieved with glucose, despite the lower biomass (P < 0.05) (Fig. 5). Formate production by UCD401 is greater than that by B. longum subsp. infantis JCM1260 when catabolizing A6 (P < 0.05). This is consistent with formate secretion while utilizing highly purified xyloglucans and suggests that UCD401 is specifically consuming this substrate in the mixed-purity preparation. Interestingly, B. longum subsp. infantis JCM1260 exhibits an acetate:lactate ratio of 2.44 ± 0.32 and 6.39 ± 0.57 mM formate secreted (Fig. 5c). Glucose flux through the bifid shunt was metabolized, as expected, to approximate the theoretical yield of 1.5 (1.82 ± 0.15 and 1.43 ± 0.01) (P < 0.05) (Fig. 5d). Again, this is consistent with utilization of the carbohydrate constituents of the A6 fraction.
FIG 5.
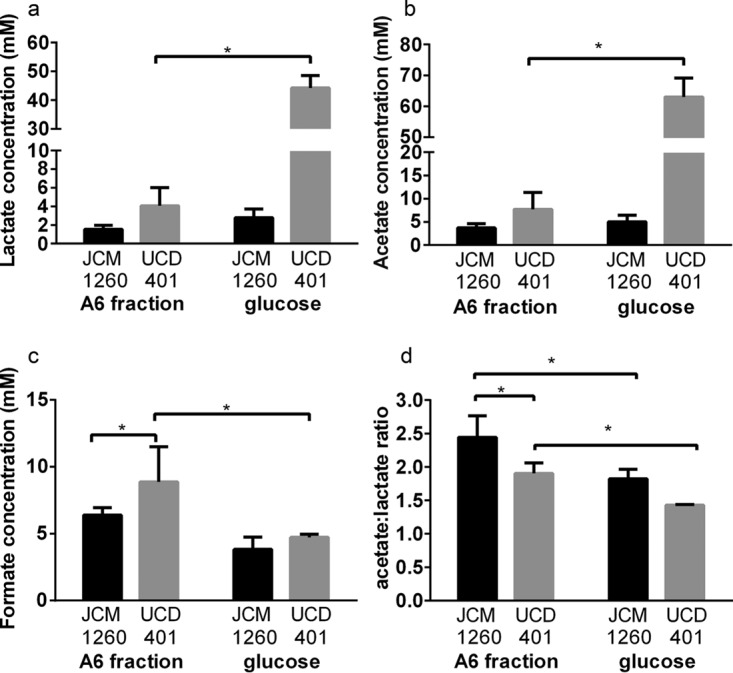
Bifidobacterial fermentative end products of cranberry fraction A6 utilization. (a to c) Lactate (a), acetate (b), and formate (c) production while B. longum subsp. infantis JCM 1260 and B. longum subsp. longum UCD401 utilize xyloglucan fraction A6. (d) Acetate:lactate ratio after fermentation. Averages of independent biological triplicates are shown, and bars represent the standard deviation of the mean. Asterisks represent significant differences evaluated by two-way ANOVA and Sidak's multiple-comparison test (P < 0.05).
Expression of arabinose utilization genes while utilizing the A6 cranberry xyloglucan fraction.
The expression of four GH family genes in the UCD401 chromosome while metabolizing A6 was evaluated. This includes an α-l-arabinofuranosidase gene (BL_0405), two arabinan endo-1,5-α-l-arabinosidase genes (BL_0404, BL_0403), and a β-xylosidase gene (BL_0402). We observed that BL_0405, BL_0404, and BL_0402 were significantly upregulated, with a minimum of a 2-fold increase during exponential phase (Fig. 6a, P < 0.05). This indicates that the expression of these genes and their products is modulated during the utilization of the A6 xyloglucan fraction. Interestingly, BL_0403 expression did not significantly change relative to that of the glucose control (P > 0.05). This indicates that the adjacent BL_0404 gene is responsible for xyloglucan hydrolysis. Moreover, differential regulation of these paralogs potentially reflects divergent enzymatic functions.
FIG 6.
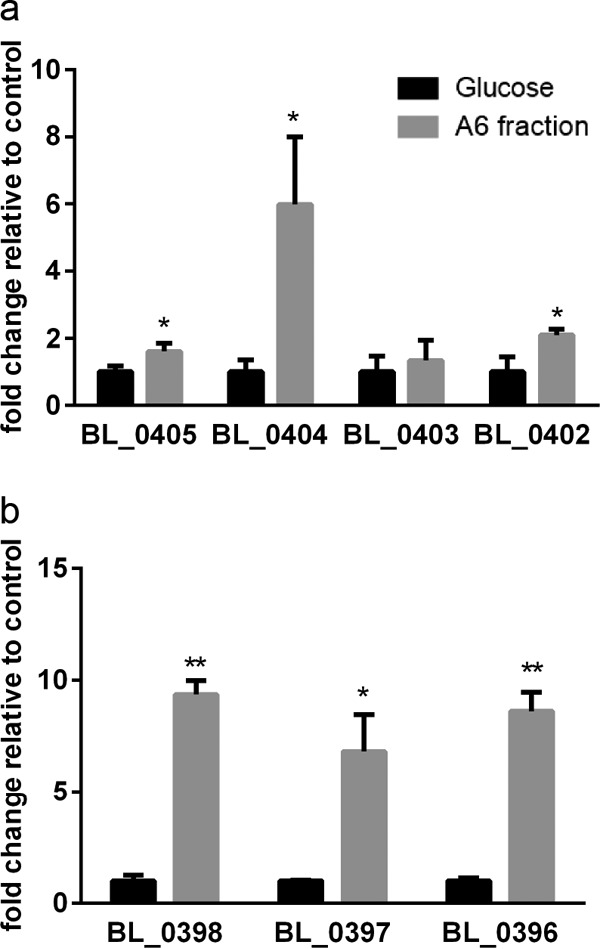
Gene expression of B. longum subsp. longum UCD401 while utilizing cranberry fraction A6 as a sole carbon source. Genes in the arabinose cluster of B. longum subsp. longum UCD401 predicted to participate in xyloglucan metabolism are depicted on the x axis. Expression of four GH family genes (those for α-l-arabinofuranosidase, C-terminal [BL_0405], arabinan endo-1,5-α-l-arabinosidase [BL_0404, BL_0403], β-xylosidase [BL_0402]) (a) and those for a carbohydrate ABC transporter substrate-binding protein (BL_0398) and carbohydrate ABC transporter membrane proteins (BL_0397, BL_0396) (b) is shown as fold change relative to the control (glucose). Averages of independent biological triplicates are shown, and bars represent the standard deviation of the mean. Asterisks (* and **) represent significant differences compared to the control (glucose), evaluated by paired t test (P values of <0.05 and <0.005, respectively).
Fermentation of the A6 fraction induces UCD401 to express transport-related proteins that may be involved in xyloglucan transport across the membrane. Upregulated transport genes, including those for a carbohydrate ABC transporter substrate-binding protein (BL_0398) and carbohydrate ABC transporter membrane proteins (BL_0397, BL_0396) exhibit >2-fold induction relative to the control (Fig. 6b, P < 0.05). This suggests recognition and a potential uptake of arabinose and xylose backbone motifs across the cell envelope.
Modeled bidirectional syntrophic interactions.
Conditioned spent medium from xyloglucan fermentations was used to assess in vitro bidirectional syntrophic interactions in microplate growth. This occurs when fermentation by a primary strain provides hydrolysis products to be used by a heterologous secondary strain that is incapable of utilizing the initial substrate. Since L. johnsonii does not grow on xyloglucans on a microplate, we included this strain as a negative control. Interestingly, B. longum subsp. infantis ATCC 15697 does not utilize purified xyloglucans as a sole carbon source; however, conditioned supernatants from L. plantarum and L. johnsonii enabled growth to final OD600s of 0.066 ± 0.008 and 0.093 ± 0.008 with growth rates of 0.940 ± 0.106 h−1 and 0.801 ± 0.077 h−1, respectively (Table 4). We interpret this as moderate growth because of the characteristic sigmoidal response curve produced during fermentation. In contrast, supernatant harvested from B. longum subsp. longum UCD401 did not enable B. longum subsp. infantis ATCC 15697 to grow (Fig. 7). This is likely due to the inability of B. longum subsp. infantis ATCC 15697 to utilize arabinosyl-reducing ends, as predicted by comparative genomics (15). UCD401 may potentially sequester and metabolize molecules that would otherwise be utilized by other bifidobacterial strains. Furthermore, it is possible that the lactobacilli secreted other products to enhance B. longum subsp. infantis growth (e.g., exopolysaccharides) (34).
TABLE 4.
Growth kinetics of strains during syntrophic interaction
| Strain | Supernatant from: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
B. longum UCD401 |
L. plantarum ATCC BAA-793 |
L. johnsonii ATCC 33200 |
|||||||
| k (h−1) | ΔODasym | tc (h) | k (h−1) | ΔODasym | tc (h) | k (h−1) | ΔODasym | tc (h) | |
| B. infantis ATCC 15697 | NDe | ND | ND | 0.940 ± 0.106a | 0.066 ± 0.008 | 10.3 ± 0.6 | 0.801 ± 0.077 | 0.093 ± 0.008 | 10.3 ± 0.6 |
| B. longum UCD401 | NAf | NA | NA | 0.632 ± 0.120b | 0.096 ± 0.005d | 12.8 ± 0.6 | 0.468 ± 0.070d | 0.162 ± 0.007c | 11.0 ± 0.0 |
| L. plantarum ATCC BAA-793 | 0.890 ± 0.158d | 0.115 ± 0.009d | 2.1 ± 0.9 | NA | NA | NA | NA | NA | NA |
Significant difference between the growth rates of B. infantis ATCC 15697 in different conditioned media determined by paired t test (P < 0.05).
Significant difference between the growth rates of B. longum UCD401 in different conditioned media determined by paired t test (P < 0.05).
Significant difference between the asymptotic OD values of B. longum UCD401 in different conditioned media determined by paired t test (P < 0.05).
Significant difference in the growth kinetics of each strain in different conditioned media and its growth kinetics in xyloglucans alone (shown in Table 2) determined by paired t test (P < 0.05).
ND, not determined.
NA, not available.
FIG 7.
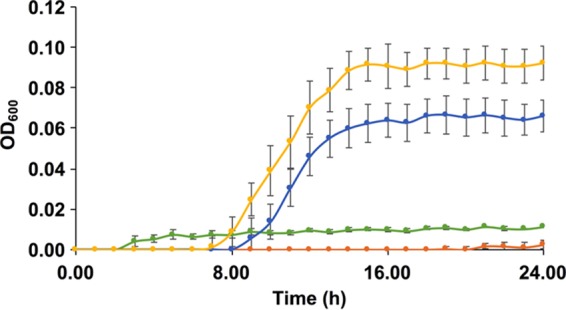
Bacterial syntrophic interactions modeled with cranberry xyloglucans. Growth curves of B. longum subsp. infantis ATCC 15697 on mMRS medium containing xyloglucans (green) and supernatants from B. longum subsp. longum UCD401 (orange), L. plantarum ATCC BAA-793 (blue), and L. johnsonii ATCC 33200 (yellow) after xyloglucan fermentation. The curves are drawn from an average of three independent experiments.
B. longum subsp. infantis ATCC 15697 utilization of carbohydrates liberated from lactobacilli and UCD401 primary fermentations were analyzed by liquid chromatography (LC)-MS (Fig. S8). Of interest, ATCC 15697 grew on the spent medium from lactobacillus primary fermentations, albeit with a stronger peak intensity observed by LC-MS. This may be due to hydrolysis of higher-molecular-weight xyloglucans or the production of exopolysaccharides, as hypothesized with primary fermentations. In addition, we determined that ATCC 15697 is incapable of utilizing the spent medium from UCD401 in a secondary fermentation.
When B. longum subsp. longum UCD401 was grown on the L. plantarum supernatant, it achieved a lower final OD600 (0.096 ± 0.005) than when it was grown on purified xyloglucans in a primary fermentation (P < 0.05), although the growth rates are similar regardless of whether UCD401 is conducting a primary or a secondary fermentation (Table 4). Conversely, the growth rate of L. plantarum on conditioned medium from B. longum subsp. longum UCD401 was significantly higher whereas its biomass production (final OD600 of 0.115 ± 0.009) was significantly lower than that of purified xyloglucans (final OD600 of 0.29 ± 0.03) (P < 0.05) (Table 4). This may indicate the production of inhibitory compounds or a preference for intact oligosaccharides.
DISCUSSION
It has been established that genotypic and phenotypic variations contribute to bifidobacterial preferences for carbon sources (12). For the former, the B. longum subsp. infantis genome has evolved to enable milk oligosaccharide utilization at the expense of plant carbohydrates. Bifidobacteria deploy transporters to capture intact oligosaccharides or their derivatives from the extracellular milieu (13, 35). Thus, the inability of B. longum subsp. infantis strains to utilize xyloglucans may be due to the absence of specific transporters. This is consistent with B. longum subsp. infantis ATCC 15697 utilization of lactobacillus hydrolysis products. These lactobacilli perform extracellular digestion to provide ATCC 15967 with fermentable substrates. In contrast, UCD401 internalizes intact oligosaccharides to withhold potential growth substrates from ATCC 15697.
It is likely that B. longum subsp. longum UCD401 utilizes xyloglucans from the terminal arabinose positioned at the reducing end. This is consistent with the expression of arabinose utilization genes (Fig. S9). This arabinose utilization cluster is conserved in B. longum subsp. longum strains with an araC transcriptional family operon upstream of α-l-arabinofuranosidase and β-xylosidase genes adjacent to an ABC transporter cassette. Importantly, B. longum subsp. longum ATCC 15707 does not grow on xylose but metabolizes arabinose as a sole carbohydrate source (36). Accordingly, B. longum subsp. longum UCD401 utilizes arabinose more efficiently than do the other B. longum subsp. longum strains tested (Fig. S10). It is noteworthy that this B. longum strain was isolated from an infant stool sample and the stronger utilization phenotype may reflect recent isolation and limited passages.
Bifidobacterial xyloglucan utilization requires the transport of arabinose following extracellular hydrolysis of higher-DP oligosaccharides. β-d-Glucopyranosyl and α-d-xylopyranosyl residues are not metabolized extracellularly. This likely causes the accumulation of lower-DP xyloglucans. Arabinose metabolism has been linked to genes encoding extracellularly secreted α-arabinofuranosidases, which are located within the UCD401 genome (Fig. S4) (37). As we predicted, B. longum subsp. longum UCD401 utilizes xyloglucan within the crude A6 fraction likely by using an extracellular arabinofuranosidase (BL_0405). Arabinose liberated from xyloglucans was completely sequestered, as it was not detected in spent medium. It has been posited that bifidobacterial strains cannot use xylose backbones longer than xylotetraose (DP of 4) (37). The B. longum subsp. longum arabinose cluster encodes predicted intracellular xylosidases, which supports the hypothesis that transport is essential for the catabolism of higher (DP of >4) xylooligosaccharides (Fig. S4). The upregulation of solute-binding transport proteins (BL_0398) and permeases (BL_0397 and BL_0396) suggests that uptake of arabinose and lower-DP xyloglucans, including xylose and glycosyl residues, occurs after the release of arabinose moieties.
In bifidobacterial carbohydrate metabolism, pyruvate is converted to lactate via lactate dehydrogenase, which does not produce additional ATP but recycles NAD+ (38). Pyruvate formate-lyase (pfl; EC 2.3.1.54) catalyzes formate and acetyl-coenzyme A (acetyl-CoA) production from pyruvate. Acetyl-CoA is subsequently metabolized to acetate or ethanol and secreted (38). This results in an increased acetate/lactate ratio and correspondingly higher ATP production (31, 38). We predicted increased acetate secretion during UCD401 xyloglucan fermentation, although the lower biomass on this substrate hinders comparisons of absolute concentrations. However, formate and ethanol secretion was previously determined to impact the acetate/lactate ratio in bifidobacteria (38). Thus, the shift toward formate production coincides with an increased need for ATP while metabolizing xyloglucans. Bifidobacterial secretion of formate while utilizing certain oligosaccharides has been previously observed (39–41). Under certain conditions, bifidobacterial oligosaccharide metabolism is initially characterized by high levels of lactate production, with a shift toward formate production occurring later in the fermentation (40, 42, 43). This suggests that formate production benefits the cell by generating additional ATP when oligosaccharides are catabolized slowly. In general, bifidobacteria secrete secondary products such as formate, succinate, and ethanol when achieving lower biomass concentrations (OD600 of <0.5) and when the carbohydrate consumption rate is diminished (31, 39, 40, 42). Our results are consistent in that formate production increases while xyloglucans are utilized slowly, ostensibly to bolster ATP production under these conditions.
Crude oligosaccharide extracts were tested, as they is of interest for the preparation of cranberry xyloglucans for prebiotic applications. The cranberry cell wall does not contain appreciable quantities of monosaccharides (44) and is not expected to influence growth (22). As the A2 fraction retains a pink color, there are likely small quantities of phenolic compounds that may contribute to the observed growth differential between the A2 fraction and the highly purified xyloglucans. Furthermore, the most abundant oligosaccharides within the A6 fraction are identical to the xyloglucans purified in this study (22). However, XSGGG-Ac and XSGGG-Ac2 structures (i.e., acetylated xyloglucan oligosaccharides) have been identified in the A6 fractions and did not remain in the highly purified xyloglucans. Thus, these oligosaccharides may be responsible for promoting L. johnsonii and B. longum subsp. infantis JCM1260 growth. B. longum typically consumes short-chain oligosaccharides, likely subsequent to intracellular transport (13, 45). Thus, it is possible that differential phenotypes are due to variation in the xyloglucan DP.
Lactobacilli are natural inhabitants of the human gastrointestinal tract and may be used as probiotics and in concert with prebiotics in synbiotic applications (26, 46). Lactobacillus strains may utilize carbohydrates as well as bifidobacteria (47). It is likely that L. plantarum uses the phosphoketolase pathway while consuming xyloglucans to secrete formate and acetate, with lactate being reduced (48, 49). This was previously observed in L. plantarum VTT E-79098 fermentation of arabino-xylooligosaccharide (33).
Our experiments suggest that syntrophic interactions following xyloglucan degradation may occur between other members of the gut consortium. The interactions among lactobacilli, bifidobacteria, and oligosaccharides are of interest in the engineering of synbiotic interventions. Moreover, lactobacillus-mediated cleavage of xyloglucans within the small intestine would impact substrates available to bifidobacterial commensals of the lower gastrointestinal tract. Syntrophic interactions during bifidobacterial in vitro cofermentations that include members of the genus Bifidobacterium on various carbohydrate sources have been previously described (12, 45, 50). Cooperative or cross-feeding between strains that colonize the gastrointestinal tract may contribute to the competitive exclusion of sensitive taxa and maintain diversity through specialist adaptation to colonize unique niches (51). Such syntrophic interactions under in vivo conditions might prompt a shift in the gut microbiome toward the metabolism of plant-derived carbohydrates with a metabolome characteristic of saccharolytic activities (52). We did not detect monosaccharides in spent medium by HPLC, leading to the hypothesis that liberated monosaccharides would be sequestered, thus rendering them undetectable. Furthermore, it is possible that the transfer of conditioned medium from the primary fermentation introduced end products into the secondary fermentation although at very low concentrations with minimal impact on growth.
Conclusions.
Among the bifidobacterial strains tested, B. longum subsp. longum UCD401 utilizes cranberry xyloglucans as a sole energy and carbon source. This is potentially significant, as it was recently isolated from an infant and subjected to limited passages. This may be due to conserved phenotypic function, as genomic or regulatory features likely remained intact in the limited generations since its isolation (53). Furthermore, it hints at the potential for strain variation within infants and adults in the capacity to utilize cranberry xyloglucans. B. longum subsp. longum colonizes both infants and postweaning individuals. The phenotypic versatility underlying the broad host range necessary to subsist on a substrate encountered in the adult diet suggests a postweaning strategy. It is notable that xyloglucan catabolism prompts a shift in the central fermentative pathway to obtain more ATP. The significance of the altered metabolic profile for microbiome function or host health remains an outstanding question.
MATERIALS AND METHODS
Isolation of xyloglucans from the cranberry cell wall.
Cranberry hulls were degraded with pectinase (Klerzyme 150; DSM Food Specialties, South Bend, IN, USA) and fractionated as previously described (24), with modifications. Briefly, 2 g of cranberry pectinase-treated powder was dissolved in 20 ml of distilled water and loaded onto a RediSep GOLD C18 reverse-phase column (Teledyne ISCO, Inc., Lincoln, NE, USA) connected to a CombiFlash Rf purification system (Teledyne ISCO, Inc.) The column was eluted sequentially with 500 ml of deionized water, 500 ml of 15% methanol-water, and 500 ml of methanol. Fractions from all of the gradients were individually pooled and lyophilized to obtain three major fractions, Cranf1W (761 mg, 38.1%) eluted with 100% deionized water, Cranf1b (476 mg, 23.8%) eluted with 15% methanol-water, and Cranf1M (562 mg, 28.1%) eluted with 100% methanol. A 100-mg sample of Cranf1b was then dissolved in 2 ml of distilled water and further purified with a size exclusion column HiPrep Sephacryl S-100 HR 16/60 (GE Healthcare Life Sciences, Pittsburgh, PA, USA). The column was isocratically eluted with deionized water at 0.5 ml/min. Eluates were collected from every 5-ml volume and evaluated for their total carbohydrate content by phenol sulfuric acid assay (54). The total carbohydrate content was assessed with a 96-well microtiter plate as previously reported (54). Briefly, in each well of a 96-well microtiter plate, 30 μl of each fraction was mixed with 100 μl of concentrated sulfuric acid and 20 μl of 5% phenol solution. The microtiter plate was than incubated at 90°C for 5 min, and the absorbance at 490 nm was recorded with a SpectraMax M2 microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Xyloglucan structural analysis.
Oligosaccharide fractions were pooled and freeze-dried to obtain 59.5 mg of cranberry xyloglucans, which was then chemically verified by a combination of MALDI-TOF MS and 1H NMR spectroscopy. Briefly, 1 μl of xyloglucan (1 mg/ml in H2O) was mixed with 1 μl of 2,3-dihydrobenzoic acid matrix solution. A 2-μl volume of this mixture was analyzed by MALDI-TOF MS (Axima Performance, Shimadzu, Kyoto, Japan) in positive reflectron mode. Five hundred profiles were collected for each experiment. Furthermore, the cranberry xyloglucans were dissolved in D2O (99.96%; Cambridge Isotope Laboratories Inc., Tewksbury, MA, USA). The 1H NMR spectrum was obtained on a 500-MHz NMR spectrometer (Varian VNMR; Agilent Technologies, Santa Clara, CA, USA) at 25°C.
Bacterial strains and propagation.
The bacterial strains used in this study are summarized in Table 1. Bifidobacterial strains were propagated in bifidobacterial selective medium or De Man-Rogosa-Sharpe (MRS; Oxoid, Hampshire, England) medium supplemented with 0.05% (wt/vol) l-cysteine (Sigma-Aldrich, St. Louis, MO) (55) at 37°C under anaerobic conditions (Coy Laboratory Products, Grass Lake, MI). Lactobacillus cultures were propagated in MRS medium supplemented with 0.05% (wt/vol) l-cysteine as a reducing agent at 37°C under anaerobic conditions. Bacterial strains were routinely verified with the bifidobacterium-specific phosphoketolase assay (56) and through microscopy. The strains used in this study have been previously confirmed by multilocus sequence typing and urease assay to distinguish B. infantis and B. longum (urease+ and urease−) (16). In addition, a PCR-based B. longum/B. infantis ratio analysis was performed to differentiate B. longum subspecies as previously described (57).
Microplate growth assay.
To evaluate growth phenotypes in a 96-well format, overnight cultures were centrifuged and washed with phosphate buffer solution that was inoculated (1% [vol/vol]) into modified MRS (mMRS) medium (without acetate and carbohydrate substrate). The sole carbon source was defined as purified xyloglucans at a final concentration of 2% (wt/vol). The growth assay was conducted anaerobically at 37°C for 72 h by assessing OD600 with an automated PowerWave HT microplate spectrophotometer (BioTek Instruments, Inc., Winooski, VT). Each strain was evaluated in biological triplicate with three technical replicates. Negative and positive controls consisted of inoculated medium in the absence of substrate and the presence of glucose (2% [wt/vol]), respectively. Bacterial growth kinetics were calculated with Wolfram Mathematica 10.3 Student Edition as described by Dai et al. (58) in accordance with the equation ΔOD(t) = ΔODasym{[1/1 + exp(ktc − t)] − [1/1 + exp(ktc)]}, where ΔODasym is the growth level at stationary phase with k representing the growth rate and tc is the inflection point indicating the time required to reach the highest growth rate.
Modeled syntrophic interactions.
To determine if degradative or secreted metabolites are utilized by a heterologous strain, supernatants from microplate-grown bacteria were analyzed. Accordingly, 800 μl of the conditioned supernatant was dissolved in 1,200 μl of mMRS medium. Bacterial strains were washed once with phosphate-buffered saline to remove the residual carbohydrates from the initial propagation, inoculated at 1% (vol/vol), and evaluated for growth in a 96-well format as already described. Bacterial growth kinetics were calculated, and the supernatant obtained after the secondary growth was analyzed by HPLC.
Xyloglucan profiling following bacterial fermentation.
Following in vitro fermentation, filtered bacterial culture supernatants were derivatized with 2-aminobenzamide (2-AB) for oligosaccharide purification. Briefly, 50 μl of cell-free supernatant was diluted with 2 ml of deionized water and loaded onto a porous graphitized carbon cartridge (1 g; Thermo Scientific, Waltham, MA, USA). The cartridge was prewashed with 5 ml of 50% (vol/vol) acetonitrile-H2O and equilibrated with 3 × 5 ml of deionized water. First, the cartridges were eluted with 5 ml of deionized water three times with oligosaccharide fractions eluted with 30% (vol/vol) acetonitrile-H2O with 0.1% trifluoroacetic acid (TFA) three times. The resulting fractions were spiked with 0.036 mg of glucose as an internal standard and freeze-dried. 2-AB at 7 mg/ml and 2-picoline borane at 3.2 mg/ml were prepared in 10% (vol/vol) acetic acid-H2O, and 200 μl was added to each mixture. The solutions were held at 40°C for 4 h, centrifuged, and dried in vacuo. The derivatized oligosaccharide mixtures were redissolved in 400 μl of deionized water for fluorescence detection (FL)-HPLC and LC-MS analyses.
FL-HPLC and LC-MS analyses.
The 2-AB-labeled xyloglucans were analyzed by FL-HPLC and LC-MS. FL-HPLC analysis was performed with a Hitachi Elite LaChrom HPLC system (Hitachi, Tokyo, Japan) connected to a fluorescence detector (L-2485; Hitachi). The samples were analyzed on a Kinetex reverse-phase C18 column (150 by 3 mm, 2.6 μm; Phenomenex, Torrance, CA, USA) at 40°C. The column was first eluted with isocratic 10% (vol/vol) methanol in H2O (with 0.1% TFA) for 10 min at 0.2 ml/min, followed by a linear gradient of 10 to 20% methanol in H2O for 90 min and then a linear gradient of 20 to 100% methanol in H2O for 15 min, and kept at 100% methanol for 30 min. Elution was monitored with a fluorescence detector with an excitation wavelength of 330 nm and an emission wavelength at 420 nm. LC-MS analysis was performed with a Shimadzu Prominence UFLC system (Shimadzu, Kyoto, Japan) coupled to an AB Sciex QTrap 4500 mass spectrometer (AB Sciex, Framingham, MA, USA) with an electrospray ionization source. The LC-MS analysis was performed with the same column and the same HPLC program as aforementioned for FL-HPLC. The mass spectrometer was operated in positive mode, and ions at m/z 200 to 2,000 were scanned. Quantification of extracted ions was normalized by 2-AB-derivatized glucose.
Characterization of bacterial organic acid production.
End products of bacterial fermentation were quantitated by HPLC. Bacterial strains were initially propagated as described above. Cell-free supernatants from microcentrifuge tubes were obtained at early stationary phase, filtered through a 0.22-μm filter following centrifugation, and stored at −80°C until analysis. Organic acids were quantified with an Agilent 1260 Infinity HPLC system (Agilent Technologies, Santa Clara, CA) equipped with a Wyatt Optilab T-rEX refractive-index detector (Wyatt Technology Corp., Santa Barbara, CA). Separation was carried out with an Aminex HPX-87H column (7.8 mm [inside diameter] by 300 mm; Bio-Rad Laboratories, Hercules, CA) at 50°C in a mobile phase of 5 mM H2SO4 at a flow rate of 0.6 ml/min with a 50-μl injection volume. Organic acids (i.e., acetic acid, lactic acid, and formic acid) were acquired from Sigma-Aldrich Co. (St. Louis, MO). Metabolite concentrations were calculated from standard curves derived from external standards for six different concentrations (0.05, 0.1, 0.5, 1, 5, and 10 mg/ml) and converted to millimolar values. Metabolite profiling was carried out in triplicate, and each measurement was performed in duplicate.
Gene expression by qRT-PCR.
Relative gene expression was performed by quantitative real-time PCR (qRT-PCR). Four-milliliter samples were collected at mid-exponential phase, pelleted at 12,000 × g for 2 min, and stored in 1 ml of Ambion RNAlater (Life Technologies, Carlsbad, CA). Total RNA was extracted with the Ambion RNAqueous minikit (Life Technologies, Carlsbad, CA) in accordance with the manufacturer's instructions. The cells were placed in lysis buffer, transferred to MP Bio Matrix E tubes, and subjected to bead beating with a FastPrep 24 instrument (MP Biomedicals, Santa Ana, CA) (2 × 5.5 m/s for 30 s). Total RNA was eluted in 50 μl of EB solution and immediately subjected to DNase treatment with the Ambion Turbo DNA-free kit (Life Technologies, Carlsbad, CA) with 1 μl of DNase I for 0.5 h. Total RNA was converted to cDNA with the High Capacity cDNA reverse transcription kit (Applied Biosystems, Carlsbad, CA) in accordance with the manufacturer's instructions. cDNA concentrations were measured with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc., Agawam, MA). qRT-PCR analysis was performed with a 7500 Fast real-time PCR system (Applied Biosystems, Singapore) with PowerUP SYBR green master mix (Applied Biosystems, Foster City, CA) and the parameters suggested by the manufacturer. Primers were designed with Primer3 (Table 5; http://frodo.wi.mit.edu). The Blon_0393 gene was used as an endogenous control as previously validated (59). Gene expression levels during growth on glucose (2%, wt/vol) were used as a reference. Results were expressed as fold changes relative to the control. Bacterial growth was performed in triplicate with triplicate measurements by qRT-PCR.
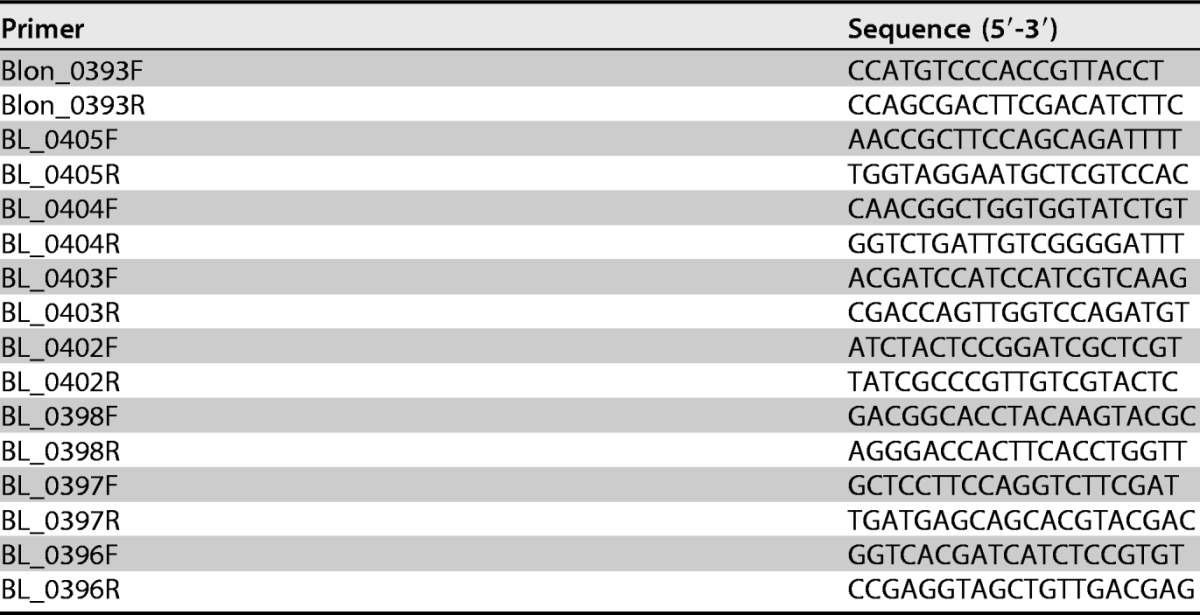
TABLE 5.
Primers used in this study
Statistical analysis.
Bacterial growth kinetics were subjected to two-way analysis of variance (ANOVA) and Tukey's honestly significant difference test for multiple comparisons of strains within a treatment compared with the positive control. Metabolite concentrations were subjected to two-way ANOVA, and Sidak's correction was used to account for multiple comparisons. Significant differences in modeled syntrophic interactions were determined by paired t test for bacterial growth kinetics. The fold change in gene expression compared to the control was analyzed by paired t test.
Supplementary Material
ACKNOWLEDGMENTS
We thank Micha Peleg and Mark D. Normand for helpful discussions on microbial growth curve kinetics and Asha Rani for critical review of the manuscript. We thank the Viticulture and Enology Culture Collection for providing B. longum UCD401.
Ocean Spray Cranberries, Inc., and the University of Massachusetts Graduate School are acknowledged for funding. In addition, the University of Massachusetts Innovation Institute is acknowledged for facilitating industrial sponsorship. Instruments used for chemical analysis of oligosaccharides were supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (2 P20 GM103430). NMR spectroscopy data were acquired at a research facility supported in part by National Science Foundation EPSCoR Cooperative Agreement EPS-1004057. The sponsors did not have any role in study design, data collection and interpretation, or the decision to submit the manuscript for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01097-17.
REFERENCES
- 1.Puertollano E, Kolida S, Yaqoob P. 2014. Biological significance of short-chain fatty acid metabolism by the intestinal microbiome. Curr Opin Clin Nutr Metab Care 17:1–16. doi: 10.1097/MCO.0000000000000025. [DOI] [PubMed] [Google Scholar]
- 2.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud D-J, Bakker BM. 2013. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. 2011. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol 9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonnenburg JL, Xu J, Leip DD, Chen C-H, Westover BP, Weatherford J, Buhler JD, Gordon JI. 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 5.Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, Flint HJ. 2006. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol 72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berni Canani R, Di Costanzo M, Leone L, Pedata M, Meli R, Calignano A. 2011. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J Gastroenterol 17:1519–1528. doi: 10.3748/wjg.v17.i12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O'Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pokusaeva K, Fitzgerald GF, van Sinderen D. 2011. Carbohydrate metabolism in bifidobacteria. Genes Nutr 6:285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fushinobu S. 2010. Unique sugar metabolic pathways of bifidobacteria. Biosci Biotechnol Biochem 74:2374–2384. doi: 10.1271/bbb.100494. [DOI] [PubMed] [Google Scholar]
- 10.Hehemann J-H, Kelly AG, Pudlo NA, Martens EC, Boraston AB. 2012. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc Natl Acad Sci U S A 109:19786–19791. doi: 10.1073/pnas.1211002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van den Broek LAM, Hinz SWA, Beldman G, Vincken J-P, Voragen AGJ. 2008. Bifidobacterium carbohydrases—their role in breakdown and synthesis of (potential) prebiotics. Mol Nutr Food Res 52:146–163. doi: 10.1002/mnfr.200700121. [DOI] [PubMed] [Google Scholar]
- 12.Milani C, Lugli GA, Duranti S, Turroni F, Mancabelli L, Ferrario C, Mangifesta M, Hevia A, Viappiani A, Scholz M, Arioli S, Sanchez B, Lane J, Ward DV, Hickey R, Mora D, Segata N, Margolles A, van Sinderen D, Ventura M. 2015. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep 5:15782. doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sela DA, Mills DA. 2010. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol 18:298–307. doi: 10.1016/j.tim.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Odamaki T, Horigome A, Sugahara H, Hashikura N, Minami J, Xiao J-Z, Abe F. 2015. Comparative genomics revealed genetic diversity and species/strain-level differences in carbohydrate metabolism of three probiotic bifidobacterial species. Int J Genomics 2015:567809. doi: 10.1155/2015/567809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A 105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LoCascio RG, Desai P, Sela DA, Weimer B, Mills DA. 2010. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl Environ Microbiol 76:7373–7381. doi: 10.1128/AEM.00675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sela DA, Garrido D, Lerno L, Wu S, Tan K, Eom H-J, Joachimiak A, Lebrilla CB, Mills DA. 2012. Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol 78:795–803. doi: 10.1128/AEM.06762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sela DA, Li Y, Lerno L, Wu S, Marcobal AM, German JB, Chen X, Lebrilla CB, Mills DA. 2011. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J Biol Chem 286:11909–11918. doi: 10.1074/jbc.M110.193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carpita NC, Gibeaut DM. 1993. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3:1–30. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh YSY, Harris PJ. 2009. Xyloglucans of monocotyledons have diverse structures. Mol Plant 2:943–965. doi: 10.1093/mp/ssp061. [DOI] [PubMed] [Google Scholar]
- 21.Brennan M, Harris PJ. 2011. Distribution of fucosylated xyloglucans among the walls of different cell types in monocotyledons determined by immunofluorescence microscopy. Mol Plant 4:144–156. doi: 10.1093/mp/ssq067. [DOI] [PubMed] [Google Scholar]
- 22.Hotchkiss AT, Nunez A, Strahan GD, Chau H, White A, Marais J, Hom K, Vakkalanka MS, Di R, Yam KL, Khoo C. 2015. Cranberry xyloglucan structure and inhibition of Escherichia coli adhesion to epithelial cells. J Agric Food Chem 63:5622–5633. doi: 10.1021/acs.jafc.5b00730. [DOI] [PubMed] [Google Scholar]
- 23.Coleman CM, Ferreira D, Howell AB, Reed JD, Krueger CG, Marais JPJ. 2010. Isolation and identification of antiadhesive urinary metabolites produced as a result of cranberry juice consumption, abstr. O-47 In 2010 Joint Annual Meeting of the American Society of Pharmacognosy & The Phytochemical Society of North America Natural Solutions to 21st Century Problems—from Discovery to Commercialization http://www.psna-online.org/ASP_2010%20Program.pdf. [Google Scholar]
- 24.Sun J, Marais JPJ, Khoo C, LaPlante K, Vejborg RM, Givskov M, Tolker-Nielsen T, Seeram NP, Rowley DC. 2015. Cranberry (Vaccinium macrocarpon) oligosaccharides decrease biofilm formation by uropathogenic Escherichia coli. J Funct Foods 17:235–242. doi: 10.1016/j.jff.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartemink R, Van Laere KMJ, Mertens A, Rombouts F. 1996. Fermentation of xyloglucan by intestinal bacteria. Anaerobe 2:223–230. doi: 10.1006/anae.1996.0031. [DOI] [Google Scholar]
- 26.Holzapfel WH, Haberer P, Snel J, Schillinger U, Huis in't Veld JH. 1998. Overview of gut flora and probiotics. Int J Food Microbiol 41:85–101. doi: 10.1016/S0168-1605(98)00044-0. [DOI] [PubMed] [Google Scholar]
- 27.McLaughlin HP, O'Connell Motherway M, Lakshminarayanan B, Stanton C, Ross RP, Brulc J, Menon R, O'Toole PW, van Sinderen D. 2015. Carbohydrate catabolic diversity of bifidobacteria and lactobacilli of human origin. Int J Food Microbiol 203:109–121. doi: 10.1016/j.ijfoodmicro.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Ward RE, Niñonuevo M, Mills DA, Lebrilla CB, German JB. 2007. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol Nutr Food Res 51:1398–1405. doi: 10.1002/mnfr.200700150. [DOI] [PubMed] [Google Scholar]
- 29.LoCascio RG, Ninonuevo MR, Freeman SL, Sela DA, Grimm R, Lebrilla CB, Mills DA, German JB. 2007. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem 55:8914–8919. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- 30.LoCascio RG, Niñonuevo MR, Kronewitter SR, Freeman SL, German JB, Lebrilla CB, Mills DA. 2009. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microb Biotechnol 2:333–342. doi: 10.1111/j.1751-7915.2008.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van der Meulen R, Adriany T, Verbrugghe K, De Vuyst L. 2006. Kinetic analysis of bifidobacterial metabolism reveals a minor role for succinic acid in the regeneration of NAD+ through its growth-associated production. Appl Environ Microbiol 72:5204–5210. doi: 10.1128/AEM.00146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borch E, Berg H, Holst O. 1991. Heterolactic fermentation by a homofermentative Lactobacillus sp. during glucose limitation in anaerobic continuous culture with complete cell recycle. J Appl Microbiol 71:265–269. doi: 10.1111/j.1365-2672.1991.tb04457.x. [DOI] [Google Scholar]
- 33.Kontula P, von Wright A, Mattila-Sandholm T. 1998. Oat bran β-gluco- and xylo-oligosaccharides as fermentative substrates for lactic acid bacteria. Int J Food Microbiol 45:163–169. doi: 10.1016/S0168-1605(98)00156-1. [DOI] [PubMed] [Google Scholar]
- 34.Salazar N, Prieto A, Leal JA, Mayo B, Bada-Gancedo JC, de los Reyes-Gavilán CG, Ruas-Madiedo P, de Bont JAM. 2009. Production of exopolysaccharides by Lactobacillus and Bifidobacterium strains of human origin, and metabolic activity of the producing bacteria in milk. J Dairy Sci 92:4158–4168. doi: 10.3168/jds.2009-2126. [DOI] [PubMed] [Google Scholar]
- 35.Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, Mangifesta M, Sanchez B, Viappiani A, Mancabelli L, Taminiau B, Delcenserie V, Barrangou R, Margolles A, van Sinderen D, Ventura M. 2014. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol 80:6290–6302. doi: 10.1128/AEM.02308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pastell H, Westermann P, Meyer AS, Päivi T, Tenkanen M. 2009. In vitro fermentation of arabinoxylan-derived carbohydrates by bifidobacteria and mixed fecal microbiota. J Agric Food Chem 57:8598–8606. doi: 10.1021/jf901397b. [DOI] [PubMed] [Google Scholar]
- 37.Rivière A, Moens F, Selak M, Maes D, Weckx S, De Vuyst L. 2014. The ability of bifidobacteria to degrade arabinoxylan oligosaccharide constituents and derived oligosaccharides is strain dependent. Appl Environ Microbiol 80:204–217. doi: 10.1128/AEM.02853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Vuyst L, Moens F, Selak M, Rivière A, Leroy F. 2014. Summer Meeting 2013: growth and physiology of bifidobacteria. J Appl Microbiol 116:477–491. doi: 10.1111/jam.12415. [DOI] [PubMed] [Google Scholar]
- 39.Palframan RJ, Gibson GR, Rastall RA. 2003. Carbohydrate preferences of Bifidobacterium species isolated from the human gut. Curr Issues Intest Microbiol 4:71–75. [PubMed] [Google Scholar]
- 40.Van der Meulen R, Makras L, Verbrugghe K, Adriany T, De Vuyst L. 2006. In vitro kinetic analysis of oligofructose consumption by Bacteroides and Bifidobacterium spp. indicates different degradation mechanisms. Appl Environ Microbiol 72:1006–1012. doi: 10.1128/AEM.72.2.1006-1012.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thum C, Roy NC, McNabb WC, Otter DE, Cookson AL. 2015. In vitro fermentation of caprine milk oligosaccharides by bifidobacteria isolated from breast-fed infants. Gut Microbes 6:352–363. doi: 10.1080/19490976.2015.1105425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van der Meulen R, Avonts L, De Vuyst L. 2004. Short fractions of oligofructose are preferentially metabolized by Bifidobacterium animalis DN-173 010. Appl Environ Microbiol 70:1923–1930. doi: 10.1128/AEM.70.4.1923-1930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Falony G, Lazidou K, Verschaeren A, Weckx S, Maes D, De Vuyst L. 2009. In vitro kinetic analysis of fermentation of prebiotic inulin-type fructans by Bifidobacterium species reveals four different phenotypes. Appl Environ Microbiol 75:454–461. doi: 10.1128/AEM.01488-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blumberg JB, Camesano TA, Cassidy A, Kris-Etherton P, Howell A, Manach C, Ostertag LM, Sies H, Skulas-Ray A, Vita JA. 2013. Cranberries and their bioactive constituents in human health. Adv Nutr 4:618–632. doi: 10.3945/an.113.004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rossi M, Corradini C, Amaretti A, Nicolini M, Pompei A, Zanoni S, Matteuzzi D. 2005. Fermentation of fructooligosaccharides and inulin by bifidobacteria: a comparative study of pure and fecal cultures. Appl Environ Microbiol 71:6150–6158. doi: 10.1128/AEM.71.10.6150-6158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibson G, Roberfroid MB. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr 125:1401–1412. [DOI] [PubMed] [Google Scholar]
- 47.Watson D, O'Connell Motherway M, Schoterman MHC, van Neerven RJJ, Nauta A, van Sinderen D. 2013. Selective carbohydrate utilization by lactobacilli and bifidobacteria. J Appl Microbiol 114:1132–1146. doi: 10.1111/jam.12105. [DOI] [PubMed] [Google Scholar]
- 48.Arsköld E, Lohmeier-Vogel E, Cao R, Roos S, Rådström P, van Niel EWJ. 2008. Phosphoketolase pathway dominates in Lactobacillus reuteri ATCC 55730 containing dual pathways for glycolysis. J Bacteriol 190:206–212. doi: 10.1128/JB.01227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okano K, Yoshida S, Tanaka T, Ogino C, Fukuda H, Kondo A. 2009. Homo-d-lactic acid fermentation from arabinose by redirection of phosphoketolase pathway to pentose phosphate pathway in l-lactate dehydrogenase gene-deficient Lactobacillus plantarum. Appl Environ Microbiol 75:5175–5178. doi: 10.1128/AEM.00573-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turroni F, Özcan E, Milani C, Mancabelli L, Viappiani A, van Sinderen D, Sela DA, Ventura M. 2015. Glycan cross-feeding activities between bifidobacteria under in vitro conditions. Front Microbiol 6:1030. doi: 10.3389/fmicb.2015.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfeiffer T, Bonhoeffer S. 2004. Evolution of cross-feeding in microbial populations. Am Nat 163:E126–135. doi: 10.1086/383593. [DOI] [PubMed] [Google Scholar]
- 52.Turroni F, Milani C, Duranti S, Mancabelli L, Mangifesta M, Viappiani A, Lugli GA, Ferrario C, Gioiosa L, Ferrarini A, Li J, Palanza P, Delledonne M, van Sinderen D, Ventura M. 2016. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J 10:1656–1668. doi: 10.1038/ismej.2015.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee J-H, Karamychev VN, Kozyavkin SA, Mills D, Pavlov AR, Pavlova NV, Polouchine NN, Richardson PM, Shakhova VV, Slesarev AI, Weimer B, O'Sullivan DJ. 2008. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics 9:247. doi: 10.1186/1471-2164-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Masuko T, Minami A, Iwasaki N, Majima T, Nishimura S-I, Lee YC. 2005. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal Biochem 339:69–72. doi: 10.1016/j.ab.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 55.Turroni F, Foroni E, Pizzetti P, Giubellini V, Ribbera A, Merusi P, Cagnasso P, Bizzarri B, de'Angelis GL, Shanahan F, van Sinderen D, Ventura M. 2009. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl Environ Microbiol 75:1534–1545. doi: 10.1128/AEM.02216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Orban JI, Patterson JA. 2000. Modification of the phosphoketolase assay for rapid identification of bifidobacteria. J Microbiol Methods 40:221–224. doi: 10.1016/S0167-7012(00)00133-0. [DOI] [PubMed] [Google Scholar]
- 57.Lewis ZT, Shani G, Masarweh CF, Popovic M, Frese SA, Sela DA, Underwood MA, Mills DA. 2016. Validating bifidobacterial species and subspecies identity in commercial probiotic products. Pediatr Res 79:445–452. doi: 10.1038/pr.2015.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dai Y, McLandsborough LA, Weiss J, Peleg M. 2010. Concentration and application order effects of sodium benzoate and eugenol mixtures on the growth inhibition of Saccharomyces cerevisiae and Zygosaccharomyces bailii. J Food Sci 75:M482–M488. doi: 10.1111/j.1750-3841.2010.01772.x. [DOI] [PubMed] [Google Scholar]
- 59.Garrido D, Kim JH, German JB, Raybould HE, Mills DA. 2011. Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS One 6:e17315. doi: 10.1371/journal.pone.0017315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.